Abstract
De novo production of RNA on RNA template, a process known as RNA-dependent RNA synthesis, RdRs, and the enzymatic activity conducting it, RNA-dependent RNA polymerase, RdRp, were initially considered to be exclusively virus-specific. Eventually, however, the occurrence of RdRs and the ubiquitous presence of conventional RdRp were demonstrated in numerous eukaryotic organisms. The evidence that the enzymatic machinery capable of RdRs is present in mammalian cells was derived from studies of viruses, such as hepatitis delta virus, HDV, that do not encode RdRp yet undergo a robust RNA replication once inside the mammalian host; thus firmly establishing its occurrence and functionality. Moreover, it became clear that RdRp activity, apparently in a non-conventional form, is constitutively present in most, if not in all, mammalian cells. Because such activity was shown to produce short transcripts, given its apparent involvement in RNA interference phenomena, and because double-stranded RNA is known to trigger cellular responses leading to its degradation, it was generally assumed that its role in mammalian cells is restricted to a regulatory function. However, at the same time, an enzymatic activity capable of generating complete antisense RNA complements of mRNAs was discovered in mammalian cells undergoing terminal differentiation. Moreover, observations of widespread synthesis of antisense RNAs initiating at the 3’poly(A) of mRNAs in human cells suggested an extensive cellular utilization of mammalian RdRp. These results led to the development of a model of RdRp-facilitated and antisense RNA-mediated amplification of mammalian mRNA. Recent detection of the major model-predicted identifiers, chimeric RNA intermediates containing both sense and antisense RNA strands covalently joined in a rigorously predicted and uniquely defined manner, as well as the identification of a putative chimeric RNA end product of this process, validated the proposed model. The results corroborating mammalian RNA-dependent mRNA amplification were obtained in vivo with cells undergoing terminal erythroid differentiation and programmed for only a short survival span. This raises a question of whether mammalian RNA-dependent mRNA amplification is a specialized occurrence limited to extreme circumstances of terminal differentiation or a general physiological phenomenon. The present study addresses this question by testing for the occurrence of RNA-dependent amplification of mRNA encoding extracellular matrix proteins abundantly produced throughout the tissue and organ development and homeostasis, an exceptionally revealing indicator of the range and scope of this phenomenon. We report here the detection of major identifiers of RNA-dependent amplification of mRNA encoding α1, β1, and γ1 chains of laminin in mouse tissues producing large quantities of extracellular matrix proteins. The results obtained warrant reinterpretation of the mechanisms involved in ubiquitous and abundant production and deposition of extracellular matrix proteins, confirm the occurrence of mammalian RNA-dependent mRNA amplification as a new mode of genomic protein-encoding information transfer, and establish it as a general physiological phenomenon.
Keywords: RNA-dependent amplification of mammalian mRNA, RNA-dependent RNA polymerase, Self-priming, Chimeric RNA intermediate, Sensestrand RNA, Antisense-strand RNA
Introduction
The process of RdRp-facilitated, antisense RNA-mediated amplification of mammalian mRNA has been studied in quite some detail in cells and tissues undergoing erythroid differentiation [1–6,38]. If this phenomenon is not limited to specialized circumstances of terminal differentiation, it is conceivable that its principles, deduced in investigations with the erythroid model [5,6,38], would apply in other physiological situations as well. The essence of the mammalian RNA-dependent mRNA amplification, expounded in the Discussion section below, is the following. It occurs in the cytoplasm and utilizes a conventional, genome-transcribed, processed and spliced mRNA as a primary template. The transcription, directed by RdRp, initiates within the 3’-terminal poly(A) of conventional mRNA and terminates at its 5′end producing a full size antisense RNA molecule, the initial intermediate of mRNA amplification and a template for the production of a new sense-orientated RNA strand. It is the mechanism of the initiation of the sense strand synthesis, also by RdRp activity, that defines a decisively identifying attribute of the amplification process. The initiation of the sense strand RNA synthesis occurs by the extension of the 3’ terminus of the antisense strand “locked” in a self-priming structure within the antisense RNA. The extension proceeds until RdRp activity reaches and transcribes the 5’poly(U) segment of the antisense strand, generating 3’-terminal poly(A). The resulting pinhead/double-stranded RNA molecule, containing covalently joined antisense and sense RNA strands, is the second intermediate of RNA-dependent mRNA amplification and the immediate precursor of amplified mRNA, which may be released by the cleavage at the loop of a hairpin structure. It is chimeric in that it contains the antisense sequence extended into the sense RNA strand. The position of the antisense/sense junction is not random but rigorously and uniquely defined by a complementarity between the 3’-terminal segment of the antisense strand and its internal segment. Such a highly regulated extension is at the core of RNA-depended mRNA amplification and defines its specificity. Therefore, the detection of the major recognizable attribute of this mechanism, an antisense molecule extended into a sense strand in a rigorously predicted and positionally uniquely defined manner, would constitute a definitive evidence for the occurrence of RNA-dependent mRNA amplification process.
Where to look for such an identifier of the occurrence of RNA-dependent mRNA amplification? Among potential cell types that may utilize mRNA amplification by the mechanism described above are cells producing extracellular matrix proteins. At multiple stages during the development, growth, repair, turnover and remodeling of connective tissues, matrix proteins are rapidly produced and, because they are secreted, the production of extraordinary amounts is not limited by cellular confines. A mechanism whereby every mRNA molecule may serve as a potential template for production of additional mRNAs may facilitate increased production of specific secreted extracellular matrix proteins at times of high demand. With this in mind, the present study utilized, as an experimental model, cells from the Engelbreth-Holm-Swarm (EHS) mouse sarcoma tumor that generate extraordinarily large quantities of basement membrane components that are collected and marketed as Matrigel [7–14]. We report below the detection, by next generation sequencing, of the definitive identifiers of RNA-dependent mRNA amplification, the chimeric antisense/sense RNA junction sequences, for mRNAs encoding α1, β1, and γ1 chains of laminin in extracellular matrix-producing EHS cells.
Results
The occurrence of chimeric junctions containing both antisense and sense sequences in mRNAs encoding extracellular matrix, ECM, components produced by EHS tumor cells was probed by next generation sequencing. Cytoplasmic RNA, isolated from tumor cells harvested ten days after tumor implantation and depleted of ribosomal RNA, was used to generate sequencing libraries, as described in the Methods section. Libraries were sequenced on an Illumina Next500 instrument. The resulting reads were aligned with or blasted against appropriate reference sequences, and sequences of fragments of interest were extracted from raw data and analyzed. Contiguous chimeric sequences containing antisense and sense components, with the junctions occurring within 5’ untranslated regions of mRNA species encoding α1, β1, and γ1 chains of laminin were detected. A selection of such chimeric junction sequences representing two different types of molecules for each of three chains of laminin is shown below:
Laminin α1, chimeric junction sequences type 1 (Figure 1, top panel): C G C C C G T G C T C G C T C A C A G G C A C T G C G C G A G T C C T T C C C C A G G A G C G C A G G G A G C G G C G G C G A C A A C A T G C G C G G C A G C G G C A C G G G A G C C G C G C T C C T G G T G C T C C T G G.
Laminin α1, chimeric junction sequences type 2 (Figure 1, bottom panel): GGTGACCCAGAGCACCGAGGCCAGGAGCACCAGGAGCGCGGCTCCCGTGCCGCTGCCGCGCATGTTGTCGCCGCCGCTCCCTGCGCTCCTGGGGAAGGTCTCGCCCGTGCTCGCTCACAGGCACTGCGCGAGTGTGCTCTTCCCCAGGAGCGCAGGGAGCGGC.
Laminin β1, chimeric junction sequences type 1 (Figure 2, top panel): G G C C C T T C C A T T T C C T G C C G C T C C C A C G G A A G C G G G G G C T G G G C C C A G G A A G G G G G T C A A G T C G C T T A A C T A C T T T G T T C T C C T C A C C C G G C T G G G C G A G C G C T C A A C C C G C T C C T G GCAGCCCACCGGGTGAGGAGAACAAAGTAGTTAAGCGAC.
Laminin β1, chimeric junction sequences type 2 (Figure 2, bottom panel): T G G G C C C A G G A A G G G G G T C A A G T C G C T T A A C T A C T T T G T T C T C C T C A C C C G G C T G G G C G A G C G C T C A A C C C G C T C C T G G C A G C C C A C C G G C C C C C G C T T C C G T G G G A G C G G C A G G A A A T G G A A G G G C C C C.
