Abstract
Background
Hypoxic preconditioning alters the biological properties of mesenchymal stem cells (MSCs). It is not known whether this process has an effect on circular RNAs (circRNAs) in MSCs.
Methods
Human placental chorionic plate‐derived MSCs (hpcpMSCs) isolated from the same placentae were classed into two groups: hypoxic pretreated (hypoxia) group and normally cultured (normoxia) group. The comparative circRNA microarray analysis was used to determine circRNAs expression and verified by quantitative reverse‐transcription polymerase chain reaction (qRT‐PCR) in the two groups.
Results
One hundred and two differentially expressed circRNAs in the hypoxia group were found compared to that in the normoxia group (fold change >1.5‐fold and P < 0.05). The expression levels of circRNAs by qRT‐PCR were consistent with those evaluated by microarray analysis. Gene ontology (GO) analysis showed that the putative function of their target genes for those differentially expressed circRNAs was primarily involved in cell development and its differentiation and regulation. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis revealed that transcriptional misregulation in cancer and mitogen‐activated protein kinase (MAPK) signaling pathway were the most significant. MAPK signaling pathway was found to be the core regulatory pathway triggered by hypoxia.
Conclusions
The results indicate that the altered expression of specific circRNAs in MSCs is associated with hypoxic preconditioning. This finding provides further exploration of underlying mechanisms of the characteristic changes of MSCs with hypoxic preconditioning.
Abbreviations
- circRNAs
circular RNAs
- ERK
extracellular signal‐regulated kinase
- ERβ
estrogen receptor β
- GO
Gene Ontology
- hpcpMSCs
human placental chorionic plate‐derived MSCs
- JNK
c‐Jun N‐terminal kinase
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MAPK
mitogen‐activated protein kinase
- MSCs
mesenchymal stem cells
- qRT‐PCR
quantitative reverse transcription‐polymerase chain reaction
1. INTRODUCTION
Mesenchymal stem cells (MSCs) are adult stem cells that are present in various tissues of the human body and are characterized by self‐renewal, multiple differentiation potential, immunosuppression, and easy transfection. These characteristics make MSCs the ideal seed cells for regenerative medicine and as gene therapy vectors.1, 2 MSCs are the second type of adult stem cells used for clinical treatment. There are many types of MSC‐based therapies for the treatment of various diseases, and additional preclinical and clinical studies are underway. Currently, there are 864 clinical trials involving MSCs registered at clinicaltrials.gov. Most studies have used bone marrow‐derived MSCs, but placenta‐derived MSCs are easy to isolate and culture. Placental tissue provides an ideal source of MSCs due to its large size and the fact that it can be acquired without any ethical issues.3 hpcpMSCs have better expansion potential and homogeneity than other sources of MSCs.4, 5 Studies have shown that the biological characteristics of MSCs are affected by various factors present during their isolation and culture. Hypoxia is one of these factors and is known to have certain effects on the cell cycle, proliferation, apoptosis, migration, and differentiation of MSCs.6, 7
CircRNAs are a class of endogenous RNA molecules that do not have a 5′‐end cap and a 3′‐end poly(A) tail. Their ends are linked by the 3′‐5′ phosphodiester covalent bonds to form the circular structures. Sanger et al8 discovered circRNAs in the 1970s. However, the number of circRNA studies has decreased after several decades. Advancement of high‐throughput sequencing technology and bioinformatics has enabled circRNAs to recently become one of the research hot spots in the field of biomedicine.9 Studies have shown that irregular expression of circRNAs is associated with an imbalance in certain homeostatic processes; moreover, circRNAs participate in the pathophysiological process of aging, development, and disease progression.10, 11 Several studies have demonstrated that circRNAs are abundant in the stem cells and can act as a molecular “sponge” to regulate the characteristics and differentiation of the stem cells.12, 13, 14 As reported by Yu et al,15 circBIRC6 competitively binds to miR‐34a and miR‐145 and is involved in the regulation of stem cell pluripotency in the embryonic stem cells. A few reports described a role played by circRNAs in physiological processes of MSCs. Maass et al16 reported expression of distinct and specific circRNAs in MSCs as well as in chondrocytes, osteoblasts, and vascular smooth muscle cells differentiated from MSCs. Li et al17 revealed that estrogen receptor β regulates osteogenic differentiation of MSCs through the 2:27713879/27755789/2:240822115/240867796‐miR‐328‐5p‐mRNA axis. Zhang et al18 uncovered numerous circRNAs differentially expressed during BMSC osteogenic differentiation. The parental genes of these circRNAs were associated with bone formation, suggesting that the differentially expressed circRNAs and their putative miRNAs were involved in the osteogenic differentiation of MSCs. The effect of hypoxic preconditioning on the expression of circRNAs in MSCs has not been reported.
