Abstract
We quantify the impact of scientific grant funding at the National Institutes of Health (NIH) on patenting by pharmaceutical and biotechnology firms. Our paper makes two contributions. First, we use newly constructed bibliometric data to develop a method for flexibly linking specific grant expenditures to private-sector innovations. Second, we take advantage of idiosyncratic rigidities in the rules governing NIH peer review to generate exogenous variation in funding across research areas. Our results show that NIH funding spurs the development of private-sector patents: a $10 million boost in NIH funding leads to a net increase of 2.3 patents. Though valuing patents is difficult, we report a range of estimates for the private value of these patents using different approaches.
Keywords: economics of science, patenting, academic research, NIH, knowledge spillovers
1. Introduction
It is often taken for granted that investments in innovation underpin economic growth (Romer 1990; Aghion and Howitt 1992). In leading models and empirical studies, these R&D investments are undertaken by private firms with the goal of creating new products or improving existing ones (Pakes and Griliches 1980). While most studies of innovation focus on a firm’s own R&D investments, and more recently on knowledge spillovers between firms (e.g., Bernstein and Nadiri 1989; Bloom, Schankerman, and Van Reenen 2013), the impact of public sector research investments has received less attention.
In many industries, private-sector innovations often have their roots in public-sector research investments. The pharmaceutical firm Novartis, for example, made use of decades of government-funded research on gene mutation and cell-signaling in the development of Gleevec, a revolutionary treatment for chronic myelogenous leukemia (Wapner 2013). In the U.S., the belief that public-sector research matters for private-sector innovation has fueled considerable federal investment in R&D for at least the past seventy years—despite the fact that economists and policymakers have acknowledged that little is known about the returns to these investments (Jaffe 2002; Marburger 2005). This paper aims to fill this gap in knowledge.
Assessing the impact of public-sector research is conceptually different from quantifying the returns to private R&D, and in many ways more difficult. There are three issues. First, while private R&D investments are typically targeted to specific applications in the hope of direct commercial payoffs, public R&D investments—especially those in basic science—are often made with the opposite goal: to produce non-rival ideas that maximize potential spillovers. As a result, traditional empirical approaches—which rely on foreseeable linkages between investments and outcomes—are ill-suited to help trace the unpredictable and often convoluted path between public expenditures and final commercial products (Griliches 1992). Second, analyses of the effects of public R&D on outcomes are beset by potential endogeneity problems: public investments may target research areas with the most potential for follow-on innovation, for example those where disease burden is rising (Acemoglu and Linn 2004) or scientific opportunities are increasing (Lichtenberg 2001). Finally, research on public R&D needs to account for the possibility that public research “crowds out” private investment (David, Hall, and Toole 2000).
This paper makes progress on each of these issues to provide causal evidence on the returns to public investments in biomedical research.1 Our empirical setting is the biopharmaceutical industry, an area where innovations are thought to be extremely important for health, productivity and welfare, and where the US National Institutes of Health (NIH) is the single largest funder of research in the world. We analyze the impact of NIH research funding on patenting by private sector firms, from 1980 through 2012.
Our first contribution is to construct new measures of the commercial output associated with publicly funded research. The most recent work in this area examines the effects of funding for a disease on outcomes relevant for that same disease, typically using pre-specified lag structures (Manton et al. 2009; Toole 2012), or selecting optimal lags based on goodness-of-fit criteria (Blume-Kohout 2012). While these papers are an important step toward understanding the relationship between public research inputs and practical outputs, a drawback to these approaches is that they do not capture the impact of funding on other diseases or with other time lags. This concern is particularly salient in our setting because the possibility of such unanticipated spillovers is among the main rationales for the public funding of science in the first place.
To capture the potentially unanticipated impact of public funding, our paper takes a different approach. We construct a dataset that uses bibliometric information to explicitly link NIH grants with the publications they support and the patents that cite those publications—even if these patent outcomes are in substantially different research areas, and regardless of the lags involved. By letting the data reveal the relevant linkages, we are able to identify patents that build on NIH-funded research without making a priori assumptions about the diffusion of scientific knowledge over time and across diseases.
Our second contribution relates to identification. Public investments may target research areas with the most potential for follow-on innovation, which could lead to a correlation between public funding and private patenting even if public investments were unproductive. To address concerns about the endogeneity of public investments, our paper begins by considering a finer-grained unit of analysis: NIH funding for a given disease (D), relying on a specific set of scientific approaches and methodologies (S), at a particular time (T). Organizing our analysis at the level of a DST captures the notion of a biomedical research area as a collection of projects focusing on a particular disease, that also share an interest in the same underlying scientific questions. Constructing funding flows for a DST is also straightforward, since every NIH grant is funded by a specific Institute (e.g., the National Cancer Institute), which tells us the disease area it is targeting, and evaluated for scientific merit by a committee (e.g., Behavioral Genetics and Epidemiology), which informs us about the science domain to which it belongs.
Using DST as the unit of analysis helps our identification strategy in two ways. First, it enables us to include detailed pairwise disease/science, disease/time, and science/time fixed effects to account for the most common potential sources of endogeneity in funding (e.g. differences in innovative potential across diseases, changes in disease burden, and changes in scientific opportunity). Second, it enables us to construct an instrument for total DST funding, by taking advantage of NIH funding rules which specify how grant applications are prioritized across disease and science areas.
As an illustration, consider a grant application related to a specific disease/science area, say cancer/cell signaling. This grant is evaluated by a group of cell signaling experts and is assigned a raw score. One might decide whether to fund this application by comparing its raw score with other cell signaling applications (its science rank) or by comparing it with other cancer applications (its disease rank). The NIH does neither. Instead, it decides whether to fund an application based on how its science rank compares with the science ranks of other applications in the same disease area (its “rank of rank”). As we explain in detail in Section 3.2, a funding threshold based on rank of ranks effectively means that DSTs can have the same number and quality of applications based on peer review evaluations, but nonetheless differ in the total funding that they receive. We operationalize this notion of “windfall” funding as the amount of funding for a DST that comes from grants that fall just above an Institute’s funding threshold—holding constant its applications’ raw scores and science ranks. Our IV approach thus compares patenting outcomes across DSTs that look similar in terms of scientific quality, but differ in windfall funding arising from their applications’ rank of ranks. We demonstrate that these windfall funds do indeed look random at the DST level and that grant applicants cannot anticipate these fluctuations and strategically submit applications in response.
The third contribution of our paper is to account for the impact of crowd-out. We develop a novel method to identify the set of private-sector patents intellectually related to a given NIH research area—even if these patents do not build explicitly on NIH-funded work. By identifying private-sector patents in areas potentially influenced by NIH funding, we are able to measure the impact of public research investments on total private-sector output in affected areas, net of potential crowd-out.
Our results show that NIH funding increases total private-sector patenting. We obtain similar estimates using both our fixed effects and IV estimation strategies. Our preferred empirical specification suggests that an additional $10 million in NIH funding for a research area generates 2.3 additional private-sector patents in that area, or roughly one patent for every two to three NIH grants. Not all patents are equally valuable; the distribution of patent value is highly skewed (Harhoff, Scherer, and Vopel 2003). In a series of back-of-the envelope calculations (Section 5.4 and Table 10) we report a range of estimates for the private value of these patents using different approaches.
Table 10:
Implied Drug Valuation of NIH Investments
| Advanced Drug Candidates |
FDA Approved |
Pre-approval | Main | Drug-level | |
|---|---|---|---|---|---|
|
Mean=0.546; SD=0.864 |
Mean=0.316; SD=0.532 |
Mean=0.212 SD=0.358 |
Mean=0.035; SD=0.084 |
Mean=0.059; SD=0.099 |
|
| (1) | (2) | (3) | (4) | (5) | |
| OLS | |||||
| DST Funding (×$10 mln.) Mean=4.06; SD=4.36 | 0.081*** (0.015) |
0.046*** (0.012) |
0.032*** (0.007) |
0.005*** (0.001) |
0.008*** (0.001) |
| Elasticity | 0.602 | 0.591 | 0.613 | 0.580 | 0.551 |
| Implied Drug Value ($ mln.) | — | $20.0 | $22.2 | $17.4 | $27.8 |
| IV | |||||
| DST Funding (×$10 mln.) Mean=4.06; SD=4.36 | 0.053 (0.040) |
0.034 (0.024) |
0.017 (0.019) |
0.001 (0.004) |
0.004 (0.005) |
| Elasticity | 0.394 | 0.437 | 0.326 | 0.116 | 0.275 |
| Implied Drug Value ($ mln.) | — | $14.7 | $11.8 | $3.5 | $13.9 |
| Observations | 14,085 | 14,085 | 14,085 | 14,085 | 14,085 |
Note: See notes to Tables 6 and 7 for sample details. The outcome variables are fractional patent counts. All specifications include disease-science FEs, disease-year FEs, science by year linear time trends, FEs for the number of applications to the DST, cubics in the average raw score and average science rank received by applications in the 25-grant radius window around the IC payline, and FEs for number of DST applicants in a 25-grant window around an IC’s funding cutoff. A patent is labelled “Private Sector” if it is assigned to a domestic US or foreign corporation (NBER assignee categories 1 and 2 minus foundations, universities, and hospitals). A patent is labeled an advanced drug candidate if it is included in IMS Patent Focus, which contains information on patents on biopharmaceutical candidates in Phase III trials or further. We do not generate an implied value for these patents since they are not necessarily associated with an approved drug/biologic. Within this set, patents are labeled as “FDA approved” if linked to an approved drug/biologic. A patent is labeled “pre-approval” if it is “FDA approved” and was filed prior to the time at which corresponding received marketing approval. A patent is labeled as “main” patent if it is the first patent ever filed associated with a marketed drug. Column 5 aggregates results to the drug level, reweighting by the number of unique drugs associated with a DST. Implied drug values are calculated assuming a mean lifetime discounted value of $3.47 billion, in 2010 dollars. This figure comes from DiMasi, Grabowski, and Vernon (2004). All estimates assume that there is one pivotal patent per drug; FDA approved patents are scaled by 8; pre-approval patents by 5; main patents and drug specific outcomes are not scaled. For instance, the OLS estimate in column (2) imply that an additional $10 mln. in NIH funding for a DST would result in $22.6 mln. in downstream pharmaceutical sales.
Standard errors in parentheses, two-way clustered at the disease and science level (*p < 0.10, **p < 0.05, ***p < 0.01).
The empirical approach we propound also sheds light on the path through which NIH investments influence private sector innovation by developing estimates of the cross-disease spillover effects of NIH funding. We show that fully half of the patents resulting from NIH funding are for disease applications distinct from the one that funded the initial research. The size of this effect underscores the importance of our approach to linking patents with funding: by looking only within the same disease area when measuring impact, the prior literature in this area appears to have missed almost half of the total impact of basic research funding.
We proceed as follows. In Section 2, we discuss institutional background and the various effects that NIH funding may have on private patenting. We describe our conceptual framework and empirical strategy in Section 3. Sections 4 and 5 present our data and main results, respectively. Section 6 concludes. Robustness checks and alternative specifications can be found in Appendices F, I, J, K, and L. Appendix M discusses the impact of NIH funding for a given research area on how firms reallocate investments to and from other areas.
2. Background
2.1. The NIH
The NIH was responsible for funding 28 percent of U.S. medical research in 2008. This compares to 37 percent of research funded by pharmaceutical firms, 15 percent by biotechnology firms, and 7 percent by medical device firms (Dorsey et al. 2013).2 The bulk of NIH funding is for “basic” research that aims to extend the frontiers of medical understanding. About one-third of NIH funding is for clinical research (including patient-oriented research, clinical trials, epidemiological and behavioral studies, as well as outcomes and health services research) that is more applied in nature. The agency also supports a range of training grants that help develop the U.S. scientific and medical workforce.
The NIH comprises 27 Institutes or Centers (ICs) that are typically organized around body systems (e.g., the National Heart, Lung, and Blood Institute), or disease areas (e.g., the National Cancer Institute). Each Institute receives its own Congressional appropriation and is responsible for funding research that is potentially relevant to its mission. Scientific evaluation of grant applications, by contrast, occurs primarily in approximately 180 standing review committees known as study sections. Each study section is organized around a scientific topic (for example, “Behavioral Genetics and Epidemiology” or “Cellular Signaling and Regulatory Systems”) and is responsible for evaluating the quality of applications in its area. Study sections review grant applications from multiple disease areas with similar scientific underpinnings. In turn, ICs fund applications evaluated by multiple study sections. As such, we construct total NIH funding for our unit of analysis, the disease/science/year (DST), by identifying the amount of funding for all grants assigned to a given NIH institute (which corresponds to a disease area) and study section (which captures the scientific area) pairing, in any given year.
Study sections assign each application a raw score. During the timespan covered in our analysis, these ranged from 5.0 (worst) to 1.0 (best). This raw score is meant to be a summary statistic for the study section’s assessment of the quality of that application. Raw scores are then normalized within a study section and converted into a percentile. We call this normalized score the application’s “science rank.” Once a study section has evaluated an application, the NIH’s funding rule is mechanical: an IC must fund the applications it is assigned in order of their science rank until its budget has been exhausted. The worst score that is still funded is known as that IC’s “payline.” In summary, the peer review process at NIH generates three separate scores for each application: (i) the “raw score” given by the study section; (ii) the within-study section “science rank” immediately derived from the raw score; and (iii) the within-IC ranking of science ranks. It is this final “rank of rank” that determines whether an application is funded. As discussed in the introduction, the structure of the NIH and its funding rules will play an important role in our empirical work. Section 3.2.2 details how we exploit these features to isolate exogenous variation in NIH investments across research areas. Appendix A provides more details about the NIH and its funding rules.
2.2. Measuring the impact of publicly funded medical research: previous research and challenges
Publicly-funded research can influence private innovation in numerous ways and through diverse channels, including through increasing the stock of knowledge (which may suggest new projects, or aid in completion of existing projects), training graduates, creating scientific instruments and tools, creating networks, and creating new firms (Salter and Martin 2001; Cohen, Nelson, and Walsh 2002; Mansfield 1995; Bekkers and Bodas Freitas 2008). Public and private sector biomedical research can be linked through all of these overlapping channels (Henderson, Orsenigo, and Pisano 1999).
One channel which has attracted considerable attention from policy makers and economists is the patenting and licensing of university inventions, which are then developed by private firms. Academic patenting and licensing have become increasingly common in recent decades, encouraged by the 1980 Bayh-Dole Act and other policies. This has led to an extensive set of studies focusing on IP-based, academic entrepreneurship (Henderson, Jaffe, and Trajtenberg 1998; Mowery et al. 2004; Azoulay, Ding, and Stuart 2009). Yet survey research (Cohen, Nelson, and Walsh 2000; Arundel and Geuna 2004; Agrawal and Henderson 2002) as well as work by economic historians (Rosenberg and Nelson 1994) suggest this channel may miss a potentially more important contribution to private-sector innovation: the informational value of scientific research (typically communicated through publication and other “open science” channels), which may suggest project ideas to firms and more generally improve the efficiency of their R&D activities.