Laminin γ1, chimeric junction sequences type 1 (Figure 3, top panel): CGGTGCGCAGCCTGCACTGTGCGCCGGGAGGTAGCGGAGGCAGCGCGATCTTGGCTCGGACGCCCACCCATCGGCTCTGCGTCCGGCTCTCGGCCTCCAGCCCGGTCCACAGCCCGGCCTCGGCCCGCAGCGGAGGATCGGCCTCGGGATACGCCGCTAG.
Laminin γ1, chimeric junction sequences type 2 (Figure 3, bottom panel): C C G A C T T C C G A G C G C G C A C T C G A G A G C G C G C T C G G A A G T C G G G G G T C G G C G C G C A G T G C A G G C T G C G C A C C G G G A G G T A G C G G A G G C A G C G.
The relevant portions of the above sequences are shown in Figures 1–3, together with the projected pathways of their generation detailed in the Figure legends and in the Discussion section below. The sequences are unique in that they are not encoded in the genome contiguously and could be generated only through the extension of the 3’ terminus of a self-primed antisense RNA, with the extension start site (ESS) and, consequently, the chimeric junction sequence defined precisely and uniquely by the position of a self-priming structure assemblies, shown in Figures 1–3. Accordingly, chimeric junction sequences serve as definitive identifiers of the occurrence of RNA-dependent mRNA amplification process.
The feasibility of the amplification process is largely decided by the sequence and structure of the 3’ termini of the antisense strands, which, in turn, is determined by the position of the transcription start site (TSS) of the gene-encoded mRNA. For all three chains of laminin, α1, β1, and γ1, the chimeric RNA sequences detected originated from mRNAs transcribed from multiple TSS positions; this explains multiple types of chimeric junctions observed. This is not surprising considering that the laminin genes and, in fact, the majority of genes encoding extracellular matrix proteins, lack the “TATA” regulatory element that rigorously defines the position of a TSS.
Consequently, due to the lack of TATA-control element, the utilization of multiple TSS positions is a normal physiological occurrence for such genes. Thus, in the top panel of Figure 1, transcription of the laminin α1 mRNA starts 70 nucleotides upstream from the AUG translation initiation codon, (line A). The 3’-terminal region of the corresponding antisense strand (line B) is then folded into a self-priming configuration (line C) and extended into the sense-oriented component of a chimeric molecule (line D). Line E shows a projected chimeric junction sequence and line F displays the detected chimeric junction sequence. In the bottom panel of Figure 1, the TSS is shifted ten nucleotides upstream. The antisense molecule forms a different self-priming structure, whose extension from a new ESS yields a distinctly differently chimeric sequence, but the end-result, as defined by the coding content of the amplified sense component, remains unchanged. Indeed, as reasoned in the Discussion section below, the translational outcome of mRNA amplification is defined by the position of self-priming, and in both cases above, as well as in the cases described below, self-priming occurs within the segment of the antisense strand corresponding to the 5’UTR of the sense strand. Therefore, the amplified mRNA retains the intact coding capacity of the conventional mRNA.
Figure 1: Sequence of a chimeric junction containing antisense and sense segments of laminin α1 mRNA and the projected pathway of its generation.

Uppercase letters – nucleotide sequence of the sense strand; lowercase letters – nucleotide sequence of the antisense strand. Highlighted in green – “AUG” translation initiation codon on the sense strand; highlighted in blue – “uac” complement of translation initiation codon on the antisense strand. In italics and highlighted in grey – detected chimeric fragments. Blue arrows: Position of antisense/sense junction. The top and bottom panels depict amplification of mRNA molecules originated from different TSSs; note that self-priming positions and, consequently, chimeric junction sequences in two panels are different. A: 5′ terminal region of laminin al mRNA. B: Antisense complement of the 5’ terminal region of α1 laminin mRNA. C: Projected folding of the antisense strand into self-priming configuration. D: Extension of selfprimed antisense strand into sense-oriented sequence. E: Projected chimeric junction sequence. F: Detected chimeric junction sequence. Complete sequences of the chimeric junctions shown in this Figure are provided in the main text above. Note that the priming occurs within the segment of antisense strand corresponding to the 5′UTR of mRNA, thus preserving the coding capacity of amplified mRNA.
A similar phenomenon is seen with mRNA for the β1 chain of laminin. Transcription from the TSS position 80 nucleotides upstream from the AUG translation initiation codon produces mRNA whose antisense counterpart is capable of self-priming its extension into a sense strand, thus producing a chimeric molecule (Figure 2, top panel). A shift of the TSS position just three nucleotides upstream results in a radically different self-priming structure assembly of the antisense strand (Figure 2, bottom panel) which is, nevertheless, extended from a new ESS into a sense strand producing a distinctly different chimeric molecule but with the unchanged protein coding content.
Figure 2: Sequence of a chimeric junction containing antisense and sense segments of laminin β1 mRNA and the projected pathway of its generation.
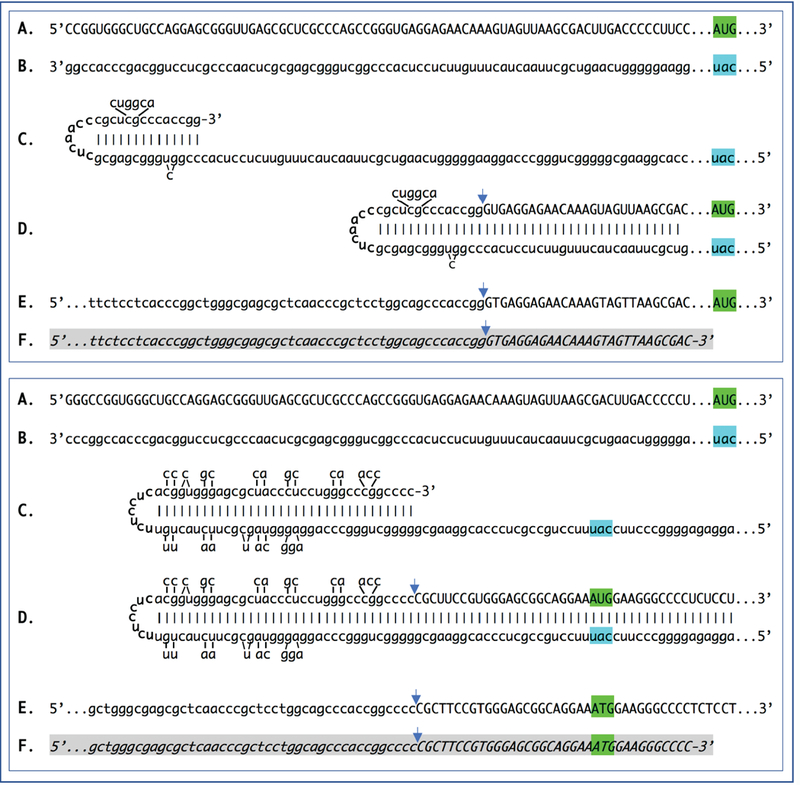
Uppercase letters – nucleotide sequence of the sense strand; lowercase letters – nucleotide sequence of the antisense strand. Highlighted in green – “AUG” translation initiation codon on the sense strand; highlighted in blue – “uac” complement of translation initiation codon on the antisense strand. In italics and highlighted in grey – detected chimeric fragments. Blue arrows: Position of antisense/sense RNA junction. The top and bottom panels depict amplification of mRNA molecules originated from different TSSs; note that self-priming positions and, consequently, chimeric junction sequences shown in two panels are different. A: 5’ terminal region of laminin pi mRNA. B: Antisense complement of the 5’ terminal region of pi laminin mRNA. C: Folding of the antisense strand into self-priming configuration. D: Extension of self-primed antisense strand into sense-oriented sequence. E: Projected chimeric junction sequence. F: Detected chimeric junction sequence. Complete sequences of the chimeric junctions shown in this Figure are provided in the main text above. Note that the priming occurs within the segment of antisense strand corresponding to the 5′UTR of mRNA, thus preserving the coding capacity of amplified mRNA.
Likewise, initiation of transcription of mRNA encoding the γ1 chain of laminin from TSS positions 210 nucleotides upstream from the AUG codon (Figure 3, top panel) or 246 nucleotides upstream from the translation start site (Figure 3, bottom panel), lead to two types of antisense RNA strands, both capable of self-priming their 3’-extension into sense-oriented molecules. The resulting composite antisense/sense molecules have distinctly different chimeric junction sequences but retain the protein coding content identical to that of the conventional mRNA.
