Abstract
Background
Rapid diagnosis of Mycobacterium tuberculosis (Mtb) is key to controlling the spread of tuberculosis, which is a global health concern. In this study, isothermal recombinase polymerase amplification (RPA) was developed to detect specific targets of Mtb, IS6110 and IS1081. Additionally, SYBR Green I was used for endpoint detection of the RPA products by the naked eye.
Method
A total of 146 genomic Mtb DNA samples and 24 genomic nontuberculous mycobacteria (NTM) DNA samples were amplified at IS6110 and IS1081 by RPA. After a complete amplification, the RPA amplicons were examined by agarose gel electrophoresis (RPA‐AGE) and SYBR Green I (RPA‐S) assays. The performance of the RPA assays was evaluated by comparing them to a conventional PCR.
Results
The RPA assay demonstrated to have a good capability to differentiate Mtb from NTM with a very short turnaround time at a constant temperature. Compared to conventional PCR, the sensitivities and specificities of RPA‐AGE for IS6110 and IS1081 were 100%. The specificity of RPA‐S was 100% for both targets; however, its sensitivities for IS6110 and IS1081 were 97.95% and 99.32%, respectively. The limits of detection of IS6110 RPA‐AGE and RPA‐S were 0.05 and 0.5 ng, respectively, while the LODs of IS1081 RPA‐AGE and RPA‐S were 0.00005 and 0.05 ng, respectively. Both RPA assays showed a satisfying diagnostic specificity, with no cross‐reaction with other bacteria.
Conclusion
A rapid, sensitive, naked eye RPA assay can be integrated into point‐of‐care diagnosis for Mtb detection, especially in remote areas where laboratory instrument resources are limited.
Keywords: IS1081, IS6110, Mycobacterium tuberculosis (Mtb), recombinase polymerase amplification (RPA), SYBR Green I
1. INTRODUCTION
Tuberculosis (TB), which is caused by Mycobacterium tuberculosis (Mtb), is the source of a major global health crisis. The World Health Organization (WHO) estimates that there are approximately 10 million people affected by TB annually, and TB is one of the top 10 causes of death worldwide.1 Although the number of people killed by TB from 2000 to 2015 was less than that in previous years, the mortality rates are still high, especially in lower‐ to middle‐income countries.1, 2 Rapid diagnosis and appropriate treatment may limit the risk of TB spreading, which is important because those are essential factors for decreasing both morbidity and mortality rates.3
TB can be diagnosed in several ways. Sputum smear microscopy, a primary screening test that is particularly used in endemic areas, is used to examine the presence of bacteria.4, 5 Even if this technique is rapid and low cost, previous studies have shown that the sensitivity of the test is variable (48%‐62%).6, 7, 8, 9 Conventional culture‐based methods, use of a reference standard, permit the growth of bacteria from clinical specimens and can be used to determine the species of Mycobacterium. Nevertheless, these methods are time‐consuming due to a requirement of TB growth and can take up to 8 weeks.10 Molecular diagnostic tests are continually being developed not only to decrease their turnaround time but also to improve the sensitivity of the TB diagnosis.11, 12 The Xpert MTB/RIF (Cepheid, Sunnyvale, CA, USA), a fully automated, walkaway, real‐time PCR‐based method with a total turnaround time of approximately 2 hours, has been endorsed by the WHO for simultaneous diagnosis of TB and rifampicin resistance.13 However, the disadvantages of the Xpert MTB/RIF include its high cost, regular maintenance, stable electric power supply requirement, and limited machine capacity (four tests every 2 hours).14 Unlike PCR‐based assays, isothermal amplification methods avoid the requirements for thermal cycling and specific power supply instruments.15 Isothermal amplification methods amplify products at a constant temperature, which can be supported with a simple heat block. This method is a major advantage for low‐resource settings and can be applied as a point‐of‐care test. Several isothermal amplification methods were recently shown to be able to detect Mtb, including transcription‐mediated amplification (TMA), loop‐mediated amplification (LAMP), and helicase‐dependent amplification (HDA).16, 17, 18, 19
Recombinase polymerase amplification (RPA) has emerged as a novel isothermal amplification method that is useful in many diagnostic fields.20 The three core enzymes required for the reaction are recombinase, single‐stranded binding protein, and DNA polymerase.21 The amplification process occurs under an optimum temperature that ranges from 37 to 42°C and takes <10‐40 minutes.22, 23 Previous studies have shown successful Mtb detection by the RPA technique via real‐time device detection or endpoint detection via bridging flocculation.24, 25 However, specific devices are required to accomplish these tests.
Hence, in this study, we developed an alternative molecular diagnostic test based on isothermal amplification methods. The aim of this study was to establish an affordable, rapid, and easy hands‐on detection tool for Mtb detection, targeting low‐resource and peripheral healthcare settings or fieldworks where specific instrumentation is limited. RPA was used to amplify target genes, specifically Mtb IS6110 and IS1081. Then, SYBR Green I was added after the amplification to observe the RPA products by the naked eye.