In this study, microarray analysis was used to compare circRNAs expressed in MSCs cultured under normoxia and hypoxia conditions. Differential expression of circRNAs was validated by qRT‐PCR. Five highly matched miRNAs were assigned to each differentially expressed circRNA, and the circRNA‐miRNA networks were constructed. The target gene of each predicted miRNA was subjected to GO analysis for functional prediction and pathway enrichment, and the core pathway was analyzed based on the interactions among pathways.
2. MATERIALS AND METHODS
2.1. Cell culture
According to routine laboratory methods, hpcpMSCs were isolated, cultured, and consistent with the MSCs as defined by the International Society for Cell Therapy. The process includes obtaining the placentas after normal cesarean section, sectioning the chorionic plate, isolation of the cells using trypsin digestion (Gibco, Gaithersburg, MD), and inoculation of the cultures in the T75 flasks (Corning, Cambridge, MA). Fibroblast‐like cells were cultured in DMEM/F12 (Gibco) containing 10% serum (Gibco). Cell surface markers were detected with corresponding antibodies (BD Biosciences, San Jose, CA). According to the manufacturer's instructions (Gibco), osteogenic and adipogenic differentiation markers were detected. Then, hpcpMSCs were proven to be purely fetal‐derived MSCs using short tandem repeat PCR (data not shown). After the P3 generation cells reached 80% confluence, cells were cooled by a programmable cooler (Planer, Sunbury, UK) and cryopreserved in liquid nitrogen at 1 × 106 cells/mL/tube. The human placenta protocol was approved by the Ethics Committee of the Guangzhou Women and Children's Medical Center, and the mothers provided the written informed consent. In this study, three tubes of P3 hpcpMSCs from different maternal placentas were rewarmed in a 37‐42°C water bath and then inoculated in the T75 flasks; these P4 cells were incubated at 37°C in a humidified chamber (Labotect, Göttingen, Germany) at 21% O2. The cells were cultured to 80% confluence, digested, passaged, and propagated into the P5 cells at a 1:2 ratio. One T75 flask was placed in a 21% O2 incubator for the normoxia group, and another T75 flask was cultured in a 1% O2 incubator for the hypoxia group.
2.2. RNA extraction
When the P5 hpcpMSCs cells in the hypoxia and normoxia groups reached 70%‐80% confluence, cells were washed with saline solution, and the total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The total RNA was purified by the RNeasy Mini Kit (Qiagen, Hilden, Germany), and the RNA was quantified and analyzed by Nanodrop ND‐1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). The purity and integrity of the RNA were detected by formaldehyde denaturing agarose gel electrophoresis.
2.3. Labeling and microarray hybridization
Microarray hybridization was performed according to the manufacturer's procedure (Arraystar, Rockville, MD). Briefly, the total RNA of 6 specimens was treated with RNase R (Epicentre, Madison, WI) to remove linear RNA molecules and enrich circRNAs. Then, circRNAs were amplified and transcribed into the fluorescently labeled cRNA with random primers using the Arraystar Super RNA Labeling Kit (Arraystar). The fluorescently labeled cRNA was purified using the RNeasy Mini Kit, and the concentration and specific activity of the labeled cRNA were detected by Nanodrop ND‐1000 spectrophotometer. A total of 1 µg of labeled cRNA, 5 μL of 10× blocking agent, and 1 μL of 25× fragmentation buffer were mixed and incubated at 60°C for 30 minutes. Finally, 25 μL of 2× hybridization buffer was added to dilute the fluorescently labeled cRNA. A total of 50 μL of the hybridization solution was loaded on the microarray and incubated at 65°C for 17 hours in an Agilent Hybridization Oven (Agilent Technologies, Santa Clara, CA). The hybridized arrays were fixed after washing and scanned with an Agilent Scanner G2505C (Agilent Technologies).
2.4. Data analysis
The microarray scan images were imported into the Agilent Feature Extraction software (Agilent Technologies). The raw data from the circRNAs microarray were extracted, and the quantile normalization of the raw data and the data analysis were performed using the R software limma package (R Foundation for Statistical Computing, Vienna, Austria). The statistical significance of differences was estimated by paired t test. CircRNAs with fold changes ≥1.5 times and P < 0.05 were defined as being differentially expressed.
