Abstract
Nav1.5 inactivation is necessary for healthy conduction of the cardiac action potential. Genetic mutations of Nav1.5 perturb inactivation and cause potentially fatal arrhythmias associated with long QT syndrome type 3. The exact structural dynamics of the inactivation complex is unknown. To sense inactivation gate conformational change in live mammalian cells, we incorporated the solvatochromic fluorescent noncanonical amino acid 3-((6-acetylnaphthalen-2-yl)amino)-2-aminopropanoic acid (ANAP) into single sites in the Nav1.5 inactivation gate. ANAP was incorporated in full-length and C-terminally truncated Nav1.5 channels using mammalian cell synthetase-tRNA technology. ANAP-incorporated channels were expressed in mammalian cells, and they exhibited pathophysiological function. A spectral imaging potassium depolarization assay was designed to detect ANAP emission shifts associated with Nav1.5 conformational change. Site-specific intracellular ANAP incorporation affords live-cell imaging and detection of Nav1.5 inactivation gate conformational change in mammalian cells.
Significance
We incorporated a fluorescent noncanonical amino acid (3-((6-acetylnaphthalen-2-yl)amino)-2-aminopropanoic acid) into single sites within the inactivation gate of human cardiac voltage-gated sodium channels (Nav1.5). We developed a fluorescence-based spectral method to monitor Nav1.5 conformational change in intact mammalian cells. This work compliments the near-atomic-level structural detail resolved in recent cryo-electron microscopy structures of full-length eukaryotic voltage-gated sodium channels and sets the foundation for measurement of voltage-gated sodium channel structural dynamics in mammalian cells.
Introduction
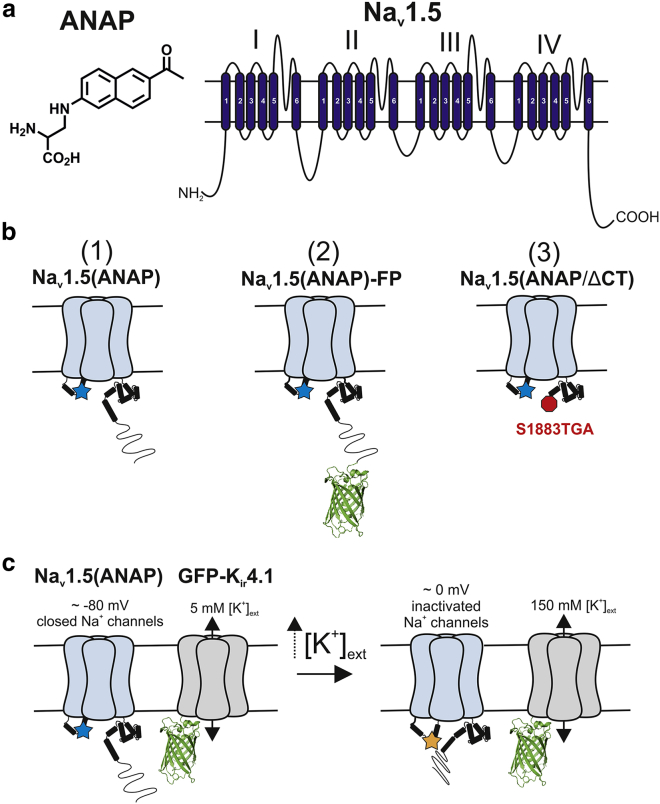
Genetic mutation in the voltage-gated sodium channel (VGSC) Nav1.5 can lead to multiple cardiac arrhythmia disorders, including Brugada Syndrome and long QT syndrome type 3 (LQT3), which can both lead to sudden cardiac death. Nav1.5 is a 260 kDa transmembrane ion channel with four semihomologous domains, each containing six transmembrane segments (1) (Fig. 1 a, right). The ∼50-amino-acid linker between domains III and IV (III-IV linker) is known as the inactivation gate. VGSC fast inactivation is thought to occur by a hinged-lid mechanism requiring the presence of a conserved hydrophobic motif, typically IFM, in the III-IV linker (1). This hydrophobic motif may stabilize a channel conformation in which the III-IV linker occludes the intracellular mouth of the pore and halts ion conduction (1, 2, 3, 4, 5). The IFM motif assumes a stable fast inactivated state by docking onto the underside of the pore via hydrophobic interactions, mainly with sites on the DIII:S4-S5 linker, DIV:S4-S5 linker, and DIV:S6 (5, 6, 7, 8, 9, 10, 11, 12, 13). The C-terminus (CT) is also important for regulating channel availability and inactivation kinetics, demonstrated by functional characterization of LQT3-associated mutations in the CT and ion channel chimera experiments (14, 15, 16, 17, 18, 19). Furthermore, the inactivated state is stabilized by an interaction between the III-IV linker and CT, forming an inactivation complex, and this requires the presence of the C-terminal part of helix 6 in the CT structured region (8, 20, 21, 22, 23).
Figure 1.
Nav1.5 constructs and K+ depolarization assay schematic. (a) Chemical structure of ANAP (left) and Nav1.5 domain structure (right) are shown. (b) Nav1.5(ANAP) constructs used in imaging and electrophysiology experiments are shown. Star = ANAP. (c) A schematic of K+ depolarization in cells expressing GFP-Kir4.1 and Nav1.5(ANAP) is shown. Net flux of K+ is zero at both membrane potentials. 5 mM [K+]ext and 150 [K+]int set the equilibrium potential at ∼ −80 mV, at which Nav channels are expected to be closed. An increase to 150 mM [K+]ext sets the equilibrium potential at ∼0 mV, at which a large population of Nav channels is expected to be inactivated. To see this figure in color, go online.
Structures of truncated regions of eukaryotic VGSCs, such as the III-IV linker, in complex with accessory proteins have been reported; however, their functional significance is unknown (24, 25). In particular, the eukaryotic III-IV linker bound to calcium/calmodulin (CaM) has been crystallized (24). Also, the crystal structure of the eukaryotic Nav1.5 CT domain in complex with fibroblast growth factor homologous factor and apo-CaM has been resolved (25). Considered together, these results raise questions of how the III-IV linker, CT, calcium, and CaM may interact in the inactivation complex of full-length eukaryotic channels. One limitation to the studies supporting the significance of a direct III-IV linker CT interaction is the measurement of interactions between purified III-IV linker and CT peptides, not in full-length channels.
A number of cryo-electron microscopy structures of full-length eukaryotic VGSCs have been published to date: insect NavPaS (26), electric eel Nav1.4 (27), human Nav1.7 (28), human Nav1.2 (29), and hybrid NavPaS/human Nav1.7 (30). Interactions between voltage sensor domain IV and the CT were visualized in the full-length hybrid channel (insect NavPaS + voltage sensor domain IV from human Nav1.7) bound to α-scorpion toxin, suggesting a possible resting state for the fast-inactivation complex (30). One limitation of structural techniques is that there is no membrane potential in a protein crystal. Therefore, it is difficult to assign the resolved structure to voltage-dependent functional ion channel states. Fluorescence imaging in mammalian cells can be a useful complement to cryo-electron microscopy studies for connecting structure with function of the Nav1.5 inactivation complex. Currently, specific fluorescent labeling of the short intracellular inactivation gate with minimal perturbation in full-length channels is a technical hurdle that must be overcome.
Options for labeling short intracellular protein linkers are limited. Fluorescent proteins are quite large, and cysteine labeling is nonspecific. Noncanonical amino acids (ncAAs) offer an alternative method for incorporating fluorophores at single sites in proteins by amber suppression. Amber suppression in mammalian cells requires expression of an orthogonal aminoacyl-tRNA synthetase-tRNA (aaRS-tRNA) pair specific for the ncAA, along with the protein of interest containing an amber stop codon at the desired incorporation site (31). 3-((6-acetylnaphthalen-2-yl)amino)-2-aminopropanoic acid (ANAP) is one of a number of solvatochromic fluorescent ncAAs whose spectrum red-shifts in increasingly polar environments (32, 33, 34) (Fig. 1 a, left). The ANAP aaRS-tRNA pair was evolved in Saccharomyces cerevisiae from the Escherichia coli leucyl aaRS-tRNA pair (32). Since the first demonstration of ANAP incorporation into model proteins in S. cerevisiae and mammalian cells, there have been a number of examples using ANAP environmental sensitivity to study ion channels in Xenopus laevis oocytes and mammalian cells (35, 36, 37, 38, 39, 40, 41, 42, 43, 44).