These potential benefits are more difficult to trace than inventions directly patented and licensed by academics. Previous research has examined the effects of public science spillovers on private innovation in different ways, including surveys (Cohen, Nelson, Walsh 2002; Mansfield 1995) and analyses relating variation in public funding (by geography, scientific area, disease area, and over time) to outcomes (Jaffe 1986, Adams 1990; in medicine see Blume-Kohout 2012; Toole 2012; Manton 2009). A common approach to measuring these spillovers from academic research is to look at citations to university patents (Trajtenberg et al. 1997). However, a high share of patent-to-patent citations comes from examiners, not applicants (Alcácer and Gittleman 2006; Sampat 2010), perhaps compromising these measures of knowledge flows. Moreover, previous research suggests that patenting is a minority activity in academia (Azoulay, Michigan, and Sampat 2007), implying that the patent-to-patent citation lens may have a narrow focus.
In the analyses below, we use patent-to-article citations instead. Building on the idea that citations in journal articles can be used to track knowledge flows, the pioneering work of Francis Narin and colleagues at CHI research in the 1970s used references on the front page of patents to scientific articles (part of the “non-patent references” cited in the patent), to examine the “science dependence” of technology (Carpenter and Narin 1983) and linkages between science and technology (Narin and Olivastro 1992, 1998). This research also found that life science patents cite non-patent references more intensively than do patents from other fields. In the economics literature, the count of non-patent references (or the share of non-patent references in all citations) has been used a proxy for the extent to which patents are science-based (e.g., Trajtenberg et al. 1997). Patent-to-article citations are less likely to come from examiners than are patent-to-patent citations, and recent validation against survey results (Roach and Cohen 2013) suggests they are more informative than the latter in measuring the intellectual influence of public sector research. Our paper builds on and extends this approach, by linking life science patents back to the articles that cite them, and the specific NIH grants funding the production of these articles.3
A long-standing challenge in evaluating spillovers from publicly funded research is that effects are realized with long and variable lags (Griliches 1992) and in potentially diverse fields. Another major advantage of linking grants to articles to patents is that this allows the data reveal where to look for impact in time and space. In addition, as discussed below, our paper goes beyond previous work trying to assess the causal impact of public funding (Blume-Kohout 2012, Toole 2007, Jaffe 1986, Adams 1990) by using plausibly exogenous sources of variation in funding. (To our knowledge the only paper to do this previously is Moretti et al. [2014]).
A research focus on spillovers implicitly assumes that NIH funding raises returns to private R&D and thus “crowds-in” private research investments. It is possible, however, that public investments may “crowd-out” private-sector efforts.4 This could happen for a variety of reasons. Public funds could simply be subsidizing the cost of a firm’s existing research. Alternatively, they could lower the costs of entry for competitors, reducing the firm’s ability to reap market rewards from its R&D investments. As explained in more detail below, our analysis also goes beyond previous work in trying to account for potential crowd-out.
3. Empirical strategy
Our approach makes progress on addressing the key measurement and inference challenges faced by the existing literature. Section 3.1 describes how we measure outputs associated with NIH funding. Section 3.2 describes our OLS and IV approaches to inference, and provides support for our identification strategy.
3.1. Measuring Biomedical Innovation Using Patents
We develop new ways to link public research investments with private patenting outcomes. Our main outcome variable is patenting by private sector biopharmaceutical firms (see Appendix B for more details on these patents). Patents may appear a surprising choice; researchers studying medical innovation have typically focused on outcomes that are more immediately welfare-relevant, such as reductions in mortality and morbidity (Manton et al. 2009), drugs entering clinical trials (Blume-Kohout 2012), or new drug approvals (Toole 2012). However, these outcomes cannot be readily linked to variation in public research expenditures without restrictive assumptions. By contrast, biomedical patents can be linked to specific grant expenditures using the bibliographic references they contain. Moreover, securing patents is the principal way that biopharmaceutical firms appropriate the returns from their R&D investments (Cohen, Nelson, and Walsh 2000).
Our key methodological innovation is in how we link patents to NIH research investments. To see this more explicitly, consider an innovation production function in which patenting output pντ in a research area ν at time τ is determined by knowledge inputs krt from research areas r, at times t. In theory, output pντ could be a function of inputs from many different research areas r ≠ ν and many different times t ≠ τ. In practice, however, previous work generally places strong restrictions on the nature of the relationship between inputs rt and outputs ντ: that investments in one area only impact outputs in that same area (r = ν) and with a fixed lag structure.5 Our approach differs in that we use bibliometric data to trace the impact of a given investment krt on patenting in a range of areas ν and time periods τ. This framework is formalized in Appendix H.
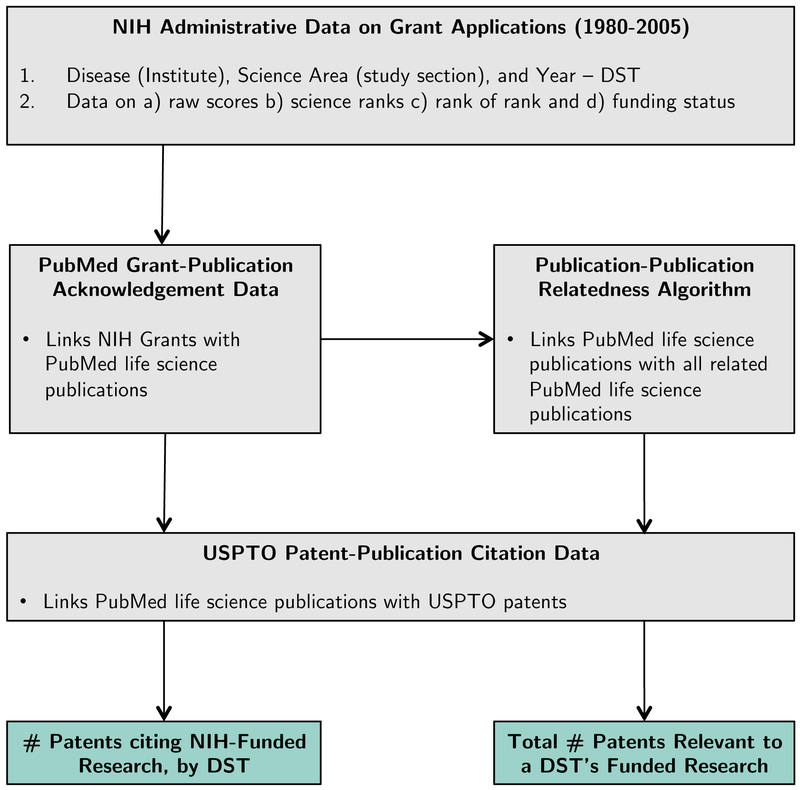
Using this approach, we construct two measures of patenting outcomes, which we describe now. Figure 1 provides an overview of this process and Appendix G provides a detailed description.
Figure 1.
Overview of Data and Construction of Patent Outcome Measures
Patents citing NIH-funded research.
We first link NIH grants to the publications they support using grant acknowledgement data.6 Second, extending bibliometric approaches surveyed in previous section, we link those publications to patents that build on their findings (Figure 1, second column).7
Taking the acknowledgment and citation data together, we define as the set of patents that cite publications that in turn acknowledge funding from that DST. These patents need not target the same disease as the original source of NIH funding with which they are linked. For example, if a patent related to cardiovascular stents cites research funded with money allocated to diabetes, we would associate this cardiovascular patent with diabetes funding. We also do not make ex ante assumptions about the nature of time lags between the date of the original grant and the date of the linked patent. A 2005 patent can be linked to a 2004 and 1994 grant if those grants produce publications cited by that patent.
This approach has two important drawbacks. First, relying on direct publication-to-patent citations limits the type of intellectual influences we can account for. We would not, for instance, credit NIH funding if it led to patenting through more complicated citation patterns (e.g., a patent that cites a publication that cites a publication that acknowledges the NIH), informal interactions (e.g., two researchers meet and exchange ideas at a conference supported by NIH funding), or the hiring of NIH-funded trainees by private-sector firms. Omitting these channels may lead us to underestimate the impact of NIH funding.
Second, by accounting only for patents that explicitly cite NIH-funded research, this measure treats patents that do not exist and patents that do exist but which cite only privately-funded research in the same way—neither are linked to a DST. As a result, if increased DST funding led to an additional linked patent, we could not tell whether this patent would otherwise have existed or not, i.e., whether private firms would have funded the necessary research instead. In other words, this first measure asks whether NIH-funded research is useful to private firms. While informative, this is not the same as asking whether NIH funding increases total private-sector innovation in a research area.
Patents related to NIH-funded research.
Our second outcome identifies all patents in the intellectual vicinity of an NIH funding area, whether or not these patents actually cite NIH-funded research. This allows us to account for a richer set of channels through which NIH funding may impact private-sector patenting. These patents, hereafter referred to as simply “related patents,” may be linked to NIH funding via a longer citation chain or belong to NIH-trained scientists who join private-sector firms. Crucially, these related patents may also be the result of private sector investments in related research areas; they need not be financially dependent on NIH at all.
Capturing the total number of private sector patents in an intellectual area is also important because it allows us to address another issue complicating previous attempts to assess the impact of science: the possibility of crowd-out. If all NIH funding did was crowd-out private research, we would not expect NIH funds to increase the total number of patents in a given research area; it would simply change the funding source for those patents. If, instead, NIH funding led to the development of patents that would not have otherwise been developed, then we should see an increase in the total amount of innovation in a research area. The impact of NIH funding on total innovation in a research area thus captures the net effect of potential crowd-in and crowd-out.
To construct this measure, we define a patent to be related to an NIH research area if it cites research similar to research that is actually funded by that area. In particular, we match each NIH grant in our sample to publications that acknowledge its support and then link these publications to a set of intellectually similar publications using a keyword-based similarity measure developed by the National Library of Medicine.8 The final step in our matching process is to identify the set of patents that cite this broader set of publications (see Figure 1). The set of patents linked to a DST in this way can be thought of as “related,” in the sense that they are part of the same intellectual area as that DST. Again, this approach does not require that “related” patents be in the same research area or issued at the same time as the original NIH disease/science area.
3.2. Estimating equation and identification
In our empirical implementation, we define a research area r at time t to be a disease/science/time combination, or DST. This is a finer-grained level of analysis than is customary in the literature, which tends to aggregate the data up to the disease level (e.g., Toole [2012]). In turn, a DST is intended to identify projects that share a similar disease application and benefit from an understanding of similar scientific methods and mechanisms at a given point in time.9 Given this unit of analysis, we estimate the following:
| (1) |
The main explanatory variable, Fundingdst, is the amount of funding allocated to grants that fall in a particular disease/science/year combination. Our outcome variable, , is the full set of private-sector patents that rely on Fundingdst as an input, even if they do not directly relate to the same disease or science area, and regardless of the lags involved.10
We address the potential endogeneity of public investments in R&D in two ways.
3.2.1. Fixed Effects Estimation
Our benchmark OLS specification is:
| (2) |
Equation (2) includes pairwise disease/science, disease/year, and science/year fixed effects that account for many common sources of endogeneity. For example, diseases that affect more people may receive more public and private interest. Further, some research topics may be more tractable than others; the genetics of breast cancer, for instance, can be studied using a variety of animal models, whereas the same is not true for the genetics of schizophrenia (Nestler and Hyman 2010). To account for time-invariant differences in innovative potential among disease/science areas, we include disease/science fixed effects (δds). The innovative or commercial potential of disease and science areas may of course also change over time. We include disease/year fixed effects γdt to control for potential confounders such as shifting disease burden or public perceptions of disease salience.11 NIH funding may also respond to scientific advances. The introduction of new DNA-sequencing technologies in the late 1990s, for instance, may have increased both public and private research funding for diseases with a genetic component. We include science/year fixed effects, νst, to control for this type of variation. Finally, in our most detailed specification, we also include fixed effects for the number of applications that a DST receives. These indicator variables proxy for time-varying interest in a particular research area that may not be captured by our other controls. In our main specifications, this regression is weighted by the average size of a DST, that is, the average yearly number of grants in a disease/science area.12 To account for serial correlation, standard errors are double-clustered at the disease and science levels (Cameron and Miller 2015).
The remaining funding variation in equation (2) comes from within-disease/year or within-science/year changes. Why is it, for instance, that cancer/cell signaling may receive more funding in 1995 than cancer/tumor physiology? After saturating our specifications with fixed effects, our identifying assumption is that NIH funding for a specific DST is not correlated with changes in the innovative or commercial potential for specific disease/science combinations.
This assumption would be violated if either Congress or NIH administrators allocated funding to DSTs on the basis of their potential. In response to the success of Gleevec, for example, the National Cancer Institute may have decided to devote a greater proportion of its budget toward the study of cell signaling or gene expression, scientific topics that are particularly relevant for targeted cancer therapies. If private firms were behaving similarly, then equation (2) would not be able to identify the impact of public funding, because we would expect changes in patenting for this area even in the absence of additional funds.
In practice it is difficult for the NIH to direct funding to DSTs on the basis of their evolving potential. As discussed in Section 3, applications are funded in order of their science ranks. This means that if cell signaling was a particularly “hot topic in a given year, the NCI could not decide to fund the top 20 cancer-related cell-signaling applications without first funding the top 19 cancer-related applications in all other science areas. Most likely, it would not have the budget to do so.13 The rigidity of this system was cited in an NIH-commissioned report from 2000, urging reform:
“…Researchers perceive that…applications describing some of the most productive, highest impact work may be assigned to too few study sections, causing too much of the ‘best science’ to compete with itself; that the scope of some study sections is restricted to research with relatively low impact, resulting in undeserved ‘entitlements’…”14
3.2.2. Instrumental Variables Estimation
Even if the NIH cannot direct funding to specific DSTs, Fundingdst could still be endogenous if study section reviewers assigned better scores to applications from DSTs with more potential. If, for instance, the cell-signaling study section decides to give better scores to cancer-related applications after the discovery of Gleevec, then the resulting funding allocation for the cancer/cell signaling DST would reflect this unobserved enthusiasm.
To address this source of endogeneity, we construct an instrument for DST funding that is not correlated with a DST’s potential. Our instrument works by isolating variation in DST funding coming from procedural rigidities in the NIH funding process that can lead equally meritorious grant applications to have different funding outcomes. These differences in grant-level funding then translate into differences in DST-level funding.
To understand how this instrument works, and the type of variation it relies on, we first consider a stylized example with two disease areas and two science areas. Having discussed how our instrument works in this setting, we then define it more generally for our entire sample.
