Abstract
Objective:
Sepsis represents an acute life-threatening disorder resulting from a dysregulated host response. For patients who survive sepsis, there remains long-term consequences, including impaired inflammation, as a result of profound immunosuppression. The mechanisms involved in this long-lasting deficient immune response are poorly defined.
Approach and Results:
Sepsis was induced using the murine model of cecal ligation and puncture (CLP). Following a full recovery period from sepsis physiology, mice were subjected to our wound healing model and wound macrophages (CD11b+,CD3-,CD19-,Ly6G-) were sorted. Post-sepsis mice demonstrated impaired wound healing and decreased reepithelization in comparison to controls. Further, post-sepsis bone marrow-derived macrophages (BMDMs) and wound macrophages exhibited decreased expression of inflammatory cytokines vital for wound repair (IL-1β, IL-12, and IL-23). To evaluate if decreased inflammatory gene expression was secondary to epigenetic modification, we conducted chromatin immunoprecipitation on post-sepsis BMDMs and wound macrophages. This demonstrated decreased expression of Mll1, an epigenetic enzyme, and impaired histone 3 lysine 4 trimethylation (activation mark) at NFkB binding sites on inflammatory gene promoters in BMDMs and wound macrophages from post-CLP mice. Bone marrow transplantation studies demonstrated epigenetic modifications initiate in bone marrow progenitor/stem cells following sepsis resulting in lasting impairment in peripheral macrophage function. Importantly, human peripheral blood leukocytes from post-septic patients demonstrate a significant reduction in MLL1 compared with non-septic controls.
Conclusion:
These data demonstrate that severe sepsis induces stable MLL1-mediated epigenetic modifications in the bone marrow, that are passed to peripheral macrophages resulting in impaired macrophage function and deficient wound healing persisting long after sepsis recovery.
Keywords: Sepsis, macrophages, epigenetics
Subject Codes: Inflammation, Vascular Biology, Epigenetics
INTRODUCTION
Sepsis is a life-threatening organ dysfunction that is caused by a dysregulated host response to infection, which despite advancements in medical therapy continues to have a high morbidity and mortality 1,2. This dysregulated systemic inflammatory response can result in hypotension, organ failure, and death. Beyond the acute pathophysiological insult, there remains additional insidious long-term consequences of patients who survive severe sepsis. In the late phases of sepsis, an output of anti-inflammatory cytokines occurs in an attempt to restrict the damage of the inflammatory reaction 3,4. This latter reaction counteracts the host’s initial response and can lead to a systemic state of immune tolerance. This so-called “sepsis-induced immunosuppression” can persist for years, rendering patients susceptible to secondary complications 1,2,5,6. Indeed, survivors of severe sepsis have a significant reduction in 5- and 8-year life expectancy compared to the unaffected age-matched population 7,8. These patients exhibit increases in a wide range of negative health outcomes such as the occurrence of cancer, infections, and impaired wound healing 9-11. Complications involving impaired tissue repair in post-septic patients include; anastomotic leaks, fascial dehiscence, and infections 12. Despite this recognized significant clinical problem, the molecular mechanism(s) that underlie impaired inflammatory response and dysfunctional healing following recovery from sepsis have not been elucidated.
A common characteristic of poorly healing wounds is an impaired initial immune response to injury. In normal wound healing, myeloid cells are mobilized from the bone marrow (BM) into circulation and migrate into peripheral tissues where they differentiate into macrophages, a key immune cell that drives wound inflammation and promotes tissue repair. This early innate inflammatory response is critical for establishing the healing cascade 13-15. During the first part of the inflammatory phase of wound healing, macrophages exist in an inflammatory phenotype where they release inflammatory cytokines and mediators, recruit additional leukocytes, and promote tissue and pathogen destruction 13. After this early inflammatory phase, macrophages undergo a phenotypic switch and begin secreting anti-inflammatory mediators as well as growth factors to promote wound resolution. Prior studies in murine models have demonstrated a decrease serum level of inflammatory cytokines during sepsis recovery 16. The ability to fully understand and control the initiation and resolution of inflammation in wound macrophages is critical to advancing the field of wound repair. At present, what specifically drives changes in wound macrophage phenotype throughout the course of healing is unknown. Thus, the examination of the molecular mechanisms underlying macrophage plasticity in wounds is necessary to address the pathologically impaired tissue repair observed following recovery from sepsis.
Evidence suggests that epigenetic regulation (e.g., DNA methylation, histone modification) of gene expression plays a key role in influencing immune cell phenotypes 17,18. Among various histone modifications, methylation of histone H3 at lysine-4 (H3K4) and at lysine-27 (H3K27), are highly correlated with transcriptional activation19 and repression,20 respectively. Interestingly, aberrant histone methylation patterns, specifically H3K9 demethylation, in human patients with malignancy and active sepsis is seen, but the impact, both acute and long-term, of H3K4 modification remains undefined 21,22. Mixed-lineage leukemia 1 (MLL1) is a histone methyltransferase with site specificity for H3K4 and has been shown to regulate cytokine expression in diabetic wound healing 23,24. Our laboratory has previously demonstrated decreased H3K4 methylation at the promoter region of the Il12 gene in lung dendritic cells in a murine model of acute sepsis 25. However, the specific contributions of epigenetic-based mechanisms on the long-term regulation of macrophage phenotypes and inflammation following sepsis recovery remain poorly defined.
Given the importance of epigenetic modification on immune cell phenotype, we investigated the role of histone methylation in directing macrophage-mediated inflammation following sepsis. Using both a murine model of sepsis and cutaneous wound healing as well as human peripheral blood leukocytes, we demonstrate that long after recovery from sepsis there remains impairment in wound healing and decreased macrophage inflammatory cytokine production in response to tissue injury. Further, this aberrant macrophage phenotype following sepsis is directed by decreased expression of the epigenetic enzyme, MLL1, and its associated histone activation mark on the NFκB binding site in promoters of key inflammatory genes critical to tissue repair. Lastly through BM transplantation experiments we show that these epigenetic modifications are set in BM myeloid progenitor cells during the initial septic insult and result in lasting impairment in peripheral macrophage function leading to dysfunctional tissue repair.
MATERIALS AND METHODS
Data available on request from the authors.
Mice
Mice were maintained in the University of Michigan pathogen-free animal facility, and all protocols were approved by and in accordance with the guidelines established by the Institutional Animal Care and Use Committee (UCUCA). Male and female mice were used for murine sepsis and wound curve experiments as detailed in the ATVB council statement 26. Mouse strains include: C57BL/6, CD45.1, and CD45.2 mice maintained on a normal diet (ND) (13.5% kcal fat; LabDiet 5001) were purchased at 10 weeks from The Jackson Laboratory (Bar Harbor, ME). Animals were housed in a barrier facility on a light:dark cycle of 14 : 10 hours (ambient temperature of 22°C) with free access to water, food (Lab diet 5001), and bedding (Andersons Lab Bedding Bed o’Cobs combo). Animals underwent all procedures at 10-14 weeks of age. Body weights were determined prior to experimentation.
Cecal Ligation and Puncture Model
To induce sepsis, the cecal ligation and puncture (CLP) model was used as previously described 27. Briefly, mice were anesthetized with ketamine and xylazine and a 1 cm midline incision was made on the anterior abdomen. The cecum was exposed and ligated below the ileocecal junction without causing total bowel obstruction. Using a 21-gauge needle, the cecum was punctured. Following this, the cecum was returned to the abdominal cavity, and the peritoneal wall and skin incision were closed with a surgical 9 mm clip. All animals received 1 mL of sterile isotonic saline subcutaneous immediately after surgery. Sham-operated animals (sham) underwent identical laparotomy but without cecal ligation or puncture. All mice undergoing CLP developed early clinical signs of sepsis, including lethargy, piloerection, and tachypnea. They were analyzed daily for 14 days to access survival rate. The average mortality rate for mice subjected to CLP in this study was 40–60% by day 4 after surgery. For experiments mice were either sacrificed during the acute sepsis physiology (days 1–7) to determine the impact of septic physiology on bone marrow function or mice were allowed to recovery for 4 weeks (post-sepsis) to determine the long-term immunological impact of sepsis survival. After 4 weeks of recovery for the post-sepsis mice, these animals underwent cutaneous wounding as described below.
Wound Healing Assessment
Before wounding, mice were anesthetized, hair was removed with Veet (Reckitt Benckiser), and skin was cleaned with sterile water. Full-thickness back wounds were created by 4-mm punch biopsy. Initial wound surface area was recorded and digital photographs were obtained daily using an Olympus digital camera. Photographs contained an internal scale to allow for standard measurement calibration. Wound area was quantified using ImageJ software (National Institutes of Health, Bethesda, MD) and was expressed as the percentage of original wound size over time. Data are representative of 5 independent experiments.
