Abstract
Focal adhesion pathway is one of the key molecular pathways affected by suboptimal culture conditions during embryonic development. The epidermal growth factor (EGF) and hyaluronic acid (HA) are believed to be involved in the focal adhesion pathway function by regulating the adherence of the molecules to the extracellular matrix. However, regulatory and molecular mechanisms through which the EGF and HA could influence the embryo development is not clear. Therefore, this study aimed to investigate the effect of continued or stage specific supplementation of EGF and/or HA on the developmental competence and quality of bovine preimplantation embryos and the subsequent consequences on the expression and DNA methylation patterns of genes involved in the focal adhesion pathway. The results revealed that, the supplementation of EGF or HA from zygote to the blastocysts stage reduced the level of reactive oxygen species and increased hatching rate after thawing. On the other hand, HA decreased the apoptotic nuclei and increased blastocyst compared to EGF supplemented group. Gene expression and DNA methylation analysis in the resulting blastocysts indicated that, combined supplementation of EGF and HA increased the expression of genes involved in focal adhesion pathway while supplementation of EGF, HA or a combination of EGF and HA during the entire preimplantation period changed the DNA methylation patterns of genes involved in focal adhesion pathway. On the other hand, blastocysts developed in culture media supplemented with EGF + HA until the 16-cell stage exhibited higher expression level of genes involved in focal adhesion pathway compared to those supplemented after the 16-cell stage. Conversely, the DNA methylation level of candidate genes was increased in the blastocysts obtained from embryos cultured in media supplemented with EGF + HA after 16-cell stage. In conclusion, supplementation of bovine embryos with EGF and/or HA during the entire preimplantation period or in a stage specific manner altered the DNA methylation and expression patterns of candidate genes involved in the focal adhesion pathway which was in turn associated with the observed embryonic developmental competence and quality.
Introduction
Suboptimal culture conditions during preimplantation period can result in long-term effects on embryo competence and pregnancy establishment. Despite many attempts to modulate the in vitro culture conditions to mimic the in vivo environment, the quality of in vitro produced embryos remains low [1]. Furthermore, suboptimal embryo culture condition decreases the quality and hinders the developmental competence of the embryo by altering the expression and DNA methylation patterns of developmentally related genes and pathways [2, 3]. Among these, focal adhesion pathway was one of the top dysregulated pathways in bovine embryos due to suboptimal culture conditions [4]. Focal adhesion is one of the cell communication mechanisms, it is vital for cell motility, differentiation, migration, proliferation and survival [5, 6]. Many of focal adhesion proteins, such as beta1 integrin, alpha4 integrin, alpha5 integrin, talin, paxillin, vinculin, focal adhesion kinase and integrin like kinase are essential for embryonic development. Furthermore, the functional loss of these proteins during embryogenesis would affect the cell-extracellular matrix (ECM) adhesion, cytoskeletal organization, polarity, migration and survivability of the embryos [7].
Adhesion to the ECM with the supplementation of growth factors is necessary for normal cell growth [8]. Moreover, supplementation of growth factors in cell-free culture media was found to improve the blastocyst rate [9]. Among these factors, epidermal growth factor (EGF) was found to improve the embryonic development in mouse [10], porcine [11] and bovine [12]. Similarly, hyaluronic acid (HA), one of the main components of ECM, is also believed to improve the blastocyst rate of in vitro produced bovine embryos [13]. However, the molecular mechanisms through which supplementation of EGF and HA influenced the embryonic development and quality remain elusive. Here, it was hypothesized that the extracellular growth factors and the extracellular components could improve the quality and development of embryos by regulating the DNA methylation and expression patterns of genes involved in focal adhesion pathway. Indeed, suboptimal in vitro culture conditions could cause DNA methylation changes during embryo development [14, 15]. We assumed that, the dynamic changes in the DNA methylation pattern of embryos during in vitro development may rely on epigenetic adaptability of embryos resulting from the persistent cellular interactions with extracellular environment via cell adhesion to ECM molecules. Therefore, this study was conducted to investigate the effect of continued or stage specific supplementation of epidermal growth factor and/or hyaluronic acid on the expression and DNA methylation patterns of the genes involved in focal adhesion pathway and its subsequent significance on the developmental competence and quality of bovine preimplantation embryos.
Materials and methods
Experimental design
Two consecutive experiments were conducted to achieve the goals of the study. The first experiment was designed to investigate the effect of EGF and/or HA on the expression and DNA methylation patterns of genes involved in focal adhesion pathway and the subsequent effects on the development and quality of bovine embryos. Therefore, supplementation of EGF and/or HA was performed from zygote to the blastocyst stage (the entire preimplantation “period”) For this, synthetic oviduct fluids (SOF) culture media supplemented with fatty acid free bovine serum albumin was considered as the basic media (BM). Accordingly, in vitro produced presumptive zygotes were cultured only in BM (control group), or BM supplemented with 10 ng/ml EGF [16–19], BM supplemented with 1mg/ml HA [13, 20–22] or BM supplemented with a combination of 10 ng/ml EGF and 1 mg/ml HA (EGF + HA), (Fig 1A). Afterwards, the effects of EGF and/or HA on the developmental competence of the day 7 preimplantation embryos were investigated. Moreover, blastocysts (four biological replicates) were subjected to total cell count, ICM/TE ratio, ROS accumulation, apoptosis, protein analysis and cryotolerance (three biological replicates) assays. Furthermore, Total RNA and genomic DNA were isolated from each four biological replicates of day 7 blastocysts group. Thereafter, the mRNA expression was quantified using qPCR and the DNA methylation level of candidate genes involved in focal adhesion pathway was analyzed using bisulfite sequencing.
Fig 1. Outline of the experimental design.
In first experiment (A), zygotes were cultured in four culture conditions until the blastocysts stage: only in basic media (BM), BM supplemented with EGF, BM supplemented with HA and BM supplemented with EGF + HA. In the second experiment (B), preimplantation embryos were cultured in BM until the blastocyst stage (BM), or supplemented with EGF + HA after 4 cell stage (After_4C), after 16 cell stage (After_16C), until 4-cell (Until_4C), until 16-cell stage (Until_16C) or from zygote to the blastocyst stage (All_stages).
The second experiment, was aimed to elucidate the effect of continued (the entire preimplantation “period”) or stage specific (before or after embryonic genome activation) supplementation of EGF + HA on the expression and DNA methylation patterns of genes involved in focal adhesion pathway (Fig 1B). Accordingly, preimplantation embryos were then cultured in six culture conditions and six blastocyst groups were generated for development assessment and molecular analysis. The first blastocyst group was generated from zygotes cultured in basic media (BM), while the second blastocyst group (After_4C) was generated from zygotes culture in BM until 4-cell stage and then transferred to culture media supplemented with 10 ng/ml EGF and 1 mg/ml HA (EGF + HA) until blastocyst stage. The third blastocyst group (After_16C) was derived from zygotes cultured until 16-cell stage in BM then cultured in media supplemented with EGF + HA until the blastocyst stage. On the other hand, the fourth blastocyst group (All_stages) was obtained from zygotes cultured until blastocyst stage in culture media supplemented with EGF + HA. The fifth group (Until_4C) was obtained from zygotes cultured in media supplemented with EGF + HA until 4-cell stage and further cultured until the blastocyst stage in BM while, the sixth blastocyst group (Until_16C) was generated from zygotes cultured in media supplemented with EGF + HA until the 16-cell stage and then cultured in BM until blastocyst stage. Accordingly, in the second experiment three embryo groups were cultured with media supplemented with EGF + HA during the embryonic genome activation period (All_stages, After_4C and Until_16C) however, three groups were cultured with the absence of EGF + HA during the embryonic genome activation period (BM, Until_4C and After_16C). Accordingly, the development data were recorded and four biological replicates of each blastocysts group (10 blastocysts / replicate) were subjected mRNA expression and DNA methylation patterns of candidate genes involved in focal adhesion were analyzed.
Oocytes collection and in vitro maturation
Bovine ovaries were collected from local slaughterhouse (Duren, Germany) and transported to the laboratory within 1–2 hr in a thermal container containing physiological saline solution (0.9% NaCl) at 37°C. After arrival, the ovaries were washed with saline solution and rinsed in 70% ethanol followed by two washes with saline solution. Cumulus oocyte complexes (COCs) were then aspirated from follicles 2–8 mm in size using a 5-ml syringe loaded with an 18- gauge needle. The COCs with more than three compacted cumulus layers and evenly granulated cytoplasm were cultured in groups of 50 in 400 μl of standard maturation medium [tissue culture media (TCM-199) (M-2154; Sigma, Munich, Germany) with Earle salts buffered with 4.43 mM HEPES (H-9136; Sigma, Munich, Germany) and 33.9 mM sodium bicarbonate (S-5761; Sigma, Munich, Germany) supplemented with 12% estrous cow serum (OCS), 0.5 mM L-glutamine, 0.2 mM pyruvate, 50 mg/ml gentamycin sulphate and 10 μg/ml FSH (Folltropin, Vetrepharm, Canada)) in four well dishes (Nunc, Roskilde, Denmark). The COCs were incubated for 22 hr at 38.7°C and 5% CO2 in air with maximum humidity.
In vitro fertilization and embryo production
A group of 50 matured COCs (n = 3550) were in vitro fertilized with 2 × 106 spermatozoa/ml using a commercial frozen-thawed semen (Borken, Germany) in Nunclon dishes in 250 μl modified Tyrode medium supplemented with 10 mM sodium lactate, 1 mM sodium pyruvate, 6 mg/ml bovine serum albumin (BSA), 1 μg/ml heparin, 10 μM hypotaurine, 20 μM penicillamine and 2 μM epinephrine at 38.7°C. The COCs and sperms were incubated in 5% CO2 in air with maximum humidity. Eighteen hours later, the presumptive zygotes were denuded by repeated pipetting, washed three times in PBS and then cultured (n = 50) in 400 μl of SOF culture medium supplemented with or without EGF and/or HA according to the experimental design (Fig 1) in four well dishes (Nunc, Roskilde, Denmark) covered with mineral oil at 38.7°C and 5% CO2 in humidified air. The cleavage and blastocyst rates, blastocyst cell number, apoptotic index, level of reactive oxygen species (ROS) and cryotolerance ability of the embryos were recorded in each treatment group of the first experiment. Moreover, in the second experiment, the cleavage rate and day 7, day 8 and day 9 blastocyst rates were recorded and the day 7 blastocyst stage embryos in both experiments were used for gene expression and DNA methylation analysis.
ROS accumulation assay
Intracellular ROS levels in four biological replicates of day 7 blastocyst stage embryos (20 blastocysts / replicate) was determined using the cell-permeant 2', 7’-dichlorodihydrofluorescein diacetate (H2DCFDA) (Life Technologies, USA) following the manufacturer’s instruction. Briefly, the four independent biological replicates of blastocysts from each group were incubated in 400 μl media containing of 5 μM H2DCFDA for 20 min at 37°C followed by washing twice with PBS-PVA 0.01%. Thereafter, the ROS level was evaluated under inverted fluorescence microscope (Leica DM IRB, Wetzlar, Germany). Afterwards, the images were captured using the green fluorescence filter which emitting 517–527 nm wave length for ROS. The background was adjusted with the negative control then the positive samples were checked using the same adjustments. The image signals were quantified using ImageJ 1.48v software (National Institutes of Health, USA, http://imagej.nih.gov).
