Abstract
Aim
To develop a new immunoassay based on the time‐resolved fluorescence immunoassay (TRFIA) system for the simultaneous measurement of IgM and IgG antibodies to HCV.
Methods
Coated recombinant HCV antigens and Eu3+‐labeled IgM and Sm3+‐labeled IgG antibodies were prepared. HCV‐IgM/IgG TRFIA was established and optimized, followed by a methodological assessment. Data were expressed as +SD and analyzed using the SPSS 13.0 software. The percentile method was used to calculate cutoff values.
Results
The detection sensitivities of HCV‐IgM and HCV‐IgG were 0.06 S/CO and 0.15 S/CO, respectively. There was a good linear response from 1:40 to 1:40 960 for HCV‐IgM and 1:20 to 1:40 960 for HCV‐IgG, when samples strongly positive for HCV‐IgM and HCV‐IgG were serially diluted from 1:10 to 1:81 920. The average intra‐assay coefficients of variation (CV) for HCV‐IgM and ‐IgG were 3.45% and 3.71% and the inter‐assay coefficients of variation (CV) were 6.49% and 6.79%, respectively. When HCV‐negative/positive sera were tested by ELISA and using established kits, the negative, positive, and total conformity rates for HCV‐IgM were 93.3% (28/30), 100% (25/25), and 96.4% (53/55), and those for HCV‐IgG were 93.3% (28/30), 100% (35/35), and 96.9% (63/65), respectively. Additionally, the established kit exhibited good stability, with declines in fluorescence values to 11.1% and 9.5%, respectively, after storage at 37°C for 7 days.
Conclusion
We established a dual‐label HCV‐IgM/IgG TRFIA assay with a wide detection range, high specificity, high sensitivity, good stability, and good clinical value for the simultaneous measurement of HCV‐IgM and HCV‐IgG titers in a single test.
Keywords: Dual‐labeling, hepatitis C virus, time‐resolved fluorescence immunoassay
1. INTRODUCTION
Hepatitis C virus (HCV) infection is a public health problem of global concern. According to the most recent report by the World Health Organization (WHO), approximately 18.5 million people have a history of HCV infection, and nearly 350 000 people die of HCV‐related diseases annually.1 China ranks first in the world, with an HCV prevalence of approximately 1% and around 13 million infected people.2, 3 The clinical symptoms of HCV infection are not easily apparent, and the spontaneous seroconversion rate is low. Approximately 75%‐85% of those who are initially infected with HCV develop chronic inflammation, which further deteriorates to liver cirrhosis or even hepatocellular carcinoma.4 The number of deaths caused by HCV infection is expected to increase threefold by 2025.5
Effective screening of HCV infection directly affects the accuracy of diagnosis and treatment, prevention of HCV infection, and the safety of blood sources. Currently, HCV is mainly identified using enzyme‐linked immunosorbent assays (ELISA) to detect HCV antibodies. Compared to HCV RNA detection, ELISA has a lower cost, is easier to operate, has a high reproducibility, and is suitable for detection using a large number of clinical serum samples. The development of third‐generation reagents has shortened the detection window period for HCV antibodies to approximately 8 weeks.6 However, improved methods for the early diagnosis of viral infections and the effective screening of blood donors are still needed. Serum HCV‐IgM antibodies appear earlier than IgG after HCV infection, with a window period of approximately 3‐7 weeks. Some studies have shown a good correlation between HCV‐IgM antibody levels and HCV RNA levels (window period of 1‐2 weeks). Despite the benefits of the joint detection of HCV‐IgM/IgG antibodies, commercial reagents for dual antibody detection are lacking. In addition, a large number of unconfirmed and false‐positive results are obtained owing to the insufficient sensitivity and stability of ELISA, which results in the missed detection of true‐positive cases and the detection of only a single target at a time. Therefore, in this study, dual‐label time‐resolved fluoroimmunoassay (TRFIA) was used to establish a method for the simultaneous detection of HCV‐IgM and IgG with high sensitivity, a wide detection range, and high stability.