Histology/Immunofluorescence
For wound histology, whole wounds were excised using a 6mm punch biopsy on day 3 post-wounding. Wound sections were fixed in 10% formalin overnight before embedding in paraffin. 5 μm sections were stained with hematoxylin and eosin for evaluation of re-epithelialization and with Masson’s Trichrome stain for collagen deposition. Images were captured using Olympus BX43 microscope and Olympus cell Sens Dimension software. Percent re-epithelialization was calculated by measuring distance traveled by epithelial tongues on both sides of wound divided by total distance needed for full re-epithelialization. For bone marrow histology, the femur and tibia were isolated at day 3 and 7 after CLP or sham procedure and fixed overnight in 3% paraformaldehyde. Bones were then decalcified in PBS + 10% EDTA (weight/volume)(pH 7.5) for 3 days following which they were embedded in paraffin. 5μm sections were stained with hematoxylin and eosin for evaluation. Slides containing 5 μm sections were deparaffinized and hydrated by incubating them in two changes of xylene for five min each, followed by 2 changes of 100% ethanol for 3 min each, 70% ethanol for 2 min, 50% ethanol for 2 min, and distilled water for 5 min. Antigen retrieval was performed by incubating the slides in 10 mM Citric acid solution (pH 6.0) in an 80 ºC oven overnight. For immunofluorescence, the slides were subsequently washed in PBS and permeabilized in 10% methanol containing 0.4% H2O2 for 30 min. After permeabilization, Fc receptors were blocked with a 2.4G2 mAb for 30 min at room temperature. The slides were stained with primary antibody, H3K4me3 (ab8580; 1.25 μg/ml), overnight at 4°C followed by a second wash with appropriated secondary (Alexa-Fluor) antibody for 1h at room temperature. Slides were then mounted using Prolong Gold Antifade Reagent with DAPI (Invitrogen). Data are representative of 2–3 independent experiments. Analysis of sections was obtained using a Nikon A1 confocal microscope system (Nikon Instruments) and Olympus BX43 Fluorescence microscope (Cell Sens Software).
Wound Digestion
Following sacrifice, wounds were collected from the backs of the mice postmortem following CO2 asphyxiation using a 6 mm wound biopsy. Sharp scissors were used to excise the full thickness dermis with a 1–2mm margin around the wound ensuring collection of granulation tissue and wounds were placed in RPMI. Wounds were then carefully minced with sharp scissors and digested by incubating in a 50 mg/ml Liberase TM (Roche) and 20U/ml DNaseI (Sigma-Aldrich) solution. Wound cell suspensions were then gently plunged and filtered through a 100μm filter to yield a single cell suspension. Cells were then either magnetic-activated cell sorted (MACs) for CD3-, CD19-, Ly6G-, CD11b+ cells for RNA studies or cultured ex-vivo for application of GolgiStop and subsequent staining for intracellular flow cytometry 28.
Wound Macrophage Isolation and Magnetic-Activated Cell Sorting
Wounds were digested as described above. Single cell suspensions were incubated with fluorescein isothiocyanate–labeled anti-CD3, anti-CD19, and anti-Ly6G (BioLegend) followed by anti–fluorescein isothiocyanate microbeads (Miltenyi Biotec). Flow-through was then incubated with anti-CD11b microbeads (Miltenyi Biotec) to isolate the non-neutrophil, non-lymphocyte, CD11b+ cells. Cells were saved in Trizol (Invitrogen) for quantitative RT-PCR analyses. For bone marrow sorting, bone marrow (lineage-/c-kit+) cells were isolated using MACs Lineage Cell Depletion Kit (Miltenyi Biotec, 130–090-858) followed by anti-CD117 microbeads (Miltenyi Biotec, 130–091-224). Cells were saved in Trizol (Invitrogen) for quantitative RT-PCR analyses
Cell Culture and Cytokine Analysis
Bone marrow (BM) cells were collected by flushing mouse femurs and tibias with RPMI. BM-derived Mϕs (BMDMs) were cultured as previously detailed 29. On day 6, the cells were replated, and after resting for 24 h, they were incubated with or without LPS (100 ng/mL) for 2–6 hours after which cells were fixed in paraformaldehyde for ChIP analysis or placed in Trizol (Invitrogen) for RNA analysis.
Colony Forming Unit Assays
2–6 mL of bone marrow was collected in sterile, preservative-free heparin tubes. CFU-C assays were performed using the MethoCultTM M3231 (STEMCELL Technologies, Vancouver, Canada.). Briefly, 1×10E+4 viable bone marrow cells were plated per 6 well plate in 2 mL of media with 10 ng/mL of GMCSF. The cells were dispersed evenly through the media by a syringe and vortexing. Cells were grown for 10 days at 37°C with 5% CO2 and >85% humidity and then counted for colonies. Data are representative of 2 independent experiments.
ChIP Assay
Chromatin immunoprecipitation (ChIP) assay was performed as described previously 21. Briefly, cells fixed in paraformaldehyde were lysed and sonicated to generate 100–300bp fragments. To immunoprecipitate, samples were incubated in anti-H3K4trimethyl antibody (Abcam, ab8580), anti-NFκB (active motif, 40916) or isotype control (rabbit polyclonal IgG) (Millipore) in parallel samples overnight followed by addition of protein A Sepharose beads (Thermo-Fisher). Bound DNA was eluted and purified using Phenol:Chloroform:Isoamyl alcohol extraction and ethanol precipitation. Primers were designed using the Ensembl genome browser to search the IL1β, IL12, and IL23 promoter for NFκB within the promoter region and then NCBI Primer-BLAST was used to design primers that flank this site. Data are representative of 2–3 independent experiments. Primer sequences are available in Supplemental Table I.
Flow Cytometry/Fluorescent Activated Cell Sorting
For surface staining, wound cell isolates were collected directly from wounds. Cells were stained with a Fixable LIVE/DEAD viability dye (Molecular Probes by Life Technologies; Ref No. L34959; 1.0 μg/ml). FcR-receptors were then blocked with anti-CD16/32 (BioXCell, Cat No. CUS-HB-197, 2.5 μg/ml) for 10 minutes. Monoclonal antibodies for surface staining included: Anti-CD3 (Biolegend, Cat No. 100204, 1.25 μg/ml), Anti-CD19 (Biolegend, Cat No. 152404, 1.25 μg/ml), Anti-Ter-119 (Biolegend, Cat No. 116206, 1.25 μg/ml), Anti-NK1.1 (Cat No. 108706, 1.25 μg/ml), Anti-Ly6G (Biolegend, Cat No. 127612, 1.25 μg/ml), Anti-CD11b (Biolegend, Cat No. 101226, 1.25 μg/ml), and Anti-Ly6C (Biolegend, Cat No. 128008, 1.25 μg/ml). After surface staining, cells were washed twice, and biotinylated antibodies were labeled with streptavidin-fluorophore (Biolegend, Cat No. 405208, 1.0 μg/ml). Next, cells were either washed and acquired for surface-only flow cytometry or were fixed with 2% formaldehyde and then washed/permeabilized with BD Perm/wash buffer (BD Biosciences, Ref No. 00–8333-56) for intracellular flow cytometry. After permeabilization, intracellular stains included: anti-IL-1β-Pro-PE Cy7 (eBioscience, Ref No. 25–7114-82, 2.5 μg/ml) and anti-tumor necrosis factor (TNF)-α-APC (Biolegend, Cat No. 506308, 2.5 μg/ml). For peripheral blood flow analysis, peripheral blood was obtained at the time of animal sacrifice. Cells were stained with a Fixable LIVE/DEAD viability dye (Molecular Probes by Life Technologies; Ref No. L34959; 1.0 μg/ml). FcR-receptors were then blocked with anti-CD16/32 (BioXCell, Cat No. CUS-HB-197, 2.5 μg/ml) for 10 minutes. Monoclonal antibodies for surface staining included: Anti-CD45.1 (eBioscience, Ref No. 25–0453-82, 1.25 μg/ml) and Anti-CD45.2 (Biolegend, Ref No. 17–0454-82, 1.25 μg/ml). For bone marrow progenitor surface staining, single cell suspensions were collected and washed two times with cold PBS and filtered into a 96-well plate for surface staining. Cells were initially stained with pacific orange LIVE/DEAD fixable viability dye (Thermofisher) and then washed two times with cold PBS. Cells were then resuspended in Flow Buffer (PBS, FBS, NaN3, and Hepes Buffer) and Fc-Receptors were blocked with anti-CD16/32 (Biolegend) prior to surface staining for hematopoietic stem cell cohort and stained with anti-CD16/32-PE (Biolegend) for myeloid progenitor cells. Monoclonal antibodies for surface staining included: Anti-CD3 (Biolegend, Cat No. 100304, 1.25 μg/ml), Anti-Ter119 (Biolegend, Cat No.116204, 2.5 μg/ml), Anti-GR.1 (Biolegend, Cat No.108404, 2.5 μg/ml), Anti-B220 (Biolegend, Cat No.103204, 2.5 μg/ml), Anti-cKit (Biolegend, Cat No. 105812, 2.5 μg/ml), Anti-Sca (eBiosciences, Cat No.56–5981-82, 2.5 μg/ml), Anti-FcgRIII (Biolegend, Cat No.101308, 2.5 μg/ml), Anti-CD41 (Biolenged, Cat No. 303713, 1.25 μg/ml), Anti-Flt3 (Biolegend, Cat No.1035405, 2.5 μg/ml), Anti-CD48 (Biolenged, Cat No. 103417, 1.25 μg/ml), Anti-CD105 (Biolegend, Cat No.120410, 1.0 μg/ml), and Anti-CD150 (Biolegend, Cat No.115922, 2.5 μg/ml). Following surface staining, cells were washed twice, and biotinylated antibodies were labeled with streptavidin APC-Cy7 or streptavidin Pacific Orange. Samples were acquired on a 3-Laser Novocyte Flow Cytometer (Acea Biosciences) or fluorescent activated cell sorting (FACS) sorted on a FACsAria III Flow Sorter. FACS was performed with FACsDiva Software (BD Biosciences), analysis was performed using FlowJo software version 10.0 (Tree Star), and data were compiled using Prism software (GraphPad). Data are representative of 2–3 independent experiments. All populations were routinely backgated to verify gating and purity.
