Abstract
Background
The H19 is a maternally expressed imprinted gene transcribing a long noncoding RNA (lncRNA), which has previously been reported to be involved in tumorigenesis and cancer progression. The aim of this study was to evaluate the associations between two lncRNA‐H19 (rs3741219 T>C and rs217727 C>T) gene polymorphisms with the risk of breast cancer (BC).
Methods
In a case‐control investigation, we evaluated 150 BC patients and 100 cancer‐free subjects in East Azerbaijan Province of Iran. To assess two gene polymorphisms, the polymerase chain reaction‐restriction fragment length polymorphism (PCR‐RFLP) method was used.
Results
The genotype frequencies of two lncRNA‐H19 (rs217727 C>T and rs3741219 T>C) gene polymorphisms TT + TC/CC and CC + CT/TT have not shown a statistically significant association with the risk of BC (P = 0.065; OR = 0.967; 95% CI, 0.938‐0.996) and (P = 0.510; OR = 1.583; 95% CI, 0.399‐6.726), respectively. In addition, our findings revealed a significant differences in allele frequencies in lncRNA‐H19 rs217727 C>T polymorphism between groups (P = 0.033; OR = 1.985; 95% CI, 1.048‐3.761).
Conclusion
Our findings suggested that rs217727 C>T polymorphism may be involved in the pathogenesis of BC, whereas rs3741219 T>C variation may not be involved in the genetic background of BC in Iranian.
Keywords: breast cancer, H19 gene, LncRNA, LncRNA‐H19
1. INTRODUCTION
Breast cancer (BC) is one of the most prominent causes of illness and death worldwide. BC is a malignant proliferation of epithelial cells that cover lacrimal ducts or lobules. Risk factors such as premenstrual meningitis, premature menopause, infertility, or age at the time of first birth may be responsible for one‐third of cases of BC. With the improvement of early diagnosis and treatment regime, the mortality rate of BC significantly decreased.1 Breast cancer is a complex disease and multifactorial process, in which a large number of epidemiological studies have been conducted to identify other risk factors for BC.2 Previous genomic research has provided evidence to support single‐nucleotide polymorphisms (SNP) in several genes associated with the risk of BC and contributed as carcinogenic agents.3, 4 The total of SNPs has been identified in cancers; nearly 10 percent were associated with a change in the amino acid sequence, while a large proportion occurred in the coding or noncoding regions.7 This highly questionable observation led to a wide range of research into the discovery of the function of noncoding sites and roles of the cancer development. These studies have led to the identification of lncRNA, which was transcribed from noncoding sites and may be increased the susceptibility of risk cancer.8, 9 The lncRNA‐H19 gene is a maternally expressed and coded a long noncoding RNA that has been reported previously involved in the tumor progression.11
LncRNA ‐H19 is a carcinogenic gene located at 11p15.5 of human chromosome, which is abnormally expressed in some types of tumors and acts as a tumor suppressor gene (TSG). According to the evidence, it suggests that genetic changes in lncRNA‐H19 play an important role in cancer development.12, 13 To date, few studies have attempted to reveal the association between lncRNA‐H19 gene polymorphism with the risk of BC.16, 17 According to the previous findings, we assume that two SNPs in lncRNA‐H19 may be related to the risk of BC in Iranian. However, the relationship between lncRNA‐H19 polymorphism and BC remained unclear. The purpose of this investigation was to examine the association between two SNPs in lncRNA‐H19 (rs3741219 T>C and rs217727 C>T) with the risk of BC.
2. MATERIAL AND METHODS
2.1. Patients
In case‐control investigation, we recruited a convenience peripheral blood sample of 150 BC women and 100 cancer‐free women, which were referral to the Tabriz International Hospital of Iran, during May 2015 to November 2017. Control group consisting of 100 healthy women without any diseases, especially chronic diseases and matched age and ethnicity, from over 2000 people, was selected. A questionnaire for women's data was filled and used. All patients were recently diagnosed with BC according to the pathological procedures.
The sample was obtained according to protocols confirmed by the ethics committee of the Tabriz University of Medical Sciences, and signed informed consent and questionnaire were received from each case. Individuals who underwent chemotherapy, radiotherapy, or proposed therapy were excluded. In addition, Ethics and Human Rights Committee in Tabriz University of Medical Sciences approved the present investigation. We have obtained 2 mL peripheral blood samples from of 100 healthy individuals and 150 patients with BC (according to inclusion and exclusion criteria) in anticoagulant tubes containing EDTA, stored at −80°C prior to DNA extraction.