2. MATERIALS AND SUBJECTS
2.1. Chemicals and reagents
HCV recombinant antigen (NS3/NS4/NS5/Core) was purchased from SYM‐BIO (Suzhou, China). The automatic DELFIA‐1235 TRFIA Analyzer was obtained from Perkin‐Elmer Life Sciences and Analytical Science/Wallac Oy (Turku, Finland). The ABI7500 Fluorescent quantitative PCR Device was purchased from Applied Biosystems (Shanghai, China). Rabbit anti‐human IgM (110M4778) and IgG (049K4803) were purchased from Sigma (St. Louis, MO, USA). Enhancer solution and Eu3+‐labeling kit (1244‐302) and Sm3+‐labeling kit (1244‐303) were purchased from PE Company (EG&G‐Wallac, Finland). Bovine serum albumin (BSA) was supplied by the Department of Health, Institute of Biological Products (OO0303A, Shanghai, China). The Hepatitis C Viral RNA Quantitative Fluorescence Diagnostic Kit was purchased from Sansure Biotech (Changsha, China). The PD‐10 column and Sepharose CL‐6B column were obtained from the Pharmacia Company (Peapack, NJ, USA). Concentrated washing solution (170213), enhancer solution (170710), and reaction buffer (170227) were supplied by Jiangsu Key Laboratory of Molecular Nuclear Medicine (Wuxi, China). ELISA kits for HCV antibody detection were purchased from Beijing Wantai BioPharm (2017080908, Beijing, China). Anti‐hepatitis A (HAV) (AM20170402B), anti‐hepatitis D (HDV) (20170503R), and anti‐hepatitis E (HEV) (EM20170101B), as well as the positive standard, were purchased from Beijing Controls & Standards Company (Beijing, China). Anti‐human immunodeficiency virus (HIV) (2017010112) and anti‐Treponema pallidum (TP) antibodies (2017102612) were purchased from Beijing Wantai BioPharm (Beijing, China). All additional chemicals and reagents were of analytical grade.
2.2. Source of samples
Serum samples were collected from 200 patients with hepatitis C infection (18 acute hepatitis C, 24 subacute hepatitis C, and 158 chronic hepatitis patients) admitted to our hospital from February 2015 to March 2016 and from 270 healthy volunteers. The inclusion criterion for the HCV‐infected patients was that their serum samples were positive for anti‐HCV antibody or HCV‐RNA. HCV‐infected individuals were excluded if they had HAV, HBV, HDV, or HEV infection. Similarly, healthy control individuals were excluded if they were infected with HAV, HBV, HCV, HDV, or HEV. Informed consent was obtained from all study participants during the medical examination, and the protocol was approved by the ethics committees of Wuxi People's Hospital Affiliated to Nanjing Medical University.
3. METHODS
3.1. Preparation of solid‐phase antigens
Recombinant HCV antigens were diluted with a coating buffer containing 50 mmol/L Na2CO3‐NaHCO3 (pH 9.6), and 100 μL coating buffer was added to each well of a 96‐well microtiter plate and incubated overnight at 4°C. The coating buffer was discarded, and each well was blocked with 100 μL of 3 g/L BSA at room temperature for 2 hours. The blocking buffer was discarded, and the plates were dried under a vacuum and stored at −20°C in a sealed plastic bag with a desiccant until use.
3.2. Preparation of labeled antibodies
Eu3+ labeling of rabbit anti‐human IgM antibodies was performed according to the instructions provided with the Eu3+ labeling kit. One milliliter of rabbit anti‐human IgM antibodies was passed through a PD‐10 column to adjust the buffering conditions using a 50 mmol/L Na2CO3‐NaHCO3 (pH 8.5) elution solution containing 0.155 mol/L NaCl. The fractions with protein peaks were collected and concentrated to 2 g/L. Then, 500 μL of the above solution was added to a bottle containing freeze‐dried 0.2 mg Eu3+‐N2‐[p‐isocyanato‐benzyl]‐diethylenetriamine tetraacetic acid and reacted for 20 hours at 25°C with a magnetic stirrer. The reaction solution was passed through an agarose CL‐6B column (1 × 30 cm) that was equilibrated with an 80 mmol/L Tris‐HCl (pH 7.8) buffer solution for chromatography with a fully automated TRFIA detector. The protein peaks were collected and packaged (1 mL/bottle) after dilution and freeze‐dried for storage. Sm3+‐labeled IgG antibodies were prepared using the same method.