Blastocyst total cell number and the inner cell mass (ICM) and trophoectoderm (TE) differentiation assessment
The day 7 blastocyst total cell number of four independent biological replicates of each group (20 blastocysts / replicate) was determined using Hoechst 33342 stain (Sigma, Munich; Germany). Blastocysts from each group were incubated for 5 min in a solution containing 2% formalin and 0.25% gluteraldehyde. Fixed blastocysts were then mounted and stained with 12.5 μg/ml solution of glycerol-based Hoechst 33342 on clean glass slides for 10 min. Stained nuclei were visualized using the epifluorescent microscope (Olympus, Tokyo, Japan), and then the cells number was recorded. Differential inner cell mass and trophoectoderm cells count was performed according to work described previously Handyside and Hunter [23]. Briefly, the blastocysts (20 blastocysts / group) were subjected to zona pellucida removal by treatment with acid tyrode (pH 2.5). Thereafter, zona-free blastocysts were incubated for 30 min at 39°C in a 1:10 dilution of rabbit antibody (Sigma, D-9656; Germany). Blastocysts were incubated for 30 min at 39°C in a solution of complement (guinea pig complement, sigma, Germany), propidiumiodide (Sigma, Germany) and 8% (v/v) FCS in PBS. Afterwards, the blastocysts were fixed in cold ethanol with bisbenzimide stain (Hoechst 33258, Sigma B2883) and kept for 20 hr at -20°C. Fixed blastocysts were then transferred to a drop of glycerol on a glass slide and covered with a cover slip and the cells were counted under a fluorescent microscope (Leica DM-IRB, Germany).
Terminal deoxynucleotidyl transferase (TdT)-mediated dUTP-biotin nick end labelling (TUNEL) assay
TUNEL assay kit (Roche®, Mannheim, Germany) was used to detect dead cells in day 7 blastocysts of different treatment groups. Four independent biological replicates blastocysts (10 blastocysts / group) from each group were fixed, permeabilized and then incubated with drops of TUNEL solutions for 1 hr in a humid chamber at 37°C in dark. Samples used as positive control were treated with DNase (Promega, WI; USA) while, the negative control ones were incubated with making solution. Thereafter, samples were washed and stained with Hoechst 33342 (Sigma, Munich; Germany), then mounted with glycerol on clean slides. The samples were observed under the fluorescence microscope (Leica, Germany) and TUNEL positive (fragmented DNA) cells were recorded.
Cryotolerance test
To investigate the effect of EGF and/or HA on embryo freezability, three biological replicates of the blastocyst stage embryos cultured in BM (n = 67), BM + EGF (n = 59), BM + HA (n = 120) and BM + EGF + HA (n = 63) were subjected to the cryotolerance test. Briefly, the day 7 blastocysts were washed with D-PBS containing 5% PVA and then transferred to a solution consisting of 1.5 M ethylene glycol and 0.1 M sucrose (BoviFreeze, Minitube GmbH, Tiefenbach, Germany). Groups of 10–15 blastocysts were loaded into an open-pulled straw (Minitube GmbH, Tiefenbach, Germany) and then immediately plunged into the programmable freezer “Freeze Control” (Consarctic, Germany). Seeding was accomplished manually with forceps cooled in liquid nitrogen at a temperature of -6°C, followed by a cooling to -30°C at a rate -0.3°C /min. Thereafter, the straws were kept in liquid nitrogen. One week later, the embryos were thawed and washed with SOF media supplemented with fatty acid free BSA. Finally, the embryos were cultured in 400 μl culture media in four well dishes (Nunc, Roskilde, Denmark) covered with mineral oil at 38.7°C in 5% CO2 and humidified air. The expansion and hatching rates were determined at 24, 32, 48 and 56 hr post thawing.
DNA and total RNA isolation and first strand cDNA synthesis
The genomic DNA (gDNA) and total RNA of each experimental group were isolated from four biological replicates of day 7 blastocysts (10 blastocysts / replicate) using RNA/DNA/protein purification plus micro kit (Norgen biotek, Cat# 51600, Canada) according to manufacturer’s instructions. The gDNA was used for DNA methylation analysis whereas the total RNA was used for the gene expression studies. Total RNA concentration was measured using NanoDrop 8000 spectrophotometer (NanoDrop technologies, USA) and the adjusted RNA (100 ng) was reverse transcribed into cDNA using thermo scientific first strand cDNA synthesis kit (Life technologies, GmbH, Germany). Briefly, total RNA of each sample was incubated with 0.5 μl oligo-dT and 0.5 μl random primers at 65°C for 5 min followed by chilling on ice. Afterwards, 1 μl ribolock RNase inhibitor, 4 μl 5x reaction buffer, 2 μl of 10 mM dNTPs mix and 2 μl reverse transcriptase were added and incubated at 25°C for 5 min, 37°C for 60 min followed by 70°C for 5 min. Samples were then stored at -20°C until use.
Gene expression analysis
To determine the effect of EGF and/or HA supplementation during in vitro embryo culture on the expression of genes involved in focal adhesion pathway, the mRNA level of candidate genes was determined using quantitative real time PCR (qPCR). For that, the mRNA expression level of 11 genes namely, actin gamma1 (ACTG1), focal adhesion kinase (FAK), phosphatase and tensin homolog (PTEN), vinculin (VCL), p21 protein (Cdc42/Rac)-activated kinase 4 (PAK4), ras-related C3 botulinum toxin substrate 1 (RAC1), collagen, type IV, alpha 1 (COL4A1), collagen, type I, alpha 2 (COL1A2), epidermal growth factor receptor (EGFR), CD44 molecule (CD44) and hyaluronan-mediated motility receptor (HMMR) were selected for analysis. In addition, the expression level of DNA (cytosine-5-)-methyltransferase 1 (DNMT1), DNA (cytosine-5-)- methyltransferase 3 alpha (DNMT3A) and DNA (cytosine-5-)-methyltransferase 3 beta (DNMT3B) was also investigated. For all samples 2 μl cDNA of four biological replicates were used and the qPCR was performed using Applied Biosystem® StepOnePlus™ System (Thermo Fisher Scientific Inc, USA) using iTaq™ Universal SYBR® Green Supermix (Bio-Rad Laboratories GmbH, Germany) with the following program; 95°C for 3 min, 40 cycles at 95°C for 15 sec, 60°C for 45 min. The data from four independent biological replicates were analyzed using a comparative threshold cycle method (2-ΔΔCT) using beta actin (β-actin) and glyceraldehyde phosphate dehydrogenase (GAPDH) expression as the normalizers. All primers (Table 1) used for qPCR were designed using the online software (http://www.ncbi.nlm.nih.gov/tools/primer-blast/).
Table 1. List of primers used for quantitative real time PCR analysis.
| Accession number | Genes | Primer sequence | Size |
|---|---|---|---|
| NM_173979 | ACTB |
F: 5´-GGCATTCACGAAACTACCTT-3´ R: 5´-CAATCCACACGGAGTACTTG-3´ |
208 |
| NM_001034034 | GAPDH |
F: 5´-ACCCAGAAGACTGTGGATGG-3´ R: 5´-ACGCCTGCTTCACCACCTT-3´ |
247 |
| NM_001033618 | ACTG1 |
F: 5´-ATCCGAAAGGACCTGTATGC-3´ R: 5´-CTTGCTGATCCACATCTGCT-3´ |
213 |
| NM_001075250 | FAK |
F 5´-ACTGGATTCAGTCAAGGCCA-3´ R 5´-CAGCCCTTGTCTGTGAGGTA-3´ |
231 |
| XM_613125 | PTEN |
F:5´-TGGGGAAGTAAGGACCAGAG-3´ R: 5´-ATTGCAAGTTCCACCACTGA-3´ |
172 |
| NM_001191370 | VCL |
F:5´-TTGCAAAGTGTGACCGAGTG-3´ R: 5´-CAACAGCTTGATGGGAGTCG-3´ |
203 |
| NM_001076184 | PAK4 |
F: 5´-TACCAGCATGAGAACGTGGT-3´ R: 5´-CTCTTGATGTCCCGGTGGAT-3´ |
212 |
| NM_174163 | RAC1 |
F: 5´-ACGGAGCTGTGGGTAAGACT-3´ R: 5´-TACATCTGTCTGCGGGTAGG-3´ |
200 |
| NM_001166511 | COL4A1 |
F: 5´-TCTGGATCGGCTACTCCTTT-3´ R: 5´-AACATCTCGCTCCGCTCTAT-3´ |
196 |
| NM_174520 | COL1A2 |
F: 5´-TGAAAAAGGTCATGCTGGTC-3´ R: 5´-TACCCCTTTCTCCTGGTTTG-3´ |
200 |
| HM749883 | EGFR |
F: 5´-GACCCGAAAGAACTGGACAT-3´ R: 5´-TGTTATATCCAGGCCGACAA-3´ |
177 |
| NM_174013 | CD44 |
F: 5´-CTGAAATGAGGGCCCAGTTA-3´ R: 5´- CCAACCCCACTTGAAAGAAA-3´ |
285 |
| NM_001206621 | HMMR |
F: 5´-TGCTTATACTCAGGCCACCC-3´ R: 5´-CGGACATCCTCTGCACTTTG-3´ |
189 |
| NM_182651 | DNMT1 |
F: 5´-TGACTACATCAAGGGCAGCA-3´ R: 5´-AGGTTGATGTCTGCGTGGTA-3´ |
192 |
| NM_001206502 | DNMT3B |
F: 5´-CTGCTGAATTACACTCGCCC-3´ R: 5´-CCAGAAGTATCGGGCTCTGT-3´ |
177 |
| NM_181813 | DNMT3A |
F: 5´-AGCACAACGGAGAAGCCTAA-3´ R: 5´-CAGCAGATGGTGCAGTAGGA-3´ |
245 |
Immunoblotting
Western blot was performed to detect the expression levels of focal adhesion pathway marker proteins (FAK and VCL) in embryos derived from zygotes cultured in media supplemented with or without EGF and/or HA. For this, (30 embryos / replicate) of day 7 blastocysts from each treatment group were boiled with 4 μl 2x SDS loading buffer at 95°C for 5 min and loaded on 4–18% gradient SDS-PAGE gel. After electrophoresis, proteins were transferred to nitrocellulose membrane (Protran®, Schleicher & Schuell Bioscience) and the later was then blocked with Roti-block solution (Carl Roth GmbH) for 1 hr at room temperature. The membrane was overnight incubated at 4°C with anti FAK goat polyclonal antibody diluted at 1:250 (Santa Cruz Biotechnology Inc, Germany). On the second day, the membrane was washed with Tween-Tris-buffer saline (TTBS) and incubated with donkey anti goat secondary antibody diluted at 1:5000 (Santa Cruz Biotechnology Inc, Germany) for 1 hr at room temperature. Thereafter, the membrane was washed with TTBS and incubated in dark with an equal amount of peroxide solution and luminal enhancer for 5 min at room temperature. Images were developed on the ChemiDoc™ XRS+ system (Bio-Rad Laboratories GmbH, Germany). Afterwards, the membrane was subjected to stripping protocol (Bio-Rad, Germany), followed by blocking with the Roti-block solution (Carl Roth GmbH). The membrane was then incubated overnight at 4°C with anti VCL rabbit polyclonal antibody (1:250) and after washing steps it incubated for 1 hr at room temperature with goat anti rabbit secondary antibody diluted at 1:5000 (Santa Cruz Biotechnology Inc, Germany). Afterwards the protein image was detected and the membrane was subjected to stripping and blocking procedure and then incubated overnight at 4°C with anti ACTB mouse monoclonal antibody (1:500) (Santa Cruz Biotechnology Inc, Germany) followed by washing steps and incubation for 1 hr at room temperature with goat anti mouse secondary antibody diluted at 1:5000 (Santa Cruz Biotechnology Inc, Germany) then subjected to image detection process.
