Abstract
Acinetobacter baumannii causes hospital-acquired infections, especially in those with impaired immune function. Biocides or disinfectants are widely used antibacterial agents used to eradicate the effect of A. baumannii on inanimate objects and health care environments. In the current study, the antimicrobial activity of chlorhexidine has been investigated against carbapenem-resistant (RS-307, RS-7434, RS-6694, and RS-122), and sensitive (ATCC-19606 and RS-10953) strains of A. baumannii. We have determined growth kinetics, antimicrobial susceptibility, ROS production, lipid peroxidation, cell viability using flow cytometry assay (FACS), and membrane integrity by scanning electron microscope (SEM). The effect of chlorhexidine on the bacterial membrane has also been investigated using Fourier transform infrared (FTIR) spectroscopy. The present study showed that 32μg/ml chlorhexidine treatment results in the decreased bacterial growth, CFU count and cell viability. The antibacterial activity of chlorhexidine is due to the elevated ROS production and higher lipid peroxidation. These biochemical changes result in the membrane damage and alteration in the membrane proteins, phospholipids, carbohydrates, nucleic acids as evident from the FTIR and SEM data. Therefore, chlorhexidine has the potential to be used in the hospital setups to remove the spread of A. baumannii.
Introduction
Acinetobacter baumannii is non-fermentative, non-motile, strictly aerobic, non-fastidious, catalase positive, oxidase negative, and Gram-negative coccobacilli [1]. In last decades, this bacteria have turned into an important nosocomial pathogen from an opportunistic bacteria [2] due to its antibacterial resistance capability [3]. A. baumannii infections are associated with devastating outcomes in terms of mortality and morbidity and contribute to higher burden on the healthcare system. Skin and mucous membrane, wounds and burns, intravascular and urinary catheters, also urinary, gastrointestinal, and respiratory tracts are the sources of its infections [4]. Hospitals sources are the patient’s bed, mattresses, pillow, table, sinks, shower units, infusion pump together with suction and resuscitation equipment. A. baumannii has gained resistance to most of the antibiotics all over the world, which is the major challenge in the treatment. Hence, availability of effective antibiotics for A. baumannii treatment is restricted because of rapid increase in drug resistance [5].
Biocides are important components in the prevention of bacterial infections, commonly in intensive care units (ICUs) where there have been considerable reports on multidrug-resistant bacteria such as carbapenem-resistant A. baumannii [6]. Chlorhexidine, first synthesized in 1954 [7], is a biocide that is used as a skin disinfectant before surgery and to sterilize surgical instruments [8] and also against nosocomial infections in shared facilities [9].
Therefore, we have investigated into the molecular mechanism of chlorhexidine on the carbapenem resistant strain of A. baumannii. Various antimicrobial molecules showed ROS dependent membrane damage as an antibacterial mechanism hence we have explored this mechanism for chlorhexidine. In the present study, we have also investigated the antibacterial effect of chlorhexidine on membrane integrity and composition using flow cytometry, scanning electron microscopy (SEM) and fourier transform infrared spectroscopy (FTIR) respectively. These results will help in developing effective biocide to control the spread of A. baumannii infections, for hospital setups where antibiotics cannot be used.
Material and methods
Reagents
For the experiments, we have used various reagents, such as MacConkey broth, Luria bertani broth, Guanidine hydrochloride, Potassium Phosphate dibasic, Potassium phosphate monobasic, Sodium dodecyl sulfate, sodium hydroxide, Nitroblue tetrazolium (NBT), potassium hydroxide, 2,4-dinitrophenylhydrazine (DNPH), 2-Thiobarbituric acid (TBA), and Dimethyl sulfoxide, that were purchased from Himedia Laboratories Ltd., India. Chlorhexidine was purchased from Sigma Aldrich, Hydrochloric acid, sodium chloride and glycine were from Merck, India. Glacial acetic acid and glycerol from Qualigens, India. Ethyl acetate purchased from Fisher scientific.2-Thiobarbituric acid (0.8%) was prepared in 1M NaOH,2,4-dinitrophenylhydrazine (10mM) was prepared in 2N HCl. For PI staining, 100mM Tris (pH 7.4), 150mM NaCl, 1mM CaCl2, 0.5mM, MgCl2, and 0.1% Triton-X were prepared, 2.5% glutaraldehyde, and ethanol.
Bacterial samples
The pre-collected bacterial strains of A. baumannii (RS307, RS6694, RS122, RS7434, RS10953 and ATCC-19606) were available in our laboratory at Central University of Rajasthan, Ajmer, India.
Growth kinetics
Growth kinetics pattern of A. baumannii in the presence and absence of chlorhexidine was analyzed by UV-Vis Spectrophotometer at 605nm. For this, A. baumannii was cultured in LB media at 37°C under shaking conditions. Readings were taken at the regular time interval of 30min. When optical density reached at 0.6, the treatment of 16μg/ml, 32μg/ml and 64μg/ml of chlorhexidine was given. Differential growth curves were plotted to compare the bacterial growth, in the presence and absence of biocides. The concentration range was selected based on the previous published articles [10,11].
Determination of bacterial cell viability using MTT assay
We have determined the bacterial cell viability using a modified MTT [3-(4, 5- Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide)] assay using published protocol [12–16]. The assay is based on conversion of yellow tetrazolium (MTT) into purple insoluble formazan crystals in the bacterial cytoplasm. This color change indicated the presence of live bacteria, whereas no color changes indicated the absence of live bacteria. In brief, firstly, 100μl cultures (OD of 0.2 at 605nm) of A. baumannii RS-307 and ATCC-19606 were transferred to 96 well-plate and treated with chlorhexidine (2μg/ml to 48μg/ml). After treatment, the multiwell plate was further incubated for 5 hours at 37°C. To determine the viable cells, 5μl of MTT solution (5mg/ml) was added to all wells and incubated for 1 hour in the dark at 37°C. After it 100μl of 100% DMSO was added to the suspension and incubated for 2hours at 37°C. Then, we measured the optical density of micro-plate at a wavelength of 570nm, and the percentage viability rate of wells was determined [17].
Nitro blue tetrazolium assay
ROS have proposed to play critical roles in the bacterial response to lethal stress like antimicrobial peptide [18]. In oxidative stress conditions, ROS levels increases and causes significant damage to bacterial cell structures. Reports showed that chlorhexidine have role in ROS elevation to produce its antimicrobial effect [19–21]. Therefore, the reactive oxygen species (ROS) was measured by nitro blue tetrazolium assay (NBT) on a multi-well scanning spectrophotometer (ELISA reader) as per published protocol [22].
Thiobarbituric acid assay
The thiobarbituric acid assay is most widely used to determine lipid peroxidation. During this process, the malondialdehyde (MDA) was formed by the decomposition of polyunsaturated fatty acids, which reacts with thiobarbituric acid. MDA (the secondary product) produce a pink-colored dimeric compound with thiobarbituric acid (TBA), which can be easily quantified using ELISA reader. We have quantified lipid peroxidation as per our published method [22].
