Abstract
In nature, most bacteria live in biofilms where they compete with their siblings and other species for space and nutrients. Some bacteria produce antibiotics in biofilms; however, since the diffusion of antibiotics is generally hindered in biofilms by extracellular polymeric substances, i.e., the biofilm matrix, their function remains unclear. The Bacillus subtilis yitPOM operon is a paralog of the sdpABC operon, which produces the secreted peptide toxin SDP. Unlike sdpABC, yitPOM is induced in biofilms by the DegS-DegU two-component regulatory system. High yitPOM expression leads to the production of a secreted toxin called YIT. Expression of yitQ, which lies upstream of yitPOM, confers resistance to the YIT toxin, suggesting that YitQ is an anti-toxin protein for the YIT toxin. The alternative sigma factor SigW also contributes to YIT toxin resistance. In a mutant lacking yitQ and sigW, the YIT toxin specifically inhibits biofilm formation, and the extracellular neutral protease NprB is required for this inhibition. The requirement for NprB is eliminated by Δeps and ΔbslA mutations, either of which impairs production of biofilm matrix polymers. Overexpression of biofilm matrix polymers prevents the action of the SDP toxin but not the YIT toxin. These results indicate that, unlike the SDP toxin and many conventional antibiotics, the YIT toxin can pass through layers of biofilm matrix polymers to attack cells within biofilms with assistance from NprB. When the wild-type strain and the YIT-sensitive mutant were grown together on a solid medium, the wild-type strain formed biofilms that excluded the YIT-sensitive mutant. This observation suggests that the YIT toxin protects B. subtilis biofilms against competitors. Several bacteria are known to produce antibiotics in biofilms. We propose that some bacteria including B. subtilis may have evolved specialized antibiotics that can function within biofilms.
Author summary
Biofilms are multicellular aggregates of bacteria that are formed on various living and non-living surfaces. Biofilms often cause serious problems, including food contamination and infectious diseases. Since bacteria in biofilms exhibit increased tolerance or resistance to antimicrobials, new agents and treatments for combating biofilm-related problems are required. In this study, we demonstrated that B. subtilis produces a secreted peptide antibiotic called the YIT toxin and its resistant proteins in biofilms. A mutant lacking the resistance genes was defective in biofilm formation. This effect resulted from the ability of the YIT toxin to pass through the biofilm defense barrier and to attack biofilm cells. Thus, unlike many conventional antibiotics, the YIT toxin can penetrate biofilms and suppress the growth of YIT toxin-sensitive cells within biofilms. Some bacteria produce antibiotics in biofilms, some of which can alter the bacterial composition in the biofilms. Taking these observations into consideration, our findings suggest that some bacteria produce special antibiotics that are effective against bacteria in biofilms, and these antibiotics might serve as anti-biofilm agents.
Introduction
In the environment, bacteria compete for space and nutrients [1]. Antibiotics are thought to play a critical role in this competition, and antibiotic-producing bacteria are indeed common in various environments [2–4]. However, in the environment, most bacteria are found in sessile multicellular bacterial communities known as biofilms, in which bacteria exhibit increased antibiotic tolerance or resistance [5, 6]. Though alternative environmental roles of antibiotics have been proposed [7], this paradox has not been explained in detail to date.
In biofilms, bacterial cells adhere to each other and to a surface via a mixture of extracellular polymeric substances called the biofilm matrix, which consists of exopolysaccharides, proteins, nucleic acids, and/or lipids [8, 9]. When encased in the biofilm matrix, cells exhibit increased tolerance or resistance to environmental stresses, antibiotics, host defense systems, and predation [8, 9]. Thus, biofilm formation enables bacteria to remain in a favorable niche and to claim territory; however, biofilms are not a utopia for bacteria. The properties of biofilms, including high cell density, decreased internal fluidity, and, in many cases, the presence of multiple species, lead to conditions of harsh competition, especially when nutrients are scarce. Many bacteria secrete biofilm formation-inhibiting molecules, such as biosurfactants, polysaccharides, and molecules that interfere with bacterial quorum sensing, and these secreted molecules help to exclude unfavorable competitors from biofilms [10]. Antibiotics might also play an important role in competition within biofilms. However, since the properties of biofilms, including the protection of member cells by the biofilm matrix, the increased expression of antibiotic resistance genes, and the decreased internal fluidity, reduce the efficacy of antibiotics against biofilm cells [11–16], little attention has been paid to the functions of antibiotics in competition within biofilms. However, some biofilms do indeed produce antibiotics, and several of these antibiotics can alter the bacterial composition of the biofilm [17–23]. These observations suggest that the functions of antibiotics produced in biofilms remain to be investigated. An understanding of how bacteria use antibiotics in biofilms will not only provide insight into bacterial survival strategies within biofilms, it will also lead to the discovery of tactics for combating biofilm-related problems, such as food and beverage safety issues, industrial contamination, and biofilm-related diseases.
The Gram-positive soil bacterium Bacillus subtilis is a model organism for biofilm formation. B. subtilis forms robust biofilms under laboratory conditions, for example, pellicles on the surface of liquid media under static culture conditions or wrinkled colonies on solid media [24]. B. subtilis biofilms are maintained by a biofilm matrix that mainly consists of exopolysaccharides, TasA amyloid fibers, and BslA hydrophobins, which are produced by proteins encoded by the epsABCDEFGHIJKLMNO operon, the tapA-sipW-tasA operon, and bslA, respectively [24–29]. These genes are directly or indirectly repressed by the transcriptional repressors AbrB and SinR [30–33]. Phosphorylation of the response regulator Spo0A induces mechanisms that antagonize these repressors, leading to the expression of the biofilm matrix synthesis genes [34, 35].
B. subtilis produces a wide array of antibiotics. Many of these antibiotics are non-ribosomally synthesized peptide compounds, such as surfactin, bacillaene, fengycin, iturin, and bacilysin, which are thought to be important in nature for competition with other organisms, including fungi [4, 36]. Furthermore, B. subtilis produces ribosomally synthesized peptide antibiotics, such as bacteriocins and other protein-derived toxins, which are generally effective against other bacteria that are genetically similar and present in similar ecological niches [4, 37–39]. One of these protein-derived toxins is the cannibalism toxin SDP [40], whose function is involved in biofilm formation. The SDP toxin is derived from the internal sequence of SdpC, and it is encoded by the sdpABC operon. SdpC is a 203 amino acid protein that contains a typical N-terminal secretion signal and a C-terminal hydrophobic domain. After secretion and cleavage of the signal sequence, SdpC is further processed into the 42 amino acid peptide known as the SDP toxin, which corresponds to the C-terminal hydrophobic domain (C141 to S182) [39–41]. SdpA and SdpB are required for the processing of SdpC to SDP, and this processing is essential for the activity of the SDP toxin [41]. The hydrophobic nature of the SDP toxin enables the SDP toxin to penetrate bacterial membranes, where it then induces cell lysis by collapsing the proton motive force [42]. Downstream of sdpABC is the sdpRI operon, which encodes its own transcriptional repressor and an anti-toxin protein to the SDP toxin [43]. SdpI is an integral membrane protein that protects cells probably by binding to the SDP toxin. Transcription of the sdpABC and sdpRI operons is directly or indirectly activated by phosphorylated Spo0A [40, 43]. Spo0A is a master regulator of stationary phase development that is phosphorylated after the onset of stationary phase [44]. However, as the phosphorylation of Spo0A is subject to a bistable regulatory mechanism, a subset of B. subtilis cells produce the SDP toxin and the SdpI anti-toxin protein [40, 43]. Consequently, the secreted SDP toxin lyses and kills a fraction of the sibling cells that do not produce the SdpI anti-toxin protein. Since phosphorylated Spo0A also induces biofilm formation in parallel, cells that produce the SDP toxin and the SdpI anti-toxin protein efficiently develop biofilms by using nutrients released from their lysed siblings [45]. Moreover, the SDP toxin is effective not only against B. subtilis, but also against many Firmicutes bacteria [39, 41, 46]. Thus, the SDP toxin likely plays an important role in the early phase of biofilm formation by eliminating unnecessary types of cells and closely related competitors in the environment.
The undomesticated B. subtilis strain NCIB3610 encodes an sdpABC paralog known as yitPOM. While transcription of sdpABC is activated by Spo0A [40], yitPOM was previously identified as a member of the group of genes regulated by the DegS-DegU two-component regulatory system [47]. Phosphorylated DegU directly induces transcription of genes for the biofilm matrix protein, BslA, several antibiotic synthetases, and many extracellular degradative enzymes, such as proteases, levansucrase, α-amylase, β-glucanases, and xylanase. These antibiotics and degradative enzymes are thought to play important roles in bacterial competitions and nutrient acquisition in nature [47, 48, and references therein]. Since DegS-DegU is required for biofilm formation, we were interested in determining whether the yitPOM-encoded toxin plays a role particularly in biofilms. In this paper, we demonstrate that yitPOM encodes a biofilm-associated secreted toxin. Unlike many conventional antibiotics, in particular positive charged antibiotics, this toxin was able to attack cells within biofilms by passing through the layers of the biofilm matrix polymers with the assistance of an extracellular protease. Given that several other bacteria produce antibiotics in biofilms, our results suggest that some bacteria may have evolved specialized antibiotics to suppress competitors in biofilms.
Results
yitPOM encodes a toxin
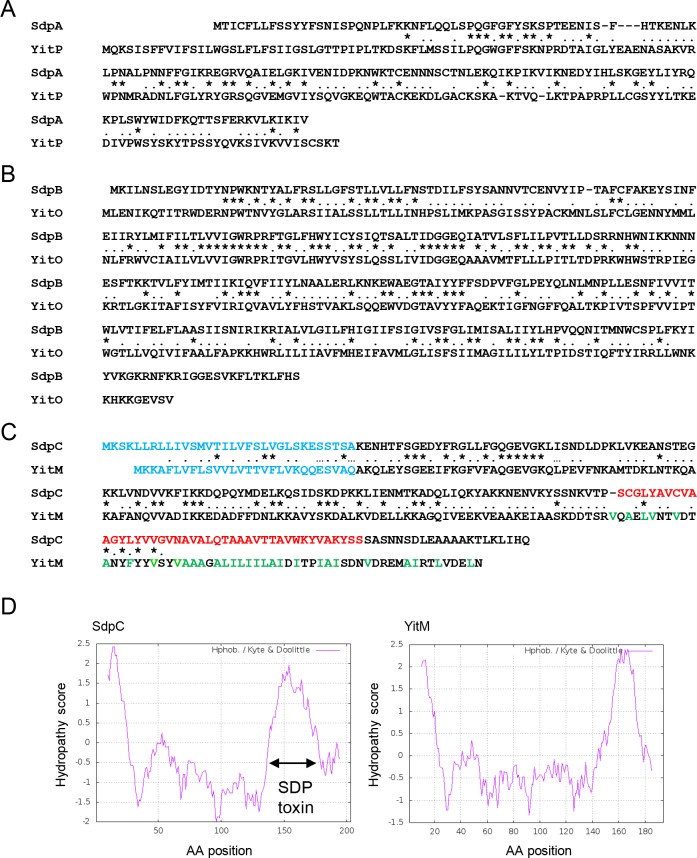
The undomesticated B. subtilis strain NCIB3610 (hereafter referred to as the wild-type strain or 3610) [24] encodes an sdpABC paralog known as yitPOM. YitP and YitO exhibit approximately 50% sequence similarity to the entire SdpA and SdpB sequences, respectively (Fig 1A and 1B). Like SdpC, YitM has an N-terminal secretion signal; however, the sequence similarity between YitM and SdpC is limited to the N-terminal three quarters of the sequence, which does not include the entire sequence corresponding to the SDP toxin (Fig 1C). Although there is no sequence similarity, like SdpC, the YitM C-terminal region contains a hydrophobic domain (Fig 1C and 1D). These observations suggest the possibility that the C-terminal hydrophobic domain of YitM might be processed to a secreted toxin via a YitP and YitO-dependent mechanism. If this is the case, then yitPOM encodes a toxin whose sequence differs from that of the SDP toxin.