3.3. Establishment of the HCV‐IgM/IgG TRFIA
The assay is summarized in Figure 1. In brief, after diluting the serum with buffer 200 times, 100 μL of test serum or positive or negative control serum was added to microplates coated with HCV antigens. After incubation at 25°C with shaking for 45 minutes, the plates were washed five times with washing solution before the addition of 100 μL/well Eu3+‐rabbit anti‐human IgM and Sm3+‐rabbit anti‐human IgG conjugates, followed by incubation at room temperature with shaking for 45 minutes. After washing twice, the enhancer solution (100 μL/well) was added. The fluorescence values were read after incubation at room temperature for 5 minutes with shaking, during which the HCV‐IgM/IgG antibodies in the serum were bound to the solid‐phase plate. HCV antigen‐HCV IgM‐Eu3+ and HCV antigen‐HCV IgG‐Sm3+ complexes formed after the addition of Eu3+‐rabbit anti‐human IgM and Sm3+‐rabbit anti‐human IgG. After the addition of a dissociation enhancer for dissociating the lanthanide ions, the fluorescence intensity was directly proportional to the levels of HCV‐IgM/IgG antibodies in the sample.
Figure 1.
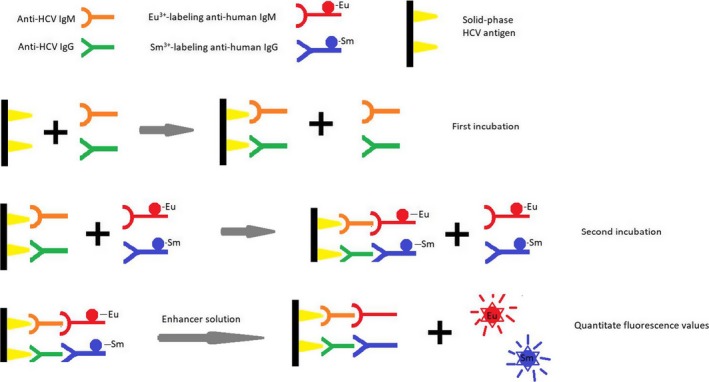
Illustration of the HCV‐IgM/IgG TRFIA detection principle
3.4. Methodological assessment and statistical analysis
The sensitivity, specificity, precision, and stability of the assay were compared with those obtained using existing methods. SPSS 13.0 was used for statistical analyses.
4. RESULTS
4.1. Labeling rate
Using the Eu3+ and Sm3+ standards provided by PerkinElmer, the labeling rates were 10.4 Eu3+ ions per anti‐human IgM antibody and 9.6 Sm3+ ions per anti‐human IgG antibody.
4.2. Optimal antibody labeling concentration
The labeled antibodies from the reaction step were diluted 1:10, 1:20, 1:50, 1:100, 1:200, 1:400, 1:800, and 1:1600. We simultaneously tested the positive and negative samples. The background signal increased as the labeling concentration increased, and the dilution with the maximum positive/negative (P/N) ratio was selected. Thus, we obtained the optimal dilutions for Eu3+‐rabbit anti‐human IgM (1:100) and Sm3+‐rabbit anti‐human IgG (1:50).
4.3. Optimal serum dilution
The negative serum was diluted 1:25, 1:50, 1:100, 1:200, 1:400, and 1:800 with the buffer to decrease nonspecific adsorption. The decline in the fluorescence value tended to plateau as the sample dilution increased, and 1:200 was the optimal serum dilution (Figure 2).
Figure 2.