Colony forming assay
The numbers of viable cells were quantified by measuring colony forming units (CFUs) for both treated and untreated bacteria as per published protocol with some modification [23]. The culture was incubated at 37°C and monitored using UV spectrophotometer at 605nm until it reached the exponential phase (at OD 0.6). Control sample remained under growth condition while treatment was given with 32μg/ml chlorhexidine and further continuing cultivation for 3 hours. Pellet of 1 ml culture was diluted in 0.9% sodium chloride solution and prepared its dilutions from 101 to 104. From this, 10μl sample was taken from different reaction mixtures and spread on MacConkey agar plates using sterile glass rods. The plates were then incubated at 37°C for 16 hours (overnight). The number of viable cells after treatment with biocides were quantified and compared with untreated control to evaluate the antibacterial effect. The log reduction was calculated by log of ‘number of viable cells before treatment’ with ‘number of viable cells after treatment’.
Flow cytometry analysis
Flow cytometry is a technique to enumerate the viable bacterial cell in the bacterial sample; here we have used treated and untreated bacterial samples. Propidium iodide is a nucleic acid dye that only leaks into the cells with ruptured cell membranes. Flow cytometry was prepared as per the method of Bankier et al. [24] with some modifications. To determine the viability status of bacterial cells, culture was grown until OD600 of 1 (109 CFU). The 10μl primary culture is suspended into 1ml of Luria bertani broth (corresponding to 2 X 106 to 1 X 107 CFU/ml). These prepared bacterial cell suspensions (at log phase) were divided into three sets. One set was used for treatment with chlorhexidine, other for heat treatment (at 70°C for 30min as positive control) and third set was untreated control. All three samples were incubated for 3hours at 37°C. The incubated samples were centrifuged, pellet was treated with 3μM from PI (stock solution 1mg/ml, 1.5mM) and incubated for 30min in dark and the samples were subjected for detection of live and dead cells by flow cytometry (FACScan, BD Bioscience). Two non-fluorescent parameters determined the bacterial acquisition gate: forward scatter (FSC), X-axis and side scatter (SSC), Y-axis, and one fluorescent parameter (red fluorescence) from PI. FSC and SSC channels help to eliminate background noise. The threshold was also set at the FSC signals of control.
Fourier transform infrared spectroscopy (FTIR)
Recently, FTIR being increasingly used to investigate the mode of action of bactericidal compounds and to determine changes in bacterial cells in response to different stresses [25]. A. baumannii strains were grown in LB broth till OD reaches 0.6 and divided into two sets. One set was treated with chlorhexidine while other was used as control. The bacterial cultures in different conditions were centrifuged at 13000RPM for 15min and the supernatant was discarded. Bacterial pellets were washed (twice) with phosphate buffer solution by centrifugation at 13000RPM for 3 minutes. All FTIR spectra were recorded in the transmission mode, by the potassium bromide (KBr) disc method. The FTIR spectra were recorded with the PerkinElmer Spectrum Bx FTIR system spectrophotometer Version 10.4 with deuterated triglycine sulfate (DTGS) detector elements. FTIR data was obtained in the wavelength range 4000–400 cm-1, 32 scans at a pixel resolution of 8 cm-1 and spectral resolution 4 cm-1. The pellets, with potassium bromide (KBr) were grinded in a mortar and pestle. The obtained background from KBr disc was automatically subtracted from the prepared KBr and sample disc spectra.
Field emission scanning electron microscopy (FESEM)
The field emission scanning electron microscope (FESEM) is a useful technique to study the morphological features of bacteria [26]. The ultrastructural changes in bacterial morphology caused by disinfectant were examined by field emission scanning electron microscopy (FESEM). The appropriate growth conditioned bacterial suspension, while was grown in LB broth until the OD reached at 0.6, was divided into two sets. One set was treated with 32μg/ml chlorhexidine while other was kept as untreated and incubated at 37°C until the OD of control reached at 1.0. 4ml bacterial cells (treated and untreated) were centrifuged at 13000 rpm for 30 minutes at 4 °C. The obtained pellets were washed with PBS to remove the remaining medium before the pellets were fixed with glutaraldehyde. A solution (100 μl) of 2.5% glutaraldehyde in PBS (pH 7) was added to the pellet and incubated at 4 °C for 2.5 hours. The pellet was washed with PBS and fixation step was repeated. After fixation, cells were washed 4 times for 30 minutes with PBS at 4 °C. Finally, 20μl PBS was added to the pellet, mixed and one drop of this suspension was placed on the glass cover slip. The smear was allowed to dry and then dehydrated with ethanol gradients (35%, 50%, 75%, 95% and 100%) followed by air dry. The fixed bacterial smears were then coated with a layer of gold (Quorum) and the conductive samples were examined with SEM (Nova NanoSEM 450).
Statistical methods
The data was analyzed for standard deviation based on the entire data using the STDEV function in Microsoft excels. The results were expressed as the mean ± the standard error of the mean (n = 3). For the statistical method, we have used graph pad prism and performed the t-test to compare the findings.
Results
In the present study, the experiments were designed to identify the biochemical alterations of biocide-treated multidrug resistance strains of A. baumannii. The focus of this study is to have a comparative difference in the A. baumannii after biocide treatment using biochemical and FTIR based signatures, which might indicate the molecular mechanism of biocide. In the present study, we have used carbapenem-resistant strains of A. baumannii which were found to be highly resistant (RS307, RS6694, and RS122), intermediate carbapenem-resistant (RS7434) and sensitive (RS-10953 and ATCC-19606) strains.
Growth kinetics showed the inhibitory effect of chlorhexidine
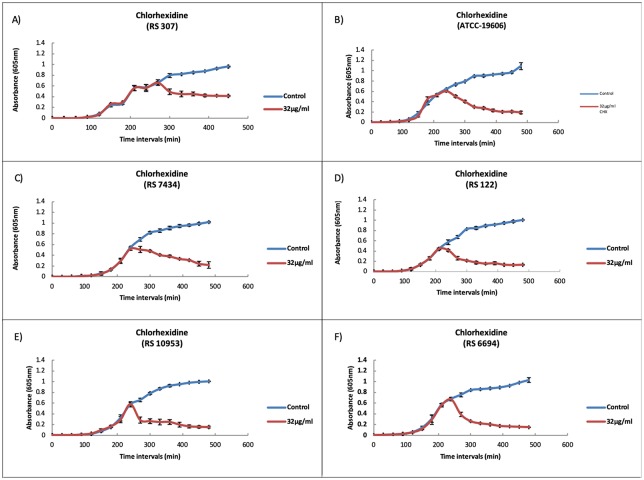
The result of growth analysis of ATCC19606 and RS307 showed that 32μg/ml and 64μg/ml showed a similar effect, therefore 32μg/ml were used in further study. The result showed that chlorhexidine has significant inhibitory effect on the strains RS307 (p-value 0.0019), ATCC19606 (p-value 0.0023), RS6694 (p-value 0.002), RS122 (p-value 0.0015), RS7434 (p-value 0.0063), and RS10953 (p-value 0.0025) (Fig 1). This result showed that the 32μg/ml chlorhexidine have inhibitory effect on different strains of A. baumannii.