Fig 1. yitPOM is a paralog of sdpABC.
(A) Alignment of SdpA and YitP. (B) Alignment of SdpB and YitO. (C) Alignment of SdpC and YitM. Identical and similar amino acid residues shared by the two proteins are indicated by asterisks and dots, respectively. The signal sequences of SdpC and YitM and the SDP toxin sequence are shown in blue and red, respectively. Hydrophobic amino acid residues in the C-terminal region of YitM are shown in green. (D) Kyte and Doolittle hydropathy plots of SdpC and YitM. The hydropathy score, representing the hydrophobic or hydrophilic properties of amino acid residues was calculated and plotted using the ExPASy website (https://web.expasy.org/protscale/) with a window size of 19.
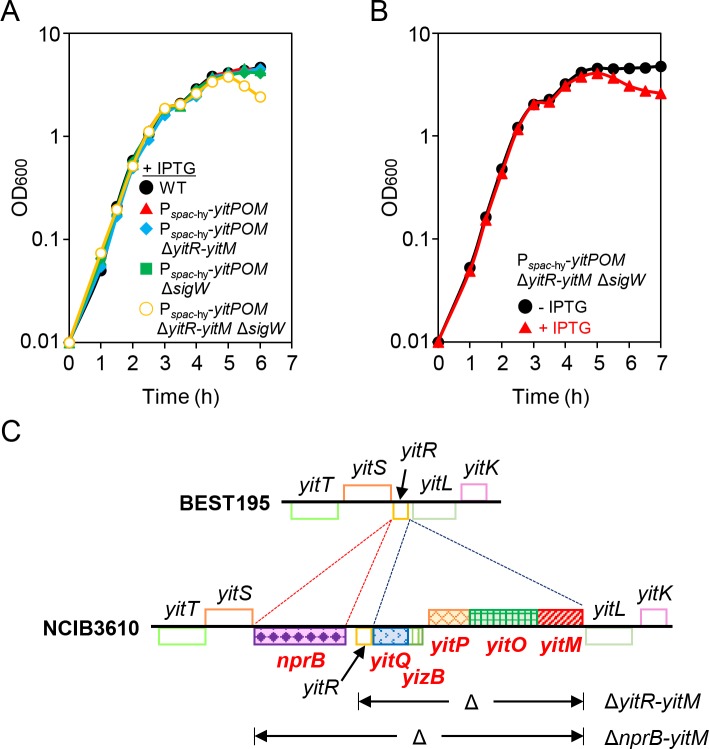
To determine whether yitPOM encodes a toxin, we examined the effect of yitPOM overexpression on growth. We constructed the strain Pspac-hy-yitPOM, which ectopically expresses yitPOM from the strong isopropyl β-D-thiogalactopyranoside (IPTG)-inducible, LacI-repressible spac-hy promoter [49] in the amyE locus on the chromosome. The wild-type and Pspac-hy-yitPOM strains were grown with vigorous shaking in 2× Schaeffer’s sporulation medium plus glucose (2×SG) [50] supplemented with 1 mM IPTG, and the optical density at 600 nm (OD600) was measured over time. These strains showed no difference in growth from exponential phase to stationary phase (Fig 2A). Toxin-producing bacteria normally express cognate anti-toxin proteins against their toxins, and the effects of the toxins do not appear unless the anti-toxin genes are deleted [2, 37, 38, 40]. Since toxin and anti-toxin genes are simultaneously inserted into the genomes as exogenous genes and are frequently located close to each other in the genome [2, 37, 38, 40], genome comparison is a powerful tool to identify toxin/anti-toxin gene sets. To identify candidates for an anti-toxin gene against the putative toxin encoded by yitPOM, we compared the genetic organization of the 3610 and B. subtilis var. natto BEST195 strains, the latter of which lacks yitPOM. This comparison revealed that yitPOM appears to be inserted between yitR and yitL in the 3610 genome, along with yizB and yitQ, which are predicted to encode a transcriptional regulator and a membrane protein, respectively (Fig 2C). Since the SdpI anti-toxin protein is a membrane protein, YitQ was a candidate for an anti-toxin protein to the putative toxin, although YitQ has no similarity to SdpI. Based on the DNA sequence, yizB and yitQ are predicted to form an operon with a downstream gene, yitR, which also encodes a membrane protein. Althogh yitR is present in BEST195, we kept it as a second candidate for the anti-toxin protein. To test whether these genes encoded anti-YIT toxin, we attempted to disrupt these genes. However, we were concerned that deleting these candidate anti-toxin genes might cause severe growth defects by releasing the activity of the toxin encoded by the genomic yitPOM operon. Therefore, we constructed a ΔyitR-yitM deletion strain that lacks the entire region from yitR to yitM, which contains the candidate anti-toxin genes yitR and yitQ, the unknown repressor gene yizB, and the putative toxin-encoding yitPOM operon (Fig 2C). Furthermore, since the expression of the secondary resistance mechanism against the SDP toxin is induced by the alternative sigma factor SigW (σW) [51], we also constructed a sigW deletion strain. Subsequently, either the ΔyitR-yitM and ΔsigW mutation alone or both mutations together were introduced into the Pspac-hy-yitPOM strain. We compared the growth of these strains in 2×SG medium supplemented with 1 mM IPTG. While the Pspac-hy-yitPOM ΔyitR-yitM and Pspac-hy-yitPOM ΔsigW mutants grew normally, the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant showed mild cell lysis 3 h after the end of exponential phase (Fig 2A). We confirmed that yitPOM expression caused this cell lysis, as cell lysis was only observed when yitPOM expression was induced with IPTG (Fig 2B). These results indicate that yitPOM expression leads to the production of a toxin that causes cell lysis in a mutant strain lacking the putative anti-toxin genes and sigW. The σW-regulated genes include multiple antibiotic resistant genes, such as a peptide exporter, an SdpI homolog, fosfomycin resistance proteins, sublancin resistance proteins, and a penicillin-binding protein [51–54]. Some of these genes might contribute to resistance to the putative toxin produced by yitPOM.
Fig 2. yitPOM encodes a toxin.
(A) Effect of yitPOM induction on cell growth. B. subtilis strains were grown at 37°C in 2×SG supplemented with 1 mM IPTG with vigorous shaking. Growth profiles were examined at least three time, and the typical examples were shown. (B) Induction of yitPOM caused mild cell lysis. The Pspac-hy-yitPOM ΔyitR-yitM ΔsigW strain was grown in 2×SG supplemented with or without 1 mM IPTG. (C) Comparison of the genetic organization in NCIB3610 and BEST195. Homologous genes are shown by boxes of the same color. Genes only present in NCIB3610 are shown in red bold. The deleted regions in the ΔyitR-yitM and ΔnprB-yitM mutants are shown below the gene map of NCIB3610.
To further confirm that yitPOM encodes a toxin, we employed a spot-on-lawn assay. We performed this assay in the ΔsdpABC-sdpIR (hereafter referred to as ΔsdpA-sdpR) ΔyitR-yitM mutant background to eliminate the effects of the endogenous sdpABC and yitPOM operons. Since the ΔsigW mutant is sensitive to multiple antibiotics and stresses, including the SDP toxin [51] and the putative toxin produced by yitPOM, we used the ΔsdpA-sdpR ΔyitR-yitM ΔsigW mutant as an antibiotic-sensitive indicator strain. When spotted on a lawn of this indicator strain, the strain expressing YitPOM (Pspac-hy-yitPOM ΔsdpA-sdpR ΔyitR-yitM) formed growth inhibition zones (halos) around its colonies (Fig 3). By contrast, a strain that does not express YitPOM (ΔsdpA-sdpR ΔyitR-yitM) formed no obvious halos around its colonies on the same lawn. These results demonstrate that, like sdpABC, yitPOM encodes a secreted toxin, which we named YIT.
Fig 3. YitQ is an anti-toxin protein to the YIT toxin.
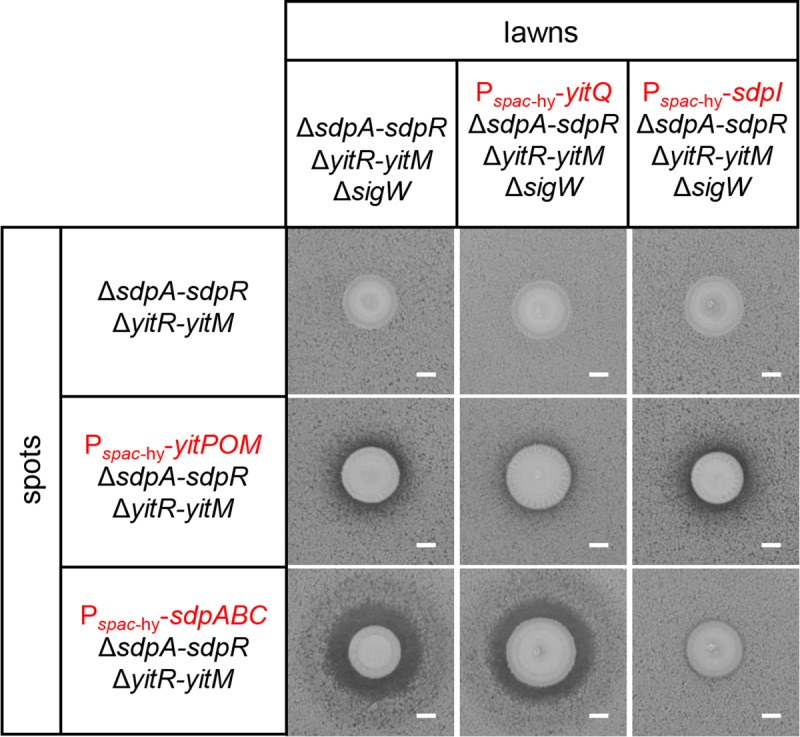
B. subtilis strains ΔsdpA-sdpR ΔyitR-yitM ΔsigW, Pspac-hy-yitQ ΔsdpA-sdpR ΔyitR-yitM ΔsigW, and Pspac-hy-sdpI ΔsdpA-sdpR ΔyitR-yitM ΔsigW were added to 2×SG agar containing 1 mM IPTG and poured into plates as lawns. Strains tested for antibiotic production (shown on the left of the figure as spots) were spotted on the lawns. Plates were incubated at 37°C. A growth inhibitory zone was observed if the lawn strain was sensitive to a compound produced by the strain spotted on it. Scale bar, 2 mm.