2. MATERIALS AND METHODS
2.1. Mtb DNA samples
A total of 170 genomic DNA samples extracted from Mycobacterium colonies were used in this study. These included 100 Thai strain Mtb DNA samples and 24 nontuberculous mycobacteria (NTM) DNA samples obtained from the Microbiology Unit, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand. Additionally, 46 Japanese strains of Mtb DNA were obtained from the Department of Bacteriology, Osaka Prefectural Institute of Public Health, Osaka, Japan. All of the Mycobacterium strains were previously cultured by a MGIT™ liquid culture system (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) and discriminated between Mtb and NTM using the SD Bioline TB Ag MPT64 assay (Standard Diagnostics, Gyeonggi‐do, Korea). DNA of the standard reference strain Mtb H37Rv genomic (kindly provided by the Microbiology Unit, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Bangkok, Thailand) was used as a positive control.
All of the genomic DNA used in this study was extracted from discarded colonies after a routine diagnostic examination, with no links to patient data. The study was exempted from ethical approval in accordance with the Research Ethics Review Committee for Research Involving Human Research Participants, Health Sciences Group, Chulalongkorn University (COA No. 036/2018).
2.2. IS6110 and IS1081 amplifications by conventional PCR
The IS6110 and IS1081 genes were amplified by conventional PCR using specific primers, as summarized in Table 1. A total of 170 samples of Mtb and NTM DNA were used as templates for both target sites. Mtb H37Rv genomic DNA and sterile distilled water were used as positive and negative controls, respectively. Each reaction contained 0.2 μmol/L forward and reverse primers, 0.1 ng of DNA, 200 μmol/L dNTPs, 1× PCR buffer, 1.5 mmol/L MgCl2, and 1.5 U of Taq DNA Polymerase (New England BioLabs, Ipswich, MA, USA). The PCR amplification conditions were predenaturing at 95°C for 5 minutes, 35 cycles of amplification (95°C 1 minute, 57°C for IS6110 or 60°C for IS1081, and 72°C 1 minute), and a final extension at 72°C 7 minutes. The PCR amplicons of both genes were analyzed by 1.5% agarose gel electrophoresis containing UltraPower DNA/RNA Safedye (Gellex, Tokyo, Japan) and visualized under UV transilluminator.
Table 1.
Primer sequences for PCR and RPA used in this study
| Primer | Sequence (5′→3′) | Product size (bp) | Reference |
|---|---|---|---|
| IS6110F PCR | GGTCGCCCGTCTACTTGGTG | 416 | 44 |
| IS6110R PCR | TGGACGCGGCTGATGTGCTC | ||
| IS1081F PCR | TCGCGTGATCCTTCGAAACG | 238 | 41 |
| IS1081R PCR | GCCGTTGCGCTGATTGGACC | ||
| IS6110F RPA | TCAGTGAGGTCGCCCGTCTACTTGGTGTTG | 423 | This study |
| IS6110R RPA | TGGACGCGGCTGATGTGCTCCTTGAGTTC | ||
| IS1081F RPA | CCTCTTCTCATCTTATCGACGCCGAGCAGC | 173 | This study |
| IS1081R RPA | CTGATTGGACCGCTCATCGCTGCGTTCGC |
2.3. RPA amplification
All 170 samples of Mtb DNA were amplified by the RPA assay targeting the IS6110 and IS1081 genes. The RPA specific primers used in this study are summarized in Table 1, and Mtb H37Rv genomic DNA and sterile distilled water were used as positive and negative controls, respectively. DNA amplification was performed with the TwistAmp® Basic kit (TwistDx Limited, Cambridge, UK) as recommended by the manufacturer. Each RPA reaction consisted of 29.5 μL rehydration buffer, 0.48 μmol/L forward and reverse primers, 0.1 ng of DNA, 14 mmol/L MgOAc, and sterile distilled water up to 50 μL. The RPA reaction mixture was incubated at 37°C for 15 minutes. After amplification, each reaction was aliquoted into two tubes to be evaluated by different detection methods: 1.5% agarose gel electrophoresis (RPA‐AGE) and SYBR Green I (RPA‐S).
2.4. Agarose gel electrophoresis
RPA products (25 μL) were purified by FavorPrep Gel/PCR Purification (Favorgen Biotech Corp., Ping‐Tung, Taiwan). The purified RPA products were visualized via 1.5% agarose gel electrophoresis containing UltraPower DNA/RNA Safedye (Gellex) under UV light. Amplifications of IS6110 and IS1081 were expected to generate visible bands of 416 and 238 bp, respectively, on the agarose gel. A 100‐bp DNA ladder was used as the molecular marker.
2.5. SYBR green I detection
Amplified IS6110 and IS1081 RPA products were directly detected by the naked eye in the natural light by adding 1 μL of 375x SYBR Green I (TaKaRa Bio, Tokyo, Japan) to the 25‐μL RPA product tube and immediately observing a color change of the solution. The solution changed from light orange to bright green in the presence of RPA amplicons, implying that Mtb DNA was detected. By contrast, the solution remained light orange in the absence of RPA amplification, indicating that Mtb DNA was not detected. RPA coupled with SYBR Green I detection was blindly interpreted to eliminate bias.