Fig 1. Growth kinetics of A. baumannii strains were determined in the presence and absence of effective concentration of chlorhexidine that measured by UV-Vis spectrophotometer at 605 nm.
Data expressed is Mean± SEM of 3 values (n = 3) (Control: Average of Control; 32μg/ml Chlorhexidine). Results of 16μg/ml and 64μg/ml chlorhexidine treated samples are provided as S1 Fig.
Microtiter plate analysis also showed the inhibitory effect of chlorhexidine
A. baumannii strains RS-307 and ATCC-19606 were treated with various concentration of chlorhexidine (2μg/ml-48μg/ml). The different concentrations of disinfectant produce a different inhibitory effect such as >50% inhibitory effect on RS307 at 2μg/ml (25.64% cell viability) and ATCC-19606 (12 μg/ml, 28.97% cell viability). Microtiter experiment also confirms the inhibitory effect of chlorhexidine on A. baumannii.
Chlorhexidine treatment decreases CFU by Acinetobacter baumannii
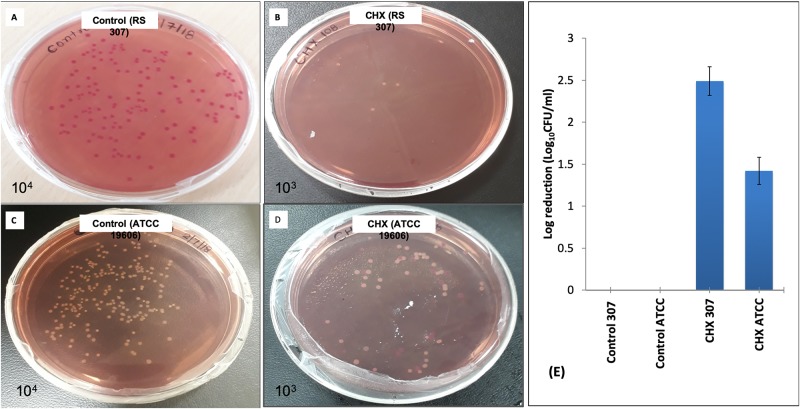
Colony forming unit (CFU) measurements indicate the antibacterial effect of chlorhexidine against the strains of A. baumannii. The viability of both bacterial strains was reduced in the presence of 32 μg/ml chlorhexidine concentration. The bacterial colonies after 103 dilutions of RS307, clearly showed that after treatment, there is a decrease in the number of CFU in the treated sample as compared to control (untreated) (Fig 2A and 2B). Similarity, bacterial colonies after 103 dilutions of ATCC-19606 showed decreases in CFU after treatment (Fig 2C and 2D). Growth of the bacterial cells was less than 5% after the incubation at 37° C for 16 hours. CFU of RS307 at 103 dilutions is 1940 x 104 while for ATCC, it is 1270 x104. The treatment with chlorhexidine results in the decrease of CFU for RS-307 (33x104) and for ATCC-19606 (340 x104). The treatment of chlorhexidine result in 2.49 and 1.42 log reduction of RS307 and ATCC-19606 strain of A. baumannii respectively as compared to the control (Fig 2E).
Fig 2. Representative culture plate of colony forming unit count of A. baumannii strains untreated and treated with 32 μg/ml Chlorhexidine: (A) Control untreated RS-307; (B) Chlorhexidine treated RS-307; (C) Control untreated ATCC-19606; (D) Chlorhexidine treated ATCC-19606; (E) Log reduction plot for chlorhexidine treatment.
Plates showing few or no bacterial growth after treatment. Experiment performed in triplicates, shown the final result observed.
Enhancement of ROS generation in A. baumannii by chlorhexidine
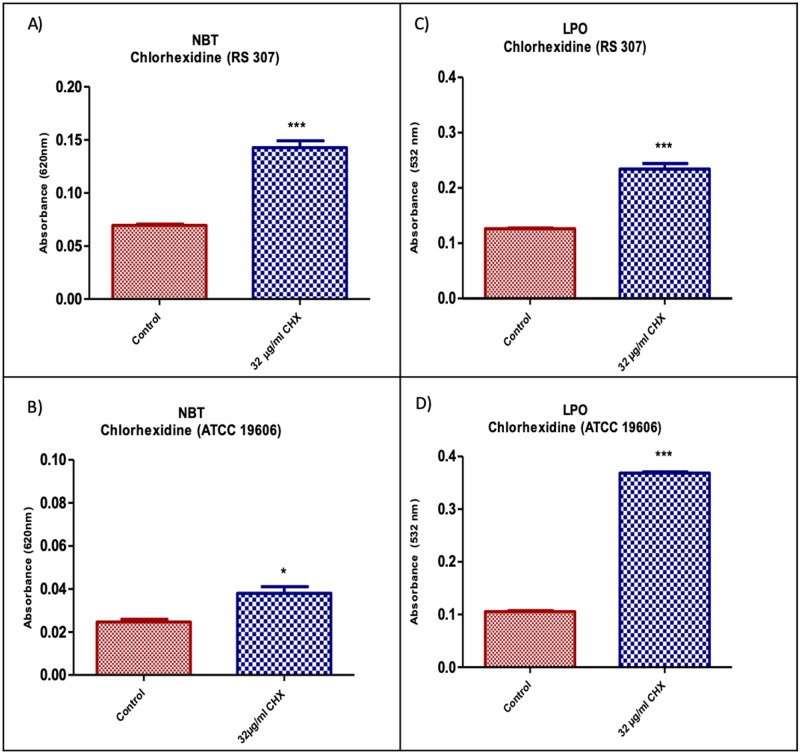
Chlorhexidine (32μg/ml) treated A. baumannii strains (RS307 and ATCC-19606) shows the significant elevated ROS generation in the treated bacterial samples as compared to the untreated bacterial samples (Fig 3A and 3B). Furthermore, 32μg/ml chlorhexidine treatment resulted in two folds ROS generation in the resistant strain.
Fig 3.
(A and B) Increase in the generation of total ROS using NBT as a substrate in treated A. baumannii strains RS307 and ATCC 19606 with chlorhexidine as compared to the control. (C and D) Absorbance values of A. baumannii strains RS307 and ATCC 19606 after treated with chlorhexidine as compared to the untreated control sample. The data expressed are mean ± SEM of at least three values (n = 3). Values were measured via the ELISA reader. ELISA, enzyme ‐linked immunosorbent assay; NBT, nitroblue tetrazolium.
Elevated lipid peroxidation after chlorhexidine treatment
Lipid peroxidation results showed that higher MDA production has seen after treatment with 32μg/ml chlorhexidine in the strain RS-307 as well as in ATCC-19606 as compared to untreated. This suggests that 32μg/ml chlorhexidine is significantly effective in lipid peroxidation of A. baumannii (Fig 3C and 3D).