2.5. Quantitative reverse transcription‐PCR validation
Nine differentially expressed circRNAs were selected, and their target miRNAs were predicted to be expressed in MSCs and influence the biological characteristics of MSCs. Using β‐actin as an internal reference, qRT‐PCR was performed by the SYBR Green method. The RNA (1 µg) was reverse‐transcribed into cDNA according to the instructions of the PrimeScript reverse transcription kit (Invitrogen), and circRNA expression levels were evaluated on a ViiA 7 real‐time PCR System (Applied Biosystems, Foster City, CA) using specific reaction primers (Appendix S1). All amplification dissolution curves had single peaks, revealing no nonspecific amplification, and the experiment was performed in triplicate. The average Cq value of each sample was recorded. The qRT‐PCR results were analyzed by the relative quantitative 2−ΔΔCq method. The target circRNA expression levels in the hypoxia group were normalized to those in the normoxia group.
2.6. Bioinformatics analysis
For each differentially expressed circRNA, circRNA and microRNA interaction predictions were performed with Arraystar's homemade miRNA target prediction software based on TargetScan & miRanda.19 The miRNA target genes were predicted using the TargetScan 7.1 and mirdb V5 databases. The target gene predictions had to be consistent across both databases. The putative genes were subjected to GO analysis (website http://www.geneontology.org) and to pathway enrichment analysis (website http://www.genome.ad.jp/kegg). The pathway‐pathway interaction network analysis was used to evaluate the interactions among pathways.20 The circRNA‐microRNA networks and the pathway‐pathway interaction networks were generated using the Cytoscape V3.6.0 software (The Cytoscape Consortium, San Diego, CA).
2.7. Statistical analysis
Data are presented as the mean ± standard deviation. All experiments were performed at least three times. The statistical significance of the differences between the groups was estimated with the paired t test. The R software limma package was used for the analysis. P < 0.05 was considered significant.
3. RESULTS
3.1. CircRNA expression profiles
In this study, the hypoxia group included three specimens, and the normoxia group also included three specimens. Out of 13 617 circRNAs on the microarray, the expression profiles of 12 114 human circRNAs were obtained by the microarray analysis using the samples from the hypoxia and the normoxia groups (Appendix S2). Differentially expressed circRNAs were identified using volcano plots and scatter plots based on fold change >1.5 and P < 0.05 (Figure 1A,B). Hierarchical cluster analysis revealed 102 circRNAs differentially expressed between the two groups. Compared with the expression levels of circRNAs in the normoxia group, the expression levels of 85 circRNAs in the hypoxia group were upregulated, and 17 circRNAs were downregulated in the hypoxia group vs the normoxia group; the changes were over twofold in 27 cases, and all these cases were upregulated (Figure 1C and Appendix S3). Among these circRNAs, hsa_circRNA_402986, hsa_circRNA_000317, and hsa_circRNA_062003 were upregulated, whereas hsa_circRNA_000310, hsa_circRNA_403519, and hsa_circRNA_103901 were downregulated with the highest fold differences.
Figure 1.
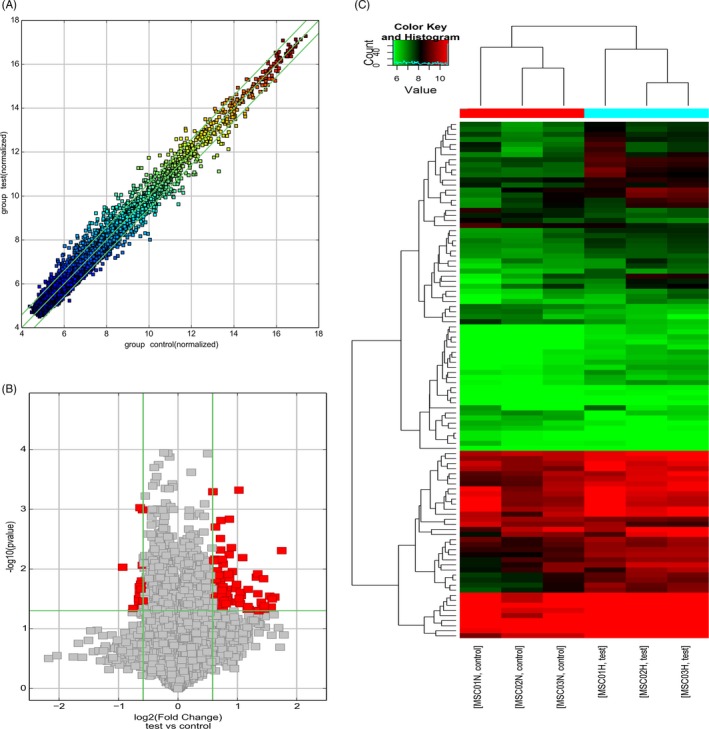
CircRNAs expression profiles in hypoxia group and normoxia group. A, Scatter plots of circRNAs expression profiles in hypoxia group and normoxia group. The values on the X and Y axes in the scatter plot are the normalized signal values of the samples (log2 scaled). The green lines correspond to 1.5‐fold changes. B, Volcano plots of circRNAs expression profiles in hypoxia group and normoxia group. Red dots represent differentially expressed circRNAs (fold change >1.5‐fold and P < 0.05). C, Heat map of circRNAs differentially expressed between hypoxia group and normoxia group
3.2. Microarray result validation
To validate the results of the microarray analysis, the expression levels of nine circRNAs (hsa_circRNA_002415, hsa_circRNA_102700, hsa_circRNA_104981, hsa_circRNA_402986, hsa_circRNA_005232, hsa_circRNA_103145, hsa_circRNA_104833, hsa_circRNA_102854, and hsa_circRNA_103859) were detected by qRT‐PCR in hpcpMSCs of the hypoxia and normoxia groups. The expression levels of circRNAs were consistent with those obtained in the comparative microarray hybridization analysis (Figure 2), confirming the reliability of the data.