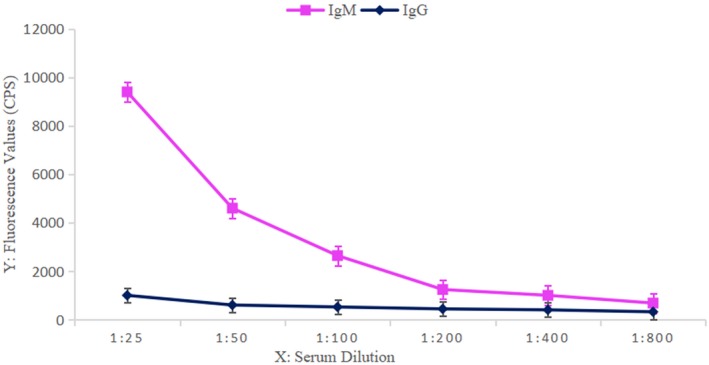
Serial dilution of negative serum samples and corresponding fluorescence values
4.4. Confirmation of positive cutoff values (CO values)
The Eu3+/Sm3+ dual‐label TRFIA was used to detect HCV in sera from 270 healthy volunteers. The HCV‐IgM and HCV‐IgG fluorescence values ( ± SD) were 4020.5 ± 1856.4 and 580.0 ± 272.3, respectively. The sample fluorescence/mean negative control fluorescence values (S/Nx) are shown in Table 1. The reference CO value was 95%, and the one‐tailed S/Nx CO values for HCV‐IgM and HCV‐IgG were 2.2 and 2.4, respectively. Thus, the following equations were derived: Cutoff IgM = negative control value × 2.2 (negative control values less than 4,021 were set to 4021); cutoff IgG = negative control value × 2.4 (negative control values less than 580 were set to 580); S/CO ≥ 1 was considered positive.
Table 1.
Serum HCV‐IgM and HCV‐IgG fluorescence values relative to the negative control fluorescence values for 270 healthy volunteers
| S/N value | 0.26~ | 0.66~ | 1.06~ | 1.46~ | 1.86~ | 2.26~ | >2.66 | |
|---|---|---|---|---|---|---|---|---|
| HCV‐IgM | n | 99 | 84 | 39 | 22 | 15 | 5 | 6 |
| Percentage (%) | 36.7 | 31.1 | 14.4 | 8.1 | 5.6 | 1.9 | 2.2 | |
| Cumulative percentage (%) | 36.7 | 67.8 | 82.2 | 90.3 | 95.9 | 97.8 | 100 |
| S/N value | 0.32~ | 0.82~ | 1.32~ | 1.82~ | 2.32~ | 2.82~ | >3.32 | |
|---|---|---|---|---|---|---|---|---|
| HCV‐IgG | n | 143 | 84 | 18 | 11 | 5 | 3 | 6 |
| Percentage (%) | 53.0 | 31.1 | 6.7 | 4.1 | 1.9 | 1.1 | 2.2 | |
| Cumulative percentage (%) | 53.0 | 84.1 | 90.7 | 94.8 | 96.7 | 97.8 | 100 |
4.5. Methodological assessment
4.5.1. Linearity
Strongly positive HCV‐IgM and HCV‐IgG samples were serially diluted from 1:10 to 1:81 920. The established TRFIA method was used to quantitate fluorescence. IgM showed a good linear response at dilutions of 1:140 to 1:40 960, and IgG showed a good linear response from 1:20 to 1:40 960 (Figure 3).
Figure 3.
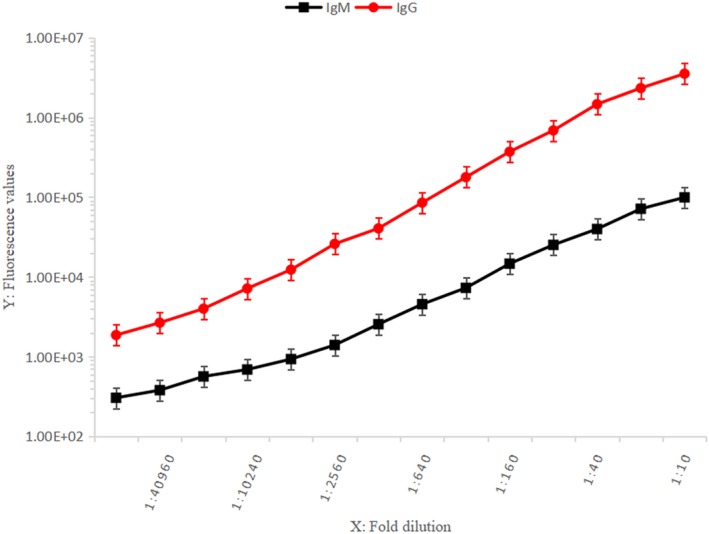
Detection range for HCV‐IgM/IgG TRFIA (1:10 to 1:81 920)
4.5.2. Specificity
The established reagent kit was used to detect reference samples positive for anti‐HAV, anti‐HDV, anti‐HEV, anti‐HIV, and anti‐TP. The test results were all negative, showing that there was no cross‐reactivity with the above antibodies.