2.2. Genotyping
For determination of genotype frequencies of two lncRNA‐H19 gene polymorphisms, we used PCR‐RFLP method and the following sequence of forward and reverse primers: 5´‐CCCCCTGCGGCGGACGGTTG‐3´ and 5´‐GGCGTAATGGAATGCTTCAA‐3´ for lncRNA‐H19 (rs3741219) polymorphism, and 5´‐ACTCACGAATCGGCTCTGGAAGGTG‐3´ and 5´‐ATGTGGTGGCTGGTGGTCAACGGT‐3´ for lncRNA‐H19 (rs217727) polymorphism.16
The PCR of lncRNA‐H19 (rs217727) gene polymorphism amplification consists of 1 µL (10 pmole) each of the primers, 2 µL genomic DNA (50 ng), 13 µL Master Mix Red 2× (Ampliqon, Odense, Denmark), and 8 µL distilled water and for lncRNA‐H19 (rs3741219) 1 µL (10 pmole) each of the primers, 1 µL genomic DNA (50 ng), 14 µL Master Mix Red 2× (Ampliqon), and 8 µL distilled water. The reaction was carried out in the final volume of 25 μL. The PCR cycle condition followed with the initial denaturation at 95°C for 5 minutes, 35 cycles for (rs3741219) and 32 cycles for (rs217727) of the PCR, consisting of denaturation at 95°C for the 20 seconds, the annealing temperature 61°C for (rs3741219) and 63°C for (rs217727) for 15 seconds, extended at 72°C for 30 seconds, and the final cycle expanded 72°C for 5 minutes. Protocol digestion was performed on the following enzymes and conditions according to the manufacturer's instructions; HhaI and RsrII (Thermo Fisher Scientific Company, Waltham, MA, USA) enzyme digestion was used for the genotyping of rs3741219 and rs217727 at 37°C for 8 hours, respectively. We obtained a PCR product with sizes of 247 and 434 bp for rs217727 and rs3741219, respectively. During product digestion, rs217727 (T allele: 247 bp, C allele: 221 + 26 bp) and rs3741219 (T allele: 342 + 92 bp, C allele: 301 + 92 + 41 bp) fragments were generated. The PCR product digested was separated on 3% agarose gel (Figures 1 and 2). We validated genotype frequencies by sequencing 10% of the examples through arbitrary selection.
Figure 1.
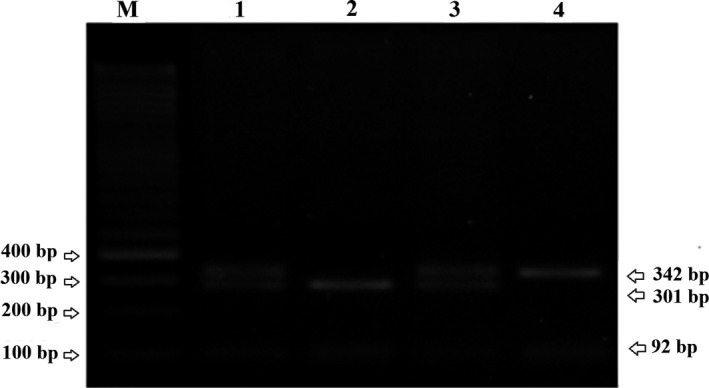
Gel electrophoresis of the PCR‐RFLP products from of lncRNA‐H19 (rs3741219) gene on 3% agarose gel electrophoresis. Lanes 1 and 3: TC genotype (342, 301, 92, and 41 bp); Lane 2: CC genotype (301, 92, and 41 bp); Lane 4: TT genotype (342 and 92 bp); M: DNA size marker 100 bp
Figure 2.
