Abstract
Background
The ratio of target cfDNA in total plasma is low. The abundant gDNA background resulting from blood cell lysis caused by improper operation has become a major obstacle to accurately measure cfDNA. In this study, we investigated the storage capacity of three blood cell collection tubes (BCTs) in the prevention of genomic DNA contamination caused by white blood cell rupture and evaluated their performance when they were utilized combining with highly sensitive mutation detection technology.
Methods
Blood samples were drawn from six healthy blood donors and stored in three types of BCTs (BD K2‐EDTA tube, Roche tube, and Streck tube). Plasma samples were isolated at specific time points (day 0, day 3, day 7, and day 14) for content analysis.
Results
Roche BCT was more capable for preventing cfDNA contamination caused by white blood cell disruption within 14 days, comparing with BD K2‐EDTA tube and Streck tube. Severe white blood cell lysis and gDNA contamination were found in the BD tube. The impacts of Roche and Streck tubes on the quantity and complexity of next‐generation sequencing (NGS) libraries did not differ significantly within 3 days, satisfying most of our daily demands. In addition, the rupture of WBC was not synchronized with hemolysis in BCTs.
Conclusion
This study showed that capacities of blood collection tubes differed considerably in preservation of blood samples. Therefore, suitable blood collection devices should be selected to minimize gDNA contamination and to standardize blood samples processing for achieving more accurate and reliable clinical analysis of cfDNA.
Keywords: blood collection tube, cfDNA for cancer diagnosis, gDNA contamination, white blood cell disruption
1. INTRODUCTION
Cell‐free DNA (cfDNA) is DNA fragments released to plasma as a consequence of apoptosis and necrosis. CfDNA was first discovered by Mandel and Metais in 1948.1 Leon and colleagues demonstrated the importance of cfDNA in clinical practice due to the significant elevation of cfDNA level in cancer patients, especially those with metastatic cancer.2 These studies shed light on the potential applications of cfDNA detection in clinical diagnosis, therapeutic response assessment, and prognosis prediction.3 Compared with healthy controls, an elevated level of cfDNA can be detected in plasma and serum of cancer patients.4, 5 Besides, in addition to the significance of cfDNA in tumor patients, fetal cfDNA can also be detected in pregnant donor blood plasma. This raised the interest of fetal DNA in prenatal diagnoses, such as sex determination test and fetal gene disorder detection.6
Although cfDNA plays an important role in clinic, the percentage of tumoral and fetal cfDNA only accounts for only a small fraction of total plasma cfDNA.7, 8 Abundant of released genomic DNA (gDNA) would mask the cfDNA of interest in subsequent clinical detection and calculation procedure.9, 10, 11 Therefore, it is necessary to minimize gDNA release for preventing cfDNA contamination in the process from phlebotomy to laboratorial DNA analysis.
One way to solve this problem is to carry out cfDNA analysis immediately after blood collection, so as to prevent the release of gDNA to the greatest extent. Nevertheless, this method may limit the scope of research and is not suitable for facilities which do not possess genetic testing and analysis instruments and capabilities. Another way to stabilize blood cell is to add traditional reagents, such as formaldehyde and glutaraldehyde, to maintain the integrity of biomolecules. However, the key problem of this method is the cross‐link of nucleic acid‐protein, which makes it difficult to isolate nucleic acid.12 Therefore, it is worthwhile to establish an optimized approach for blood sample preservation.
In this study, characterization of three types of commercial blood collection tubes (BCTs) was investigated. We evaluated the ability of three different BCTs to prevent cfDNA contamination caused by gDNA release through quantitative real‐time PCR, and compared impacts of the three tubes on next‐generation sequencing (NGS) library construction. In addition, we also evaluated whether the WBC rupture is synchronized with hemolysis. The purpose of this study was to provide a valuable guidance for the selection of BCTs in specific situation.
