Abstract
Three new species, closely related to Sphaerostilbella broomeana, are described from the USA and India. These species form septate conidia from simple conidiophores with individual branches terminating in a single phialide and chlamydospores. Teleomorphs, known for S. broomeana and S. appalachiensis, are characterised by hairy perithecia and fusiform, apiculate, and conspicuously warted ascospores. This combination of characters distinguishes the S. broomeana-group from other members of Sphaerostilbella that all form gliocladium-type anamorphs and mostly grow on basidiomata of Stereum spp. Like in other species of the genus, the majority of hosts of the species described in this paper belong to wood-inhabiting taxa of Russulales. Sphaerostilbella broomeana had been recorded from a few regions in Europe and exclusively on Heterobasidion annosum. Herein, it is reported also from H. parviporum in many other localities and on H. insulare s.l. at the foothills of the Himalayas. Its sister species, found in the same region in northern India on another member of Russulales (Dichostereum effuscatum), is described as S. himalayensis. The two species described from North America colonize polypores from various taxa. Whereas S. appalachiensis occurs in eastern USA, with H. irregulare among its hosts, S. toxica is so far known only from two locations in eastern Texas, growing on Gloeophyllum striatum (Polyporales). Despite their great similarity in morphology and ITS rDNA, TEF1 sequences clearly distinguish these two North-American species. Moreover, the two strains of S. toxica appeared metabolically distinct as their organic extracts strongly inhibited the growth of human pathogenic microbes grown in vitro. Phylogenetic analysis of rDNA sequences supports monophyly of the genus Sphaerostilbella and the included S. broomeana-group, established here.
Keywords: Anamorphic fungi, Antagonism, Fungicolous fungi, Host specialization, Hypocreaceae, Parasites, New taxa
Introduction
Most members of Hypocreaceae that grow on wood-decaying basidiomycetes belong to the genus Hypomyces and are characterised by forming comparatively large and apiculate ascospores in light- to bright-coloured perithecia on an effused subiculum. Such species produce cladobotryum-type anamorphs characterised by multi-celled conidia that are formed either singly or in dry chains at one or several loci of the conidiogenous cell (Rogerson and Samuels 1993; Põldmaa and Samuels 1999). Species of Trichoderma (teleomorphs until recently classified in Hypocrea), several of which grow on polypores, form much smaller, nonapiculate ascospores that disarticulate inside asci in perithecia immersed in a stroma, and aseptate conidia that are produced from the single locus of the phialide where they are accumulating in heads. Fungicolous fungi, closely related to these two species-rich genera, combine these features in different ways but are all non-stromatic and produce conidia in a drop of liquid at the tips of phialides. Among these, species of Protocrea and Sphaerostilbella form gliocladium-type anamorphs, characterised by usually aseptate conidia held in liquid heads at the top of penicillately branched conidiophores (Seifert 1985; Jaklitsch et al. 2008; Zare and Gams 2016). In most cases, these penicillate conidiophores are accompanied by verticillium-like conidiophores with phialides arranged in divergent whorls. In contrast, anamorphs of Sporophagomyces produce simple, unbranched conidiophores and septate conidia, held in liquid droplets (Rogerson and Samuels 1993; Põldmaa et al. 1999).
Sphaerostilbella broomeana is characterised by a unique combination of apiculate warted ascospores, nearly penicillate conidiophores, and the occurrence on basidiomata of devastating forest pathogens from the Heterobasidion annosum-group (Bondarzewiaceae, Russulales). During the last two decades, morphologically similar fungi have been collected on various wood-decaying fungi outside Europe, from where all previous S. broomeana reports originate. Several of the hosts belong to the Russulales, yet not to the genus Stereum, included in it, that represents the main host of majority of other Sphaerostilbella species. These encompass S. aurifila (W.R. Gerard) Rossman, L. Lombard & Crous (=S. lutea (Henn.) Sacc. & D. Sacc., the type species of the genus), S. berkeleyana (Plowr. & Cooke) Samuels & Cand., S. penicilloides (Corda) Rossman, L. Lombard & Crous, (=S. aureonitens (Tul. & C. Tul.) Seifert, Samuels & W. Gams), and Gliocladium polyporicola (Seifert 1985; Candoussau and Magni 1995), the taxonomy and description of which will be amended in a future publication.
The aim of this paper was to resolve the taxonomy of recent collections that are similar to S. broomeana but have been found outside the known distribution and host range of this species. For that purpose, we studied the morphology as well as sequenced ITS-LSU rDNA and parts of the TEF1 gene of 18 specimens and cultures isolated from these. Analyses of these data along similar species and sequences available in GenBank led to the delineation of the S. broomeana-group and its four members. The paper presents detailed descriptions of S. broomeana and three new species.
Materials and methods
Morphology and cultures
Twenty-five or more ascospores and conidia were measured from each specimen/isolate in 3% aqueous KOH solution. In the given ranges of measurements of spores, the innermost number(s) represents (the range of) the mean values of the specimen(s) and the two outermost numbers the limits of the 90% range of estimated normal distribution observed in the single or the most divergent specimens rounded to the nearest 0.5 μm. Absolute ranges are presented for all other structures. Two size ranges are provided for the length and width of ascospores and their length/width ratio (Q). One provides the dimensions of whole ascospores, while the other excludes apiculi and ornamentation. All specimens reported in the study have been deposited at the mycological collection of the Natural History Museum of the University of Tartu (TU) or the Estonian University of Life Sciences (TAAM).
Ascospores or conidia were isolated and colonies grown on 1.5% malt extract agar (MEA, Oxoid, Cambridge, UK) in darkness at 24 °C. Colony growth was measured from 9 cm diam plastic Petri dishes inoculated with a 4 × 4-mm plug taken from the edge of an actively growing colony. Characters were evaluated also from cultures grown on cornmeal agar (CMA, Oxoid, Cambridge, UK) and potato dextrose agar (PDA, Merck KGaA, Darmstadt, Germany). All isolates reported in the study have been deposited at the Tartu Fungal Culture Collection (TFC) of the University of Tartu.
DNA extraction, PCR, and sequencing
Genomic DNA was extracted from mycelium grown for 4–7 days on a sheet of cellophane on top of a MEA surface. Material, stored at −20 °C, was soaked in 100 μl of buffer containing 0.015 U/μl proteinase K (Thermo Fisher Scientific, Waltham, MA, USA), 0.8 M Tris–HCl, 0.2 M (NH4)2SO4, and 0.2% w/v Tween-20 (Solis BioDyne, Tartu, Estonia) and incubated at 56 °C for 15 h. After inactivation of proteinase K at 98 °C for 15 min, the lysate was centrifuged at 6010×g for 2 min. The supernatant was diluted by 10 times, of which up to 2.5 μl were used as a template for PCR. PCR amplicons of ITS and the 5′-end of the LSU rDNA were generated using primers ITS0F (Tedersoo et al. 2008) and ITS4 (White et al. 1990) or LR5 (Vilgalys and Hester 1990), with annealing temperature of 55 °C. TEF1 amplicons including introns 5, 6 and exons 5, 6 were generated with primers EF1–728F (Carbone and Kohn 1999) and TEFLLErev (Jaklitsch et al. 2005) or the region of introns 5, 6 and exon 5 was amplified with primers EF1–728F and EF2 (O’Donnell et al. 1998), and exon 6 with primers EF1–983fand EF1–2218r (Rehner 2001). All TEF regions were amplified with annealing temperature of 66 °C for the first 10 cycles followed by annealing temperature of 57 °C for the next 35 cycles. PCR was performed using 5 × HOT FIREPol® Blend Master Mix (Solis BioDyne, Tartu, Estonia) in a 25-μ1 reaction volume. The PCR products were purified using Exo-FastAP enzymes (Thermo Fisher Scientific, Waltham, MA, USA). Amplicons were sequenced at Macrogen Inc. (Amsterdam, The Netherlands). Sequences were edited and assembled with Sequencher 5.4.6 (Gene Codes, Ann Arbor, MI, USA) and deposited in INSD under accession numbers MH795080–MH795107.