RNA Analysis
Total RNA extraction was performed using Trizol (Invitrogen) or Trizol LS (for human samples) according to manufacturer’s instructions. RNA was then reversed transcribed to cDNA using iScript (Biorad). PCR was performed with 2X Taqman PCR mix using the 7500 Real-Time PCR System. Primers for Arg1 (Mm00475988), Col1a1 (01302050_m1), Col1a3 (01254476_m1), Il-1β (Mm00434228_m1), Il-12 (Mm00434165_m1, Mm01288992_m1), Il-23 (Mm00518984_m1), Mll1(Mm01179235_m1), Nos2 (Mm00440502_m1), Tgfb1 (00441724_m1), Tnf⍺ (Mm00443258_m1), human IL-1β (Hs00233688_m1), human IL-23 (Hs00372324_m1) and human MLL1 (Hs00610538_m1) were purchased (Applied Biosystems). 18S or GAPDH was used as the internal control. Data were then analyzed relative to 18s ribosomal RNA or GAPDH (2ΔCt). All samples were assayed in triplicate. The threshold cycle values were used to plot a standard curve. Data are representative of 2–3 independent experiments were compiled in Microsoft Excel and presented using Prism software (GraphPad).
IL-1β ELISA
Mouse IL-1β concentration was measured by the Quantikine mouse IL-1β ELISA kit (R&D Systems DY401) per the manufacturer’s protocol. Color intensity was measured at 450 nm. The ELISA kit has a detection limit of 15.6 pg/mL.
Bone Marrow Transplant
Recipient mice underwent CLP procedure and then were allowed to recover for 4 weeks. Following the 4 week recovery period, recipient mice received 8.0 Gy TBI (orthovoltage X-ray source) split in two fractions, 3 h apart. Donor mice were randomized to undergo either sham or CLP procedure and then were allowed to recover for 4 weeks. Following the 4 week recovery period, bone marrow cells were harvested from donor mice and resuspended in serum-free medium (DMEM, 0.1% BSA,1% penicillin-streptomycin, 1% L-glutamine, and 0.1% amphotericin B). Bone marrow cells (5 × 106) were administered by tail vein injection into TBI recipient mice. All experiments with BMT mice were performed 5–6 wk post-BMT when mice were fully donor-cell reconstituted. Spleen cells were >87% donor derived at this time point 30. Data are representative of 2 independent experiments.
Human Peripheral Blood Analysis
Human peripheral blood leukocytes were obtained from patients enrolled in the Michigan Genomics Institute under the direction of institutional review board protocol (HUM00126299). The electronic medical record was reviewed to determine patient samples who satisfied an inclusion criteria of a patient with history of sepsis as defined by Sepsis-3 criteria with a Sequential Organ Failure Assessment (SOFA) score ≥2 and a documented urinary tract infection 31. Of note, electronic medical record-based case definitions using the sepsis criteria have been found to be more accurate than the use of claims based data 32. Demographic and clinical data can be found in Supplemental Table II. The average days of patient recovery since last documented SOFA score ≥2 was 1280 days (range 14–2431 days). Once our sepsis patient cohort was obtained, patients were then matched based on age, sex, and medical history to blood samples that had no documented history of sepsis physiology to obtain the control population. For both patient cohorts, exclusion criteria included patients <18 years of age and those with a documented history of leukemia and/or lymphoma given the potential confounding impact of leukemia/lymphoma on the myeloid cell population.
Statistical Analysis
Data were analyzed using GraphPad Prism software version 6. We expressed the results as means ±SEM. Shapiro-Wilk test was used to determine normality of data and Brown-Forsythe or F test to determine equality of variances, and then a parametric or nonparametric test was performed accordingly. Parametric statistical analysis was performed using unpaired Student’s t test (2 tailed) between 2 groups and 1-way ANOVA followed by post hoc analysis (Bonferroni, Dunnett, or Newman-Keuls Multiple Comparison test) for analysis of differences between >2 groups. Nonparametric statistical analysis was performed using Mann-Whitney U test. For human peripheral blood leukocytes, statistical significance in expression of MLL1 was determined using conditional logistic regression. This was performed using a Cox Regression analysis with Time Dependent Covariates test in SPSS v25.0. Individuals diagnosed with a SOFA score of 2 or greater were paired based on age, sex, and medical history with an individual without a diagnosis of sepsis based on SOFA score using the information available in the database. P values <0.05 were considered significant.
Study approval
Animal experiments were conducted following the NIH guidelines and were approved by the IACUC of the University of Michigan. Human blood specimens were collected with the approval of the institutional review committee of University of Michigan Medical Center. All patients gave written informed consent.
RESULTS
Cutaneous Wound Healing is Impaired Following Recovery from Sepsis and Wound Macrophages Demonstrate Decreased Inflammatory Cytokine Expression
Previous human and murine studies have demonstrated innate immune dysfunction occurs following sepsis, however the molecular mechanisms remain poorly defined 3,6,33-35. Increasing evidence suggests that proper wound healing requires the establishment of a regulated inflammatory response following tissue injury 13,36,37. Therefore, we examined the impact of sepsis survival on wound repair. Polymicrobial abdominal sepsis was induced in 10–14 week old C57BL/6 mice by cecal ligation and puncture. No statistical differences were found between groups with respect to sex. Consistent with previous studies, CLP had a mortality of 40–60% secondary to septic physiology 27. Following full recovery from sepsis, post-septic and control mice were subjected to 4-mm full thickness wounds as previously described 36. Wound repair was monitored daily and we identified that post-septic mice had impaired wound healing throughout the entirety of the wound course (Figure 1A). Histological assessment showed that the wounds from post-septic mice had less reepithelialization (Figure 1B).
Figure 1. Cutaneous Wound Healing is Impaired Following Recovery from Sepsis and Wound Macrophages Demonstrate Decreased Inflammatory Cytokine Expression.
A: Wounds were created by 4-mm punch biopsy on the backs of post-sepsis or Sham mice 4 weeks after recovery. The change in wound area was recorded daily with Image J software until complete healing was observed. Representative photographs of the wounds of post-sepsis and sham mice are shown on days 0 and 4 are shown (n=20/group, repeated 2×). *p<0.05 by Mann-Whitney U test. B: Wounds were harvested on day 3, paraffin embedded and sectioned. 5 µM sections were stained with hematoxylin and eosin. Percent re-epithelialization was calculated by measuring distance traveled by epithelial tongues on both sides of wound divided by total distance for full re-epithelialization. Representative images are shown in 2× magnification (n=5/group). Black bar over tissue represents overall distance of the wound, arrowheads identify epithelial tongue ingrowth, scale bar equals 300 μm. *p<0.05 by Student t test with Welch’s correction. C: Wound myeloid cells CD11b+[CD3−CD19−Ly6G−] were isolated from post-sepsis and control mice. Gene expression of Il1b, Nos2, Il23, Il12, and Arginase 1 was quantified using qPCR (n=5/group). **p<0.01 and ***p<0.001 by Student t test with Welch’s correction. D: Post-sepsis and control wound cell isolates were processed for intracellular flow cytometry. The gating strategy used for intracellular flow cytometry selecting live, lineage−, Ly6G−, CD11b+ cells is shown. Flow cytometry quantification of IL1β+ cells (% of Live, Lineage−, Ly6G−, CD11b+, Ly6CHi Cells) in wounds (n = 10/group). *p<0.05 by Student t test with Welch’s correction. E: Levels of IL1β in supernatant of wound macrophages from DIO and control mice following treatment with LPS for 24 hours measured by ELISA (n = 3/group). **p<0.01 by Student t test with Welch’s correction. FMO, fluorescence minus one; FSC, forward scatter; FSC-A, forward scatter area; FSC-H, forward scatter height; SSC, side scatter.
Since progression through the initial inflammatory phase of wound healing is largely directed by macrophage plasticity, we theorized that the impaired wound healing observed in post-septic mice was secondary to macrophage dysfunction. To study this, in vivo wound monocyte/macrophages (CD11b+[CD3-CD19-Ly6G-]) were sorted from wounds of post-septic and control mice at day 3 post injury as we have previously described 38,39. Resident F4/80 macrophages are known to be a minor percentage of the monocyte/macrophage population during tissue repair and were thereby not separately analyzed (Supplemental Figure IA). Expression of inflammatory (Il1b, Nos2, Il12, and Il23) and anti-inflammatory (Arginase1 and Tgfβ1) cytokines known to play a major role in healing were examined. Inflammatory cytokine levels were markedly reduced in post-sepsis wound macrophages with a reciprocal increase in anti-inflammatory cytokine levels (Figure 1C and Supplemental Figure IB-D). To investigate if these altered inflammatory transcript levels correlated with decreased cytokine production by wound monocyte/macrophages, we performed flow cytometry on day 3 wounds and found that the monocyte/macrophages (live, lineage−, Ly6G−, CD11b+, Ly6CHi) from post-septic wounds made significantly less IL-1β than controls (Figure 1D). This was further supported with ELISA analysis of IL1β levels demonstrating a significant reduction in cytokine production in post-septic wound macrophages (Figure 1E). We examined day 3 post-wounding to allow circulating monocytes adequate time to enter the tissues, transform into macrophages, and assume a functional role 40,41.
BM Stem/Progenitor Cells from Post-Septic Mice Demonstrate Reduced MLL1 and Decreased H3K4me3 at NFκB Inflammatory Gene Promoters.