Immunohistochemistry
The localization of VCL protein was performed using an immunohistochemistry assay. Briefly, four biological replicates (15 blastocysts / replicate) from each group were washed three times in phosphate- buffer saline (PBS), and then fixed overnight at 4°C in 4% paraformaldehyde in PBS. Fixed samples were washed twice with glycine in PBS and then permeabilized with 0.5% (v/v) Triton-X100 (Sigma, Munich, Germany) in PBS for 4 hr at room temperature. The permeabilized blastocysts were incubated in 3% normal donkey serum (Sigma, Munich, Germany) in PBS for 1 hr at room temperature. Samples were incubated overnight at 4°C with anti VCL rabbit polyclonal antibody diluted at 1:250 (Santa Cruz Biotechnology Inc, Germany). Afterwards, blastocysts were incubated at 37°C for 2 hr in the dark with FITC-conjugated goat anti-rabbit secondary antibody diluted at 1:200 (Lifespan Biosciences, Seattle, WA). Blastocysts were mounted in mounting medium containing DAPI on a clean slid glass and were then visualized under the CLSM LSM-780 confocal laser scanning microscope (Zeiss, Germany). For that, the confocal microscope was adjusted using the suitable wavelength for FITC (excitation/emission, 495/517) and DAPI dyes at 40x magnification then the background was adjusted using the negative control samples (blastocysts incubated with secondary antibody only) thereafter, images were acquired for all samples. The images were analyzed using ImageJ 1.48v software (National Institutes of Health, USA, http://imagej.nih.gov). The images were analyzed separately and the intensity of the signal from each embryo was recorded and the data were subjected to statistical analysis.
Bisulfite sequencing
The genomic DNA (gDNA) isolated from four biological replicates of day 7 blastocysts was subjected to bisulfite treatment using EZ DNA methylation direct kit (Zymo Research, USA) and amplified using gene specific primers (Table 2) according to the manufacturer’s recommendation. The primers for bisulfite sequencing were designed using the online software (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi). The PCR amplification was performed using 2 μl of bisulfite treated gDNA, 1 μl of forward primer, 1 μl of reverse primer, 12.5 μl of 2x reaction buffer, 0.3 μl of 10 mM dNTP mix, 0.2 μl of Taq polymerase, Zymo Taq™ DNA polymerase (Zymo Research, USA) and nuclease free water was added up to 25 μl and the mix was then incubated in thermocycler using PCR touchdown protocol. The PCR thermocycler was adjusted using touch down program as following 95°C 5 min 1 cycle, 95°C 30 sec and touch down 0.5°C each cycle till reach the annealing TM followed by 72°C 1 min for 10 cycles, 95°C 30 sec, 55°C 30 sec followed by 72°C 1 min for 35 cycles and 72°C 10 min 1 cycle. The presence of the PCR product was confirmed after loading 5 μl of the PCR product on 2% agarose gel electrophoreses run on 1% TE buffer. The remaining PCR product was purified using the QIAquick PCR purification (Qiagen, Germany), cloned to pGEM®-T Easy Vector Systems (Promega, WI, USA) and transformed to E. coli competent cells. The bacterial culture was then plated onto the LB agar/ampicillin/IPTG/X-gal plate and incubated overnight at 37°C. Independent clear white colonies were selected for sequencing. For that, the colonies were lysed at 95°C for 15 min and 10 μl of the lysate was used for PCR amplification using M13 primer. The samples (10 μl) in addition to 0.5 μl of M13 forward primer, 0.5 μl of M13 reverse primer, 2 μl of 10x reaction buffer, 0.5 μl of 10 mM dNTP mix, 0.2 μl of Taq polymerase and nuclease free water was added up to 20 μl were incubated in thermocycler using the following PCR protocol, 95°C 30 sec, 60°C 30 sec followed by 72°C 1 min for 40 cycles and 72°C 10 min 1 cycle The PCR product was then subjected to sequencing protocol and sequenced using the GenomeLab™ GeXP Genetic Analysis System (Beckman Coulter). The bisulfite sequencing DNA methylation analysis software (BISMA) (http://services.ibc.uni-stuttgart.de/BDPC/BISMA/) was used to analyze the sequencing data.
Table 2. List of primers used for DNA methylation analysis.
| Accession number | Genes | Primer sequence | Number of CpGs in the product |
|---|---|---|---|
| ENSBTAG00000013472 | COL1A2 |
F: 5´-GGGATTTTAAGTTTATTTTTTAATAAA-3´ R: 5´-AACCCTACCTACCTTATACCCTAC-3´ |
16 |
|
F: 5´- TGGTTTGTTGGTAAAGTTTATTTTTTT -3´ R: 5´-CCAACCCTACCTACCTTATACCCTAC-3´ |
22 | ||
| ENSBTAG00000012849 | COL4A1 |
F 5´-GGGTAGGATTTTATATTAGTTTTTGATGTT-3´ R 5´-ACCTTCTCTTAAATTCCCCTTCAAT-3´ |
7 |
|
F: 5´-GATAGAGGAGAAGGGGAATTTAGGAT-3´ R: 5´-AACTCCCACCAAAAACCCTATTT-3´ |
9 | ||
| ENSBTAG00000009233 | RAC1 |
F 5´-AAGTGAATGGTAGTTTTTAGGAATTT-3´ R 5´-AAAAAAATTATTTACCTCCCATTAATA-3´ |
8 |
|
F 5´-TTTTTGGGGTTTGGATATTTGTAT-3´ R 5´-AAACCAAACACCCCTAAAACTAAAC-3´ |
14 |
Statistical analysis
The data of this study were analyzed using Statistical Analysis System (SAS) version 9.1 software (SAS Institute Inc., Cary, NC, USA). One-way analysis of variance (ANOVA) followed by Tukey multiple pairwise comparison was performed between treatment groups. Number of blastocysts survived after cryopreservation was analyzed using the chi square test. Differences were considered significant when P < 0.05.
Ethics approval and consent to participate
The study was conducted on bovine in vitro produced blastocysts using oocytes collected from slaughterhouse ovaries and thus special approval of this experiment was not required.
Results
Hyaluronic acid supplementation influenced embryo development and apoptotic index
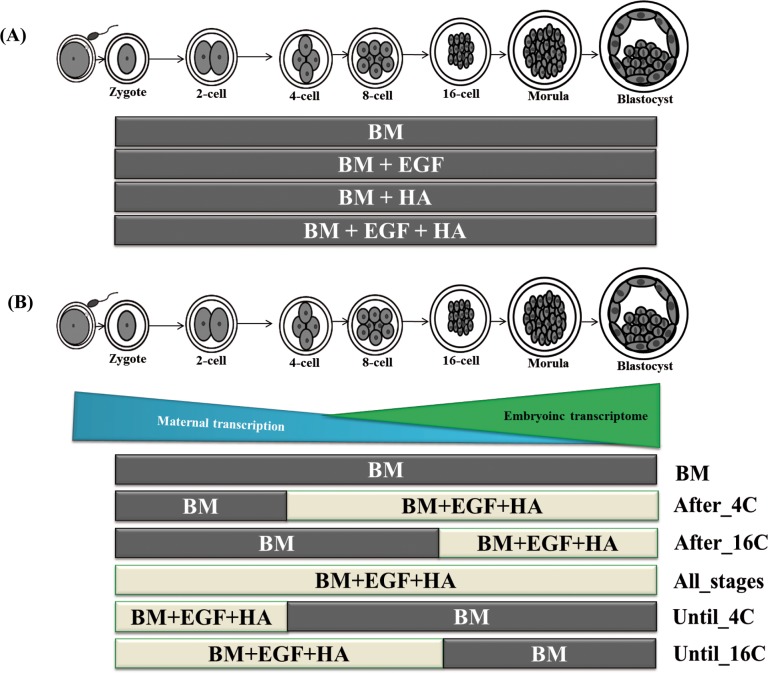
Supplementation of culture media with EGF, HA or EGF + HA did not affect the cleavage rate and blastocyst formation compared to the control group. Nevertheless, the day 7 blastocyst rate was significantly higher in the HA supplemented group compared to the EGF group (Table 3). Moreover, supplementation of HA resulted in insignificant increases in blastocyst total cell number and the ICM: TE ratio (Table 3). On the other hand, supplementation of HA reduced the apoptotic index in the resulting blastocysts compared to EGF counterparts (Fig 2).
Table 3. The development rate and blastocysts cell number of embryos cultured in presence of EGF, HA or EGF + HA throughout preimplantation period.
| BM | BM + EGF | BM + HA | BM + EGF + HA | |
|---|---|---|---|---|
| No. zygotes | 425 | 416 | 466 | 446 |
| Cleavage rate | 81.3 ± 4.9 | 82.2 ± 5.9 | 84.4 ± 6.2 | 82.1 ± 3.8 |
| Day 7 blastocyst rate | 28.9 ± 7.1ab | 23.8 ± 7.08a | 32.1 ± 6.1b | 28.7 ± 5.3ab |
| Day 8 blastocyst rate | 42.7 ± 8.1 | 37 ± 8.2 | 42.1 ± 5.6 | 38.6 ± 5.6 |
| Day 9 blastocyst rate | 46.1 ± 6.4 | 41.5 ± 7.4 | 44.9 ± 5.9 | 39.7 ± 7.4 |
| Blastocyst total cell number | 127±7 | 123±13 | 131±4 | 118±6 |
| ICM:TE ratio | 1.73 | 1.74 | 1.87 | 1.81 |
Data represented as mean ± SEM and different letters in the same raw indicate significantly different (p < 0.05.).
Fig 2. TUNEL assay in blastocysts of different blastocysts group.
(A) Representative blastocysts showing TUNEL positive cells. (B) The percentage of apoptotic nuclei is presented as mean ± SEM. Bars with different letters are statistically significant different at p < 0.05.