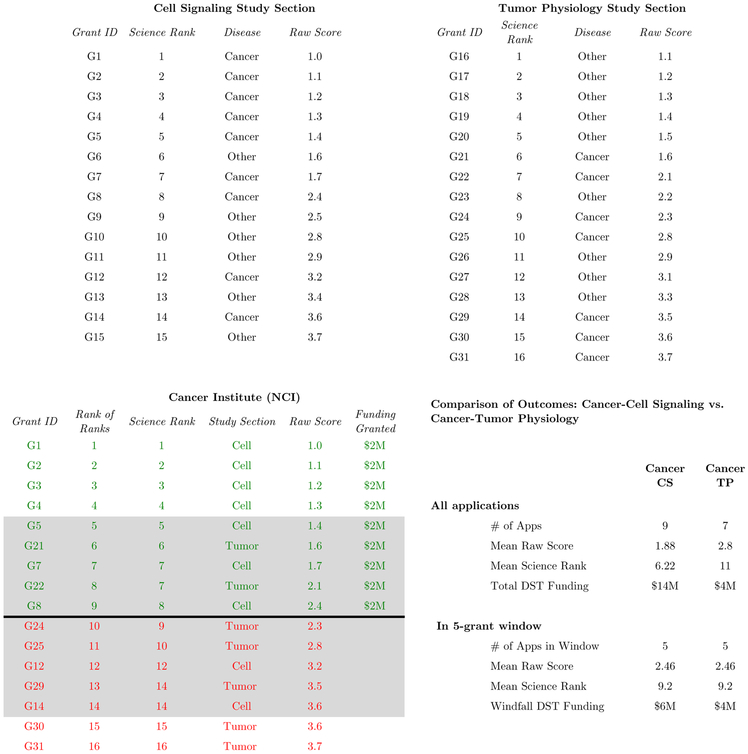
Stylized example.
Figure 2 illustrates our identifying variation. We focus on the National Cancer Institute (NCI) and label grants assigned to other disease areas as “Other.” The NCI is responsible for funding grant applications from two study sections: Cell Signaling and Tumor Physiology. We focus on the following two DSTs: Cancer/Cell Signaling and Cancer/Tumor Physiology (the time dimension is fixed in a given year and suppressed for expositional convenience).
Figure 2.
Example of Windfall DST Funding
The top two panels of Figure 2 describe the scores of grant applications to study sections. Each row represents a grant application. Study sections are science-based evaluation committees that score grant applications, potentially from many disease areas. In the top left panel, the cell signaling study section reviews applications related to cancer and other disease areas. In the top right panel, the tumor physiology study section reviews cancer and other applications as well.
Recall from Section 2.2 that the NIH implicity assigns three scores to each grant application: (i) a cardinal raw score directly given by peer evaluators in a science-based study section; (ii) an ordinal science rank, which describes how an application’s raw score compares to other applications evaluated in the same science-based study section; and (iii) another ordinal “rank of ranks” that describes how an application’s science rank comes to the science ranks other applications evaluated by different study sections but which share the same disease area. The top left panel of Figure 2 lists raw scores and science ranks for all 15 applications evaluated by the cell signaling study section. Similarly, the top right panel does so for applications evaluated by the tumor physiology study section. For completeness, we have also included the dollar amount requested by each grant, which we assume for simplicity is $2 million for all grants (requested funding amounts do not impact an application’s score).
Study sections score grant applications within science areas, but do not fund them. Funding is provided by NIH Institutes at the disease area. The bottom panel of Figure 2 illustrates funding allocations for cancer-related grant applications. In order to decide which applications to fund, the NCI ranks grant applications on the basis of their “rank of rank.” For example, G1-G5 have the five highest science ranks of all cancer applications, so in this example the five top cancer applications come from cell signaling. The cancer application with the next highest science rank is G21, a tumor physiology application with a science rank of 6. Next, both G7 and G22 have science ranks of 7; we list G7 first then G22, using their raw score as a tiebreaker. Application G8 has the next highest science rank, 8, so it receives the next best priority rank, 9. The bottom left panel of this Figure illustrates the completed rankings for all 16 cancer applications across both the cell signaling and tumor physiology study sections.
Such a priority ranking of applications is created for each NIH Institute and funding is allocated based on this ranking. In our example, we assume that the NCI has enough resources to fund the top 9 applications only. The bottom right section of this figure illustrates statistics for the grant applications associated with our two DSTs of interest: cancer-cell signaling and cancer-tumor physiology. Overall, research in the cancer-cell signaling DST is designed to potentially be of higher quality: there are more applications, perhaps signaling more interest and on average these applications receive both better raw scores and better within-study section science ranks. As a result, the cancer-cell signaling DST receives more total funding: $14 million compared to $4 million.
In general, this funding difference could be correlated with unobserved scientific potential or other factors that may also impact follow on private-sector patenting: cancer-cell signaling might have more scientific potential, which leads to both more total DST funding and, independently, more patenting.
We address this by using variation in DST funding within a tight window around an IC’s payline, rather than using variation in total DST funding. In this example, we consider the 5 grant window above and below the NCI’s payline: these are shaded darkly in Figure 2. Within this window, the grant applications associated with cancer-cell signaling and cancer-tumor physiology appear similar in quality: both DSTs have the same number of applications within this window, and those applications receive the same average raw score and have the same average science rank. Nonetheless, however, even within this window, our DSTs receive different amounts funding. Cancer-cell signaling receives funding associated with G5, G7, and G8 for a total of $6 million while cancer-tumor physiology receives funding for G21 and G22, for a total of $4 million.
This difference of $2 million comes from differences not in raw scores or rank scores (both of which might plausibly be thought of as reasonable measures of application quality), but in “rank of ranks” score. Holding constant average raw scores and rank scores as we do in this example, the remaining variation in rank of ranks is driven by the relative quality of applications in other disease areas. For example, the cancer-tumor physiology application, G24, is not funded because the cancer-cell signaling application G8 has a higher science rank. This happens not because of the quality of G24 relative to G8 (in fact, G24 receives a higher raw score), but because G8 faces weaker competition from cell-signaling applications in other disease areas (for example, G9-G11, G13, and G15).
We believe that this type of variation is not correlated with underlying innovative potential: the additional $2 million that cancer-cell signaling receives in this +/− 5 grant window around the NCI’s payline can be thought of as a “windfall” because it is not correlated with the quality of applications in this window. Our strategy is to instrument total DST funding (the $14 million that cancer-cell signaling receives) with this windfall amount.
Generalization: Instrument construction for the entire sample.
As in our motivating example above, our main IV specification also examines funding for DSTs that have the same number and quality of grant applications near an IC’s payline, but which receive different amounts of “windfall” funding. Specifically, we estimate:
| (3) |
instrumenting Fundingdst with
| (4) |
WindfallFundingdst is the amount of funding for a DST that comes from the set of grants, Wdt, that are within a window around its IC’s payline. In our simple example from the previous section, Wdt was the 5-application window on either side of the NCI’s payline. In our main specifications, we define Wdt to be the set of 25 grant applications on either side of the funding threshold for disease area d in year t; we construct the windfall funding amount to be the sum of funding for grants within this set that are actually funded. On average, windfall funding accounts for 5.6% of a DST’s total funding in that year. The median IC receives 750 applications in a given year (the mean is 1,100), making this a relatively tight window. Our results are robust to a variety of other bandwidths.
In general, WindfallFundingdst, as currently defined, may still be endogenous. This is because what we call windfall funding is simply the marginal funding that a DST barely gets. Better DSTs may have more applications that are highly scored and those DSTs would have a greater representation of grants in the set Wdt of applications near an IC’s payline; if this were the case, these better DSTs would also be likely to have more funded grants within this set. Similarly, even if two DSTs have the same number of grant applications near an IC’s payline, applications from better DSTs may justifiably receive better scores and, as a result, better DSTs may have a greater number of grants that are actually funded.
To address these concerns, we use WindfallFundingdst as an instrument for Fundingdst only after including additional variables controlling for the quality of a DST’s applications. Specifically, Equation (3) includes a full set of indicator variables for the number of grant applications any given DST has near the threshold set Wdt (i.e., the function ϒ in equation (3)), as well as separate cubics in the average raw score and average science ranks of all DST applications within the threshold set W (i.e., the functions Φ and Ψ in equation (3)). Controlling for both the raw score and science rank accounts for any differences in quality among applications, meaning that the remaining variation comes only from how science ranks translate into rank of ranks.15 In our earlier example, we show how it is possible for two DSTs to have the same number of applications within a window, for those applications to have the same average raw scores and science ranks, and for DSTs to nonetheless receive different amounts of windfall funding.
3.2.3. Identification Checks
In this section, we investigate the extent to which DST funding may be correlated with other factors that may also impact private sector innovation.
We first show that the NIH does not appear to be as sensitive to the scientific potential of research areas as one might expect. If the NIH were directing funding to DSTs on the basis of their time-varying scientific potential, we would expect that the amount of funding for DSTs that share the same scientific interests should be correlated; for example, if the NIH were allocating more money to genetics because of increased potential in that area, then we should weakly expect funding for genetics–related research to be positively correlated across disease areas. Table 4 tests this by examining the correlation between own-disease funding for a science area, Fundingdst, and funding for that same science area from other diseases Funding−d,st. Column 1, which includes only year fixed effects, shows a strong negative correlation between own and other funding. This, however, is likely due to the mechanical relationship between the size of one’s own disease area in a given science area, and the size of other disease areas. After controlling for disease by science fixed effects in Columns 2 and 3, we find no remaining correlation. Columns 4 through 6 repeat this exercise using the proportion of a disease area’s funding devoted to a particular science area as the variable of interest; we find no correlation in these specifications either.
Table 4:
Relationship Between Own DST Funding and Funding by Other Diseases for the Same Science Area
| DST Funding ($10 mln.) | |||
|---|---|---|---|
| (1) | (2) | (3) | |
| DST Funding, Other Diseases, Same Science (×$10 mln.) | −0.446*** (0.017) |
0.009 (0.063) |
−0.008 (0.060) |
| R2 | 0.134 | 0.732 | 0.771 |
| Observations | 14,085 | 14,085 | 14,085 |
| Year FEs | Incl. | Incl. | Incl. |
| Disease × Science FEs | Incl. | Incl. | |
| Disease × Year FEs | Incl. | ||
Note: Each cell is a study section/IC/year. Funding is defined by the sum of project-cycle allocations for all Type I and II grants reviewed by that study section. See notes to Tables 1 and 2 for additional details about this sample.
Standard errors in parentheses, two-way clustered at the disease and science level (*p < 0.10, **p < 0.05, ***p < 0.01).
Our next tests deal with our IV specification in particular. Table 5 tests alternative first stages using past or future windfalls as an instrument. If windfall funding for a DST is correlated with time-varying observed potential in that disease/science area after conditioning on the number of applications around the payline and their raw scores and science ranks, then we might expect past or future windfalls to still be predictive of current funding; excitement about targeted cancer therapies in the wake of Gleevec might, for instance, drive funding for cancer/cell-signaling for several years. The results in Table 5 show, however, that this is not the case. While current windfalls (Column 2) are strongly predictive of total DST funding, past and future windfalls are not.
Table 5:
Alternative First Stages, Past and Future Windfalls
| Dependent variable: Total DST Funding | |||
|---|---|---|---|
| Past Windfall | Current Windfall | Future Windfall | |
| (1) | (2) | (3) | |
| Windfall Funding | 0.067 (0.243) |
1.251*** (0.232) |
0.085 (0.205) |
| R2 | 0.927 | 0.921 | 0.927 |
| Observations | 9,326 | 14,085 | 9,326 |
Note: This table presents alternative first stages using past and future windfall funding. Current windfall funding is the total amount of funding for awarded DST grants within 25 grants of an Institute specific award cutoff in the same year T. Future windfall is this same amount, but defined for DS,T+1. Past windfall funding is similarly defined, for DS,T-1. Controls include disease-science and disease-year fixed effects, linear science-year time trends, as well as fixed effects for the number of applicants to a DST, the number of applicants within a 25-grant radius window around the IC payline, as well as cubics in the average raw and rank scores of applications in the funding window. The outcome variables are fractional patent counts.
Standard errors in parentheses, clustered at the disease/science level (*p < 0.10, **p < 0.05, ***p < 0.01).
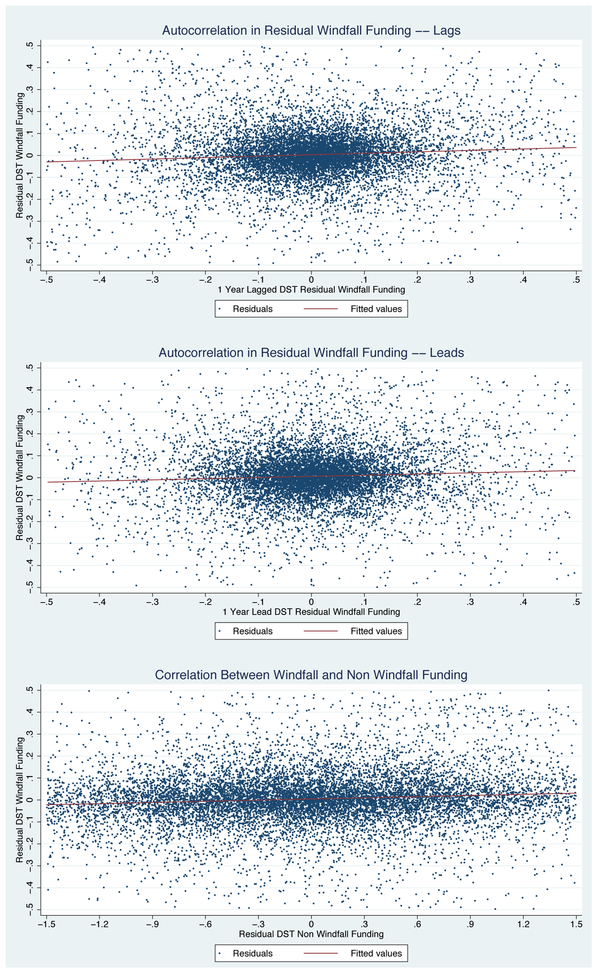
Figure 3 illustrates this point graphically. The first panel of Figure 3 plots past windfall funding on the x-axis against current windfall funding on the y-axis and finds no evidence of a relationship. The second panel does the same for current and future windfall funding. The final panel examines the relationship between windfall funding and “non-windfall” funding, i.e., Fundingdst – WindfallFundingdst. If windfall funding were truly random, then it should not be correlated with the overall quality of the DST as given by the amount of non-marginal funding it receives. Again, we find no relationship.
Figure 3.
Correlation Between Windfall DST Funding and Other DST Funding
These results show that a DST’s windfall funding, controlling for these variables, is uncorrelated with non-windfall funding, previous and future windfall funding, and other measures of DST output. Appendix J reports additional specification and robustness checks.
4. Data Construction and Descriptive Statistics
Our analysis combines data from several primary sources: (i) Administrative data on NIH grant applications from the IMPAC II database; (ii) publication data from PubMed including information on grant acknowledgements; (iii) patent data from the USPTO; and (iv) information on patents related to FDA-approved drugs from the FDA’s “Orange Book” and IMS-Health. Our final analytic sample captures linkages between the universe of NIH-funded grants from 1980-2005 at both the individual grant and DST levels, and the universe of biomedical patents granted between 1980 and 2012.16
4.1. Grant-level Patent Match
We begin with data on all 153,076 NIH grants from 1980-2005 that were evaluated in chartered study sections (those that are associated with a specific science area, rather than convened on an ad hoc basis). These grants were evaluated by 624 such study sections and funded by 17 Institutes.17 The characteristics of these grants are described in Table 1. In total, we have grant-level data that aggregate up to the activities of 14,085 DSTs. This is a only a small fraction of the 624 × 17 × 25 = 265, 200 potential DSTs. Many potential DSTs do not exist because they do not represent intellectually coherent D-S combinations. Appendix F provides details about our disease-science panel dataset and shows that our results are robust to restricting to a panel of disease-science areas that receive non-zero funding for all years for which it is in existence.