2. MATERIALS AND METHODS
2.1. Recruitment of blood donors
Six volunteer donors enrolled by Burning Rock Dx provided informed consents. All the donors were healthy, and blood samples were obtained through venipuncture.
2.2. Information of blood collection tubes
We compared three types of blood collection tubes in this study: Vacutainer Plastic K2EDTA Tube (BD, 367527), Cell‐Free DNA Collection Tube (Roche, 7785674001), and Cell‐Free DNA tube (Streck, 218997). All the three types of tube share same volume of 8 mL.
2.3. Sample processing
A total of 24 mL blood was drawn from each blood donor using disposable venous blood lancet and stored directly in three types of blood collection tubes (8 mL per tube). Blood samples were stored at room temperature and separated at specific time points at indicates (day 0, day 3, day 7, and day 14). Before each separation process, blood collection tubes were mixed upside down gently for several times. Specific aliquots of blood were transferred to new vials gently (100 μL for quantitative real‐time PCR and hemoglobin absorbance of each time point, 7.5 mL for NGS library construction). Tubes with blood were centrifuged at 4°C for 10 minutes at 2000 g using a balanced, swing‐out centrifuge. The supernatants (plasma) were transferred into new tubes. After that, plasma samples were further centrifuged at 4°C for 10 minutes at 16 000 g to remove the potential contaminating cells. The supernatant plasma sample was removed into another new tubes again and stored in refrigerator at −80°C for further analysis.
2.4. Quantitative Real‐Time PCR
Human L1PA2 primers were prepared based on a previous study, and the sequences were listed as follows13: Forward primer: 5′‐TGC CGC AAT AAA CAT ACG TG‐3′; reverse primer: 5′‐GAC CCA GCC ATC CCA TTA C‐3′. This pair of primers produces a 90‐bp amplicon of L1PA2, which can be used to indicate the overall DNA level in plasma, including target cfDNA and degraded gDNA released by WBC rupture.
Plasma samples were taken out of −80°C refrigerator and thawed on ice, followed by real‐time PCR for plasma cfDNA quantification using KAPA2G Fast PCR Kits (Kapa Biosystem, KK5500). For each well in PCR plate, ten microliters reaction volume contained 0.1 μL of plasma, 1 μL SYBR Green (1.5X), 1 μL ROX (50X) reference dye, 2 μL KAPA2G buffer A, 2 μL dNTP mix (10 mmol/L), 0.04 μL KAPA2G Fast HotStart DNA polymerase (5 U/μL), 1 μL of each primer (1 μmol/L), and PCR‐grade water. PCR amplification reaction was initiated with denaturation for 10 minutes at 98°C, followed by 39 cycles of 30 seconds at 95°C, 30 seconds at 60°C, and 30 seconds at 72°C. All samples were amplified in triplicates to diminish variations.
2.5. Absorbance spectra of free hemoglobin in 414 nm
Plasma samples were thawed on ice before hemoglobin absorbance measurement. The hemoglobin absorbance spectrum was measured by NanoDrop 2000 UV‐Vis Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) according to manufacturer's instruction following a well‐established and validated protocol.14 Wavelengths of 414 nm were preset in the software (NanoDrop 2000) before measurement. Instrument was primarily washed and adjusted to zero with nuclease‐free water. 1.5 μL plasma liquid was used for hemoglobin absorbance measurement. Three repeats were measured for each patient and at each time point to minimize variation.
2.6. Evaluation of quantity and complexity of NGS library
Plasma was separated from 7.5 mL blood storing in the three types of BCTs by centrifugation (K2EDTA tube, Day 0, as control; Streck and Roche tubes, Day 3). CfDNA (about 20~30 ng, 55 μL) was isolated from plasma (3~4 mL) using the QIAamp Circulating Nucleic Acid Kit (Qiagen, Valencia, CA, USA) following manufacturer's protocol. A total of 50 μL cfDNA was used for NGS library construction. DNA shearing was performed using Covaris M220 (Covaris Inc., Woburn, MA, USA). End repair and A tailing were performed followed by adaptor ligation. The ligated fragments with size of 170 bp were then selected by beads (Agencourt AMPure XP Kit; Beckman Coulter, Brea, CA, USA), hybridized with probe baits, and amplified by polymerase chain reaction. Indexed samples were sequenced on Nextseq500 sequencer (Illumina, Inc., San Diego, CA, USA) with pair‐end reads.