Epidermal growth factor and/or hyaluronic acid supplementation reduced the oxidative stress and increased the cryotolerance ability in the resulting blastocysts
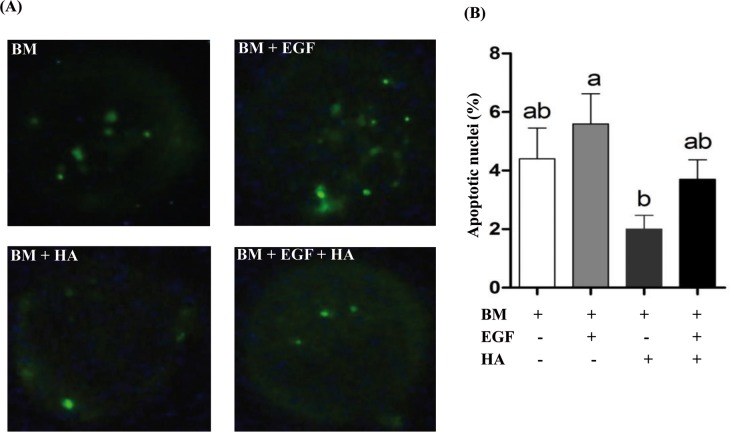
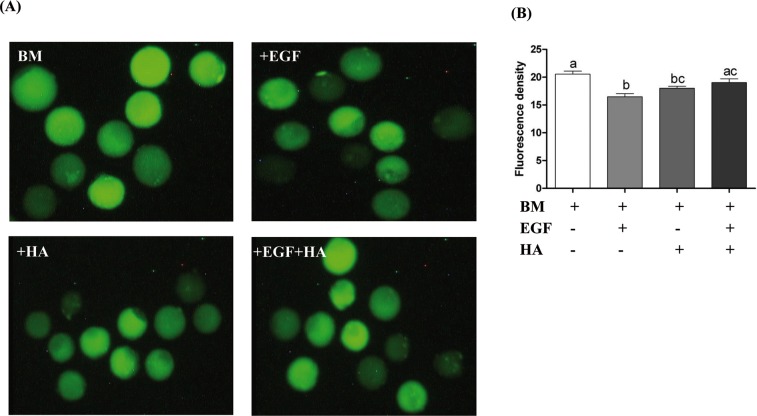
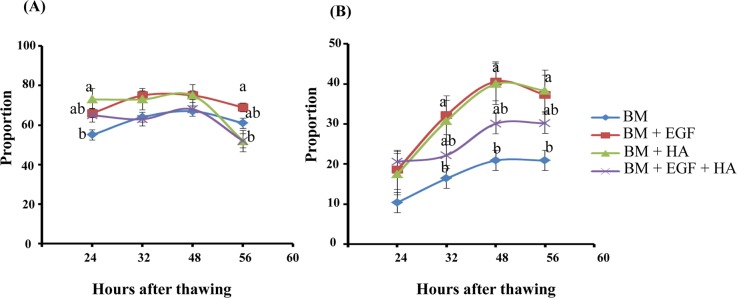
In this study, the reactive oxygen species (ROS) levels were significantly reduced in the blastocysts derived from zygotes cultured in culture media supplemented with EGF or HA compared to control ones. However, blastocysts derived from zygotes supplemented with EGF + HA did not show a significant difference in ROS levels compared to control and HA groups (Fig 3). In addition, the blastocysts derived from zygotes cultured in media supplemented with HA exhibited higher re-expansion rates at 24 hr after thawing compared to the control group. However, at 56 hr post thawing, the re-expansion rate was significantly reduced in the HA group compared to the EGF group (Fig 4A). Moreover, the hatching rate was significantly higher in EGF and HA groups at 32, 48 and 56 hr after thawing compared to the control counterparts (Fig 4B).
Fig 3. Intracellular ROS accumulation in blastocysts derived from zygotes cultured in media supplemented with EGF and/or HA.
(A) Representative figures displaying the ROS accumulation levels in the blastocysts. (B) The fluorescent density (mean ± SEM) showing the level of ROS in each blastocyst group. Bars with different letters are statistically significant (p < 0.05). Scale bars represent 100 μm.
Fig 4. Effects of EGF and/or HA supplementation on the cryotolerance ability of the embryos.
Expansion (A) and hatching rates (B) of blastocysts at 24, 32, 48 and 56 hr after thawing of embryos supplemented with EGF and/or HA. Data are presented as mean ± SEM and different letters on the bars indicate statistical significant differences (p < 0.05).
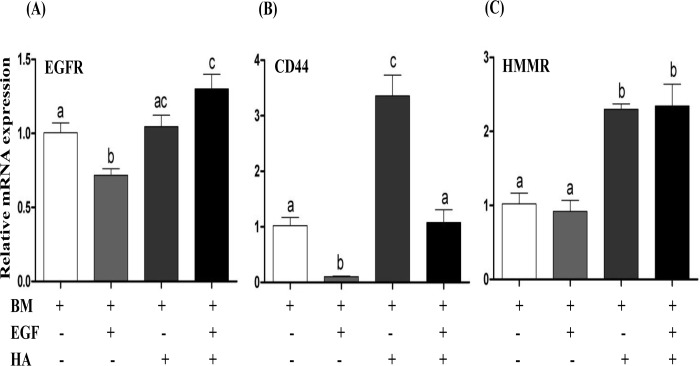
Epidermal growth factor and/or hyaluronic acid supplementation altered the expression level of epidermal growth factor and hyaluronic acid receptor genes
In order to identify the association of EGF and/or HA supplementation and the activity of their receptors, the mRNA abundance level of EGF and/or HA receptors were quantified in blastocysts derived from embryos developed in the culture media supplemented with EGF and/or HA during the entire preimplantation period. While supplementation of HA increased the expression level of its receptor (CD44 and HMMR) genes, supplementation of EGF significantly reduced the mRNA expression level of its receptor (EGFR). However, when EGF was combined with HA, the expression level of EGFR was significantly increased compared to control and EGF groups (Fig 5).
Fig 5.
Supplementation of EGF and/or HA on the expression level of EGF (A) and HA receptors (B and C). Data are presented as mean ± SEM. Bars with different letters are statistically significant (p < 0.05).
Epidermal growth factor and hyaluronic acid supplementation induced the expression of genes involved in focal adhesion pathway
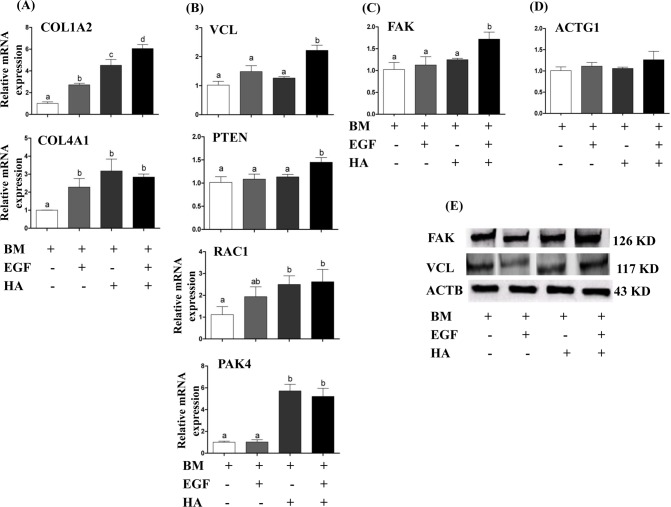
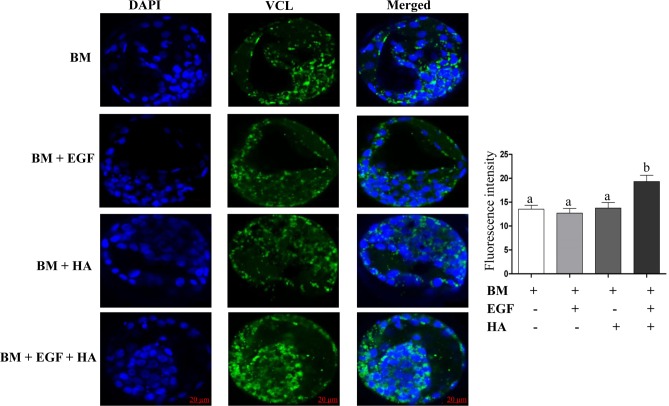
The expression level of candidate genes involved in focal adhesion pathway was analyzed in blastocysts derived from treatment of different groups. For this, the mRNA level of the genes representing the ligands (COL1A2 and COL4A1), cytoplasmic structures (ACTG1), adaptors and focal adhesion marker (VCL), enzymatic regulation (FAK), inhibitor of focal adhesion kinase (PTEN) and actin regulators (RAC1 and PAK4) was quantified using qPCR. The results revealed that except ACTG1, the expression level of the candidate genes was increased in the blastocysts derived from embryos cultured in the presence of EGF + HA during the entire preimplantation period (Fig 6). Furthermore, immunohistochemical analysis indicated higher VCL protein in the cell membrane of the blastocysts derived from embryos cultured in media supplemented with EGF + HA (Fig 7).
Fig 6.
The mRNA expressions level of ligands (A), adaptor (B), regulator (C) and structure linker (D) of focal adhesion pathway genes in blastocysts of different treatment groups. (E) Western blot analysis of FAK and VCL proteins in blastocysts derived from zygotes supplemented with or without EGF and/or HA (E). Bars represent mean ± SEM. Different letters on bars are statistically significant (p < 0.05).
Fig 7. Immunohistochemical analysis of VCL, focal adhesion protein marker gene, in blastocysts derived from zygotes cultured in different treatment groups.
The green colour indicates the expression of protein, while the blue colour indicates the nuclear staining using 4′,6-diamidino-2-phenylindole (DAPI).
Combined supplementation of epidermal growth factor and hyaluronic acid altered expression of genes involved in focal adhesion pathway in a stage specific manner
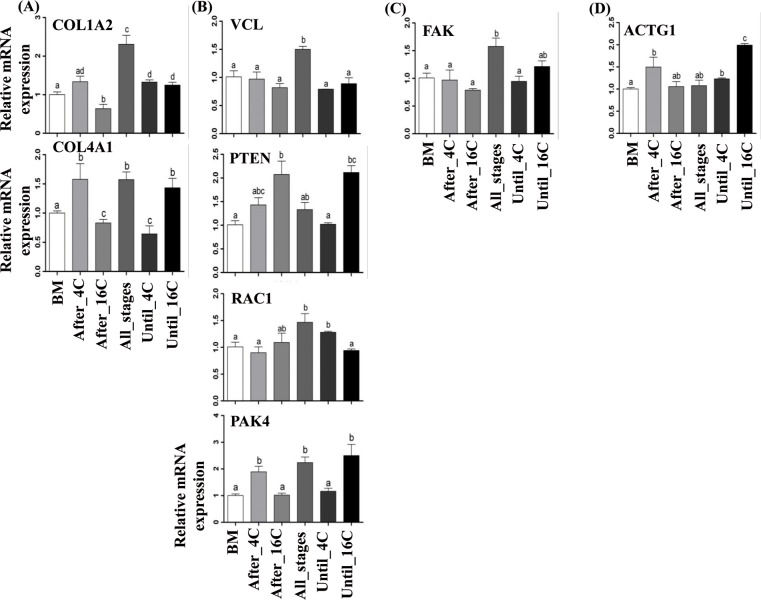
The results from the first experiment indicated that the preimplantation embryos cultured from zygote to blastocyst stage in the presence of EGF + HA showed a significant increase in the mRNA and protein expression level of genes involved in focal adhesion pathway. However, in the first experiment, supplementation of EGF and/or HA was performed through the preimplantation period and it was not clear whether the effect of EGF + HA was prominent before, during or after the embryonic genome activation. Therefore, to address this, embryos were supplemented with EGF + HA after 4-cell (After_4C) or after 16- cell stage (After_16C). Other embryo groups were cultured in BM supplemented with EGF + HA until 4-cell (Until_4C) or until 16-cell stage (Until_16C) and then cultured only in BM until the blastocysts stage. Although the number of embryos reached day7 blastocyst stage was higher in Until_16C compared to Until_4C or After_16C while was higher at day8 and day9 blastocyst stage of After_4C group, however, there was absence of significant differences between all experimental groups (Table 4). On the other hand, the gene expression analysis indicated that the expression level of the candidate genes (COL4A1, PAK4and ACTG1) was higher in the blastocysts of After_4C and Until_16C supplemented groups compared to After_16C supplemented group (Fig 8). However, unlike other genes, the expression level of RAC1 was increased in the blastocysts of Until_4C group.
Table 4. The correlation between the DNA methylation and expression patterns of the candidate genes in blastocysts of different groups.
| Expression = E | After_4C | After_16C | All_stages | Until_4C | Until_16C | BM + EGF | BM + HA | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Methylation = M | E | M | E | M | E | M | E | M | E | M | E | M | E | M | |
| COL1A2 | Promoter | ↑ | ↑ | ↓ | ↑ | ↑ | ↓ | ↑ | ↓ | ↑ | ↑ | ↑ | ↑ | ↑ | ↓ |
| Distal Promoter | ↑ | ↑ | ↓ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | |
| COL4A1 | Promoter | ↑ | ↑ | ↓ | ↑ | ↑ | ↑ | ↓ | ↑ | ↑ | ↑ | ↑ | ↑ | ↑ | ─ |
| Distal Promoter | ↑ | ↓ | ↓ | ↓ | ↑ | ↑ | ↓ | ↑ | ↑ | ↓ | ↑ | ↓ | ↑ | ↓ | |
| RAC1 | Promoter | ─ | ─ | ─ | ↑ | ↑ | ↑ | ↑ | ↑ | ─ | ─ | ─ | ↑ | ↑ | ↑ |
| Intron | ─ | ─ | ─ | ─ | ↑ | ↑ | ↑ | ↑ | ─ | ↑ | ─ | ↓ | ↑ | ─ | |
Symbols ↑ and↓ indicate up and downregulation of gene expression, respectively while ↑ and ↓ indicate the hypermethylation and hypomethylation of the gene, respectively.