Table 1:
Grant Characteristics, 1980-2005
| Grants Linked to Private-sector Patents |
|||
|---|---|---|---|
| Full Sample | Cited by Patents | Related to Patents | |
| Sample Coverage | |||
| # Grants | 153,076 | 66,085 | 123,872 |
| # Disease Areas (Institutes) | 17 | 17 | 17 |
| # Science Areas (Study Sections) | 624 | 548 | 598 |
| # DSTs | 14,085 | 9,951 | 13,092 |
| Grant Characteristics | |||
| % R01 equivalent Grants | 73.74 | 77.46 | 74.33 |
| % Center Grants | 3.26 | 4.79 | 3.20 |
| % Teaching or Fellowship Grants | 11.43 | 10.12 | 11.27 |
| % New | 59.50 | 51.08 | 58.55 |
| Funding Amount (total project allocation, 2010 dollars; mean & s.d.) | $1,556,969 (2,198,506) |
$1,875,779 (2,783,272) |
$1,568,881 (2,215,371) |
| Number of Acknowledged Publications | 1.41 (3.58) |
3.27 (4.86) |
1.75 (3.91) |
| Number of Related Publications | 84.80 (194.36) |
166.10 (271.34) |
104.90 (211.24) |
| # of Patents Citing Grant (weighted counts) | 0.43 (2.36) |
1.00 (3.51) |
0.54 2.62 |
| # of Patents Related to Grant (weighted counts) | 0.84 (2.21) |
1.60 (3.05) |
1.04 (2.41) |
Note: Sample is the set of all NIH-funded grants from 1980-2005, excluding NINR, NLM, and NIMHD grants (see Appendix A for a full list of ICs in the sample) and evaluated by chartered study sections. The sample is restricted to new and competitive renewal grants so that there is one observation per successful grant application cycle. A grant is defined as cited by patents if there exists a patent that cites a publication that acknowledges funding from that grant. A grant is matched with a publication if it acknowledges the project number of the grant and is published within 5 years of the grant’s funding year. A patent is citation-linked to a grant if it cites a publication that is linked to a grant. A grant is considered related to a patent if that grant produces a publication that is similar (as defined by the PubMed Relatedness Matching Algorithm) to a publication that is cited by a patent. In this paper, we require that similar publications be published within 5 years of each other. A grant is an R01 equivalent (e.g. a large project-based grant) if its NIH funding mechanism is either an R01, R23, R29, or R37. Center grants are those grants whose mechanism starts with a “P” (e.g., a P01 grant containing multiple projects). A teaching or fellowship grant is one whose grant mechanism designation begins with a “T” or an “F.” New grants are projects that have not previously received NIH funding. Acknowledged publications are the unique count of PubMed publications which acknowledge the grant′s main project number and which are published within five years of grant receipt. Related publications include directly acknowledged publications, in addition to all publications related to them, according to the PMRA algorithm discussed in the text, and published within a 5 year window.
The average award size for grants in our sample is approximately $1.6 million. Seventy four percent of grants are R01s—the R01 is a renewable, project-based grant that constitutes the majority of NIH’s grant spending—and most (60%) are for new research projects (as opposed to renewals of existing projects).
Table 2 describes the patents in our sample and show how they are linked to NIH funding. We begin with the universe of 315,982 life-science patents granted by the USPTO between 1980 and 2012. Of these, 232,276 (74%) are private-sector patents and 83,394 (26%) are what we call public-sector patents, meaning those assigned to governments, universities, hospitals, and other institutions (see Appendix B for a description of patent types and definitions). Despite the large number of patents we examine, Table 2 shows that only 4,718 private-sector patents (2%) are associated with advanced drug candidates—drugs and biologics in Phase III trials and beyond—and even fewer, 1,999 (<1%) are associated with FDA-approved new chemical entities and new biological entities.
Table 2:
Patent Characteristics, 1980-2012
| Patents Linked to NIH Funding | |||
|---|---|---|---|
| Full Sample | % Citing NIH Funded Research |
% Related to NIH Funded Research |
|
| Sample Coverage | |||
| # Patents | 315,982 | 44.00 | 84.10 |
| Patent Characteristics: General | |||
| Private Sector | 232,276 | 39.38 | 82.33 |
| Public Sector | 83,394 | 56.91 | 89.07 |
| Patent Characteristics: Private Sector Only | |||
| Advanced Drug Candidates | 4,718 | 49.92 | 88.22 |
| FDA Approved Drugs | 1,999 | 42.47 | 86.79 |
| Large Asssignee | 164,431 | 36.23 | 80.37 |
| Small Asssignee | 29,183 | 51.37 | 87.89 |
Note: Sample is the set of all USPTO granted patents from 1980-2012 that meet the following criteria: (i) they are either in NBER Patent Categories 1 (“Chemicals”) or 3 (“Drugs and Medical”) and (ii) they cite at least one publication in the PubMed database. A patent is defined as citing NIH-funded research if it cites a publication that acknowledges the project number of an NIH grant and is published within 5 years of that grant’s funding year. A patent is considered related to NIH funding if it cites a publication that is similar (as defined by the PubMed Relatedness Matching Algorithm) to a publication that acknowledges NIH funding. We require that similar publications be published within 5 years of each other. A patent is labelled “Private Sector” if it is assigned to a domestic US or foreign corporation (NBER assignee categories 1 and 2 minus foundations, universities, and hospitals). A patent is labelled “Public Sector” if it is assigned to a US or foreign goverment (NBER categories 5 and 6) or if it is assigned to a foundation, university, or hospital. A patent is labeled an advanced drug candidate if it is associated with a drug or biologic in Phase III clinical trials or beyond (these are listed in Orange Book and/or IMS Patent Focus); A patent is associated with an FDA approved drug if that patent is associated with a marketed treatment accoding to IMS Health. A patent is associated with a large assignee if its assignee employs over 500 employees; it is considered small otherwise.
Table 2 also shows that NIH funding is relevant for organizations seeking patents. Forty-four percent of life-science patents in our sample directly cite NIH-funded research. Among the subset of private-sector patents, this figure is 39%. For public-sector patents, this figure is 57%. We further document a greater role of NIH-funded research in the development of high value patents: 50% of patents associated with advanced drug candidates—those that have entered clinical trials—cite NIH-funded research (Sampat and Lichtenberg 2011).
Table 2 also shows that the vast majority of patents—265,741 patents or about 84% of the universe—cite research that is similar to research funded by an NIH DST. This is true, moreover, for private- and public-sector patents, as well as high value patents, and those from both large and small firms.
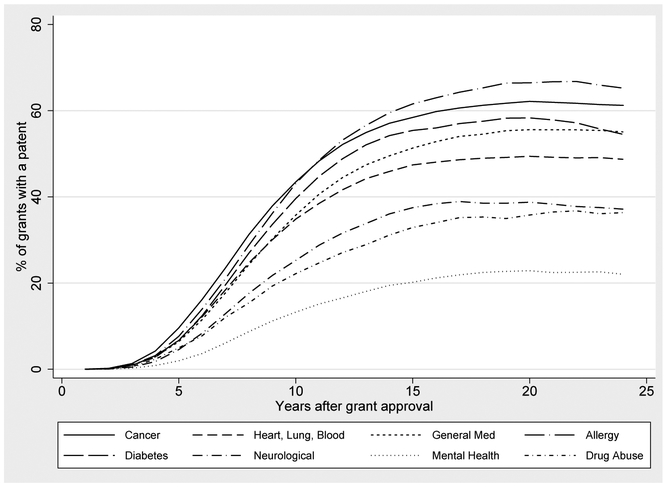
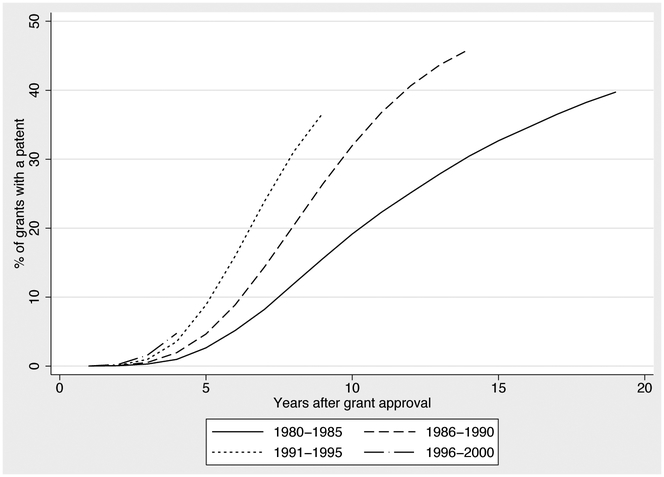
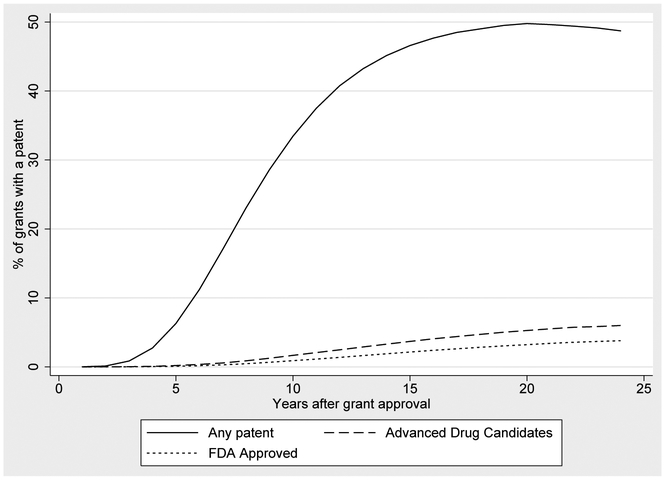
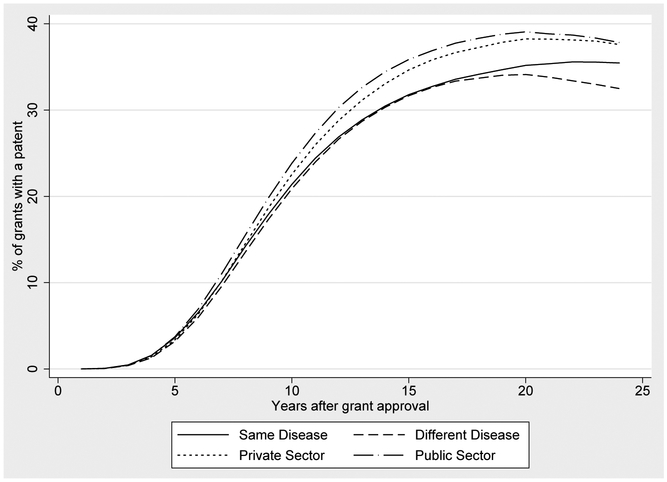
According to Table 1, 66,085 or 43% of the NIH grants in our sample produce a publication that is directly cited by a patent. This figure is a lower bound because our publication and patent data are truncated in 2012. Figures 4, 5, 6 and 7 describe the lag times between NIH funding and follow-on patenting. Each figure displays a curve graphing the cumulative probability that a grant is linked to follow on patenting, over time. At a given point t on the x-axis, we plot the proportion of t year old grants that have produced a publication that is cited by a patent. The curve is generally increasing because a grant’s likelihood of being linked to a patent increases with age. In some cases, these curves turn downward in later years because of changes in cohort composition: to compute the proportion of grants linked to a patent at t, we exclude grants that are not yet t years old, meaning that our calculations for higher t do not include more recent grants. This provides a graphical way to examine the diffusion of knowledge stemming from NIH expenditures, and how this diffusion process varies over time and across diseases.
Figure 4.
Grant-Patent Lags by Disease Area — Top 10 ICs
Figure 5.
Grant-Patent Lags by Grant Cohort
Figure 6.
Grant-Patent Lags by Patent Quality
Figure 7.
Grant-Patent Lags by Patent Type
Figure 4 documents substantial variation in the relevance of NIH funding for patenting across diseases. Approximately 15 years after funding, over 60% of grants funded by the National Institutes for Allergy and Infectious Diseases have produced research that has been cited by a patent. By contrast, this is true of only 20% of grants funded by the National Institutes of Mental Health. We caution that these differences should not be interpreted as comparisons of the efficacy of NIH funds, as they also reflect differences in the ease of biomedical innovation across disease areas and the types of research funded by different Institutes.
Figure 5, meanwhile, shows that time-to-patent has been decreasing over time. Only 20% of grants awarded between 1980 and 1985 produced research that is relevant for a patent in the ten years following. For grants awarded between 1991 and 1995, this figure is on track to be almost 40%. One interpretation of this finding is that NIH efforts to encourage “translational research” have been successful. An alternative view is that patentability has steadily moved upstream along the biopharmaceutical R&D value chain (Eisenberg and Nelson 2002; Jensen and Murray 2005).
Figure 6 underscores the fact that although 43% of grants are associated with patents, “important” patents—those pertaining to advanced drug candidates, or to FDA-approved treatments—are still relatively rare. Even twenty years after approval, less than 5% of NIH grants produce research cited by a patent associated with an FDA-approved drug; this figure is only slightly higher for advanced drug candidates, those at or beyond Phase 3 clinical trials.
Finally, Figure 7 shows that a grant is just as likely to produce research relevant for patents primarily associated with other disease areas as it is for patents associated with its own disease area. Our matching process allows a patent to be associated with more than one Institute (conditional on being linked to a DST, the average patent is linked to 7 different ICs). For each patent, we define its primary disease area as the IC responsible for funding the plurality of the publications that it cites. Then we categorize each patent-to-grant linkage as being for the same disease or for a different disease, where the reference disease is simply given by the funding IC for the focal grant. Figure 7 also shows that both private- and public-sector entities take advantage of NIH-funded research.
From here on, we focus on the impact of NIH funding on private-sector patents. This designation excludes patents to universities, governments, hospitals, and other non-profit institutions. Appendix Table K5 reports our main results with public-sector patents instead. Appendix N presents results that circumvents the use of publication data by restricting the patent data to the set of “Bayh-Dole” patents, i.e., patents held by the PIs of NIH grants and reported to NIH as products of these grants. OLS estimates imply an elasticity only approximately half as large as that observed in Table 7. The corresponding IV estimates are negative and imprecisely estimated. To summarize, despite its prominence in policy discussion, academic entrepreneurship (as proxied by academic patenting) corresponds to only a small fraction of the impact of NIH-funded research on patenting more generally.