Sequence alignments and phylogenetic analyses
ITS-LSU and TEF1 sequences were aligned using MAFFT v7 (Katoh and Standley 2013), followed by manual adjustment in AliView 1.20 (Larsson 2014). The final rDNA dataset included 683 sites for ITS, of which an ambiguously aligned region of 28 bp was excluded, and 817 sites for LSU. Bayesian analyses were run in MrBayes v. 3.2.6 (Ronquist et al. 2012) for 10 million generations under the GTR+I + G evolutionary model at the CIPRES Science Gateway v3.3 (http://www.phylo.org), and sampled at each 1000th generation. By the end of the run, the average standard deviation of split frequencies attained 0.003. The first 25% of the trees were discarded as a burn-in and the posterior probabilities (PP) were calculated from the remaining trees. The TEF1 dataset was divided into two partitions, distinguishing coding (1140 sites) and non-coding regions (424 sites) and analysed in MrBayes using settings as described above. Maximum likelihood analysis of the rDNA dataset was run in RAxML (Stamatakis 2014) at the CIPRES Science Gateway v3.3 (http://www.phylo.org) applying the gamma model of rate heterogeneity, with automatic halting of bootstrapping.
Results and discussion
ITS-LSU rDNA-based phylogenetic analyses supported the monophyly of the ingroup and eight subgroups comprised in it (Fig. 1). The most specimen-rich subgroup included S. broomeana and all the morphologically similar collections. Among these, an isolate from the Himalayas represented the sister-group of S. broomeana, while the remaining six isolates formed another subgroup that was supported in the Bayesian analysis. This, in turn, comprised two further subgroups, one of which obtained maximum Bayesian posterior probability and bootstrap support in maximum likelihood analysis. In the Bayesian analysis of TEF1 sequences, however, both subgroups received maximum support (supplementary Fig. 1). Altogether, the analyses of molecular data distinguished S. broomeana and three closely related lineages representing putative new species.
Fig. 1.
Bayesian phylogeny of Sphaerostilbella species based on ITS and LSU rDNA sequences. Sporophagomyces chrysotomus represents an outgroup. Numbers on the branches indicate posterior probability scores and bootstrap support. Scale bar presents substitutions per site
Regarding morphology, the anamorphs of the three putative new species, albeit clearly distinct from S. broomeana, are difficult to distinguish among. However, the aggregation and abundance of chlamydospores vary among the four, each of which also has a characteristic growth rate on MEA. The only new teleomorph can easily be delimited from that of S. broomeana through ascospore size and ornamentation as well as width of apiculi. Based on morphological and molecular evidence, we consider the lineages, closely related to S. broomeana, to represent three new species of Sphaerostilbella that will be described below, next to a detailed description of S. broomeana.
Teleomorphs, known in two of the Sphaerostilbella species treated here, are characterised by a tomentose layer of perithecia formed in a scanty subiculum. Such a morphological aspect results from the many free ends of hyphae originating from the subiculum and cells at the base of the papillae. Among species growing on polypores, hairy perithecia also occur in Sporophagomyces spp., Hypomyces villosus Rogerson and Samuels (Rogerson and Samuels 1993), and H. viridigriseus K. Põldmaa & Samuels (Põldmaa et al. 1997). Anamorphic collections of the two Sphaerostilbella species often produce perithecia when host fruitbodies are incubated in moist conditions and/or on agar media in culture.
The anamorphs of S. broomeana and the closely related species described below differ from those in Hypocreaceae in forming septate conidia from phialides borne on simple unbranched or nearly penicillate conidiophores and chlamydospores. Chlamydospores are formed in large irregular clusters in two species from North America or in intercalary chains in cultures of S. broomeana and its sibling, while not being observed in other Sphaerostilbella species. Synnematous anamorphs and yellow pigments, shared by most members of the genus, are not formed by any of the four species treated below. However, like typical gliocladium-type anamorphs of Sphaerostilbella, S. broomeana tends to form penicillate conidiophores with adpressed branches bearing phialides, which, however, differ in their larger dimensions. By contrast, in the three new species, individual conidiophores cannot clearly be distinguished because they consist of simple septate lateral branches of aerial hyphae that typically terminate in a single phialide and form septate conidia. Accordingly, the anamorphs resemble that of Sporophagomyces chrysostomus (Berk. & Broome) K. Põldmaa & Samuels, previously classified in Acremonium, Moeszia, and Septocylindrium (Arnold 1970; Rogerson and Samuels 1993). The somewhat similar acremonium-like anamorphs in Trichoderma sect. Hypocreanum Bissett, e.g., T. pulvinatum (Fuckel) Jaklitsch & Voglmayr, differ in more robust structures and aseptate conidia.
Sphaerostilbella broomeana was thus far known from a few countries in Europe but is shown here to be of a wider and regionally frequent occurrence. Its easternmost location in the Himalayas hosted also an undescribed species, most closely related to S. broomeana. Like most other species of the genus, both grow on wood-decaying members of the Russulales, albeit from different genera. Similar collections originating from the eastern United States on different polypore taxa proved to represent two additional new species with supposedly distinct host and geographic ranges but minor morphological differences. One of these has twice been collected on Gloeophyllum striatum (Polyporales), Although phylogenetically not closely related to Heterobasidion, that includes the main hosts of the S. broomeana-group. However, the two polypore genera share several features to which the ancestor of these parasites might have become adapted, with later speciation events accompanied by host jumps across a shared substratum. Namely, both genera comprise pioneer species on coniferous wood that form perennial, leathery, dimitic basidiomata.
The four species treated in this paper form a morphologically and genetically distinct group, referred to as the S. broomeana-group. It is considered to belong to Sphaerostilbella based on previously published phylogenies that have included its core species, S. broomeana (Põldmaa et al. 1999; Põldmaa 2000; Jaklitsch et al. 2008; Zare and Gams 2016), and the rDNA-based phylogeny presented herein. The incorporation of the S. broomeana-group expands the concept of Sphaerostilbella, originally including species with <12 μm long, spinulose, naviculate and nonapiculate ascospores, penicillate conidiophores, and aseptate conidia but no chlamydospores (Seifert 1985). Species of the S. broomeana-group, in contrast, have >12 μm long, warted, fusiform, and apiculate ascospores, rather undifferentiated conidiophores, septate conidia, and chlamydospores. While the rDNA ITS and LSU regions analysed in this study provided strong support for the monophyly of the S. broomeana-group, phylogenetic relationships with and among other members of the genus did not receive adequate support. Resolving the generic status of the S. broomeana-group has thus to await phylogenetic analyses of a multigene dataset of Hypocreaceae, including several undescribed species, preliminarily identified in Sphaerostilbella.