Fig 8.
Supplementation of EGF + HA before or after embryonic genome activation on the expression pattern of ligands (A), adaptor (B), regulator (C) and structure linker (D) of focal adhesion pathway genes. Data presented as mean ± SEM. Different letters are statistically significant (p < 0.05).
Supplementation of epidermal growth factor and/or hyaluronic acid altered the DNA methylation patterns of genes involved in focal adhesion pathway
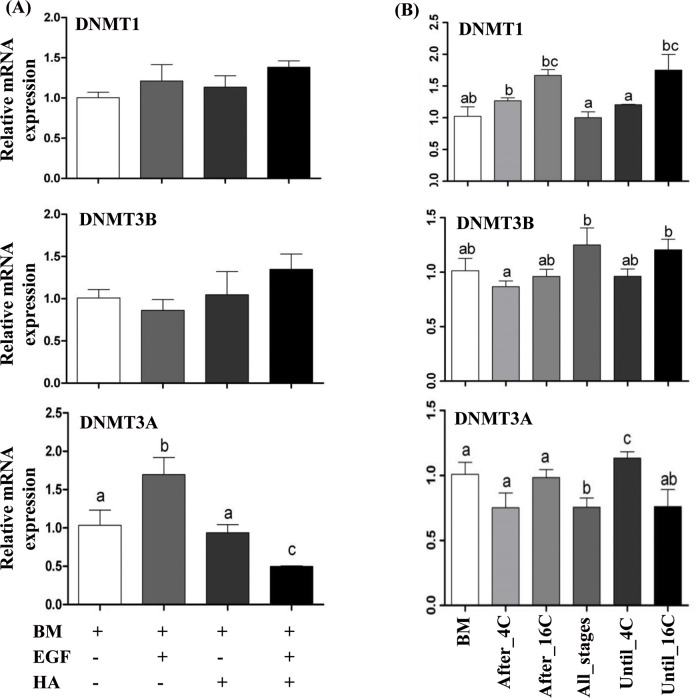
Since supplementation of EGF and/or HA affected both the blastocyst quality and the expression of genes involved in focal adhesion pathway, we speculated that EGF and/or HA may affect the embryo quality by epigenetically regulating the genes involved in focal adhesion pathway. For this, first the expression levels of genes associated with DNA methylation namely, the DNA methyltransferase related genes (DNMT1, DNMT3A and DNMT3B) were investigated in the blastocysts obtained from embryos cultured in media supplemented with EGF, HA and EGF + HA from zygote to blastocyst stage or with EGF + HA before or after embryonic genome activation. Results showed that the expression level of DNMT3A was significantly increased in the blastocysts derived from culture media supplemented with EGF and decreased in the EGF + HA group. However, HA alone or in a combination with EGF did not show any significant effects on the expression levels of DNMT1 and DNMT3B (Fig 9A). Nevertheless, the expression level of DNMT1 was significantly higher in After_4C supplemented group compared to Until_4C supplemented ones. In addition, the expression level of DNMT3B was significantly higher in blastocysts of Until_16C compared to After_4C group, whereas the mRNA level of DNMT3A was significantly increased in the Until_4C supplemented group (Fig 9B).
Fig 9.
Relative mRNA expression level of DNMT1, DNMT3B and DNMT3A in blastocysts derived from zygotes cultured in media supplemented with or without EGF and/or HA until blastocyst stage (A) or blastocysts derived from embryo supplemented with or without EGF + HA before or after embryo genome activation (B). Data are presented as mean ± SEM. Bars with different letters are statistical significant (p < 0.05).
Following the expression analysis, we have performed DNA methylation analysis in three candidate genes, as their expression was affected by continued or stage specific supplementation of EGF + HA and their critical role in focal adhesion pathway. The three selected genes are namely COL4A1 (Chr. 12, 11623958–11624257 with 7 CpG sites at the promoter region and 9 CpG sites at distal promoter region 11623579–116239544), COL1A2 (Chr. 12, 89010212–89010447 with 16 CpG sites at the promoter region and 22 CpG sites at the distal promoter region 890106686–890106949) and RAC1 (Chr. 25, 38828881–38829079 with 8 CpG sites at the prompter region and 14 CpG sites at the intronic region 38834424–38834685). DNA methylation analysis was performed using bisulfate sequencing from four biological pools of embryos in each experimental group. About 16 clones from each experimental group were subjected for sequencing and 8 clones with accurate sequencing results were subjected for final analysis.
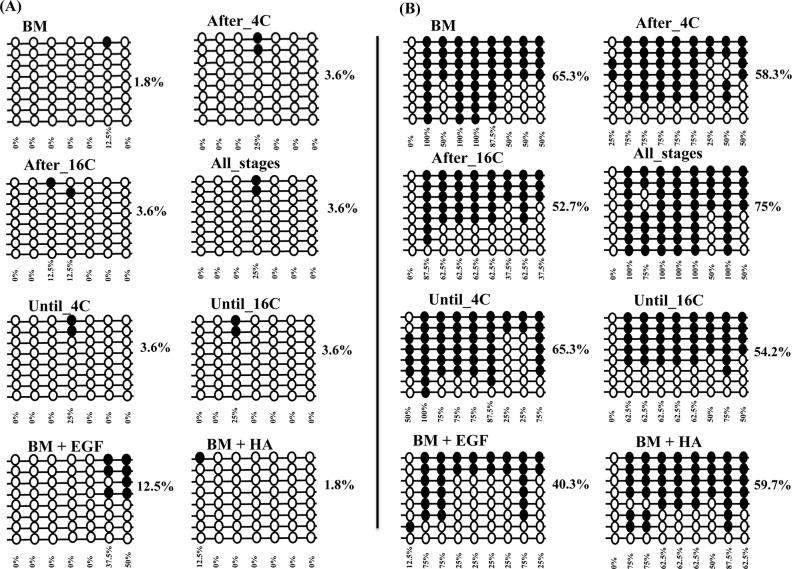
Results revealed that, unlike HA supplemented group, in vitro culture of embryos in the presence of EGF alone or in a combination with HA increased and decreased the DNA methylation level of the COL4A1 gene at promoter and distal promoter regions, respectively compared to the control group. On the other hand, except Until_4C group, the DNA methylation level of COL4A1 gene was reduced in the stage specific supplemented groups (EGF + HA before, during or after embryo genome activation) compared to control one (Fig 10).
Fig 10.
The DNA methylation level of COL4A1 at the promoter (A) or distal promoter (B) region of blastocysts of different treatment groups. The rows indicate the sequence of different colonies while, the circles indicate the CpG sites within the sequence. The black and white circles indicate the methylated and unmethylated CpG sites, respectively.
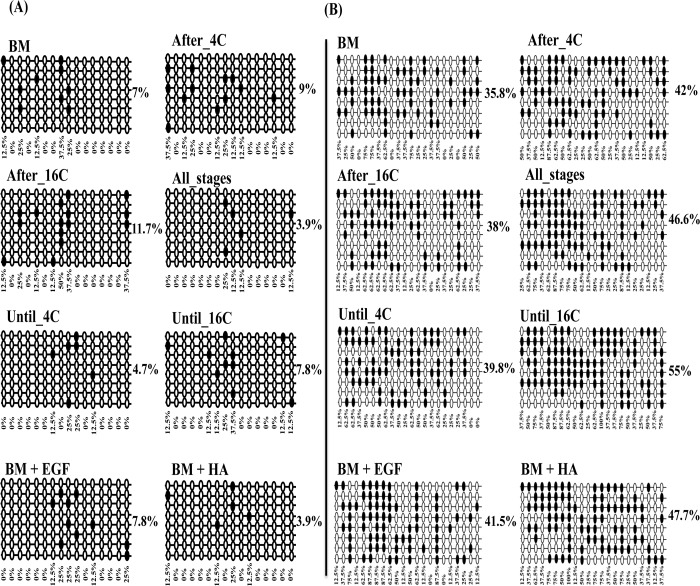
Similarly, results showed that supplementation of HA alone or in combination with EGF through the preimplantation period decreased and increased the promoter and distal promoter DNA methylation level of COL1A2 gene in the resulting blastocysts. While, supplementation of EGF + HA after the 16-cell stage (After_16C) increased the DNA methylation at the promoter region of COL1A2 gene compared to Until_4C or Until_16C groups. However, higher DNA methylation at distal promoter region of COL1A2 was detected in blastocysts derived from Until_16C group (Fig 11).
Fig 11.
The DNA methylation level of COL1A2 at the promoter (A) or distal promoter (B) region of blastocysts of different treatment groups. The rows indicate the sequence of different colonies while, the circles indicate the CpG sites within the sequence The black and white circles indicate the methylated and unmethylated CpG sites, respectively.
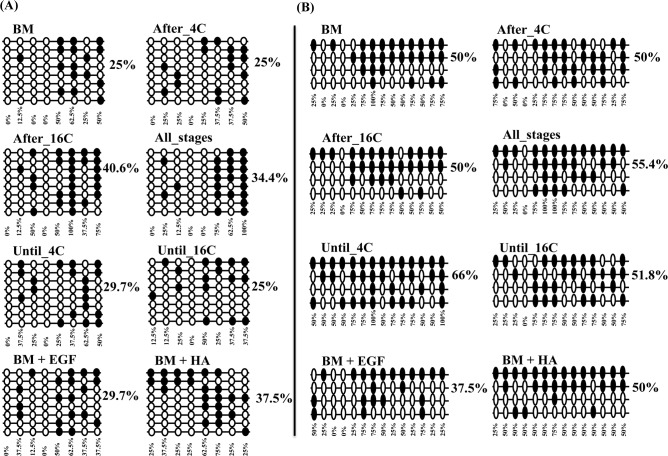
In addition, the results showed that supplementation of embryo during the entire preimplantation period with EGF and/or HA increased DNA methylation level of RAC1 at the promoter region. However, EGF supplementation resulted in reduction of DNA methylation level at intronic region of the same gene. Moreover, the results from stage specific supplementation of EGF + HA showed that blastocysts obtained from After_16C group had the highest DNA methylation level at promoter region of RAC1 gene (Fig 12). At the intronic region of RAC1 gene, higher DNA methylation was detected in the blastocysts of the Until_4C group compared to blastocysts of all groups.
Fig 12.
The DNA methylation pattern of RAC1 gene at promoter (A) or gene body region (B) of blastocysts of different treatment groups. The rows indicate the sequence of different colonies while, the circles indicate the CpG sites within the sequence The black and white circles indicate the methylated and unmethylated CpG sites, respectively.