Figure 3: Sequence of a chimeric junction containing antisense and sense segments of laminin γ1 mRNA and the projected pathway of its generation.
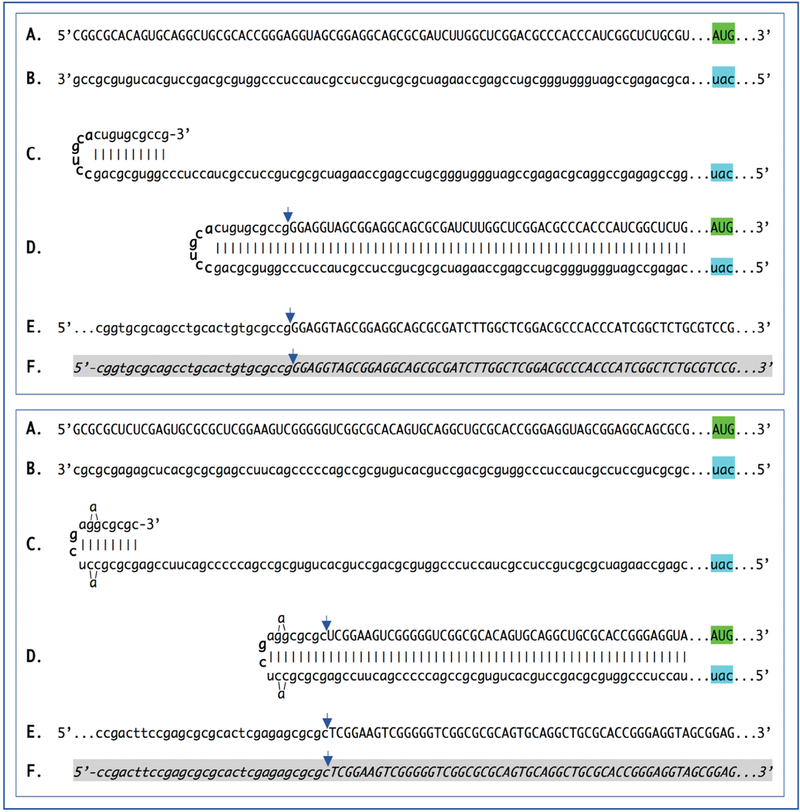
Uppercase letters – nucleotide sequence of the sense strand; lowercase letters – nucleotide sequence of the antisense strand. Highlighted in green – “AUG” translation initiation codon on the sense strand; highlighted in blue – “uac” complement of translation initiation codon on the antisense strand. In italics and highlighted in grey – detected chimeric fragments. Blue arrows: Position of antisense/sense RNA junction. The top and bottom panels depict amplification of mRNA molecules originated from different TSSs; note that self-priming positions and, consequently, chimeric junction sequences shown in two panels are different. A: 5’ terminal region of laminin γ1 mRNA. B: Antisense complement of the 5’ terminal region of γ1 laminin mRNA. C: Projected folding of the antisense strand into self-priming configuration. D: Extension of self-primed antisense strand into sense-oriented sequence. E: Projected chimeric junction sequence. F: Detected chimeric junction sequence. Complete sequences of the chimeric junctions shown in this Figure are provided in the main text above. Note that the priming occurs within the segment of antisense strand corresponding to the 5′UTR of conventional mRNA, thus preserving the coding capacity of amplified mRNA.
As was mentioned above, chimeric junction RNA sequences for α1, β1, and γ1 chains of laminin were detected in the EHS cells ten days after tumor implantation. Interestingly and importantly, in identical multiple searches, no chimeric junction sequences for α1, β1, and γ1 laminin RNA were detected in EHS cells at an earlier stage, six days after tumor implantation. This was not for the lack of laminin genes expression. Abundant transcripts of α1, β1, and γ1 laminin genes were identified in sequencing libraries prepared six days post-implantation, but none of the chimeric RNA junction sequences were found. The significance and implications of these observations are discussed below.
Discussion
Cellullar origin of the detected chimeric junction sequences
The essence of the present study is the detection of a new type of RNA molecule. Its structure is evident: A chimeric RNA molecule containing 3’-terminal segment of the antisense strand covalently connected, at a precisely determined position, to the sense strand segment of the same mRNA species. Whereas numerous mechanistic aspects of the overall cellular mechanism involving this type of biomolecule remain to be elucidated and require further investigations, the mechanism of generation of the chimeric sequences observed in the present study is very clear: The extension of the antisense strand, self-primed by its 3’-terminal segment. The position of the junction between the antisense and the sense components of a chimeric molecule is uniquely defined by the folding of the antisense into self-priming configuration. As shown here, this, in turn, is specified by the position of transcription start site(s) of a conventional genome-transcribed mRNA molecule. Moreover, multiple TSSs may lead to multiple chimeric RNAs, with junctions at different positions, for a single mRNA species, an outcome observed in the present study. Of crucial importance for the conclusions of this study are considerations pointing to the cellular origin of the observed chimeric junctions, i.e. that they were present in the initial RNA preparations rather than inadvertently generated during the construction of sequencing libraries. The chimeric junction sequences described above were identified by next generation sequencing. This procedure not only provides the nucleotide sequence of a single molecule but contains safeguards against mis-exposure of chimeric fragments that could be possibly generated during sequencing library preparation. As described below, even if such fragments were generated during the procedure, they could not be sequenced. Thus, the procedure used in the present study substantiates cellular origin of the detected chimeric sequences.
Previously, it was shown that self-priming can occur and a fragment containing both sense and antisense components may be generated during reverse transcription, provided the antisense strand (cDNA) contains the required complementary elements [3]. For several reasons, it can be stated with a high degree of certainty that chimeric laminin RNA junction sequences seen in the present study did not arise during the reverse transcription or the enrichment stages of sequencing libraries construction but were present in the initial cytoplasmic RNA preparations. First, any possible synthesis of the second cDNA strand during the stage of first strand synthesis was suppressed by the addition of the NEB strand specificity reagent, as described in the Methods section. Second, even if a hairpin structure were created during the reverse transcription stage, it would represent a dead end in the library preparation process because no adaptor could be ligated. Thirdly, due to the intrinsic geometry of the observed chimeric reads, the structure of many of the chimeric fragments detected in the present study decisively rules out the possibility of their generation during the reverse transcription stage of library preparation. This is because they have, when folded by matching the complementary elements discussed above, 3’-protruding sense-orientation segments. Since the length of the antisense component 5’ of the position of self-priming structure in these fragments is significantly smaller than that of the sense-oriented segment, the former could not serve as a template for the latter during the library preparation process thus corroborating the occurrence of the observed chimeric molecules in the initial RNA preparation. Forth, the inadvertent generation of a chimeric fragment, either by intramolecular or intermolecular processes, during the PCR enrichment of a sequencing library can also be ruled out. For this to occur, the 3′-terminal portion of the antisense strand should anneal to the internal portion of the same or another antisense strand and be extended into a sense strand. In sequencing library preparation, the PCR enrichment step follows the adaptor ligation step. If antisense strand contains an adaptor, its 3′-terminal complementary element cannot be extended, even if it anneals either intramolecularly or intermolecularly. If an antisense strand does not contain an adaptor, it potentially can anneal and be extended but cannot be sequenced. Finally, the strongest indication of the cellular origin of the chimeric antisense/sense molecules is that they were observed to occur in a developmental stage-specific manner. Indeed, whereas numerous chimeric junction reads were obtained with RNA from ten day post-implantation cells, none were seen with RNA from an earlier, six day post-implantation stage, a stage where the abundant expression of α1, β1, and γ1 laminin genes already occurs but the RNA-dependent mRNA amplification process is apparently not yet activated. These observations decisively rule out origination of chimeric sequences during library construction process.
The chimeric pathway of mammalian RNA-dependent mRNA amplification
The results of the present study show the occurrence of chimeric molecules containing covalently bound sense and antisense strands of RNA encoding all three chains of laminin in a tissue that produces extraordinary quantities of this protein. While limited to the demonstration of a new type of RNA molecules, these results are indisputable. Likewise is their origin: these molecules are clearly produced by the 3’ extension of the antisense RNA self-primed in a sequence-specific manner. These results can be interpreted within the framework of the mammalian RNA-dependent mRNA amplification model developed in the previous studies [1–6,38], reviewed in detail elsewhere [6], diagrammed in Figure 4 below and briefly summarized as follows.