YitQ is an anti-toxin protein to the YIT toxin
To determine whether YitQ is an anti-toxin protein to the YIT toxin, we examined the effect of yitQ overexpression on the YIT toxin activity. To this end, the Pspac-hy-yitQ construct was introduced into the amyE locus of the indicator strain (i.e., the ΔsdpA-sdpR ΔyitR-yitM ΔsigW mutant). When spotted on a lawn of the indicator strain expressing YitQ (Pspac-hy-yitQ ΔsdpA-sdpR ΔyitR-yitM ΔsigW), the strain expressing YitPOM (Pspac-hy-yitPOM ΔsdpA-sdpR ΔyitR-yitM) only formed weak halos around its colonies (Fig 3). Thus, yitQ expression confered resistance to the YIT toxin.
We were interested in whether there is crosstalk between yitPOM/yitQ and sdpABC/sdpI. To explore this possibility, a strain expressing SdpABC (Pspac-hy-sdpABC ΔsdpA-sdpR ΔyitR-yitM) was spotted onto lawns of the control indicator strain (ΔsdpA-sdpR ΔyitR-yitM Δspo0A) and the indicator strain expressing YitQ (Pspac-hy-yitQ ΔsdpA-sdpR ΔyitR-yitM ΔsigW) (Fig 3). The SdpABC-expressing strain formed clear halos around its colonies on both types of lawns. Thus, yitQ expression did not confer resistance to the SDP toxin. We also tested whether SdpI expression confers resistance to the YIT and SDP toxins. A strain expressing YitPOM (Pspac-hy-yitPOM ΔsdpA-sdpR ΔyitR-yitM) formed halos around its colonies on a lawn of the indicator strain expressing SdpI (Pspac-hy-sdpI ΔsdpA-sdpR ΔyitR-yitM ΔsigW), whereas the strain expressing SdpABC (Pspac-hy-sdpABC ΔsdpA-sdpR ΔyitR-yitM) did not. These results indicate that YitQ and SdpI are anti-toxin proteins specific to the YIT and SDP toxins, respectively. Thus, the two toxin/anti-toxin gene pairs yitPOM/yitQ and sdpABC/sdpI most likely function independently.
Expression of yitPOM and yitQ
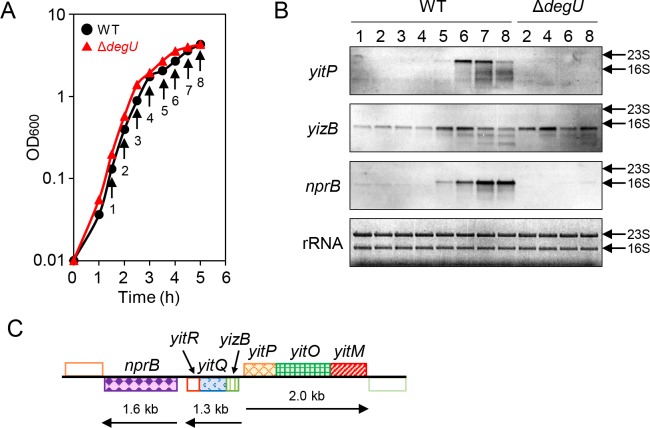
The yitPOM operon was previously identified as a member of the DegS-DegU-regulated genes via a DNA microarray analysis using another B. subtilis strain, ATCC6051 [47]. To confirm this property in strain 3610, we carried out a Northern blot analysis. RNA samples were isolated from wild-type and ΔdegU mutant cells grown for various lengths of time in 2×SG with vigorous shaking (Fig 4A). We detected a single band at a position between the 23S rRNA (2904 nt) and 16S rRNA (1541 nt) on Northern blots with a yitP-specific probe (Fig 4B). The size of the band was consistent with the length of the entire yitPOM locus (2031 bp), confirming that the yitPOM locus is transcribed as an operon (Fig 4C). On the Northern blots, the yitPOM transcript was observed in the stationary phase samples from the wild-type strain but not in those from the ΔdegU mutant (Fig 4B). These results indicate that DegS-DegU directly or indirectly induces yitPOM transcription in stationary phase.
Fig 4. Transcription of yitPOM and yitQ in the ΔdegU mutant.
(A) Growth profiles of the wild-type and ΔdegU mutant strains. Strains were grown in 2×SG with vigorous shaking. Arrows indicate the time points at which samples were taken for RNA isolation. (B) Northern blot analysis of yitPOM, yizB-yitQ-yitR, and nprB. Transcripts were detected with gene-specific DIG-labeled RNA probes. Lane numbers (time points) under the strain names correspond to the time points shown in panel A. rRNA stained with methylene blue is shown as a loading control. The positions of 23S rRNA and 16S rRNA are indicated by arrows. (C) The transcription map of the yitPOM region. The transcripts are represented as lines with arrows below the gene map, and their estimated lengths are indicated.
yitQ is predicted to form an operon with its upstream and downstream genes, yizB and yitR. We detected a band below the position of 16S rRNA on Northern blots with a yizB-specific probe (Fig 3B). The size of the band was consistent with the length of the yizB-yitQ-yitR locus (1244 bp), supporting the conclusion that yizB, yitQ, and yitR are transcribed as an operon (Fig 4C). Based on the Northern blots, the yizB-yitQ-yitR operon was transcribed at low levels during exponential phase and then induced during stationary phase in the wild-type strain (Fig 4B). The ΔdegU mutation had no significant effect on the transcription of the yizB-yitQ-yitR operon. The SDP toxin mediates cannibalism between “Spo0A ON” and “Spo0A OFF” cells [40]. The finding that DegS-DegU regulates yitPOM but not yitQ rules out the possibility that the YIT toxin mediates cannibalism between “DegU ON” and “DegU OFF” cells.
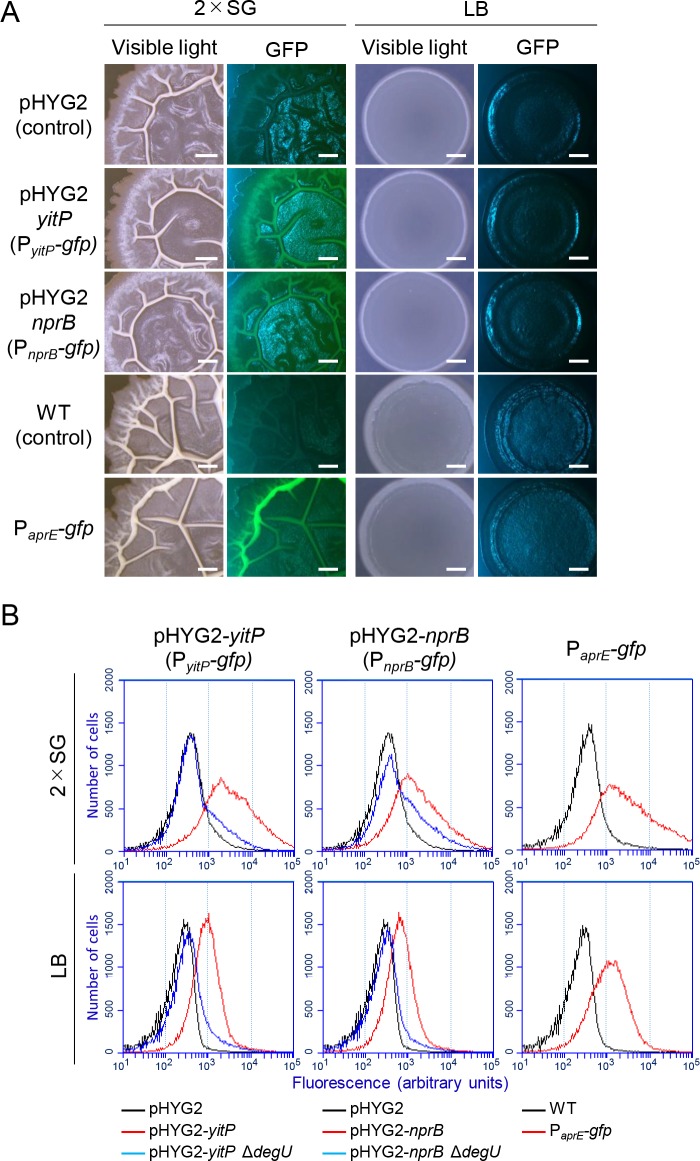
To explore the function of the YIT toxin, we asked under what conditions yitPOM expression is induced. The DegS-DegU-regulated gene bpr, which encodes an extracellular protease, is expressed in biofilms [55]. We therefore speculated that yitPOM is also expressed in biofilms. To visualize yitPOM expression in biofilms, the yitP promoter was fused to the green fluorescent protein (GFP) reporter, and the resulting PyitP-gfp reporter construct was introduced to the amyE locus on the chromosome of the wild-type strain. B. subtilis biofilms are wrinkled structures on the surfaces of colonies grown on solid media that support biofilm formation, such as 2×SG [24]; therefore, we attempted to examine the expression level of the PyitP-gfp reporter in colonies grown on 2×SG solid medium. However, we did not detect any fluorescent GFP signal on these colonies, probably because the yitP promoter activity was too low to detect signals in our microscopy. We next examined the expression of the PyitP-gfp reporter inserted into the multi-copy plasmid pHYG2. The expression of the PyitP-gfp reporter on the plasmid was examined at 37°C because the plasmid pHYG2-yitP negatively affected biofilm formation at 30°C. The wild-type strain carrying the multi-copy plasmid pHYG2-yitP (PyitP-gfp) formed wrinkled structures on the surfaces of colonies grown on 2×SG solid medium as the biofilms developed. Weak green GFP signal was observed in these wrinkles with a color digital camera (Fig 5A). By contrast, this strain formed flat colonies on LB medium, which does not support biofilm formation, and produced no detectable green GFP signal. Under the same conditions, the wild-type strain carrying the parental plasmid pHYG2 (promoterless gfp) produced no green fluorescent signal on either 2×SG or LB.
Fig 5. yitPOM expression is induced in biofilms by DegS-DegU.