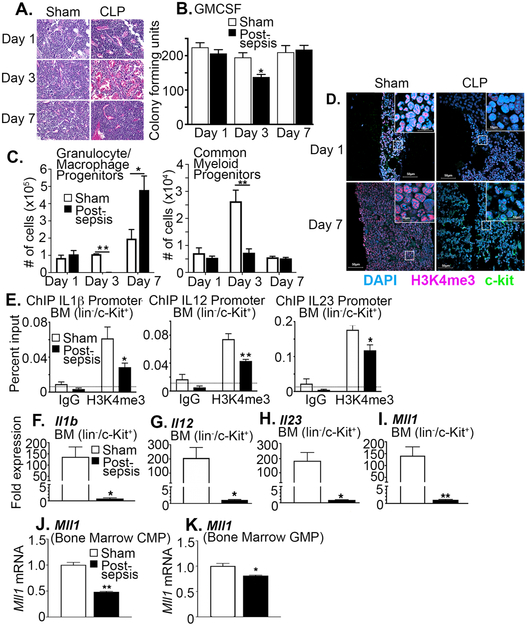
Peripheral tissue monocyte/macrophages arise from myeloid progenitors in the BM and differentiate in response to stimuli. Previous studies have demonstrated that systemic pathological conditions, such as chronic hyperglycemia, can alter bone marrow progenitor cells 3,36. To determine if altered bone marrow progenitor cells impact the peripheral monocyte/macrophage phenotype during and following sepsis, we first analyzed BM stem/progenitor cells during acute sepsis physiology. Histological analysis demonstrated that immediately following CLP during a period of active sepsis there is a reduction in bone marrow progenitor cells at day 3 in comparison to sham operated controls (Figure 2A). Further, BM myeloid cells exhibited reduced growth potential immediately after sepsis; however, by day 7 after sepsis this was restored (Figure 2A, B). Analysis of progenitor subtypes within the bone marrow demonstrated that at day 3 following sepsis induction, post-sepsis bone marrow had a reduction in granulocyte/monocyte progenitors as well as common myeloid progenitors. This was normalized by day 7, thereby suggesting that a prolonged reduction in BM myeloid progenitor cell growth or progenitor subtypes in septic mice is unlikely to explain impaired immunological function after recovery (Figure 2C and Supplemental Figure IIA-G).
Figure 2. BM Stem/Progenitor Cells from Post-Septic Mice Demonstrate Decreased H3K4me3 at NFκB Inflammatory Gene Promoters.
A: BM was isolated on days 1, 3, and 7 following induction of CLP or sham surgery paraffin embedded and sectioned. 5 µM sections were stained with hematoxylin and eosin to analyze cellularity. Representative images are shown in 40×. B: BM growth was accessed by colony forming unit assays on days 1, 3, and 7 in post-sepsis and sham mice (n = 5/group, repeated 3 times). *p<0.05 by Mann-Whitney U test. C: Post-sepsis and sham bone marrow was processed for flow cytometry to identify granulocyte/macrophage and common myeloid progenitor populations at day 1, 3, and 7 after surgical procedure. The gating strategy can be viewed in Supplemental Figure II. *p<0.05 and **p<0.01 Mann-Whitney U test. D: Immunofluorescence was performed on days 1 and 7 in post-sepsis and sham bone marrow to analyze H3K4me3 expression. Representative images are shown in 10× and 200× magnification. E: BM stem/progenitor cells (lin−/c-Kit+) were isolated in vivo using MACS and analyzed by ChIP for H3K4me3 levels on the promoter of Il1b, Il12 and Il23 (n = 3/group, plated in triplicate). *p<0.05 by ANOVA test with Bonferroni pos hoc test. F, G, H: BM stem/progenitor cells (lin−/c-Kit+) were isolated using MACS isolation and gene expression of Il1b, Il12 and Il23 was quantified by qPCR (n = 5/group). *p<0.05 by Student t test with Welch’s correction. I: BM stem/progenitor cells (lin−/c-Kit+) were isolated using MACS isolation and gene expression of Mll1 was quantified by qPCR (n = 5/group). **p<0.01 by Student t test with Welch’s correction J-K: Bone marrow common myeloid progenitors (CMP) and granulocyte/macrophage progenitors (GMP) were isolated using FACs and gene expression of Mll1 was quantified by qPCR (n = 5/group). *p<0.05 by Student t test with Welch’s correction. Data are presented as the mean±SEM. Data are representative of 2-3 independent experiments.
Previous studies have demonstrated that epigenetic reprograming, most notably histone methylation, is a key mechanism that can regulate transcriptional reprogramming during sepsis 42,43. To determine if epigenetic mechanisms impact BM stem/progenitor cells during sepsis we analyzed histone modifications and found that H3K4 trimethylation (H3K4me3) was significantly reduced in post-sepsis BM on day 3 and remained depressed at day 7 even after restoration of absolute BM cellularity (Figure 2D).
We next assessed if this alteration in the H3K4me3 was preserved over time in animals surviving experimental sepsis. In this set of studies, we analyzed BM from post-sepsis mice after 4 weeks of recovery using chromatin immunoprecipitation (ChIP) analysis. No difference in weight or behavior was observed between the 2 groups of mice at this time point. We identified that H3K4me3 and NFκB binding remained significantly decreased on the Il1b, Il12, and Il23 gene promoters in the BM stem/progenitor cells in comparison to controls (Figure 2E and Supplemental Figure IIH). Consistent with the removal of an activating epigenetic mark, post-septic BM stem/progenitor cells exhibited a significant decrease in inflammatory mediator production (Figure 2F-H). The epigenetic enzyme, MLL1, has site specificity for H3K4 and since we identified that H3K4me3 was decreased in post-septic BM myeloid cells and we have previously published that MLL1 plays a vital role in regulating macrophage phenotype 44-46, we analyzed Mll1 expression. In BM stem/progenitor cells (including lineage-/c-kit+ cells, common myeloid progenitors, and granulocyte-macrophage progenitors) we found a significant decrease in Mll1 expression in post-septic mice compared to controls (Figure 2I-K). Collectively, severe sepsis results in decreased H3K4me3 on inflammatory cytokine promoters in BM stem/progenitor cells that is mediated by reduced Mll1 expression and H3K4me3 which is maintained long-term following sepsis recovery.
Human Peripheral Blood from Recovered Post-Septic Patients Demonstrates Decreased MLL1
To evaluate whether our findings are translatable to humans, we isolated peripheral blood leukocytes from recovered post-septic patients and age matched controls. MLL1 expression was significantly decreased in the peripheral blood of post-septic patients in comparison to aged matched controls who had no documented history of sepsis (Figure 3A). We did not see a difference in inflammatory cytokine expression, likely secondary to analysis of human peripheral blood with different cell types having variable chromatin structure and inflammatory cytokine expression (Supplemental Figure IIIA). However, the reduction in MLL1 expression highlights the fact that this change likely happens in the bone marrow, and bone marrow derived cells maintain a low level of expression. Additionally, we found the reduction in MLL1 expression was independent of patient age or time since sepsis (Supplemental Figure IIIB-D). This may indicate that in patients that survive sepsis, an absolute reduction in MLL1 expression and the decrease in its activating histone mark drives the myeloid phenotype in peripheral tissues and promotes an aberrant response to further injury.
Figure 3. Human Peripheral Blood from Recovered Post-Septic Patients Demonstrates Decreased MLL1.
A: Peripheral blood leukocytes (30 mL) were collected from post-sepsis patients and control subjects without documented history of sepsis. The average time of recovery since sepsis was 1280 days (range 14-2431 days). No statistical differences were found between groups with respect to sex, age, or comorbid conditions. MLL1 gene expression was measured by qPCR (n = 33 patients in post-sepsis cohort and 61 patients in control). Data are presented as the mean±SEM. *p<0.05. For human peripheral blood leukocytes Cox regression analysis with a fixed covariate (sepsis vs non-septic) was used in which gene expression data from each septic donor was paired with expression data from a healthy age and sex matched control.
Wound Monocyte/Macrophages from Post-Septic Mice Demonstrate Decreased MLL1 and H3K4me3 at NFκB Binding Sites on Inflammatory Gene Promoters
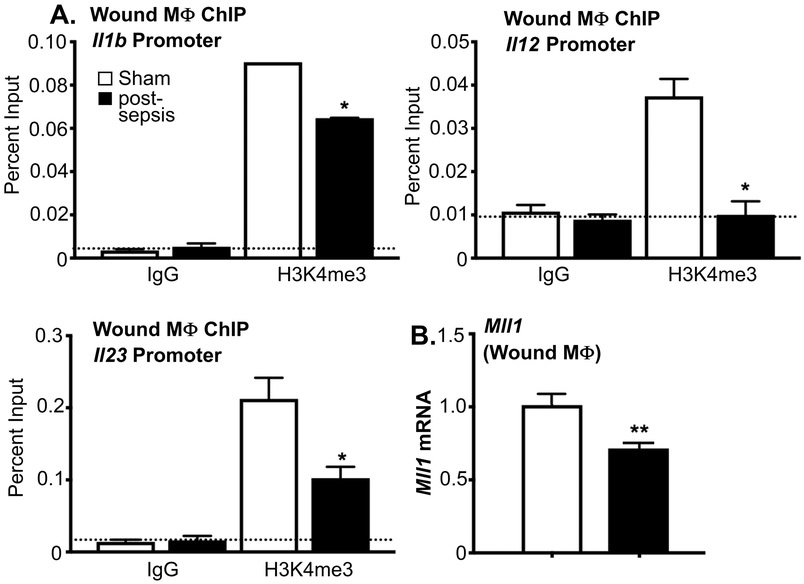
Given the long-term persistence of epigenetic modifications in the BM progenitor/stem cells following recovery from sepsis, we next sought to determine the impact of these BM changes on peripheral monocytes/macrophages. As such, in vivo wound monocyte/macrophages (CD11b+[CD3-CD19-Ly6G-]) were sorted from wounds of post-septic and control mice on day 3 post injury and a ChIP analysis was performed at the NFkB binding sites on the Il1b, Il12, and Il23 promoters. NFkB binding sites on Il1b, Il12, and Il23 promoters demonstrated a significant decrease in H3K4me3 in the post-septic wound macrophages compared with controls (Figure 4A). In addition, Mll1 was found to be decreased in post-septic wound macrophages (CD11b+[CD3-CD19-Ly6G-]) in comparison to controls (Figure 4B). No difference in Mll1 expression was noted in wound F4/80+ resident cells (Supplemental Figure IV). These findings suggest that decreased Mll1 and absence of the activating histone methylation mark, H3K4me3, on inflammatory gene promoters in recovered post-septic mice is a direct result of the changes in MLL1 in the BM stem/progenitor cells.