Flow cytometry also confirmed the bacterial membrane disruption after chlorhexidine
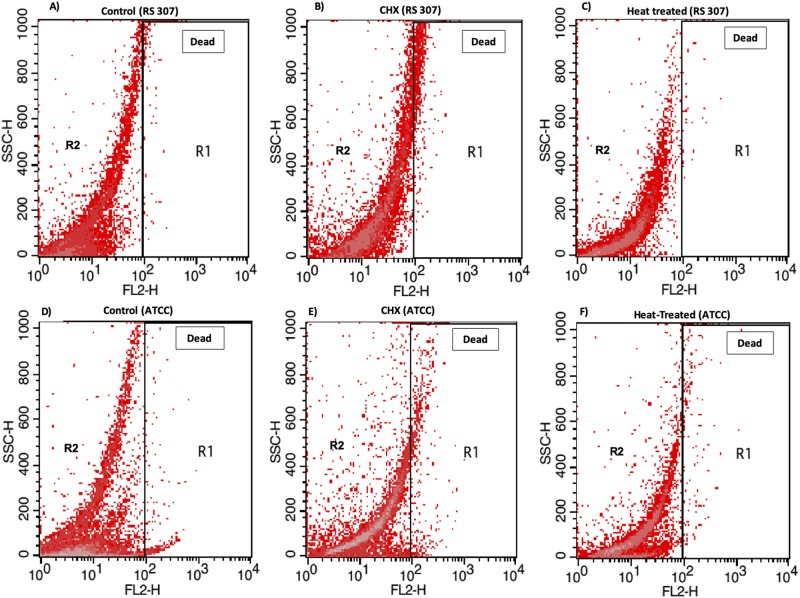
Flow cytometric analysis of A. baumannii viability control showed that the bacteria were more viable as compared to the treated bacteria (Fig 4). The heat-treated dead cells showed less viable cells as compared to control. Chlorhexidine treatment showed more inhibitory effect as compared to control and heat-treated cells (PI-positive cells). Cells coming in the range of R2 zone of autofluorescence from unstained cells are set within the first log decade (100 to 101). The result showed that there is a difference in the Geo-mean of untreated RS307 (6.19), heat treated RS307 (6.66), chlorhexidine-treated RS307 (23.46). Similarly, Geo-mean of ATCC-19606 strain also showed a difference in untreated (9.11), heat-treated ATCC-19606 (9.61), chlorhexidine-treated ATCC 19606 (14.94). The events count for untreated, chlorhexidine-treated and heat treated was found to be 247, 1904 and 202 for RS307 while 300, 515 and 135 events for ATCC-19606 respectively. These results state that there are more dead cells observed in the treated samples as compared to the untreated samples. Flow cytometry was also used to monitor the cell membrane integrity or disruption, and a positive PI cells in flow cytometry showed membrane disruption [27,28]. The significant difference shown between control, and treated bacterial cells suggests changes in A. baumannii cell membrane integrity after chlorhexidine treatment.
Fig 4. Flow cytometry dot plots of A. baumannii strains (RS 307 and ATCC): (A) Control (RS 307) (B) CHX treated (RS 307) (C) Heat-treated (RS 307)) (D) Control (ATCC) (E) CHX treated (ATCC) (F) Heat-treated (ATCC).
Histogram graphs of flow cytometry are provided as S2 Fig.
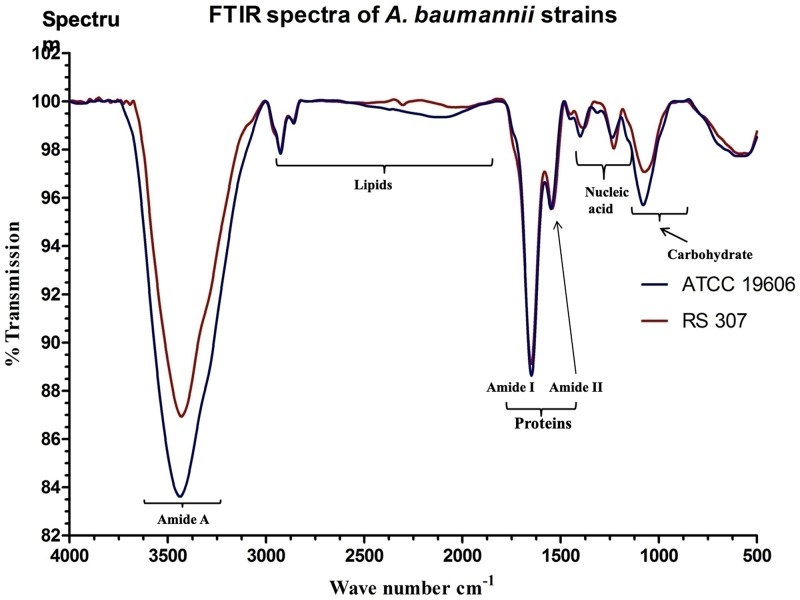
FTIR signature in the control and resistant strain of A. baumannii
FTIR spectra of the control ATCC and resistant strain RS307, showed biomolecules characteristics IR bands: lipids (3000–2800 cm-1), protein/amides I and II (1700–1500 cm-1), phospholipids/DNA/RNA (1500–1185 cm-1), polysaccharides (1185–900 cm-1) and fingerprint region (900–600 cm-1) [29]. The phospholipids/DNA/RNA and carbohydrate region are previously used to discriminate different clones of A. baumannii [30]. FTIR spectra of both the strains are very similar, but some differences are also observed in both the spectra (Fig 5). The comparative spectra showed the minor shift in spectra from around 1400 cm-1 in ATCC-19606 to 1382cm-1 in RS-307, this showed the difference in the fatty acid chains of phospholipids and amino acid side chain [31]. Similarly, there is a minor shift in FTIR spectra form around 1240cm-1 in ATCC-196906 to 1230 cm-1 in RS-307 which showed changes in phospholipids and some nucleic acids of two strains [32]. Comparative FTIR result around 600nm also showed that there is a minor change in the carbohydrate composition of these two strains. The resistant strain also has more proteins (Amide A band at 3430cm-1) and phospholipid (1080cm-1) as compare to sensitive strain. These changes in the carbohydrate and phospholipid may be associated with the higher drug tolerance of the resistant strain.
Fig 5. Comparative FTIR spectra (500–4000 cm-1) of A. baumannii strains (ATCC 19606 and RS 307).