Figure 2.
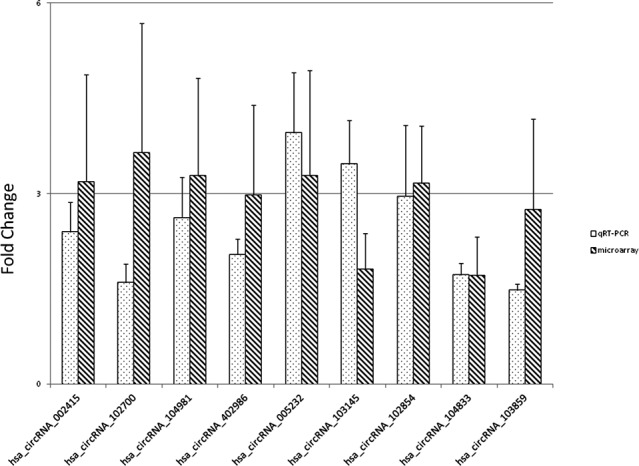
qRT‐PCR validation of expression levels of circRNAs identified by microarray analysis. The error bar stands for standard deviations
3.3. Bioinformatics analysis
Five high‐valued miRNA targets were predicted for each differentially expressed circRNAs using Arraystar's homemade miRNA target prediction software according to specific base pairing. A circRNA‐miRNA network was constructed for nine circRNAs (Appendix S4). The Gene Ontology (GO) analysis of the predicted target genes detects the putative functions of the target genes (Appendix S5). In the biological process of GO analysis, the most enriched GO terms are “developmental process” and “nervous system development.” Other enriched terms include “multicellular organism development,” “cellular developmental process,” and “positive regulation of cellular process” (Figure 3). The KEGG analysis showed four meaningful enrichment signal pathways (P < 0.05 and false discovery rate <0.05; Figure 4, Appendix S6), with the pathways “transcriptional misregulation in cancer” and “mitogen‐activated protein kinase (MAPK) signaling” being the most significant. The constructed pathway interaction network was evaluated for pathway‐pathway interactions (Figure 5). The MAPK signaling pathway was at the core of these interactions.
Figure 3.
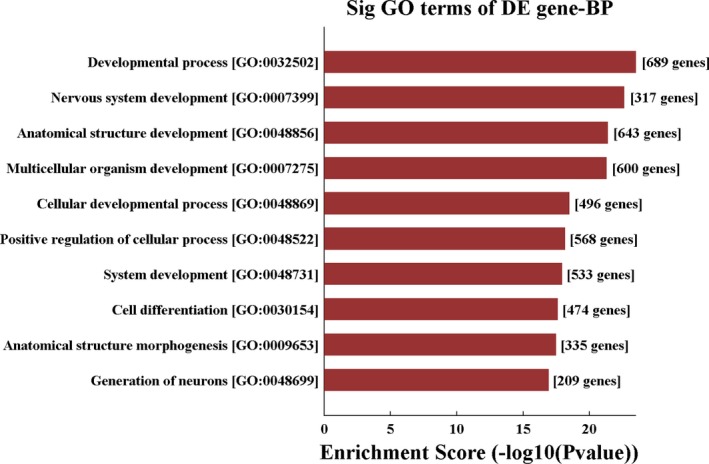
Top 10 significant GO biological processes of the target genes of miRNAs interacted with differentially expressed circRNAs
Figure 4.