Figure 4. Wound Monocyte/Macrophages from Post-Septic Mice Demonstrate Decreased MLL1 and H3K4me3 at NFκB Binding Sites on Inflammatory Gene Promoters.
A: Wound myeloid cells were isolated at day 3 postinjury from post-sepsis and sham control mice by MACS for CD11b+[CD3-CD19-Ly6G-] cells. ChIP analysis for H3K4me3 at Il1b, Il12, and Il23 promoter was performed (n=5/group). For all ChIP experiments, isotype control antibody to IgG was run in parallel. Dotted line represents isotype control. *p<0.05 by ANOVA test with Newman-Keuls Multiple Comparison test. B: Wound myeloid cells CD11b+[CD3-CD19-Ly6G-] were isolated from post-sepsis and control mice and Mll1 expression was quantified using qPCR (n=5/group). **p<0.01 by Student t test with Welch’s correction.
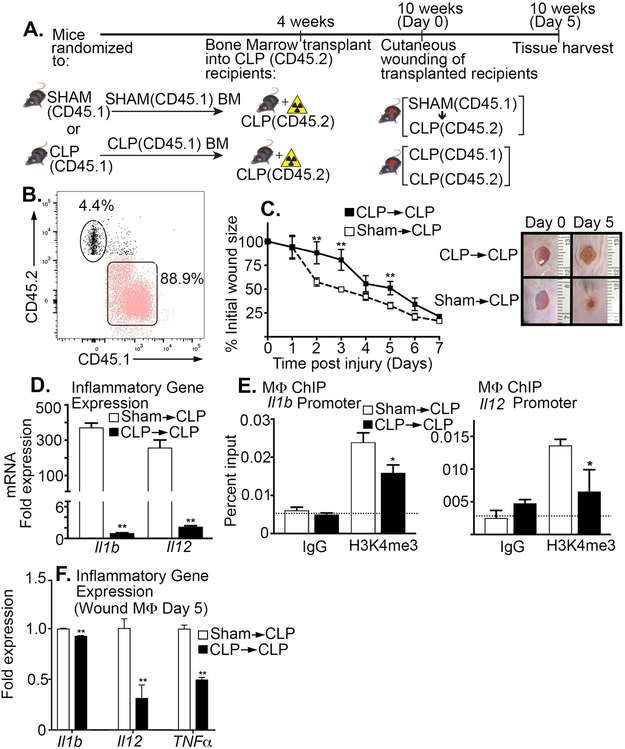
Chimeric Mice Receiving BM Transplant Display Improved Wound Healing and Restored Wound Monocyte/Macrophage Phenotype
To evaluate whether the peripheral monocyte/macrophage phenotype is controlled by BM stem/progenitor cells following sepsis recovery, we first performed a congenic bone marrow transplant. Following recovery from sepsis, bone marrow was isolated from Sham or CLP donor mice expressing CD45.1 and injected into recipient mice, expressing CD45.2, that had previously undergone a CLP procedure. Degree of chimerism was found to be 88% at 8 weeks posttransplant using peripheral blood flow cytometry (Figure 5A,B). Wound healing was significantly improved in the CLP mice receiving sham BM (Figure 5C). Expression of inflammatory cytokines in peripheral blood monocytes was increased in the chimeric mice receiving Sham BM (Figure 5D). Further, using ChIP assays, we found H3K4me3 was significantly increased on the NFκB binding sites of Il1b and Il12 in peripheral monocytes that received Sham BM (Figure 5E). Finally, analysis of wound monocytes/macrophages demonstrated increased inflammatory cytokine production at day 5 post wounding in CLP mice receiving Sham BM in comparison to control bone marrow (Figure 5F). Taken together, these results suggest that MLL1-mediated H3K4 trimethylation in BM cells may play a key role in promoting the impaired peripheral macrophage inflammatory phenotype and delayed wound healing following full recovery from sepsis.
Figure 5. Chimeric Mice Receiving BM Transplant Display Improved Wound Healing and Restored Wound Monocyte/Macrophage Phenotype.
A: BM chimeras were created using a congenic bone marrow transplant. Following either CLP or sham surgery mice were allowed to recover and then BM from donor sham or CLP mice expressing CD45.1 was transplanted into irradiated CLP recipient mice expressing CD45.2. B: Peripheral blood analysis was performed at 8 weeks confirming 88.9% donor chimerism. C: Wounds were created by 4-mm punch biopsy on the backs of CLP→CLP and Sham→CLP mice 6 weeks after recovery. The change in wound area was recorded daily with Image J software until complete healing was observed. Representative photographs of the wounds of post-sepsis and sham mice are shown on days 0 and 5 are shown (n=7/group). **p<0.01 by Mann-Whitney U test. D: Myeloid cells CD11b+[CD3-CD19-Ly6G-] were isolated from CLP→CLP and Sham→CLP at day 5 post injury mice. Gene expression of Il1b and IL12 was quantified using qPCR (n=5/group). **p<0.01 by Mann-Whitney U test. E: Myeloid cells were isolated at day 5 post injury from CLP→CLP and Sham→CLP mice by MACS for CD11b+[CD3-CD19-Ly6G-] cells. ChIP analysis for H3K4me3 at Il1b and Il12 promoter was performed (n = 5/group). For all ChIP experiments, isotype control antibody to IgG was run in parallel. *p<0.05 by ANOVA test Newman-Keuls Multiple Comparison test. F: Wound myeloid cells CD11b+[CD3-CD19-Ly6G-] were isolated from CLP→CLP and Sham→CLP mice and Il1b, Il12, and Tnfa expression was quantified using qPCR (n=5/group). **p<0.01 by Student t test with Welch’s correction. Data are presented as the mean±SEM. Data are representative of 2 independent experiments.
DISCUSSION
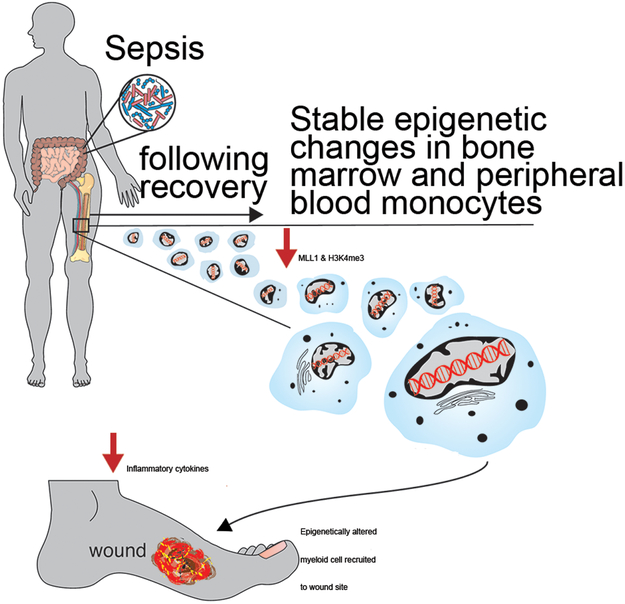
Sepsis represents an acute life-threatening disorder resulting from a dysregulated host response to infection. Beyond the acute insult, there remains additional long-term consequences after sepsis recovery such as impaired wound healing for which the molecular mechanisms remain unknown. Herein, we identify that following recovery from sepsis, peripheral monocyte/macrophages are poised for an impaired inflammatory phenotype, resulting in significantly delayed cutaneous wound repair following injury. This aberrant wound macrophage phenotype in post-septic mice is, at least partially, directed by low levels of the epigenetic enzyme, MLL1, and its associated histone H3K4me3 activation mark on the NFkB binding sites on promoters of inflammatory genes. Further, human blood samples corroborate these findings in patients who survive sepsis exhibiting decreased MLL1 in comparison to non-sepsis controls. Finally, bone marrow transplant experiments demonstrate that these epigenetic modifications post-sepsis initiate in BM progenitor/stem cells resulting in impairment in peripheral macrophage function (Figure 6).
Figure 6. Schematic of epigenetic Modifications to Macrophages Following Survival from Sepsis.