4.5.3. Sensitivity
The fluorescence values for blank controls in 10 wells were measured according to + 2SD. The detection limits for HCV‐IgM and HCV‐IgG were 0.06 S/CO and 0.15 S/CO, respectively.
4.5.4. Precision
Samples with high, medium, and low concentrations were tested 10 times under similar experimental conditions within 8 days (Table 2). The mean intra‐assay Circulation Volume (CV) for IgM and IgG were 3.45% and 3.71%, respectively, and the mean inter‐assay CV were 6.49% and 6.79%, respectively. The mean intra‐assay CV was less than 10% and the mean inter‐assay CV was less than 15%, showing that our method has good precision.
Table 2.
Intra‐assay and inter‐assay precision
| Group | Low | Medium | High | ||||
|---|---|---|---|---|---|---|---|
| IgM | IgG | IgM | IgG | IgM | IgG | ||
| Intra‐assay precision (n = 20) | Concentration (S/CO) | 1.75 | 1.87 | 5.91 | 18.36 | 20.54 | 88.63 |
| CV (%) | 4.23 | 5.12 | 3.25 | 3.48 | 2.87 | 2.52 | |
| Inter‐assay precision (n = 10) | Concentration (S/CO) | 1.68 | 1.73 | 5.84 | 18.65 | 20.42 | 87.45 |
| CV (%) | 7.71 | 8.54 | 6.63 | 6.46 | 5.13 | 5.38 | |
4.5.5. Comparative tests
The established TRFIA method and ELISA for HCV‐IgM and IgG detection were simultaneously used to test 55 serum samples from 200 patients with HCV and 65 serum samples from 270 healthy volunteers collected from February 2015 to March 2016. The comparative results are summarized in Table 3. The IgM positive, negative, and total coincidence rates were 100% (25/25), 93.3% (28/30), and 96.4% (53/55), respectively. Similarly, the IgG positive, negative, and total coincidence rates were 100% (35/35), 93.3% (28/30), and 96.9% (63/65), respectively.
Table 3.
Comparison of HCV‐IgM and HCV‐IgG results
| TRFIA | ELISA | Total | ||
|---|---|---|---|---|
| Positive (n) | Negative (n) | |||
| HCV‐IgM | Positive (n) | 25 | 2 | 27 |
| Negative (n) | 0 | 28 | 28 | |
| Total | 25 | 30 | 55 | |
| HCV‐IgG | Positive (n) | 35 | 2 | 37 |
| Negative (n) | 0 | 28 | 28 | |
| Total | 35 | 30 | 65 | |
4.5.6. Stability
Reagent kits stored at 37°C for 7 days and those stored under normal conditions were used to simultaneously quantitate six HCV‐positive samples. The quantitation results showed that the HCV IgM fluorescence values obtained using the reagents stored at 37°C decreased by 12.2%, 13.8%, 11.0%, 10.2%, 9.8%, and 9.6% compared to those obtained using the reagents stored under normal conditions, while the IgG fluorescence values decreased by 10.4%, 8.4%, 9.6%, 10.7%, 9.7%, and 8.4%. The mean decreases for IgM and IgG were 11.1% and 9.5%, respectively.
4.6. Clinical applications
The prepared HCV‐IgM/IgG TRFIA reagent kit was used to analyze sera from 200 patients with HCV and compared with the results of the HCV‐RNA and HCV‐IgG and IgM. The joint testing of HCV‐IgM and IgG increased the positive rate of HCV detection (Table 4).
Table 4.