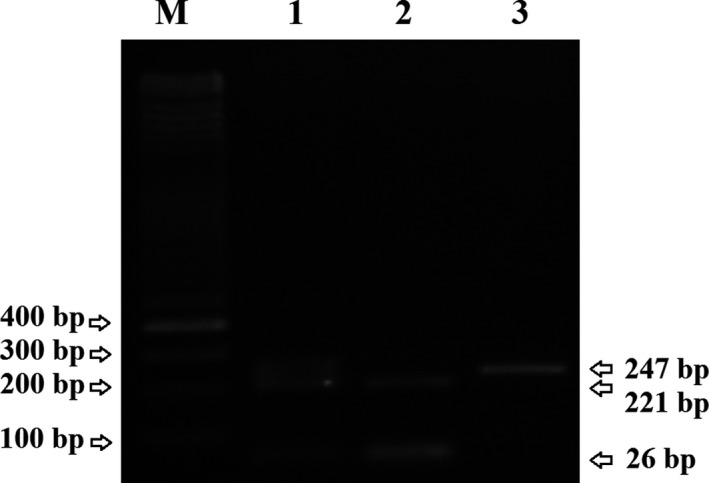
Gel electrophoresis of the PCR‐RFLP products from of lncRNA‐H19 (rs217727) gene on 3% agarose gel electrophoresis. Lane 1: CT genotype (247, 221, and 26 bp); Lane 2: CC genotype (221 and 26 bp); Lane 3: TT genotype (247 bp); M: DNA size marker 100 bp
2.3. Statistical analysis
In order to determine the allele and genotype frequencies of lncRNA‐H19 in two samples, the multivariate logistic regression and chi‐square test were used, respectively. In addition, deductive variables and the difference ratio (OR) and confidence interval (CI = 95%) were used to estimate the association between the lncRNA‐H19 polymorphism and probability of BC. Statistical analysis was performed by using SPSS 23.0 software package (SPSS Inc, Chicago, IL, USA). All the tests were two‐sided, and a P‐value under 0.05 was considered to be statistically significant.
3. RESULTS
3.1. Population characteristics
The mean age characteristics of the subjects investigated were 46.59 ± 11.18 for women with BC (range: 22‐78 years) and 43.91 ± 9.99 for healthy women (range: 25‐75 years). For overwhelming the influences of disturbing factors, the case and control groups were matched regarding the age and ethnicity. The genotype frequencies of lncRNA‐H19 (rs3741219 T>C) polymorphism were revealed (TT = 119, TC = 24, and CC = 7 and TT = 80, TC = 17, and CC = 3) in case and control, respectively (P = 0.796).
The genotype frequencies of lncRNA‐H19 (rs217727 C>T) polymorphism have shown (CC = 116, CT = 29, and TT = 5 and CC = 86, CT = 14, and TT = 0) in both groups (P = 0.087). However, our findings did not reveal a significant association between two SNPs of lncRNA‐H19 with the risk of BC. In our investigation, in order to assess a statistically significant difference in genotype frequencies in co‐dominant, dominant, and recessive heredity models, three genotypes were analyzed in two groups. In recessive genetically model, the TT + CT/CC genotype frequencies showed a borderline association between lncRNA‐H19 (rs217727 C>T) gene polymorphisms with the risk of BC (P = 0.065; OR = 0.967; 95% CI, 0.938‐0.996). In addition, our findings revealed a significant differences in allele frequencies in lncRNA‐H19 rs217727 C>T polymorphism between groups (P = 0.033; OR = 1.985; 95% CI, 1.048‐3.761). The genotype frequencies of lncRNA‐H19 (rs217727 C>T) gene polymorphisms TC + CC/TT and CC + CT/TT did not showed a statistically significant association with the risk of BC (P = 0.308; OR = 1.472; 95% CI, 0.735‐2.950) and (P = 0.088; OR = 1.800; 95% CI, 0.910‐3.561), respectively (Table 1). Of note, in homozygous co‐dominant model (TT vs CC), genotype frequencies showed a borderline association between lncRNA‐H19 (rs217727 C>T) gene polymorphisms with the risk of BC (P = 0.056; OR = 0.959; 95% CI, 0.924‐0.995).
Table 1.