2.6. Limit of detection determination
To determine the limit of detection (LOD) of the RPA assay, genomic DNA extracted from Mtb H37Rv was tenfold serially diluted from 5 to 0.00005 ng and used as a template for each pair of primer sets for amplification. Sterile distilled water was used as a negative control. After amplification, each reaction was aliquoted for SYBR Green I detection and compared to 1.5% agarose gel electrophoresis detection.
2.7. Specificity testing
The specificities of RPA‐S and RPA‐AGE with each pair of primer sets were analyzed with genomic DNA from Acinetobacter baumannii, Haemophilus influenzae, Klebsiella pneumoniae, Moraxella catarrhalis, Pseudomonas aeruginosa, Streptococcus pneumonia, Streptococcus pyogenes, M. avium, and M. intracellulare. Sterile distilled water was used as a negative control. After amplification, each reaction was aliquoted for SYBR Green I detection and compared to the 1.5% agarose gel electrophoresis detection results.
3. RESULTS
3.1. PCR amplifications
A total of 146 Mtb and 24 NTM isolates, confirmed by liquid culture and the SD Bioline TB Ag MPT64 assay, were correctly identified at the species level by conventional PCR using IS6110 and IS1081 as targets. The amplified PCR products for IS6110 and IS1081 were 416 and 238 bp, respectively. Both PCR primer sets were able to detect DNA from all 100 Thai and 46 Japanese Mtb strains via PCR amplification. On the other hand, neither IS6110 nor IS1081 products were detected in the 24 NTM strains. The PCR amplification results for each target are summarized in Table 2.
Table 2.
Mycobacterium strains detected by conventional PCR
| 100 Thai Mtb strains | 46 Japanese Mtb strains | 24 NTM strains | |
|---|---|---|---|
| IS6110 positive | 100 | 46 | 0 |
| IS6110 negative | 0 | 0 | 24 |
| IS1081 positive | 100 | 46 | 0 |
| IS10081 negative | 0 | 0 | 24 |
3.2. RPA amplification
Amplification by RPA generated IS6110 (423 bp) and IS1081 (173 bp) products. To detect RPA products of the 146 Mtb and 24 NTM genomic DNA samples, two different DNA product analyses were applied. For RPA‐AGE, the purified RPA products were analyzed by 1.5% agarose gel electrophoresis (Figure 1). All of the 146 Mtb strains, but not the NTM strains, were able to be detected by RPA‐AGE using both the IS6110 and IS1081 targets. The rest of nonpurified RPA products were directly added to 1 μL of 375x SYBR Green I, and an immediate color change of the solution was observed by the naked eye. Figure 2 shows the final color of the RPA solution after SYBR Green I was added. A green color solution indicates a positive result (Mtb DNA was detected), while an orange color solution indicates a negative result (Mtb DNA was not detected). RPA‐S was able to detect 143 Mtb strains using the IS6110 target and 145 Mtb strains using the IS1081 target. Three samples for the IS6110 target and one sample for the IS1081 target gave indefinite results; borderline orange‐green colors observed for RPA‐S using IS6110 and IS1081 as the targets, respectively. However, in this study, IS6110 and IS1081 were able to differentiate NTM from Mtb. None of the NTM strains were able to be detected by either RPA‐AGE or RPA‐S. The RPA amplifications for each target are summarized in Table 3.
Figure 1.
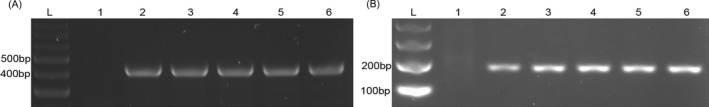
Agarose gel electrophoresis of RPA amplification of IS6110 and IS1081. A, shows 423‐bp bands of IS6110. B, shows 173‐bp bands of IS1081. In each figure, lane L is a 100‐bp DNA ladder, lane 1 is a negative control, lanes 2‐5 are Mtb DNA, and lane 6 is Mtb H37Rv DNA positive control. Each figure is the representative of triplicate on three separate experiments
Figure 2.
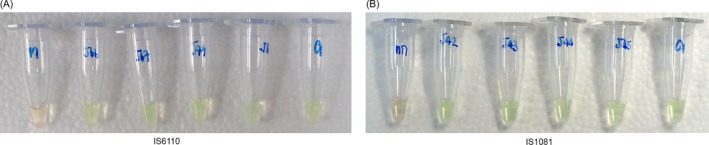
SYBR Green I detection of RPA amplification of IS6110 and IS1081. RPA products of IS6110 (A) and IS1081 (B) were endpoint‐detected by SYBR Green I. For both figures, the leftmost tubes are negative control, showing orange color. The rightmost tubes are Mtb H37Rv DNA positive control, showing a green color. The other tubes are Mtb DNA, showing green color, implying the positive result of amplification. Each figure is the representative of triplicate on three separate experiments
Table 3.