(A) Expression of yitPOM and nprB in biofilms. The wild-type strain 3610 carrying the multi-copy plasmid pHYG2 (promoterless gfp), pHYG2-yitP containing the PyitP-gfp reporter, or the single copy of aprE-gfp was grown at 37°C for 24 h on 2×SG or LB. GFP fluorescence was analyzed with a digital color camera. GFP fluorescence was observed as green light signals. Some excitation light reflections on the surfaces of colonies and media were observed as blue light signals on GFP images. Strains 3610 pHYG2 (promoter-less gfp) and 3610 were used as negative controls. Scale bar, 1 mm. (B) Flow cytometry analysis of gfp reporter strains. B. subtilis strains were grown at 37°C for 24 h on 2×SG or LB. Expression of gfp reporters in the colonies was analyzed using stains 3610 pHYG2 and 3610 as negative controls.
The expression of PyitP-gfp in these colonies was also analyzed at the single-cell level. Flow cytometry analysis revealed that the expression of PyitP-gfp was heterogeneous in the population in colonies grown on 2×SG and that the considerable portion of PyitP-gfp cells exhibited stronger fluorescence in 2×SG than in LB (Fig 5B). Combined with the microscopic observation, these results indicate that biofilms contains cells that highly express the yitPOM operon. Moreover, we confirmed that the expression of PyitP-gfp in these colonies was DegU-dependent as the level of fluorescence decreased to the background level in the ΔdegU mutant (Fig 5B).
To eliminate potential artifacts resulting from multi-copy plasmid-based experiments, we examined the expression of aprE, which is one of the most highly expressed genes among the DegS-DegU-regulated genes [47]. We used a strain carrying a single copy of the PaprE-gfp reporter inserted into the amyE locus on the chromosome. The PaprE-gfp strain produced GFP fluorescent signal in the wrinkles of colonies grown on 2×SG (Fig 5A). By contrast, no detectable GFP signal was observed when the PaprE-gfp strain was grown on LB. Flow cytometry analysis revealed that the considerable portion of PaprE-gfp cells exhibited stronger fluorescence in 2×SG than in LB (Fig 5B). Thus, the expression profiles of PaprE-gfp were quite similar to those of PyitP-gfp. These results indicate that DegS-DegU strongly induces its regulatory target genes, including yitPOM, in biofilms and that the YIT toxin may play a role in biofilms.
The YIT toxin inhibits colony biofilm formation
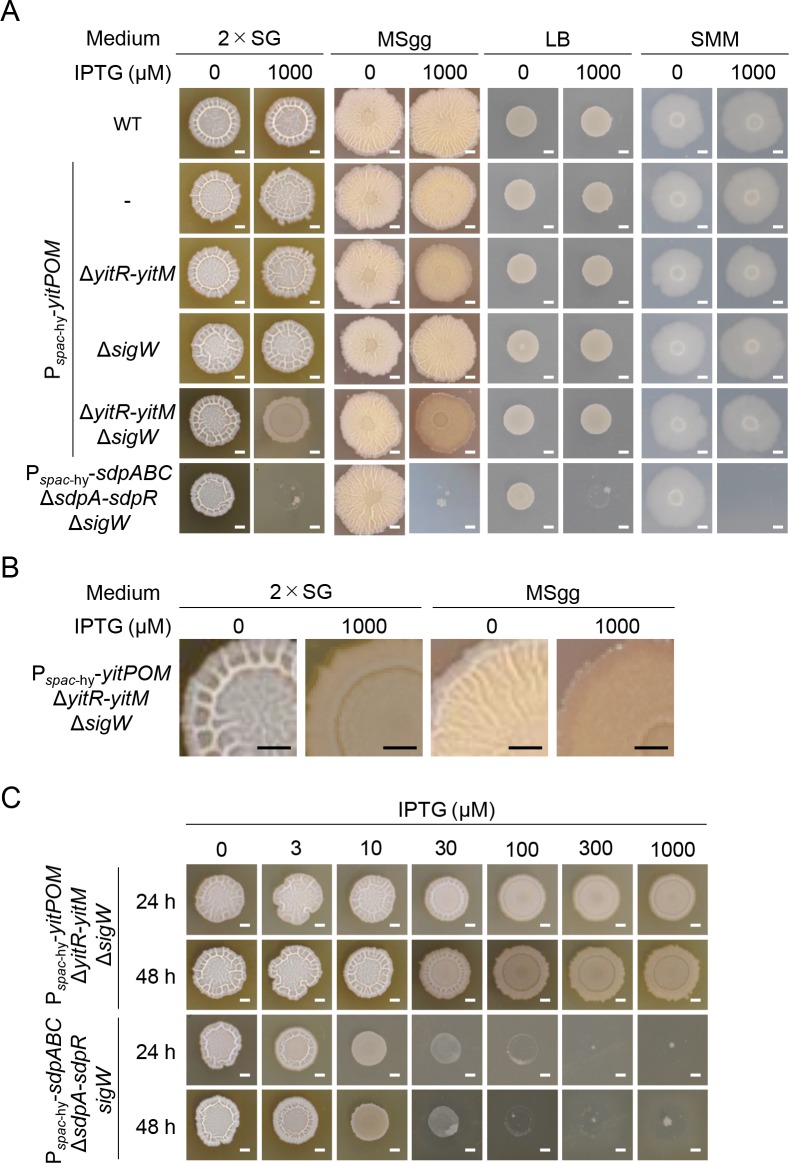
We hypothesized that if the YIT toxin has an effect on biofilm formation, we expected to detect this effect on biofilm supporting media but not on the media that do not support biofilm formation. First, we examined whether yitPOM overexpression from the spac-hy promoter affects colony biofilm formation on biofilm-supporting media. We used two biofilm-supporting media, the rich complex medium 2×SG and the synthetic medium MSgg [24]. On 2×SG solid medium, the wild-type strain formed whitish wrinkled colonies (Fig 6A). Induction of yitPOM did not affect the colony morphologies of the wild-type, ΔyitR-yitM mutant, or ΔsigW mutant strains; these Pspac-hy-yitPOM strains formed similar whitish wrinkled colonies in the presence or absence of IPTG. By contrast, yitPOM induction altered the colony morphology of the ΔyitR-yitM ΔsigW mutant; the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant formed brown flat colonies in the presence of IPTG (Fig 6A). Magnified images showed that the whitish wrinkled layers (biofilms) were completely absent on the surfaces of the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant colonies in the presence of IPTG (Fig 6B). Similar results were obtained on MSgg medium. The wild-type strain formed light brown wrinkled colonies on MSgg (Fig 6A). Induction of yitPOM altered the colony morphology of the ΔyitR-yitM ΔsigW mutant. The Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant formed brown flat colonies in the presence of IPTG (Fig 6A), and these colonies completely lacked the light brown wrinkled layers (biofilms) on their surfaces (Fig 6B). Unlike on rich 2×SG medium, induction of yitPOM also altered the colony morphology of the ΔyitR-yitM mutant when it was grown on MSgg (Fig 6A). In the presence of IPTG, the Pspac-hy-yitPOM ΔyitR-yitM mutant formed colonies covered with attenuated wrinkles at 96 h post-inoculation; however, these wrinkles faded over time (S1 Fig). This phenotype suggests that YitQ may play a major role in resistance to the YIT toxin under low nutrient conditions, such as B. subtilis natural habitats, soils. We next examined the colony morphologies on the complex medium LB and on the synthetic medium Spizizen minimal medium (SMM) [56]. B. subtilis forms flat colonies rather than biofilms on these media. On these media, yitPOM induction had little or no effect on colony morphology, even in the ΔyitR-yitM ΔsigW mutant (Fig 6A). We compared the effect of yitPOM overexpression on colony morphology with that of sdpABC overexpression. For this, sdpABC was expressed under the control of the same promoter (spac-hy) in the ΔsdpA-sdpR ΔsigW mutant. The Pspac-hy-sdpABC ΔsdpA-sdpR ΔsigW mutant formed normal colonies in all four media in the absence of IPTG, but it did not form colonies in the presence of IPTG (Fig 6A). Thus, the SDP toxin inhibited overall cell growth independently of the medium conditions.
Fig 6. Expression of yitPOM inhibits biofilm formation.
(A) Pspac-hy-yitPOM strains with the indicated mutations were grown at 30°C for 48 h on biofilm formation media (2×SG and MSgg) and non-biofilm formation media (LB and SMM) with or without 1000 μM IPTG. Colonies of the wild-type and Pspac-hy-sdpABC ΔsdpA-sdpR ΔsigW strains are also shown as references. (B) Magnified images of the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant colonies shown in panel A. (C) Comparison of the effects of yitPOM and sdpABC overexpression on colony morphology. The Pspac-hy-yitPOM ΔyitR-yitM ΔsigW and Pspac-hy-sdpABC ΔsdpA-sdpR ΔsigW mutant strains were grown at 30°C for 48 h on 2×SG with various IPTG concentrations. Colony morphology analysis was done at least three time, and the typical examples were shown in the figure. Scale bar, 2 mm.
We investigated the relationship between the expression levels of yitPOM and sdpABC and colony morphology. For this purpose, Pspac-hy-yitPOM ΔyitR-yitM ΔsigW and Pspac-hy-sdpABC ΔsdpA-sdpR ΔsigW mutants were grown on 2×SG medium supplemented with various IPTG concentrations (0 to 1000 μM) (Fig 6C). The effect of yitPOM expression on colony morphology appeared when the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant was grown in the presence of 30 μM or higher IPTG concentrations. At 30 μM IPTG, attenuated wrinkles appeared on the colonies at 24 h post-inoculation; however, these wrinkles failed to grow further. At 100 μM IPTG, the colonies completely lacked the whitish wrinkled layers on their surface and became flat. Higher IPTG concentrations did not further alter the colony morphology. Despite having an obvious effect on colony morphology, yitPOM induction did not affect colony size. By contrast, when grown on 2×SG medium supplemented with various IPTG concentrations, the Pspac-hy-sdpABC ΔsdpA-sdpR ΔsigW mutant formed small colonies in the presence of 10 or 30 μM IPTG but did not form colonies at 100 μM or higher IPTG concentrations (Fig 6C). Thus, sdpABC expression exerted a stronger effect on colony formation as its expression levels increased. These results demonstrate that the YIT and SDP toxins have different effects on colony growth and that the YIT toxin specifically inhibits biofilm formation in the absence of its resistance genes.
We considered how the YIT toxin inhibits biofilm formation. As described above, yitPOM induction caused mild cell lysis only in ΔyitR-yitM ΔsigW mutant cells grown in 2×SG medium with shaking. Induction of yitPOM in the ΔyitR-yitM ΔsigW mutant did not cause cell lysis in cultures grown in LB medium with shaking (S2 Fig). Thus, yitPOM induction caused cell lysis and inhibition of biofilm formation only in the ΔyitR-yitM ΔsigW mutant cells grown on biofilm formation media, indicating that these two phenotypes represent different aspects of one phenomenon. Moreover, induction of yitPOM led to formation of halos in the spot-on-lawn assays. We propose that the YIT toxin likely inhibits biofilm formation by killing biofilm-forming cells rather than by preventing expression of biofilm formation genes.