Comparative analysis of gene expression and DNA methylation
Once, we have analyzed the expression and DNA methylation patterns in blastocysts of different groups, we performed comparative analysis between the expression and DNA methylation levels. For this, the DNA methylation data of COL4A1, COL1A2 and RAC1 were superimposed to the expression level. Accordingly, the results indicated that the DNA methylation pattern of COL4A1 at the promoter and distal promoter regions was negatively correlated with the gene expression patterns in blastocysts of EGF supplemented group. Moreover, at distal promoter of COL4A1, the DNA methylation pattern was negatively correlated with the corresponding gene expression in all treatment groups except After_16C, All_stages groups (Table 4). However, the DNA methylation level in the distal promoter region of COL1A2 was positively correlated with its gene expression in all blastocysts groups except in After_16 group. On the other hand, the DNA methylation of COL1A2 at the promoter region was negatively correlated with the corresponding gene expression in blastocysts of After_16C, All_stages, Until_4C and HA groups. Moreover, at the intronic region of the RAC1 gene, the DNA methylation pattern was positively correlated with gene expression in all blastocyst groups except After_16C, EGF and HA (Table 4).
Discussion
Robust embryo-maternal communication permits normal development, high quality embryos, proper implantation and maintenance of pregnancy [24]. Thus, understanding the mechanism through which the preimplantation embryos communicate with in vivo or in vitro extracellular micro-environment is one of the most vital aspects to be considered in order to unravel the effects of suboptimal environmental conditions during embryogenesis. In this regard, our previous results demonstrated that, in vivo and in vitro alternative culture conditions altered the expression and DNA methylation pattern of genes involved in focal adhesion pathway in the resulted blastocysts [2, 3].
Focal adhesion is one of the essential molecular signaling pathways that regulate interaction and communication between the cell and the extracellular micro environment [25]. Focal adhesion is required for normal cell growth [26] and cellular functions [6, 25]. Indeed, previous studies indicated that normal cell growth is associated with stimulation of cell adhesion in the presence of growth factors [8]. In this regard, supplementation of epidermal growth factor modulated cell growth [27] and improved embryo development [28–34]. The potential effects of EGF may regulate by redistributing focal adhesion constituents to adhesion sites and by amplifying the levels of focal adhesion protein members [35]. Additionally, hyaluronic acid is an extracellular matrix component which is highly abundant in follicular, oviduct and uterine fluids of different species [36–39], is involved in multicellular functions [40–44] by increasing cell-to-cell communication and cell-to-extracellular matrix adhesion [45].Interestingly, supplementation of HA to in vitro culture media improved bovine and porcine oocyte maturation and embryo developmental competence [46–48]. Therefore, in the current study, bovine preimplantation embryos were in vitro cultured in the presence or absence of EGF and/or HA with to elucidate the potential effect of EGF and HA on bovine embryonic development and epigenetic landscape of genes involved in focal adhesion pathway. Results revealed that EGF and/or HA did not significantly affect the cleavage and the blastocyst rates. These results were in consistent with previous reports [19, 49, 50] which indicated that, EGF alone or in combination with HA did not influence the bovine blastocyst rate while other studies indicated that HA increased bovine blastocyst rate [46, 47, 51]. However, there was a positive effect of HA on reducing the percentage of apoptotic cells, suggesting the potential role of hyaluronic acid in regulation of cell proliferation and apoptosis compared to EGF group [44].
One of the critical cellular phenotypes that might mark the development and quality of in vitro produced embryos is the level of intracellular reactive oxygen species (ROS) [52]. In line to this, the current study revealed that EGF or HA significantly reduced the ROS levels in the resulted blastocysts. Indeed, EGF [53] and HA are believed to reduce the embryo intracellular ROS levels by regulating Nrf2 and Akt genes [54]. Nevertheless, the interaction between EGF and HA receptors is believed to increase the ROS levels [55]. Furthermore, cryo-preservability of the embryo is one of the main quality indicators of embryos developed under different culture conditions [56–58]. In the current study, HA was found to improve the blastocyst cryotolerance which was revealed by significantly higher blastocyst hatching rate at different time points after thawing. Previous studies also showed that, HA improved embryo cryotolerance and increased pregnancy rate after embryo transfer [13, 59, 60].
Following the phenotypic assessments, the effect of EGF and HA on the expression level of EGF receptor (EGFR) and HA receptors (CD44 and HMMR) was determined. EGF resulted in a significant decrease in the expression level of EGFR compared to other groups. These findings were in agreement with the previous studies which indicated that, excessive binding of EGF with its receptors led to reduction in mRNA level of EGFR [61]. On the other hand, a combined supplementation of EGF and HA induced the expression level of EGFR, this may suggest the potential role of HA in EGFR activation through specific receptors of HA namely, CD44 and HMMR [51, 62–65]. Moreover, CD44 is regulated by EGFR and the inhibition of EGFR resulted in a reduction of CD44 mRNA expression level [66]. In the present study HA induced the expression level of CD44 and HMMR receptors. HA is believed to promote the rapid appearance of HMMR expression [67] and the number of HMMR receptors increased significantly at morula stage of bovine embryos and then reduced when embryos are cultured in serum-containing medium [68].
The mRNA expression analysis revealed that, a combined supplementation of EGF and HA increased the expression level of candidate focal adhesion genes including the focal adhesion kinase (FAK) gene. FAK is a non-receptor tyrosine kinase focal adaptor protein which is localized in integrin-focal sites [69] to link the growth factors and integrins [35]. The expression of FAK is induced either by direct effect of growth factors such as EGF [35] or by the action of extracellular signal-regulated kinase1/2 (ERK1/2) which is activated by EGF and HA stimulation [35, 43]. The active FAK induces the phosphatidylinositol 3-kinase (PI3K) expression [69], ultimately leading to the induction of the RAC1 and PAK family genes [70, 71]. In line with this, our results revealed a higher expression level of RAC1 and PAK4 genes with the presence of HA. In fact, FAK and RAC1 are believed to induce focal adhesion complex proteins such as vinculin (VCL), a focal adhesion marker protein that controls focal adhesion [72, 73]. Our results also revealed a higher mRNA and protein expression levels of vinculin in blastocysts of EGF + HA compared to other groups.
Apart from this, considering the fact that major bovine EGA occurs around the 8- to 16-cell stages [74, 75] and the minor genome around the 4-cell embryonic stage of development, blastocysts were generated according to experimental design (Fig 1B). In this study there was absence of significant differences in embryonic development rates between all groups. On the other hand, the supplemented with EGF + HA from 4-cell stage until blastocyst stage (After_4C) or from zygote stage until 16-cell stage (Until_16C) increased the expression level of focal adhesion genes, indicating the importance of EGF + HA during the time of embryonic genome activation.
Additionally, the expression pattern of DNA methyltransferase genes (DNMT1, DNMT3A and DNMT3B) was investigated to get insight about the effects of EGF and/or HA on the expression pattern of genes involved in DNA methylation mechanism during embryonic development. The expression level of DNMT3A, which is one of the key genes involved in de novo DNA methylation, was significantly increased by the supplementation of EGF. However, it was significantly reduced with the supplementation of EGF + HA compared to control or HA groups. Similarly, previous studies also indicated higher expression of DNMT3A in bovine embryos cultured under different in vitro culture conditions [76]. On the other hand, the presence or absence of EGF + HA in culture media during the entire embryo genome activation period altered the expression level of DNA methyltransferase genes. These findings suggested that destabilization of embryos by the addition or removing of a single molecule particularly during entire genome activation period may lead to the disturbance of the DNA methylome of genes related to development. Therefore, we assumed that, the alteration of DNA methyltransferases particularly DNMT3A may influence the DNA methylation pattern of genes involved in focal adhesion pathway. For this, COL1A2, COL4A1 and RAC1 genes were selected for DNA methylation analysis based on their mRNA expression pattern. Bisulfite sequencing data revealed that the DNA methylation pattern of these candidate genes was changed by continued or stage specific supplementation of EGF and/or HA at the promoter, distal promoter or gene body regions. These results were found to be in agreement with our previous findings reported of in vitro-in vivo alternative embryo culture conditions [3]. Moreover, the DNA methylation level at the promoter regions of COL4A1, COL1A2 and RAC1 genes was induced in the EGF group compared to the HA one, but the gene expression tended to be lower in the EGF group compared to the HA group. Furthermore, the DNA methylation pattern of the candidate genes was inversely or positively correlated to the gene expression in the promoter, distal promoter or in the gene body regions. Previous studies reported a negative correlation between the DNA methylation level at promoter region and gene expression pattern [77] while, methylation at gene body correlates with gene activity [78]. Although our results did not show specific trend of the overall DNA methylation percentage of the selected genes however, the loss or gain of DNA methylation at specific CpG sites within the CpG island of candidate regions may result in different expression pattern of candidate gene. It was reported that, differentially DNA methylation patterns at a single CpG site could influence the transcription factors access leading to activation or inactivation of the gene [79, 80].
In conclusion, this was the first study that demonstrated the effect of culture media supplemented with EGF and/or HA before, during or after embryonic genome activation on development and quality of bovine preimplantation embryos, expression and DNA methylation pattern of genes involved in focal adhesion pathway. The presence of EGF and/or HA during bovine preimplantation embryo development altered the expression and DNA methylation patterns of genes involved in focal adhesion pathway associated with embryonic competence. Further studies are needed to investigate the effect of those components on embryo differentiated cells with the corresponding DNA methylome changes of specific CpG sites.
Acknowledgments
We are grateful to Affairs Sector and missions (Egyptian Ministry of Higher Education) for its support to Dr. Mohammed Saeed-Zidane during the study. We also thankful to Mrs. Birgit Koch-Fabritius at ITW, Bonn University for her help during bisulfite sequencing.
Abbreviations
- EGF
Epidermal growth factor
- HA
Hyaluronic acid
- DNA
Deoxyribonucleic acid
- ECM
extracellular matrix
- BM
Basic media
- SOF
synthetic oviduct fluids
- After_4C
Blastocyst group that was generated from zygotes culture in BM until 4-cell stage and then supplemented with 10 ng/ml EGF and 1 mg/ml HA (EGF + HA) until blastocyst stage
- After_16C
Blastocyst group that was derived from zygotes cultured until 16-cell stage in BM followed by EGF + HA supplementation until the blastocyst stage
- All_stages
Blastocyst group that was obtained from zygotes cultured until blastocyst stage in BM supplemented with EGF + HA
- Until_4C
Blastocysts group that was obtained from zygotes cultured in BM supplemented with EGF + HA until 4-cell stage and further cultured until the blastocyst stage in BM
- Until_16C
Blastocyst group that was generated from zygotes cultured in BM supplemented with EGF + HA until the 16-cell stage and then cultured in BM until blastocyst stage
- HAmRNA
Messenger RNA
- COCs
Cumulus oocyte complexes
- PBS
Phosphate buffer saline
- ROS
Reactive oxygen species
- ICM
Inner cell mass
- TE
Trophoectoderm
- TUNEL
Terminal deoxynucleotidyl transferase (TdT)-mediated dUTP-biotin nick end labelling
- qPCR
quantitative real time PCR
Data Availability
All relevant data are within the paper.