Table 7:
Effect of NIH Investments on Total Related Private-Sector Patenting
| # of Patents Related to NIH-Funded Research | |||||
|---|---|---|---|---|---|
| (1) | (2) | (3) | (4) | (5) | |
| Fractional Patent Counts: Mean=24.8; SD=28.0 | |||||
| DST Funding (×$10 mln.) Mean=4.06; SD=4.36 | 4.516*** (0.210) |
3.593*** (0.512) |
3.590*** (0.537) |
3.712*** (0.601) |
3.239*** (0.372) |
| Elasticity | 0.738 | 0.588 | 0.587 | 0.607 | 0.530 |
| R2 | 0.536 | 0.759 | 0.783 | 0.965 | 0.974 |
| Unit Patent Counts: Mean=3,969; SD=3,918 | |||||
| DST Funding (×$10 mln.) Mean=4.06; SD=4.36 | 603.082*** (26.714) |
456.685*** (53.002) |
453.133*** (56.424) |
504.728*** (80.237) |
445.983*** (41.404) |
| Elasticity | 0.696 | 0.527 | 0.523 | 0.583 | 0.515 |
| R2 | 0.561 | 0.843 | 0.861 | 0.978 | 0.983 |
| Observations | 14,085 | 14,085 | 14,085 | 14,085 | 14,085 |
| Year FEs | Incl. | Incl. | Incl. | Incl. | Incl. |
| Disease × Science FEs | Incl. | Incl. | Incl. | Incl. | |
| Disease × Year FEs | Incl. | Incl. | Incl. | ||
| Science × Year FEs | Incl. | Incl. | |||
| Application Count FEs | Incl. | ||||
Note: Each observation is Disease/Science/Time (DST) combination. A patent is considered to be in the same area as an NIH grant if it cites a publication that is similar (as defined by the PubMed Relatedness Matching Algorithm) to a publication that is linked to a patent. For more details on this sample, See the notes to Tables 1 and 2. Funding is defined by the sum of project-cycle allocations for all new and competing renewal grants that are associated with that DST. The patent sample is restricted to those with private sector assignees, and weighted by average DST size, i.e., the average yearly number of grants in a Disease/Science research area. See Table 2 for more details. Year FEs are fixed effects for the fiscal year associated with a DST. NIH Institutes are taken to represent diseases and NIH study sections (review committees) are taken to represent science areas. Elasticities are evaluated at sample means. Application count FEs are indicator variables for the number of applications that a DST receives.
Standard errors in parentheses, two-way clustered at the disease and science level (*p < 0.10, **p < 0.05, ***p < 0.01).
4.2. DST-level Patent Match
Recall that our analysis is at the DST level: each observation is an Institute-study section pairing at a point in time, and we are interested in how funding for this DST relates to later patenting. Table 3 describes the characteristics of the DSTs in our sample. The average DST supports 11 grants totaling $47 million in funding (weighted by DST size). Table 3 also indicates that 13,027 or over 80% of DSTs produce research that is potentially relevant for patenting. In contrast, 8,886 DSTs (63%) can be linked to patents through a direct citation link.
Table 3:
NIH Research Area (DST) Characteristics, 1980-2005
| DSTs Linked to Patents | |||
|---|---|---|---|
| Full Sample | Cited by Patents | Related to Patents | |
| Average # of Grants | 10.85 (16.58) |
15.60 (19.05) |
11.62 (17.01) |
| Output Characteristics | |||
| Funding Amount (DST) | $40,631,460 (43,611,800) |
$45,556,350 (44,448,260) |
$41,397,230 43,683,690 |
| # of Patents Citing NIH-Funded Research (Fractional counts) | 12.82 (19.17) |
14.71 (19.85) |
13.07 (19.28) |
| # of Patents Citing NIH-Funded Research (Unit counts) | 101.7 (153.6) |
116.8 (159.1) |
103.7 (154.4) |
| # of Patents Related to NIH-Funded Research (Fractional counts) | 24.84 (27.95) |
28.33 (28.31) |
25.30 (28.00) |
| # of Patents Related to NIH-Funded Research (Unit counts) | 3,520 (3,742) |
4,023 (3,755) |
3,589 (3,745) |
| N | 14,085 | 8,886 | 13,027 |
Note: Sample is the same as that in Table 1, except aggregated to the NIH Disease/Science/Time level. See the notes to Table 1 for additional definitions. The funding and patent variables are weighted by average DST size, i.e., the average yearly number of grants in a Disease/Science research area. In fractional patent counts, a patent matched to N distinct DSTs counts as 1/Nth of a patent for each DST. In unit patent counts, a single patent matched to N distinct DSTs counts as one patent for each DST. Funding amounts are expressed in 2010 dollars (deflated by the Biomedical R&D Producer Price Index).
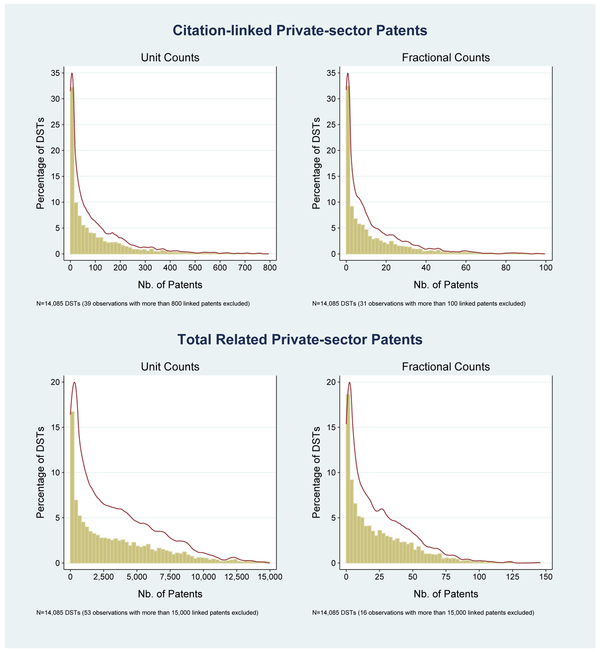
The correct attribution of patents to DSTs depends on the innovation production function and the degree to which any particular piece of knowledge is instrumental in generating the patent. If DSTs are pure substitutes in the production of patents and if a patent is linked to N DSTs, then each DST should receive credit for 1/Nth of that patent. Table 3 shows that the average DST in our sample produces research that is directly cited by 12.8 private-sector patents and is intellectually related to a total of 24.8 patents, using this “fractional” patent count. If, instead, the contributions of various DSTs are complements, then a patent should count for more than ; in the extreme, support from each DST is critical such that production is Leontief. In this case, DSTs should receive full credit for each patent it is linked to, which we designate as a “unit” patent count. Applying this assumption to our data, we find that the average DST is directly cited by 102 unit patents. The distribution of patent counts at the DST level exhibits skewness, as can be observed in the histograms displayed in Figure 8.
Figure 8.
Outcome Measures by DST
5. Main Results
Tables 6 and 7 present the fixed effects estimates of the impact of NIH funding on our two measures of patent outcomes. The top panel of Table 6 describes the impact of NIH funding on the number of patents that cite NIH-funded work, using fractional patent counts. Without any controls, we find that a $10 million increase in funding for a research area (DST) is associated with 2.6 more patents. Adding fixed effects for research areas (disease/science groupings) reduces this coefficient to 2.3. We add increasingly detailed fixed effects in each successive column; interestingly, our estimates remain relatively stable. One explanation for this is consistency is that, at the time it makes funding decisions, the NIH may not be able to anticipate which DSTs have greater future innovative potential. In this case, the amount of funding that a DST receives may be relatively uncorrelated with its future patent output. With our full set of controls, we estimate that a $10 million increase in funding contributes to 2.5 additional patents. With an average grant size of $1.6 million, this is equivalent to about one patent for every 2 to 3 NIH grants.
Table 6:
Effect of NIH Investments on Follow-On Patenting by Private-Sector Firms
| # of Patents Citing NIH-Funded Research | |||||
|---|---|---|---|---|---|
| (1) | (2) | (3) | (4) | (5) | |
| Fractional Patent Counts: Mean=12.82; SD=19.17 | |||||
| DST Funding (×$10 mln.) Mean=4.06; SD=4.36 | 2.595*** (0.220) |
2.281*** (0.356) |
2.242*** (0.359) |
2.550*** (0.654) |
2.450*** (0.568) |
| Elasticity | 0.822 | 0.723 | 0.71 | 0.808 | 0.777 |
| R2 | 0.417 | 0.600 | 0.641 | 0.918 | 0.933 |
| Unit Patent Counts: Mean= 101.7; SD= 153.6 | |||||
| DST Funding (×$10 mln.) Mean=4.06; SD=4.36 | 21.830*** (1.731) |
17.831*** (2.068) |
17.842*** (2.067) |
18.626*** (4.308) |
18.412*** (3.648) |
| Elasticity | 0.872 | 0.712 | 0.713 | 0.744 | 0.735 |
| R2 | 0.447 | 0.674 | 0.710 | 0.944 | 0.956 |
| Observations | 14,085 | 14,085 | 14,085 | 14,085 | 14,085 |
| Year FEs | Incl. | Incl. | Incl. | Incl. | Incl. |
| Disease × Science FEs | Incl. | Incl. | Incl. | Incl. | |
| Disease × Year FEs | Incl. | Incl. | Incl. | ||
| Science × Year FEs | Incl. | Incl. | |||
| Application Count FEs | Incl. | ||||
Note: Each observation is Disease/Science/Time (DST) combination. A patent is citation-linked to a DST if it cites research that acknowledges funding from that DST. For more details on this sample, see the notes to Tables 1 and 3. Funding is defined by the sum of project-cycle allocations for all new and competing renewal grants that are associated with that DST. The patent sample is restricted to those with private sector assignees, and weighted by average DST size, i.e., the average yearly number of grants in a Disease/Science research area. See Table 2 for more details. Year FEs are fixed effects for the fiscal year associated with a DST. NIH Institutes are taken to represent diseases and NIH study sections (review committees) are taken to represent science areas. Elasticities are evaluated at sample means. Application count FEs are indicator variables for the number of applications that a DST receives.
Standard errors in parentheses, two-way clustered at the disease and science level (*p < 0.10, **p < 0.05, ***p < 0.01).
The bottom panel presents an equivalent set of results using unit patent counts. Here, we estimate that $10 million leads to 18.4 more patents in the specification that is saturated with fixed effects (column 5). The difference in estimates between the top and bottom panels of Table 6 are substantial and arise because using unit count assumes that publications are perfect complements in patent production, as discussed in Section 4.2. Yet, the corresponding elasticities are very similar in both cases. Since patents can cite many publications (14 on average), it may not be reasonable to assume that all publications are required to produce a given patent. As such, in the remainder of the manuscript we focus on the smaller and more conservative fractional counts as our preferred outcome variable.
The estimates in Table 6 would not reflect the true value of NIH funding if public support for science either crowds out private investment or if it spurs patenting in ways that cannot be captured by a direct grant-publication-patent link. The top panel of Table 7 reports the impact of NIH expenditures on the total amount of private-sector patenting in areas related to a DST, whether or not these patents directly cite NIH-funded research. This specification is designed to assess the net impact of NIH funding on private-sector innovation in an area, accounting for both the possibility of crowd-out and the possibility that not all patents spurred by NIH funding can be linked via direct citations. Column 5 of Table 7 finds that a $10 million increase in DST funding results in a 3.2 net increase in the number of related private-sector patents, or about one patent for every two NIH grants.
If NIH funding fully crowded out industry investments, we would expect the coefficients reported in Table 7 to be zero. In fact, the magnitude of the impact of NIH funding on total patenting is slightly larger than its effect on patenting that can be directly linked to NIH funds (see Table 6). This is consistent with the absence of crowd-out. Alternatively, even if NIH funding crowds out some private investment, it is offset by increases in the number of patents related to NIH funding through indirect citation channels, or by increases in the productivity of private R&D investments.18
The bottom panel of Table 7 reports these results with fractional patent counts, yielding effect sizes that are an order of magnitude larger. These results, however, are unlikely to reflect the true effect of NIH funding. Recall that this final outcome measure is designed to capture the influence that NIH funding may have on patenting that does not require a direct citation linkage between funding and patents. In this measure, patents are linked to study sections through shared intellectual foci, reflecting the notion that public funding in a particular area produces knowledge that enhances productivity of others working in that area. Each DST is associated with many more patents in this way, thus driving a large wedge between fractional and unit impacts. Unlike the direct method which connect patents to a small number of study sections, our indirect method often yields connections to hundreds of study sections in related intellectual realms. While all linkages may be important, it is harder to imagine that each unit of knowledge is instrumental, and thus we favor the more conservative fractional approach in this case. Going forward, we will discuss estimates of the effect of funding on overall patent production using only the more conservative fractional counts.
Table 8 displays IV estimates using our instrumental variable for funding. Column 1 reports the first stage estimate of the relationship between total DST funding and windfall DST funding, controlling flexibly for raw scores and science ranks, as well as the number of applications that a disease/science paring has in a 25-grant window surrounding that IC’s funding threshold for that year. Because our IV strategy requires that we control for these additional variables, which we do not use in Tables 6 and 7, we report both our IV estimates as well as OLS estimates using the same set of first stage controls. Table 8 also reports tests of the strength of our windfall funding instrument. We obtain a Cragg-Donald Wald F-statistic of 478 and a Kleibergen-Paap Wald F-statistic of 37.5; both reject the null hypothesis that our instrument is weak. Comparing OLS and 2SLS specifications, we find similar effects of NIH funding on the number of directly cited patents (2.5 vs. 2.0) and a slightly smaller effect for the total number of patents related to an NIH research area (3.6 vs. 2.3). We take the 2.3 figure in Column 5 as our preferred estimate of the impact of NIH funding on private-sector patenting. Appendix Table J2 reports reduced-form estimates using windfall funding as the explanatory variable; we find similar, or even slightly larger results.
Table 8:
Effect of NIH Investments on Private-Sector Patenting Windfall Funding IV
| First Stage | Citation Linked | Total Related | ||||
|---|---|---|---|---|---|---|
| DST Funding (× $10 mln.) |
Mean=12.82; SD=19.17 | Mean=24.8; SD=28.0 | ||||
| OLS | IV | OLS | IV | |||
| (1) | (2) | (3) | (4) | (5) | ||
| Windfall Funding (×$10 mln.) | 1.251*** (0.194) |
DST Funding (×$10 mln.) Mean=4.06; | 2.478*** (0.658) |
2.002* (1.106) |
3.615*** (0.817) |
2.329** (1.159) |
| Elasticity | 0.785 | 0.634 | 0.592 | 0.381 | ||
| Cragg-Donald Wald F-stat | 478 | |||||
| Kleibergen-Paap Wald F-stat | 37.51 | |||||
| Observations | 14,085 | 14,085 | 14,085 | 14,085 | 14,085 | |
| Year FEs | Incl. | Incl. | Incl. | Incl. | Incl. | |
| Disease × Science FEs | Incl. | Incl. | Incl. | Incl. | Incl. | |
| Disease × Year FEs | Incl. | Incl. | Incl. | Incl. | Incl. | |
| Science × Year Linear Trends | Incl. | Incl. | Incl. | Incl. | Incl. | |
| Application Controls | Incl. | Incl. | Incl. | Incl. | Incl. | |
Note: See notes to Tables 6 and 7 for details about the sample. The outcome variables are fractional patent counts. The instrument is the total amount of funding (2010 dollars) for the subset of grants funded by a DST whose rank of rank scores were marginal, i.e., were within 25 applications of the award cutoff for their specific disease area (Institute). Application controls include (i) FEs for the number of applications that a DST receives; (ii) FEs for the number of applications associated with a DST that are also in a 50-grant window around the relevant IC payline, as well as (iii) cubics in the average raw and rank scores of applications associated with a DST that are also in a 50-grant window around the payline. Elasticities are evaluated at the sample means.