Comparison of HCV‐RNA detection by FQ‐PCR and HCV‐IgM and IgG detection by TRFIA
| HCV‐RNA | n | HCV‐IgM | HCV‐IgG | ||
|---|---|---|---|---|---|
| − | + | − | + | ||
| − | 93 | 93 | 0 | 0 | 93 |
| + | 107 | 59 | 48 | 8 | 99 |
| Total | 200 | 152 | 48 | 8 | 192 |
5. DISCUSSION
HCV ELISA reagent kits have undergone multiple rounds of improvement, and their sensitivity and specificity have been continuously optimized since their initial production in 1989, when Choo et al (Chiron Corporation, Emeryville, CA, USA) established the first generation of HCV antibody diagnosis reagent kits.6, 7 However, as ELISA uses active enzymes, many factors may affect the labeling process, and issues with the linear range and stability are common drawbacks. Hence, a large number of unconfirmed and false‐positive results are obtained in low‐risk populations, and true HCV‐positive cases may sometimes be missed, which is a matter of immense concern.8 In addition, there is variation in results depending on the kit manufacturer. Currently, the clinical detection of HCV antibodies is mainly based on IgG detection by ELISA, but the single detection of HCV IgG cannot be used to distinguish between acute and chronic hepatitis, and the window period is long. Thus, ELISA has limitations for the detection of early acute infections. The HCV‐IgM titer, which is positive at about 4 weeks of HCV acute infection, starts to decrease when the HCV‐IgG titer reaches its peak at weeks 12‐16. Thus, HCV‐IgM is more appropriate for the early diagnosis of HCV infection and the determination of antiviral therapeutic efficacy, liver disease activity, and correlations with viral replication.9, 10
For suspected cases of HCV‐IgG‐negative samples based on ELISA, HCV‐RNA can be detected by fluorescence quantitative PCR (FQ‐PCR), but the positive detection rate is low because of instrument‐associated problems, a low detection threshold, and low levels of replicating virus in patients after treatment.
Commonly used immunoassays, such as radioimmunoassays, ELISA, chemiluminescent immunoassays, and electrochemiluminescence immunoassays have various limitations. For example, the use of FQ‐PCR is limited by its complexity, and ELISA is only able to detect single antibodies. TRFIA is a novel technology that uses lanthanide elements for labeling, and it provides various benefits, such as a high sensitivity, wide linear range, and good tracer stability; in addition, it is not easily affected by sample autofluorescence.11, 12 Moreover, multiple labeling analyses can be performed as the detection windows and wavelengths of lanthanide elements vary. Eu3+ and Sm3+, which are used extensively in clinical and scientific research, use the same enhancer system containing β‐naphthoyl trifluoroacetone (β‐NTA) and a pair of common labels.13, 14
The Eu3+ and Sm3+ dual‐labeled HCV‐IgM/IgG TRFIA method established in this study can be used to simultaneously detect serum HCV‐IgM and HCV‐IgG in a single experiment. In this system, the detection sensitivity, mean intra‐assay CV, and inter‐assay CV of HCV‐IgM were 0.06 S/CO, 3.45%, and 6.49%, respectively, and those of HCV‐IgG were 0.15 S/CO, 3.71%, and 6.79%, respectively, conforming to clinical requirements. ELISA was used as a reference to detect HCV‐positive and ‐negative samples; the HCV‐IgM positive coincidence rate was 100%, negative coincidence rate was 93.3%, and total coincidence rate was 96.4%. The HCV‐IgG positive, negative, and total coincidence rates were 100%, 93.3%, and 96.9%, respectively. In addition, samples strongly positive for HCV‐IgM and HCV‐IgG were serially diluted from 1:10 to 1:81 920. HCV‐IgM and HCV‐IgG showed a wide linear range, with good linear responses from 1:40 to 1:40 960 and 1:20 to 1:40 960, respectively. The stability of the assay also satisfies clinical reagent requirements. When reagent kits stored at 37°C for 7 days and those stored under normal conditions were used to simultaneously quantitate six HCV‐positive samples, the mean HCV IgM and IgG fluorescence values for the reagent box stored at 37°C decreased by only 11.1% and 9.5%, respectively.
In conclusion, the HCV‐IgM/IgG TRFIA method established in this study does not cause radioactive pollution, requires 2‐3 hours for complete detection, has a wide detection range, and can simultaneously generate results for both HCV‐IgM and HCV‐IgG in a single experiment. This reduces the clinical workload, labor, and reagent costs, and increases the accuracy of detection by avoiding false‐positive and false‐negative results and limits the differences resulting from variation in experimental conditions. The detection levels for TRFIA were better than those of other methods, reaching 10−18 mol/L (chemiluminescence: 10−15 mol/L, ELISA: 10−9 mol/L), and the results were reproducible. In addition, the instruments required for TRFIA are easy to operate and are widely used in China. We therefore expect this method to improve the early diagnosis of HCV infection and the effective screening of blood donors.