Mycobacterium strains detected by RPA‐AGE and RPA‐S
| 100 Thai Mtb strains | 46 Japanese Mtb strains | 24 NTM strains | |
|---|---|---|---|
| IS6110 | |||
| RPA‐AGE positive | 100 | 46 | 0 |
| RPA‐AGE negative | 0 | 0 | 24 |
| RPAS positive | 97 | 46 | 0 |
| RPAS negative | 3a | 0 | 24 |
| IS1081 | |||
| RPA‐AGE positive | 100 | 46 | 0 |
| RPA‐AGE negative | 0 | 0 | 24 |
| RPAS positive | 99 | 46 | 0 |
| RPAS negative | 1a | 0 | 24 |
Indefinite results are interpreted as negative results.
3.3. Performance of RPA
Among these 170 samples, compared to conventional PCR, the sensitivity and specificity of the IS6110 RPA‐AGE assay were 100% (95% CI: 97.51, 100) and 100% (95% CI: 85.75, 100) and of the IS6110 RPA‐S assay were 97.95% (95% CI: 94.11, 100) and 100% (95% CI: 85.75, 100), respectively. The sensitivity and specificity of the IS1081 RPA‐AGE assay were 100% (95% CI: 97.51, 100) and 100% (95% CI: 85.75, 100) and of the IS1081 RPA‐S assay were 99.32% (95% CI: 96.24, 99.98) and 100%, respectively.
3.4. LOD of RPA‐AGE and RPA‐S
A limit of detection was determined according to the lowest DNA concentration that could be detected by either RPA‐AGE or RPA‐S. The LODs of RPA‐AGE were found to be 100 and 1000 times lower than those RPA‐S when targeting IS6110 and IS1081, respectively. When using the IS1081 primers, only 0.00005 and 0.05 ng of Mtb H37Rv DNA were required for the RPA‐AGE and RPA‐S reactions, respectively, to be positive. However, when using the IS6110 primers, 0.005 and 0.5 ng of the DNA templates were required for RPA‐AGE and RPA‐S reactions, respectively, to be consistently positive. IS1081 was found to be the more sensitive target compared to IS6110 in this study.
3.5. Specificity of RPA
The specificity of the RPA assay was assessed for both target sequences, IS6110 and IS1081. Samples of genomic DNA from A. baumannii, H. influenzae, K. pneumoniae, M. catarrhalis, P. aeruginosa, S. pneumonia, S. pyogenes, M. avium, and M. intracellulare were tested by RPA‐AGE and RPA‐S. Neither assay detected DNA from these bacteria. Figure 3 shows the detection of RPA products of IS1081 (data not shown for IS6110 detection).
Figure 3.
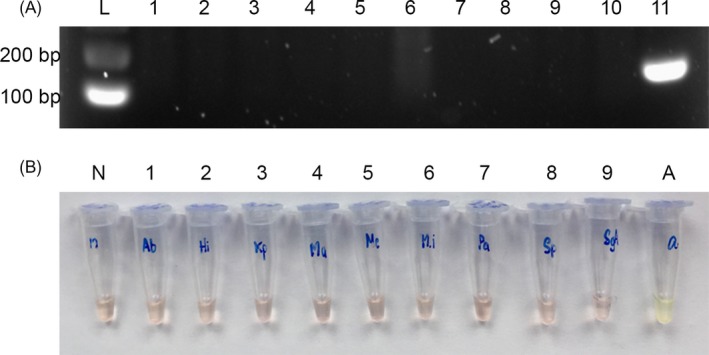
Specificity testing of RPA. A, RPA products of IS1081 were detected by agarose gel electrophoresis. Lane L is a 100‐bp DNA ladder, lane 1 is a negative control, and lanes 2‐10 are DNA from A. baumannii, H. influenzae, K. pneumoniae, M. catarrhalis, M. avium, M. intracellulare, P. aeruginosa, S. pneumonia, and S. pyogenes, respectively, showing no visible band. Lane 11 is Mtb H37Rv DNA, showing a positive 173‐bp band of IS1081. Each figure is the representative of triplicate on three separate experiments. B, RPA products of IS1081 were endpoint‐detected by SYBR Green I. Tube N is a negative control, and tubes 1‐9 are DNA from A. baumannii, H. influenzae, K. pneumoniae, M. catarrhalis, M. avium, M. intracellulare, P. aeruginosa, S. pneumonia, and S. pyogenes, respectively, showing an orange color. Tube A is Mtb H37Rv DNA, showing a green color, implying the positive result of amplification. Each figure is the representative of triplicate on three separate experiments
4. DISCUSSION
TB is a global healthcare concern worldwide due to its high mortality and morbidity. A rapid, accurate diagnosis and promptly appropriate treatment could be effective for decreasing death rates and disease transmission. Sputum microscopy is a frontline diagnostic test for TB used in developing countries, particularly in low‐resource and peripheral healthcare settings. However, the low sensitivity of sputum microscopy endangers the control of TB diseases.26 Isolation of M. tuberculosis, the causative agent of TB, is the most sensitive diagnostic method, but it is time‐consuming and requires sophisticated laboratory facilities.15 Despite the fact that nucleic acid amplification‐based diagnosis enables sensitive detection of TB at low density with a rapid turnaround time, the utility of current conventional and commercial amplification methods is limited by their costs and complexity.15 Previously, applications of isothermal amplification for Mtb detection have been reported.17, 27, 28 Isothermal amplification techniques do not require thermocycling instrumentation, which is a major advantage for point‐of‐care devices as the whole amplification processes take place under isothermal conditions.29 Furthermore, when isothermal amplification is combined with direct visual detection, a complete instrument‐free nucleic acid amplification is possible.