Standard errors in parentheses, two-way clustered at the disease and science level (*p < 0.10, **p < 0.05, ***p < 0.01).
Finally, we note that although we take our slightly smaller IV estimates as our preferred specification, our OLS fixed effect and IV approaches should be considered complementary because they identify slightly different sources of funding variation. In particular, our OLS estimates will capture the impact of both anticipated and unanticipated changes in NIH funding. Increases in funding for a research area may lead to more total patenting in this area both by providing support for existing research ideas that would not have been funded otherwise, or by encouraging scientists to enter or extend their research in this area. The latter effect depends on scientists being aware of funding changes in advance. Our OLS estimates allow us to capture both these effects, especially in specifications that control for fewer fixed effects. The downside of these estimates is that such variation is also potentially endogenous to scientific potential although, in practice, our estimates are not very sensitive to the inclusion of more fixed effects, suggesting that the impact we estimate in the OLS is less likely to be purely driven by endogenous factors correlated with our fixed effects.
Our IV estimates, on the other hand, are driven by differences in windfall funding coming as a result of the relative ranking of grant applications that have already been submitted. As such, they only capture the impact of unanticipated increases in NIH funding for a given research area. Such variation is more likely to be exogenous, but the tradeoff is that we identify a less comprehensive source of variation. This may be another potential reason we find a slightly smaller impact of funding using our instrument.
5.1. Patents related to NIH-funded research: stable keyword approach
In Table 7 and in Columns 4 and 5 of Table 8, we examine the impact of NIH funding on the total number of intellectually related patents, whether or not these patents actually cite NIH-funded research. We define a patent as intellectually related to an NIH DST if that patent cites any publications that are intellectually similar (according to keyword overlap) to publications funded by that DST (see Appendix E for details). A potential drawback of this approach is that our definition of a DST’s “intellectual area” can vary over time. If funding allows a disease/science area to expand the set of topics that it supports, then we may associate increased funding with more patents simply because higher levels of grant expenditures leads us to credit DSTs with patents over a wider slice of technological space.
To ensure that our results are not driven by this phenomenon, we also reestimate our results restricting to a definition of intellectual area that is stable for each disease/science (DS) area. To do this, we categorize all MeSH keywords associated with a publication funded by a DS combination into one of two types: “stable” keywords appear in publications funded by that DS across all years in the observation window, whereas “peripheral” keywords appear only in a subset of years in the data. We then restrict the set of related publications to those that match to a DS on stable keywords only. This fixes the boundaries of an intellectual area over time and therefore breaks any mechanical relationship that might exist between funding and the number of indirectly linked patents.
Appendix Table L1 examines the impact of NIH funding on the number of intellectually related patents, using a variety of ways to standardize the keywords that define a stable intellectual area. The details of this approach are discussed in Appendix L. In general, two features of the results presented in Appendix Table L1 deserve mention. First, the magnitudes of the coefficients are slightly smaller than those observed in Table 8. This is to be expected since our “stable” linking strategy shrinks the number of opportunities to associate patents with DSTs. The IV estimates are more imprecisely estimated (statistically significant at the 10% level for three out of four specifications). Second, the elasticities are comparable in magnitude to those computed in Columns 4 and 5 of Table 8. We believe these results provide evidence that our main conclusions are not driven by a potential mechanical linkage between DST funding and the size of its related intellectual area.
5.2. Additional Robustness Checks
We probe the robustness of our results using a variety of approaches, described in more detail in Appendices F, I, J, and K.
Appendix F discusses the idea of “missing” DSTs, i.e., those DST observations that are absent in our sample of 14,085 DSTs. Appendix Table F1 repeats our analysis on a balanced panel of 7,966 contiguous DSTs—those DS combinations that receive funding in all years between the first and last year in which the DS is observed. Our estimates are almost numerically identical. Appendix I compares traditional production function estimation with “fixed lags” to the estimates generated by our approach. Appendix J provides additional tests of our identifying assumptions. For example, the NIH occasionally funds grant applications out of the order in which they are scored. If DSTs that receive more out-of-order funding also have unobservably higher innovative potential, then this may bias our estimates. We discuss a variety of specification checks that together demonstrate that this threat to identification is not a concern empirically. Appendix J also provides evidence for the plausibility of the exclusion restriction for the instrument, in addition to the tests already presented in Section 3.2.3. We show that WindfallFundingdst is not correlated with past patent output in a DS.
Appendix K considers alternative specifications and samples. We show that our results are robust to not using weights in our regressions, so that each DST contributes to the same extent to the results, regardless of how many grants it supports. We estimate non-linear specifications using logs of funding and patenting, as well as a Poisson parametrization. Our main results also hold when restricting our sample to NIH Institutes that are the most directly identified with disease and body system areas and we also examine the impact of NIH funding on public sector patenting. Finally, we also examine the impact of NIH funding on “embodied” versus “disembodied linkages by separating the effect of funding on patenting by the same research team that receives the grant from its impact on patenting by different research teams.
5.3. Heterogeneity
In addition to quantifying the impact of NIH funding on overall patenting, we also examine which type of patents are most responsive to NIH expenditures. The impact of NIH funding on the development of high-value patents need not be similar to its impact on overall patenting; if firms direct their resources to the most promising projects, then the marginal patent that is created because of NIH funding may be relatively low quality. Conversely, if it is unprofitable for firms to invest in risky or early-stage research, then the marginal patent supported by the NIH may be of high quality. Column 1 of Table 9 reproduces the estimates of the impact of funding on total private-sector patenting from Table 8. Column 2 focuses on “important” patents, those that either pertain to advanced drug candidates or to FDA-approved biopharmaceuticals (traditional “small molecule” drugs as well as vaccines and biologics).
Table 9:
Effect of NIH Investments on Private-Sector Patenting Heterogeneity by Patent Type
| All Private Sector |
Advanced Drug Candidates |
Highly Cited | Same Area | Different Area | Large Assignee | Small Assignee | |
|---|---|---|---|---|---|---|---|
|
Mean=24.8; SD=28.0 |
Mean=0.546; SD=0.864 |
Mean=1.28; SD=1.76 |
Mean=18.9; SD=23.8 |
Mean=15.9; SD=19.0 |
Mean=17.5; SD=20.7 |
Mean=3.47; SD=4.18 |
|
| −1 | (2) | (3) | (4) | (5) | (6) | (7) | |
| OLS | |||||||
| DST Funding (×$10 mln.) Mean=4.06; SD=4.36 | 3.615*** (0.817) |
0.081*** (0.014) |
0.175*** (0.052) |
2.699*** (0.450) |
2.297*** (0.711) |
2.562*** (0.620) |
0.506*** (0.102) |
| Elasticity | 0.592 | 0.602 | 0.555 | 0.580 | 0.587 | 0.594 | 0.592 |
| IV | |||||||
| DST Funding (×$10 mln.) Mean=4.06; SD=4.36 | 2.329** (1.159) |
0.053 (0.040) |
0.111** (0.067) |
1.202* (0.731) |
1.894** (0.939) |
1.658** (0.778) |
0.362 (0.230) |
| Elasticity | 0.381 | 0.394 | 0.352 | 0.258 | 0.484 | 0.385 | 0.424 |
| Observations | 14,085 | 14,085 | 14,085 | 14,085 | 14,085 | 14,085 | 14,085 |
Note: See notes to Tables 6 and 7 for sample details. The outcome variables are fractional patent counts. All specifications include disease-science FEs, disease-year FEs, science by year linear time trends, FEs for the number of applications to the DST, cubics in the average raw score and average science rank received by applications in the 25-grant radius window around the IC payline, and FEs for number of DST applicants in a 25-grant window around an IC’s funding cutoff. A patent is labelled “Private Sector” if it is assigned to a domestic US or foreign corporation (NBER assignee categories 1 and 2 minus foundations, universities, and hospitals). A patent is labeled an advanced drug candidate if it is included in IMS Patent Focus, which has information on patents on drugs in Phase III trials or further. A patent is in the same disease area as a DST if the majority of NIH research areas that it is linked are also associated with that same “D” disease area. A patent is associated with a large assignee if its first assignee employs more than 500 employees; it is considered small otherwise.
Standard errors in parentheses, two-way clustered at the disease and science level (*p < 0.10, **p < 0.05, ***p < 0.01).
The OLS and IV estimates reported in Column 2 of Table 9 show that a $10 million increase in DST funding leads to a net increase of 0.05 to 0.08 patents associated with advanced drug candidates (those that have entered clinical trials) and FDA-approved drugs. While this figure is small in magnitude, it translates into an elasticity of patenting with respect to funding of between 0.4 to 0.6, comparable to the elasticity we estimate for private-sector patents in general. We will discuss alternative measures of patent value in the next section, when we discuss the economic magnitude of our results.
Our next set of results consider the impact of spillovers from funding in one disease area on innovation in others. Many studies document cases in which existing medical treatments have been successfully used to treat new conditions (Gelijns et al. 1998; Wurtman and Bettiker 1994). Similarly, drug development efforts often build on research originally intended for other diseases, reflecting the importance of knowledge spillovers across diseases (Henderson and Cockburn 1996). Our results provide evidence on the magnitude of these cross-disease knowledge spillovers. To measure spillovers, we assign a primary disease affiliation to each patent in our data by finding the NIH Institute that is responsible for funding the plurality of publications cited by that patent. We find that NIH funding directed toward one disease area is as likely—if not more likely—to translate into patents that are primarily affiliated with other disease areas as it is to translate into patents affiliated with its own. The IV estimate in Column 4 of Table 9 indicates that a $10 million increase in funding for a DST generates 1.20 additional patents with the same primary disease affiliation. Column 5, however, shows that this same funding also generates 1.89 additional patents with a different primary disease affiliation. Part of the reason for such large cross-disease funding spillovers may be due to the fact that much of the research that the NIH supports centers on scientific questions that are relevant to many disease areas. The National Cancer Institute may, for instance, fund a study of cell division in frog embryos; this research may also be relevant for the study of tissue regeneration and aging-related disorders. These findings highlight the importance of using a patent-linking strategy that does not assume that funding only impacts innovation in its intended area. Had we made this assumption, we would have failed to account for over half of the relevant innovative outputs.
Finally, Table 9 also shows that NIH investments increase patenting for both large and small assignees. While larger assignees produce a larger number of patents in response to increases in NIH funding, the response of small assignees is equally elastic. This finding is consistent with our summary statistics in Table 2, which show that a greater proportion of patents assigned to small firms cite NIH-funded research.
5.4. Valuing the Impacts of NIH Investments
Our results suggest that a $10 million increase in NIH funding leads to a net increase of 2.3 weighted private-sector patents. Putting a dollar value on these patents is difficult, for several reasons. It is well known that patent value distributions are highly skewed (Harhoff, Scherer, and Vopel 2003). Moreover, only the private value of patents is typically calculated, and the social value can be much larger. As such, we utilize a variety of approaches to monetize this return.
One approach to valuing the returns to NIH funding in dollars, rather than patents, is to rely on estimates for the market value of patents taken from the literature. Bessen (2009) quantifies the effect of patent stocks on Tobin’s q, and uses these estimates to derive the market value of a patent across sectors of the economy. In the biopharmaceutical sector, his estimates imply that an additional patent is valued by the stock market at about $11.2 million (2010 dollars). Combined with our estimate in Table 6, Column 5, a back-of-the-envelope calculation indicate that a $10 million dollar increase in NIH funding would yield $34.7 million in firm market value. As Bessen (2009) notes, a problem with this approach is that patents may be picking up the effects of other factors correlated with market value; accordingly this figure probably represents an upper bound.
A different approach is to focus on patents associated with marketed drugs. Very few of the patents in our sample are for drugs, let alone marketed drugs. However, for this set we have another measure of private value, drug sales. DiMasi, Grabowski, and Vernon (2004) report that the mean present discounted value (PDV) of lifetime sales for new drugs approved by the FDA between 1990 and 1994 was approximately $3.47 billion (2010 dollars). More recent research (Berndt et al. 2015) shows similar orders of magnitude, although the returns appear to have been declining over time.
Table 10 presents implied drug valuation estimates of our results based on the DiMasi et al. figure reported above. Column 1 reproduces our findings from Table 9 with respect to all advanced drug candidates. Another variation is to restrict the outcome to patents associated with FDA-approved drugs. Column 2 reports OLS and IV estimates using only these patents to construct the outcome variables at the DST level and finds that a $10 million dollar increase in funding results in approximately 0.034 more such patents. In this definition, we include all patents we can link to a drug (including those listed in the Orange Book, as well as additional patents from IMS Patent Focus); there are approximately eight patents associated with every FDA-approved drug on average (see Appendix B). If the inventions associated which each of these eight patents are essential to the development of the corresponding drug, then we should fully credit each with the value of that drug. In this case, we would expect $10 million dollar increase in funding to generate an expected PDV of 0.034 × $3.47 billion = $149.2 million dollars in sales.
If we instead assumed that the invention underlying each patent contributes equally to the drug, we would expect this funding amount to translate into 0.034/8 = 0.004 drugs, with an expected PDV of 0.004 × $3.47 billion = $14.7 million.
However, even within drug, there may be heterogeneity in patent importance.19 Many “secondary” Orange Book patents are not even filed until well after the product is launched (Kapcynski et al. 2012; Hemphill and Sampat 2013); IMS patents may be even more peripheral.20 Attributing the same share of product sales to these patents as to the “main patent” associated with that drug may lead to overstating the effect of NIH funding. To explore this heterogeneity, we ran several additional models. The first looks only at “pre-approval” patents (from the Orange Book and/or IMS), those filed before drug approval (on average, there are five such patents per drug). In Column 4, we are more conservative, limiting the outcome variable to the first patent associated with a marketed drug, on the assumption that this is the main patent. (No scaling is required in this case since we are only looking at one patent per drug.) Finally, Column 5 examines drug level outcomes: in this case, we match the number of discrete drugs associated with a DST, rather than the number of patents. In all three of these columns, the OLS estimates are statically significant and similar in magnitude to those reported for FDA approved drugs, from Column 2, but the IV estimates are smaller and statistically insignificant.21
There exists a vast literature estimating the rate of return to private R&D. These estimates are highly variable, ranging between 0 and 100% (see Hall, Mairesse, and Mohnen (2001) for a comprehensive summary). Two caveats must be kept in mind when comparing our results with those previously reported. First, the level of analysis employed in our study (the research area) is very different from that typically encountered in the literature, which tends to analyze data collected at the industry-, firm-, or plant-level. Second, we focus on a single industry, the biopharmaceutical industry, rather than a wide cross-section of industries. That said, our implied rate of return (based on the $14.7 million implied drug value of a $10 million investment seen in Table 10, column 2) is quite similar to the middle of the range of estimates reported in the literature.