In recent years, ANAP incorporation has been used increasingly for studying protein dynamics in live cells. In 2013, Kalstrup and Blunck demonstrated voltage-clamp fluorometry of ANAP-incorporated Shaker potassium channels in X. laevis oocytes and have published video protocols for reproducing ANAP voltage-clamp fluorometry experiments (35, 45). ANAP spectral shifts were used to monitor protein misfolding of luciferase in S. cerevisiae (46). In X. laevis oocytes, ANAP has been incorporated into glycine receptors, CNGA1 channels, voltage-sensing phosphatases, KCNH channels, and ASIC channels (36, 37, 38, 39, 40, 41). ANAP incorporated in CNGA1 and KCNH channels was used to measure intracellular interactions by transition metal Förster resonance energy transfer (FRET; 36). However, there is a need for characterization of ion-channel structural dynamics in the mammalian cell context. A few studies have answered to that need. Zagotta et al. demonstrated transition metal FRET between ANAP incorporated in TRPV1 channels and the plasma membrane of unroofed mammalian cells (42). In Chinese hamster ovary cells, ANAP was incorporated into hASIC1a, and increasing concentrations of the channel toxin mambalgin-1 were detected spectrally by a red shift in ANAP fluorescence (43). ANAP was demonstrated as a FRET donor to enhanced green fluorescent protein (EGFP) in an FP-reporter fusion protein expressed in mammalian cells (47). Recently, Puljung et al. interrogated activation of ATP-sensitive K+ channels in unroofed human embryonic kidney (HEK)293T cells via FRET between ANAP and fluorescent nucleotides (44).
We hypothesized that ANAP can be incorporated into the Nav1.5 inactivation gate using mammalian cell synthetase-tRNA technology. Once incorporated, ANAP would act as a fluorescent reporter of human Nav1.5 conformational change. An ANAP spectral assay was developed to address whether ANAP responds to 1) the local environment of the inactivation gate when incorporated at different sites (Fig. 1 b), 2) the absence of the distal CT (Fig. 1 b), and 3) conformational rearrangement elicited by a potassium (K+) depolarization (Fig. 1 c).
Materials and Methods
Chemical reagents
All reagents were purchased commercially. A 1.7 mM stock solution of ANAP (3-((6-acetylnaphthalen-2-yl)amino)-2-aminopropanoic acid) (AsisChem, Waltham, MA) was made in anhydrous dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO). A 50 μM tetradotoxin (TTX; Abcam, Cambridge, UK) solution was made in ddH2O from a 5 mM stock solution. Reagents were aliquoted and stored at −20°C. Chemicals for physiological solutions were purchased from Sigma-Aldrich and stored at room temperature unless otherwise noted. Stock physiological solutions were stored at 4°C.
Plasmids, cell lines, and molecular biology
AnapRS-LeutRNACUA, pcDNA4-EGFP(Y40TAG) “pSWAN-GFP37TAG,” and pCMV-MmPylRS-PylT were gifts from Peter G. Schultz (33, 48). Template plasmids pH2B-EGFP and pcDNA3-SCN5A hh1c were available in-house. Template plasmid pH2B-mCherry was a gift from Robert Benezra (Addgene plasmid # 20972; Watertown, MA). HEK293T cells were available in-house. The GFP-Kir4.1 HEK293T cell line was a gift from Christopher Ahern. QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA) and/or Gibson Assembly (New England Biolabs, Ipswich, MA) was performed according to the manufacturer’s instructions with modifications as needed.
Mammalian tissue culture and transfection
HEK293T cells were grown in a medium containing minimum essential medium (MEM) (Gibco, Gaithersburg, MD) or Dulbecco’s modified Eagle’s medium containing 4.5 g/L glucose, L-glutamine, and sodium pyruvate (Corning 10-013-CM; Corning, NY), 10% fetal bovine serum, 1% penicillin-streptomycin, and 2 mM GlutaMAX (Gibco) supplement at 37°C in a humidified atmosphere of 5% CO2. Transfections of all cell lines for Western blot and live-cell imaging (six-well tissue-culture-treated plates for Western blot, LabTek eight-well chambered coverglass tissue-culture-treated plates for imaging) were performed using Xtremegene HP (Roche, Basel, Switzerland) or Lipofectamine 2000 (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. A typical imaging experiment involved transfection of 1 μg total DNA, including 0.6 μg Nav1.5 reporter construct and 0.4 μg AnapRS-LeutRNACUA (1.5:1 target protein/aaRS-tRNA mass ratio). Plasmids were co-transfected with reagent (3 μL reagent to 1 μg DNA) in Opti-MEM (Gibco). In +ANAP conditions, ANAP was added to the well at a final concentration of 10 μM ∼30 min before adding transfection reaction. This recipe was scaled up to 2 μg total DNA for electrophysiology experiments, transfected in T25 flasks, then split 24 h later and seeded into 35 mm plates. EGFP or EGFP(Y40TAG) was co-transfected as a marker of cells to patch as needed (1:1 with Nav1.5(TAG)). GFP-Kir4.1 HEK293T were cultured in Dulbecco’s modified Eagle’s medium (Gibco), 10% fetal bovine serum, 1% penicillin-streptomycin, 2 mM GlutaMAX, and Zeocin (Gibco) at 37°C in a humidified atmosphere of 5% CO2.
Western blot
HEK293T cells were seeded at 3 × 105 cells per well in a six-well tissue-culture-treated plate, transfected according to manufacturer’s instructions. 1.2 μg of DNA encoding Nav1.5(TAG)-EGFP, Nav1.5-EGFP, EGFP(Y40TAG), or EGFP was co-transfected with 0.8 μg of AnapRS-LeutRNACUA with Xtremegene (Roche) at a ratio of 3:1 in Opti-MEM (Gibco) supplemented with 2 mM GlutaMAX. 10 μM ANAP was added to the media ∼30 min before adding transfection reaction. Transfected cells were cultured at 37°C, 5% CO2 36–48 h before harvesting and lysis. Before lysis, cells were washed once with Tris-HCl-buffered saline (TBS), then lysed with Cell Lysis Buffer (Cell Signaling Technology, Danvers, MA) with 1:100 proteasome inhibitor complex on ice for 5 min. Lysis buffer contained (mM) 20 Tris-HCl (pH 7.5), 150 NaCl, 1 Na2EDTA, 1 EGTA, 1% Triton, 2.5 sodium pyrophosphate, 1 β-glycerophosphate, 1 Na3VO4, and 1 μg/mL leupeptin. Cells were scraped and resuspended in lysis buffer and incubated at 4°C for 30 min. Samples were centrifuged 10 min at 12,000 × g, then heated (55°C). Samples were loaded onto a 5 and 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and electrophoresed for 45 min at 10 mA/gel, then 90 min at 20 mA/gel. Protein gels were wet-transferred onto polyvinylidene difluoride membranes overnight at 4°C (15 V). The membranes were washed briefly with TBS, then blocked with 5% w/v skim milk in 1× TBS with Tween 20 (TBS-T) for 30 min at room temperature. Membranes were then incubated with 1:8000 primary antibody (rabbit polyclonal anti-GFP) in TBS-T for 1 h at room temperature. Membranes were washed 3 × 10 min at room temperature in TBS-T and incubated with secondary antibody (anti-rabbit horseradish peroxidase) 30 min at room temperature. Membranes were washed 2 × 10 min at room temperature with TBS-T, then 1 × 10 min at room temperature with TBS and developed using ECL Western Blot Detection Reagents (Bio-Rad, Hercules, CA). Images were taken by exposing light sensitive films at 30 s, 1, 2, 3, and 5 min on a developer (Kodak, Rochester, NY) in a dark room. Membranes were stripped, stained with anti-β-tubulin, and imaged as above.
Whole-cell patch-clamp electrophysiology
Sodium channel electrophysiology
Electrophysiological recordings of sodium channels expressed in HEK293T were carried out in a whole-cell patch-clamp configuration using physiological solutions. Pipette resistances ranged from 1.5 to 4 MOhm. The INa internal solution contained (mM) 50 aspartic acid, 60 CsCl, 5 Na2-ATP, 11 EGTA, 10 HEPES, 1 CaCl2, and 1 MgCl2, with pH 7.4 adjusted with CsOH. The INa external solution contained (mM) 130 NaCl, 2 CaCl2, 5 CsCl, 1.2 MgCl2, 10 HEPES, 5 glucose, with pH 7.4 adjusted with CsOH. Whole-cell sodium currents were recorded under voltage-clamp conditions. A voltage-step protocol was used, holding at −100 mV, step depolarization to −20 mV for 200 ms, then back to holding. Cells were pulsed at a frequency of 0.1 Hz. Steady-state inactivation (SSI) voltage protocol was as follows: holding at −100 or −90 mV, step to the conditioning pulse for 500 ms, depolarization to −10 mV for 20 ms, return to holding. Conditioning pulses ranged from −130 to −20 mV. Late sodium current was measured as TTX-sensitive current 200 ms after depolarization to −20 mV. Percent late current was measured as the average current between 195 and 200 mV divided by the peak current. Data was collected using an Axopatch 200B amplifier (Axon Instruments, San Jose, CA) and Digidata 1440A digitizer. Data was recorded with pClamp 8, 10, or 10.5 (Molecular Devices, San Jose, CA). Capacitive current and series resistance compensation were carried out using analog techniques according to the amplifier manufacturer (Axon Instruments). All measurements were obtained at room temperature (25°C). Statistical significance was determined using single-factor analysis of variance (ANOVA) when comparing multiple groups and Student’s t-test assuming equal or unequal variances based on a prior F-test of variance. p < 0.05 was considered statistically significant.