Following recovery from sepsis, peripheral monocyte/macrophages are poised toward an impaired inflammatory phenotype, resulting in significantly delayed cutaneous wound repair following injury. This aberrant wound macrophage phenotype in post-septic mice is directed by low levels of the epigenetic enzyme, MLL1, and its associated H3K4me3, histone activation mark.
The process of wound healing involves a programmed local inflammatory reaction dictated mainly by macrophages in response to tissue damage. During the first phase of wound healing, an initial inflammatory response ensues that is characterized by macrophage tissue destruction, release of inflammatory cytokines, and clearance of invading pathogens 47. This is followed by cessation of inflammation and fibrin/collagen deposition resulting in initiation of tissue repair. The precise timing of both the initiation and resolution of inflammation is essential for restoring tissue integrity following injury. Although macrophage phenotype/function in tissue repair post-sepsis has not been well-studied, recent work has demonstrated that monocytes/macrophages display decreased capacity to release proinflammatory cytokines in response to endotoxin [lipopolysaccharide (LPS)], TLR agonists and various other bacterial compounds 48,49. In clinical studies, the magnitude and the persistent nature of this refractory state is associated with increased mortality and nosocomial infections 50. Within tissue repair, the local inflammatory response to injury may be influenced by this dysregulation of systemic inflammation 12,16,51. Prior studies investigating the association of wound healing and sepsis have focused on either the acute sepsis episode 16, connective tissue synthesis 52,53, or epidermal barrier function 12, however, few investigations have focused on the protracted immunosuppression and its impact on wound repair following sepsis recovery. Our results demonstrate that bone marrow progenitor/stem cells and local wound monocyte/macrophages continue to exhibit an impaired inflammatory response long after recovery from sepsis. This results in decreased production of inflammatory cytokines that are vital for the first phase of wound healing. This inability to generate an initial inflammatory response negatively impacts downstream orchestration of subsequent phases 47.
These current findings support the concept that the severe sepsis alters monocyte/macrophage phenotypes through epigenetics 6,21,54. These epigenetic changes can contribute to a phenomenon known as ‘innate immune memory’ where chromatin modifications persist even after a normal physiological environment is restored 3,55. Innate immune cells have classically been thought to lack immunological memory. However, recent studies have challenged this dogma by demonstrating that innate immune cells can also develop persistent phenotypic alterations following infection, BCG vaccination, or even β-glucan challenge. Considering the nature of monocyte/macrophage (Mo/Mac) differentiation, as well as their relatively short lifespan investigations have shown that the innate immune memory arises from epigenetic imprinting of hematopoietic stem cells56,57. This response is considered to be independent of adaptive immunity and driven by epigenetic modifications that influence gene expression in mononuclear phagocytes58,59. Our results are consistent with the development of innate immune memory after sepsis, however unlike prior investigations which demonstrate a trained robust innate response to secondary infection, our investigation suggests the development of an immune tolerance in macrophages which hinders tissue repair. Profound changes in histone methylation and acetylation, as well as in DNA methylation, have been described after sepsis 60. In models of endotoxin tolerance, epigenetic modification of pro-inflammatory gene loci plays a prominent part in the regulation of their transcription 58,61. Similarly, the role of epigenetic modifications during sepsis has previously been demonstrated in lung-resident macrophages 25. Septic mice (24 h post-surgery/sepsis induction) exhibit significantly decreased expression of TNF, IL12p40 and iNOS when restimulated ex vivo with LPS as compared to sham surgery controls 54. In addition, these post-septic lung macrophages exhibit decreased acetylation of histone 4 (H4Ac) which is a chromatin mark known to increase gene transcription. In our study, we demonstrate for the first time that BM progenitor/stem cells and wound myeloid cells display decreased expression of the epigenetic enzyme MLL1 and its associated histone activation mark, H3K4me3, following prolonged recovery from sepsis. Although we have previously shown, that MLL1, and its associated H3K4 trimethylation, is involved in the regulation of inflammatory cytokine production in diabetic wound macrophages 23, to date no studies have investigated the role of MLL1 in sepsis. MLL1 has been shown to be recruited by NFkB to the promoters of inflammatory genes, tumor necrosis factor-α and matrix metalloproteinase-9, suggesting a role for MLL1 in NFkB–mediated signaling 62,63. This is likely relevant in human tissues because MLL1 expression in human monocyte–derived macrophages has been shown to influence inflammatory gene transcription 64. Since the NFkB pathway is critical for inflammatory gene transcription in wound macrophages, alterations in MLL1 expression are likely to influence macrophage inflammatory response. These results suggest that the “cytokine storm” of sepsis may be directing the loss of specific activating epigenetic marks at promoters of proinflammatory genes in macrophages. Lastly, recent investigations have suggested there exists multiple sepsis phenotypes within the classically defined clinical definitions (Sequential Organ Failure Assessment)65,66. These variable clinical phenotypes correlated with host-response patterns and clinical outcomes and may be driven by differences in epigenetic modifications.
This study offers insight into the epigenetic regulation of macrophage phenotype and wound healing following sepsis; however some limitations must be addressed. Although the H3K4me3/MLL1 down regulation in these post-sepsis mice suggests a potential mechanism for the impaired inflammatory reaction, it is unlikely that MLL1 is the sole contributor to macrophage inflammatory cytokine production. Indeed, other epigenetic enzymes have been shown to result in aberrant myeloid cell function in pathological states 61,67. Further, as variation in progenitor cells during the course of sepsis recovery could ultimately impact other processes happening within wound healing that are independent of epigenetic changes. Thus, further studies assessing the role of other specific epigenetic enzymes in the regulation of macrophage phenotype and wound healing following sepsis would allow insight into the full regulation of post-septic macrophages.
In summary, we demonstrate that following recovery from sepsis, bone marrow progenitors and peripheral monocytes/macrophages are poised toward an impaired inflammatory phenotype resulting in a delay in cutaneous wound healing. This is driven by an MLL1-mediated epigenetic modification and its associated activating histone mark on NFkB inflammatory gene promoters. Further, post-septic human patients display consistent reduction in MLL1 expression in comparison to controls without a history of sepsis. These results suggest that the physiological stress imparted on the immune system during severe sepsis can have significant long-term effects on gene expression and that epigenetic regulatory “signatures” in macrophages can be modulated resulting in impaired response to tissue injury.
Supplementary Material
Highlights.
Using both a murine model of sepsis and cutaneous wound healing we demonstrate that long after recovery from sepsis there remains impairment in wound healing and decreased macrophage inflammatory cytokine production in response to tissue injury.
This post-sepsis immunosuppression is mediated by decreased expression of Mll1, an epigenetic enzymes, and impaired histone H3 lysine 4 trimethylation (activation mark) at NFkB binding sites on inflammatory gene promoters.
Bone marrow transplantation studies demonstrated epigenetic modifications initiate in bone marrow progenitor/stem cells following sepsis resulting in lasting impairment in peripheral macrophage function.
Human peripheral blood leukocytes from post-septic patients demonstrate a significant reduction in MLL1 compared with non-septic controls.
Acknowledgements:
We thank Robin Kunkel for her assistance with the graphical illustrations.
Funding Sources: This work is supported in part by National Institutes of Health grants R01-HL137919 (KG), K08-DK102357 (KG), F32-DK117545 (FD), American College of Surgeons Resident Fellowship (FD), T32-HL076123 (AK), R01-HL031237 (SK), HL35HL144481 (BBM), the Michigan Diabetes and Research Center (P30-DK020572), and the Doris Duke Foundation (KG).