Assigning value to individual patents is notoriously difficult, and the different approaches above yield different magnitudes for the effects of NIH funding. Accordingly, beyond presenting a range of implied drug valuations, we are not in a position to report a specific rate of return. Any such estimate would only capture the private (rather than social) value of the patented technologies.22 Finally, as we will emphasize in the conclusion, there are many effects of NIH funding that do not result in patentable research at all.
6. Conclusion
Modern growth theory highlights the importance of knowledge spillovers for long-run economic growth. These spillovers mean that private firms will under-invest in the production of knowledge. Two types of policies aim to ameliorate this “market failure”: patent policy and public funding of research. While there is now a significant body of empirical research on the former, the effects of public funding, and public funding of science in particular, have received less attention.
One reason for this paucity of evidence on the impacts of public research investments is that it is difficult to measure the effects of knowledge that is both non-rival and difficult to appropriate (Griliches 1992). While the idea that public science has large effects is central to U.S. policy—going back to Vannevar Bush’s 1945 assertion that basic research is “the pacemaker of technological progress”—economists emphasize that evidence in support of this claim is rather limited (Garber and Romer 1996; Cockburn and Henderson 1998).
In this paper, we examine the effects of public science on private sector innovation in the life sciences, focusing on funding by the largest funder of research in the world, the National Institutes of Health. Our results show that NIH investments in a research area increase subsequent private-sector patenting in that area; a $10 million increase in funding for an area leads to 2.3 additional patents or, equivalently, we expect one private-sector patent generated for every two to three NIH-funded grants. This result holds across a variety of OLS and IV specifications. This positive impact, moreover, does not appear to be associated with lower private investments in other research areas. We cannot perform a formal rate of return calculation since our analysis focuses on only one aspect of the effect of NIH funding, that of sales associated with patented drugs. One rough calculation suggests that $1 dollar in NIH funding generates around $1.40 in drug sales.
We find that over half of the patents that result from NIH funding flow across disease areas. This has implications for measurement: had we looked only at patents in the same disease area, we would have missed half the output. This finding speaks to a long-standing question in postwar medical research policy: the feasibility and desirability of targeting research to diseases. Claims that scientific research often flows across disease areas have been common from NIH Directors since the agency’s founding, especially during Congressional debates about whether particular diseases are over/underfunded or in response to advocates lobbying for a new Institute for “their” disease (Sampat 2012). Our results support the view that there are strong cross-disease spillovers. The organization of the agency around disease-specific Institutes, though useful for mobilizing funding, may not reflect the importance of the interplay of ideas from different disease areas and fields in shaping biomedical research progress.
Throughout the text, we emphasized numerous caveats. We highlight several here. First, we are examining only one type of return to NIH funding, those that flow through patented innovations. This neglects a number of other socially important benefits of publicly-funded medical research, including applied epidemiological and clinical research that changes medical practice or health behaviors. Previous research (Cutler and Kadiyala 2003; Heidenreich and McClellan 2003) suggests this research has high value. Ignoring these outcomes could lead to large underestimates of the value of NIH funding.
A second potential limitation is the assumption that patent-to-publication citations reflect real linkages between the cited grant/publications and citing patents. For the goal of measuring knowledge spillovers from public research, these citations are much more meaningful than patent-to-patent citations, for reasons already discussed. However, articles are cited in patents for legal reasons, to denote “prior art” material to patentability, and decisions about how much to cite are influenced by factors including patent importance and applicant patent strategy (Sampat 2010). Not all articles cited are crucial for the development of the citing patent. Citations that are not real intellectual influences would lead to overestimates of the effects of NIH funding. (At the same time there are false negatives—not all knowledge firms “build on” must be cited—which would lead to underestimates of the effects of NIH funding.)
Third, our implied drug valuations were based on publicly available estimates on the distribution of drug sales, and assumptions about how to divide a drug’s value across its many patents. There is likely considerable heterogeneity in the private and social value of drugs (Garthwaite and Duggan 2012), and individual patents (Hemphill and Sampat 2011), which our back-of-the-envelope calculations could not fully incorporate.
Finally, our analysis implicitly assumes a “linear” flow from science to technology, and does not account for the complementary investments made by other actors (e.g., the NSF, or venture capital firms) in the path from laboratory to marketplace, or the feedbacks from technology to the progress of science. This “linear model” of research is well known to be an oversimplification, but even its detractors acknowledge that it is more reasonable in the life sciences than in other fields, and that alternative models would be far less empirically tractable (Balconi et al. 2010).
Despite these limitations, our analysis uses novel data and a new source of identification to provide estimates on an important but understudied component of the innovation production function: spillovers from public research. In future work, this framework could be extended to examine a range of other questions of interest to economists and policymakers, including heterogeneity in types of research (whether more or less targeted research has higher impact) and how the presence or absence of intellectual property rights affects returns to public research investments.
Supplementary Material
Acknowledgments
We acknowledge the financial support of the National Science Foundation through its SciSIP Program (Award SBE-0738142). We are grateful to Jason Abaluck, David Autor, Jim Bessen, Alex Frankel, David Genesove, Gordon Hanson, Ben Jones, Naomi Hausman, Kelly Shue, Scott Stern, John Van Reenen, Heidi Williams, and numerous seminar participants for helpful comments and suggestions. Azoulay and Graff Zivin are currently supported by the NIH for a project examining life cycle events, innovation, and the evolution of scientific fields (P01-AG039347). Azoulay, Graff Zivin, and Sampat are all prior recipients of NIH funding. Azoulay and Li have a current special volunteer agreement with NIH to facilitate access to peer review data. All errors are our own.
Footnotes
Adams (1990) uses distributed lags and panel data to shed light on the effect of scientific knowledge stocks on productivity growth at the industry level. Moretti, Steinwender, and Van Reenen (2014) use shocks to defense R&D induced by the end of the cold war to identify the impact of government expenditures on TFP growth, once again at the industry level.
Other funders include foundations, accounting for 4 percent, other federal funders, about 5 percent, and state and local governments, also about 5 percent.
The practical challenges encountered in order to systematically track and catalog patent-to-publication citation linkages are described in Appendix D2.
This concern is especially salient in the life sciences, since the organization of drug discovery research in the biopharmaceutical industry has been greatly transformed to mimic that of academic labs in terms of size, intellectual autonomy granted to researchers, and rewards linked to the production of high-impact publications (Henderson 1994). Many biomedical scientists also search for positions in academe and industry simultaneously (Stern 2004), and the patterns of mobility between the private and the public sector have been extensively documented (Zucker, Darby, and Torero 2002).
One notable exception is Bloom, Schankerman, and Van Reenen, who consider spillovers associated with private R&D investments.
This is relatively straightforward because PubMed started capturing this information systematically starting in 1980. Appendix D1 provides more detail, and discusses the issues that may arise in our design if researchers inflate their publication accomplishments to improve their odds of getting a grant renewed.
In previous work, Sampat and Lichtenberg (2011) looked at marketed drugs citing NIH publications, finding that over 40 percent of the drugs approved between 1988 and 2005 cite an NIH-funded publication. This paper builds on the strategy of linking drugs to patents to publications to grants, but extends it in several ways. Most importantly, rather than a retrospective approach examining what share of drug development can be linked back to NIH funding, our analysis is prospective, examining how variation in NIH funding relates to subsequent innovation. This approach allows for “failure” (grants that do not generate any innovation), and is relevant for policy makers considering changes to NIH funding.
The PubMed Related Article (PMRA) algorithm analyzes keywords and keyword combinations that are assigned to all life-science publications by the National Library of Medicine and defines similarity on the basis of how many of these keywords overlap. This is discussed in detail in Appendix E.
We discuss the practical details involved in assigning grants to particular DSTs in Section 4.1.
An alternative approach would be to define a research area narrowly, for example at the level of the individual grant. In Appendix C, we explain why we believe that exploiting grant-level variation in the funding process is less useful to shed light on the main questions of policy interest.
For instance, Congress may allocate more money to the National Cancer Institute in order to fight the “war on cancer” (Mukherjee 2010), and the private sector may make similar investments, suggesting a causal relationship that may in fact be spurious.
Unweighted results are presented in Appendix K, Table K1.
The main way that ICs get around these rules is to either fund an application out of scoring order or to issue a request for proposals (RFPs) or applications (RFAs) on a specific topic. RFPs and RFAs account for only a small portion of NIH grant spending. Grants responding to these are evaluated in specially empaneled study sections, which we exclude from our analysis. See Appendix J for a discussion of out-of-order grant funding.
“Recommendations for Change at The NIH Center For Scientific Review,” Final Phase 1 Report, Jan 14, 2000.
Jacob and Lefgren (2011) investigate the impact of receiving NIH funding on the publication output of individual scientists using a regression discontinuity design and compare outcomes for grant applications just above and just below an Institute’s payline. We cannot use the same design because the running variable—rank of rank—applies to individual grants but not to DSTs. There is no DST-level discontinuity. Instead, we compare DSTs with similar quality applications as judged by their raw and science rank scores, but which receive different levels of windfall funding.
A patent is part of our universe if (i) it is in a relevant patent class and (ii) cites at least one article indexed by PubMed. The relevant patent classes are the 92 classes belonging to categories 1 and 3 in the NBER USPTO database (see Appendix B for a complete list). Note that in practice, the second requirement is almost always satisfied for patents in these classes.
The list of the included Institutes is described in Appendix A, Table A1. Briefly, we exclude three small ICs (the National Institute on Minority Health and Health Disparities, the National Institute of Nursing Research, and the National Library of Medicine), as well as six NIH centers which serve mainly administrative functions. Our primary analyses do include three ICs that are not oriented towards a particular disease: the National Institute of General Medical Sciences (NIGMS), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and the National Human Genome Research Institute (NHGRI). Note, however, that these Institutes review grant applications from several study sections, which is all that our identification strategy requires. In a robustness test, we show that our results are robust to excluding ICs that are not primarily devoted to the study of specific diseases or body-systems (Appendix K, Table K4).
This may occur, inter alia, because researchers trained with NIH funds find jobs in the private sector where they go on to patent in the same area, or because NIH investments clarify the scientific potential of different research areas, allowing biopharmaceutical firms to target their investments more efficiently. In both cases, total private patenting in an area may still increase even if overall private investment decreases.
The active ingredient patent is typically thought to be more important than other Orange Book-listed patents (on average there is a single active ingredient patent per drug, and three total Orange Book patents). As an illustration of this, generics typically are able to enter after the expiration of the active ingredient patent: later Orange Book patents are often found to be irrelevant or invalid (Hemphill and Sampat 2012).
On average, 5 of the 8 patents for each drug were in IMS only. These were patents that did not meet the FDA’s standards for being relevant to the marketed drugs. Nevertheless, as discussed in Appendix B, we include IMS patents since the Orange Book has very limited coverage for biologic drugs, even though it does introduce many peripheral patents for traditional, “small molecule” drugs.
In our data, there are only 332 drugs and 270 “main” patents that can be matched to NIH grants over the course of our 25 year sample. Because the IV estimates rely on limited variation around an IC’s funding payline, there may not be enough data to obtain reliable IV estimates when these extremely rare patents are used to construct outcome variables at the DST level.
For biopharmaceuticals, some estimates suggest that the social value of an innovation can exceed its private value by a factor ranging from 4 to 20 (Lakdawalla et al. 2010; Philipson and Jena 2005, Goldman et al. 2010). Other authors strike a more skeptical note, emphasizing that the enormous costs of adopting certain medical technologies can sometimes drive social benefits far below the level of the surplus captured by their manufacturers (Murphy and Topel 2003; Chandra and Skinner 2012).
Contributor Information
Pierre Azoulay, MIT & NBER.
Danielle Li, Harvard University.
Joshua S. Graff Zivin, UCSD & NBER.