Taxonomy
Sphaerostilbella broomeana (Tul. & C. Tul.) K. Põldmaa, Canad. J. Bot. 77: 1764. 1999. Fig. 2
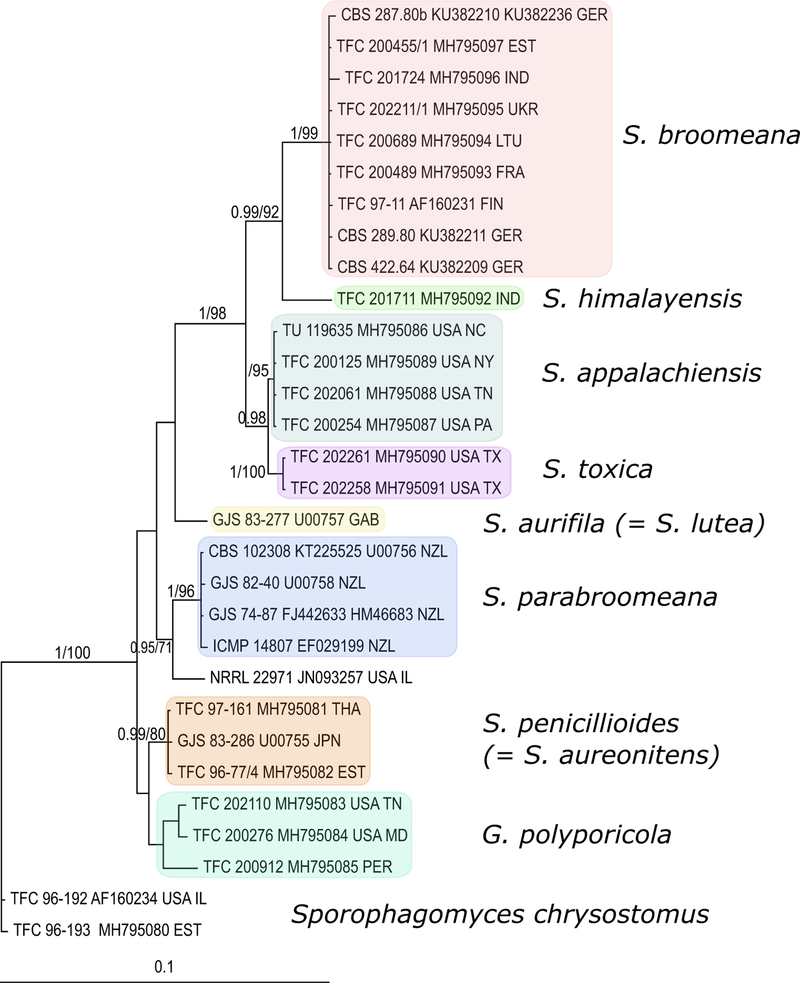
Fig. 2. Sphaerostilbella broomeana.
a effused layer of perithecia; b hairy appearance of perithecia; c anamorph next to teleomorph; d, e perithecia covered by hyphae with free ends; f asci; g, h (part)ascospores; i loose fascicles of conidiophores; j–n conidiophores with conidia; o conidia; p chlamydospores. a, c–f TAAM169844; b TU131906; g, h TAAM161513; i TAAM169547; j, k TFC200455; l, n TFC200398; m, o TFC94–62; p TFC97–167. a–i on host hymenophore, j–p on MEA. Scale bars: a 1 mm; b 200 μm; c 300 μm; d, i, m 50 μm; e, j–l, n 20 μm; f, g, and o 10 μm; h 5 μm; p 15 μm
≡Hypomyces broomeanus Tul. & C. Tul., Sel. Fung. Carpol. 3: 108. 1865
≡Nectriopsis broomeana (Tul.)W. Gams, Neth. J. Pl. Path. 88: 73. 1982
Anamorph known as Gliocladium microspermum (Sacc.) W. Gams, Neth. J. Pl. Path. 88: 73. 1982
≡Verticillium microspermum Sacc., Syll. Fung. 4: 153. 1886
=Gliocladium strictum Petch, Trans. Brit. mycol. Soc. 21: 274. 1938
Holotype: PC?. Epitype, designated by Zare and Gams (2016), MBT374110: Germany, Schonbuch near Tübingen, 2 km north of Bebenhausen, on Heterobasidion annosum, leg. R.J. Bandoni, August 1979, isol. C.T. Rogerson from single ascospore (CBS H-22417); ex-type culture, CBS 287.80B (=C.T.R. 79–315). Ex-type culture derived sequences, KU382210 (rDNA ITS), KU382236 (LSU)
Subiculum white, consisting of a cottony mat of hyaline hyphae effused over host hymenophore, hyphae loosely aggregated, compacted below perithecia, 1.5–3.0 μm wide, hyaline, smooth-walled. Perithecia gregarious, semi-immersed in subiculum, yellowish white to ochraceous, not changing colour in KOH, 250–460 μm high, venter 200–260 μm in diam; papilla 50–140 μm high, 80–130 μm wide at base, with obtuse apex 30–50 μm wide; perithecial wall around 20 μm thick, composed of one region of flattened cells with wall ca 1 μm thick; white mycelial weft covering perithecia with apex of papilla protruding, composed of loosely interwoven hyphae 1.5–3 μm in diam, of which many terminate in straight, septate, 1.5–2.5 μm wide, smooth-walled free ends. Asci cylindrical, 110–140 × 4–5 μm, apex thickened up to 2 μm, containing eight uniseriately placed ascospores with ends overlapping. Ascospores fusiform to naviculate, apiculate, with a median or rarely submedian septum, wall covered by warts ca 0.5 (im high and wide, 10.5–(11.6–13.2)–15.0 × 2.5–(3.0–3.7)–4.0 μm, Q = 2.9–(3.6–3.9)–4.5, without apiculi and warts 9.0–(10.6–11.3)–13.0 × 2.0–(2.6–2.8)–3.5 μm, Q = 2.7–(3.5–4.0)–5.0; apiculi 0.5–(0.8–0.9)–1.2 × 0.4–(0.6– 0.7)–1.0 μm, Q = 0.9–(1.4)–1.8, usually of equal size at both ends of the spore, apex blunt.
Conidiophores erect, single to fasciculate; stipe 120230 μm long, 3–4 μm wide near base, tapering to 2–3 μm, hyaline, smooth–walled, bearing few, mostly two adpressed phialides or few, mostly two, 15–30 μm long and 1.5–2.5 μm wide branches bearing phialides. Phialides almost cylindrical, 30–50 μm long, 1.5–2.0 μm wide at base, tapering gradually to 1–1.5 μm width at apex. Conidial mass translucent, globose, 20–40 μm diam. Conidia variable in size and shape, ellipsoidal to oblong or allantoid, 0–(1)-septate, base flattened, hilum indistinguishable, 3.0–(7.1)–11.0 × 2.0–(2.7)–3.5 μm, Q = 1.5–(2.6)–3.7. Chlamydospores not observed on host fruitbodies.
Colonies on MEA reaching 25–35 mm diam in 7 days, cottony or floccose, uniform or concentrically zonate; aerial mycelium scarse to abundant, white, densely covered by watery droplets containing conidia; margin even or fasciculate; conidiation abundant; reverse whitish.
Conidiophores ascending, formed mostly at right angle from aerial hyphae, not clearly differentiated from these; stipe (35)70–350(500) μm long, 2–3 μm wide near base, hyaline, smooth-walled, branched in the middle or upper part or unbranched and ending in two phialides; branches formed by one or two at one to three levels, 20–40(80) μm long and 1.5–3 μm wide. Phialides on uppermost branches of conidiophores, typically in groups of two or three, adpressed, almost cylindrical, 25–50(70) μm long, 1.5–2.0 μm wide at base, tapering gradually to 1–1.5 μm width at apex. Conidial mass translucent, globose, 10–35 μm diam, often merging with others from neighbouring conidiophores.