ABBREVIATIONS
- BM
Bone Marrow
- BMDM
Bone marrow derived macrophages
- CLP
Cecal ligation and puncture
- H3K4
Histone 3 Lysine 4
- H3K4me3
Trimethylation on histone 3 lysine 4
- IL
Interleukin
- MLL1
Mixed-lineage leukemia 1
- NFκB
Nuclear factor kappa-light-chain-enhancer of activated B cells
Footnotes
Disclosures: The authors have no conflicts of interest
References
- 1.Vincent J-L, Marshall JC, Namendys-Silva SA, François B, Martin-Loeches I, Lipman J, Reinhart K, Antonelli M, Pickkers P, Njimi H, Jimenez E, Sakr Y, ICON investigators. Assessment of the worldwide burden of critical illness: the intensive care over nations (ICON) audit. Lancet Respir Med. 2014;2(5):380–386. doi: 10.1016/S2213-2600(14)70061-X. [DOI] [PubMed] [Google Scholar]
- 2.Fleischmann C, Scherag A, Adhikari NKJ, Hartog CS, Tsaganos T, Schlattmann P, Angus DC, Reinhart K, International Forum of Acute Care Trialists. Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Current Estimates and Limitations. Am J Respir Crit Care Med. 2016;193(3):259–272. doi: 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- 3.Hotchkiss RS, Monneret G, Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat Rev Immunol. 2013;13(12):862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hotchkiss RS, Monneret G, Payen D. Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Infect Dis. 2013;13(3):260–268. doi: 10.1016/S1473-3099(13)70001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arens C, Bajwa SA, Koch C, Siegler BH, Schneck E, Hecker A, Weiterer S, Lichtenstern C, Weigand MA, Uhle F. Sepsis-induced long-term immune paralysis – results of a descriptive, explorative study. Crit Care. 2016;20(1):93. doi: 10.1186/s13054-016-1233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carson WF, Cavassani KA, Dou Y, Kunkel SL. Epigenetic regulation of immune cell functions during post-septic immunosuppression. Epigenetics. 2011;6(3):273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quartin AA, Schein RM, Kett DH, Peduzzi PN. Magnitude and duration of the effect of sepsis on survival. Department of Veterans Affairs Systemic Sepsis Cooperative Studies Group. JAMA. 1997;277(13):1058–1063. [PubMed] [Google Scholar]
- 8.Perl TM, Dvorak L, Hwang T, Wenzel RP. Long-term survival and function after suspected gram-negative sepsis. JAMA. 1995;274(4):338–345. [PubMed] [Google Scholar]
- 9.Maley JH, Mikkelsen ME. Short-term Gains with Long-term Consequences: The Evolving Story of Sepsis Survivorship. Clin Chest Med. 2016;37(2):367–380. doi: 10.1016/j.ccm.2016.01.017. [DOI] [PubMed] [Google Scholar]
- 10.Jones TK, Fuchs BD, Small DS, Halpern SD, Hanish A, Umscheid CA, Baillie CA, Kerlin MP, Gaieski DF, Mikkelsen ME. Post–Acute Care Use and Hospital Readmission after Sepsis. Ann Am Thorac Soc. 2015;12(6):904–913. doi: 10.1513/AnnalsATS.201411-504OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ortego A, Gaieski DF, Fuchs BD, Jones T, Halpern SD, Small DS, Sante SC, Drumheller B, Christie JD, Mikkelsen ME. Hospital-based acute care use in survivors of septic shock. Crit Care Med. 2015;43(4):729–737. doi: 10.1097/CCM.0000000000000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koskela M, Gäddnäs F, Ala-Kokko TI, Laurila JJ, Saarnio J, Oikarinen A, Koivukangas V. Epidermal wound healing in severe sepsis and septic shock in humans. Crit Care. 2009;13(3):R100. doi: 10.1186/cc7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood S, Jayaraman V, Huelsmann EJ, Bonish B, Burgad D, Sivaramakrishnan G, Qin S, DiPietro LA, Zloza A, Zhang C, Shafikhani SH. Pro-inflammatory chemokine CCL2 (MCP-1) promotes healing in diabetic wounds by restoring the macrophage response Liu G, ed. PLoS One. 2014;9(3):e91574. doi: 10.1371/journal.pone.0091574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maruyama K, Asai J, Ii M, Thorne T, Losordo DW, D’Amore PA. Decreased macrophage number and activation lead to reduced lymphatic vessel formation and contribute to impaired diabetic wound healing. Am J Pathol. 2007;170(4):1178–1191. doi: 10.2353/ajpath.2007.060018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snyder RJ, Lantis J, Kirsner RS, Shah V, Molyneaux M, Carter MJ. Macrophages: A review of their role in wound healing and their therapeutic use. Wound Repair Regen. 2016;24(4):613–629. doi: 10.1111/wrr.12444. [DOI] [PubMed] [Google Scholar]
- 16.Sommer K, Sander AL, Albig M, Weber R, Henrich D, Frank J, Marzi I, Jakob H. Delayed wound repair in sepsis is associated with reduced local pro-inflammatory cytokine expression. Zhou Z, ed. PLoS One. 2013;8(9):e73992. doi: 10.1371/journal.pone.0073992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15(2):163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 18.Stability Reik W. and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447(7143):425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 19.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6(1):73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 20.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14(2):155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Ishii M, Wen H, Corsa CAS, Liu T, Coelho AL, Allen RM, Carson WF, Cavassani KA, Li X, Lukacs NW, Hogaboam CM, Dou Y, Kunkel SL. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114(15):3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang L, Wang Y, Zhu D, Xue Z, Mao H. Alteration of histone H3 lysine 9 dimethylation in peripheral white blood cells of septic patients with trauma and cancer. Mol Med Rep. 2016;14(6):5467–5474. doi: 10.3892/mmr.2016.5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimball A, Joshi A, Carson WF, Boniakowski A, Davis F, Schaller M, Allen R, Bermick J, Henke PK, Burant CF, Kunkel SL, Gallagher KA. The Histone Methyltransferase, MLL1, Directs Macrophage-Mediated Inflammation in Wound Healing and is Altered in a Murine Model of Obesity and Type 2 Diabetes. Diabetes. June 2017:db170194. doi: 10.2337/db17-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yamashita M, Hirahara K, Shinnakasu R, Hosokawa H, Norikane S, Kimura MY, Hasegawa A, Nakayama T. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 2006;24(5):611–622. doi: 10.1016/j.immuni.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 25.Wen H, Dou Y, Hogaboam CM, Kunkel SL. Epigenetic regulation of dendritic cell-derived interleukin-12 facilitates immunosuppression after a severe innate immune response. Blood. 2008;111(4):1797–1804. doi: 10.1182/blood-2007-08-106443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinet P, Milewicz DM, Cassis LA, Leeper NJ, Lu HS, Smith JD. Consideration of Sex Differences in Design and Reporting of Experimental Arterial Pathology Studies-Statement From ATVB Council. Arterioscler Thromb Vasc Biol. 2018;38(2):292–303. doi: 10.1161/ATVBAHA.117.309524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen H, Hogaboam CM, Gauldie J, Kunkel SL. Severe sepsis exacerbates cell-mediated immunity in the lung due to an altered dendritic cell cytokine profile. Am J Pathol. 2006;168(6):1940–1950. doi: 10.2353/ajpath.2006.051155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirza RE, Fang MM, Ennis WJ, Koh TJ. Blocking Interleukin-1 Induces a Healing-Associated Wound Macrophage Phenotype and Improves Healing in Type 2 Diabetes. Diabetes. 2013;62(7):2579–2587. doi: 10.2337/db12-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii M, Wen H, Corsa CAS, Liu T, Coelho AL, Allen RM, Carson WF, Cavassani KA, Li X, Lukacs NW, Hogaboam CM, Dou Y, Kunkel SL. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114(15):3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubbard LLN, Ballinger MN, Wilke CA, Moore BB. Comparison of conditioning regimens for alveolar macrophage reconstitution and innate immune function post bone marrow transplant. Exp Lung Res. 2008;34(5):263–275. doi: 10.1080/01902140802022518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singer M, Deutschman CS, Seymour CW, Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche J-D, Coopersmith CM, Hotchkiss RS, Levy MM, Marshall JC, Martin GS, Opal SM, Rubenfeld GD, van der Poll T, Vincent J-L, Angus DC. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA. 2016;315(8):801. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhee C, Dantes R, Epstein L, Murphy DJ, Seymour CW, Iwashyna TJ, Kadri SS, Angus DC, Danner RL, Fiore AE, Jernigan JA, Martin GS, Septimus E, Warren DK, Karcz A, Chan C, Menchaca JT, Wang R, Gruber S, Klompas M, CDC Prevention Epicenter Program. Incidence and Trends of Sepsis in US Hospitals Using Clinical vs Claims Data, 2009–2014. JAMA. 2017;318(13):1241–1249. doi: 10.1001/jama.2017.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wen H, Dou Y, Hogaboam CM, Kunkel SL. Epigenetic regulation of dendritic cell-derived interleukin-12 facilitates immunosuppression after a severe innate immune response. Blood. 2008;111(4):1797–1804. doi: 10.1182/blood-2007-08-106443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fattahi F, Ward PA. Understanding Immunosuppression after Sepsis. Immunity. 2017;47(1):3–5. doi: 10.1016/j.immuni.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Schaack D, Siegler BH, Tamulyte S, Weigand MA, Uhle F. The immunosuppressive face of sepsis early on intensive care unit-A large-scale microarray meta-analysis. Ahuja SK, ed. PLoS One. 2018;13(6):e0198555. doi: 10.1371/journal.pone.0198555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gallagher K a., Joshi A, Carson WF, Schaller M, Allen R, Mukerjee S, Kittan N, Feldman EL, Henke PK, Hogaboam C, Burant CF, Kunkel SL. Epigenetic changes in bone marrow progenitor cells influence the inflammatory phenotype and alter wound healing in type 2 diabetes. Diabetes. 