Potassium channel electrophysiology
Electrophysiological recordings of GFP-Kir4.1 HEK293T were recorded under voltage-clamp or current-clamp conditions. Internal solution contained (mM) 150 KCl, 3 MgCl2, 5 EGTA, 10 HEPES, with pH 7.4 adjusted with KOH. Extracellular solution contained (mM) 150 NaCl, 5 KCl, 1 MgCl2, 1.8 CaCl2, 10 HEPES, with pH 7.4 adjusted with NaOH. Changes in membrane potential were recorded over time once patch stability and Kir4.1 channel expression were confirmed by applying a voltage-ramp protocol: cells were held at −80 mV, and a 500 ms voltage ramp was applied once every 3 s, with voltage increasing linearly from −120 to +60 mV, then returned to holding. Using pClamp software, a 50× data reduction of recordings was performed for data transfer compatibility to Microsoft Excel and Origin (Microcal Software) software. All measurements were obtained at room temperature (25°C). Where appropriate, as in comparing reversal potentials between low K+ and high K+, statistical significance was determined using Student’s t-test assuming equal or unequal variances based on a prior F-test of variance. p < 0.05 was considered statistically significant.
Live-cell imaging
ANAP incorporation and spectral imaging of different sites in Nav1.5
HEK293T cells were plated at a density of 10,000–20,000 cells per well in LabTek eight-well chambered coverglass tissue-culture-treated plates (Nunc, 1.5 borosilicate) 12–24 h before transfection. Transfections were performed as described previously. Fluorescence microscopy and spectral imaging of ANAP incorporated at different sites in Nav1.5 were performed on a Zeiss LSM 700 confocal microscope (Zeiss, Oberkochen, Germany). ANAP was excited at 405 nm and spectral emission collected from 410 to 600 nm in 10 nm intensity bins (Zeiss). To match the laser intensity as accurately as possible, intensity over wavelength was measured for each manually chosen region of interest (ROI), normalized to ROI area, and subtracted by a laser-line background-area-normalized spectrum measured near the original ROI, which did not contain a visible cell. Laser-subtracted spectra were then normalized to the sum of the intensities under the peak. Resulting normalized spectra were averaged and errors determined as mean ± standard error (SE). Peak centroids of each averaged spectrum were calculated as
ANAP spectral imaging with K+ depolarization
Spectral imaging for the K+ depolarization experiment was performed on a Nikon A1RMP confocal microscope with spectral detector (Nikon, Tokyo, Japan). Cells were maintained at 37°C in a 5% CO2 humidified atmosphere on the microscope with a stage-top incubator in live-cell imaging solution (Molecular Probes, Eugene, OR) or electrophysiological solutions. Live-cell imaging solution contained (mM) 140 NaCl, 2.5 KCl, 1.8 CaCl2, 1.0 MgCl2, 20 HEPES (pH 7.4). In K+ depolarization experiments, low K+ external solution contained (mM) 150 NaCl, 5 KCl, 1 MgCl2, 1.8 CaCl2, 10 HEPES, with pH 7.4 adjusted with NaOH. High K+ external solution contained (mM) 150 KCl, 5 NaCl, 1 MgCl2, 1.8 CaCl2, 10 HEPES, with pH 7.4 adjusted with KOH. During imaging, cells were maintained in low K+ external solution before buffer exchange to high K+. ANAP was excited at 405 nm and spectral emission collected from 402 to 588 nm in 6 nm intensity bins using the spectral detector. The three shortest wavelength bins were affected by a mechanical filter inside the spectral detector—a metal finger blocker just after the grating that prevents excitation light from entering the photomultiplier arrays. Unmixed ANAP or GFP spectra had negligible intensities at the affected wavelengths (Fig. S2). Data analysis was performed as follows. Composite ANAP and GFP spectra were fitted and unmixed using a MATLAB (The MathWorks, Natick, MA) linear unmixing algorithm built in-house. Three spectral components were modeled to fit and unmix experimental spectra: 1) ANAP, 2) 405 nm laser line, and 3) GFP. First, the model ANAP spectrum was extracted from ANAP-only control HEK293T expressing Nav1.5(Q1475ANAP) and fitted as the linear combination of two Gaussians to obtain a smooth spectrum. Second, a model spectrum of the peak 405 nm laser light background was determined by measuring spectra from ROIs drawn in which there were no visible cells. Third, the GFP model spectrum was extracted from GFP-only control GFP-Kir4.1 co-transfected with Nav1.5(Q1475TAG) and AnapRS-LeutRNACUA in the absence of ANAP. The GFP model spectrum was laser-subtracted using the model spectrum of the laser light background determined in (2). The best GFP fit was a linear combination of two log-normal distributions (49). Once the model spectra were obtained, least-squares regression was performed over the entire wavelength range (402–588 nm) to fit how bright each of the peaks needed to be to explain the observed spectrum at each wavelength. To account for ANAP shifts, the least-squares fit was calculated at one particular wavelength of ANAP, and the residual was recorded. The code iterated 20 nm to the left and right of the peak in 1 nm steps, downsampled to the instrument resolution, until residuals were minimized, resulting in the best fit overall.
Results
Fluorescent ncAA ANAP was incorporated into the Nav1.5 inactivation gate in mammalian cells
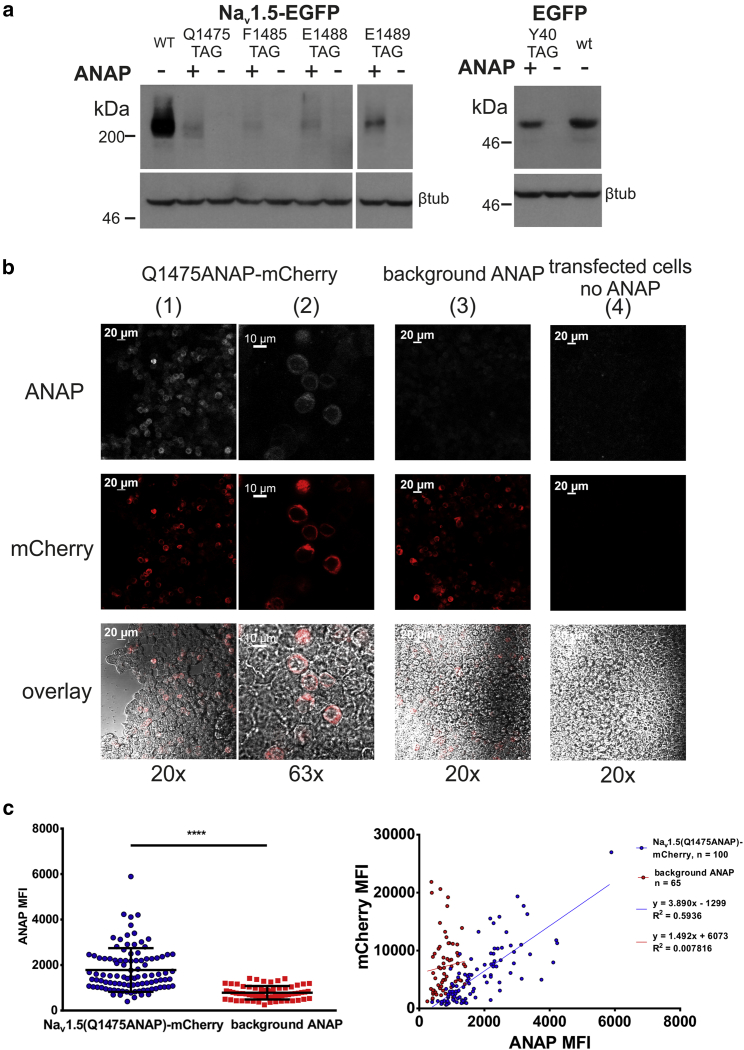
Can fluorescent ncAA ANAP be incorporated into the human Nav1.5 inactivation gate in mammalian cells? In proof-of-principle ANAP incorporation experiments, EGFP or mCherry was fused to the human Nav1.5 CT as a reporter of full-length channel expression (Fig. 1 b, construct 2). ANAP-dependent expression of full-length Nav1.5-EGFP (∼275 kDa) was measured by Western blot for four sites: Q1475TAG, F1485TAG, E1488TAG, and E1489TAG (Fig. 2 a). In the presence of ANAP, a band was observed above 200 kDa, corresponding to the more intense positive control band of the same size, representing full-length Nav1.5-EGFP. The positive control for ncAA incorporation was to incorporate ANAP into EGFP(Y40TAG). ANAP-dependent full-length expression of EGFP (∼27 kDa) was observed at ∼50 kDa, corresponding to the band from cells expressing wild-type EGFP (Fig. 2 a). In both cases, no band was observed in the absence of ANAP. β-tubulin normalized band intensities in the ncAA-incorporation conditions were 16% for Q1475ANAP, 7% for F1485ANAP, 11% for E1488ANAP, and 31% for E1489ANAP of that in the wild-type condition. These results demonstrate ANAP-dependent expression of full-length sodium channel reporter protein in mammalian cells and emphasize the high variability in ncAA-incorporation efficiency depending on target incorporation site.