Conidia emerging straight or obliquely from conidiogenous locus, variable in size and shape, ellipsoidal to oblong or (sub)allantoid, sometimes narrow obclavate, straight, curved or bent in lower part, widest in the middle or in the lower or the upper part, sometimes slightly attenuated towards the base, 0–1(3)-septate, base flattened or rounded, hilum mostly indistinguishable, 3.0–(8.2–10.5)–17.5 × 1.5–(2.5–3.0)–4.0 μm, Q =1.5–(3.3–3.9)–5.8. Chlamydospores formed in simple or divergent intercalary chains, sometimes forming irregular clusters, subglobose 7–13 × 5–8 μm, wall thickened, 0.5–1 μm, smooth, abundant on submerged mycelium.
Hosts: Heterobasidion annosum s.l. (incl. H. parviporum) and H. insulare s.l.
Geographical distribution: Europe (Estonia, Finland, France, Germany, Lithuania, Norway, Ukraine, United Kingdom) and Asia (India, at foothills of the Himalayas).
Specimens examined. Estonia, Pärnu Co., Mereäärse, on Heterobasidion parviporum, leg. K. Põldmaa, 27 Sep. 1994 (TAAM161513, culture TFC94–62); Ida–Virumaa Co., Kaukvere primaeval forest, on H. parviporum, leg. K. Põldmaa, 3 Sep. 1996 (TAAM169547, culture TFC190254); Tartu Co., Alam-Pedja Nature Reserve, SE part of forest Suur Peenar, on H. parviporum on Picea abies, leg. K. Põldmaa, 10 Sep. 1997 (TAAM169844, culture TFC97–167); on H. parviporum, leg. M. Hanso, 2003 (TAAM170777, culture TFC200455); JärvamaaCo., on H. parviporum on Picea abies, leg. K. Põldmaa, 2005 (TAAM170902, culture TFC200733). Finland, Varsinais-Suomi Prov., Nousiainen Comm., Pukkipalo virgin forest, on H. parviporum on Picea abies, leg. K. Põldmaa, 4 June 1997 (TAAM169659, culture TFC97–11). France, Rimont, Ariege, on Heterobasidion sp., leg. J. Fournier, 2004 (TAAM170845, culture TFC200489). India, Uttarakhand, Dehradun distr., 4 km W from Dhanaulti, on H. insulare s.l., growing together with anamorph of Hypomyces aurantius (Pers.: Fr.) Tul. & C. Tul., leg. K. Põldmaa, 9 Oct. 2012 (TU119036B, culture TFC201724). Lithuania, Kaunas County, Prienas distr., Nemunas Loops Regional Park, Drobungis botanical preserve, leg. G.R.W. Arnold & K. Põldmaa, 29 Sep. 1995 (TAAM170702, culture TFC200689). Norway, on Heterobasidion sp., leg. L. Ryvarden & K.–H. Larsson (TU131909). Ukraine, Ivano-Frankivsk region, Kosiv distr., Hutsulschyna National Nature Park, Korovyak stow, near Sheshory village, on H. parviporum on Picea abies (L.) Karst., leg. A. Akulov, 5 Aug. 2017 (CWU (MYC) AS 6523, TU131906; culture TFC202211).
Notes: Sphaerostilbella broomeana was originally described as a species of Hypomyces, most similar to H. aureonitens Tul. & C. Tul. (Tulasne and Tulasne 1865). However, later on the former species was combined in Nectriopsis (Gams and van Zaayen 1982) but the latter in Sphaerostilbella (Seifert 1985). The anamorph, reported by the Tulasne brothers as being similar to Mariannaea (=Penicillium) elegans (Corda) Samson, was later described as a species of Verticillium or Gliocladium by Saccardo and Petch, respectively. Molecular evidence supported the transfer of the species to Sphaerostilbella (Põldmaa et al. 1999). All holotype collections originated from the UK, and only a century later additional material was reported from Germany and Estonia (Arnold 1963; Gams and van Zaayen 1982; Helfer 1991; Põldmaa 1999). The new records provided here suggest that S. broomeana is more widely distributed in Europe but can easily be overlooked because of its cryptic growth on the hymenophore of host basidiomata, which themselves are often also hidden under the decaying tree trunks or stumps. In the literature, it has only been reported to grow on H. annosum (Fr.) Bref, that can be regarded as a species complex. In fact, most of the specimens listed above are from H. parviporum Niemelä & Korhonen, segregated from H. annosum in 1998, and were collected at the base of decaying trunks of Picea abies. The specimen from the Himalayas suggests a broader geographic and host range for S. broomeana than previously recognized. The host is a member of the H. insulare-group with its ITS sequence (UNITE: UDB018604) belonging to UNITE species hypothesis SH201110.07FU (Kõljalg et al. 2013) that includes H. cf. linzhiense with the most similar (99.5%) sequence. The identification of S. broomeana from a fallen leaf of Guazuma ulmifolia in Cuba (Zare and Gams 2016) is doubtful and needs verification.
Sphaerostilbella broomeana stands out in its readiness to form perithecia abundantly in culture as well as on host fruitbodies naturally infected with the anamorph and incubated in a container at adequate moisture. In some collections, the (over)mature ascospores were disarticulated, as observed also in Sporophagomyces chrysostomus (Põldmaa et al. 1999) and some species of Hypomyces. The anamorph of S. broomeana deviates from the gliocladium-type in forming a less distinguishable conidiophore stipe, 0- and 1-septate conidia and chlamydospores. Sphaerostilbella parabroomeana Zare & Gams, in which some of the conidia also are reported to be 1-septate, is only distantly related to S. broomeana (Fig. 1).
The ITS rDNA sequences that we obtained from five specimens from Western to Eastern Europe were identical, whereas a few base pair differences were found among sequences derived from Genbank and originating from Germany. Similarly, TEF1 sequences, obtained from two European specimens, were identical. The collection from India, however, was more distinct, differing at three positions and having some longer homopolymer repeats in the ITS region. Their TEF1 sequences were even more distinct, differing at 21 positions and having three indels.
Sphaerostilbella himalayensis K. Põldmaa, sp. nova Fig. 3
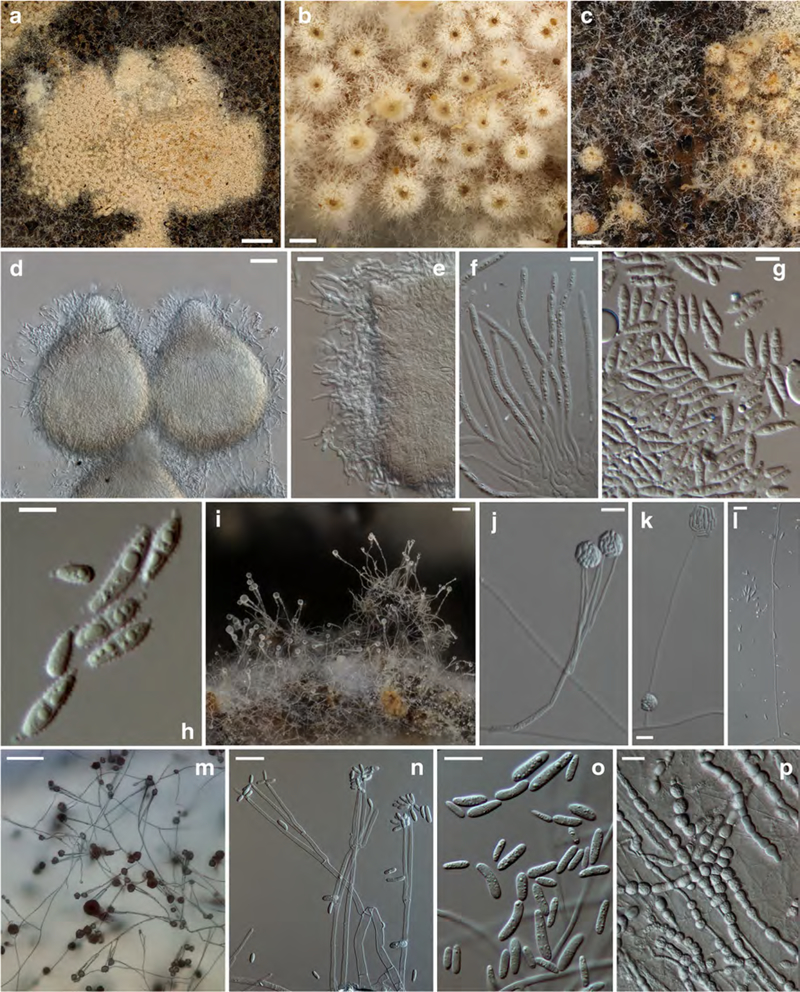
Fig. 3. Sphaerostilbella himalayensis.