Figure 4: Projected stages of of RdRp-directed, antisense RNA-mediated amplification of mammalian mRNA.
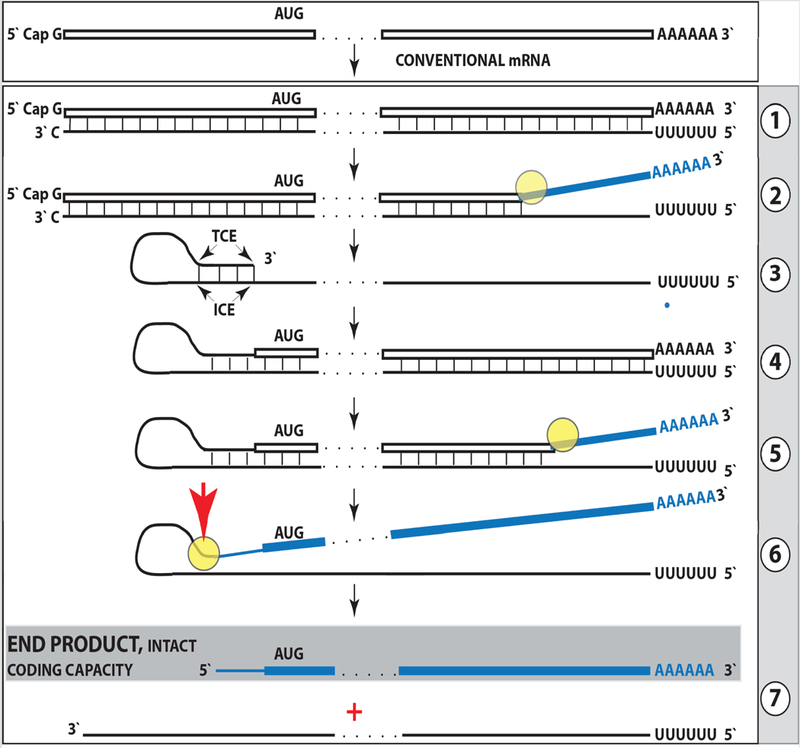
Top panel: Conventional, genome-originated mRNA molecule. Bottom panel: Projected stages of antisense RNA-mediated mRNA amplification. Boxed line – sense strand RNA. Single line – antisense strand RNA. “AUG” – functional translation initiation codon (could be other than “AUG”). “TCE”– 3′-terminal complementary element; “ICE”– internal complementary element, both on the antisense RNA strand. Yellow circle -helicase/modifying activity complex. Blue lines (both single and boxed) – RNA strand modified and separated from its complement by a helicase complex. Red arrowhead – position of cleavage of the chimeric intermediate. Step 1: Synthesis of antisense strand; step 2: Strand separation; step 3: Folding of antisense strand into self-priming configuration; step 4: Extension of self-primed antisense RNA; step 5: Strand separation; step 6: Cleavage of the chimeric intermediate; step 7: End-products of amplification. Note that chimeric RNA end product retains the intact coding capacity of conventional mRNA.
The amplification process occurs in the cytoplasm and starts with transcription of the antisense complement from a conventional spliced mRNA template, initiating at the 3’poly(A), possibly with the help of a uridilated protein, as seen in viral RdRs [15] (Figure 4, Step 1). Generation of a complete antisense transcript requires the presence of an eligible RNA template and a compatible polymerase activity. The only major prerequisite for a potential RNA template appears to be the presence of the poly(A) segment at its 3’ terminus [4–6]. The compatible polymerase activity is the RNA-dependent RNA polymerase. The RdRp activity in mammalian cells appears to be non-conventional; two possible candidates for this role are the RNA polymerase II complex or its components [16,17] and RdRp activity of the TERT complex [18], both ubiquitously present in all cells. Under regular circumstances, the RdRp activity in mammalian cells produces only short antisense RNA transcripts. For example, a widespread synthesis of diverse short antisense RNA transcripts initiating at the 3’poly(A) of mRNA was observed in human cells [19]. On the other hand, RdRp activity isolated from rabbit reticulocytes [20] was able to produce, in assays, long antisense RNA transcripts. Subsequent studies identified full-length antisense transcripts of globin mRNA in erythroid cells [4–6]. It could be argued that the component responsible for the production of long antisense transcripts in mammalian cells is a processivity conferring co-factor of RdRp activity that is induced under special circumstances when overproduction of specific proteins is required. The notion of a processivity co-factor is strongly supported by studies of RdRp-deficient HDV replication in “normal” (i.e. apparently lacking processivity co-factor) mammalian cells, [21,22]. Within the framework of the above considerations, the ability of RdRp-deficient viruses to use RdRp activity of mammalian cell for their replication implies that they should encode a processivity co-factor of cellular RdRp. In case of HDV, it appears to be hepatitis delta antigen HDAg, the only protein encoded by HDV. HDAg is essential [21] both for production of long transcripts by cellular RdRp, and for viral replication. In its absence only short transcripts are generated [21]. These observations provide a proof of concept for the notion of RdRp processivity co-factor, central to our understanding of mammalian mRNA amplification. Identification of a cellular homolog of HDAg, DIPA [22,23] suggests directions for a search for the cellular RdRp processivity co-factor.
The resulting double-stranded sense/antisense structure is then separated into single-stranded molecules by a helicase activity that mounts the poly(A) segment of the 3’poly(A)-containing strand (the sense-oriented strand) of the double helical structure and proceeds along this strand modifying, on average, every fifth nucleotide in the process [5,6] (Figure 4, Step 2). Only purines, “A” and “G”, appear to be modified in the separation/modification process [5,6]. The 5’ poly(U)-containing antisense strand remains unmodified during and after the separation [4–6]; this being essential for the production of a new sense strand since modifications were shown to interfere with complementary interactions required in this process [5,6,38].
The vast majority of mammalian mRNA species contains 3’-terminal poly(A) segments. The notion that many, or possibly most, of them could be eligible templates for RdRp was suggested in our previous studies [4–6]. Subsequent observations by Kapranov et al. showed a widespread synthesis of antisense RNAs initiating, apparently indiscriminately, at the 3’ poly(A) of mRNA in human cells [19]. This, seemingly undiscerning, RdRp template eligibility of the bulk of mammalian mRNA species raises questions with regard to mechanisms underlying the manifestly stringent specificity of the mRNA amplification process as seen, for example, in erythropoietic differentiation [4–6]. The specificity of mRNA amplification appears to be determined at the 3’ terminus of an antisense transcript by its ability or inability to support production of a complementary sense-strand molecule.
The generation of a sense strand on an antisense template occurs via the extension of the 3’ terminus of a self-primed antisense template and requires the presence within the antisense transcript of two spatially independent complementary elements [3]. One of these is the strictly 3’-Terminal Complementary Element (TCE), the other is the Internal Complementary Element (ICE). These elements (Figure 4, Step 3) must be sufficiently complementary to form a priming structure but may contain mismatches and utilize unconventional G/U pairings [3–6]. The generation of a sense strand also requires the thermodynamic feasibility, enhanced/enabled by the occurrence of two complementary and topologically compatible elements, of the antisense strand folding into a self-priming configuration.
Provided that a self-priming structure is formed, the 3′ end of the folded antisense strand is extended by RdRp into a sense-oriented molecule terminating with the poly(A) at the 3′ end (Figure 4, Step 4), thus generating a hairpin-structured chimeric intermediate consisting of covalently joined sense and antisense strands. The double stranded portion of the resulting structure is separated by a helicase activity invoked above, which mounts the 3′poly(A) of a newly synthesized sense strand component of the chimeric intermediate and proceeds along this strand in the 5′ direction modifying the molecule as it advances (Figure 4, Step 5). When the helicase activity reaches a single stranded portion of the hairpin structure, it, or associated activities, cleave the molecule either within the TCE, at a TCE/ICE mismatch, or immediately upstream of the TCE. The cleavage occurs between the 5’ hydroxyl group and the 3’ phosphate [5,6] (red arrowhead, Figure 4, Step 6).