Loop‐mediated amplification is an isothermal amplification recommended by the WHO for detection of pulmonary TB.19 However, in our study, we did not perform LAMP assay to compare efficiency with the developed RPA assay. To the best of our knowledge, there is no evidence comparing RPA to LAMP to detect Mtb from the direct specimens. RPA and LAMP showed performances equivalent to the PCR to directly detect pathogens, including virus30 and fungus,31 in clinical samples. Both RPA and LAMP had short turnaround times coupled with satisfactory sensitivity and specificity. However, RPA was more simple to operate with a lower risk of contamination than with LAMP.31 Additionally, RPA has great feasibility of primer design and multiplexing.20
Recombinase polymerase amplification is a promising isothermal molecular tool that has been increasingly used in clinical microbiology laboratories because it has a simple, affordable, rapid, and sensitive method for identifying pathogens.20 Recently, RPA has been developed to detect Mtb DNA in combination with a variety of detection formats, including a fluorescence detection device24 and bridging flocculation.25 However, specific or electrical instruments are still required for these detection formats. Here, we attempted to develop RPA combined with SYBR Green I to obtain instrument‐free nucleic acid amplification and detection for Mtb diagnosis. With the RPA system, the manufacturer recommends general guidelines for designing RPA primers for a successful amplification.32 These include avoiding long tracks of one particular nucleotide, especially guanine, or a large number of small repeats at the 5′ terminus. GC content higher than 70% or lower than 30% must be avoided. RPA primers should be 30‐35 bases long, while more extended primer over 45 bases are not recommended because they are like to form the secondary structures.32 In our study, two to three forward and reverse primers for each specific sequence (IS6110 and IS1081) were designed. These RPA primer candidates contained 29‐30 bases and had GC content ranging from 56% to 62%. All primer candidates were analyzed for a possibility to form secondary structure (such as hairpin loop and self‐dimer) using OligoAnalyzer 3.1 program (IDT, Skokie, IL, USA). Each forward and reverse primer candidates were paired and preliminarily screened with the TwistAmp® Basic Kit as recommended by the manufacturer. Once optimal primers were identified, they were subsequently used in our developed RPA assay for specific amplification. SYBR Green I (250, 375, 500, 750, and 1000× in a final volume of 25 μL RPA products) was also validated to obtain the appropriate concentration. In our study, 375x of SYBR Green I showed the most distinguishable positive and negative results when detecting RPA products and was selected in the development processes. During RPA assay development, amount of DNA template, primer concentration, MgOAc concentration, and incubation temperature and period were optimized until gaining the most appropriate condition. In every optimization, the RPA amplicons were inspected by SYBR Green I compared with agarose gel electrophoresis. The only condition that produced clearly readout results (when observed with SYBR Green I) without nonspecific bands in agarose gel electrophoresis was selected.
SYBR Green I has been previously used for endpoint DNA product detection in several works. Cyanine dye preferentially binds to double‐stranded DNA, including PCR or RPA amplicons, resulting in a DNA‐dye complex that emits green light and can be read by the naked eye.33 Due to the simple colorimetric signal detection property of SYBR Green I, when combined with our RPA assay, this platform offers an intermediate outcome within only 15‐20 minutes. Additionally, no further specific instruments are required, permitting the development of point‐of‐care devices with fully portable test platforms.
Compared to conventional PCR, the RPA technique demonstrated a good capability to amplify Mtb DNA using IS6110 and IS1081 as targets. All of the Mtb isolates were correctly differentiated from NTM isolates via RPA‐AGE (100% sensitivity and specificity for either IS6110 or IS1081 targets). Our study showed that RPA‐S had reduced sensitivity to detect Mtb via either IS6110 (97.95% sensitivity) or IS1081 (99.32% sensitivity); however, it still had 100% specificity for both targets. There were 3 (for IS6110) and 1 (for IS1081) RPA‐S reactions that exhibited an indefinite orange‐green color after adding SYBR Green I, which were interpreted as negative for Mtb detection. A previous study showed that there was no significant difference between the LAMP results detected by agarose gel electrophoresis or naked eye detection of LAMP products after SYBR Green I addition.34 Although SYBR Green I exhibits good sensitivity for DNA detection, false negatives somehow occur when the DNA content is low.35 DNA from 300 ng to 2 μg has been suggested to be used as the template amount for amplification, followed by the detection by SYBR Green I; otherwise, the experimental results cannot be visualized by the naked eye.36 In our RPA‐S assay, the final amount of template DNA used was only 0.1 ng, which may have led to the false negatives observed in the four negative RPA‐S reactions mentioned above. Despite the disadvantages of SYBR Green I, such as extensive optimization or preferential binding to certain DNA sequences,37 it remains one of the most widely used dyes for nonspecific DNA detection due to its cost efficiency and easy setup.38
Our RPA assay targeted two different insertion sequences that are highly specific for Mtb, IS6110 and IS1081. In this study, RPA‐AGE and RPA‐S showed satisfying diagnostic specificity, with no cross‐reaction with other bacterial genera or species. While IS6110 is present in Mtb at a copy number of up to 25 per genome and has high sensitivity for Mtb diagnosis by PCR,39 Mycobacterium tuberculosis complex isolates with no IS6110 have been reported.40 On the contrary, IS1081 is present in all MTBC species, with a stable copy number of 5‐7 repeats per genome.41 When testing with IS1081, RPA‐AGE and RPA‐S were able to detect Mtb DNA as low as 0.00005 ng and 0.05 ng, respectively. However, IS6110 required a slightly higher amount of DNA 0.005 ng and 0.5 ng for the RPA‐AGE and RPA‐S assays, respectively. The DNA product length has been shown to impact the LOD of SYBR Green I. The optimal DNA product length recommended for SYBR Green I detection is generally less than 200 bp.33 Amplification of IS6110 by RPA generated products of 423 bp, which is larger than IS1081, which is173 bp. The reduced LOD of IS6110 RPA‐S may be due to the loss of efficiency of SYBR Green I when detecting the longer product.