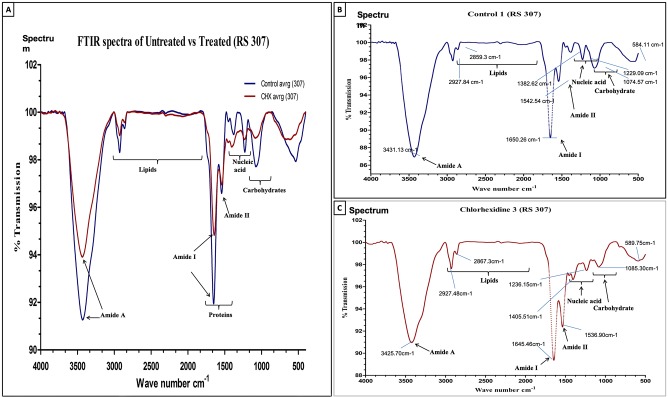
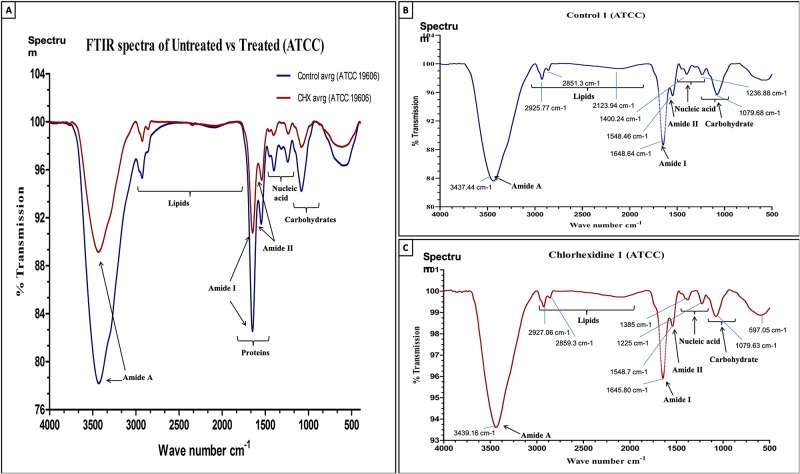
FTIR spectra confirm the alteration in membrane proteins, phospholipids, nucleic acids and carbohydrate by chlorhexidine
To investigate the effect of chlorhexidine on the Acinetobacter baumannii, we have recorded the FTIR spectra for chlorhexidine treated and untreated strains of A. baumannii (ATCC-19606 and RS-307). The experiments were performed in triplicates and have obtained individual spectra of each sample. This experiment was performed using the whole bacterial culture, and we found out a significant spectral difference in the untreated and treated samples, associated with absorbance of macromolecules such as proteins, nucleic acids, carbohydrates and phospholipids. Changes in the shape and frequencies are noticeable (Figs 6 and 7). It was observed that after chlorhexidine treatment, there is a change in the transmittance or absorbance of IR signal for nucleic acids, carbohydrates, protein and phospholipids in the treated sample as compared to the untreated sample. Results implicate the significant shift in the IR spectra for stretching modes of phosphates of phospholipids (1240cm-1), bending of CH2 backbone of phospholipids (1460cm-1), rocking of CH2 (720cm-1) deformation of asymmetric CH3 (1405cm-1), phospholipids (1191cm-1) and lipids esters (1740cm-1) after treatment with chlorhexidine. These finding suggests damage of phospholipid and degradation of lipid esters of the membrane. Chlorhexidine treatment also changes the protein structures that are evident from the amide I and amide II bands. It was observed that chlorhexidine treatment resulted into shift in the alpha-helix in range of 1648–1660cm-1 (1652cm-1 in amide I and 1548cm-1 in amide II), shift in beta-sheet (1630cm-1 for anti-parallel beta-sheet and 1640cm-1 for parallel beta-sheet)in the range of 1620–1640cm-1, main component) and 1670–1695cm-1, second frequency, and turn (1680cm-1)in the range of 1620–1640cm-1, main component and 1650–1695cm-1, second frequency. This suggests that the Acinetobacter membrane disruption and protein degradation, or denaturation is the target of the chlorhexidine.
Fig 6.
FTIR spectroscopy-based spectra (500–4000 cm-1) of (A) Combined spectra of Control and 32 μg/ml CHX treated A. baumannii (RS 307). (B) Control. and (C) 32 μg/ml CHX Treated, in transmission mode. The experiment was performed in triplicates. Results of two sets are provided as S3 Fig.
Fig 7.
FTIR spectroscopy-based spectra (500–4000 cm-1) of (A) Combined spectra of Control and 32 μg/ml CHX treated A. baumannii (ATCC). (B) Control. and (C) 32 μg/ml CHX Treated, in transmission mode. The experiment was performed in triplicates. Results of two sets are provided as S4 Fig.
Similarly, further analysis of chlorhexidine on nucleic acid of ATCC-19606 showed that there is a shift in the FTIR signature peak of PO2- from 1229cm-1 in B-DNA spectrum to 1239cm-1 in the A-DNA spectrum [33]. This showed that chlorhexidine also causes conformational changes in the DNA to inactivate the Acinetobacter baumannii. In resistant strain RS307, amide I peak of untreated sample is located at 1650 cm-1 (%T 89.122502), while in case of chlorhexidine treated sample, the peak observed at around 1634.78 cm-1 (% T 98.898112). This showed that chlorhexidine also induce some alpha-helix (1650cm-1) to beta sheet (1635cm-1) transition in protein structure [32]. These two observations need further analysis before confirmation.
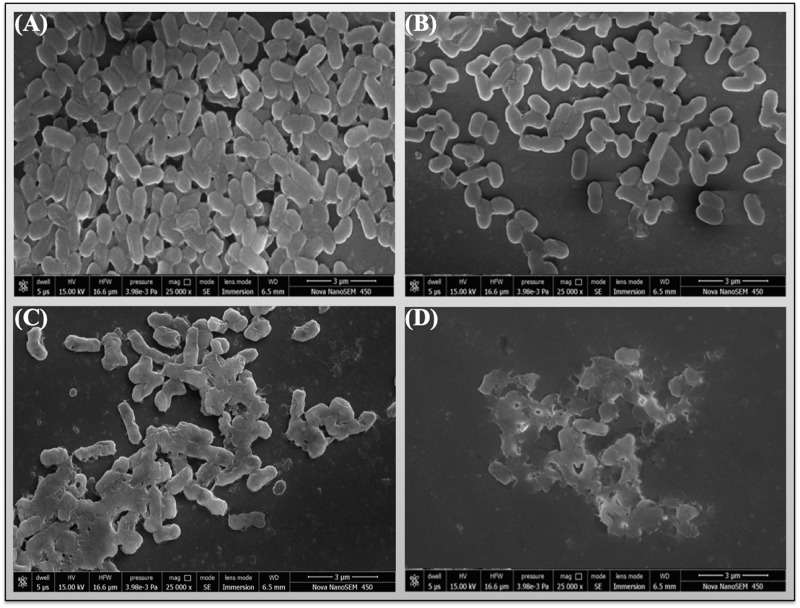
Scanning electron microscopy confirms membrane disruption by chlorhexidine
FESEM analysis of A. baumannii (strains RS-307 and ATCC-19606) bacterial cells showed observable change in cellular morphology post exposure to chlorhexidine (Fig 8C and 8D) as compared to control (Fig 8A and 8B). The figure also showed that the treatment of chlorhexidine resulted into the loss of cellular integrity and membrane rupture, which further supports the results from other techniques like FTIR, flow cytometry etc.