NprB allows the YIT toxin to attack cells within biofilms
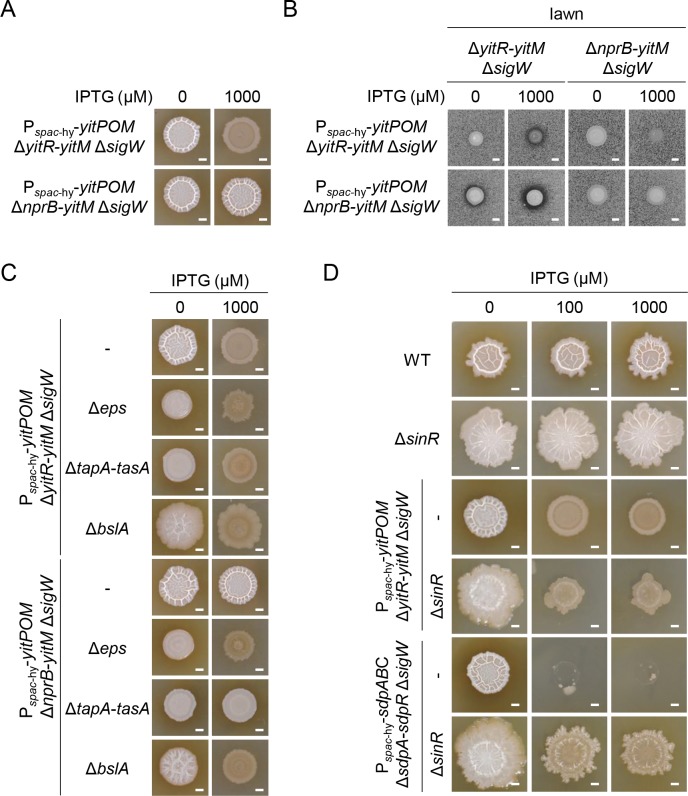
Induction of yitPOM exerted its effects only in cells grown on biofilm formation media. However, the spac-hy promoter is active in rich and poor media, including LB and SMM, as observed for the Pspac-hy-sdpABC ΔsdpA-sdpR ΔsigW mutant (Fig 6A). These observations suggest the involvement of other factor(s) in the functions of the YIT toxin. A comparison of the genetic organization of the 3610 and BEST195 strains revealed that, in addition to yizB and yitQ, nprB appears to be inserted into the 3610 genome along with yitPOM (Fig 2C). Like yitPOM, nprB, which encodes an extracellular neutral protease, was transcribed in a DegU-dependent manner (Fig 4B), and its expression was induced in biofilms (Fig 5A and 5B). To determine whether nprB is involved in the YIT toxin function, we introduced a deletion of the nprB-yitM region (Fig 2C) into the Pspac-hy-yitPOM ΔsigW mutant and examined the colony morphology of the resulting strain. Unlike in the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant, yitPOM induction did not inhibit biofilm formation in the Pspac-hy-yitPOM ΔnprB-yitM ΔsigW mutant. This strain formed whitish wrinkled colonies like those of the wild-type strain in the presence or absence of IPTG (Fig 7A). Because nprB deletion was the only genetic difference between Pspac-hy-yitPOM ΔyitR-yitM ΔsigW and Pspac-hy-yitPOM ΔnprB-yitM ΔsigW mutants (Fig 2C), this result suggests that the NprB protease is required for the production or function of the YIT toxin.
Fig 7. The NprB protease is required for the YIT toxin to inhibit biofilm formation.
(A) The ΔnprB mutation prevents the YIT toxin from inhibiting biofilm formation. Pspac-hy-yitPOM ΔyitR-yitM ΔsigW and Pspac-hy-yitPOM ΔnprB-yitM ΔsigW cells were grown on 2×SG at 30°C for 48 h with or without 1000 μM IPTG. (B) Production of the YIT toxin. ΔyitR-yitM ΔsigW and ΔnprB-yitM ΔsigW cells were added to 2×SG 1.2% agar with or without 1000 μM IPTG, and the mixtures were poured into plates. Pspac-hy-yitPOM ΔyitR-yitM ΔsigW and Pspac-hy-yitPOM ΔnprB-yitM ΔsigW cells were spotted on these lawn plates. The plates were then incubated at 37°C for 24 h. (C) The Δeps and ΔbslA mutations bypass the requirement for NprB in the ability of the YIT toxin to inhibit biofilm formation. (D) Overproduction of biofilm matrix polymers interfered with the action of the SDP toxin but not with that of the YIT toxin. Scale bar, 2 mm.
To distinguish these possibilities, we examined the production of the YIT toxin in these mutants via spot-on-lawn assays. The Pspac-hy-yitPOM ΔyitR-yitM ΔsigW and Pspac-hy-yitPOM ΔnprB-yitM ΔsigW mutants were spotted on the lawn of the ΔyitR-yitM ΔsigW mutant. Although both mutants formed halos around their colonies in the presence of IPTG, the Pspac-hy-yitPOM ΔnprB-yitM ΔsigW mutant formed clearer halos than did the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant (Fig 7B). The Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant formed smaller colonies on the lawn than did the Pspac-hy-yitPOM ΔnprB-yitM ΔsigW mutant, likely due to loss of biofilm formation. Therefore, we compared the YIT toxin production between the Pspac-hy-yitPOM ΔyitR-yitM and Pspac-hy-yitPOM ΔnprB-yitM mutants. Although these mutants formed similar colonies on the lawn of the ΔyitR-yitM ΔsigW mutant, the Pspac-hy-yitPOM ΔnprB-yitM mutant formed clearer halos than did the Pspac-hy-yitPOM ΔyitR-yitM mutant (S3 Fig). Thus, the ΔnprB mutation increased YIT toxin production or activity. These results suggest that NprB is not required for YIT toxin production. Given that NprB is an extracellular neutral protease, these results suggest that the YIT toxin is probably a substrate for NprB.
We next examined the alternative possibility that NprB might be required for the function of the YIT toxin. To test this idea, we spotted the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW and Pspac-hy-yitPOM ΔnprB-yitM ΔsigW mutants on the lawn of the ΔnprB-yitM ΔsigW mutant. Both mutants failed to form clear halos around their colonies (Fig 7B), supporting this idea.
We explored why the ΔnprB mutation impaired the function of the YIT toxin. Cells in biofilms are covered with and protected by biofilm matrix polymers, a key reason why cells in biofilms exhibit increased antibiotic tolerance or resistance [5, 6]. We hypothesized that a similar mechanism might work against the YIT toxin and that the NprB protease might enable the YIT toxin molecules to pass through the layers of the biofilm matrix polymers to attack cells within the biofilms. If this were true, then disrupting the biofilm matrix would enable the YIT toxin to inhibit biofilm formation even in the ΔnprB mutant. The biofilm matrix of B. subtilis biofilms mainly consists of exopolysaccharides (synthesized by the products of the eps operon) and polymers of the TasA (produced by the tapA-tasA operon) and BslA proteins [24–29]. To test our hypothesis, we introduced Δeps, ΔtapA-tasA, and ΔbslA deletion mutations into the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW and Pspac-hy-yitPOM ΔnprB-yitM ΔsigW mutants and examined their colony morphologies. The Pspac-hy-yitPOM ΔyitR-yitM ΔsigW Δeps mutant formed whitish mucoid colonies in the absence of IPTG, while it formed flat brown colonies in the presence of IPTG (Fig 7C). The difference in colony morphology depending on the presence or absence of IPTG indicates that the induced YIT toxin can function in these colonies even though the Δeps mutation impaired biofilm formation and led to the formation of mucoid colonies. Unlike the Pspac-hy-yitPOM ΔnprB-yitM ΔsigW mutant, the Pspac-hy-yitPOM ΔnprB-yitM ΔsigW Δeps mutant also formed whitish mucoid colonies in the absence of IPTG and flat brown colonies in the presence of IPTG (Fig 7C). Thus, the ΔnprB mutation did not interfere with the function of the YIT toxin in the Δeps mutant. Similar results were obtained with the ΔbslA mutant. The Pspac-hy-yitPOM ΔyitR-yitM ΔsigW ΔbslA and Pspac-hy-yitPOM ΔnprB-yitM ΔsigW ΔbslA mutants formed whitish mucoid colonies in the absence of IPTG and flat brown colonies in the presence of IPTG (Fig 7C). These results demonstrate that the Δeps and ΔbslA mutations eliminate the requirement for NprB in the function of the YIT toxin. Thus, our idea that NprB enables the YIT toxin to pass through the layers of the biofilm matrix polymers to attack cells in the biofilm is very likely. On the other hand, the ΔtapA-tasA mutation did not restore the YIT toxin activity in the ΔnprB-yitM mutant. The Pspac-hy-yitPOM ΔnprB-yitM ΔsigW ΔtapA-tasA mutants formed whitish mucoid colonies in the presence or absence of IPTG (Fig 7C). These results indicate that exopolysacchrides, BslA polymers or molecules associated with these polymers probably trap the YIT toxin in ΔnprB mutant biofilms.
We further examined the effect of overexpression of biofilm matrix polymers on the YIT toxin activity. SinR is a major repressor of the biofilm matrix synthesis genes [32], and a ΔsinR mutant formed large swollen colonies due to overproduction of biofilm matrix polymers (Fig 7D). We introduced the ΔsinR mutation into the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant and examined the colony morphology of the resulting strain. In the absence of IPTG, the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW ΔsinR mutant formed large swollen colonies, like those of the ΔsinR mutant. Induction of yitPOM inhibited biofilm formation in this mutant. The Pspac-hy-yitPOM ΔyitR-yitM ΔsigW ΔsinR mutant formed flat brown colonies in the presence of 100 or 1000 μM IPTG (Fig 7D), as was also observed in the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant (Fig 6C). We also examined the effect of the ΔsinR mutation on the SDP toxin. The Pspac-hy-sdpABC ΔsdpA-R ΔsigW ΔsinR mutant formed large swollen colonies in the absence of IPTG. Induction of sdpABC did not inhibit colony formation and only partly suppressed the swollen colony phenotype even in the presence of 1000 μM IPTG (Fig 7D). These results indicate that overproduction of biofilm matrix polymers interferes with the activity of the SDP toxin but not with that of the YIT toxin. Based on these results, we conclude that the YIT toxin can function within mature biofilms with the assistance of the extracellular neutral protease NprB.
We examined the colony morphology of the ΔnprB mutant. The ΔnprB mutant formed wrinkled colonies on 2×SG medium similar to those of the wild-type strain (Fig 8A). We extracted the extracellular proteins and cell surface-associated proteins from these colony biofilms and analyzed them via SDS-PAGE. We detected little or no difference in the protein composition between the wild-type and ΔnprB mutant strains in the gels after Coomassie brilliant blue (CBB) staining (Fig 8B). These results indicate that, despite a clear effect on the function of the YIT toxin, the ΔnprB mutation does not significantly alter biofilm structure.