The different LODs between RPA‐AGE and RPA‐S may be due to the efficiency of the dye used in the individual system. In the RPA‐AGE assay, the UltraPower DNA/RNA fluorescent dye was used to detect RPA amplicons when electroporated, and it is able to detect at least 20 pg of double‐stranded DNA in an agarose gel.42 SYBR Green I was used to detect RPA amplicons in the RPA‐S assay. However, SYBR Green I also has a high sensitivity for double‐stranded DNA detection, as low as 60 pg, when coupled with agarose gel electrophoresis with visualization via an UV transilluminator.43 In our study, the RPA‐S reaction was observed for a SYBR Green I color change; this change was observed by the naked eye. This detection method probably limits the efficiency of SYBR Green I when performing the LOD test via RPA‐S compared with RPA‐AGE in this study.
Conforming to a previous study that provided evidence for using RPA as a tool for the rapid and sensitive detection of DNA from Mtb,24 our developed RPA‐S assay provides a rapid (~15‐20 min from sampling to readout), sensitive (0.5‐0.05 ng for LOD), convenient molecular method for Mtb detection at an affordable price (~5 USD per reaction). Moreover, RPA‐S does not require expensive or specific laboratory devices, making it a possible candidate for development as a point‐of‐care diagnostic device for TB infection in future. Although a rapid amplification is now available, extraction of nucleic acid from direct specimens remains a laboratory‐based practice. Adoption of DNA tests at the point‐of‐care setting still requires the development of simplified sample‐handling procedures.
In conclusion, our study demonstrates the application of SYBR Green I with the RPA assay for Mtb detection by the naked eye. This assay can be implemented as an alternative molecular diagnostic test for the screening of TB infection, which could help control the spread of TB, especially in low‐resource and peripheral healthcare settings.
ACKNOWLEDGMENTS
The authors would like to acknowledge the assistance from the Clinical Microbiology Laboratory, Faculty of Medicine Ramathibodi Hospital, Mahidol University, Thailand, for providing Mtb H37Rv and Thai Mtb strains. We also would like to acknowledge the Department of Bacteriology, Osaka Prefectural Institute of Public Health, Osaka, Japan, for providing Japanese Mtb strains. This study was partially supported by the scholarship from the Graduate School, Chulalongkorn University, to commemorate the 72th anniversary of his Majesty King Bhumibol Adulyadej, the 90th Anniversary Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund), and the Chulalongkorn University Office of International Affairs Scholarship for Short‐term Research.
Singpanomchai N, Akeda Y, Tomono K, Tamaru A, Santanirand P, Ratthawongjirakul P. Naked eye detection of the Mycobacterium tuberculosis complex by recombinase polymerase amplification—SYBR green I assays. J Clin Lab Anal. 2019;33:e22655 10.1002/jcla.22655
REFERENCES
- 1. WHO . Global Tuberculosis Report 2017. Geneva: World Health Organization; 2017. http://www.who.int/tb/publications/global_report/en/. Accessed December 22, 2016. [Google Scholar]
- 2. WHO . Global Tuberculosis Report 2016. Geneva: World Health Organization; 2016. http://www.who.int/tb/publications/global_report/en/. Accessed December 22, 2016. [Google Scholar]
- 3. Uys PW, Warren R, van Helden PD, Murray M, Victor TC. Potential of rapid diagnosis for controlling drug‐susceptible and drug‐resistant tuberculosis in communities where Mycobacterium tuberculosis infections are highly prevalent. J Clin Microbiol. 2009;47(5):1484‐1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. CDC . Core Curriculum on Tuberculosis: What the Clinician Should Know, 6th ed Atlanta, GA: Centers for Disease Control and Prevention; 2013. [Google Scholar]
- 5. WHO . Internaltional standards for tuberculosis care. http://www.who.int/tb/publications/ISTC_3rdEd.pdf. Accessed May 15, 2017.