Fig 8. Scanning electron microscopy (SEM) images of A. baumannii cells after chlorhexidine treatment.
SEM image of untreated RS-307 strain (A); untreated ATCC-19606 (B); RS-307 treated with chlorhexidine (C); ATCC-19606 treated with chlorhexidine (D). (magnification: 25000 X; SE mode; Bars: 3μm).
Discussion
Various studies have investigated that bacteria can persist in the hospital environment from several days to a few months. Due of this, the hospital environment becomes a reservoir for hospital-associated bacteria and contributes to their transmission. There is an increasing evidence that effective biocide of the hospital environment plays an essential role in controlling the transfer of multi-resistant bacteria. Acinetobacter have developed resistance mechanism to most of the antibiotics [5,34–37] and different approaches [38–41] have been investigated to control this pathogen. Studies have been done to demonstrate the antimicrobial activity of biocides. Still, the mechanism behind the antibacterial activity is a matter of intensive research [42]. Biocides are the chemical agents inactivate virtually all recognized pathogenic bacteria. A. baumannii is a critical nosocomial pathogen with resistance to various antimicrobial agent [43]. It is an opportunistic and multidrug-resistant pathogen [44]. When they build up their biological community in a hospital, it is tough to get them out, since they thrive in respirator tubing, as well as on inanimate surfaces, for example, tabletops and bed guardrails [45]. Therefore, biocides play a very important role to irradiate it. It is generally accepted that biocides have multiple target sites within the bacterial cell, and overall damage to these target sites results in the bactericidal effect; but insusceptibility mechanism of biocides may be similar, whereas not necessarily identical [46]. Chlorhexidine has been used to eradicate the nosocomial infections; present on the hospital environment or inanimate objects [47]. In a recent study, the effect of chlorhexidine bathing as compared to routine bathing has been tested in multi-drug resistant organisms [48]. Increased usage of disinfectants has not been matched by an increase in surveillance [42]. This might be because of the reasons for the lack of surveillance is the lack of both standardized testing methods and definitions for reduced susceptibility.
Previously it was shown that 32μg/ml chlorhexidine is effective biocide against MRSA [11]; therefore we have screened different concentrations of chlorhexidine (16μg/ml, 32μg/ml and 64μg/ml). The results state that 32μg/ml and 64μg/ml chlorhexidine showed similar antibacterial activity; hence, we had selected 32μg/ml for further analysis. Our result demonstrated that chlorhexidine effectively reduces the number of multi-drug resistance strains of A. baumannii. This was confirmed by CFU and growth kinetics assay. To investigate the molecular mechanism, we have also monitored the involvement of reactive oxygen species by nitro blue tetrazolium assay and thiobarbituric acid assay. Higher production of ROS may damage varieties of cellular macromolecules, for example, RNA, DNA, protein and lipids etc which leads to loss of function, an increased rate of mutagenesis and ultimately bacterial cell death [49]. Result of NBT assay has shown increases ROS in treated bacteria with 32μg/ml chlorhexidine in both the strains, also increased in lipid peroxidation. Colony forming unit was found to be less in the Acinetobacter baumannii treated with chlorhexidine as compared to untreated.
We performed flow cytometry for determination of the dead cells present in the treated and untreated bacterial culture. For the experiment, we have used Propidium iodide (PI), a red-fluorescent nuclear staining dye that is used to analyze the dead cells in a population [50]. The result of the flow cytometry assay in the strain RS 307 and ATCC 19606 demonstrated that the control sample appeared with the less dead cell as compared to the chlorhexidine treated and heat-treated cells.
FTIR spectroscopic analysis has observed as a label-free characterization [51]. Each bacterial species has a complex cell wall composition, which gives a unique FTIR metabolic fingerprint. This distinctiveness is because of the stretching and bending vibrations of molecular bonds or functional groups present in its biomolecules, such as proteins, nucleic acid, lipids, sugars, and lipopolysaccharides within the cell wall. The molecular compositions can alter from species to species and at the level of strain. Therefore, all the bacteria will have their unique and characteristic spectrum, and single microorganisms could be identified from an FTIR spectrum. It was measured for intact bacterial cells that are complex, and the peaks are broad because of superposition of contributions from all the biomolecules present in the cells of bacteria. The results showed similarities in the FTIR signature of ATCC-19606 and resistant strain RS-307 but some difference in the phospholipids and carbohydrate reason. The treatment of chlorhexidine to the ATCC-19606 and RS-307 result in different FTIR pattern but both FTIR spectra confirms the damage in the membrane, alteration in phospholipids, protein and nucleic acids. SEM results also confirmed the membrane disruption by chlorhexidine. The overall results indicate that chlorhexidine is an effective disinfectant against the A. baumannii strains and leads to the membrane damage to exert its antibacterial effects.
Conclusion
The present result concludes that the growth kinetics graph and CFU counting after 32μg/ml chlorhexidine treatment result in bacterial cell death. Based on flow cytometry, FTIR and SEM analysis, the probable mechanism of action of chlorhexidine involves a ROS dependent membrane damage and alterations in membrane compositions. The membrane damage is a result of oxidative stress and lipid peroxidation in the bacterial cell. FTIR signature shows similarly in the Acinetobacter baumannii ATCC-19606 and RS-307 while the FTIR spectra also pointed out some differences after chlorhexidine treatment. The results highlighted that the FTIR spectra change attributed mainly to phospholipids and proteins as well as minor changes in the nucleic acids. They also enlighten the use of FTIR to analyze the molecular mechanism of disinfectants against Acinetobacter baumannii. Further studies are required to determine the precise molecular mechanism of disinfectants. Further, a dose-dependent study may also be needed to enhance the outcomes of this study.
Supporting information
(TIFF)
(TIFF)
(TIFF)
(TIFF)
Acknowledgments
Deepika Biswas would like to thank UGC for her PhD fellowship. MT would like to thank the Central University of Rajasthan for her Ph.D. fellowship. VT would like to thank MNIT, Jaipur for SEM analysis on payment basis.
Data Availability
All data is provided along with MS.