a, b anamorph on host hymenophore; c, e conidiogenous branch; d, f–h conidia; i chain of chlamydospores. a–d TU119041; e–i ex-type culture TFC 201711, e–g on MEA, h–i on PDA. Scale bars: a 1 mm; b 250 μm; c, f, h, and i 10 μm; d, g 5 μm, e 20 μm
MycoBank: MB 827698
Etymology: Epithet refers to the geographic origin of the holotype
Holotype: India, Uttarakhand, Dehradun distr., Dhanaulti, next to Crystal View Hotel, 30.42897° N 78.23483° E on Dichostereum effuscatum, leg. K. Põldmaa, 9 Oct. 2012, (TU119041, ex-type culture TFC201711). Sequences deposited in GenBank: MH795092 (rDNA ITS and LSU); MH795106 (TEF1)
Teleomorph unknown. Scarce whitish mycelium effused at margins of host fruitbodies. Simple branches of aerial hyphae terminating in a single phialide bearing several conidia. Conidiogenous branches septate, 40–80 μm long (and probably longer), 1.5–2.5 μm wide near base, and ca 1 μm at apex. Conidia oblong, straight, equilateral, 0–1(2)–septate, 9.5–(13.1)–16.5 × 2.5–(3.0)–3.5 μm, and Q =3.4–(4.4)–5.4; hilum not visible or low and flat, rarely pronounced, 0.5–0.7 × 0.7–1.3 μm. Chlamydospores not observed.
Colonies on MEA reaching 15–17 mm diam in 7 days, white, margin even, central part compact cottony, margin floccose with sparser aerial mycelium, densely covered by watery droplets containing conidia. Aerial hyphae hyaline, 1–2(3) μm wide, septate; submerged hyphae forming dense mat of narrow hyphae, ca 1 μm in diam. Conidiophores not distinct from aerial hyphae, ascending at right angle from upright aerial hyphae forming simple lateral branches, 1–2 from one point, conidiogenous branches with up to eight septa, 95–300 μm long or longer, of constant width 1.2–1.8 μm throughout their length, terminating in a single phialide. Phialides tapering slightly to 0.8–1.2 μm at the apex, bearing masses of conidia that adhere in translucent droplets of liquid.
Conidia formed in upright position at the apex of the phialide, of variable shape and size, ellipsoidal to oblong or narrow clavate, occasionally widest near base, straight and equilateral but often bent near base; some constricted in the middle, (0–)1(–3)-septate, often slightly attenuated towards base, base flattened, or rounded; some with a hilum 0.5–1.2 μm high and ca 1.3 μm wide, 9–(14.1)–19.5 × 2.0–(3.0)–4.0 μm, and Q = 3.3–(4.7)– 6.1. Chlamydospores not observed among aerial mycelium but chlamydospore-like cells scattered among submerged hyphae, hyaline, multiguttulate, 4–8 μm in diam, formed singly or in chains in intercalary position.
Hosts: Dichostereum effuscatum (Cooke & Ellis) Boidin & Lanq.
Geographical distribution: Asia (India, at foothills of the Himalayas), known only from the type collection
Notes: Sphaerostilbella himalayensis represents the sister species of S. broomeana both in rDNA and TEF1-based phylogenies (Fig. 1, supplementary Fig. 1) with numerous nucleotide differences observed in these gene regions. Both species parasitize wood-inhabiting members of Russulales, yet from different genera and families. Additional collections are needed to confirm their distinct host ranges. The new species is distinguished from S. broomeana by much less differentiated conidiophores ending in a single phialide and on average longer, slenderer, and mostly septate conidia. Accordingly, S. himalayensis is similar to Sporophagomyces chrysostomus and the two new species described below.
Sphaerostilbella appalachiensis K. Põldmaa, sp. Nova Fig. 4
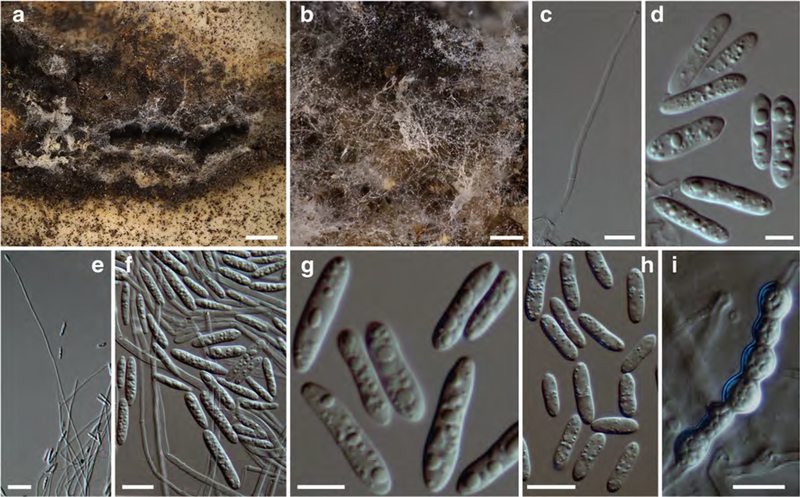
Fig. 4. Sphaerostilbella appalachiensis.
a perithecia and subiculum on host hymenophore; b–d perithecia covered by hyphae with free ends (d at papilla); e ascospores; f, g, and l conidiogenous branches and conidia; h–j anamorph on host hymenophore; k translucent globose mass of conidia; m conidia; n chlamydospores in clusters. a–c, e holotype TU119727; d TAAM170269; f–j TAAM 170565; k–n ex-type culture TFC202061. a–j on host, k–m on MEA, n on PDA. Scale bars: a 500 μm; b, i–k 100 μm; c, l 40 μm; d, g, n 10 μm; e, m 5 μm; f 20 μm; h 200 μm
MycoBank: MB 827699
Etymology: In reference to the geographic origin of the described specimens
Holotype: USA, Tennessee, Sevier County, Ramsey Cascades trail, 35.703394° N 83.353538° W, on Heterobasidion irregulare, leg. K. Põldmaa, 30 Sep. 2015 (TU119727, ex-type culture TFC202061). Sequences deposited in GenBank: MH795088 (rDNA ITS and LSU); MH795101 (TEF1)
Subiculum white, loosely cottony, effused over host hymenophore, composed of hyaline hyphae, densely compacted below perithecia; hyphae 1.5–3.0 μm wide, mostly smooth-walled, some spinulose. Perithecia gregarious, semiimmersed in subiculum, ivory, not changing colour in KOH, 300–400 μm high, venter 166–250 μm in diam, papilla pronounced, 85–145 μm high, 70–105 μm wide at base, with obtuse apex, 30–55 μm wide; wall 18–20 μm thick, composed of one region of flattened cells, with wall 0.5–1 μm thick; perithecia covered with white mycelial weft, from which usually the papilla or its apex protrude, composed of loosely interwoven hyphae 2–4 μm in diam with some cells inflated to 6 μm diam and with numerous free ends; free ends of hyphae reaching the surface straight, septate, 1.5–2.5 μm wide, smooth-walled. Asci cylindrical, 110–160 × 5.5–7 μm, apex slightly thickened, up to 2 μm, containing eight uniseriately placed ascospores with ends overlapping. Ascospores fusiform, apiculate, with a median or rarely submedian) septum, wall grossly warted-tuberculate, warts 0.3–0.6 μm high and 0.3–1 μm wide, 15.5–(17.5)–19.5 × 4.5–(5.0)–5.5 μm, Q =3.1–(3.5)–4.0, without apiculi and warts 13.5–(15.2)–17.0 × 3.5–(4.1)–4.5 μ, Q = 3.1–(3.8)– 4.4); apiculi 0.7–(1.1)–1.5 × 1.0–(1.7)–2.2 μm, Q = 0.3–(0.7)–1.1, of equal size at both ends, apex blunt.