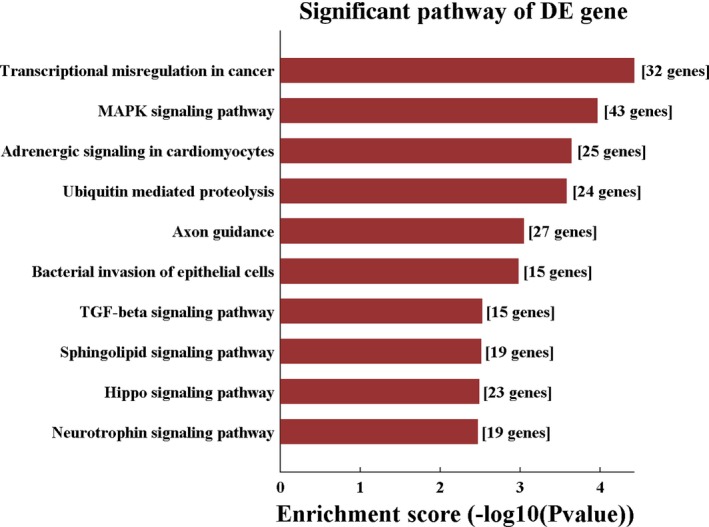
Top 10 significant KEGG pathway of the target genes of miRNAs interacted with differentially expressed circRNAs
Figure 5.
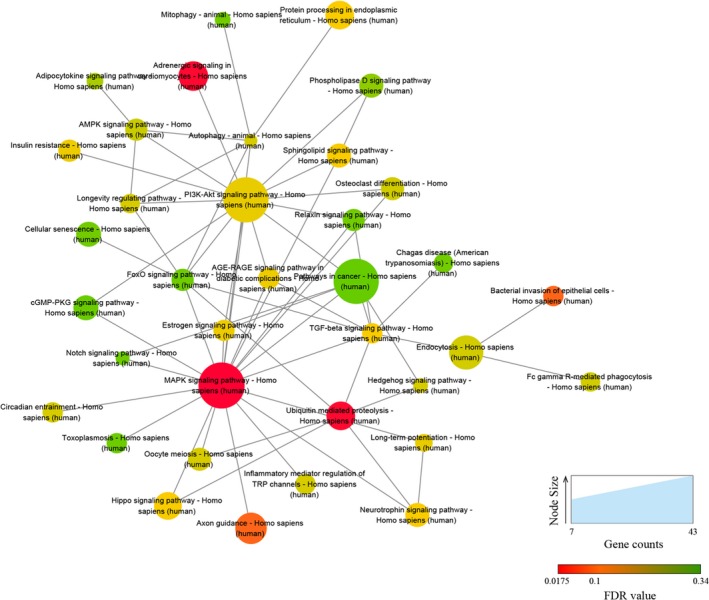
Pathway‐pathway interaction network. A higher number of edges for a pathway indicate a greater degree of interconnection and a more central role within the network
4. DISCUSSION
Oxygen maintains metabolism and function of the cells and is essential for almost all life forms. The effect of oxygen tension on cellular biological behavior is the most prominent of a series of influencing factors, and it is simple and easy to control the biological behavior of the cells by controlling oxygen tension.21 The discovery of hypoxia‐inducing factors in the 1990s gave rise to a new understanding of the effects of hypoxia in the body.22 The effect of hypoxia on the biological characteristics of MSCs and its application in regenerative medicine are not negligible.6, 7 Studies have shown that hypoxia affects cell cycle, apoptosis, migration, proliferation, and differentiation of MSCs. Hypoxic preconditioning can be used as an effective means to overcome the shortcomings of the biological characteristics of MSCs, such as slow proliferation, low mobility after transplantation, and gene instability, and can improve the effectiveness and safety of clinical application of MSCs. It is very important for studies of applications of MSCs in regenerative medicine.23, 24
Competitive endogenous RNAs (pseudogene transcripts, lncRNAs, and circRNAs) are one of the hot spots of biomedical research, functioning to regulate gene expression by competitively binding the miRNA response elements.25 The studies of circRNAs mainly focus on the field of cancer, and circRNAs are involved in the development of tumors.26 There are few reports on the physiological processes involving circRNAs in MSCs. Li et al17 reported that estrogen receptor β regulates osteogenesis of MSCs through the 2:27713879/27755789/2:240822115/240867796‐miR‐328‐5p‐mRNA axis. CircRNAs and their interacting miRNAs play an important role in the osteogenic differentiation of BMSCs.18 Numerous studies reported the effects of miRNAs on MSCs that involve cell cycle, proliferation, differentiation, and migration of MSCs.27, 28 It has been reported that miR‐21 is strongly expressed in MSCs and promotes osteogenic and adipogenic differentiation by regulating BMPR2, SPRY1, TGFBR2, and BMP, inhibits cell proliferation by regulating Sox2, and regulates PDCD4, SPRY1, SPRY2, and PTEN to enhance cell survival and to protect mitochondrial function from mitochondrial apoptosis.29, 30 miR‐27b inhibits the migration of MSCs by regulating the expression of SDF‐1α, inhibits adipogenesis by targeting transcription factors PPARγ and C/EBPα, and affects osteogenic differentiation by regulating APC, SATB2, and GCA.31, 32
In this study, comparison of the expression levels of circRNAs in the hypoxia and the normoxia groups identified 102 differentially expressed circRNAs; 85 circRNAs were upregulated, 17 were downregulated, and 27 were upregulated by more than twofold. These differentially expressed circRNAs may be involved in various biological processes and molecular regulatory mechanisms of hpcpMSCs under hypoxic conditions. Bioinformatics analyses revealed the target binding sites of differentially expressed circRNAs and predicted the