2.7. Statistical analysis
Statistical analysis was processed by R studio and Microsoft Office Excel 2010. All data were calculated and presented using paired, two‐tailed Student's t test in P value.
3. RESULTS
3.1. Study design and workflow overview
Twenty‐four milliliters of blood was drawn from each of the six healthy donors with disposable venous blood needle and stored directly into three different types of blood collection tubes (8 mL per tube). The tubes containing blood samples were stored in a relatively stationary room temperature (20‐30°C). Specific volumes of blood samples were separated at desired time points (day 0, day 3, day 7, and day 14). Plasma samples were isolated and stored in −80°C refrigerator for L1PA2 gene level assessment, hemoglobin absorption at optical density (OD) 414 nm, and NGS library evaluation (Figure 1).
Figure 1.
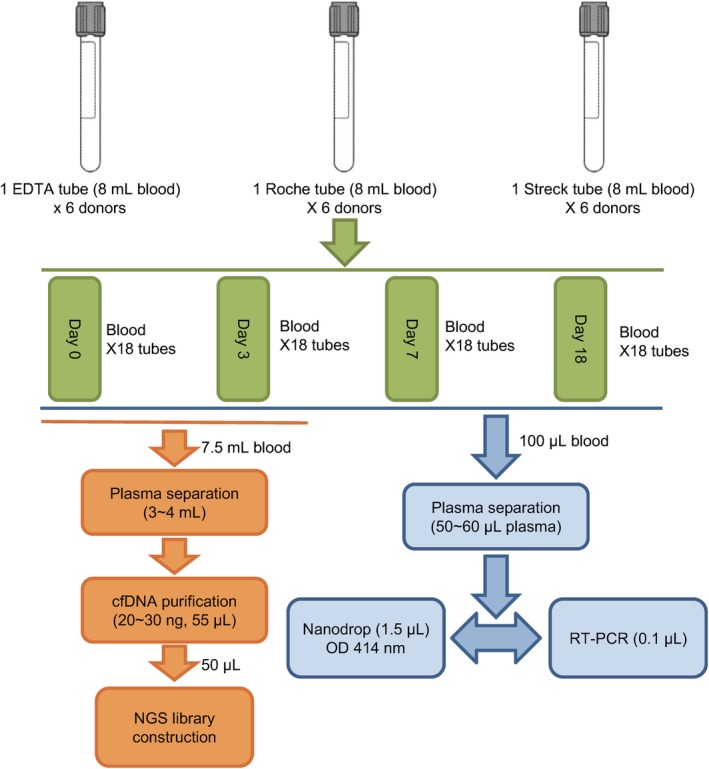
Experimental design and workflow of the blood collection tubes evaluation
3.2. Evaluation of different BCTs for preventing white blood cell lysis
To evaluate gDNA contamination caused by WBC lysis, we use L1PA2 level to measure the total plasma DNA level. L1PA2 is an abundant gene which intersperses throughout the human genome.
In this study, delta C t was calculated by negative C t (Day 0) minus plasma C t (Day 3, Day 7, or Day 14) to reflect the total DNA level in the plasma (10delta Ct) and to demonstrate the ability of tubes for WBC stabilization. After 7‐day storage at room temperature, blood samples in Roche (1.30 at day 3; 2.50 at day 7) and Streck blood cell collection tubes (BCT) (2.18 at day 3; 2.87 at day 7) showed marginal gDNA releasing from WBC lysis, comparing to those in K2‐EDTA tubes (5.35 at day 3; 118.26 at day 7) which had a plenty of gDNA contamination (Figure 2A‐D). As the storage time prolonged to 14 days, Roche BCT showed a relatively low DNA (2.73) level than Streck one (11.49), indicating that Roche tube was more effective in preventing release of gDNA from WBC.