2015;64(4):1420–1430. doi: 10.2337/db14-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koh TJ, DiPietro LA. Inflammation and wound healing: the role of the macrophage. Expert Rev Mol Med. 2011;13:e23. doi: 10.1017/S1462399411001943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimball A, Schaller M, Joshi A, Davis FM, denDekker A, Boniakowski A, Bermick J, Obi A, Moore B, Henke PK, Kunkel SL, Gallagher KA. Ly6CHi Blood Monocyte/Macrophage Drive Chronic Inflammation and Impair Wound Healing in Diabetes Mellitus. Arterioscler Thromb Vasc Biol. 2018;38(5):1102–1114. doi: 10.1161/ATVBAHA.118.310703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davis FM, Kimball A, denDekker A, Joshi AD, Boniakowski AE, Nysz D, Allen RM, Obi A, Singer K, Henke PK, Moore BB, Kunkel SL, Gallagher KA. Histone Methylation Directs Myeloid TLR4 Expression and Regulates Wound Healing following Cutaneous Tissue Injury. J Immunol. 2019;202(6):1777–1785. doi: 10.4049/jimmunol.1801258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fathke C, Wilson L, Hutter J, Kapoor V, Smith A, Hocking A, Isik F. Contribution of bone marrow-derived cells to skin: collagen deposition and wound repair. Stem Cells. 2004;22(5):812–822. doi: 10.1634/stemcells.22-5-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y, Wang J, Scott PG, Tredget EE. Bone marrow-derived stem cells in wound healing: a review. Wound Repair Regen. 2007;15 Suppl 1(s1):S18–26. doi: 10.1111/j.1524-475X.2007.00221.x. [DOI] [PubMed] [Google Scholar]
- 42.Davenport EE, Burnham KL, Radhakrishnan J, Humburg P, Hutton P, Mills TC, Rautanen A, Gordon AC, Garrard C, Hill AVS, Hinds CJ, Knight JC. Genomic landscape of the individual host response and outcomes in sepsis: a prospective cohort study. Lancet Respir Med. 2016;4(4):259–271. doi: 10.1016/S2213-2600(16)00046-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cazalis M- A, Lepape A, Venet F, Frager F, Mougin B, Vallin H, Paye M, Pachot A, Monneret G. Early and dynamic changes in gene expression in septic shock patients: a genome-wide approach. Intensive care Med Exp. 2014;2(1):20. doi: 10.1186/s40635-014-0020-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaller M, Ito T, Allen RM, Kroetz D, Kittan N, Ptaschinski C, Cavassani K, Carson WF, Godessart N, Grembecka J, Cierpicki T, Dou Y, Kunkel SL. Epigenetic regulation of IL-12-dependent T cell proliferation. J Leukoc Biol. 2015;98(4):601–613. doi: 10.1189/jlb.1A0814-375RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamashita M, Hirahara K, Shinnakasu R, Hosokawa H, Norikane S, Kimura MY, Hasegawa A, Nakayama T. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 2006;24(5):611–622. doi: 10.1016/j.immuni.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 46.Carson WF, Cavassani KA, Soares EM, Hirai S, Kittan NA, Schaller MA, Scola MM, Joshi A, Matsukawa A, Aronoff DM, Johnson CN, Dou Y, Gallagher KA, Kunkel SL. The STAT4/MLL1 Epigenetic Axis Regulates the Antimicrobial Functions of Murine Macrophages. J Immunol. 2017;199(5):1865–1874. doi: 10.4049/jimmunol.1601272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis FMFM, Kimball A, Boniakowski A, Gallagher K. Dysfunctional Wound Healing in Diabetic Foot Ulcers: New Crossroads. Curr Diab Rep. 2018;18(1):2. doi: 10.1007/s11892-018-0970-z. [DOI] [PubMed] [Google Scholar]
- 48.Cavaillon J-M, Adib-Conquy M. Bench-to-bedside review: endotoxin tolerance as a model of leukocyte reprogramming in sepsis. Crit Care. 2006;10(5):233. doi: 10.1186/cc5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30(10):475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Monneret G, Lepape A, Voirin N, Bohé J, Venet F, Debard A- L, Thizy H, Bienvenu J, Gueyffier F, Vanhems P. Persisting low monocyte human leukocyte antigen-DR expression predicts mortality in septic shock. Intensive Care Med. 2006;32(8):1175–1183. doi: 10.1007/s00134-006-0204-8. [DOI] [PubMed] [Google Scholar]
- 51.Rico RM, Ripamonti R, Burns AL, Gamelli RL, DiPietro LA. The effect of sepsis on wound healing. J Surg Res. 2002;102(2):193–197. doi: 10.1006/jsre.2001.6316. [DOI] [PubMed] [Google Scholar]
- 52.Jacinto A, Martinez-Arias A, Martin P. Mechanisms of epithelial fusion and repair. Nat Cell Biol. 2001;3(5):E117–23. doi: 10.1038/35074643. [DOI] [PubMed] [Google Scholar]
- 53.Greenhalgh DG, Gamelli RL. Is impaired wound healing caused by infection or nutritional depletion? Surgery. 1987;102(2):306–312. [PubMed] [Google Scholar]
- 54.Lyn-Kew K, Rich E, Zeng X, Wen H, Kunkel SL, Newstead MW, Bhan U, Standiford TJ. IRAK-M regulates chromatin remodeling in lung macrophages during experimental sepsis. Jeyaseelan S, ed. PLoS One. 2010;5(6):e11145. doi: 10.1371/journal.pone.0011145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bomans K, Schenz J, Sztwiertnia I, Schaack D, Weigand MA, Uhle F. Sepsis Induces a Long-Lasting State of Trained Immunity in Bone Marrow Monocytes. Front Immunol. 2018;9:2685. doi: 10.3389/fimmu.2018.02685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kaufmann E, Sanz J, Dunn JL, Khan N, Mendonça LE, Pacis A, Tzelepis F, Pernet E, Dumaine A, Grenier J- C, Mailhot-Léonard F, Ahmed E, Belle J, Besla R, Mazer B, King IL, Nijnik A, Robbins CS, Barreiro LB, Divangahi M. BCG Educates Hematopoietic Stem Cells to Generate Protective Innate Immunity against Tuberculosis. Cell. 2018;172(1–2):176–190.e19. doi: 10.1016/j.cell.2017.12.031. [DOI] [PubMed] [Google Scholar]
- 57.Mitroulis I, Ruppova K, Wang B, Chen L-S, Grzybek M, Grinenko T, Eugster A, Troullinaki M, Palladini A, Kourtzelis I, Chatzigeorgiou A, Schlitzer A, Beyer M, Joosten LAB, Isermann B, Lesche M, Petzold A, Simons K, Henry I, Dahl A, Schultze JL, Wielockx B, Zamboni N, Mirtschink P, Coskun Ü, Hajishengallis G, Netea MG, Chavakis T. Modulation of Myelopoiesis Progenitors Is an Integral Component of Trained Immunity. Cell. 2018;172(1–2):147–161.e12. doi: 10.1016/j.cell.2017.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Novakovic B, Habibi E, Wang S- Y, Arts RJW, Davar R, Megchelenbrink W, Kim B, Kuznetsova T, Kox M, Zwaag J, Matarese F, van Heeringen SJ, Janssen-Megens EM, Sharifi N, Wang C, Keramati F, Schoonenberg V, Flicek P, Clarke L, Pickkers P, Heath S, Gut I, Netea MG, Martens JHA, Logie C, Stunnenberg HG. β-Glucan Reverses the Epigenetic State of LPS-Induced Immunological Tolerance. Cell. 2016;167(5):1354–1368.e14. doi: 10.1016/j.cell.2016.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kleinnijenhuis J, Quintin J, Preijers F, Joosten LAB, Jacobs C, Xavier RJ, van der Meer JWM, van Crevel R, Netea MG. BCG-induced trained immunity in NK cells: Role for non-specific protection to infection. Clin Immunol. 2014;155(2):213–219. doi: 10.1016/j.clim.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leentjens J, Kox M, van der Hoeven JG, Netea MG, Pickkers P. Immunotherapy for the adjunctive treatment of sepsis: from immunosuppression to immunostimulation. Time for a paradigm change? Am J Respir Crit Care Med. 2013;187(12):1287–1293. doi: 10.1164/rccm.201301-0036CP. [DOI] [PubMed] [Google Scholar]
- 61.El Gazzar M, Yoza BK, Chen X, Garcia BA, Young NL, McCall CE. Chromatin-specific remodeling by HMGB1 and linker histone H1 silences proinflammatory genes during endotoxin tolerance. Mol Cell Biol. 2009;29(7):1959–1971. doi: 10.1128/MCB.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang X, Zhu K, Li S, Liao Y, Du R, Zhang X, Shu H- B, Guo A- Y, Li L, Wu M. MLL1, a H3K4 methyltransferase, regulates the TNFα-stimulated activation of genes downstream of NF-κB. J Cell Sci. 2012;125(Pt 17):4058–4066. doi: 10.1242/jcs.103531. [DOI] [PubMed] [Google Scholar]
- 63.Robert I, Aussems M, Keutgens A, Zhang X, Hennuy B, Viatour P, Vanstraelen G, Merville M- P, Chapelle J- P, de Leval L, Lambert F, Dejardin E, Gothot A, Chariot A. Matrix Metalloproteinase-9 gene induction by a truncated oncogenic NF-kappaB2 protein involves the recruitment of MLL1 and MLL2 H3K4 histone methyltransferase complexes. Oncogene. 2009;28(13):1626–1638. doi: 10.1038/onc.2009.6. [DOI] [PubMed] [Google Scholar]
- 64.Kittan NA, Allen RM, Dhaliwal A, Cavassani KA, Schaller M, Gallagher KA, Carson WF, Mukherjee S, Grembecka J, Cierpicki T, Jarai G, Westwick J, Kunkel SL, Hogaboam CM. Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. Zissel G, ed. PLoS One. 2013;8(10):e78045. doi: 10.1371/journal.pone.0078045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Knaus WA, Marks RD. New Phenotypes for Sepsis: The Promise and Problem of Applying Machine Learning and Artificial Intelligence in Clinical Research. JAMA. 2019;321(20):1981. doi: 10.1001/jama.2019.5794. [DOI] [PubMed] [Google Scholar]
- 66.Seymour CW, Kennedy JN, Wang S, Chang C-CH, Elliott CF, Xu Z, Berry S, Clermont G, Cooper G, Gomez H, Huang DT, Kellum JA, Mi Q, Opal SM, Talisa V, van der Poll T, Visweswaran S, Vodovotz Y, Weiss JC, Yealy DM, Yende S, Angus DC. Derivation, Validation, and Potential Treatment Implications of Novel Clinical Phenotypes for Sepsis. JAMA. 2019;321(20):2003. doi: 10.1001/jama.2019.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chan C, Li L, McCall CE, Yoza BK. Endotoxin tolerance disrupts chromatin remodeling and NF-kappaB transactivation at the IL-1beta promoter. J Immunol. 2005;175(1):461–468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.