interactions of miRNAs with their putative genes. GO analysis indicated that cell development and regulation of cell development are two of the main functions of differentially expressed circRNAs. hsa_circRNA_005232 and hsa_circRNA_102700 act on miR‐27a‐3p and miR‐27b‐3p, respectively. hsa_circRNA_102854 acts on miR‐21‐3p. Signal pathway enrichment analysis revealed that “transcriptional misregulation in cancer” and “mitogen‐activated protein kinase signaling pathway” were the most significant pathways, suggesting that these differentially expressed circRNAs may affect the genome stability and biological characteristics of hypoxia‐preconditioned MSCs. The results of bioinformatics analysis of differentially expressed circRNAs are consistent with the reported effects of hypoxic preconditioning on the biological characteristics of MSCs.6, 7
In this study, 43 genes are associated with the MAPK signaling pathway; the MAPK signaling pathway is at the core of the pathway‐pathway interaction network. The MAPK signaling pathway is involved in various cellular functions and physiological processes of MSCs, such as cell movement, apoptosis, differentiation, growth, and proliferation.33, 34 ERK, p38, and JNK are the MAPK family members involved in signal transduction during MSC differentiation.35 ERK‐MAPK is activated during cell development and is closely related to cell proliferation and initiation of differentiation, while the p38 and JNK signaling pathways are likely to play a role in the late stages of cell differentiation or apoptosis.33, 36 Recent studies have shown that the effects of circular RNAs involve the MAPK signaling pathway, such as TNF‐α promoting circ‐4099 expression through the MAPK and NF‐κB pathways and overexpression of circ‐4099 increasing the expression of collagen II and aggrecan in the nucleus pulposus tissue of degenerated human intervertebral disks.37 CircRNA BANP and circRNA ITCH can interact with miRNA34a and miRNA146a to regulate osteogenic differentiation of the periodontal ligament stem cells via the MAPK pathway.38 Promotion of osteogenic differentiation of the periodontal ligament stem cells related to circRNA CDR1as is modulated through the Smad‐dependent and p38 MAPK signaling pathways.39
In summary, we identified 102 differentially expressed circRNAs in hypoxia‐pretreated MSCs. GO analysis and pathway enrichment analysis showed that the differentially expressed circRNAs affect the biological characteristics of MSCs, such as cell development, differentiation, cellular processes, and the stability of the genome, in agreement with previous experimental studies. Our findings indicate that specific circRNAs regulate the biological characteristics of MSCs in hypoxic preconditioning.
Supporting information
ACKNOWLEDGMENTS
The work was supported by the National Natural Science Foundation of China (81601004 and 81102248) and the Natural Science Foundation of Guangdong Province (2016A030310139) and Paediatric Institute Foundation of Guangzhou women and children's Medical Center (YIP‐2016‐014) and Guangdong Provincial Key Laboratory for Diagnosis and Treatment of Major Neurological Diseases (2017B030314103) and the Southern China International Cooperation Base for Early Intervention and Functional Rehabilitation of Neurological Diseases (2015B050501003) and Guangdong Provincial Engineering Center for Major Neurological Disease Treatment.