Fig 8. The ΔnprB mutation had no significant effect on biofilm formation.
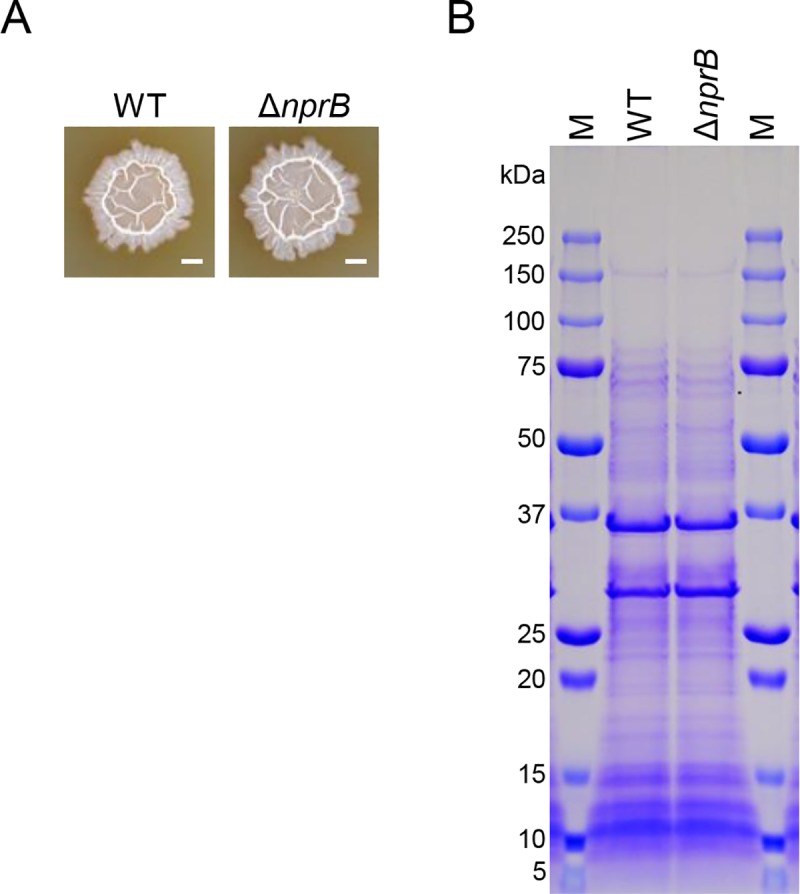
(A) Colony biofilms of the wild-type and ΔnprB mutant strains. These strains were grown at 30°C for 48 h on 2×SG. Scale bar, 2 mm. (B) The ΔnprB mutation had no significant effect on the composition of the extracellular proteins of colony biofilms. Colonies grown at 30°C for 48 h on 2×SG were suspended in SDS-PAGE sample buffer (62.5 mM Tris-HCl (pH 6.8), 1% SDS, 10% glycerol, 2.5% 2-mercaptoethanol, 2 mM PMSF, and 5 mM EDTA) and boiled for 2 min. After centrifugation, the supernatants were subjected to SDS-PAGE. Protein bands were visualized via Coomassie brilliant blue (CBB) staining. The size of protein molecular weight markers (lanes M) is indicated on the left side. The experiment was done twice, and the typical example was shown.
The YIT toxin is present in biofilms of the wild-type strain
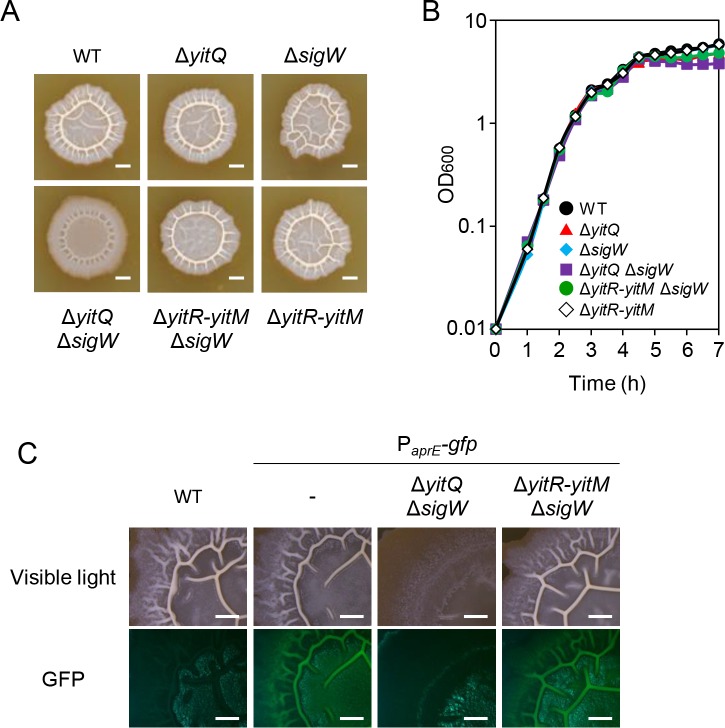
So far, we have reported the results of experiments designed to uncover the function of the YIT toxin via yitPOM expression from the strong spac-hy promoter. We asked whether yitPOM expression from its own promoter is sufficiently high to exhibit the phenotypes observed above. As described above, the action of the YIT toxin was antagonized by YitQ and unidentified σW-regulated gene product(s) in the wild-type strain. If the YIT toxin is present in biofilms of the wild-type strain, its effect should appear in yitQ and sigW mutants. Therefore, we examined the colony morphologies of mutants lacking yitQ and/or sigW on 2×SG medium (Fig 9A). While the ΔyitQ and ΔsigW single mutants formed whitish wrinkled colonies like those of the wild-type strain, the ΔyitQ ΔsigW double mutant formed colonies with attenuated wrinkles (biofilms). The ΔyitR-yitM ΔsigW mutant, which lacks both the toxin and anti-toxin genes, formed whitish wrinkled colonies like those of the wild-type strain. Thus, the phenotype of the ΔyitQ ΔsigW mutant was caused by the YIT toxin. However, the phenotype of the ΔyitQ ΔsigW mutant was slightly less noticeable than that of the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant in the presence of IPTG. Likewise, when grown in shaking culture, the ΔyitQ ΔsigW mutant did not display the culture lysis phenotype as observed for the Pspac-hy-yitPOM ΔyitR-yitM ΔsigW mutant; instead, the ΔyitQ ΔsigW mutant reached slightly lower OD600 in the stationary phase than that of the wild-type strain (Fig 9B).
Fig 9. The YIT toxin is expressed and functions in the wild-type strain.
(A) Colony morphologies of mutants lacking the resistance genes to the YIT toxin. The strains were grown at 30°C for 48 h on 2×SG. Scale bar, 2 mm. (B) Growth profiles in 2×SG shaking cultures. (C) PaprE-gfp expression was reduced in the ΔyitQ ΔsigW mutant. Scale bar, 1 mm.
As mentioned above, the PaprE-gfp reporter is induced in biofilms by DegS-DegU. The wild-type strain and the ΔyitR-yitM ΔsigW mutant with the PaprE-gfp reporter displayed bright GFP signal at wrinkles on colonies. However, the ΔyitQ ΔsigW mutant with the PaprE-gfp reporter displayed no detectable GFP signal on its colonies with attenuated wrinkles (Fig 9C). Thus, the ΔyitQ ΔsigW mutant showed decreased biofilm formation and decreased aprE expression. These results indicate that the YIT toxin reduces the number of biofilm-forming cells that express DegS-DegU-regulated genes, including yitPOM, in the ΔyitQ ΔsigW mutant. In other words, the action of the YIT toxin reduces the number of cells producing the YIT toxin in the ΔyitQ ΔsigW mutant. This effect can explain why the phenotype of the ΔyitQ ΔsigW mutant was slightly less obvious. These results demonstrate that the YIT toxin is present within biofilms of the wild-type strain.
The YIT toxin can mediate intercellular competition within biofilms
We hypothesized that the YIT toxin might mediate intercellular competition within biofilms. To test this hypothesis, we designed the following experiment. Dilutions of cultures of the wild-type and ΔyitR-yitM ΔsigW mutant strains were mixed, and the mixtures were spotted on 2×SG solid medium. The inoculated cells grew and formed biofilms in which the wild-type cells were expected to produce the YIT toxin. The YIT toxin then exerted its effect in biofilms with the assistance of NprB, which expressed in wild-type and ΔyitR-yitM ΔsigW mutant biofilms. If the YIT toxin suppressed the growth of ΔyitR-yitM ΔsigW mutant cells within biofilms, the ratio of these strains within the biofilms would change from the initial ratio. To estimate the ratio of two strains within biofilms, the PaprE-gfp reporter was introduced into one strain to detect its cells within biofilms.
First, wild-type cells carrying the PaprE-gfp reporter (the PaprE-gfp strain) were mixed with wild-type cells at various ratios from 10:0 to 0:10, and the mixtures were spotted on 2×SG solid medium. After 2 days of incubation, we observed the colonies with a fluorescence stereomicroscope. A bright GFP fluorescent signal was detected on colonies grown from the 10:0 mixture, and the GFP signal decreased as the proportion of the PaprE-gfp strain decreased (Fig 10A). When PaprE-gfp-expressing cells were mixed with ΔyitR-yitM ΔsigW mutant cells and grown on 2×SG medium, the GFP signal on the colonies also decreased as the proportion of the PaprE-gfp strain decreased; however, its decrease was moderate compared with that in the former experiment. For example, at a ratio of 3:7, obvious GFP signal was observed on colonies grown from the mixture of the aprE-gfp and ΔyitR-yitM ΔsigW mutant strains but not on colonies grown from the mixture of the aprE-gfp and wild-type strains. We also introduced the PaprE-gfp reporter into the ΔyitR-yitM ΔsigW mutant. When ΔyitR-yitM ΔsigW PaprE-gfp mutant cells were mixed with wild-type cells, no GFP signal was observed, even on the colonies grown from the 9:1 mixture (Fig 10A). By contrast, when ΔyitR-yitM ΔsigW PaprE-gfp mutant cells were mixed with ΔyitR-yitM ΔsigW mutant cells, the GFP signal on the colonies decreased as the proportion of the ΔyitR-yitM ΔsigW PaprE-gfp mutant cells decreased, as was also the case with the mixture of the aprE-gfp and wild-type strains. Thus, the YIT toxin is likely to mediate intercellular competition within biofilms.
Fig 10. The YIT toxin mediates competition within biofilms.