Conidiophores/conidiogenous branches arising singly or in small fascicles from hyphae on host hymenophore, sparingly branched, branches formed singly or in pairs at opposite position, >100 μm long and 2 μm wide, with up to five septa and ending in a single phialide. Conidial mass translucent, globose, 20–30 μm in diam. Conidia ellipsoidal to oblong, straight, equilateral, occasionally wider in basal part, or constricted in the middle, (0–)1(–3)-septate, often attenuated near base, 8.0–(10.9–12.6)–17.0 × 2.5–(3.2–3.4)–4.0 μm, Q = 2.5–(3.4–3.7)–4.9, base truncate, hilum not distinguished. Chlamydospores forming irregular clusters among subicular hyphae, (sub)globose, 4.0–9.0 μm in diam, hyaline to pale ochraceous, wall 0.3–0.7 μm thick, smooth.
Colonies on MEA reaching 17–22 mm diam in 7 days, white, margin even or crenate; cottony or floccose, aerial mycelium scarce to abundant, homogenous and compact or with zones of different mycelium texture, white, and densely covered by watery droplets containing conidia. Conidiophores indistinct, formed laterally on aerial hyphae at 45–90°, unbranched or with a supporting branch of 25–32 × 2–2.5 μm bearing further two or three conidiogenous branches; terminal branches with up to seven septa, 50–420 μm long (or longer), 2–3 μm wide near the base, terminating in a single phialide. Phialides tapering slightly, ca 1.5 μm wide at tip. Conidia held in a translucent drop of liquid that joins conidia from one or several adjacent conidiogenous branches.
Conidia emerging straight or obliquely from the conidiogenous locus, of regular shape, ellipsoidal to oblong, straight or bent in lower part, sometimes constricted in the middle, (0–)1(–3)-septate, often slightly attenuated towards base, 8.5–(10.2–14.7)–18.5 × 2.5–(3.0–3.1)–4.0 μm, Q = 2.4–(3.3–4.9)–6.1; base flattened or rounded, without a distinct hilum. Chlamydospores observed only on submerged hyphae, in short chains or forming irregular clusters, hyaline, subglobose, 7.513 × 6.5–12 μm, wall smooth, ca 0.5 μm thick.
Colonies on CMA reaching 15–20 mm diam in 7 days. Aerial mycelium scarce. Conidiogenous structures as those observed on MEA. Chlamydospores abundant, submerged or covering surface of agar medium, forming large clusters, 7.5–9.5 × 6.5–8.0. Conidia 7.5–(10.0)–13.0 × 2.5–(3.1)–3.5–μm, Q = 2.5–3.3–4.2.
Hosts: Heterobasidion irregulare Garbel. & Otrosina, Rigidoporus crocatus (Pat.) Ryvarden, Trichaptum biforme (Fr.) Ryvarden
Geographical distribution: North America, Appalachian mountain range
Specimens examined: USA. New York, Oswego County, near Cleaveland, Vanderkamp Station, on Rigidoporus crocatus, on a decaying trunk of Pinus sp., leg. K. Põldmaa, 2 Oct. 1999 (TAAM170269, ex ascospore culture TFC200125). North Carolina, Blue Ridge Mountains, Blue Ridge Assembly, at start of the California trail, on Trichaptum biforme, leg. K. Põldmaa, 24 Sep. 2015 (TU119635). Pennsylvania, Westmoreland County, Laurel Summit Picnic Area, trail to a bog, on T. biforme, leg. K. Põldmaa, 16 Sep. 2000 (TAAM170565, anamorph with juvenile perithecia; ex conidium culture, TFC200254).
Notes: Indistinguishable in gross appearance, S. appalachiensis and S. broomeana differ in micromorphology of both the teleo- and the anamorph. Namely, ascospores of the former species are larger, more grossly warted, and bear more prominent (wider) apiculi than in the latter. Ascospores could be measured only in the holotype, as in TAAM170269 most of the ascospores were overmature and some disarticulated into partspores. Like in some collections of S. broomeana, the holotype of S. appalachiensis included only the anamorph when collected but produced abundant perithecia on the host after kept in a container with adequate moisture content. In contrast to the more or less penicillate conidiophores as well as often irregularly shaped and aseptate conidia of S. broomeana, S. appalachiensis typically produces homogeneously shaped and consistently septate conidia on solitary phialides formed on simple, septate lateral branches of aerial hyphae. However, in all these anamorph characters, S. appalachiensis cannot be distinguished from S. toxica, described below. Both species form abundant chlamydospores in irregular clusters, while in cultures of S. broomeana these are infrequent and held in intercalary chains. Similar, however, larger chlamydospores are characteristic of several species of Hypomyces with cladobotryum-type anamorphs (Rogerson and Samuels 1993; Põldmaa and Samuels 1999; Põldmaa 2011) but are not observed in other species of Sphaerostilbella.
ITS-LSU rDNA and TEF1-based phylogenies revealed S. appalachiensis as part of the sister clade of S. broomeana and S. himalayensis (Fig. 1, supplementary Fig. 1) with more than 25 nucleotide substitutions in ITS regions distinguishing S. appalachiensis from the other two species. Whereas the exon regions of TEF1 in S. appalachiensis and S. broomeana differed in many positions, the divergence of their intron regions did not allow unambiguous alignment. Despite a considerable latitudinal gradient among the geographic locations of the four collections of S. appalachiensis and their host fungi belonging to different orders (Hymenochaetales, Polyporales, Russulales), their ITS sequences were identical, while differences were encountered at three positions in the TEF1 sequences. Differences from S. toxica are discussed under that species.