Sun X, Jin Y, Liang Q, et al. Altered expression of circular RNAs in human placental chorionic plate‐derived mesenchymal stem cells pretreated with hypoxia. J Clin Lab Anal. 2019;33:e22825 10.1002/jcla.22825
Funding information
The National Natural Science Foundation of China, Grant/Award Number: 81601004 and 81102248; the Natural Science Foundation of Guangdong Province, Grant/Award Number: 2016A030310139; Paediatric Institute Foundation of Guangzhou Women and Children's Medical Center, Grant Number/Award: YIP‐2016‐014; Guangdong Provincial Key Laboratory for Diagnosis and Treatment of Major Neurological Diseases, Grant/Award Number: 2017B030314103; the Southern China International Cooperation Base for Early Intervention and Functional Rehabilitation of Neurological Diseases, Grant/Award Number: 2015B050501003; Guangdong Provincial Engineering Center for Major Neurological Disease Treatment.
REFERENCES
- 1. Mo M, Wang S, Zhou Y, et al. Mesenchymal stem cell subpopulations: phenotype, property and therapeutic potential. Cell Mol Life Sci. 2016;73:3311‐3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mohammadian M, Abasi E, Akbarzadeh A. Mesenchymal stem cell‐based gene therapy: a promising therapeutic strategy. Artif Cells Nanomed Biotechnol. 2016;44:1206‐1211. [DOI] [PubMed] [Google Scholar]
- 3. Pelekanos RA, Sardesai VS, Futrega K, et al. Isolation and expansion of mesenchymal stem/stromal cells derived from human placenta tissue. J Vis Exp. 2016;112:54204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Passipieri JA, Kasai‐Brunswick TH, Suhett G, et al. Improvement of cardiac function by placenta‐derived mesenchymal stem cells does not require permanent engraftment and is independent of the insulin signaling pathway. Stem Cell Res Ther. 2014;5:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim MJ, Shin KS, Jeon JH, et al. Human chorionic‐plate‐derived mesenchymal stem cells and Wharton's jelly‐derived mesenchymal stem cells: a comparative analysis of their potential as placenta‐derived stem cells. Cell Tissue Res. 2011;346:53‐64. [DOI] [PubMed] [Google Scholar]
- 6. Li L, Jaiswal PK, Makhoul G, et al. Hypoxia modulates cell migration and proliferation in placenta‐derived mesenchymal stem cells. J Thorac Cardiovasc Surg. 2017;154:543‐552. [DOI] [PubMed] [Google Scholar]
- 7. Ejtehadifar M, Shamsasenjan K, Movassaghpour A, et al. The effect of hypoxia on mesenchymal stem cell biology. Adv Pharm Bull. 2015;5:141‐149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sanger HL, Klotz G, Riesner D, et al. Viroids are single‐stranded covalently closed circular RNA molecules existing as highly base‐paired rod‐like structures. Proc Natl Acad Sci USA. 1976;73:3852‐3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71:428‐442. [DOI] [PubMed] [Google Scholar]
- 10. Li M, Ding W, Sun T, et al. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. FEBS J. 2018;285:220‐232. [DOI] [PubMed] [Google Scholar]
- 11. Lu C, Sun X, Li N, et al. CircRNAs in the tree shrew (Tupaia belangeri) brain during postnatal development and aging. Aging (Albany NY). 2018;10:833‐852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kristensen LS, Okholm T, Venø MT, Kjems J. Circular RNAs are abundantly expressed and upregulated during human epidermal stem cell differentiation. RNA Biol. 2018;15:280‐291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yan N, Xu H, Zhang J, et al. Circular RNA profile indicates circular RNA VRK1 is negatively related with breast cancer stem cells. Oncotarget. 2017;8:95704‐95718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lei W, Feng T, Fang X, et al. Signature of circular RNAs in human induced pluripotent stem cells and derived cardiomyocytes. Stem Cell Res Ther. 2018;9:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yu CY, Li TC, Wu YY, et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun. 2017;8:1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maass PG, Glažar P, Memczak S, et al. A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl). 2017;95:1179‐1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X, Peng B, Zhu X, et al. Changes in related circular RNAs following ERβ knockdown and the relationship to rBMSC osteogenesis. Biochem Biophys Res Commun. 2017;493:100‐107. [DOI] [PubMed] [Google Scholar]
- 18. Zhang M, Jia L, Zheng Y. circRNA expression profiles in human bone marrow stem cells undergoing osteoblast differentiation. Stem Cell Rev. 2018; 10.1007/s12015-018-9841-x [DOI] [PubMed] [Google Scholar]