AUTHOR CONTRIBUTIONS
Z.G. Hu conceived the study and designed the experiments. X. Yang and Y. Ye conducted the experiments and wrote the manuscript. T.T. Wang, M. Li, L. Yu, M. Xia, and J. Qian analyzed the results.
ACKNOWLEDGMENTS
Funding: This work was supported by the Key Project of Nanjing Medical University (JMUZD057), Wuxi Key Medical Talents (zdrc006), Wuxi Young Medical Talents (QNRC045), and Jiangsu Province Science and Education Project (CXTDB2017016).
Yang X, Ye Y, Wang T, et al. Eu3+/Sm3+ dual‐label time‐resolved fluoroimmunoassay for measurement of hepatitis C virus antibodies. J Clin Lab Anal. 2019;33:e22659 10.1002/jcla.22659
Yang, Ye and Wang contributed equally to this work and should be considered as first authors.
REFERENCES
- 1. World Health Organization . Guidelines for the Screening Care and Treatment of Persons with Chronic Hepatitis C Infection (Updated Version). Geneva: WHO Document Production Services; 2016:43‐47. [PubMed] [Google Scholar]
- 2. Cui Y, Jia J. Update on epidemiology of hepatitis B and C in China. J Gastroenterol Hepatol. 2013;28:7‐10. [DOI] [PubMed] [Google Scholar]
- 3. Hajarizadeh B, Grebely J, Dore GJ. Epidemiology and natural history of HCV infection. Nat Rev Gastroenterol Hepatol. 2013;10:553‐562. [DOI] [PubMed] [Google Scholar]
- 4. Mohamed AA, Elbedewy TA, El‐Serafy M, et al. Hepatitis C virus: a global view. World J. Hepatol. 2015;7:2676‐2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Thomas DL. Global control of hepatitis C: where challenge meets opportunity. Nat Med. 2013;19:850‐858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alborino F, Burighel A, Tiller FW, et al. Multicenter evaluation of a fully automated third‐generation anti‐HCV antibody screening test with excellent sensitivity and specificity. Med Microbiol Immunol. 2011;200:77‐83. [DOI] [PubMed] [Google Scholar]
- 7. Maity S, Nandi S, Biswas S, et al. Performance and diagnostic usefulness of commercially available enzyme linked immunosorbent assay and rapid kits for detection of HIV, HBV and HCV in India. Virol J. 2012;9:290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pondé RA. Enzyme‐linked immunosorbent/chemiluminescence assays, recombinant immunoblot assays and nucleic acid tests in the diagnosis of HCV infection. Eur J Clin Microbiol Infect Dis. 2013;32:985. [DOI] [PubMed] [Google Scholar]
- 9. Irshad M, Mankotia DS, Irshad K. An insight into the diagnosis and pathogenesis of hepatitis C virus infection. World J Gastroenterol. 2013;19:7896‐7909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chevaliez S, Feld J, Cheng K, et al. Clinical utility of HCV core antigen detection and quantification in the diagnosis and management of patients with chronic hepatitis C receiving an all‐oral, interferon‐free regimen. Antivir Ther. 2016;23:211‐217. [DOI] [PubMed] [Google Scholar]
- 11. Ye Y, Hu Z, Liu J, et al. A new Eu(3 + )‐labeled method for anticardiolipin antibody IgM. J Clin Lab Anal. 2014;28:335‐340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liang RL, Yang YS, Zhou JW, et al. Dual‐labeled time‐resolved immunofluorometric assay for the simultaneous quantitative detection of hepatitis B virus antigens in human serum. J Fluoresc. 2016;27:1‐8. [DOI] [PubMed] [Google Scholar]
- 13. Li M, Xiao HL, Liu J, et al. The establishment and application of a time‐resolved fluoroimmunoassay in detection of HBV large surface protein. Chin J Nucl Med. 2014;34:362‐365. [Google Scholar]
- 14. De Pauw PE, Vermeulen I, Ubani OC, et al. Simultaneous measurement of plasma concentrations of proinsulin and C‐peptide and their ratio with a trefoil‐type time‐resolved fluorescence immunoassay. Clin Chem. 2008;54:1990‐1998. [DOI] [PubMed] [Google Scholar]