Figure 2.
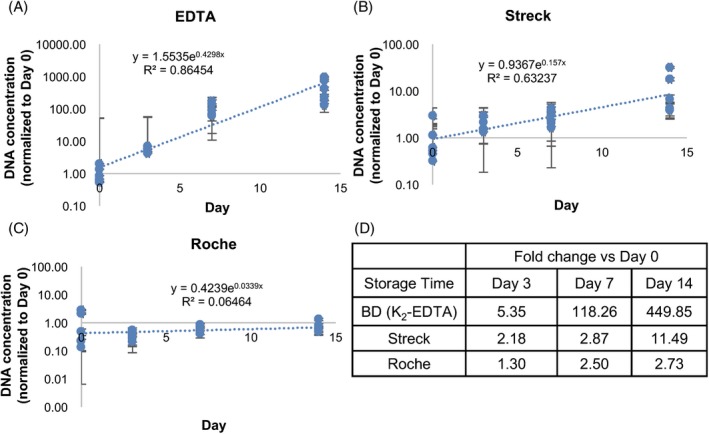
Quantitative Real‐Time PCR of L1PA2 gene level for indication of white blood cell disruption. Y‐axis means delta relative cfDNA concentration normalized to Day 0 point. Line slope presents the L1PA2 level increasing rate due to WBC lysis and showed as R 2 in the graph. Delta C t was calculated by negative C t (nuclease‐free water as control) minis plasma C t. A, cfDNA concentration in BD K2‐EDTA blood cell collection tubes (BCT) for specific storage periods as indicated. B, cfDNA level change in Streck tube with the prolonged storage time points. C, cfDNA level in Roche tube. D, Summary of relative cfDNA concentration for indicated storage time periods and each BCT which was normalized to Day 0
3.3. Evaluation of different BCTs in terms of NGS library quantity and complexity
In most cases, the process from blood draw to laboratorial NGS library construction in our facility can be completed within 3 days. Therefore, we further evaluated the impacts of different BCTs (Roche and Streck) on blood samples within 3 days in terms of NGS library quantity and complexity. Plasma collected in K2EDTA tubes and isolated within 2 hours (Day 0) after phlebotomy was used as control. Quantity of NGS library was measured by Qubit® 3.0 Fluorometer (Thermo Fisher Scientific) and LabChip GXII Touch (PerkinElmer) before and after baits capture. We found that there was no significant difference in terms of NGS library quantity among the three groups (K2EDTA, Day 0; Roche, Day 3; Streck, Day 3; Figure 3A). Library complexity is defined as the expected number of distinct molecules sequenced in a given set of reads produced in a sequencing experiment, which reflects the high fidelity the original complexity of the source material. The results showed that there was also no significant difference among K2EDTA, Roche, and Streck groups in terms of complexities of NGS library (Figure 3B). These results indicated the blood storage applicability of the two tubes for NGS within 3 days.
Figure 3.