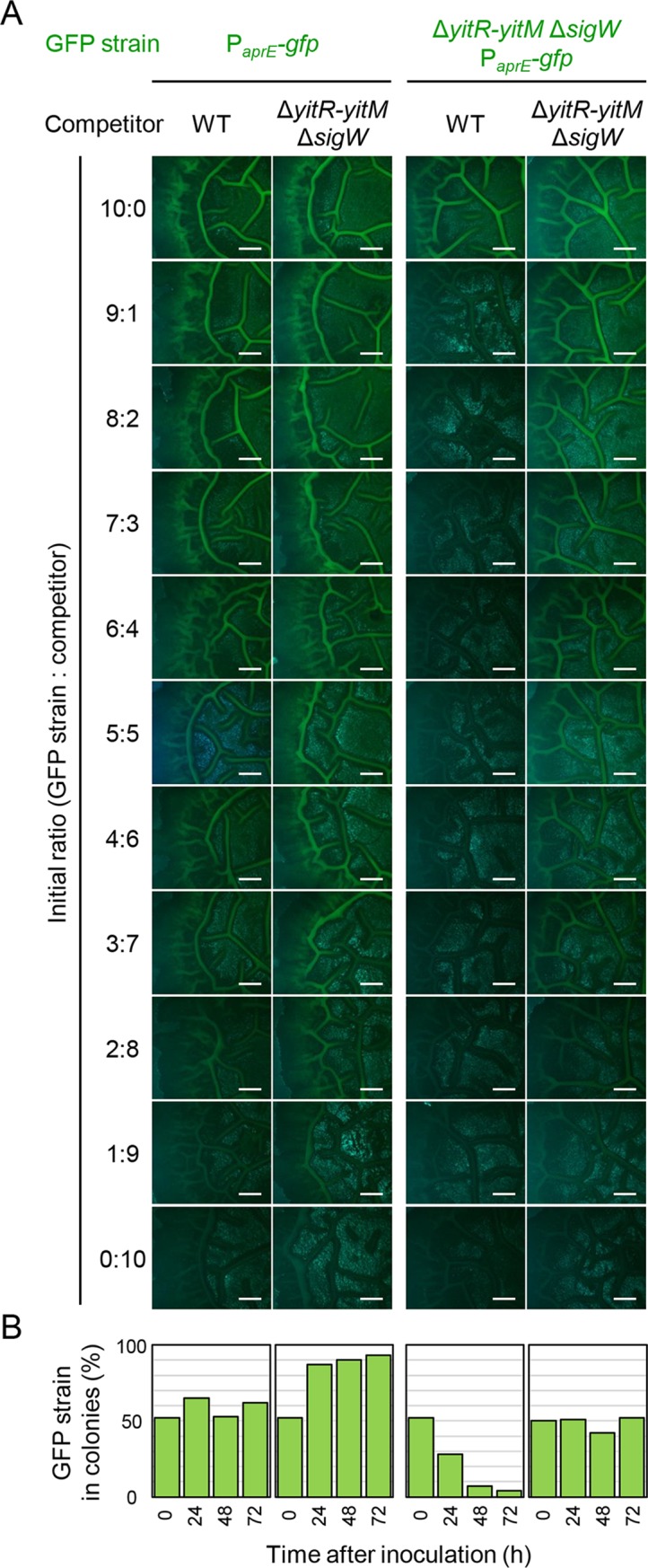
(A) PaprE-gfp expression in mixed colonies. Dilutions of cultures of the indicated strains were mixed at various ratios (10:0 to 0:10) and spotted on 2×SG. PaprE-gfp expression in the resultant colonies was analyzed 48 h after inoculation. The fluorescent images of colonies are shown. Scale bar, 1 mm. (B) The population ratios of PaprE-gfp cells in mixed colonies. Dilutions of cultures of the indicated strains on the top of the panel (A) were mixed at the ratio of 1:1, and spotted on 2×SG. The population ratios (%) of PaprE-gfp cells (Cmr) in the resultant colonies were analyzed over time by determining the numbers of Cmr CFUs and total CFUs in colonies. The values are the average of 4 independent colony measurements.
To confirm these results, we analyzed the population of cells carrying the aprE-gfp reporter in those colonies over time by determining colony forming units. In this experiment, aprE-gfp reporter strains were mixed with competitors at the ratio of 1:1. In the colonies of aprE-gfp and wild-type cells, the population ratio of the aprE-gfp cells did not change drastically from the initial ratio by 72 h after inoculation (Fig 10B). By contrast, in the colonies of PaprE-gfp and ΔyitR-yitM ΔsigW cells, a drastic increase in the population ratio of the aprE-gfp cells was observed 24 h after inoculation, and its ratio increased to 93% 72 h after inoculation. In colonies of PaprE-gfp ΔyitR-yitM ΔsigW and wild-type cells, the population ratio of the PaprE-gfp ΔyitR-yitM ΔsigW cells decreased to 4% by 72 h after inoculation. However, such a decrease was not observed in colonies of PaprE-gfp ΔyitR-yitM ΔsigW and ΔyitR-yitM ΔsigW cells. These results demonstrate that the YIT toxin can mediate intercellular competition within biofilms. In conclusion, we propose that the YIT toxin functions within B. subtilis biofilms without being obstructed by the biofilm matrix polymers with the assistance of NprB, thus protecting the biofilms from YIT toxin-sensitive unfavorable competitors.
The variety of SDP toxin homologs
Genome comparison revealed that sdpABC homologs are widely conserved among B. subtilis strains (S1 Table). sdpABC homologs, including sdpABC itself and yitPOM, can be classified into five groups based on their genome positions and SdpC homolog sequences (S1 Table, S4 and S5 Figs). Many B. subtilis strains have one or two sdpABC homologs, and yitPOM appears to be more widely conserved among B. subtilis strains than the other sdpABC homologs (S1 Table). Homologous genes encoding membrane proteins are found downstream of sdpABC homologs 2 and 4 (S4 Fig). The sdpIR homolog are found upstream of the sdpABC homolog 3. Some strains possess only sdpRI homologs in the sdpABC homolog 3 locus. These observations suggest functional similarities between the sdpABC homolog 2 and the sdpABC homolog 4 and between sdpABC and the sdpABC homolog 3. However, each group of SdpC homologs has a unique C-terminal hydrophobic domain, the sequence of which differs from those of the others (S5 Fig); therefore, each group of SdpC homolog-derived toxins may have different sequences. If the differences in these sequences impart their functional differences, as observed for SdpC and YitM, then SdpC homolog-derived toxins are likely to play more diverse roles.
Discussion
Biofilms often contain a high-density bacterial community consisting of a mixture of various species, and under these conditions, the bacteria compete with their own siblings and other species for limited space and nutrients. Since cells in biofilms exhibit increased antibiotic tolerance or resistance, the functions of antibiotics in the competition between biofilm bacteria remain unclear. In this study, we demonstrated that B. subtilis produces a biofilm-associated toxin, and that this toxin can attack toxin-sensitive cells within the biofilm by passing through the protective barriers of the biofilm with assistance from an endogenously produced extracellular protease.
The yitPOM operon is a paralog of the sdpABC operon; however, these operons are under different control and play distinct roles. Transcription of sdpABC is induced by low levels of Spo0A-P [44]. Given that low levels of Spo0A-P also induce the expression of biofilm matrix synthesis genes including the eps operon [44, 45], it is likely that the SDP toxin is induced during the early phase of biofilm formation. By contrast, yitPOM is induced by DegS-DegU in biofilms. Induction of these operons had different effects on B. subtilis cells lacking genes whose products confer resistance to the SDP and YIT toxins, i.e., sdpABC induction prevented colony formation independently of the medium conditions, whereas yitPOM specifically inhibited biofilm formation. Like SdpC, YitM contains a C-terminal hydrophobic domain that seems to be processed to produce the YIT toxin. The hydrophobic nature of the YIT toxin probably enables the YIT toxin to penetrate bacterial membrane, as observed for the SDP toxin. However, the sequence of the hydrophobic domain of YitM is different from that of SdpC. We assume that this difference contributes to the differences in the roles of these toxins. The genomes of many B. subtilis strains have both the sdpABC and yitPOM operons (S1 Table), suggesting that having both sdpABC and yitPOM may provide survival advantages in the environment.
Overproduction of biofilm matrix polymers interfered with the function of the SDP toxin but not with that of the YIT toxin, suggesting that the YIT toxin has a mechanism to pass through the layers of the biofilm matrix polymers, and we showed that this mechanism involves the extracellular neutral protease NprB. The YIT toxin could not inhibit biofilm formation in the absence of NprB even though the ΔnprB mutation increased the production of the YIT toxin. The requirement of NprB was eliminated by Δeps and ΔbslA mutations, either of which impairs biofilm matrix formation and, thus, biofilm formation [24, 28]. Cells in biofilms are encased in the biofilm matrix, which functions as a physical and chemical barrier against antibiotics by limiting their penetration [11–16]. Our results suggest that similar mechanisms might contribute to resistance to the YIT toxin in B. subtilis biofilms and that the extracellular protease NprB is required for the YIT toxin to pass through these defense barriers. The ΔnprB mutation had no significant effect on the composition of the extracellular and cell surface-associated proteins of biofilms nor on biofilm formation. We speculate that NprB may degrades the exopolysaccharides- or BslA polymer-associated protein that interacts with the YIT toxin, or that NprB may digest the YIT toxin smaller. By either or both actions, NprB may enable the YIT toxin molecules to pass through the layers of the biofilm matrix polymers. Moreover, we showed that the ΔnprB mutation increased activity of the YIT toxin, suggesting the possibility that the YIT toxin is a substrate for NprB protease. We speculate that NprB may also play a role in controlling YIT toxin levels in biofilms to avoid self-intoxication.
If NprB mediates structural changes in biofilms, that can increase the risk that biofilms become susceptible to antibiotics produced by other bacteria. However, the capability of the YIT toxin to attack sensitive cells within biofilms must be important for maintaining biofilm communities. Biofilms often consist of multiple types and multiple species of bacterial cells, some of which exploit others as free-loaders or cheaters that do not produce biofilm matrix polymers and other public goods [57]. The production of biofilm matrix polymers is metabolically costly; however, biofilm matrix polymers are extracellular products that are accessible even to non-producing cheater cells from which they receive protection [57]. An increase in the number of cheater cells, therefore, can disturb the cooperative relationships within biofilm communities and lead to instability within biofilm communities. The production of antibiotics that can diffuse through the biofilm offers the great advantage of being able to eliminate cheater cells and other unfavorable competitors present in the biofilm. Further work is required to determine which types of B. subtilis cells or what bacterial species are susceptible to the YIT toxin.
Previous studies showed that positively charged antibiotics interact with negatively charged matrix components, such as extracellular DNA and exopolysaccharides, and impede their penetration into biofilms [12, 15]. The SDP toxin contains two positively charged amino acid residues, whereas the hydrophobic region of YitM contains four negatively charged amino acid residues but no positively charged amino acid residues. These observations suggest that both NprB function and the amino acid sequence of the YIT toxin may be important for the ability of the YIT toxin to pass through the layers of biofilm matrix polymers; however, we have not yet determined the relevant sequence of the YIT toxin.