Strand separation, in conjunction with the cleavage, produces two single-stranded molecules (Figure 4, Step 7) one of which is a chimeric mRNA, the functional mRNA end product of amplification and the basis for defining this pathway as the “chimeric”. The chimeric nature of this end product is due to the presence at its 5’ end of a 3’-terminal segment of the antisense strand, consisting of either the entire TCE or a portion thereof, depending on the site of cleavage of the chimeric intermediate, and covalently attached, in a 5’ to 3’ orientation, to the 5’-truncated sense strand. This chimeric molecule is modified and 3’ polyadenylated. In contrast to conventional mRNA that can be repeatedly used as RdRp template [5,6], it cannot be further amplified because its antisense complement would be lacking the TCE, but can be translated into either the conventional mRNA-encoded polypeptide [5,6] or its C-terminal fragment [5,6,24–27] depending on the extent of 5’truncation. The extent of 5’truncation of the sense strand component of the chimeric end product is defined by the intramolecular position of the internal complementary element, ICE, within the antisense template, allowing for conceptually distinct outcomes. Figure 4 illustrates the situation whereby the ICE of the antisense strand is located within its segment corresponding to the 5’ untranslated region, 5′UTR, of a conventional progenitor mRNA. Consequently, the chimeric end product contains the entire protein coding region of a conventional mRNA and can be translated into the original, conventional mRNA-encoded, polypeptide [5,6]. Additional translational outcomes reflecting alternative positions of the internal complementary element are discussed elsewhere [6].
In the chimeric pathway of mRNA amplification, the cleavage of the chimeric intermediate following the strand separation and the associated modification of its poly(A)-containing component of the double stranded structure is the ultimate step in the generation of the chimeric mRNA end product. Consequently, it is formed already modified and is never present in the unmodified form. Therefore, because modified amplified RNA is resistant to reverse transcription [5,6], it cannot be detected by conventional sequencing methods. This explains the striking absence of chimeric RNA end product sequences among detected chimeric reads. Indeed, in chimeric junction sequences detected in the present study, antisense components extend upstream from the junction site well beyond the loop portion, defining them as segments of the chimeric hairpin intermediate. These intermediates remains unmodified until the extension of a self-primed antisense strand into the sense-orientated molecule is concluded by the synthesis of the 3’ poly(A) segment transcribed from the 5’ poly(U) of the antisense strand and required for the commencement of strand separation/modification. Moreover, following the completion of the extension and the commencement of separation, the sense/antisense chimeric junction is not modified for the duration of the strand separation process downstream from the junction thus generating a certain steady-state level and a temporal window of opportunity enabling the detection of the yet unmodified junction segment by the conventional reverse transcription-based methods.
The functions of nucleotide modifications occurring during mRNA amplification remain to be elucidated. They could include the facilitation of RNA strand separation, stimulation of the cap-independent translation of the amplification end product, and regulation of its stability [5,6]. There may also be a connection between nucleotide modifications of amplified mRNA and the activation of the mRNA amplification process. The amplified and heavily modified mRNA could behave in ways that are different spatially, qualitatively and quantitatively from those of conventional mRNA. For example, the amplification of mRNA species encoding secreted proteins, such as laminin or other extracellular matrix proteins, could overwhelm the ER, cause ER stress and trigger cell death. One cellular response to ER stress appears to be a shift of translation and secretion outside the ER [35]. It could be suggested, therefore, that nucleotide modifications of amplified mRNA may direct its translation and secretion of the resulting protein via pathways bypassing the ER, despite the presence of a signal peptide sequence. The initial ER stress resulting from increased transcription and subsequent translation of conventional mRNA encoding a secreted protein could be one of potentially multiple cellular events, probably certain types of stresses, which may trigger mRNA amplification processes. In this case, one can envision that conventional overproduction of a secreted protein induces ER stress and activates multiple transcription factors [36,37], which, in turn, activate mRNA amplification pathway thus facilitating overproduction and at the same time relieving the ER stress, since in the mRNA amplification process, a portion of conventionally produced mRNA molecules, used as templates for the production of antisense RNA, is apparently modified during strand separation (Figure 4, step 2) and would be translationally processed outside the ER, alongside the modified chimeric RNA end product of mRNA amplification.
Genes for α1, β1, and γ1 chains of laminin have multiple TSS positions, most of which are inconsistent with the eventual generation of antisense molecules capable of self-priming within their segments corresponding to the 5’UTRs of mRNA because none of the complementary elements on the antisense strand is 3’-terminal [28,29]. Observations described above suggest a possibility that a shift in TSS positions can play an important regulatory role in defining the eligibility of a transcript for amplification, i.e. the ability of the antisense strand to fold into a self-priming configuration. The concepts of such a regulation are summarized in Figure 5.
Figure 5: TSS shift as a potential regulator of the eligibility of an mRNA for the amplification process.
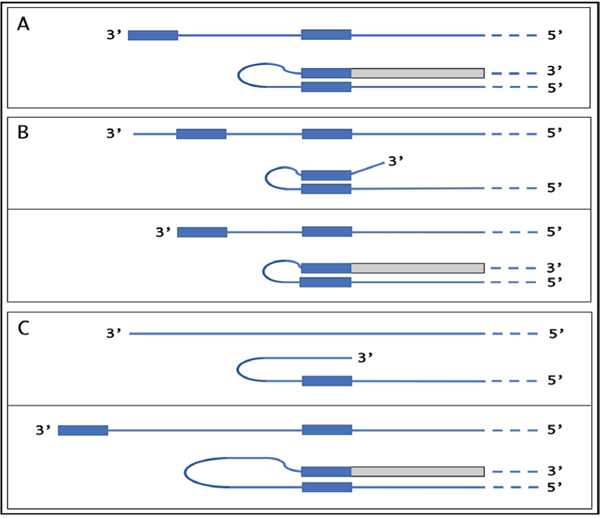
Single line – 3′ terminus of the antisense strand. Filled grey boxes -sense strand. Filled blue boxes – topologically compatible complementary elements on the antisense strand. A: One of the complementary elements is 3’-terminal; folding results in a self-priming structure which is extended into the sense strand. B: Both complementary elements are internal, no self-priming is possible; TSS shift in the downstream direction makes one of the elements 3’-terminal and allows self-priming and extension into the sense strand. C: There are no complementary elements, no self-priming can occur; TSS shift in the upstream direction generates complementary elements one of which is 3’-terminal and thus enables self-priming and extension. Note that processes depicted in panels B and C can occur in reverse resulting in a loss, rather than the acquisition, of the eligibility.
If the 3’-distant complementary element of the antisense strand is terminal (Figure 5, panel A), it can form a self-priming structure. If, however, both elements are internal (Figure 5, panel B), a downstream shift of the TSS position can make one of them 3’-terminal and thus enable self-priming. When, on the other hand, the 3’ segment of the antisense strand has no topologically compatible complementary sequences, an upstream shift of the TSS position (Figure 5, panel C) can generate such an element and make the transcript eligible for amplification. The events diagrammed in panels B and C of Figure 5 can also occur in reverse, making previously eligible mRNA ineligible for the amplification process. As a possible example of such regulation, antisense/sense chimeric RNA junctions, the identifiers of RNA-dependent mRNA amplification, were observed in the present study for all three chains of laminin at ten days post-implantation of EHS cells, but were absent at six days post-implantation despite the abundant presence of conventional transcripts. One interpretation of this observation, suggested above, is that the mRNA amplification machinery or some of its components are not yet activated at the early developmental stage. Another interpretation, in light of the above considerations, is that at the early developmental stage mRNA molecules encoding all three laminin chains are initiated at TSS positions that prevent self-priming of antisense RNA strands because of the lack or non-3’-terminal position of one of complementary elements. In such a case, the activation of RNA-dependent amplification of laminin mRNA at the later stage would occur by shifts of corresponding TSS positions in a developmental stage-specific manner. Alternatively, regulation may occur on both, enzymatic as well as transcriptional levels.
Our previous results indicated that RdRp can transcribe the cap “G” of mRNA, despite its inverted orientation [5,38]. Observations of the present study confirm this conclusion. The underlying arguments are based on the results with the α1 chain of laminin shown in the top panel of Figure 1 and are illustrated in Figure 6. If the cap “G” is not transcribed, the antisense strand terminates with the “u” and its folding configuration would be as shown in Figure 6, panel “A”. After the extension, the sense/antisense junction would consist of the “uC” as depicted in Figure 6, panel “B”. The experimental results, however, are different. They show that the sequence of the sense/antisense junction is, in fact, “ucC” (Figure 6, panel “C”). Since the genomic sequence upstream of the TSS cannot account for the additional 3’-terminal “C” in the antisense strand [28], the only remaining possibility is that the “C” in question is a transcript of the cap “G” of the sense strand and that the antisense folding into a self-priming configuration occurs as shown in Figure 6, panel “D”.