Funding Statement
This work was funded by SERB, India for EMR grant (SERB/EMR/2017/1854) to VT. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Falah F, Shokoohizadeh L, Adabi M (2019) Molecular identification and genotyping of Acinetobacter baumannii isolated from burn patients by PCR and ERIC-PCR. Scars, burns & healing 5: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agard MJ, Ozer EA, Morris AR, Piseaux R, Hauser AR (2019) A Genomic Approach to Identify Klebsiella pneumoniae and Acinetobacter baumannii Strains with Enhanced Competitive Fitness in the Lungs During Multi-Strain Pneumonia. Infection and Immunity 87(6):e00871 10.1128/IAI.00871-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sousa C, Silva L, Grosso F, Lopes J, Peixe L (2014) Development of a FTIR-ATR based model for typing clinically relevant Acinetobacter baumannii clones belonging to ST98, ST103, ST208 and ST218. J Photochem Photobiol B 133: 108–114. 10.1016/j.jphotobiol.2014.02.015 [DOI] [PubMed] [Google Scholar]
- 4.Al-Anazi KA, Al-Jasser AM (2014) Infections Caused by Acinetobacter baumannii in Recipients of Hematopoietic Stem Cell Transplantation. Front Oncol 4: 186 10.3389/fonc.2014.00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tiwari V, Vashistt J, Kapil A, Moganty RR (2012) Comparative proteomics of inner membrane fraction from carbapenem-resistant Acinetobacter baumannii with a reference strain. PLoS One 7: e39451 10.1371/journal.pone.0039451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, Lou J, Zhang W, Wu C, Jin Z (2018) Bacteria killing in ICU associated infections: antibacterial nanosheets as disinfectant. RSC Advances 8: 278–283. [Google Scholar]
- 7.Davies GE, Francis J, Martin AR, Rose FL, Swain G (1954) 1:6-Di-4'-chlorophenyldiguanidohexane (hibitane); laboratory investigation of a new antibacterial agent of high potency. Br J Pharmacol Chemother 9: 192–196. 10.1111/j.1476-5381.1954.tb00840.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sompornrattanaphan M, Kreetapirom P, Srinoulprasert Y, Kanistanon D, Klinniyom A, Wongsa C, et al. (2019) Severe anaphylaxis after pelvic examination: a case report of dual latex and chlorhexidine allergies. Allergy, Asthma & Clinical Immunology 15: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson J, Suwantarat N, Colantuoni E, Ross TL, Aucott SW, Carroll KC, et al. (2019) The impact of chlorhexidine gluconate bathing on skin bacterial burden of neonates admitted to the Neonatal Intensive Care Unit. J Perinatol 39: 63–71. 10.1038/s41372-018-0231-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saleem HG, Seers CA, Sabri AN, Reynolds EC (2016) Dental plaque bacteria with reduced susceptibility to chlorhexidine are multidrug resistant. BMC Microbiol 16: 214 10.1186/s12866-016-0833-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hardy K, Sunnucks K, Gil H, Shabir S, Trampari E, Hawkey P, et al. (2018) Increased Usage of Antiseptics Is Associated with Reduced Susceptibility in Clinical Isolates of Staphylococcus aureus. MBio 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raut U, Narang P, Mendiratta DK, Narang R, Deotale V (2008) Evaluation of rapid MTT tube method for detection of drug susceptibility of Mycobacterium tuberculosis to rifampicin and isoniazid. Indian J Med Microbiol 26: 222–227. [DOI] [PubMed] [Google Scholar]
- 13.Stevens MG, Olsen SC (1993) Comparative analysis of using MTT and XTT in colorimetric assays for quantitating bovine neutrophil bactericidal activity. J Immunol Methods 157: 225–231. 10.1016/0022-1759(93)90091-k [DOI] [PubMed] [Google Scholar]
- 14.Grela E, Kozlowska J, Grabowiecka A (2018) Current methodology of MTT assay in bacteria—A review. Acta Histochem 120: 303–311. 10.1016/j.acthis.2018.03.007 [DOI] [PubMed] [Google Scholar]
- 15.Mshana RN, Tadesse G, Abate G, Miorner H (1998) Use of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide for rapid detection of rifampin-resistant Mycobacterium tuberculosis. J Clin Microbiol 36: 1214–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haase H, Jordan L, Keitel L, Keil C, Mahltig B (2017) Comparison of methods for determining the effectiveness of antibacterial functionalized textiles. PLoS One 12: e0188304 10.1371/journal.pone.0188304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yalcin HT, Ozen MO, Gocmen B, Nalbantsoy A (2014) Effect of Ottoman Viper (Montivipera xanthina (Gray, 1849)) Venom on Various Cancer Cells and on Microorganisms. Cytotechnology 66: 87–94. 10.1007/s10616-013-9540-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Drlica K (2014) Reactive oxygen species and the bacterial response to lethal stress. Curr Opin Microbiol 21: 1–6. 10.1016/j.mib.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mageshwaran T, Ebenezar AR, Madhanamadhubala M, Kavitha S, Mahalaxmi S (2012) Counteraction of reactive oxygen species and determination of antibacterial efficacy of proanthocyanidin and lycopene when mixed with calcium hydroxide and chlorhexidine mixture: An in vitro comparative study. J Conserv Dent 15: 337–341. 10.4103/0972-0707.101893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeung SY, Huang CS, Chan CP, Lin CP, Lin HN, Lee PH, et al. (2007) Antioxidant and pro-oxidant properties of chlorhexidine and its interaction with calcium hydroxide solutions. Int Endod J 40: 837–844. 10.1111/j.1365-2591.2007.01271.x [DOI] [PubMed] [Google Scholar]
- 21.Trastoy R, Manso T, Fernandez-Garcia L, Blasco L, Ambroa A, Perez Del Molino ML, et al. (2018) Mechanisms of Bacterial Tolerance and Persistence in the Gastrointestinal and Respiratory Environments. Clin Microbiol Rev 31:e00023–18. 10.1128/CMR.00023-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biswas D, Tiwari M, Tiwari V (2018) Comparative mechanism based study on disinfectants against multidrug-resistant Acinetobacter baumannii. J Cell Biochem 119:10314–10326. 10.1002/jcb.27373 [DOI] [PubMed] [Google Scholar]