Figure 2.
Full-length Nav1.5 expression depends on presence of ANAP and its synthetase-tRNA pair. (a) Western blot of Nav1.5(Q1475TAG)-EGFP and EGFP(Y40TAG) grown in HEK293T ± 10 μM ANAP, probed with rabbit pAb-GFP, is shown. All conditions were co-transfected with AnapRS-tRNALeuCUA. (Left) Nav1.5-EGFP, 5% SDS-PAGE, 5 min exposure is shown. (Right) EGFP, 10% SDS-PAGE, 1 min exposure is shown. Each image in its entirety was adjusted for contrast and brightness enhancement. Full-length EGFP is ∼27 kDa and was observed ∼50 kDa in this blot. Full-length Nav1.5-EGFP is ∼275 kDa and was observed above 200 kDa in this blot. (b) Fluorescence imaging of HEK293T expressing Nav1.5(Q1475ANAP)-mCherry (columns 1, 2), Nav1.5-mCherry+10 μM ANAP (background ANAP, column 3), or Nav1.5(Q1475TAG)-mCherry in the absence of ANAP (transfected cells, no ANAP, column 4) is shown. ANAP (ex405, em445/50 nm) and mCherry (ex555, em647/70 nm) fluorescence were imaged. Overlay image includes ANAP (white), mCherry (red), and brightfield channels. All except brightfield images were equally adjusted in their entirety for contrast enhancement. Columns 1, 3, and 4 scale bars represent 20 μm. Column 2 scale bars represent 10 μm. (c) Quantification of the 20× images in (b) is shown. (Left) ANAP mean fluorescence intensity (MFI) was significantly increased when the target codon for incorporation was present. Error bars are ± standard deviation. ∗∗∗∗p < 0.0001, two-tailed unpaired t-test with Welch’s correction. (Right) mCherry MFI plotted as a function of ANAP MFI is shown. To see this figure in color, go online.
Nav1.5 was tested for permissibility to ANAP incorporation. A site in DIII:S6 Q1475TAG was chosen initially for ANAP incorporation because we hypothesized that this site being close to the membrane-cytosol interface would elicit detectable spectral shifts representing changes in hydrophobicity in the K+ depolarization assay (26, 30). HEK293T cells expressing Nav1.5(Q1475ANAP)-mCherry were imaged by confocal microscopy (Fig. 2 b). In the +ANAP condition, ANAP and mCherry fluorescence was observed at the plasma membrane and localized to intracellular compartments (Fig. 2 b, columns 1, 2). This was expected because of the absence of β-subunit overexpression, which can cause defective plasma membrane trafficking (50). ANAP fluorescence intensity correlated (R2 = 0.6) with mCherry fluorescence intensity in the Nav1.5(Q1475ANAP)-mCherry condition (Fig. 2 c, right). As a negative control representing background ANAP fluorescence, wild-type Nav1.5-mCherry and AnapRS-LeutRNACUA were coexpressed in HEK293T+10 μM ANAP (Fig. 2 b, column 3). Bright mCherry fluorescence and dim ANAP fluorescence were observed, but ANAP signal was significantly decreased compared to the Nav1.5(Q1475ANAP)-mCherry condition (Fig. 2 c, left).
As a negative control to evaluate ANAP-independent expression of the reporter protein, Nav1.5(Q1475TAG)-mCherry was expressed in the presence of AnapRS-LeutRNACUA in HEK293T grown without ANAP (Fig. 2 b, column 4). Cell autofluorescence and dim mCherry signal were observed, but both signals were decreased compared to the Nav1.5(Q1475ANAP)-mCherry condition. Together, these data demonstrated ANAP-dependent full-length Nav1.5 expression, suggesting that ANAP can be incorporated into the inactivation gate. Furthermore, ANAP fluorescence signal can be visualized and distinguished from background by routine confocal imaging.
Electrophysiology of ANAP incorporated full-length and ΔCT truncated Nav1.5 in mammalian cells
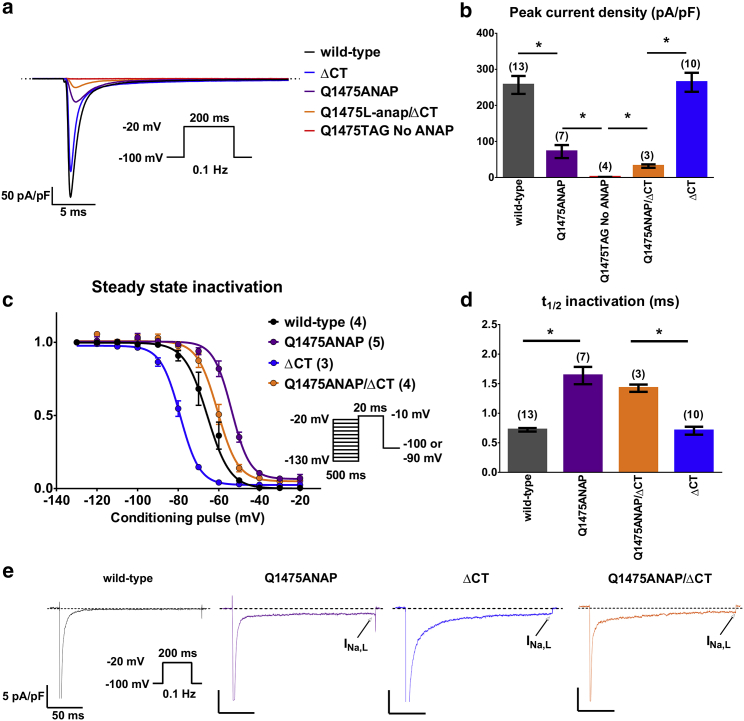
We asked whether functional channel expression at the plasma membrane depended on the presence of ANAP and its incorporation machinery. Furthermore, because the Nav1.5 inactivated state is stabilized by helix 6 in the CT structured region, we sought to measure the effect of truncating helix 6 and the entire distal CT on Nav1.5(ANAP) function (8, 20, 21, 22, 23). We used the previously characterized truncation mutation S1883TGA (ΔCT) (8). Patch-clamp electrophysiology was used to characterize Nav1.5(Q1475ANAP), Nav1.5(ΔCT), or Nav1.5(Q1475ANAP/ΔCT) coexpressed with EGFP(Y40ANAP) in HEK293T cells. EGFP(Y40ANAP) was a marker of cells able to carry out ncAA incorporation. In +ANAP conditions, TTX-sensitive sodium currents were observed in EGFP-positive cells (Fig. 3 a), although peak current density was significantly decreased compared to wild-type channels (Fig. 3 b). Decreased peak current density of ANAP-incorporated channels could be due to defects in channel trafficking to the plasma membrane, lower channel expression, or a decrease in single-channel conductance. Notably, small or no currents were observed in cells expressing all components for ANAP incorporation but grown in the absence of ANAP (Fig. 3, a and b). This suggests that sodium currents observed in the presence of ANAP are dependent on ANAP incorporation and represent full-length Nav1.5(Q1475ANAP) or truncated Nav1.5(Q1475ANAP/ΔCT) channels.
Figure 3.
Electrophysiology of ANAP incorporated full-length and ΔCT truncated Nav1.5 in HEK293T cells. (a) Representative TTX-sensitive capacitance-normalized current traces of wild-type Nav1.5, Nav1.5(ΔCT), Nav1.5(Q1475ANAP), or Nav1.5(Q1475ANAP/ΔCT) expressed in HEK293T is shown. All conditions were co-transfected with AnapRS-LeutRNACUA. Inset is the voltage protocol. (b) Average peak current density (pA/pF) of all conditions is shown. Error bars are ± SEM. Numbers above bars are the number of cells. ∗(left to right) two-tailed p = 8.5 × 10−5, 1.7 × 10−5, 0.02, 5.6 × 10−6, Student’s t-test. Peak currents and cell capacitances are in Fig. S1. (c) SSI of all constructs ± 10 μM ANAP is shown. Error bars are ± SEM. Inset is the voltage protocol. Numbers in legend refer to number of cells. V1/2: WT −64.6 ± 0.4 mV, Q1475ANAP −51.8 ± 0.7 mV, ΔCT −79.2 ± 0.4 mV, Q1475ANAP/ΔCT −59.7 ± 0.6 mV (d) Time to half-complete inactivation (t1/2, ms) of all constructs is shown. Error bars are ± SEM. Numbers above bars are the number of cells. ∗(left to right) two-tailed p = 0.0009, 0.0002, Student’s t-test. (e) High-gain TTX-sensitive capacitance-normalized traces of all constructs using same voltage protocol as in (a) are shown. Arrow indicates late sodium current (INa,L). INa,L (% peak current, late current density): WT (0.3%, 1 pA/pF), Q1475ANAP (1.3%, 0.9 pA/pF), ΔCT (0.46%,1.2 pA/pF), Q1475ANAP/ΔCT (3.1%, 0.8 pA/pF). To see this figure in color, go online.