Figure 6: RNA-dependent RNA polymerase can transcribe the cap “G” of mRNA.

Data shown is adapted from Figure 1, top panel. Uppercase letters – nucleotide sequence of the sense strand; lowercase letters – nucleotide sequence of the antisense strand. “u” highlighted in blue – 3′-terminal nucleotide of the antisense strand corresponding to the transcription start site of mRNA; “uC” highlighted in blue –the projected junction structure in the absence of the cap “G” transcription. “c” highlighted in green – transcript of the cap”G”; “ucC” highlighted in green – the resulting junction structure when cap “G” is transcribed. A: Projected self-priming configuration of the antisense strand in the absence of the cap “G” transcription. B: Projected nucleotide sequence of the sense/antisense junction in the absence of the cap “G” transcription. C: Detected nucleotide sequence of the sense/ antisense junction. D: Self-priming configuration of the antisense strand as defined by experimental results. Note that the genomic sequence upstream of the TSS cannot account for the additional 3′-terminal “C” in the antisense strand.
Novel experimental strategies
The detection of chimeric antisense/sense RNA junctions in the present study provides decisive evidence in support of the occurrence of RNA-dependent amplification of mRNA encoding all three chains of laminin in EHS cells. Multiple lines of reasoning articulated above indicate that these junction sequences are of cellular origin. Yet, very strong arguments for the cellular origin of these chimeric RNA molecules notwithstanding, their possible inadvertent and unforeseen generation during multiple experimental manipulations involved in sequencing libraries construction cannot be ruled out with absolute certainty. Ideally, the definitive and unquestionable proof would be a direct measurement, without any outside experimental manipulations, of a key cellular product of this process. Is it feasible?
The experimental strategy to obtain such a proof is suggested by a possible scenario of RNA-dependent mRNA amplification where the antisense segment of the chimeric RNA end product contains a functional translation initiation codon [6]. In such a case, if the folding/self-priming of the antisense RNA occurs within its segment corresponding to the 5’UTR of conventional mRNA and if the initiation codon within the antisense portion of the chimeric RNA end product is in-frame with the mRNA coding sequence, translation will result in a highly distinct chimeric protein non-contiguously encoded in the genome. It will contain the polypeptide encoded by a conventional mRNA, enhanced at its N-end by additional amino acids encoded by the antisense portion of the chimeric RNA end product and by a segment of the 5’UTR of conventional mRNA. Such a protein could be probed for directly in cell lysate and its detection would constitute an undisputable proof of the occurrence of RNA-dependent mRNA amplification. This scenario can be enacted experimentally.
The experimental model, based on results described above, is the in vitro amplification of mRNA encoding the murine alpha1 chain of laminin. The experimental design, outlined in Figure 7, starts with the elucidated pathway shown in the bottom panel of Figure 1 and modifies it, by editing the alpha1 laminin gene, to achieve the desired outcome. In the top panel of Figure 7, line A shows the 5’terminal portion of conventional mRNA and line B shows the 5’terminal portion of the projected chimeric RNA end product of amplification (capital letters-sense RNA; lower case letters-antisense RNA); it should be noted that both share a portion of the 5’UTR and contain the same coding regions. Line C shows the polypeptide (highlighted in blue) resulting from translation of either conventional or amplified chimeric laminin mRNA. In the middle panel, line A shows the 5’terminal portion of edited mRNA and line B shows its antisense counterpart. “CAU” highlighted in blue (line A) is transcribed into 5’-aug-3’ highlighted in green (shown as 3’-gua-5’) in the antisense strand (line B).
Figure 7: Novel experimental design: in vitro generation of a chimeric polypeptide containing murine alpha 1 laminin chain, initiated from the antisense RNA and non-contiguously encoded in the genome.
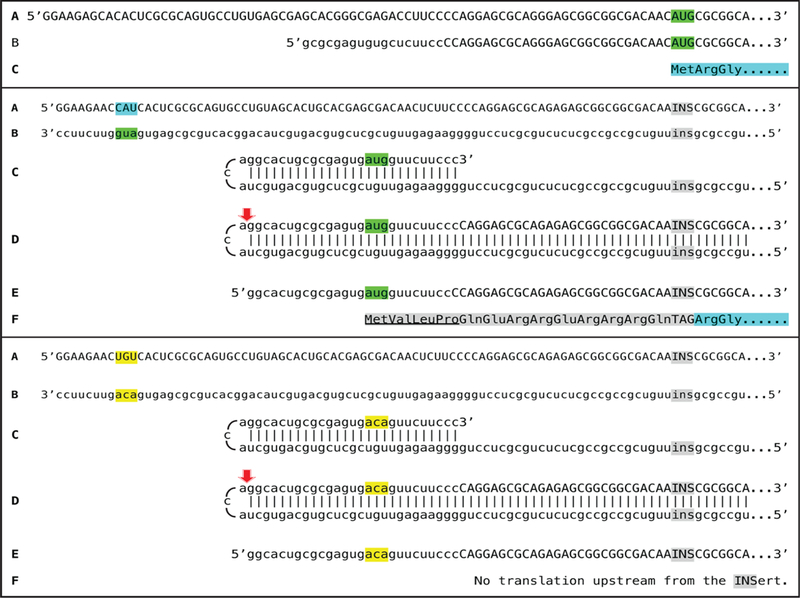
Uppercase letters: sense strand RNA. Lowercase letters: antisense strand RNA. Highlighted in green: AUG translation initiation codon. 5′-CAU-3′ (highlighted in blue) on the sense RNA: Complement of 5’-aug-3’ (highlighted in green) on the antisense RNA. “INS” highlighted in grey: Insert encoding a Tag peptide and lacking initiation codon; “TAG” highlighted in grey: Peptide encoded by “INS”. “ins” highlighted in grey: “INS” complement on the antisense RNA. Amino acid sequence highlighted in blue: Polypeptide encoded by conventional mRNA or by the amplified mRNA. Amino acid sequence highlighted in grey: The N-end extension of conventional polypeptide; underlined portion: Amino acids encoded by the antisense RNA. Highlighted in yellow: Editing changes resulting in replacement of the “aug” by the “aca” on the antisense RNA. Red arrowhead: Position of cleavage of the chimeric intermediate. Top panel: A-conventional mRNA encoding alpha 1 chain of laminin. B-chimeric RNA end product of amplification (adapted from Figure 1) encoding the same polypeptide as “A”. C-amino acid sequence encoded by either “A” or “B”. Middle panel: A-projected edited “dormant” mRNA originated from edited alpha 1 laminin gene. B-antisense complement of edited alpha 1 laminin mRNA. C-folding of the antisense strand into self-priming configuration; 3’terminal “c” is a transcript of the 5’capG of mRNA. D-extension of self-primed antisense strand into sense-oriented sequence followed by strand separation and cleavage of the chimeric intermediate. E-chimeric RNA end product of RNA-dependent amplification of edited alpha 1 laminin mRNA. F-projected translational outcome. Bottom panel: Same as the middle panel with the exception of editing changes resulting in replacement of the “aug” by the “aca” (highlighted in yellow) on the antisense RNA.
The major objectives of the editing are the following: (1) To delete the translation initiation AUG codon of the conventional mRNA; (2) To replace it with an insert (marked “INS” in Figure 7) lacking the translation initiation codon and encoding a Tag peptide so as to enable detection and isolation of the resulting polypeptide; (3) To eliminate all potential translation initiation codons [30,31] upstream from and in-frame with the “INS”/protein-encoding sequence in the edited mRNA; (4) To introduce within the TCE of the projected antisense strand an AUG codon in the 5’ to 3’ orientation, in the optimal translation initiation context, and in-frame with the “INS”/ protein-encoding sequence in the projected chimeric RNA end product of amplification; (5) To minimize TCE/ICE mismatches so as to assure that the cleavage of a chimeric intermediate does not occur downstream from the “AUG” and to maximize the length of the 5’UTR in the projected chimeric RNA end product; (6) To ascertain that the “AUG” of the antisense RNA is followed by a codon encoding Val so as to confer to the resulting protein the maximum half-life, in accordance with the N-end rule pathway of protein degradation [32], if the N-terminal Met is removed by the N-terminal methionyl aminopeptidase [33]; (7) To minimally interfere with the 5’terminus of the sense strand transcript so as to preserve the position of the TSS.