The allele and genotype frequencies of SNPs lncRNA‐H19 in case and control subjects in three heredity models
| Gene polymorphism |
Case n = 150 (%) |
Control n = 100 (%) |
Total n = 250 |
OR | CI 95% | P‐value | |
|---|---|---|---|---|---|---|---|
| Down | Up | ||||||
| lncRNA‐H19 (rs217727 C>T) | |||||||
| Co‐dominant | |||||||
| CC | 116 (57%) | 86 (43%) | 202 | ||||
| CT | 29 (67%) | 14 (33%) | 43 | 0.651 | 0.325 | 1.306 | 0.225 |
| TT | 5 (100%) | 0 (—) | 5 | 0.959 | 0.924 | 0.995 | 0.056 |
| Recessive | |||||||
| TT | 5 (100%) | 0 (—) | 5 | 0.967 | 0.938 | 0.996 | 0.065 |
| TC + CC | 145 (59%) | 100 (41%) | 245 | ||||
| Dominant | |||||||
| CC | 116 (57%) | 86 (43%) | 202 | 1.800 | 0.910 | 3.561 | 0.088 |
| TC + TT | 34 (71%) | 14 (29%) | 48 | ||||
| Frequency of C allele | 261 (58%) | 186 (42%) | 447 | 1.985 | 1.048 | 3.761 | 0.033 |
| Frequency of T allele | 39 (74%) | 14 (26%) | 53 | ||||
| lncRNA‐H19 (rs3741219 T>C) | |||||||
| Co‐dominant | |||||||
| TT | 119 (60%) | 80 (40%) | 199 | ||||
| TC | 24 (59%) | 17 (42%) | 41 | 1.054 | 0.532 | 2.806 | 0.881 |
| CC | 7 (70%) | 3 (30%) | 10 | 0.638 | 0.160 | 2.539 | 0.520 |
| Recessive | |||||||
| CC | 7 (70%) | 3 (30%) | 10 | 1.583 | 0.399 | 6.727 | 0.510 |
| CT + TT | 143 (60%) | 97 (40%) | 240 | ||||
| Dominant | |||||||
| TT | 119 (60%) | 80 (40%) | 199 | 1.042 | 0.555 | 1.956 | 0.898 |
| CT + CC | 31 (61%) | 20 (39%) | 51 | ||||
| Frequency of T allele | 262 (60%) | 177 (40%) | 439 | 1.116 | 0.643 | 1.938 | 0.696 |
| Frequency of C allele | 38 (62%) | 23 (38%) | 61 | ||||
The genotype and allele frequencies of lncRNA‐H19 (rs3741219 T>C) gene polymorphism did not reveal a statistically significant difference between the two groups in three heredity models (P > 0.05; Table 1).
We also compared genotype frequencies for each polymorphism in case and control groups with clinicopathological parameters including, age, tumor stage (Table 2), estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) receptor status (Table 3).
Table 2.
Correlation between of two SNPs lncRNA‐H19 gene with the clinicopathological parameters of BC patients
| Genotype lncRNA‐H19 rs3741219 T>C | Genotype lncRNA‐H19 rs217727 C>T | |||||||
|---|---|---|---|---|---|---|---|---|
| Normal TT | Heterozygote TC | Homozygote CC | P‐values | Normal CC | Heterozygote CT | Homozygote TT | P‐values | |
| Age < 65 | 82 | 14 | 4 | 0.353 | 79 | 17 | 4 | 0.818 |
| Age > 65 | 70 | 30 | 0 | 80 | 20 | 0 | ||
| Stage 1 | 78 | 19 | 3 | 0.585 | 76 | 20 | 4 | 0.642 |
| Stage 2 | 86 | 8 | 6 | 79 | 17 | 4 | ||
| Stage 3 | 74 | 21 | 5 | 79 | 21 | 0 | ||
| Stage 4 | 81 | 15 | 4 | 92 | 0 | 8 | ||
Table 3.
The associations between two SNPs lncRNA‐H19 and ER, PR, and HER2 status of BC patients
| Genotype | ER‐PR | P‐value | HER2 | P‐value | ||
|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | |||
| LncRNA‐H19 rs3741219 T>C | ||||||
| TT | 82 | 82 | 0.835 | 86 | 78 | 0.185 |
| TC | 16 | 15 | 0.814 | 12 | 17 | 0.347 |
| CC | 2 | 3 | 0.394 | 2 | 5 | 0.344 |
| TT + TC | 98 | 97 | 0.394 | 98 | 95 | 0.344 |
| LncRNA‐H19 rs217727 C>T | ||||||
| CC | 82 | 78 | 0.628 | 73 | 83 | 0.168 |
| CT | 14 | 19 | 0.492 | 25 | 13 | 0.071 |
| TT | 4 | 3 | 0.772 | 2 | 4 | 0.483 |
| CT + TT | 18 | 22 | 0.722 | 27 | 17 | 0.483 |
4. DISCUSSION
We examined the performance of two polymorphisms rs217727 and rs3741219 in lncRNA‐H19 on susceptibility to BC in populations in East Azerbaijan Province, Iran. To get the best information, this study looked up at the role of nucleotide changes in lncRNA‐H19 in BC carcinogenesis. Our findings revealed a significant differences in allele frequencies in lncRNA‐H19 rs217727 C>T polymorphism between groups. On this basis, it can be suggested that the presence of rs217727 C>T polymorphism poses a risk of increased BC.