Bhaven N. Sampat, Columbia University & NBER
References
- Acemoglu Daron, and Linn Joshua. 2004. “Market Size in Innovation: Theory and Evidence from the Pharmaceutical Industry.” Quarterly Journal of Economics 119(3): 1049–1090. [Google Scholar]
- Adams James D. 1990. “Fundamental Stocks of Knowledge and Productivity Growth.” Journal of Political Economy 98(4): 673–702. [Google Scholar]
- Aghion Philippe, and Howitt Peter. 1992. “A Model of Growth through Creative Destruction.” Econometrica 60(2): 323–351. [Google Scholar]
- Agrawal Ajay, and Henderson Rebecca. 2002. “Putting Patents in Context: Exploring Knowledge Transfer from MIT.” Management Science 48(1): 44–60. [Google Scholar]
- Alcácer Juan, and Gittelman Michelle. 2006. “Patent Citations as a Measure of Knowledge Flows: The Influence of Examiner Citations.” Review of Economics and Statistics 88(4): 774–779. [Google Scholar]
- Arundel Anthony, and Geuna Aldo. 2004. “Proximity and the Use of Public Science by Innovative European Firms.” Economics of Innovation and New Technology 13(6): 559–580. [Google Scholar]
- Azoulay Pierre, Michigan Ryan, and Sampat Bhaven. 2007. “The Anatomy of Medical School Patenting.” New England Journal of Medicine 357(20): 2049–2056. [DOI] [PubMed] [Google Scholar]
- Azoulay Pierre, Ding Waverly, and Stuart Toby. 2009. “The Effect of Academic Patenting on the Rate, Quality, and Direction of (Public) Research Output.” Journal of Industrial Economics 57(4): 637–676. [Google Scholar]
- Balconi Margherita, Brusoni Stefano, and Orsenigo Luigi. 2010. “In Defence of the Linear Model: An essay.” Research Policy 39(1): 1–13. [Google Scholar]
- Bekkers Rudi, and Bodas Freitas Isabel Maria. 2008. “Analysing Knowledge Transfer Channels Between Universities and Industry: To What Degree Do Sectors Also Matter?” Research Policy 37(10): 1837–1853. [Google Scholar]
- Bernstein Jeffrey I., and Nadiri M. Ishaq. 1989. “R&D and Intra-industry Spillovers: An Empirical Application of Dynamic Duality.” Review of Economic Studies 56(2): 249–267. [Google Scholar]
- Berndt Ernst R., Nass Deanna, Kleinrock Michael, and Aitken Murray. 2015. “Decline in Economic Returns from New Drugs Raises Questions About Sustaining Innovations.” Health Affairs 34(2): 245–252. [DOI] [PubMed] [Google Scholar]
- Bessen James. 2009. “Estimates of Patent Rents from Firm Market Value.” Research Policy 38(10): 1604–1616. [Google Scholar]
- Bloom Nicholas, Schankerman Mark, and Van Reenen John. 2013. “Identifying Technology Spillovers and Product Market Rivalry.” Econometrica 81(4): 1347–1393. [Google Scholar]
- Blume-Kohout Margaret E. 2012. “Does Targeted, Disease-Specific Public Research Funding Influence Pharmaceutical Innovation?” Journal of Policy Analysis and Management 31(3): 641–660. [DOI] [PubMed] [Google Scholar]
- Brooks Harvey. 1995. “The Evolution of U.S. Science Policy” In Smith Bruce L.R., and Barfield Claude E. (Eds.), Technology, R&D, and, the Economy, pp. 15–48. Washington, D.C.: The Brookings Institution and American Enterprise Institute. [Google Scholar]
- Bush Vannevar. 1945. Science: The Endless Frontier. Washington, DC: US General Printing Office. [Google Scholar]
- Colin Cameron, A., and Miller Douglas L.. 2015. “A Practitioner’s Guide to Cluster- Robust Inference.” Journal of Human Resources 50(2): 317–372. [Google Scholar]
- Carpenter Mark P., and Narin Francis. 1983. “Validation Study: Patent Citations as Indicators of Science and Foreign Dependence.” World Patent Information 5(3): 180–185. [Google Scholar]
- Chandra Amitabh, and Skinner Jonathan. 2012. “Technology Growth and Expenditure Growth in Health Care.” Journal of Economic Literature 50(3): 645–680. [Google Scholar]
- Cockburn Iain, and Henderson Rebecca. 1996. “Public-Private Interaction in Pharmaceutical Research.” Proceedings of the National Academy of Sciences 93(23): 12725–12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockburn Iain M., and Henderson Rebecca M.. 1998. “Absorptive Capacity, Coauthoring Behavior, and the Organization of Research in Drug Discovery.” Journal of Industrial Economics 46(2): 157–182. [Google Scholar]
- Cohen Wesley M., Nelson Richard R., and Walsh John P.. 2000. “Protecting Their Intellectual Assets: Appropriability Conditions and Why U.S. Manufacturing Firms Patent (or Not).” NBER Working Paper #7552. [Google Scholar]
- Cohen Wesley M., Nelson Richard R., and Walsh John P.. 2002. “Links and Impacts: The Influence of Public Research on Industrial R&D.” Management Science 48(1): 1–23. [Google Scholar]
- Cutler David M., and Kadiyala Srikanth. 2003. “The Economic Value of Medical Research” In Murphy Kevin M., and Topel Robert H. (Eds.), Measuring the Gains from Medical Research: An Economic Approach, pp. 110–162. Chicago: University of Chicago. [Google Scholar]
- David Paul A., Hall Bronwyn H., and Toole Andrew A.. 2000. “Is Public R&D a Complement or Substitute for Private R&D? A Review of the Econometric Evidence.” Research Policy 29(4–5): 497–529. [Google Scholar]
- DiMasi Joseph A., Grabowski Henry G., and Vernon John. 2004. “R&D Costs and Returns by Therapeutic Category.” Drug Information Journal 38(3): 211–223. [Google Scholar]
- Ray Dorsey, E., de Roulet Jason, Thompson Joel P., Reminick Jason I., Thai Ashley, White-Stellato Zachary, Beck Christopher A., George Benjamin P., and Moses Hamilton III. 2013. “Funding of US Biomedical Research, 2003–2008.” JAMA 303(2): 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg Rebecca S., and Nelson Richard R.. 2002. “Public vs. Proprietary Science: A Fruitful Tension?” Academic Medicine 77(12): 1392–1399. [DOI] [PubMed] [Google Scholar]
- Fourrier François, Chopin Claude, Goudemand Jenny, Hendrycx Sylvie, Caron Claudine, Rime Alain, Marcy Anne, and Lestavel Philippe. 1992. “Septic Shock, Multiple Organ Failure, and Disseminated Intravascular Coagulation.” Chest 101(3): 816–823. [DOI] [PubMed] [Google Scholar]
- Garber Alan M., and Romer Paul M.. 1996. “Evaluating the Federal Role in Financing Health-Related Research.” Proceedings of the National Academy of Sciences 93(23): 12717–12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garthwaite Craig, and Duggan Mark. 2012. “Empirical Evidence on the Value of Pharmaceuticals” In Danzon Patricia M., and Nicholson Sean (Eds.), The Oxford Handbook of the Economics of the Biopharmaceutical Industry, pp. 463–492. New York: Oxford University Press. [Google Scholar]
- Gelijns Annetine C., Rosenberg Nathan, and Moskowitz Alan J.. 1998. “Capturing the Unexpected Benefits of Medical Research.” New England Journal of Medicine 339(10): 693–698. [DOI] [PubMed] [Google Scholar]
- Goldman Dana P., Jena Anupam B., Lakdawalla Darius N., Malin Jennifer L., Malkin Jesse D., and Sun Eric. 2010. “The Value of Specialty Oncology Drugs.” Health Services Research 45(1): 115–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griliches Zvi. 1992. “The Search for R&D Spillovers.” Scandinavian Journal of Economics 94(Suppl): S29–S47. [Google Scholar]
- Hall Bronwyn H., Griliches Zvi, and Hausman Jerry A.. 1986. “Patents and R&D: Is There a Lag?” Econometrica 27(2): 265–283. [Google Scholar]
- Hall Bronwyn H., Mairesse Jacques, and Mohnen Pierre. 2010. “Measuring the Returns to R&D” Chapter 24 in Hall Bronwyn H., and Rosenberg Nathan (Eds.), Handbook of The Economics of Innovation, 2:1033–1082. Amsterdam: North-Holland. [Google Scholar]
- Harhoff Dietmar, Scherer Frederic M., and Vopel Katrin. 2003. “Exploring the Tail of Patented Invention Value Distributions” In Grandstrand Ove (Ed.), Economics, Law, and Intellectual Property: Seeking Strategies for Research and Teaching in a Developing Field, pp. 279–309. Boston/Dordrecht/London: Kluwer Academic Publisher. [Google Scholar]
- Heidenreich Paul, and McClellan Mark. 2003. “The Economic Value of Medical Research” In Murphy Kevin M., and Topel Robert H. (Eds.), Biomedical Research and Then Some: The Causes of Technological Change in Heart Attack Treatment, pp. 163–205. Chicago: University of Chicago. [Google Scholar]
- Scott Hemphill, C., and Sampat Bhaven N.. 2011. “When Do Generics Challenge Drug Patents?” Journal of Empirical Legal Studies 8(4): 613–649. [Google Scholar]
- Scott Hemphill, C., and Sampat Bhaven N.. 2012. “Evergreening, Patent Challenges, and Effective Market Life in Pharmaceuticals.” Journal of Health Economics 31(2): 327–339. [DOI] [PubMed] [Google Scholar]
- Scott Hemphill, C., and Sampat Bhaven. 2013. “Drug Patents at the Supreme Court.” Science 339(6126): 1386–1387. [DOI] [PubMed] [Google Scholar]
- Henderson Rebecca. 1994. “The Evolution of Integrative Capability: Innovation in Cardiovascular Drug Discovery.” Industrial & Corporate Change 3(3): 607–630. [Google Scholar]
- Henderson Rebecca, and Cockburn Iain. 1996. “Scale, Scope, and Spillovers: The Determinants of Research Productivity in Drug Discovery.” The RAND Journal of Economics 27(1): 32–59. [PubMed] [Google Scholar]
- Henderson Rebecca, Jaffe Adam, and Trajtenberg Manuel. 1998. “Universities as a Source of Commercial Technology: A Detailed Analysis of University Patenting 1965–1988.” Review of Economics and Statistics 80(1): 119–127. [Google Scholar]
- Henderson Rebecca, Orsenigo Luigi, and Pisano Gary P.. 1999. “The Pharmaceutical Industry and the Revolution in Molecular Biology: Interactions Among Scientific, Institutional, and Organizational Change” In Mowery David C., and Nelson Richard R. (Eds.), Sources of Industrial Leadership, pp. 267–311. New York: Cambridge University Press. [Google Scholar]
- Hunter Tony. 2007. “Treatment for Chronic Myelogenous Leukemia: The Long Road to imatinib.” Journal of Clinical Investigation 117(8): 2036–2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob Brian A., and Lefgren Lars. 2011. “The Impact of Research Grant Funding on Research Productivity.” Journal of Public Economics 95(9–10): 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe Adam B. 1986. “Technological Opportunity and Spillovers from R&D: Evidence from Firms’ Patents, Profits, and Market Value.” American Economic Review 76(5): 984–1001. [Google Scholar]
- Jaffe Adam B. 2002. “Building Program Evaluation Into the Design of Public Research Support Programs.” Oxford Review of Economic Policy 18(1): 22–34. [Google Scholar]
- Jensen Kyle, and Murray Fiona. 2005. “Intellectual Property Landscape of the Human Genome.” Science 310(5746): 239–240. [DOI] [PubMed] [Google Scholar]
- Kapczynski Amy, Park Chan, and Sampat Bhaven. 2012. “Polymorphs and Prodrugs and Salts (Oh My!): An Empirical Analysis of ‘Secondary’ Pharmaceutical Patents.” PLoS one 7(12): e49470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakdawalla Darius N., Sun Eric C., Jena Anupam B., Reyes Carolina M., Goldman Dana P., and Philipson Tomas J.. 2010. “An Economic Evaluation of the War on Cancer.” Journal of Health Economics 29(3): 333–346. [DOI] [PubMed] [Google Scholar]
- Lemley Mark A., and Sampat Bhaven. 2012. “Examiner Experience and Patent Office Outcomes.” Review of Economics and Statistics 94(3): 817–827. [Google Scholar]
- Lichtenberg Frank R. 2001. “The Allocation of Publicly Funded Biomedical Research” In Berndt Ernst, and Cutler David (Eds.), Medical Care Output and Productivity, pp. 565–589. Washington, DC: University of Chicago Press. [Google Scholar]
- Mansfield Edwin. 1995. “Academic Research Underlying Industrial Innovations: Sources, Characteristics, and Financing.” Review of Economics and Statistics 77(1): 55–65. [Google Scholar]
- Manton Kenneth G., Gu Xi-Liang, Lowrimore Gene, Ullian Arthur, and Tolley H. Dennis. 2009. “NIH Funding Trajectories and their Correlations with US Health Dynamics from 1950 to 2004.” Proceedings of the National Academy of Sciences 106(27): 10981–10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marburger III, John H. 2005. “Wanted: Better Benchmarks.” Science 308(5725): 1087. [DOI] [PubMed] [Google Scholar]
- Moretti Enrico, Steinwender Claudia, and Van Reenen John. 2014. “The Intellectual Spoils of War? Defense R&D, Productivity and Spillovers.” Working Paper, London School of Economics. [Google Scholar]
- Mowery David C., Nelson Richard R., Sampat Bhaven N., and Ziedonis Arvids A.. 2004. Ivory Tower and Industrial Innovation University-Industry Technology Transfer Before and After the Bayh-Dole Act. Palo Alto, CA: Stanford University Press. [Google Scholar]
- Mukherjee Siddhartha. 2010. The Emperor of All Maladies: A Biography of Cancer. New York: Scribner. [Google Scholar]
- Murphy Kevin M., and Topel Robert H.. 2003. “The Economic Value of Medical Research” In Murphy Kevin M., and Topel Robert H. (Eds.), Measuring the Gains from Medical Research: An Economic Approach, pp. 41–73. Chicago: University of Chicago. [Google Scholar]
- Murray Fiona. 2002. “Innovation as Co-Evolution of Scientific and Technological Networks: Exploring Tissue Engineering.” Research Policy 31(8–9): 1389–1403. [Google Scholar]
- Narin Francis, and Olivastro Dominic. 1992. “Status Report: Linkage between Technology and Science.” Research Policy 21(3): 237–249. [Google Scholar]
- Narin Francis, and Olivastro Dominic. 1998. “Linkage Between Patents and Papers: An Interim EPO/US Comparison.” Scientometrics 41(1–2): 51–59. [Google Scholar]
- Nelson Richard R. 1982. “The Role of Knowledge in R&D Efficiency.” Quarterly Journal of Economics 97(3): 453–470. [Google Scholar]
- Nestler Eric J., and Hyman Steven E.. 2010. “Animal Models of Neuropsychiatric Disorders.” Nature Neuroscience 13(10): 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell Peter C., and Hungerford David A.. 1960. “A Minute Chromosome in Human Chronic Granulocytic Leukemia.” Science 132(3438): 1497. [Google Scholar]
- Pakes Ariel, and Griliches Zvi. 1980. “Patents and R&D at the Firm Level: A First Report.” Economic Letters 5(4): 377–381. [Google Scholar]
- Philipson Tomas J., and Jena Anupam B.. 2005. “Who Benefits from New Medical Technologies? Estimates of Consumer and Producer Surpluses for HIV/AIDS Drugs.” NBER Working Paper #11810. [Google Scholar]
- Roach Michael, and Cohen Wesley M.. 2013. “Lens or Prism? Patent Citations as a Measure of Knowledge Flows from Public Research.” Management Science 59(2): 504–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romer Paul M. 1990. “Endogenous Technological Change.” Journal of Political Economy 98(5): S71–S102. [Google Scholar]
- Rosenberg Nathan, and Nelson Richard R.. 1994. “American Universities and Technical Advance in Industry.” Research Policy 23(3): 323–348. [Google Scholar]
- Salter Ammon J., and Martin Ben R.. 2001. “The Economic Benefits of Publicly Funded Basic Research: A Critical Review.” Research Policy 30(3): 509–532. [Google Scholar]
- Sampat Bhaven N. 2010. “Lessons from Bayh-Dole.” Nature 468(7325): 755–756. [DOI] [PubMed] [Google Scholar]
- Sampat Bhaven N. 2012. “Mission-oriented Biomedical Research at the NIH.” Research Policy 41(10): 1729–1741. [Google Scholar]
- Sampat Bhaven N., and Lichtenberg Frank R.. 2011. “What Are the Respective Roles of the Public and Private Sectors in Pharmaceutical Innovation?” Health Affairs 30(2): 332–339. [DOI] [PubMed] [Google Scholar]
- Stern Scott. 2004. “Do Scientists Pay to Be Scientists?” Management Science 50(6): 835–853. [Google Scholar]
- Toole Andrew A. 2007. “Does Public Scientific Research Complement Private Investment in R&D in the Pharmaceutical Industry?” Journal of Law & Economics 50(1): 81–104. [Google Scholar]
- Toole Andrew A. 2012. “The Impact of Public Basic Research on Industrial Innovation: Evidence from the Pharmaceutical Industry.” Research Policy 41(1): 1–12. [Google Scholar]
- Trajtenberg Manuel, Henderson Rebecca M., and Jaffe Adam B.. 1997. “University vs. Corporate Patents: A Window on the Basicness of Innovations.” Economics of Innovation and New Technology 5(1): 19–50. [Google Scholar]
- Varmus Harold. 2009. The Art and Politics of Science: W. W. Norton & Company. [PubMed] [Google Scholar]
- Wapner Jessica. 2013. The Philadelphia Chromosome. New York: The Experiment. [Google Scholar]
- Wurtman Robert J., and Bettiker Robert L.. 1995. “The Slowing of Treatment Discovery, 1965–1995.” Nature Medicine 1(11): 1122–1125. [DOI] [PubMed] [Google Scholar]
- Zucker Lynn G., Darby Michael R., and Torero Maximo. 2002. “Labor Mobility from Academe to Commerce.” Journal of Labor Economics 20(3): 629–660. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.