Stages C and D of the middle panel of Figure 7 show the folding of the antisense RNA (stage C) and its extension into chimeric intermediate followed by strand separation and cleavage (red arrow, stage D). Line E shows the projected edited chimeric RNA end product. In its antisense portion it contains an AUG codon (“aug” highlighted in green) in an optimal translation initiation context (purine in position -3 and “G” in position +4 relative to the “A” of the AUG) and in-frame with the INS/laminin coding sequence. Translation of the edited chimeric RNA end product would result in a chimeric polypeptide shown in line F of the middle panel and comprised of a laminin portion (highlighted in blue and identical to that shown in the top panel, line C), a Tag peptide, a portion (highlighted in grey) encoded by a segment of the 5’UTR upstream of the “INS”, and the N-end portion encoded by the antisense RNA component of the chimeric RNA end product (highlighted in grey and underlined); it would be non-contiguously encoded in the genome. The detection of a Tag peptide would indicate that a chimeric polypeptide has been produced and the isolation and sequencing of this polypeptide would provide a definitive and irrefutable proof of the occurrence of the RNA-dependent mRNA amplification process.
Identification/sequencing of the projected chimeric polypeptide would be sufficient to make a determination of the occurrence of RNA-dependent mRNA amplification, but the above experimental design allows a number of additional controls. One of these is illustrated in the bottom panel of Figure 7. The AUG codon of the antisense strand can be mutated into a codon (ACA) not known to initiate translation, without changing the complementary relationship between the TCE and the ICE components of the antisense RNA. In such a case, the amplification would proceed similarly to that depicted in the middle panel, but the resulting chimeric RNA end product (bottom panel, line E) would lack a functional in-frame translation initiation codon and no translation of INS/laminin sequence would occur.
Other possible controls are frame-shifts of the edited RNA shown in the middle panel of Figure 7. Effecting a frame-shift of one, two but not three nucleotides in the region between the AUG of the antisense portion and the “INS” of the chimeric RNA end product (middle panel, line E) should abolish translation of the INS/laminin whereas a frame-shift of either one or two or three nucleotides upstream from the AUG of the antisense RNA segment within the same molecule should have no such effect.
In the framework of the above experimental design, the edited gene would produce translationally “silent” mRNA. The protein-coding content could be activated in a rigorously predicted “enhanced” manner only when mRNA amplification process is induced. Such an experiment can be carried out in a murine astrocyte cell line known to elevate the expression of laminin under hypoxic conditions [34]. There are good reasons to believe that severe chronic hypoxia would induce the mRNA amplification process in these cells and enable the testing of the above experimental design [6].
Significance and directions of future investigations
Mammalian RNA-dependent mRNA amplification was discovered, investigated, and its principles were established in the erythroid model system with cells undergoing terminal erythroid differentiation and programmed for only a very short life span [1–6]. The present study was initiated to distinguish between the narrowly specialized, terminal differentiation-restricted, and the general physiological nature of this phenomenon. The results obtained demonstrate the occurrence of RNA-dependent amplification of mRNA encoding all three chains of laminin, a major component of extracellular matrix abundantly produced throughout the tissue and organ development and homeostasis, and an exceptionally revealing indicator of the range and scope of this phenomenon. These findings suggest reinterpretation of the mechanisms involved in ubiquitous and abundant production and deposition of extracellular matrix proteins and establish RNA-dependent mRNA amplification as a general physiological phenomenon in mammalian cells.
Malfunctions of this process may be involved in multiple pathologies associated either with the deficiency of a protein normally produced by this mechanism or with the overproduction of a protein normally not involved in such a process. One of the experimental approaches to assess its involvement in such pathologies would be to interfere in vivo with the extent of complementarity between the two elements involved in the antisense RNA self-priming. In fact, multiple experiments of this sort have been carried out by nature. Thus, at least four different types of familial beta-thalassemia, characterized by the reduced production of beta-globin chains, are associated with different point mutations in the 5’UTR of human beta-globin mRNA that clearly impede the amplification-associated complementary TCE/ICE relationship within the antisense beta-globin RNA and appear to solely account for the disease [6]. A possible example of an opposite impact, the mRNA amplification-driven pathology that is due to the abnormal overproduction of a polypeptide, is the increased generation of beta-amyloid in sporadic Alzheimer’s disease discussed in detail elsewhere [6,24–27].
Multiple directions of investigation would be instrumental in elucidating the molecular processes involved in RNA-dependent amplification of mammalian mRNA. Importantly, both genetic/molecular biology research and biochemical studies should be included in these investigations. In the first category, a well targeted DNA editing and the assessment of its consequences in a variety of experimental models could be crucial in eliciting additional information. In the second category, the choice of the starting material could be decisive in studying enzymatic activities involved in the amplification process and their networking and regulation. In this respect, anemia-driven splenic erythropoiesis appears to be, for reasons discussed elsewhere [5,6,38], the best and most suitable currently available experimental model. The increased understanding of components and mechanisms involved in RNA-dependent mammalian mRNA amplification could open up new ways and approaches not only for therapeutic interference in a number of pathologies but also for novel and powerful forms of bioengineering.
Conclusion
In conclusion, the results of the present study suggest reinterpretation of the mechanisms involved in ubiquitous and abundant production and deposition of extracellular matrix proteins, confirm RNA-dependent mRNA amplification as a new mode of genomic protein-encoding information transfer, and establish it as a general physiological phenomenon in mammalian cells. Its potential physiological impact is substantial and it appears relevant to multiple pathologies. Its increased understanding would open new venues of therapeutic interference, it suggests powerful novel experimental and bioengineering approaches and its further rigorous investigations are highly warranted.
Materials and Methods
EHS tumor tissue, removed from mice six or ten days postimplantation, was provided by MuriGenics. Inc. (CA). Tissue was reduced to a cell suspension by rubbing against and washing through a 70μm cell strainer. Pelleted cells were lysed in a buffer containing 30 mM Hepes, pH8.2; 50mM NacCl; 5 mM MgCL2; 1% NP40; 10% sucrose; 5 mM DTT and 1.5 units/μl RNase inhibitor (NEB). Nuclei were removed by centrifugation for 3 min at maximum speed in an Eppendorf centrifuge and the supernatant was vigorously mixed with 10 volumes of Trizol reagent. RNA was isolated from the Trizol mix as suggested by the manufacturer. Note that RNA intended for studies of RNA amplification was extracted from an isolated cytoplasmic fraction, after removal of cell nuclei. This is because the bulk of amplified RNA, both end product and intermediates, is at least partially modified. It tends to bind to DNA [5,6] and would be removed if the isolation is attempted from total cell lysates. Since, in the absence of DNA, modified RNA binds to a predominant RNA species [5], cytoplasmic RNA was fragmented as described below prior to the rRNA depletion in preparation for the construction of sequencing libraries. For rRNA depletion, total cytoplasmic RNA was fragmented by incubation for 12 minutes at 94°C in first strand synthesis buffer (NEB, kit E7770). Following rRNA depletion with NEBNext rRNA depletion kit (E6310), RNA was purified with SPRI magnetic beads and used for preparation of sequencing libraries. Sequencing libraries were prepared using NEBNext Ultra II RNA library preparation kit for Illumina (NEB E7770) as suggested by the manufacturer with the following modifications: Step 1 (fragmentation and priming) was for 3 minutes; in step 2 (first strand synthesis) 8ul of the strand specificity reagent (NEB) was added instead of water and an additional incubation at 50° for 15 minutes was included; after adaptor ligation and before PCR enrichment, an additional round of purification on SPRI beads was included; PCR enrichment was carried on for 7 cycles. After quality control, libraries were sequenced by the Harvard Biopolymers Facility using Illumina Next500 instrument. The initial analysis of the antisense folding patterns was carried out with the help of IDT oligonucleotides analizing software. Sequencing data obtained were converted into blasting databases with the help of the Harvard Research Computing department and blasted against reference sequences at the stringency of word size 7. Sequences of interest were then extracted from the raw data and analyzed.