- 19. Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271‐282. [DOI] [PubMed] [Google Scholar]
- 20. Wen Y, Alshikho MJ, Herbert MR. Pathway network analyses for autism reveal multisystem involvement, major overlaps with other diseases and convergence upon MAPK and calcium signaling. PLoS One. 2016;11:e0153329‐e153423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muz B, de la Puente P, Azab F, Azab AK. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl). 2015;3:83‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Semenza GL. Hypoxia‐inducible factor 1: master regulator of O2 homeostasis. Curr Opin Genet Dev. 1998;8:588‐594. [DOI] [PubMed] [Google Scholar]
- 23. Haque N, Rahman MT, Abu Kasim NH, Alabsi AM. Hypoxic culture conditions as a solution for mesenchymal stem cell based regenerative therapy. Sci World J. 2013;2013:632972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buravkova LB, Andreeva ER, Gogvadze V, Zhivotovsky B. Mesenchymal stem cells and hypoxia: where are we? Mitochondrion. 2014;19:105‐112. [DOI] [PubMed] [Google Scholar]
- 25. Yang C, Wu D, Gao L, et al. Competing endogenous RNA networks in human cancer: hypothesis, validation, and perspectives. Oncotarget. 2016;7:13479‐13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Han C, Seebacher NA, Hornicek FJ, et al. Regulation of microRNAs function by circular RNAs in human cancer. Oncotarget. 2017;8:64622‐64637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Clark EA, Kalomoiris S, Nolta JA, Fierro FA. Concise review: microRNA function in multipotent mesenchymal stromal cells. Stem Cells. 2014;32:1074‐1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hamam D, Ali D, Kassem M, et al. microRNAs as regulators of adipogenic differentiation of mesenchymal stem cells. Stem Cells Dev. 2015;24:417‐425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sekar D, Saravanan S, Karikalan K, et al. Role of microRNA 21 in mesenchymal stem cell (MSC) differentiation: a powerful biomarker in MSCs derived cells. Curr Pharm Biotechnol. 2015;16:43‐48. [DOI] [PubMed] [Google Scholar]
- 30. Kang H, Hata A. The role of microRNAs in cell fate determination of mesenchymal stem cells: balancing adipogenesis and osteogenesis. BMB Rep. 2015;48:319‐323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yuan Z, Li Q, Luo S, et al. PPARγ and Wnt signaling in adipogenic and osteogenic differentiation of mesenchymal stem cells. Curr Stem Cell Res Ther. 2016;11:216‐225. [DOI] [PubMed] [Google Scholar]
- 32. Fang S, Deng Y, Gu P, Fan X. MicroRNAs regulate bone development and regeneration. Int J Mol Sci. 2015;16:8227‐8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Majidinia M, Sadeghpour A, Yousefi B. The roles of signaling pathways in bone repair and regeneration. J Cell Physiol. 2018;233:2937‐2948. [DOI] [PubMed] [Google Scholar]
- 34. Mei Y, Bian C, Li J, et al. miR‐21 modulates the ERK‐MAPK signaling pathway by regulating SPRY2 expression during human mesenchymal stem cell differentiation. J Cell Biochem. 2013;114:1374‐1384. [DOI] [PubMed] [Google Scholar]
- 35. Jadlowiec J, Koch H, Zhang X, et al. Phosphophoryn regulates the gene expression and differentiation of NIH3T3, MC3T3‐E1, and human mesenchymal stem cells via the integrin/MAPK signaling pathway. J Biol Chem. 2004;279:53323‐53330. [DOI] [PubMed] [Google Scholar]
- 36. Jaiswal RK, Jaiswal N, Bruder SP, et al. Adult human mesenchymal stem cell differentiation to the osteogenic or adipogenic lineage is regulated by mitogen‐activated protein kinase. J Biol Chem. 2000;275:9645‐9652. [DOI] [PubMed] [Google Scholar]
- 37. Wang H, He P, Pan H, et al. Circular RNA circ‐4099 is induced by TNF‐α and regulates ECM synthesis by blocking miR‐616‐5p inhibition of Sox9 in intervertebral disc degeneration. Exp Mol Med. 2018;50:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gu X, Li M, Jin Y, et al. Identification and integrated analysis of differentially expressed lncRNAs and circRNAs reveal the potential ceRNA networks during PDLSC osteogenic differentiation. BMC Genet. 2017;18:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li X, Zheng Y, Zheng Y, et al. Circular RNA CDR1as regulates osteoblastic differentiation of periodontal ligament stem cells via the miR‐7/GDF5/SMAD and p38 MAPK signaling pathway. Stem Cell Res Ther. 2018;9:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