Next, the inactivation properties of Nav1.5(Q1475ANAP) and Nav1.5(Q1475ANAP/ΔCT) channels were characterized. Macroscopically, currents resembled Nav channels, with rapid opening and inactivation in response to a step change in membrane depolarization. There was lengthened time to half-complete inactivation (t1/2) and increased late current (INa,L) in cells expressing Nav1.5(Q1475ANAP), Nav1.5(ΔCT), or Nav1.5(Q1475ANAP/ΔCT) (Fig. 3, d and e). The SSI curve of Nav1.5(Q1475ANAP), a measure of voltage-dependent transitions between the closed and closed-inactivated states, was shifted toward more depolarized potentials relative to the wild type, suggesting destabilized inactivation and thus, an increase in availability of these channels to open and inactivate (Fig. 3 c). The SSI curve of Nav1.5(ΔCT) was shifted toward hyperpolarized potentials relative to the wild type (Fig. 3 c). Incorporation of ANAP at Q1475 resulted in a depolarizing shift of the SSI curve relative to the wild type, and the combination of Q1475ANAP/ΔCT counteracted the depolarizing shift (Fig. 3 c). Together, electrophysiological data suggest that ANAP can be incorporated at Q1475 in the Nav1.5 inactivation gate to make functional channels. Although channel function is different from the wild type, it is within pathophysiological range (51, 52, 53).
Spectral imaging of Nav1.5ANAP enables sensing of hydrophobicity in intracellular environment
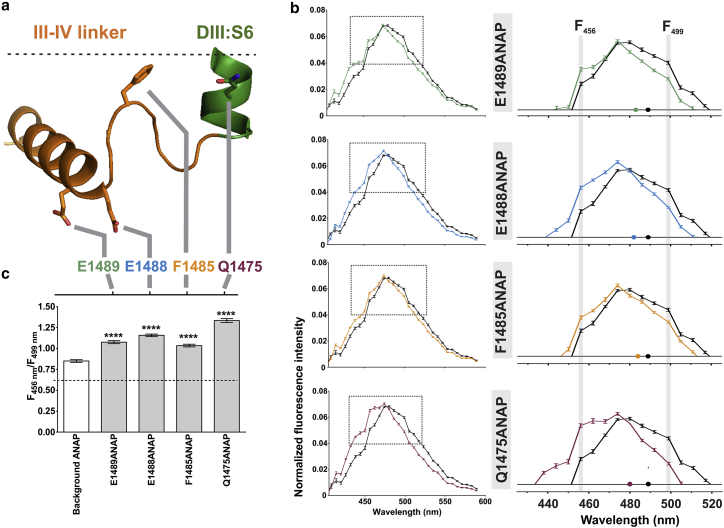
We then asked if ANAP environmentally sensitive fluorescence could be measured in mammalian cells. Spectral imaging was used to measure ANAP fluorescence emission and spectral shifts in mammalian cells. Four different sites were tested for ANAP incorporation into Nav1.5(TAG) expressed in HEK293T (Fig. 4 a). ANAP spectra associated with each site demonstrated unique emission peaks: Nav1.5(Q1475ANAP) (centroid 480 nm), Nav1.5(F1485ANAP) (centroid 484 nm), Nav1.5(E1488ANAP) (centroid 482 nm), and Nav1.5(E1489ANAP) (centroid 483 nm) (Fig. 4 b). Background ANAP in the presence of wild-type Nav1.5 was red-shifted with a peak centroid of 489 nm. The fluorescence ratio F456 nm/F499 nm was significantly different between sites, with Nav1.5(Q1475ANAP) being the most blue-shifted (high F456 nm/F499 nm) and F1485ANAP (low F456 nm/F499 nm) the most red-shifted (Fig. 4 c). As a test for the reliability of observations using spectral imaging and separation of signal spectrum from laser light, results were compared between biological replicates, and the resulting shifts were not significantly different (Fig. S3). A blue shift represents a more hydrophobic environment. Thus, Nav1.5(Q1475ANAP) was situated in the most hydrophobic environment; the location of this site at the end of DIII-S6 lends itself to an expectation of a more hydrophobic environment (26, 30). Nav1.5(F1485ANAP) was in a relatively less hydrophobic environment. Background ANAP was red-shifted and therefore was situated in the least hydrophobic environment. These data represent sensing of hydrophobicity in the local environment of ANAP incorporated in the inactivation gate of full-length Nav1.5 expressed in mammalian cells.
Figure 4.
ANAP spectral imaging afforded detection of hydrophobicity in intracellular environment. (a) A schematic of the inactivation gate depicting the location of the four sites tested, Q1475, F1485, E1488, and E1489is given. F1485 is a part of the IFM motif, known as the inactivation particle. Approximate location of the membrane is depicted by the dashed line. Structure adapted from Protein Data Bank: 5XSY (27). (b) ANAP spectra, laser line background-subtracted and normalized to the sum of intensities under each peak, are shown. ANAP at each of the four sites is compared with background ANAP control (wild-type Nav1.5+10 μM ANAP, black). Dashed box shows inset in which spectra are zoomed in on the peak. n = 12 cells for each condition, representative of two experiments, error bars are mean ± SE. (c) Fluorescence ratio F456 nm/F499 nm measured for each condition, error bars are mean ± SE. ∗∗∗∗p < 0.0001, ANOVA with Tukey’s multiple comparisons test. To see this figure in color, go online.
A spectral imaging assay allows measurement of Nav1.5 conformational change in mammalian cells
A spectral imaging assay was developed to detect ANAP fluorescence changes correlated with inactivated states of Nav1.5 channels expressed in mammalian cells. Q1475ANAP was chosen as a proof of principle for the spectral imaging assay because we hypothesized that this site being close to the membrane-cytosol interface would elicit detectable spectral shifts representing changes in hydrophobicity. We explored the effect of the ΔCT mutation on ANAP spectra with K+ depolarization because truncation of the distal CT destabilizes the inactivated state, causing a marked shift in the voltage-dependence of SSI (8, 23). We hypothesized that these changes would cause a conformational change that ANAP fluorescence shifting would reflect.
Spectral imaging of ANAP was combined with K+ depolarization such that the sodium channel state could be changed from closed to inactivated during imaging. To control membrane potential without voltage-clamp, we used Kir4.1, which is sensitive to [K+]ext and thus enables 1) establishment of the initial RMP at a hyperpolarized potential such that VGSCs will be closed and available and 2) steady-state depolarization of the RMP upon addition of increased [K+]ext to drive the transition between VGSC closed and inactivated states (54, 55, 56). Based on SSI curves of Nav1.5(Q1475ANAP) (Fig. 3 c), when the resting membrane potential (RMP) is set to a hyperpolarized potential, channels at the membrane are expected to be in the resting, closed-available state (Fig. 1 c). With a depolarization of the membrane, channels are expected to open and quickly inactivate or undergo transitions to slow-inactivated and/or closed-inactivated states, resulting in a mixed population of inactivated channels at steady state (Fig. 1 c) (57).
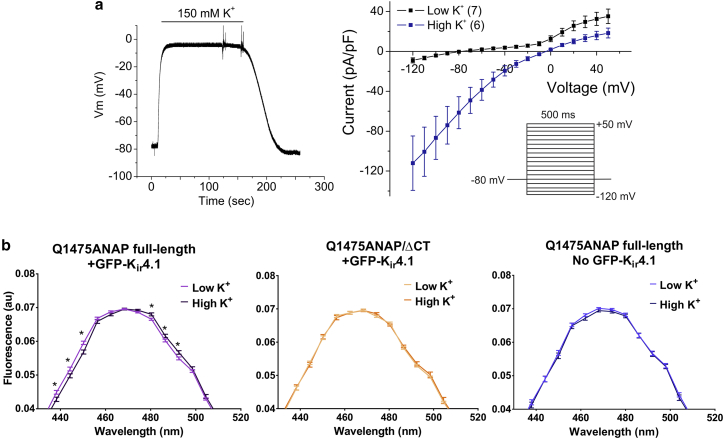
A stable HEK293T cell line expressing GFP-Kir4.1 was obtained to control RMP during spectral imaging experiments. A representative current-clamp trace demonstrates that transition from 5 mM [K+]ext (low K+) to 150 mM [K+]ext (high K+) changed the membrane potential from −76 to −5 mV (Fig. 5 a, left). The reversal potential of these cells was shifted from approximately −80 to 0 mV with increased [K+]ext (Fig. 5 a, right). Expression of Nav1.5(ANAP) in the GFP-Kir4.1 HEK293T cell line resulted in composite ANAP-GFP spectra that were unmixed and fitted to measure shifting ANAP peaks (Fig. S4). Dual-fluorescent ANAP- and GFP-positive cells were manually chosen for spectral analysis. The spectrum from each cell was individually unmixed from laser-line background and GFP.