Sphaerostilbella toxica K. Põldmaa, Bills & D.P. Lewis, sp. nova Fig. 5
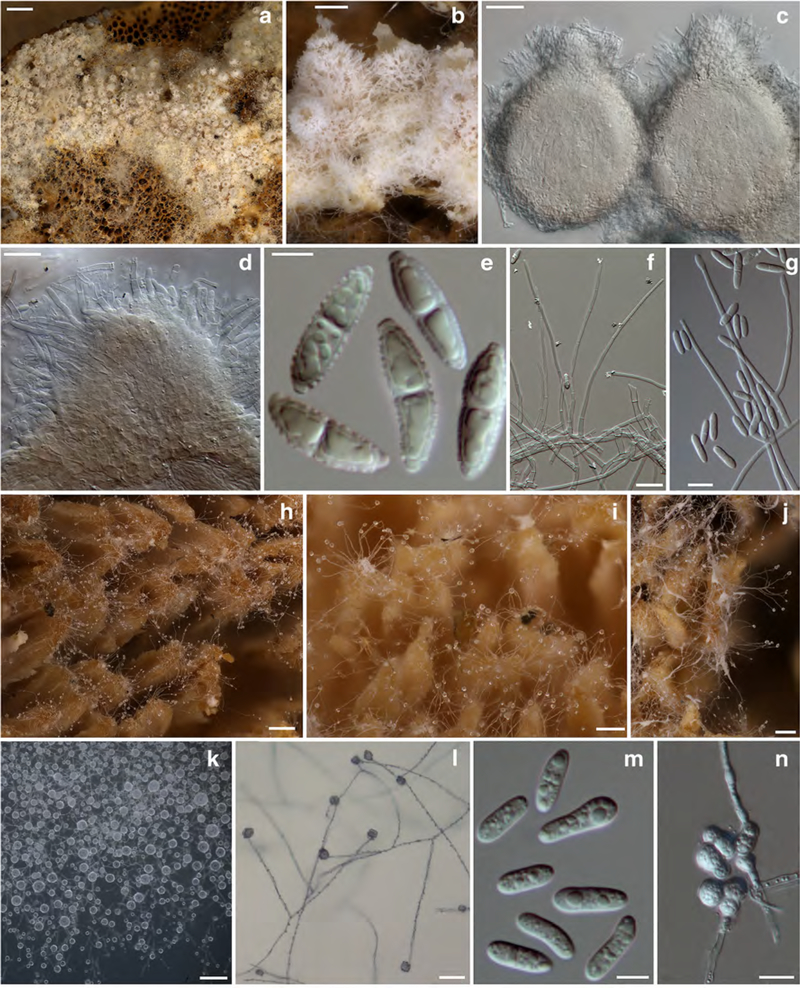
Fig. 5. Sphaerostilbella toxica.
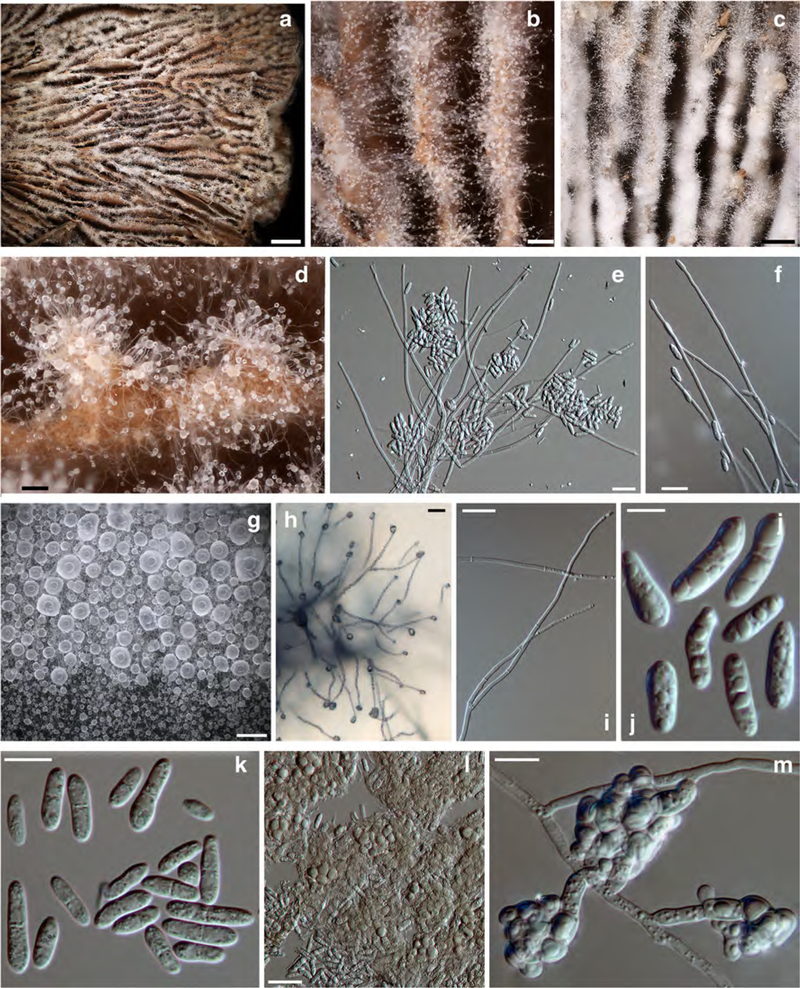
a–d anamorph on host hymenophore; e, f, h, and i conidiogenous branches and conidia; g translucent globose mass of conidia; j, k conidia; l, m clusters of chlamydospores. a, b, d, e, and l holotype TU131905; c, f TU131904; g, h, k, m TFC202061; i, j ex–type culture TFC202258. a–f, and l on the host; h on CMD; g, i–k, m on MEA. Scale bars: a 2 mm; b, d, g 100 μm; c 300 μm; e, h 25 μm; f, i, l 20 μm; j 5 μm; k, m 10 μm
MycoBank: MB 827700
Etymology: In reference to the ability of the isolates to suppress the growth of other fungi and bacteria due to potent antibiotic activity of its organic solvent extracts
Holotype: USA, Texas, Newton County, near Bleakwood, 30.7073° N 93.8269° W, on Gloeophyllum striatum, leg. David P. Lewis, Feb. 2018 (TU131905; ex-type culture TFC202258, Texas Therapeutics Institute DPL-12808). Sequences deposited in GenBank: MH795091 (rDNA ITS and LSU), MH795102 (TEF1)
Colonies effused on host basidiomata forming a palisadelike layer on hymenophore and sometimes also upper surface of pileus, composed of sparse to dense white cottony mycelium with erect branches bearing liquid drops of conidia. Conidiophores/conidiogenous branches arising from hyphae on host hymenophore singly or in small fascicles that are composed of a few adherent, hyaline to pale ochraceous, frequently septate hyphae with cells swollen, 8–20×3.5–5.5. μm, giving frequent lateral branches that terminate in phialides. Conidiogenous branches straight, seldom slightly curved, with up to six septa, terminating in a single phialide. Conidial mass translucent, (sub)globose, 15–30 μm in diam. Conidia oblong, straight, mostly equilateral, (0–)1(–2)–septate, septum median, 8.0–(10.5–11.0)–14.0 × 2.5–(3.1–3.3)–4.0, Q = 2.4–3.3–3.5–4.3 μm, with a flat or truncate base. Chlamydospores abundant among conidiophores and embedded in host tissue, globose, hyaline to pale ochraceous, 4.5–7.5 μm in diam, wall 0.5–1 μm thick, smooth; in irregular clusters of 15–22 μm in diam.
Colonies on MEA reaching 22–25 mm in 7 days; cottony or floccose, white, margin even or crenate; aerial mycelium scarce to abundant, homogenous and compact or concentrically zonate, densely covered by watery droplets containing conidia. Conidiophores indistinct, formed laterally on aerial hyphae at 45–90°, unbranched or with basal part 25–32 × 22.5 μm, that bears further two or three branches; terminal branches with up to seven septa, 50–180 μm long, 1.5–2.5 μm wide near base, tapering to 1.0–2.0 μm at tip, terminating in a single phialide.
Conidia formed in upright position and held in a translucent drop of liquid that can merge with those of neighbouring conidiogenous branches; oblong, ellipsoidal to suballantoid, (0–)1(–2)-septate, lower cell often wider than the upper one, straight or bent in lower part, 7.5–(11.2–11.6)–16.0 × 2.5–(3.4–3.6)–4.5 μm, Q = 2.1–(3.2–3.4)–4.5; base mostly truncate, rarely rounded or with a flat low hilum. Chlamydospores abundant on submerged mycelium, forming irregular clusters, hyaline, subglobose, 5.5–9.0 × 5.5–8.0 μm, wall smooth, ca 0.5 μm thick.