According to the latest knowledge by Xia et al,16 reported that the nucleotide changes in lncRNA‐H19 was associated with the risk of BC. The results showed that nonsignificant deviations from Hardy‐Weinberg equilibrium for two polymorphisms were found among a total of 467 healthy women. Contradictory, our findings revealed that the rs217727 C>T polymorphism may be involved in BC carcinogenesis, whereas the genotyping results showed that the rs217727 was associated with a reduction in the risk of BC.
Previous studies have shown that lncRNAs can control the stages of the cell cycle, such as progression, cell death, invasion, and migration. The genetic variation in lncRNAs may alter the structures and stability of lncRNAs and involved in carcinogenesis. Wu et al reported that the A allele compared to the rs11752942 G allele significantly improved lincRNA‐uc003opf.1 by binding with microRNA‐149 and affects the cellular proliferation of the esophageal cells, thereby causing cancer. They also noted that the nucleotide change in rs12325489 C to T could affect the transplant sites for miRNA‐370 and affected the transcriptional activity of lncRNA‐ENST00000515084 and cell proliferation of BC.18
The lncRNA‐H19 gene plays an important role in controlling fetal growth and development, and it is used to indicate the role of insulin‐like growth factor 2 (IGF2). However, lncRNA‐H19 and IGF2 are regulated by the internal control of the gene (ICR) and most commonly used augmentation regions. In humans, the CTCF split protein binding to the ICR promotes the expression of lncRNA‐H19 from maternal alleles.19, 20 Recently, it has been shown that lncRNA‐H19 gene polymorphisms are associated with several disorders. Gao et al21 also demonstrated that the nucleotide change in rs217727 C to T is associated with an increased risk of coronary artery disease, while the rs2067051 G to A is associated with a reduction in the risk of coronary artery disease. Hewage et al,22 in a case‐control investigation, assessed 177 patients with bladder cancer and 204 controls and reported that rs2839698 T>C was associated with a reduction in the risk of bladder cancer. In addition, findings showed a significant reduction in the bladder cancer risk in carriers of the T rs217727 allele compared with carrier C allele.
In addition, Li et al23 have been assessed the frequencies of three lncRNA‐H19 SNPs such as rs217727, rs2735971, and rs3024270 in the 200 bladder cancer patients and 200 healthy subjects. The findings suggested that rs217727 was correlated with increased risk of bladder cancer and may be involved in the background genetics of bladder cancer susceptibility. Yuan et al24 examined four functional SNPs in lncRNA‐H19 to evaluate the influence of the risk of oral squamous cell carcinoma (OSCC) in a Chinese population. The results revealed that the two SNPs rs217727 and rs2839701 were correlated with the risk of OSCC and proposed the critical roles in genetic susceptibility to OSCC. The recent investigation was performed on the 555 lung cancer patients (LC) and 618 healthy individuals to evaluate the relationship between rs217727 polymorphisms with the susceptibility of LC. They noted that the homozygous genotype of rs217727 was notably correlated with an elevated LC risk.25
Yang et al12 studied the effect of four independent lncRNA‐H19 SNPs (rs217727, rs2839698, rs3741216, and rs3741219) with gastric cancer risk in China population and showed that the T allele of rs217727 was associated with a higher risk of gastric cancer. Individuals with rs2839698 TT genotype showed a significant increase in gastric cancer susceptibility. Similarly, in the present study, we found a positive relationship between the rs217727 polymorphism and BC susceptibility. A nucleotide variation of lncRNA may be changing the structure and affects miRNA‐lncRNA interaction. Increasing evidence suggests that lncRNAs can be directly regulated by miRNAs.24 The effect of rs3741219 T>C lncRNA‐H19 gene polymorphism and interaction of miRNA‐lncRNA in BC cells remained unclear, and so further studies are needed to find the mechanism. Xia et al16 showed that the release of miR‐675 with the lncRNA‐H19 prevented estrogen proliferation of ERa of cancerous cells. Furthermore, the SNPs in the lncRNA‐H19 may have a relationship with the risk of BC.