Figure 5.
A K+-depolarization ANAP spectral imaging assay of Nav1.5 conformational change. (a) Establishment of K+ depolarization to control RMP is shown. (Left) Representative current-clamp trace of GFP-Kir4.1 HEK293T stable cells is shown. Range in Vm is −80 to 0 mV with increasing [K+]ext from 5 to 150 mM. (Right) Reversal potential of GFP-Kir4.1 cells can be changed from −80 mV in low K+ (5 mM [K+]ext) to ∼0 mV in high K+ (150 mM [K+]ext). Error bars are ± SEM. Inset is the voltage protocol. (b) Spectra of full-length Nav1.5(Q1475ANAP) (left) and Nav1.5(Q1475ANAP/ΔCT) (middle) in cells expressing GFP-Kir4.1 and Nav1.5(Q1475ANAP) in cells with no GFP-Kir4.1 (AGFP/SAR < 0.3) (right), at low K+ and high K+, are shown. Q1475ANAP low K+, n = 36 cells; Q1475ANAP high K+, n = 30 cells; Q1475ANAP/ΔCT low K+, n = 21 cells; Q1475ANAP/ΔCT high K+, n = 19 cells; Q1475ANAP no GFP-Kir4.1 low K+, n = 18 cells; Q1475ANAP no GFP-Kir4.1 high K+, n = 16 cells. Error bars are ± SE of the average relative brightness of the channel. Spectra are zoomed in on the peak. ∗p-values (left to right) full-length Nav1.5(Q1475ANAP): p = 0.02, 0.02, 0.0009, 0.008, 0.04, 0.01, Student’s t-test. To see this figure in color, go online.
To enhance the reliability of unmixing of spectra and to isolate cells expressing GFP-Kir4.1, selection criteria were introduced in postprocessing. First, as a goodness-of-fit measure, the summed absolute residuals (SARs) after the fit were compared to the total intensity (TI) in the spectrum. A threshold in the SAR/TI ratio was identified empirically as one at which the SD of the shift in the fitted ANAP peak rapidly started to increase (Fig. S6 a). Cells with SAR/TI < 0.04 were excluded from further processing. Second, spectra from reliably expressing GFP-Kir4.1 cells were selected by comparing the amplitude of the fitted GFP peak (AGFP) to the SAR. The AGFP/SAR ratio below which the GFP signal was comparable to the residuals was deemed the threshold for accepting a positive identification of GFP-Kir4.1 expression, at AGFP/SAR > 0.3 (Fig. S6, b and c). Because certain spectra showed high expression of GFP with relatively low ANAP signals, a further criterion was introduced to ensure that a well-resolved ANAP signal existed—ANAP peak amplitude (APA)/SAR ratio above APA/SAR > 1.
Spectra of Nav1.5(Q1475ANAP) show ANAP fluorescence shifts depending on [K+]ext and presence of the distal CT (Fig. 5 b). GFP peaks were unaltered in the same experiments (Fig. S5). K+ depolarization caused a small but significant red shift in the ANAP spectra of full-length Nav1.5(Q1475ANAP) (Fig. 5 b, left). Notably, K+ depolarization caused no change in ANAP spectra of ΔCT channels (Fig. 5 b, center). The red shift was not observed in cells expressing Nav1.5(Q1475ANAP) but failing the GFP threshold test, validating that the spectral shift depends on the presence of GFP-Kir1.4 and thus on the membrane potential (Fig. 5 b, right). These data suggest the distal CT plays a role in mediating inactivation gate conformational change associated with inactivation. A general ANAP spectral imaging K+ depolarization assay for probing Nav1.5 conformational change in channels expressed in mammalian cells has been established.
Discussion
Site-specific incorporation of the environmentally sensitive fluorescent ncAA ANAP into the Nav1.5 intracellular inactivation gate was achieved using synthetase-tRNA technology in mammalian cells. ANAP incorporated into a fluorescent reporter Nav1.5(Q1475TAG)-mCherry was imaged, demonstrating ANAP and mCherry fluorescence localization at the cell membrane and intracellular compartments (Fig. 2 b). In Nav1.5(Q1475ANAP)-mCherry imaging experiments, ANAP background fluorescence was observed and may result from incomplete washout of unincorporated ANAP. Also, ANAP incorporation could potentially occur at nontarget amber codons. In the absence of ANAP, autofluorescence was observed in the ANAP channel, and dim mCherry fluorescence was observed in the mCherry channel. Collected ANAP emission was in the blue visible light spectrum in which autofluorescence is expected because of natural amino acids and other biomolecules such as NADH and NADPH (58, 59). mCherry background fluorescence may result from ANAP-independent readthrough of the target stop codon or initiation of mCherry expression. It is notable that autofluorescence and background mCherry fluorescence signals were negligible compared to fluorescence signals when ANAP and its target site of incorporation were present (Fig. 2, b and c).
Patch-clamp electrophysiology revealed ncAA-dependent sodium currents in cells expressing Nav1.5(Q1475ANAP) or Nav1.5(Q1475ANAP/ΔCT), whereas few to no sodium currents were observed in the absence of ANAP (Fig. 3), further corroborating successful incorporation of the fluorescent ncAA ANAP into functional channels. Electrophysiological characterization of Nav1.5(Q1475ANAP) and Nav1.5(Q1475ANAP/ΔCT) demonstrated decreased peak current density, and for ANAP incorporation, imaging data suggest a considerable population of fluorescent channels was localized in intracellular compartments (Fig. 2 b). Decreased peak current density could represent defective channel trafficking, lower channel expression, or decreased single-channel conductance. Single-channel functional characterization was not performed in this work and would clarify the possible effect of each ncAA on single-channel conductance. However, defective trafficking and lower channel expression are expected to be primary causes of decreased peak current density. Lower expression of ncAA-incorporated channels is expected because of competition with endogenous release factors at the site of incorporation (31). Defects in channel trafficking to the membrane are known to occur in the absence of β-subunit overexpression and could be compounded by an unknown trafficking effect associated with ncAA-incorporated channels (50); coexpression with β-subunit would likely improve Nav1.5(ANAP) expression at the membrane and should be employed in future work. Single-amino-acid mutation of this region is sufficient to alter channel function, evidenced by LQT3 mutations and the work herein (51). Although channel function was altered by ANAP incorporation into the inactivation gate, properties remained within pathophysiological range and most closely mimicked those of a characterized LQT3 mutant channel F1473C (51). The method developed herein facilitates experiments with LQT3 mutant channels that could elucidate Nav1.5 structural dynamics in a pathological context.
Spectral imaging was used to measure ANAP emission in Nav1.5 in live mammalian cells. Unique ANAP spectra were observed with ANAP incorporated at four different sites in Nav1.5 Q1475, F1485, E1488 and E1489, and spectra were red-shifted in the absence of a target stop codon for incorporation, indicating sensing of hydrophobicity in local environment of the inactivation gate (Fig. 4). It should be noted that fluorescence from background ANAP (unincorporated or in a channel not in the membrane) cannot be separated from target-incorporated ANAP fluorescence at this point. Therefore, the interpretation of absolute shifts in ANAP spectra cannot be clear, and we restrict our conclusions to relative shifts. Although imaging data suggest a considerable portion of ANAP was localized to intracellular compartments, electrophysiology data showed that a detectable population of functional channels reached the plasma membrane in an ANAP-dependent fashion. Spectra from the membrane-only versus membrane and cytosol exhibited no observable difference in our setup, so spectra from the whole cell were analyzed to maximize ANAP signal (Fig. S2).
This study provided proof-of-principle evidence of a K+ depolarization ANAP spectral imaging assay that was developed to detect effects on conformational rearrangement of the inactivation gate in response to a change in membrane potential without patch-clamp. For this purpose, experiments were performed to test the specific effects of the ΔCT and channel state. K+ depolarization allowed sodium channel state at the plasma membrane to be changed from closed to a mixture of open- and closed-inactivated states in a steady-state fashion during spectral imaging (Fig. 1 c). The timescale of the K+ depolarization assay was significantly longer than that of fast inactivation; therefore, it is more likely the assay detected intermediate- or slow-inactivated states (57). When channels are inactivated and the CT is intact, a change in the position of DIII-S6 (relative to resting) may increase hydrophobicity in the environment of Q1475ANAP, leading to a subtle red shift in the ANAP spectra (Fig. 5 b, left). The change in position of DIII-S6 may lead to a change in the affinity of the III-IV linker-pore complex, destabilizing the inactivation gate. ANAP spectra were unshifted in ΔCT channels with K+ depolarization, which may suggest that the absence of the distal CT can prevent entry into inactivated states, although it is possible there is a conformational change that is undetectable with this experimental setup.