Funding Statement
Dr. Mohammed Saeed-Zidane was funded by Affairs Sector and missions (Egyptian Ministry of Higher Education). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Lonergan P, Rizos D, Gutiérrez-Adán A, Fair T, Boland MP. Effect of culture environment on embryo quality and gene expression—experience from animal studies. Reprod Biomed Online. 2003;7(6):657–63. 10.1016/s1472-6483(10)62088-3 [DOI] [PubMed] [Google Scholar]
- 2.Gad A, Hoelker M, Besenfelder U, Havlicek V, Cinar U, Rings F, et al. Molecular mechanisms and pathways involved in bovine embryonic genome activation and their regulation by alternative in vivo and in vitro culture conditions. Biol Reprod. 2012;87(4):100 10.1095/biolreprod.112.099697 [DOI] [PubMed] [Google Scholar]
- 3.Salilew-Wondim D, Fournier E, Hoelker M, Saeed-Zidane M, Tholen E, Looft C, et al. Genome-Wide DNA Methylation Patterns of Bovine Blastocysts Developed In Vivo from Embryos Completed Different Stages of Development In Vitro. PLoS One. 2015;10(11):e0140467 10.1371/journal.pone.0140467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salilew-Wondim D, Saeed-Zidane M, Hoelker M, Gebremedhn S, Poirier M, Om Pandey H, et al. Genome-wide DNA methylation patterns of bovine blastocysts derived from in vivo embryos subjected to in vitro culture before, during or after embryonic genome activation. BMC Genomics. 2018;19:424 10.1186/s12864-018-4826-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wrighton KH. Cell adhesion: the 'ins' and 'outs' of integrin signalling. Nat Rev Mol Cell Biol. 2013;14(12):752. [DOI] [PubMed] [Google Scholar]
- 6.Kaneko Y, Lecce L, Day ML, Murphy CR. Focal adhesion kinase localizes to sites of cell-to-cell contact in vivo and increases apically in rat uterine luminal epithelium and the blastocyst at the time of implantation. J Morphol. 2012;273(6):639–50. 10.1002/jmor.20010 [DOI] [PubMed] [Google Scholar]
- 7.Bladt F, Aippersbach E, Gelkop S, Strasser GA, Nash P, Tafuri A, et al. The murine Nck SH2/SH3 adaptors are important for the development of mesoderm-derived embryonic structures and for regulating the cellular actin network. Mol Cell Biol. 2003;23(13):4586–97. 10.1128/MCB.23.13.4586-4597.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renshaw MW, Price LS, Schwartz MA. Focal adhesion kinase mediates the integrin signaling requirement for growth factor activation of MAP kinase. J Cell Biol. 1999;147(3):611–8. 10.1083/jcb.147.3.611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinclair KD, Rooke JA, McEvoy TG. Regulation of nutrient uptake and metabolism in pre-elongation ruminant embryos. Reprod Suppl. 2003;61):371–85. [PubMed] [Google Scholar]
- 10.Adamson ED. Activities of growth factors in preimplantation embryos. J Cell Biochem. 1993;53(4):280–7. 10.1002/jcb.240530403 [DOI] [PubMed] [Google Scholar]
- 11.Wei Z, Park KW, Day BN, Prather RS. Effect of epidermal growth factor on preimplantation development and its receptor expression in porcine embryos. Mol Reprod Dev. 2001;60(4):457–62. 10.1002/mrd.1110 [DOI] [PubMed] [Google Scholar]
- 12.Cebrian-Serrano A, Salvador I, Silvestre MA. Beneficial effect of two culture systems with small groups of embryos on the development and quality of in vitro-produced bovine embryos. Anat Histol Embryol. 2014;43(1):22–30. 10.1111/ahe.12043 [DOI] [PubMed] [Google Scholar]
- 13.Block J, Bonilla L, Hansen PJ. Effect of addition of hyaluronan to embryo culture medium on survival of bovine embryos in vitro following vitrification and establishment of pregnancy after transfer to recipients. Theriogenology. 2009;71(7):1063–71. 10.1016/j.theriogenology.2008.11.007 [DOI] [PubMed] [Google Scholar]
- 14.Reis e Silva AR, Bruno C, Fleurot R, Daniel N, Archilla C, Peynot N, et al. Alteration of DNA demethylation dynamics by in vitro culture conditions in rabbit pre-implantation embryos. Epigenetics. 2012;7(5):440–6. 10.4161/epi.19563 [DOI] [PubMed] [Google Scholar]
- 15.Katari S, Turan N, Bibikova M, Erinle O, Chalian R, Foster M, et al. DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum Mol Genet. 2009;18(20):3769–78. 10.1093/hmg/ddp319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park YS, Lin YC. Effect of epidermal growth factor (EGF) and defined simple media on in vitro bovine oocyte maturation and early embryonic development. Theriogenology. 1993;39(2):475–84. 10.1016/0093-691x(93)90390-q [DOI] [PubMed] [Google Scholar]
- 17.Mtango NR, Varisanga MD, Dong YJ, Rajamahendran R, Suzuki T. Growth factors and growth hormone enhance in vitro embryo production and post-thaw survival of vitrified bovine blastocysts. Theriogenology. 2003;59(5–6):1393–402. 10.1016/s0093-691x(02)01163-9 [DOI] [PubMed] [Google Scholar]
- 18.Oyamada T, Iwayama H, Fukui Y. Additional effect of epidermal growth factor during in vitro maturation for individual bovine oocytes using a chemically defined medium. Zygote. 2004;12(2):143–50. [DOI] [PubMed] [Google Scholar]
- 19.Ahumada CJ, Salvador I, Cebrian-Serrano A, Lopera R, Silvestre MA. Effect of supplementation of different growth factors in embryo culture medium with a small number of bovine embryos on in vitro embryo development and quality. Animal. 2013;7(3):455–62. 10.1017/S1751731112001991 [DOI] [PubMed] [Google Scholar]
- 20.Furnus CC, Matos DG de, Martinez AG. Effect of hyaluronic acid on development of in vitro produced bovine embryos. Theriogenology. 1998;49(8):1489–99. 10.1016/s0093-691x(98)00095-8 [DOI] [PubMed] [Google Scholar]
- 21.Palasz AT, Rodriguez-Martinez H, Beltran-Brena P, Perez-Garnelo S, Martinez MF, Gutierrez-Adan A, La Fuente J de. Effects of hyaluronan, BSA, and serum on bovine embryo in vitro development, ultrastructure, and gene expression patterns. Mol Reprod Dev. 2006;73(12):1503–11. 10.1002/mrd.20516 [DOI] [PubMed] [Google Scholar]
- 22.Palasz AT, Brena PB, Martinez MF, Perez-Garnelo SS, Ramirez MA, Gutierrez-Adan A, La Fuente J de. Development, molecular composition and freeze tolerance of bovine embryos cultured in TCM-199 supplemented with hyaluronan. Zygote. 2008;16(1):39–47. 10.1017/S0967199407004467 [DOI] [PubMed] [Google Scholar]
- 23.Handyside AH, Hunter S. A rapid procedure for visualising the inner cell mass and trophectoderm nuclei of mouse blastocysts in situ using polynucleotide-specific fluorochromes. J Exp Zool. 1984;231(3):429–34. 10.1002/jez.1402310317 [DOI] [PubMed] [Google Scholar]
- 24.Wolf E, Arnold GJ, Bauersachs S, Beier HM, Blum H, Einspanier R, et al. Embryo-maternal communication in bovine—strategies for deciphering a complex cross-talk. Reprod Domest Anim. 2003;38(4):276–89. [DOI] [PubMed] [Google Scholar]
- 25.Wu C. Focal adhesion: a focal point in current cell biology and molecular medicine. Cell Adh Migr. 2007;1(1):13–8. 10.4161/cam.1.1.4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 2004;145(12):5439–47. 10.1210/en.2004-0959 [DOI] [PubMed] [Google Scholar]
- 27.Goodsell DS. The molecular perspective: epidermal growth factor. Stem Cells. 2003;21(6):702–3. 10.1634/stemcells.21-6-702 [DOI] [PubMed] [Google Scholar]
- 28.Fujihara M, Comizzoli P, Keefer CL, Wildt DE, Songsasen N. Epidermal growth factor (EGF) sustains in vitro primordial follicle viability by enhancing stromal cell proliferation via MAPK and PI3K pathways in the prepubertal, but not adult, cat ovary. Biol Reprod. 2014;90(4):86 10.1095/biolreprod.113.115089 [DOI] [PubMed] [Google Scholar]
- 29.Prochazka R, Petlach M, Nagyova E, Nemcova L. Effect of epidermal growth factor-like peptides on pig cumulus cell expansion, oocyte maturation, and acquisition of developmental competence in vitro: comparison with gonadotropins. Reproduction. 2011;141(4):425–35. 10.1530/REP-10-0418 [DOI] [PubMed] [Google Scholar]
- 30.Lorenzo PL, Illera MJ, Illera JC, Illera M. Enhancement of cumulus expansion and nuclear maturation during bovine oocyte maturation in vitro by the addition of epidermal growth factor and insulin-like growth factor I. J Reprod Fertil. 1994;101(3):697–701. 10.1530/jrf.0.1010697 [DOI] [PubMed] [Google Scholar]
- 31.Shabankareh HK, Zandi M. Developmental potential of sheep oocytes cultured in different maturation media: effects of epidermal growth factor, insulin-like growth factor I, and cysteamine. Fertil Steril. 2010;94(1):335–40. 10.1016/j.fertnstert.2009.01.160 [DOI] [PubMed] [Google Scholar]
- 32.Sirisathien S, Hernandez-Fonseca HJ, Brackett BG. Influences of epidermal growth factor and insulin-like growth factor-I on bovine blastocyst development in vitro. Anim Reprod Sci. 2003;77(1–2):21–32. [DOI] [PubMed] [Google Scholar]
- 33.Song HJ, Kang EJ, Maeng GH, Ock SA, Lee SL, Yoo JG, et al. Influence of epidermal growth factor supplementation during in vitro maturation on nuclear status and gene expression of canine oocytes. Res Vet Sci. 2011;91(3):439–45. 10.1016/j.rvsc.2010.09.003 [DOI] [PubMed] [Google Scholar]
- 34.Neira JA, Tainturier D, Pena MA, Martal J. Effect of the association of IGF-I, IGF-II, bFGF, TGF-beta1, GM-CSF, and LIF on the development of bovine embryos produced in vitro. Theriogenology. 2010;73(5):595–604. 10.1016/j.theriogenology.2009.10.015 [DOI] [PubMed] [Google Scholar]
- 35.Eberwein P, Laird D, Schulz S, Reinhard T, Steinberg T, Tomakidi P. Modulation of focal adhesion constituents and their down-stream events by EGF: On the cross-talk of integrins and growth factor receptors. Biochim Biophys Acta. 2015;1853(10 Pt A):2183–98. [DOI] [PubMed] [Google Scholar]
- 36.Lee CN, Ax RL. Concentrations and composition of glycosaminoglycans in the female bovine reproductive tract. J Dairy Sci. 1984;67(9):2006–9. 10.3168/jds.S0022-0302(84)81536-2 [DOI] [PubMed] [Google Scholar]
- 37.Suchanek E, Simunic V, Juretic D, Grizelj V. Follicular fluid contents of hyaluronic acid, follicle-stimulating hormone and steroids relative to the success of in vitro fertilization of human oocytes. Fertil Steril. 1994;62(2):347–52. 10.1016/s0015-0282(16)56890-3 [DOI] [PubMed] [Google Scholar]
- 38.Ohta N, Saito H, Kaneko T, Yoshida M, Takahashi T, Saito T, et al. Soluble CD44 in human ovarian follicular fluid. J Assist Reprod Genet. 2001;18(1):21–5. 10.1023/A:1026494528415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Archibong AE, Petters RM, Johnson BH. Development of porcine embryos from one- and two-cell stages to blastocysts in culture medium supplemented with porcine oviductal fluid. Biol Reprod. 1989;41(6):1076–83. 10.1095/biolreprod41.6.1076 [DOI] [PubMed] [Google Scholar]