- 6. Reechaipichitkul W, Suleesathira T, Chaimanee P. Comparison of GeneXpert MTB/RIF assay with conventional AFB smear for duagnosis of pulmonary tuberculosis in northeastern Thailand. Southeast Asian J Trop Med Public Health. 2017;48:313‐321. [PubMed] [Google Scholar]
- 7. Pinyopornpanish K, Chaiwarith R, Pantip C, et al. Comparison of Xpert MTB/RIF assay and the conventional sputum microscopy in detecting Mycobacterium tuberculosis in northern Thailand. Tuberc Res Treat. 2015; 10.1155/2015/571782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dhingra VK, Aggarwal N, Rajpal S, Aggarwal JK, Gaur SN. Validity and reliability of sputum smear examination as diagnostic and screening test for tuberculosis. Indian J Allergy Asthma Immunol. 2003;17(2):67‐69. [Google Scholar]
- 9. Ganoza CA, Ricaldi JN, Chauca J, et al. Novel hypertonic saline‐sodium hydroxide (HS‐SH) method for decontamination and concentration of sputum samples for Mycobacterium tuberculosis microscopy and culture. J Med Microbiol. 2008;57(Pt 9):1094‐1098. [DOI] [PubMed] [Google Scholar]
- 10. Simner PJ, Doerr KA, Steinmetz LK, Wengenack NL. Mycobacterium and aerobic Actinomycete culture: are two medium types and extended incubation times necessary? J Clin Microbiol. 2016;54(4):1089‐1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park S‐H, Kim C‐K, Jeong H‐R, Son H, Kim S‐H, Park M‐S. Evaluation and comparison of molecular and conventional diagnostic tests for detecting tuberculosis in Korea, 2013. Osong Public Health Res Perspect. 2014;5(Suppl):S3‐S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Causse M, Ruiz P, Gutierrez‐Aroca JB, Casal M. Comparison of two molecular methods for rapid diagnosis of extrapulmonary tuberculosis. J Clin Microbiol. 2011;49(8):3065‐3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. WHO . Rapid Implementation of the Xpert MTB/RIF Diagnostic Test. World Health Organization; 2011. http://apps.who.int/iris/bitstream/10665/44593/1/9789241501569_eng.pdf. Accessed March 8, 2016. [Google Scholar]
- 14. Teo J, Jureen R, Chiang D, Chan D, Lin R. Comparison of two nucleic acid amplification assays, the Xpert MTB/RIF assay and the amplified Mycobacterium tuberculosis Direct assay, for detection of Mycobacterium tuberculosis in respiratory and nonrespiratory specimens. J Clin Microbiol. 2011;49(10):3659‐3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McNerney R, Daley P. Towards a point‐of‐care test for active tuberculosis: obstacles and opportunities. Nat Rev Microbiol. 2011;9(3):204‐213. [DOI] [PubMed] [Google Scholar]
- 16. Miller N, Hernandez SG, Cleary TJ. Evaluation of Gen‐probe amplified Mycobacterium tuberculosis direct test and PCR for direct detection of Mycobacterium tuberculosis in clinical specimens. J Clin Microbiol. 1994;32(2):393‐397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwamoto T, Sonobe T, Hayashi K. Loop‐mediated isothermal amplification for direct detection of Mycobacterium tuberculosis complex, M. avium, and M. intracellulare in sputum samples. J Clin Microbiol. 2003;41(6):2616‐2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. WHO . The Use of a Commercial Loop‐Mediated Isothermal Amplification Assay (TB‐LAMP) for the Detection of Tuberculosis: Expert Group Meeting Report Geneva. World Health Oraganization; 2013. apps.who.int/iris/bitstream/10665/…/WHO_HTM_TB_2013.05_eng.pdf. Accessed January 15, 2017. [Google Scholar]
- 19. WHO . The Use of Loop‐Mediated Isothermal Amplification (TB‐LAMP) for the Diagnosis of Pulmonary Tuberculosis: Policy Guidance. World Health Organization; 2016. http://www.who.int/tb/publications/lamp-diagnosis-molecular/en/. Accessed January 15, 2017. [PubMed] [Google Scholar]
- 20. Daher RK, Stewart G, Boissinot M, Bergeron MG. Recombinase polymerase amplification for diagnostic applications. Clin Chem. 2016;62(7):947‐958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Piepenburg O, Williams CH, Stemple DL, Armes NA. DNA detection using recombination proteins. PLoS Biol. 2006;4(7):e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Crannell ZA, Rohrman B, Richards‐Kortum R. Equipment‐free incubation of recombinase polymerase amplification reactions using body heat. PLoS One. 2014;9(11):e112146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang R, Zhang F, Wang L, et al. Instant, visual, and instrument‐free method for on‐site screening of GTS 40‐3‐2 soybean based on body‐heat triggered recombinase polymerase amplification. Anal Chem. 2017;89(8):4413‐4418. [DOI] [PubMed] [Google Scholar]