Simultaneous electrophysiology and fluorescence measurements in mammalian cells would definitively link ANAP spectral shifts to Nav1.5 inactivation dynamics, and characterization of all the ANAP-incorporated sites in the K+ depolarization assay would further validate the method. The ability to label the inactivation gate and measure fluorescence spectral shifts associated with VGSC conformational change is an invaluable method setting the foundation for such future studies. Notably, the presented method could be used to screen environmentally sensitive positions in Nav1.5 or other channels in mammalian cells as a first-pass substitute for patch-clamp fluorometry.
Conclusions
ANAP was incorporated into the Nav1.5 inactivation gate in mammalian cells, resulting channel function was modestly perturbed, and ANAP fluorescence was sensitive to the intracellular environment, as well as to channel state. Site-specific fluorescent incorporation of ANAP combined with K+ depolarization enabled direct monitoring of Nav1.5 inactivation gate conformational rearrangement in channels expressed in live mammalian cells. The K+ depolarization ANAP spectral assay is a general assay for studying conformational dynamics of the Nav1.5 inactivation gate and regulators thereof in mammalian cells. It could provide insight on outstanding questions of VGSC regulation, for instance, by calcium, kinases (PKA, CAMKII, Fyn), and growth factors (fibroblast growth factor homologous factor) (60). Furthermore, the development of such an assay can similarly be applied to other ion channels and membrane proteins to gain site-specific knowledge of their conformational dynamics in mammalian cells.
Author Contributions
M.A.S., V.W.C., and R.S.K. devised the project. M.A.S. designed and performed experiments and wrote the manuscript. J.R.Q. contributed to experimental design and performed experiments. M.Y. contributed to experimental design. V.W.C. and R.S.K. designed experiments and supervised research. All authors reviewed the final results and edited the manuscript.
Acknowledgments
We thank Istvan Cziegler for assistance with imaging data analysis, Cecile Terrenoire and Seth Robey for assistance with electrophysiology experiments, Kevin Sampson and Manu Ben-Johny for assistance with the manuscript, and Zhixing Chen, Casey Brown, and Chaoran Jing for insightful conversations. We thank Christopher Ahern for the GFP-Kir4.1 stable cell line. We thank Jenny Rao for technical assistance.
M.A.S. was supported by a Columbia University Research Initiatives in Science and Engineering award. Images were collected in the Confocal and Specialized Microscopy Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University, supported by National Institutes of Health grant #P30 CA013696 (National Cancer Institute). The confocal microscope was purchased with National Institutes of Health grant #S10 RR025686. R.S.K. was supported by National Institutes of Health grant HL 123453.
Editor: Christopher Ahern.
Footnotes
Mia A. Shandell’s present address is York Biomedical Research Institute, Hull York Medical School, University of York, York, UK.
Supporting Material can be found online at https://doi.org/10.1016/j.bpj.2019.08.028.
Contributor Information
Mia A. Shandell, Email: mia.shandell@york.ac.uk.
Virginia W. Cornish, Email: vc114@columbia.edu.
Robert S. Kass, Email: rsk20@cumc.columbia.edu.
Supporting Material
References
- 1.Catterall W.A. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- 2.Kellenberger S., West J.W., Scheuer T. Molecular analysis of potential hinge residues in the inactivation gate of brain type IIA Na+ channels. J. Gen. Physiol. 1997;109:607–617. doi: 10.1085/jgp.109.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kellenberger S., West J.W., Catterall W.A. Molecular analysis of the putative inactivation particle in the inactivation gate of brain type IIA Na+ channels. J. Gen. Physiol. 1997;109:589–605. doi: 10.1085/jgp.109.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kellenberger S., Scheuer T., Catterall W.A. Movement of the Na+ channel inactivation gate during inactivation. J. Biol. Chem. 1996;271:30971–30979. doi: 10.1074/jbc.271.48.30971. [DOI] [PubMed] [Google Scholar]
- 5.Smith M.R., Goldin A.L. Interaction between the sodium channel inactivation linker and domain III S4-S5. Biophys. J. 1997;73:1885–1895. doi: 10.1016/S0006-3495(97)78219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West J.W., Patton D.E., Catterall W.A. A cluster of hydrophobic amino acid residues required for fast Na(+)-channel inactivation. Proc. Natl. Acad. Sci. USA. 1992;89:10910–10914. doi: 10.1073/pnas.89.22.10910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaholtz G., Scheuer T., Catterall W.A. Restoration of inactivation and block of open sodium channels by an inactivation gate peptide. Neuron. 1994;12:1041–1048. doi: 10.1016/0896-6273(94)90312-3. [DOI] [PubMed] [Google Scholar]
- 8.Motoike H.K., Liu H., Kass R.S. The Na+ channel inactivation gate is a molecular complex: a novel role of the COOH-terminal domain. J. Gen. Physiol. 2004;123:155–165. doi: 10.1085/jgp.200308929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McPhee J.C., Ragsdale D.S., Catterall W.A. A critical role for transmembrane segment IVS6 of the sodium channel α subunit in fast inactivation. J. Biol. Chem. 1995;270:12025–12034. doi: 10.1074/jbc.270.20.12025. [DOI] [PubMed] [Google Scholar]
- 10.Lerche H., Peter W., Lehmann-Horn F. Role in fast inactivation of the IV/S4-S5 loop of the human muscle Na+ channel probed by cysteine mutagenesis. J. Physiol. 1997;505:345–352. doi: 10.1111/j.1469-7793.1997.345bb.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filatov G.N., Nguyen T.P., Barchi R.L. Inactivation and secondary structure in the D4/S4-5 region of the SkM1 sodium channel. J. Gen. Physiol. 1998;111:703–715. doi: 10.1085/jgp.111.6.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McPhee J.C., Ragsdale D.S., Catterall W.A. A critical role for the S4-S5 intracellular loop in domain IV of the sodium channel α-subunit in fast inactivation. J. Biol. Chem. 1998;273:1121–1129. doi: 10.1074/jbc.273.2.1121. [DOI] [PubMed] [Google Scholar]
- 13.Tang L., Chehab N., Kallen R.G. Glutamine substitution at alanine1649 in the S4-S5 cytoplasmic loop of domain 4 removes the voltage sensitivity of fast inactivation in the human heart sodium channel. J. Gen. Physiol. 1998;111:639–652. doi: 10.1085/jgp.111.5.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Veldkamp M.W., Viswanathan P.C., Balser J.R. Two distinct congenital arrhythmias evoked by a multidysfunctional Na(+) channel. Circ. Res. 2000;86:E91–E97. doi: 10.1161/01.res.86.9.e91. [DOI] [PubMed] [Google Scholar]
- 15.Bezzina C., Veldkamp M.W., Wilde A.A. A single Na(+) channel mutation causing both long-QT and Brugada syndromes. Circ. Res. 1999;85:1206–1213. doi: 10.1161/01.res.85.12.1206. [DOI] [PubMed] [Google Scholar]
- 16.Rivolta I., Abriel H., Kass R.S. Inherited Brugada and long QT-3 syndrome mutations of a single residue of the cardiac sodium channel confer distinct channel and clinical phenotypes. J. Biol. Chem. 2001;276:30623–30630. doi: 10.1074/jbc.M104471200. [DOI] [PubMed] [Google Scholar]
- 17.An R.H., Wang X.L., Kass R.S. Novel LQT-3 mutation affects Na+ channel activity through interactions between α- and β1-subunits. Circ. Res. 1998;83:141–146. doi: 10.1161/01.res.83.2.141. [DOI] [PubMed] [Google Scholar]
- 18.Tateyama M., Liu H., Kass R.S. Structural effects of an LQT-3 mutation on heart Na+ channel gating. Biophys. J. 2004;86:1843–1851. doi: 10.1016/S0006-3495(04)74251-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mantegazza M., Yu F.H., Scheuer T. Role of the C-terminal domain in inactivation of brain and cardiac sodium channels. Proc. Natl. Acad. Sci. USA. 2001;98:15348–15353. doi: 10.1073/pnas.211563298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glaaser I.W., Bankston J.R., Kass R.S. A carboxyl-terminal hydrophobic interface is critical to sodium channel function. Relevance to inherited disorders. J. Biol. Chem. 2006;281:24015–24023. doi: 10.1074/jbc.M605473200. [DOI] [PubMed] [Google Scholar]
- 21.Glaaser I.W., Osteen J.D., Kass R.S. Perturbation of sodium channel structure by an inherited Long QT Syndrome mutation. Nat. Commun. 2012;3:706. doi: 10.1038/ncomms1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bankston J.R., Sampson K.J., Kass R.S. A novel LQT-3 mutation disrupts an inactivation gate complex with distinct rate-dependent phenotypic consequences. Channels (Austin) 2007;1:273–280. doi: 10.4161/chan.4956. [DOI] [PubMed] [Google Scholar]