- 23.Arakha M, Pal S, Samantarrai D, Panigrahi TK, Mallick BC, Pramanik K, et al. (2015) Antimicrobial activity of iron oxide nanoparticle upon modulation of nanoparticle-bacteria interface. Sci Rep 5: 14813 10.1038/srep14813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bankier C, Cheong Y, Mahalingam S, Edirisinghe M, Ren G, Cloutman-Green E, et al. (2018) A comparison of methods to assess the antimicrobial activity of nanoparticle combinations on bacterial cells. PLOS ONE 13: e0192093 10.1371/journal.pone.0192093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Booyens J, Thantsha MS (2014) Fourier transform infra-red spectroscopy and flow cytometric assessment of the antibacterial mechanism of action of aqueous extract of garlic (Allium sativum) against selected probiotic Bifidobacterium strains. BMC Complement Altern Med 14: 289 10.1186/1472-6882-14-289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Golding CG, Lamboo LL, Beniac DR, Booth TF (2016) The scanning electron microscope in microbiology and diagnosis of infectious disease. Sci Rep 6: 26516 10.1038/srep26516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Brien-Simpson NM, Pantarat N, Attard TJ, Walsh KA, Reynolds EC (2016) A Rapid and Quantitative Flow Cytometry Method for the Analysis of Membrane Disruptive Antimicrobial Activity. PLoS One 11: e0151694 10.1371/journal.pone.0151694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gui S, Li R, Feng Y, Wang S (2014) Transmission electron microscopic morphological study and flow cytometric viability assessment of Acinetobacter baumannii susceptible to Musca domestica cecropin. ScientificWorldJournal 2014: 657536 10.1155/2014/657536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sousa C, Silva L, Grosso F, Nemec A, Lopes J, Peixe L, et al. (2014) Discrimination of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex species by Fourier transform infrared spectroscopy. Eur J Clin Microbiol Infect Dis 33: 1345–1353. 10.1007/s10096-014-2078-y [DOI] [PubMed] [Google Scholar]
- 30.Sousa C, Grosso F, Meirinhos-Soares L, Peixe L, Lopes J (2014) Identification of carbapenem-resistant Acinetobacter baumannii clones using infrared spectroscopy. J Biophotonics 7: 287–294. 10.1002/jbio.201200075 [DOI] [PubMed] [Google Scholar]
- 31.Tang M, McEwen GD, Wu Y, Miller CD, Zhou A (2013) Characterization and analysis of mycobacteria and Gram-negative bacteria and co-culture mixtures by Raman microspectroscopy, FTIR, and atomic force microscopy. Anal Bioanal Chem 405: 1577–1591. 10.1007/s00216-012-6556-8 [DOI] [PubMed] [Google Scholar]
- 32.Talari ACS, Martinez MAG, Movasaghi Z, Rehman S, Rehman IU (2017) Advances in Fourier transform infrared (FTIR) spectroscopy of biological tissues. Applied Spectroscopy Reviews 52: 456–506. [Google Scholar]
- 33.Aboualizadeh E, Bumah VV, Masson-Meyers DS, Eells JT, Hirschmugl CJ, Enwemeka CS (2017) Understanding the antimicrobial activity of selected disinfectants against methicillin-resistant Staphylococcus aureus (MRSA). PLoS One 12: e0186375 10.1371/journal.pone.0186375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiwari V, Rajeswari MR (2013) Effect of Iron Availability on the Survival of Carbapenem-Resistant Acinetobacter baumannii: a Proteomic Approach. Journal of Proteomics & Bioinformatics 06: 125–131. [Google Scholar]
- 35.Tiwari V, Moganty RR (2014) Conformational stability of OXA-51 beta-lactamase explains its role in carbapenem resistance of Acinetobacter baumannii. J Biomol Struct Dyn 32: 1406–1420. 10.1080/07391102.2013.819789 [DOI] [PubMed] [Google Scholar]
- 36.Tiwari V, Kapil A, Moganty RR (2012) Carbapenem-hydrolyzing oxacillinase in high resistant strains of Acinetobacter baumannii isolated from India. Microb Pathog 53: 81–86. 10.1016/j.micpath.2012.05.004 [DOI] [PubMed] [Google Scholar]
- 37.Tiwari V, Rajeswari MR, Tiwari M (2019) Proteomic analysis of iron-regulated membrane proteins identify FhuE receptor as a target to inhibit siderophore-mediated iron acquisition in Acinetobacter baumannii. International Journal of Biological Macromolecules 125: 1156–1167. 10.1016/j.ijbiomac.2018.12.173 [DOI] [PubMed] [Google Scholar]
- 38.Tiwari V, Mishra N, Gadani K, Solanki PS, Shah NA, Tiwari M (2018) Mechanism of Anti-bacterial Activity of Zinc Oxide Nanoparticle Against Carbapenem-Resistant Acinetobacter baumannii. Front Microbiol 9: 1218 10.3389/fmicb.2018.01218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tiwari V, Tiwari M, Solanki V (2017) Polyvinylpyrrolidone-Capped Silver Nanoparticle Inhibits Infection of Carbapenem-Resistant Strain of Acinetobacter baumannii in the Human Pulmonary Epithelial Cell. Front Immunol 8: 973 10.3389/fimmu.2017.00973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiwari V, Tiwari M, Biswas D (2018) Rationale and design of an inhibitor of RecA protein as an inhibitor of Acinetobacter baumannii. J Antibiot (Tokyo) 71: 522–534. [DOI] [PubMed] [Google Scholar]
- 41.Tiwari V, Tiwari M (2015) Phosphoproteomics as an emerging weapon to develop new antibiotics against carbapenem resistant strain of Acinetobacter baumannii. Journal of Proteomics 112: 336–338. 10.1016/j.jprot.2014.09.008 [DOI] [PubMed] [Google Scholar]
- 42.Harbarth S, Tuan Soh S, Horner C, Wilcox MH (2014) Is reduced susceptibility to disinfectants and antiseptics a risk in healthcare settings? A point/counterpoint review. J Hosp Infect 87: 194–202. 10.1016/j.jhin.2014.04.012 [DOI] [PubMed] [Google Scholar]
- 43.Ye J Jr., Huang C-T, Shie S-S, Huang P-Y, Su LH, Chiu CH, et al. (2010) Multidrug Resistant Acinetobacter baumannii: Risk Factors for Appearance of Imipenem Resistant Strains on Patients Formerly with Susceptible Strains. PLOS ONE 5: e9947 10.1371/journal.pone.0009947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48: 1–12. 10.1086/595011 [DOI] [PubMed] [Google Scholar]
- 45.Sepkowitz KA (2002) Antibiotic prophylaxis in patients receiving hematopoietic stem cell transplant. Bone Marrow Transplant 29: 367–371. 10.1038/sj.bmt.1703366 [DOI] [PubMed] [Google Scholar]
- 46.Sanchez-Vizuete P, Orgaz B, Aymerich S, Le Coq D, Briandet R (2015) Pathogens protection against the action of disinfectants in multispecies biofilms. Front Microbiol 6: 705 10.3389/fmicb.2015.00705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simon AY, Ihejirika KA, Ogunkoya AB, Hassan AZ, Adawa DAY, Adeyanju JB (2007) Comparison of the antimicrobial efficacy oftopical antiseptic creams on canine wounds(preliminary communication). Bulgarian Journal of Veterinary Medicine 10: 273–281. [Google Scholar]
- 48.Septimus E, Huang SS, Septimus E, Kleinman K, Moody J, Hickok J, et al. (2019) Chlorhexidine versus routine bathing to prevent multidrug-resistant organisms and all-cause bloodstream infections in general medical and surgical units (ABATE Infection trial): a cluster-randomised trial. Lancet 393: 1205–1215. 10.1016/S0140-6736(18)32593-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brynildsen MP, Winkler JA, Spina CS, MacDonald IC, Collins JJ (2013) Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat Biotechnol 31: 160–165. 10.1038/nbt.2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pischel D, Buchbinder JH, Sundmacher K, Lavrik IN, Flassig RJ (2018) A guide to automated apoptosis detection: How to make sense of imaging flow cytometry data. PLOS ONE 13: e0197208 10.1371/journal.pone.0197208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lotfollahi M, Berisha S, Daeinejad D, Mayerich D (2019) Digital Staining of High-Definition Fourier Transform Infrared (FT-IR) Images Using Deep Learning. Applied Spectroscopy 73: 556–564. 10.1177/0003702818819857 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIFF)
(TIFF)
(TIFF)
(TIFF)
Data Availability Statement
All data is provided along with MS.