Colonies on CMA reaching 13 mm in 7 days. Conidiogenous structures like those observed on MEA but chlamydospores even more abundant. Conidiophores arising from aerial hyphae, both giving abundant, typically solitary lateral conidiogenous branches; conidia unexceptionally 1-septate, shape variable; chlamydospores formed only among submerged mycelium, in large clusters that fill in most of the agar medium.
Hosts: Gloeophyllum striatum (Fr.) Murrill
Geographical distribution: Southern North America, known only from two localities in eastern Texas, USA
Other specimen examined: USA, Texas, Hardin Co., Saratoga, Lance Rosier Unit, Big Thicket National Preserve, Teel Road area, on Gloeophyllum striatum, leg. Gulf States Mycological Society members, Nov. 12, 2016 (TU131904; culture TFC202061, Texas Therapeutics Institute TTI-0467)
Notes: This new species is morphologically very similar to S. appalachiensis. Like in the latter, the mostly oblong, 1-septate conidia are formed on long septate branches, each of which bears a single phialide. Chlamydospores, produced in both species, are even more abundant in S. toxica, being frequently formed also among conidiophores and tissue of host basidiomata. Despite the lack of distinguishing morphological characters, the isolates of the two species were clearly different in their growth rate and in the capacity of their organic extracts to suppress the growth of human pathogenic fungi and bacteria such as Candida albicans, Cryptococcus neoformans, and Staphylococcus aureus. Only isolates of S. toxica showed the antagonistic effects; the toxic constituents of these strains have undergone preliminary identification and will be characterised in a forthcoming article.
ITS rDNA sequences from the two isolates of S. toxica are identical and differ from those of S. appalachiensis at six positions. However, TEF1 exon and intron regions provide ample characters (127 substitutions and 22 indels) for their distinction (Supplementary Fig. 1). Moreover, the available collections, albeit few, indicate that the two species might have distinct host and geographic ranges. Whereas S. appalachiensis has been collected in eastern USA from New York to Tennessee on unrelated hosts, S. toxica may have a more southern distribution and preference for Gloeophyllum striatum basidiomata. However, more collections are needed to verify that these species have non–overlapping habitats.
Supplementary Material
Acknowledgements
KP and GB are grateful to the late Walter Gams for all his help and guidance in the world of conidial fungi, for sending and commenting on many isolates, and assistance with editing manuscripts. Thanks are also due to Jacques Fournier, Leif Ryvarden, and Oleksandr Akulov for providing specimens for this study and to Vjacheslav Spirin, who checked or provided identifications and discussion on the characters of the hosts. We are grateful to Irma Zettur and Rasmus Puusepp (University of Tartu) for the assistance in the fungal culture and molecular lab, respectively. Two reviewers and the section editor, in particular, are acknowledged for revising the manuscript.
Funding information The study was supported by the Estonian Science Agency (project IUT20–30) and the European Regional Development Fund (Centre of Excellence EcolChange). GB has been supported by University of Texas Health Science Center at Houston new faculty start-up funds, the Kay and Ben Fortson Endowment, and a grant from the NIH (R01GM121458).
Footnotes
Publisher’s Note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s11557–018-01468-w) contains supplementary material, which is available to authorized users.
References
- Arnold G (1963) Einige seltene Pilze auf Hymenomyceten. Z Pilzk 29: 33–36 [Google Scholar]
- Arnold G (1970) Über Hypomyces porotheliiformis und seine Nebenfruchtform. Westfälische Pilzbriefe 8:11–13 [Google Scholar]
- Candoussau F, Magni J–F (1995) New French records. Mycologist 9(1): 12–14 [Google Scholar]
- Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556 [Google Scholar]
- Gams W, van Zaayen A (1982) Contribution to the taxonomy and pathogenicity of fungicolous Verticillium species I: taxonomy. Neth J Plant Pathol 88:57–78 [Google Scholar]
- Helfer W (1991) Pilze auf Pilzfruchtkörpern. Untersuchungen zur Ökologie, Systematik und Chemie. Libri Botanici 1:1–157 [Google Scholar]
- Jaklitsch WM, Komon M, Kubicek CP, Druzhinina IS (2005) Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97(6):1365–1378. 10.1080/15572536.2006.11832743 [DOI] [PubMed] [Google Scholar]
- Jaklitsch WM, Põldmaa K, Samuels (2008) Reconsideration of Protocrea (Hypocreales, Hypocreaceae). Mycologia 100:962–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson-Palme J, Callaghan TM, Douglas B, Drenkhan T, Eberhardt U, Dueñas M, Grebenc T, Griffith GW, Hartmann M, Kirk PM, Kohout P, Larsson E, Lindahl BD, Lücking R, Martín MP, Matheny PB, Nguyen NH, Niskanen T, Oja J, Peay KG, Peintner U, Peterson M, Põldmaa K, Saag L, Saar I, Schüßler A, Scott JA, Senés C, Smith ME, Suija A, Taylor DL, Telleria MT, Weiß M, Larsson K– H (2013) Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology, doi: 10.1111/mec.12481 [DOI] [PubMed] [Google Scholar]
- Larsson A (2014) AliView: a fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30(22):3276–3278. 10.1093/bioinformatics/btu531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC (1998) Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci U S A 95:2044–2049. 10.1073/pnas.95.5.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Põldmaa K (1999) The genus Hypomyces and allied fungicolous fungi in Estonia. I. Species growing on aphyllophoralean basidiomycetes. Folia Cryptogamica Estonica 34:15–31 [Google Scholar]
- Põldmaa K (2000) Generic delimitation of fungicolous Hypocreaceae. Stud Mycol 45:83–94 [Google Scholar]
- Põldmaa K (2011) Tropical species of Cladobotryum and Hypomyces producing red pigments. Stud Mycol 68:1–34. 1e34. 10.3114/sim.2011.68.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Põldmaa K, Larsson E, Köljalg U (1999) Phylogenetic relationships in Hypomyces and allied genera, with emphasis on species growing on wood-decaying homobasidiomycetes. Can J Bot 77:1756–1767 [Google Scholar]
- Põldmaa K, Samuels GJ, Lodge DJ (1997) Three new polyporicolous species of Hypomyces and their Cladobotryum anamorphs. Sydowia 49:80–93 [Google Scholar]
- Põldmaa K, Samuels GJ (1999) Aphyllophoricolous species of Hypomyces with KOH-negative perithecia. Mycologia 91:177–199 [Google Scholar]
- Rehner SA (2001) Primers for Elongation Factor 1-alpha (EF1-alpha). http://www.aftol.org/pdfs/EF1primer.pdf
- Rogerson CT, Samuels GJ (1993) Polyporicolous species of Hypomyces. Mycologia 85:231–272 [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert KA (1985) A monograph of Stilbella and some allied Hyphomycetes. Stud Mycol 27:1–235 [Google Scholar]
- Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L, Jairus T, Horton BM, Abarenkov A, Suvi T, Saar I, Kõljalg U (2008) Strong host preference of ectomycorrhizal fungi in a Tasmanian wet sclerophyll forest as revealed by DNA barcoding and taxon-specific primers. New Phytol 180:479–490. 10.1111/j.1469-8137.2008.02561.x [DOI] [PubMed] [Google Scholar]
- Tulasne LR, Tulasne C (1865) Selecta fungorum carpologia 3. Jusseau, Paris [Google Scholar]
- Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics In: Innis MA, Gelfand H, Sninsky JS, White TJ (eds) PCRprotocols: a guide to methods and applications. Academic Press, San Diego, pp 315–322 [Google Scholar]
- Zare R, Gams W (2016) More white verticillium-like anamorphs with erect conidiophores. Mycol Prog 15:993–1030. 10.1007/s11557-016-1214-8 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.