Butt et al26 have been revealed that the T vs C allele in lncRNA‐H19 rs2107425 polymorphism decreased the risk of BC. They reported that no significant association was found between rs2107425 changes with the risk of BC in Swedish population.
In addition, Barnholtz‐Sloan et al27 assessed the relationship between lncRNA‐H19 rs2107425 changes in African American and White cases. The findings did not reveal a significant association between this polymorphism and risk of BC in African American.
Hassanzarei et al,28 in case‐control study was performed on 230 BC patients and 240 ages adjusted healthy women with no history of any cancer in Southeast Iranian population. The findings revealed an association between lncRNA‐H19 gene polymorphism and BC risk. In addition, it suggested that the nucleotide changes in lncRNA‐H19 may be considered as a potential biomarker for predisposition to BC.
Our findings involve several significant points that should be mentioned. The present investigation assessed the initial nucleotide variations of lncRNA‐H19 in relationship to BC susceptibility in Azeri‐Turk Iranian population. In addition, in our survey, the all individuals matched on age and the racial characteristic identical. In our investigation, we have evaluated the patient samples which all newly pathological diagnosed BC disease without obtaining any remedy regime. However, our consideration had several limitations including the analytical strength, the sample composition, and a large percentage of loss factors for some variables. In addition, we did not incorporate the effects of the lncRNA‐H19 nucleotide changes in gene expression alteration and activity. Therefore, somewhat investigations are profitable to confirm whether lncRNA‐H19 variations could influence the pathogenesis of BC.
In conclusion, our findings demonstrate that the presence of rs217727 C>T polymorphism may play crucial roles in the pathogenesis of BC, whereas rs3741219 T>C variation cannot be promoting risk of BC susceptibility.
AUTHOR CONTRIBUTIONS
Safa Abdollahzadeh and Saeid Ghorbian designed the study; Safa Abdollahzadeh and Saeid Ghorbian contributed to the sample collection; Safa Abdollahzadeh and Saeid Ghorbian performed the study; Safa Abdollahzadeh and Saeid Ghorbian analyzed the data; and Safa Abdollahzadeh and Saeid Ghorbian drafted the manuscript.
Abdollahzadeh S, Ghorbian S. Association of the study between LncRNA‐H19 gene polymorphisms with the risk of breast cancer. J Clin Lab Anal. 2019;33:e22826 10.1002/jcla.22826
REFERENCES
- 1. Even‐Sapir E, Inbar M. PET in women with high risk for breast or ovarian cancer. Lancet Oncol. 2010;11(9):899‐905. [DOI] [PubMed] [Google Scholar]
- 2. Key TJ, Verkasalo PK, Banks E. Epidemiology of breast cancer. Lancet Oncol. 2001;2(3):133‐140. [DOI] [PubMed] [Google Scholar]
- 3. Bradbury AR, Olopade OI. Genetic susceptibility to breast cancer. Rev Endocr Metab Disord. 2007;8(3):255‐267. [DOI] [PubMed] [Google Scholar]
- 4. Sapkota Y, Mackey JR, Lai R, et al. Assessing SNP‐SNP interactions among DNA repair, modification and metabolism related pathway genes in breast cancer susceptibility. PLoS One. 2014;8(6):e64896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ghorbian S, Nargesian M, Talaneh S, Asnaashar O, Sharifi R. Association of genetic variations in XRCC1 and ERCC1 genes with sporadic breast cancer. Gene Cell Tissue. 2018;5(2):e80166. [Google Scholar]
- 6. Kian R, Moradi S, Ghorbian S. Role of components of microRNA machinery in carcinogenesis. Exp Oncol. 2018;40(1):2‐9. [PubMed] [Google Scholar]
- 7. Haemmerle M, Gutschner T. Long non‐coding RNAs in cancer and development: where do we go from here? Int J Mol Sci. 2015;16(1):1395‐1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jendrzejewski J, He H, Radomska HS, et al. The polymorphism rs944289 predisposes to papillary thyroid carcinoma through a large intergenic noncoding RNA gene of tumor suppressor type. Proc Natl Acad Sci USA. 2012;109(22):8646‐8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Can Res. 2014;74(11):2913‐2921. [DOI] [PubMed] [Google Scholar]