- 40.Laurent TC, Fraser JR. Hyaluronan. FASEB J. 1992;6(7):2397–404. [PubMed] [Google Scholar]
- 41.Qhattal HSS, Hye T, Alali A, Liu X. Hyaluronan polymer length, grafting density, and surface poly(ethylene glycol) coating influence in vivo circulation and tumor targeting of hyaluronan-grafted liposomes. ACS Nano. 2014;8(6):5423–40. 10.1021/nn405839n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oommen OP, Duehrkop C, Nilsson B, Hilborn J, Varghese OP. Multifunctional Hyaluronic Acid and Chondroitin Sulfate Nanoparticles: Impact of Glycosaminoglycan Presentation on Receptor Mediated Cellular Uptake and Immune Activation. ACS Appl Mater Interfaces. 2016;8(32):20614–24. 10.1021/acsami.6b06823 [DOI] [PubMed] [Google Scholar]
- 43.Donejko M, Rysiak E, Galicka E, Terlikowski R, Glazewska EK, Przylipiak A. Protective influence of hyaluronic acid on focal adhesion kinase activity in human skin fibroblasts exposed to ethanol. Drug Des Devel Ther. 2017;11):669–76. 10.2147/DDDT.S125843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaneko T, Saito H, Toya M, Satio T, Nakahara K, Hiroi M. Hyaluronic acid inhibits apoptosis in granulosa cells via CD44. J Assist Reprod Genet. 2000;17(3):162–7. 10.1023/A:1009470206468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Turley E, Moore D. Hyaluronate binding proteins also bind to fibronectin, laminin and collagen. Biochem Biophys Res Commun. 1984;121(3):808–14. 10.1016/0006-291x(84)90750-2 [DOI] [PubMed] [Google Scholar]
- 46.Marei WF, Ghafari F, Fouladi-Nashta AA. Role of hyaluronic acid in maturation and further early embryo development of bovine oocytes. Theriogenology. 2012;78(3):670–7. 10.1016/j.theriogenology.2012.03.013 [DOI] [PubMed] [Google Scholar]
- 47.Yokoo M, Shimizu T, Kimura N, Tunjung WAS, Matsumoto H, Abe H, et al. Role of the hyaluronan receptor CD44 during porcine oocyte maturation. J Reprod Dev. 2007;53(2):263–70. 10.1262/jrd.18047 [DOI] [PubMed] [Google Scholar]
- 48.Opiela J, Romanek J, Lipinski D, Smorag Z. Effect of hyaluronan on developmental competence and quality of oocytes and obtained blastocysts from in vitro maturation of bovine oocytes. Biomed Res Int. 2014;2014):519189 10.1155/2014/519189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Keefer CL, Stice SL, Paprocki AM, Golueke P. In vitro culture of bovine IVM-IVF embryos: Cooperative interaction among embryos and the role of growth factors. Theriogenology. 1994;41(6):1323–31. 10.1016/0093-691x(94)90491-z [DOI] [PubMed] [Google Scholar]
- 50.Rios GL, Buschiazzo J, Mucci NC, Kaiser GG, Cesari A, Alberio RH. Combined epidermal growth factor and hyaluronic acid supplementation of in vitro maturation medium and its impact on bovine oocyte proteome and competence. Theriogenology. 2015;83(5):874–80. 10.1016/j.theriogenology.2014.11.022 [DOI] [PubMed] [Google Scholar]
- 51.Furnus CC, Valcarcel A, Dulout FN, Errecalde AL. The hyaluronic acid receptor (CD44) is expressed in bovine oocytes and early stage embryos. Theriogenology. 2003;60(9):1633–44. 10.1016/s0093-691x(03)00116-x [DOI] [PubMed] [Google Scholar]
- 52.Bedaiwy MA, Mahfouz RZ, Goldberg JM, Sharma R, Falcone T, Abdel Hafez MF, Agarwal A. Relationship of reactive oxygen species levels in day 3 culture media to the outcome of in vitro fertilization/intracytoplasmic sperm injection cycles. Fertil Steril. 2010;94(6):2037–42. 10.1016/j.fertnstert.2009.12.020 [DOI] [PubMed] [Google Scholar]
- 53.Kurzawa R, Glabowski W, Baczkowski T, Wiszniewska B, Marchlewicz M. Growth factors protect in vitro cultured embryos from the consequences of oxidative stress. Zygote. 2004;12(3):231–40. [DOI] [PubMed] [Google Scholar]
- 54.Onodera Y, Teramura T, Takehara T, Fukuda K. Hyaluronic acid regulates a key redox control factor Nrf2 via phosphorylation of Akt in bovine articular chondrocytes. FEBS Open Bio. 2015;5):476–84. 10.1016/j.fob.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim Y, Lee Y-S, Choe J, Lee H, Kim Y-M, Jeoung D. CD44-epidermal growth factor receptor interaction mediates hyaluronic acid-promoted cell motility by activating protein kinase C signaling involving Akt, Rac1, Phox, reactive oxygen species, focal adhesion kinase, and MMP-2. J Biol Chem. 2008;283(33):22513–28. 10.1074/jbc.M708319200 [DOI] [PubMed] [Google Scholar]
- 56.Rizos D, Ward F, Boland MP, Lonergan P. Effect of culture system on the yield and quality of bovine blastocysts as assessed by survival after vitrification. Theriogenology. 2001;56(1):1–16. 10.1016/s0093-691x(01)00538-6 [DOI] [PubMed] [Google Scholar]
- 57.Rizos D, Gutierrez-Adan A, Perez-Garnelo S, La Fuente J de, Boland MP, Lonergan P. Bovine embryo culture in the presence or absence of serum: implications for blastocyst development, cryotolerance, and messenger RNA expression. Biol Reprod. 2003;68(1):236–43. 10.1095/biolreprod.102.007799 [DOI] [PubMed] [Google Scholar]
- 58.Kuzmany A, Havlicek V, Wrenzycki C, Wilkening S, Brem G, Besenfelder U. Expression of mRNA, before and after freezing, in bovine blastocysts cultured under different conditions. Theriogenology. 2011;75(3):482–94. 10.1016/j.theriogenology.2010.09.016 [DOI] [PubMed] [Google Scholar]
- 59.Gardner DK, Rodriegez-Martinez H, Lane M. Fetal development after transfer is increased by replacing protein with the glycosaminoglycan hyaluronan for mouse embryo culture and transfer. Hum Reprod. 1999;14(10):2575–80. 10.1093/humrep/14.10.2575 [DOI] [PubMed] [Google Scholar]
- 60.Dattena M, Mara L, Bin T AA, Cappai P. Lambing rate using vitrified blastocysts is improved by culture with BSA and hyaluronan. Mol Reprod Dev. 2007;74(1):42–7. 10.1002/mrd.20576 [DOI] [PubMed] [Google Scholar]
- 61.Thongkittidilok C, Tharasanit T, Songsasen N, Sananmuang T, Buarpung S, Techakumphu M. Epidermal growth factor improves developmental competence and embryonic quality of singly cultured domestic cat embryos. J Reprod Dev. 2015;61(4):269–76. 10.1262/jrd.2014-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jackson RL, Busch SJ, Cardin AD. Glycosaminoglycans: molecular properties, protein interactions, and role in physiological processes. Physiol Rev. 1991;71(2):481–539. 10.1152/physrev.1991.71.2.481 [DOI] [PubMed] [Google Scholar]
- 63.Roy F, DeBlois C, Doillon CJ. Extracellular matrix analogs as carriers for growth factors: in vitro fibroblast behavior. J Biomed Mater Res. 1993;27(3):389–97. 10.1002/jbm.820270312 [DOI] [PubMed] [Google Scholar]
- 64.Papakonstantinou E, Karakiulakis G, Roth M, Block LH. Platelet-derived growth factor stimulates the secretion of hyaluronic acid by proliferating human vascular smooth muscle cells. Proc Natl Acad Sci U S A. 1995;92(21):9881–5. 10.1073/pnas.92.21.9881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tirone E, D'Alessandris C, Hascall VC, Siracusa G, Salustri A. Hyaluronan synthesis by mouse cumulus cells is regulated by interactions between follicle-stimulating hormone (or epidermal growth factor) and a soluble oocyte factor (or transforming growth factor beta1). J Biol Chem. 1997;272(8):4787–94. 10.1074/jbc.272.8.4787 [DOI] [PubMed] [Google Scholar]
- 66.Xu H, Wu K, Tian Y, Liu Q, Han N, Yuan X, et al. CD44 correlates with clinicopathological characteristics and is upregulated by EGFR in breast cancer. Int J Oncol. 2016;49(4):1343–50. 10.3892/ijo.2016.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang S, Chang MC, Zylka D, Turley S, Harrison R, Turley EA. The hyaluronan receptor RHAMM regulates extracellular-regulated kinase. J Biol Chem. 1998;273(18):11342–8. 10.1074/jbc.273.18.11342 [DOI] [PubMed] [Google Scholar]
- 68.Stojkovic M, Krebs O, Kolle S, Prelle K, Assmann V, Zakhartchenko V, et al. Developmental regulation of hyaluronan-binding protein (RHAMM/IHABP) expression in early bovine embryos. Biol Reprod. 2003;68(1):60–6. 10.1095/biolreprod.102.007716 [DOI] [PubMed] [Google Scholar]
- 69.Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005;6(1):56–68. 10.1038/nrm1549 [DOI] [PubMed] [Google Scholar]
- 70.Yang Y, Du J, Hu Z, Liu J, Tian Y, Zhu Y, et al. Activation of Rac1-PI3K/Akt is required for epidermal growth factor-induced PAK1 activation and cell migration in MDA-MB-231 breast cancer cells. J Biomed Res. 2011;25(4):237–45. 10.1016/S1674-8301(11)60032-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manser E, Huang HY, Loo TH, Chen XQ, Dong JM, Leung T, Lim L. Expression of constitutively active alpha-PAK reveals effects of the kinase on actin and focal complexes. Mol Cell Biol. 1997;17(3):1129–43. 10.1128/mcb.17.3.1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang F, Lemmon CA, Park D, Romer LH. FAK potentiates Rac1 activation and localization to matrix adhesion sites: a role for betaPIX. Mol Biol Cell. 2007;18(1):253–64. 10.1091/mbc.E06-03-0207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carisey A, Ballestrem C. Vinculin, an adapter protein in control of cell adhesion signalling. Eur J Cell Biol. 2011;90(2–3):157–63. 10.1016/j.ejcb.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Memili E, First NL. Zygotic and embryonic gene expression in cow: a review of timing and mechanisms of early gene expression as compared with other species. Zygote. 2000;8(1):87–96. [DOI] [PubMed] [Google Scholar]
- 75.Graf A, Krebs S, Heininen-Brown M, Zakhartchenko V, Blum H, Wolf E. Genome activation in bovine embryos: review of the literature and new insights from RNA sequencing experiments. Anim Reprod Sci. 2014;149(1–2):46–58. 10.1016/j.anireprosci.2014.05.016 [DOI] [PubMed] [Google Scholar]
- 76.Sagirkaya H, Misirlioglu M, Kaya A, First NL, Parrish JJ, Memili E. Developmental and molecular correlates of bovine preimplantation embryos. Reproduction. 2006;131(5):895–904. 10.1530/rep.1.01021 [DOI] [PubMed] [Google Scholar]
- 77.Wang Y, Jadhav RR, Liu J, Wilson D, Chen Y, Thompson IM, et al. Roles of Distal and Genic Methylation in the Development of Prostate Tumorigenesis Revealed by Genome-wide DNA Methylation Analysis. Sci Rep. 2016;6):22051 10.1038/srep22051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doerfler W. On the biological significance of DNA methylation. Biochemistry (Mosc). 2005;70(5):505–24. [DOI] [PubMed] [Google Scholar]
- 79.Long MD, Smiraglia DJ, Campbell MJ. The Genomic Impact of DNA CpG Methylation on Gene Expression; Relationships in Prostate Cancer. Biomolecules 2017. 10.3390/biom7010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2(9):1136–43. 10.1101/gad.2.9.1136 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.