- 23.Cormier J.W., Rivolta I., Kass R.S. Secondary structure of the human cardiac Na+ channel C terminus: evidence for a role of helical structures in modulation of channel inactivation. J. Biol. Chem. 2002;277:9233–9241. doi: 10.1074/jbc.M110204200. [DOI] [PubMed] [Google Scholar]
- 24.Sarhan M.F., Tung C.C., Ahern C.A. Crystallographic basis for calcium regulation of sodium channels. Proc. Natl. Acad. Sci. USA. 2012;109:3558–3563. doi: 10.1073/pnas.1114748109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C., Chung B.C., Pitt G.S. Crystal structure of the ternary complex of a NaV C-terminal domain, a fibroblast growth factor homologous factor, and calmodulin. Structure. 2012;20:1167–1176. doi: 10.1016/j.str.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen H., Zhou Q., Yan N. Structure of a eukaryotic voltage-gated sodium channel at near-atomic resolution. Science. 2017;355:eaal4326. doi: 10.1126/science.aal4326. [DOI] [PubMed] [Google Scholar]
- 27.Yan Z., Zhou Q., Yan N. Structure of the Nav1.4-β1 complex from electric eel. Cell. 2017;170:470–482.e11. doi: 10.1016/j.cell.2017.06.039. [DOI] [PubMed] [Google Scholar]
- 28.Shen H., Liu D., Yan N. Structures of human Nav1.7 channel in complex with auxiliary subunits and animal toxins. Science. 2019;363:1303–1308. doi: 10.1126/science.aaw2493. [DOI] [PubMed] [Google Scholar]
- 29.Pan X., Li Z., Yan N. Molecular basis for pore blockade of human Na+ channel Nav1.2 by the μ-conotoxin KIIIA. Science. 2019;363:1309–1313. doi: 10.1126/science.aaw2999. [DOI] [PubMed] [Google Scholar]
- 30.Clairfeuille T., Cloake A., Payandeh J. Structural basis of α-scorpion toxin action on Nav channels. Science. 2019;363:eaav8573. doi: 10.1126/science.aav8573. [DOI] [PubMed] [Google Scholar]
- 31.Chin J.W. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 32.Lee H.S., Guo J., Schultz P.G. Genetic incorporation of a small, environmentally sensitive, fluorescent probe into proteins in Saccharomyces cerevisiae. J. Am. Chem. Soc. 2009;131:12921–12923. doi: 10.1021/ja904896s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chatterjee A., Guo J., Schultz P.G. A genetically encoded fluorescent probe in mammalian cells. J. Am. Chem. Soc. 2013;135:12540–12543. doi: 10.1021/ja4059553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen B.E., McAnaney T.B., Jan L.Y. Probing protein electrostatics with a synthetic fluorescent amino acid. Science. 2002;296:1700–1703. doi: 10.1126/science.1069346. [DOI] [PubMed] [Google Scholar]
- 35.Kalstrup T., Blunck R. Dynamics of internal pore opening in K(V) channels probed by a fluorescent unnatural amino acid. Proc. Natl. Acad. Sci. USA. 2013;110:8272–8277. doi: 10.1073/pnas.1220398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aman T.K., Gordon S.E., Zagotta W.N. Regulation of CNGA1 channel gating by interactions with the membrane. J. Biol. Chem. 2016;291:9939–9947. doi: 10.1074/jbc.M116.723932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Soh M.S., Estrada-Mondragon A., Lynch J.W. Probing the structural mechanism of partial agonism in glycine receptors using the fluorescent artificial amino acid, ANAP. ACS Chem. Biol. 2017;12:805–813. doi: 10.1021/acschembio.6b00926. [DOI] [PubMed] [Google Scholar]
- 38.Sakata S., Jinno Y., Okamura Y. Voltage-dependent motion of the catalytic region of voltage-sensing phosphatase monitored by a fluorescent amino acid. Proc. Natl. Acad. Sci. USA. 2016;113:7521–7526. doi: 10.1073/pnas.1604218113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dai G., James Z.M., Zagotta W.N. Dynamic rearrangement of the intrinsic ligand regulates KCNH potassium channels. J. Gen. Physiol. 2018;150:625–635. doi: 10.1085/jgp.201711989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai G., Zagotta W.N. Molecular mechanism of voltage-dependent potentiation of KCNH potassium channels. eLife. 2017;6:e26355. doi: 10.7554/eLife.26355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wulf M., Pless S.A. High-sensitivity fluorometry to resolve ion channel conformational dynamics. Cell Reports. 2018;22:1615–1626. doi: 10.1016/j.celrep.2018.01.029. [DOI] [PubMed] [Google Scholar]
- 42.Zagotta W.N., Gordon M.T., Gordon S.E. Measuring distances between TRPV1 and the plasma membrane using a noncanonical amino acid and transition metal ion FRET. J. Gen. Physiol. 2016;147:201–216. doi: 10.1085/jgp.201511531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wen M., Guo X., Tian C. Site-specific fluorescence spectrum detection and characterization of hASIC1a channels upon toxin mambalgin-1 binding in live mammalian cells. Chem. Commun. (Camb.) 2015;51:8153–8156. doi: 10.1039/c5cc01418b. [DOI] [PubMed] [Google Scholar]
- 44.Puljung M., Vedovato N., Ashcroft F. Activation mechanism of ATP-sensitive K+ channels explored with real-time nucleotide binding. eLife. 2019;8:e41103. doi: 10.7554/eLife.41103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalstrup T., Blunck R. Voltage-clamp fluorometry in xenopus oocytes using fluorescent unnatural amino acids. J. Vis. Exp. 2017;123:e55598. doi: 10.3791/55598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsieh T.Y., Nillegoda N.B., Kramer G. Monitoring protein misfolding by site-specific labeling of proteins in vivo. PLoS One. 2014;9:e99395. doi: 10.1371/journal.pone.0099395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitchell A.L., Addy P.S., Chatterjee A. A unique genetically encoded FRET pair in mammalian cells. ChemBioChem. 2017;18:511–514. doi: 10.1002/cbic.201600668. [DOI] [PubMed] [Google Scholar]
- 48.Chen P.R., Groff D., Schultz P.G. A facile system for encoding unnatural amino acids in mammalian cells. Angew. Chem. Int. Engl. 2009;48:4052–4055. doi: 10.1002/anie.200900683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bacalum M., Zorilă B., Radu M. Fluorescence spectra decomposition by asymmetric functions: laurdan spectrum revisited. Anal. Biochem. 2013;440:123–129. doi: 10.1016/j.ab.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 50.Kruger L.C., Isom L.L. Voltage-gated Na+ channels: not just for conduction. Cold Spring Harb. Perspect. Biol. 2016;8:a029264. doi: 10.1101/cshperspect.a029264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bohnen M.S., Peng G., Kass R.S. Molecular pathophysiology of congenital long QT syndrome. Physiol. Rev. 2017;97:89–134. doi: 10.1152/physrev.00008.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bankston J.R., Yue M., Kass R.S. A novel and lethal de novo LQT-3 mutation in a newborn with distinct molecular pharmacology and therapeutic response. PLoS One. 2007;2:e1258. doi: 10.1371/journal.pone.0001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Popa M.O., Alekov A.K., Lerche H. Cooperative effect of S4-S5 loops in domains D3 and D4 on fast inactivation of the Na+ channel. J. Physiol. 2004;561:39–51. doi: 10.1113/jphysiol.2004.065912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kirkton R.D., Bursac N. Engineering biosynthetic excitable tissues from unexcitable cells for electrophysiological and cell therapy studies. Nat. Commun. 2011;2:300. doi: 10.1038/ncomms1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dhamoon A.S., Jalife J. The inward rectifier current (IK1) controls cardiac excitability and is involved in arrhythmogenesis. Heart Rhythm. 2005;2:316–324. doi: 10.1016/j.hrthm.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 56.Chang H.K., Lee J.R., Shieh R.C. The extracellular K+ concentration dependence of outward currents through Kir2.1 channels is regulated by extracellular Na+ and Ca2+ J. Biol. Chem. 2010;285:23115–23125. doi: 10.1074/jbc.M110.121186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Silva J.R., Goldstein S.A. Voltage-sensor movements describe slow inactivation of voltage-gated sodium channels II: a periodic paralysis mutation in Na(V)1.4 (L689I) J. Gen. Physiol. 2013;141:323–334. doi: 10.1085/jgp.201210910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blacker T.S., Duchen M.R. Investigating mitochondrial redox state using NADH and NADPH autofluorescence. Free Radic. Biol. Med. 2016;100:53–65. doi: 10.1016/j.freeradbiomed.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Teale F.W., Weber G. Ultraviolet fluorescence of the aromatic amino acids. Biochem. J. 1957;65:476–482. doi: 10.1042/bj0650476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abriel H., Kass R.S. Regulation of the voltage-gated cardiac sodium channel Nav1.5 by interacting proteins. Trends Cardiovasc. Med. 2005;15:35–40. doi: 10.1016/j.tcm.2005.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.