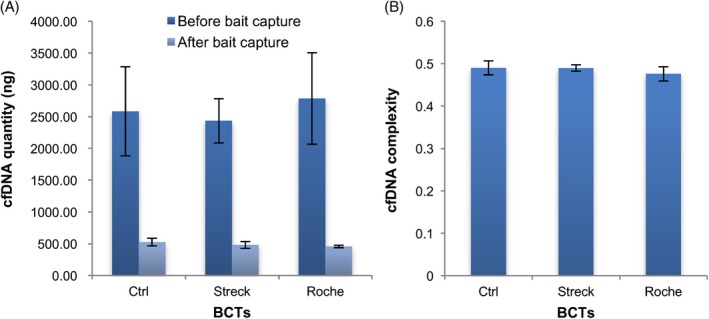
Evaluation of quantity and complexity of plasma cfDNA from Streck and Roche tubes by next‐generation sequencing (NGS). A, Quantity comparison of cfDNA from different blood cell collection tubes (BCTs) before and after baits capture. B, Complexity evaluation of cfDNA in different BCTs. cfDNA from BD K2‐EDTA tube was used as negative control
3.4. Rupture of WBC was not synchronous with hemolysis among the three types of BCTs
Hemoglobin is a metalloprotein in red blood cells, which accounted for about 96% of the red blood cells dry content (by weight).15 This gave us the implication that hemoglobin concentration can be utilized as an indicator for red blood cell disruption.
Hemoglobin has an absorption peak at OD 414 nm region. Therefore, absorption value of 414 nm was used to indicate the hemoglobin concentration in this study. Compared to K2‐EDTA tube (P = 6.99E‐12), the absorbance of hemoglobin at 414 nm in Roche (P = 8.80E‐08) and Streck (P = 3.55E‐06) tubes was relatively low in short storage period of less than 7 days, and the Streck tube exhibited a slightly lower absorption than Roche one. This observation demonstrated that Streck and Roche BCT can prevent hemolysis and maintain red blood cells in a more stable status, especially the Streck tube. In addition, when the storage period was extended to 14 days, Roche (P = 1.59E‐13) tube showed significant effective hemolytic prevention, compared to the Streck tube (P = 4.16E‐21) (Figure 4). This result indicated that the rupture of WBC in the three types of BCTs was not synchronized with hemolysis. Data analysis was conducted by paired, two‐tailed Student's t test.
Figure 4.
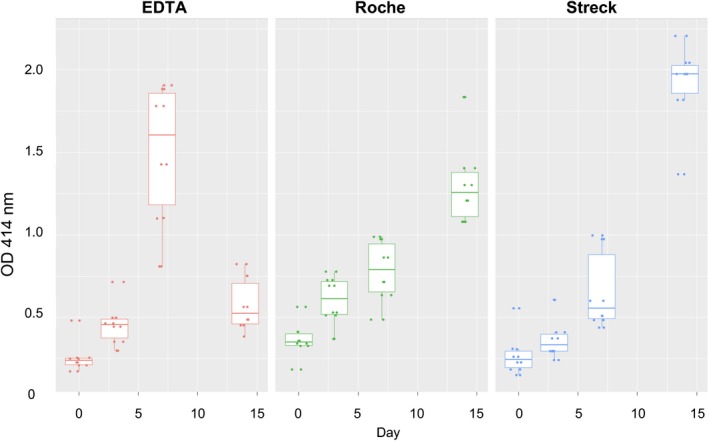
Measurements of absorption value at optical density (OD) 414 nm for plasma hemoglobin in three blood collection tubes. Specific volume of blood samples which were stored for different time periods and in different blood cell collection tubes (BCTs) was transferred to a new vial for plasma isolation
4. DISCUSSION
It is greatly recommended to minimize gDNA contamination caused by WBC rupture, so as to get a relatively accurate and reliable cfDNA level throughout the whole blood sample preanalytic process. As far as we know, most of previous studies focused on transportation condition, such as physical shock, shipping temperature, chemical cocktail for maintaining blood samples stabilization.16, 17 In this study, we evaluated three types of commonly used blood collection tubes for their capability of preventing WBC rupture and impacts on quantity and complexity of NGS library. Blood samples stored for different periods were used to assess the blood stabilizing capability of blood collection tubes. The three types of BCTs showed different performance in preservation of blood samples, which provided guidance for BCT selection in specific studies.