- 24. Boyle DS, McNerney R, Teng Low H, et al. Rapid detection of Mycobacterium tuberculosis by recombinase polymerase amplification. PLoS One. 2014;9(8):e103091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ng BYC, Wee EJH, West NP, Trau M. Rapid DNA detection of Mycobacterium tuberculosis‐towards single cell sensitivity in point‐of‐care diagnosis. Sci Rep. 2015;5:15027. [Google Scholar]
- 26. Keeler E, Perkins MD, Small P, et al. Reducing the global burden of tuberculosis: the contribution of improved diagnostics. Nature. 2006;444(Suppl 1):49‐57. [DOI] [PubMed] [Google Scholar]
- 27. Moon SH, Kim EJ, Tomono J, et al. Detection of Mycobacterium tuberculosis complex in sputum specimens using a loop‐mediated isothermal amplification assay in Korea. J Med Microbiol. 2015;64(11):1335‐1340. [DOI] [PubMed] [Google Scholar]
- 28. Mori Y, Notomi T. Loop‐mediated isothermal amplification (LAMP): a rapid, accurate, and cost‐effective diagnostic method for infectious diseases. J Infect Chemother. 2009;15(2):62‐69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao Y, Chen F, Li Q, Wang L, Fan C. Isothermal amplification of nucleic acids. Chem Rev. 2015;115(22):12491‐12545. [DOI] [PubMed] [Google Scholar]
- 30. Escadafal C, Paweska JT, Grobbelaar A, et al. International external quality assessment of molecular detection of Rift Valley fever virus. PLoS Negl Trop Dis. 2013;7(5):e2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ahmed SA, van de Sande WW, Desnos‐Ollivier M, Fahal AH, Mhmoud NA, de Hoog GS. Application of isothermal amplification techniques for identification of Madurella mycetomatis, the prevalent agent of human mycetoma. J Clin Microbiol. 2015;53(10):3280‐3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. TwistDx . Appendix to the TwistAmp™ reaction kit manuals. https://www.twistdx.co.uk/docs/default-source/twistamp-manuals/newappendix_2_.pdf?sfvrsn=8. Accessed December 25, 2016.
- 33. Debode F, Marien A, Janssen É, Bragard C, Berben G. The influence of amplicon length on real‐time PCR results. Biotechnol Agron Soc Environ. 2017;21(1):3‐11. [Google Scholar]
- 34. Birmpa A, Kalogeropoulos K, Kokkinos P, Vantarakis A. Evaluation of two loop‐mediated isothermal amplification methods for the detection of Salmonella Enteritidis and Listeria Monocytogenes in artificially contaminated ready‐to‐eat fresh produce. Ital J Food Saf. 2015;4(3):5383 10.4081/ijfs.2015.5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang Y, Yang K, Bai C, et al. Development of a SYBR Green I real‐time PCR for the detection of the orf virus. AMB Express. 2017;7:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu Y, Lei T, Liu Z, Kuang Y, Lyu J, Wang Q. A Novel technique to detect EGFR mutations in lung cancer. Int J Mol Sci. 2016;17(5):792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gudnason H, Dufva M, Bang D, Wolff A. Comparison of multiple DNA dyes for real‐time PCR: effects of dye concentration and sequence composition on DNA amplification and melting temperature. Nucleic Acids Res. 2007;35(19):e127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Negura L, Negura A. Limits and pitfalls of SYBR Green detection in quantitative PCR. Analele Ştiinţifice Ale Universităţii Alexandru Ioan Cuza din Iași,Sectiunea II A: Genetica si Biologie Moleculara; 2012.
- 39. Flores LL, Pai M, Colford JM, Riley LW. In‐house nucleic acid amplification tests for the detection of Mycobacterium tuberculosis in sputum specimens: meta‐analysis and meta‐regression. BMC Microbiol. 2005;5:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huyen MN, Tiemersma EW, Kremer K, et al. Characterisation of Mycobacterium tuberculosis isolates lacking IS6110 in Viet Nam. Int J Tuberc Lung Dis. 2013;17(11):1479‐1485. [DOI] [PubMed] [Google Scholar]
- 41. Collins DM, Stephens DM. Identification of an insertion sequence, IS1081, in Mycobacterium bovis . FEMS Microbiol Lett. 1991;67(1):11‐15. [DOI] [PubMed] [Google Scholar]
- 42. Gallex . UltraPower DNA/RNA safe dye. http://www.gellex.jp/item/ultrapower_safedye. Accessed February 24, 2018.
- 43. TaKaRa . SYBR green I nucleic acid gel stain. http://catalog.takara-bio.co.jp/PDFS/5760A_5761A_5770A_5771A_j.pdf. Accessed February 26, 2018.
- 44. Nghiem MN, Nguyen BV, Nguyen ST, Vo TT, Nong HV. A Simple, single triplex PCR of IS6110, IS1081, and 23S ribosomal DNA targets, developed for rapid detection and discrimination of Mycobacterium from clinical samples. J Microbiol Biotechnol. 2015;25(5):745‐752. [DOI] [PubMed] [Google Scholar]