Acknowledgements
Authors are grateful to George Martin, Henry Lopez and MuriGenics Inc. for providing EHS tumor tissues, to New England Biolabs for gift of valuable reagents, and to Kathleen Keating and Harvard Research Computing for assisting with the data analysis.
Authors Contributions
V.V. developed the underlying concepts, designed and performed experiments, analyzed data and wrote the manuscript. S.R. and B.O. contributed to conceptualization, discussions, data analysis and preparation of the manuscript. B.O. suggested the study model.
Funding
NIH R01 AR036819 “Biogenesis of Extracellular Matrix”.
References
- 1.Volloch V. Cytoplasmic synthesis of globin RNA in differentiated murine erythroleukemia cells: possible involvement of RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 1986;83(5):1208–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Volloch V, Schweitzer B, Rits S. Synthesis of globin RNA in enucleated differentiating murine erythroleukemia cells. J Cell Biol. 1987;105(1):137–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volloch VZ, Schweitzer B, Rits S. Evolutionarily conserved elements in the 5’ untranslated region of beta globin mRNA mediate site-specific priming of a unique hairpin structure during cDNA synthesis. Nucleic Acids Res. 1994;22(24):5302–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volloch V, Schweitzer B, Rits S. Antisense globin RNA in mouse erythroid tissues: structure, origin, and possible function. Proc Natl Acad Sci U S A. 1996;93(6):2476–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rits S, Olsen B, Volloch V. RNA-dependent synthesis of mammalian mRNA: Identification of chimeric intermediate and putative end-product. BioRxiv. 2016. [Google Scholar]
- 6.Volloch V. Protein-encoding RNA to RNA information transfer in mammatian cells: principles of RNA dependent mRNA amplification. Ann Integr Mol Med. 2019;1(1):1002. [PMC free article] [PubMed] [Google Scholar]
- 7.Orkin RW, Gehron P, McGoodwin EB, Martin GR, Valentine T, Swarm R. A murine tumor producing a matrix of basement membrane. J Exp Med. 1977;145(1):204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleinman HK, McGarvey ML, Liotta LA, Robey PG, Tryggvason K, Martin GR. Isolation and characterization of type IV procollagen, laminin, and heparan sulfate proteoglycan from the EHS sarcoma. Biochemistry. 1982;21(24):6188–93. [DOI] [PubMed] [Google Scholar]
- 9.Kleinman HK, McGarvey ML, Hassell JR, Star VL, Cannon FB, Laurie GW, et al. Cannon FB. Laurie GW. Martin GR. Basement membrane complexes with biological activity. Biochemistry. 1986;25(2):312–8. [DOI] [PubMed] [Google Scholar]
- 10.Martin GR, Timpl R. Laminin and other basement membrane components. Annu Rev Cell Biol. 1987;3:57–85. [DOI] [PubMed] [Google Scholar]
- 11.Engbring JA, Kleinman HK. The basement membrane matrix in malignancy. J Pathol. 2003;200(4):465–70. [DOI] [PubMed] [Google Scholar]
- 12.Kleinman HK, Koblinski J, Lee S, Engbring J. Role of basement membrane in tumor growth and metastasis. Surg Oncol Clin N Am. 2001;10(2):329–38, ix. [PubMed] [Google Scholar]
- 13.Kleinman HK, Martin GR. Matrigel: basement membrane matrix with biological activity. Semin Cancer Biol. 2005;15(5):378–86. [DOI] [PubMed] [Google Scholar]
- 14.Benton G, Kleinman HK, George J, Arnaoutova I. Multiple uses of basement membrane-like matrix (BME/Matrigel) in vitro and in vivo with cancer cells. Int J Cancer. 2011;128(8):1751–7. [DOI] [PubMed] [Google Scholar]
- 15.Richards OC, Ehrenfeld E. Poliovirus RNA replication. Curr Top Microbiol Immunol. 1990;161:89–119. [DOI] [PubMed] [Google Scholar]
- 16.Lehmann E, Brueckner F, Cramer P. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007;450(7168):445–9. [DOI] [PubMed] [Google Scholar]
- 17.Wagner SD, Yakovchuk P, Gilman B, Ponicsan SL, Drullinger LF, Kugel JF, et al. RNA polymerase II acts as an RNA-dependent RNA polymerase to extend and destabilize a non-coding RNA. EMBO J. 2013;32(6):781–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maida Y, Yasukawa M, Masutomi K. De Novo RNA Synthesis by RNA-Dependent RNA Polymerase Activity of Telomerase Reverse Transcriptase. Mol Cell Biol. 2016;36(8):1248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapranov P, Ozsolak F, Kim SW, Foissac S, Lipson D, Hart C, et al. New class of gene-termini-associated human RNAs suggests a novel RNA copying mechanism. Nature. 2010;466(7306):642–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Downey KM, Byrnes JJ, Jurmark BS, So AG. Reticulocyte RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 1973;70(12):3400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tseng CH, Lai MM. Hepatitis delta virus RNA replication. Viruses. 2009;1(3):818–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brazas R, Ganem D. A cellular homolog of hepatitis delta antigen: implications for viral replication and evolution. Science. 1996;274(5284):90–4. [DOI] [PubMed] [Google Scholar]
- 23.Huang CR, Lo SJ. Evolution and diversity of the human hepatitis d virus genome. Adv Bioinformatics. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volloch V. A mechanism for beta-amyloid overproduction in Alzheimer’s disease: precursor-independent generation of beta-amyloid via antisense RNA-primed mRNA synthesis. FEBS Lett. 1996;390(2):124–8. [DOI] [PubMed] [Google Scholar]
- 25.Volloch V. Mechanism for ß-amyloid overproduction in Alzheimer’s Disease: Possible antisense RNA-mediated generation of a 5’-truncated ßAPP mRNA encoding 12 kDa C-terminal fragment of ßAPP, the immediate precursor of Aß In Molecular Mechanisms of Dementia. 1997, Wasco W, Tanzi R, Eds.; Humana Press. [Google Scholar]
- 26.Volloch V. Possible mechanism for resistance to Alzheimer’s disease (AD) in mice suggests a new approach to generate a mouse model for sporadic AD and may explain familial resistance to AD in man. Exp Neurol. 1997;144(1):214–8. [DOI] [PubMed] [Google Scholar]
- 27.Volloch V, Rits S. Results of Beta Secretase-Inhibitor Clinical Trials Support Amyloid Precursor Protein-Independent Generation of Beta Amyloid in Sporadic Alzheimer’s Disease. Med Sci (Basel). 2018;6(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piccinni SA, Bolcato-Bellemin AL, Klein A, Yang VW, Kedinger M, Simon-Assmann P, et al. Kruppel-like factors regulate the Lama1 gene encoding the laminin alpha1 chain. J Biol Chem. 2004;279(10):9103–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okano R, Mita T, Matsui T. Characterization of a novel promoter structure and its transcriptional regulation of the murine laminin B1 gene. Biochim Biophys Acta. 1992;1132(1):49–57. [DOI] [PubMed] [Google Scholar]
- 30.Juba AN, Chaput JC, Wellensiek BP. Exploring the Role of AUG Triplets in Human Cap-Independent Translation Enhancing Elements. Biochemistry. 2018;57(44):6308–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kearse MG, Wilusz JE. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev. 2017;31(17):1717–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varshavsky A. The N-end rule pathway of protein degradation. Genes Cells. 1997;2(1):13–28. [DOI] [PubMed] [Google Scholar]
- 33.Giglione C, Boularot A, Meinnel T. Protein N-terminal methionine excision. Cell Mol Life Sci. 2004;61(12):1455–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halder SK, Kant R, Milner R. Chronic mild hypoxia increases expression of laminins 111 and 411 and the laminin receptor i±6i21 integrin at the blood-brain barrier. Brain Res. 2018;1700:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabouille C. Pathways of unconventional protein secretion. Trends Cell Biol. 2017;27(3):230–40. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107(7):881–91. [DOI] [PubMed] [Google Scholar]
- 37.Jiang S, Zhang E, Zhang R, Li X. Altered activity patterns of transcription factors induced by endoplasmic reticulum stress. BMC Biochem. 2016;17:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rits S, Olsen BR, Volloch V. Protein-encoding RNA to RNA information transfer in mammalian cells:RNA-dependent mRNA amplification. Identification of chimeric RNA intermediates and putative RNA end products. Ann Integr Mol Med. 2019;1(1):1003. [PMC free article] [PubMed] [Google Scholar]