In previous studies, the indicators of gDNA contamination were mainly focused on the level of relative abundant genes such as beta‐Actin, which was used as genomic equivalents.16, 17 Digital PCR is the commonly used technique to estimate DNA level of genomic equivalents to indicate impacts of factors during blood shipping and storage. In this study, we chose the widespread gene L1PA2 as genomic equivalent and performed PCR to estimate gDNA contamination in three types of BCTs. After 7‐day storage at room temperature, Roche and Streck BCTs had a similar prevention effect on the release of gDNA, while K2‐EDTA tube showed a plenty of gDNA release, which was consistent with previous studies.18, 19, 20, 21 When the storage period was extended to 14 days, Roche BCT showed better gDNA contamination prevention effect than the other two BCTs.
Rare studies evaluated the performance of BCTs for their applicability using highly sensitive mutation detection technology.19 One novelty of our study is that we used NGS to evaluate the impact of BCT on quantity and complexity of NGS library. With the development of genomic sequencing for liquid biopsies using NGS, complexity and quality of NGS library were important factors determining the quality of NGS. The complexity of NGS library can reflect the bias of NGS result caused by cfDNA quality. The ideal library complexity can reflect the high fidelity of original sample molecular complexity. We evaluated the impacts of BCTs on complexity and quality of NGS library. Our result showed that complexity and quality of NGS library constructed from cfDNA stored in Roche and Streck tubes for 3 days were similar to each other and had no significant difference to control group. This indicated that the impacts of Roche and Streck BCTs on NGS library construction were very marginal after 3 days of storage.
Beside the evaluation of gDNA contamination by L1PA2 gene level, we also detected the cell‐free hemoglobin concentration to indicate the difference of RBC hemolysis in the three types of BCTs. Our result showed that Roche tube exhibited less gDNA release than Streck one after 7 days of storage, especially after 14 days. However, Roche tube resulted in more hemoglobin release in day 7 than Streck one. This observation can be interpreted as different protection ability for gDNA release and RBC rupture of the three BCTs. In another aspect, gDNA release and RBC rupture may happen asynchronously. Overall, the results gave us the indication that it was necessary to select suitable BCT for specific blood storage needs.
There are a few limitations associated with this study. It is worth noting that plasma samples after 14 days of storage in BD K2‐EDTA tubes displayed an abnormal absorbance at OD 414 nm, which was even lower than Roche and Streck tubes. This observation may be resulted from the conflict between high absorbance and limited detection range/sensitivity of the analytic technology. We also observed that the Roche tubes had unexpectedly high OD readings at day 0 and at day 3, and this phenomenon was likely due to the fixative used in Roche tubes which have absorbance at 414 nm.
In summary, the three types of BCTs exhibited diverse performances for blood sample storage, which provided guidance for BCT selection in specific study use. Therefore, it is of great importance to standardize the blood samples processing and choose the most suitable BCT to achieve reliable and valuable individual blood analysis within a comparable experimental group.
CONSENT FOR PUBLICATION
All blood donors provided written informed consent for this study.
AUTHORS’ CONTRIBUTIONS
All authors read and approved the final manuscript.