- 10. Rossi S, Sevignani C, Nnadi SC, Siracusa LD, Calin GA. Cancer‐associated genomic regions (CAGRs) and noncoding RNAs: bioinformatics and therapeutic implications. Mamm Genome. 2008;19(7‐8):526‐540. [DOI] [PubMed] [Google Scholar]
- 11. Noori‐Daloii MR, Eshaghkhani Y. LncRNAs: new approach in cancer therapy. Med Sci J. 2015;25(4):249‐256. [Google Scholar]
- 12. Yang C, Tang R, Ma X, et al. Tag SNPs in long non‐coding RNA H19 contribute to susceptibility to gastric cancer in the Chinese Han population. Oncotarget. 2015;6(17):15311‐15320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raveh E, Matouk IJ, Gilon M, Hochberg A. The H19 Long non‐coding RNA in cancer initiation, progression and metastasis – a proposed unifying theory. Mol Cancer. 2015;14:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matouk IJ, Halle D, Raveh E, Gilon M, Sorin V, Hochberg A. The role of the oncofetal H19 lncRNA in tumor metastasis: orchestrating the EMT‐MET decision. Oncotarget. 2016;7(4):3748‐3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang L, Zhou Y, Huang T, et al. The interplay of LncRNA‐H19 and its binding partners in physiological process and gastric carcinogenesis. Int J Mol Sci. 2017;18(2):450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xia Z, Yan R, Duan F, Song C, Wang P, Wang K. Genetic polymorphisms in long noncoding RNA H19 are associated with susceptibility to breast cancer in Chinese population. Medicine. 2016;95(7):e2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lin Y, Fu F, Chen Y, et al. Genetic variants in long noncoding RNA H19 contribute to the risk of breast cancer in a southeast China Han population. Onco Targets Ther. 2017;10:4369‐4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu HC, Zheng J, Deng JQ, et al. A genetic polymorphism in lincRNA‐uc003opf.1 is associated with susceptibility to esophageal squamous cell carcinoma in Chinese populations. Carcinogenesis. 2013;34:2908‐2917. [DOI] [PubMed] [Google Scholar]
- 19. Takai D, Gonzales FA, Tsai YC, et al. Large scale mapping of methylcytosines in CTCF‐binding sites in the human H19 promoter and aberrant hypomethylation in human bladder cancer. Hum Mol Genet. 2001;10:2619‐2626. [DOI] [PubMed] [Google Scholar]
- 20. Wilkin F, Paquette J, Ledru E, et al. H19 sense and antisense transgenes modify insulin‐like growth factor‐II mRNA levels. Eur J Biochem. 2000;267:4020‐4027. [DOI] [PubMed] [Google Scholar]
- 21. Gao W, Zhu M, Wang H, et al. Association of polymorphisms in long non‐coding RNA H19 with coronary artery disease risk in a Chinese population. Mutat Res. 2015;772:15‐22. [DOI] [PubMed] [Google Scholar]
- 22. Hewage AS, Jayanthiny P, Tennekoon KH, et al. H19 rs217727 genotype and IGF‐1/intron‐2 dinucleotide CT repeat polymorphism are independently associated with birth weight. Endocrine. 2015;48:1010‐1012. [DOI] [PubMed] [Google Scholar]
- 23. Li Z, Niu Y. Association between lncRNA H19 (rs217727, rs2735971 and rs3024270) polymorphisms and the risk of bladder cancer in Chinese population. Minerva Urol Nefrol. 2018. [Epub ahead of print]. 10.23736/S0393-2249.18.03004-7 [DOI] [PubMed] [Google Scholar]
- 24. Yuan Z, Yu Y, Zhang B, et al. Genetic variants in lncRNA H19 are associated with the risk of oral squamous cell carcinoma in a Chinese population. Oncotarget. 2018;9(35):23915‐23922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li L, Guo G, Zhang H, et al. Association between H19 SNP rs217727 and lung cancer risk in a Chinese population: a case control study. BMC Med Genet. 2018;19(1):136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Butt S, Harlid S, Borgquist S, et al. Genetic predisposition, parity, age at first childbirth and risk for breast cancer. BMC Res Notes. 2012;5(1):414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barnholtz‐Sloan JS, Shetty PB, Guan X, et al. FGFR2 and other loci identified in genome‐wide association studies are associated with breast cancer in African‐American and younger women. Carcinogenesis. 2010;31(8):1417‐1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hassanzarei S, Hashemi M, Sattarifard H, Hashemi SM, Bahari G. Genetic polymorphisms in long noncoding RNA H19 are associated with breast cancer susceptibility in Iranian population. Meta Gene. 2017;14:1‐5. [Google Scholar]