Zhao Y, Li Y, Chen P, Li S, Luo J, Xia H. Performance comparison of blood collection tubes as liquid biopsy storage system for minimizing cfDNA contamination from genomic DNA. J Clin Lab Anal. 2019;33:e22670 10.1002/jcla.22670
REFERENCES
- 1. Mandel P, Metais P. Les acides nucléiques du plasma sanguin chez l'homme. C R Seances Soc Biol Fil. 1948;142:241‐243. [PubMed] [Google Scholar]
- 2. Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37(3):646‐650. [PubMed] [Google Scholar]
- 3. Sorenson GD, Pribish DM, Valone FH, Memoli VA, Bzik DJ, Yao SL. Soluble normal and mutated DNA sequences from single‐copy genes in human blood. Cancer Epidemiol Biomarkers Prev. 1994;3(1):67‐71. [PubMed] [Google Scholar]
- 4. Kohler C, Barekati Z, Radpour R, Zhong XY. Cell‐free DNA in the circulation as a potential cancer biomarker. Anticancer Res. 2011;31(8):2623‐2628. [PubMed] [Google Scholar]
- 5. Schwarzenbach H, Hoon DS, Pantel K. Cell‐free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11(6):426‐437. [DOI] [PubMed] [Google Scholar]
- 6. Lo YM, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485‐487. [DOI] [PubMed] [Google Scholar]
- 7. Lo YM, Tein MS, Lau TK, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62(4):768‐775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hung EC, Chiu RW, Lo YM. Detection of circulating fetal nucleic acids: a review of methods and applications. J Clin Pathol. 2009;62(4):308‐313. [DOI] [PubMed] [Google Scholar]
- 9. Skvortsova TE, Rykova EY, Tamkovich SN, et al. Cell‐free and cell‐bound circulating DNA in breast tumours: DNA quantification and analysis of tumour‐related gene methylation. Br J Cancer. 2006;94(10):1492‐1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barrett AN, Zimmermann BG, Wang D, Holloway A, Chitty LS. Implementing prenatal diagnosis based on cell‐free fetal DNA: accurate identification of factors affecting fetal DNA yield. PLoS ONE. 2011;6(10):e25202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fernando MR, Chen K, Norton S, et al. A new methodology to preserve the original proportion and integrity of cell‐free fetal DNA in maternal plasma during sample processing and storage. Prenat Diagn. 2010;30(5):418‐424. [DOI] [PubMed] [Google Scholar]
- 12. Das K, Dumais J, Basiaga S, Krzyzanowski GD. Carbon‐13 nuclear magnetic resonance analysis of formaldehyde free preservatives. Acta Histochem. 2013;115(5):481‐486. [DOI] [PubMed] [Google Scholar]
- 13. Breitbach S, Tug S, Helmig S, et al. Direct quantification of cell‐free, circulating DNA from unpurified plasma. PLoS ONE. 2014;9(3):e87838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shah JS, Soon PS, Marsh DJ. Comparison of methodologies to detect low levels of hemolysis in serum for accurate assessment of serum microRNAs. PLoS ONE. 2016;11(4):e0153200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Weed RI, Reed CF, Berg G. Is hemoglobin an essential structural component of human erythrocyte membranes? J Clin Invest. 1963;42:581‐588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Norton SE, Lechner JM, Williams T, Fernando MR. A stabilizing reagent prevents cell‐free DNA contamination by cellular DNA in plasma during blood sample storage and shipping as determined by digital PCR. Clin Biochem. 2013;46(15):1561‐1565. [DOI] [PubMed] [Google Scholar]
- 17. Norton SE, Luna KK, Lechner JM, Qin J, Fernando MR. A new blood collection device minimizes cellular DNA release during sample storage and shipping when compared to a standard device. J Clin Lab Anal. 2013;27(4):305‐311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Parpart‐Li S, Bartlett B, Popoli M, et al. The effect of preservative and temperature on the analysis of circulating tumor DNA. Clin Cancer Res. 2017;23(10):2471‐2477. [DOI] [PubMed] [Google Scholar]
- 19. Medina Diaz I, Nocon A, Mehnert DH, Fredebohm J, Diehl F, Holtrup F. Performance of streck cfDNA blood collection tubes for liquid biopsy testing. PLoS ONE. 2016;11(11):e0166354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Warton K, Yuwono NL, Cowley MJ, McCabe MJ, So A, Ford CE. Evaluation of streck BCT and PAXgene stabilised blood collection tubes for cell‐free circulating DNA studies in plasma. Mol Diagn Ther. 2017;21(5):563‐570. [DOI] [PubMed] [Google Scholar]
- 21. Wong D, Moturi S, Angkachatchai V, et al. Optimizing blood collection, transport and storage conditions for cell free DNA increases access to prenatal testing. Clin Biochem. 2013;46(12):1099‐1104. [DOI] [PubMed] [Google Scholar]