Bacterial competition is mediated by multiple factors [58]. In addition to the YIT toxin, DegS-DegU directly or indirectly induces non-ribosomally synthesized peptide antibiotics, e.g., bacilysin, fengycin, iturin, difficidin, and bacillomycin, although the repertoire of antibiotic synthesis genes differs from strain to strain and no B. subtilis strain produces all of these [59, 60, 61, 62]. DegS-DegU also induces a wide array of extracellular degradative enzymes, including six extracellular proteases [47 and references therein]. Although these proteases have been thought to play roles in nutrient acquisition from the surrounding environment, our results suggest that these degradative enzymes may also play roles in competition within biofilms. Indeed, several proteases were previously shown to disrupt biofilms of heterologous bacteria by degrading critical protein components in biofilms [63, 64, 65, 66]. Simultaneously producing multiple antibiotics and degradative enzymes within biofilms affords B. subtilis the ability to attack competitor cells protected by their own biofilm matrixes and to exclude them from biofilm communities. We expect that antibiotics and degradative enzymes cooperate extensively in B. subtilis biofilms.
The YIT toxin/NprB system seems to have evolved to be specifically adapted to the B. subtilis biofilm environment. Some bacteria produce specific antibiotics in biofilms [17–21]. Among them, the Escherichia coli ROAR029 strain produces the bacteriocin colicin R in biofilms, and colicin R is more active against biofilms than against planktonic cultures, as is the YIT toxin [20]. Pseudomonas aeruginosa produces bacteriocins pyocins and can suppress the growth of pyocin-sensitive bacteria in biofilms [21, 22]. Based on our results and these previous observations, we propose that bacteria may have evolved specialized antibiotics that function in biofilms as biofilm-specific competition mechanisms. The properties of these antibiotics may differ from those of conventional antibiotics. Biofilm-associated antibiotics might serve as anti-biofilm agents, especially in combination with degradative enzymes.
Materials and methods
Bacterial strains and culture condition
B. subtilis strain NCIB3610 and its derivatives used in this study are listed in Table 1. Construction of the B. subtilis mutants is described in S1 File. Primers used for the strain construction are listed in S2 Table. B. subtilis strains were maintained in LB (LB Lennox; BD Difco, Franklin Lakes, NJ, USA). For colony morphology observation, B. subtilis strains were grown at 30°C on LB plates overnight. A small single colony was suspended in 100 μl of LB, and 2 μl of the suspension was spotted onto 2×SG [49], MSgg [24], LB, and SMM media [55]. The plates were incubated at 30°C. Colony morphology was observed after 48 h of incubation on 2×SG and LB, after 72 h on SMM, and after 144 h on MSgg. Colony morphology observation was carried out at least three times and typical examples were shown in figures.
Table 1. B. subtilis strains used in this study.
| Strain name | Genotypes | References or constructiona |
|---|---|---|
| NCIB3610 | prototroph | 24 |
| N1285 | amyE::Pspac-hy-yitPOM (erm) | This study |
| N1263 | ΔyitR-yitM::tet | This study |
| N1286 | amyE::Pspac-hy-yitPOM (erm) ΔyitR-yitM::tet | N1263 → N1285 |
| NTF88 | ΔsigW::cat | 67 |
| N1356 | amyE::Pspac-hy-yitPOM (erm) ΔsigW::cat | NTF88 → N1285 |
| N1357 | amyE::Pspac-hy-yitPOM (erm) ΔyitR-yitM::tet ΔsigW::cat | NTF88 → N1286 |
| N1333 | amyE::Pspac-hy-sdpABC (erm) | This study |
| N1458 | ΔsdpA-sdpR::spc | This study |
| N1335 | amyE::Pspac-hy-sdpABC (erm) ΔsdpA-sdpR::spc | N1458 → N1333 |
| N1337 | amyE::Pspac-hy-sdpABC (erm) ΔsigW::cat | NTF88 → N1333 |
| N1340 | amyE::Pspac-hy-sdpABC (erm) ΔsdpA-sdpR::spc ΔsigW::cat | N1458 → N1335 |
| N741 | ΔsdpA-sdpR::spc ΔyitR-M::tet | N1458 → N1263 |
| N764 | ΔsdpA-sdpR::spc ΔyitR-M::tet ΔsigW::cat | NTF88 → N741 |
| N1498 | amyE::Pspac-hy-yitQ (erm) ΔsdpA-sdpR::spc ΔyitR-M::tet ΔsigW::cat | amyE::Pspac-hy-yitQ (erm) → N764 |
| N1497 | amyE::Pspac-hy-sdpI (erm) ΔsdpA-sdpR::spc ΔyitR-M::tet ΔsigW::cat | amyE::Pspac-hy-sdpI (erm) → N764 |
| N942 | amyE::Pspac-hy-yitPOM (erm) ΔsdpA-sdpR::spc ΔyitR-yitM::tet | N1285 → N741 |
| N776 | amyE::Pspac-hy-sdpABC (erm) ΔsdpA-sdpR::spc ΔyitR-yitM::tet | N1333 → N741 |
| NTF28 | ΔdegU::cat | 62 |
| N1443 | pHYG2 (promoter-less gfp, tet) | pHYG2 → NCIB3610 |
| N1444 | pHYG2-yitP (PyitP-gfp, tet) | pHYG2-yitP → NCIB3610 |
| N1446 | pHYG2-nprB (PnprB-gfp, tet) | pHYG2-nprB → NCIB3610 |
| N345 | ΔdegU::kan | This study |
| N1515 | pHYG2-yitP (PyitP-gfp, tet) ΔdegU::kan | N345 → N1444 |
| N1516 | pHYG2-nprB (PnprB-gfp, tet) ΔdegU::kan | N345 → N1446 |
| N1382 | amyE::PaprE-gfp (cat) | W740 (amyE::PaprE-gfp (cat)) [68] → NCIB3610 |
| N1268 | ΔsigW::neo | This study |
| N355 | ΔepsA-O::spc | This study |
| N1230 | amyE::Pspac-hy-yitPOM (erm) ΔyitR-yitM::tet ΔsigW::neo | N1268 → N1286 |
| N1253 | amyE::Pspac-hy-yitPOM (erm) ΔyitR-yitM::tet ΔsigW::neo ΔepsA-O::spc | N355 → N1230 |
| N1500 | amyE::Pspac-hy-yitPOM (erm) ΔyitR-yitM::tet ΔsigW::neo ΔtapA-tasA::cat | N11 (ΔtapA-tasA::cat) [62] → N1230 |
| N1255 | amyE::Pspac-hy-yitPOM (erm) ΔyitR-yitM::tet ΔbslA::spc | N254 (ΔbslA::spc) [28] → N1230 |
| N924 | ΔnprB-yitM::tet | This study |
| N1290 | amyE::Pspac-hy-yitPOM (erm) ΔnprB-yitM::tet | N924 → N1285 |
| N1238 | amyE::Pspac-hy-yitPOM (erm) ΔnprB-yitM::tet ΔsigW::neo | N1268 →N1290 |
| N1293 | amyE::Pspac-hy-yitPOM (erm) ΔnprB-yitM::tet ΔsigW::neo ΔepsA-O::spc | N355 →N1238 |
| N1503 | amyE::Pspac-hy-yitPOM (erm) ΔnprB-yitM::tet ΔsigW::neo ΔtapA-tasA::cat | N11 (ΔtapA-tasA::cat) [62] →N1238 |
| N1294 | amyE::Pspac-hy-yitPOM (erm) ΔnprB-yitM::tet ΔsigW::neo ΔbslA::spc | N254 (ΔbslA::spc) [28] →N1238 |
| N1358 | amyE::Pspac-hy-yitPOM (erm) ΔyitR-yitM::tet ΔsigW::neo ΔsinR::cat | WTF92 (ΔsinR::cat) [68] → N1238 |
| N999 | amyE::Pspac-hy-sdpABC (erm) ΔsdpA-sdpR::spc ΔsigW::neo | N1268 →N1335 |
| N1334 | amyE::Pspac-hy-sdpABC (erm) ΔsdpA-sdpR::spc ΔsigW::neo ΔsinR::cat | WTF92 (ΔsinR::cat) [68] → N999 |
| N1264 | nprB::cat | W115 (nprB::cat) [47] → NCIB3610 |
| N1287 | ΔyitQ::cat | This study |
| N1288 | ΔyitQ::cat ΔsigW::neo | N1268 → N1287 |
| N1234 | ΔyitR-yitM::tet ΔsigW::neo | N1268 → N1263 |
| N1413 | ΔyitQ::spc ΔsigW::neo amyE::PaprE-gfp (cat) | N1382 → N1288 |
| N1388 | ΔyitR-yitM::tet ΔsigW::neo amyE::PaprE-gfp (cat) | N1382 → N1234 |
aArrows indicate B. subtilis transformation: donor strain name → recipient strain name.
Comparison of genetic organization and protein sequence analysis
A comparison of the genetic organization in different B. subtilis strains was carried out using the MBGD website (http://mbgd.genome.ad.jp/) [67]. Protein alignments were constructed using the Protein BLAST program on the NCBI website (https://www.nlm.nih.gov/) and GENETYX ver.14 (GENETYX, Tokyo, Japan). Kyte &Doolittle hydropathy plots were constructed using the ExPASy website (https://web.expasy.org/protscale/) with a window size of 19 and default settings (hydropathy scale values of amino acids; A,1.8; R, -4.5; N, -3.5; D,-3.5; C 2.5; Q, -3.5; G, -0.4; H, -3.2; I, 4.5; L, 3.8; K, -3.9; M, 1.9; F, 2.8; P, -1.6;S, -0.8; T, -0.7; W, -1.3; V, 4.2).
Comparison of growth profiles
B. subtilis strains grown at 30°C on LB plates overnight were inoculated into 5 ml of 2×SG and were grown at 37°C to the mid-exponential phase with vigorously shaking. These cultures were then added to 50 ml of warm 2×SG in a 500 ml baffled flask to give an OD600 of 0.01. These cultures were shaking at 37°C, and OD600 of these cultures was measured over time. The experiments were performed at least three times, and the typical results were shown in figures.
Spot-on-lawn assay
Indicator strains (lawn strains) were grown at 28°C overnight in LB with vigorous shaking. Culture (1 μl) was mixed with 12 ml of 50°C 2×SG 1.2% agar with brief vortexing, and the mixture was immediately poured into a ϕ9 cm plate. The lawn plates were dried for 20 min in a laminar flow cabinet. The strains tested for antibiotic production were grown at 28°C overnight in LB with vigorous shaking. These cultures were diluted 10 times with LB, and 2 μl of the dilutions were spotted onto the dried lawn plates. The plates were incubated at 37°C for 24 to 30 h until halos appeared around the colonies. The experiments were performed at least three times, and the typical example was shown in figures.
Northern blot analysis
Wild-type and ΔdegU mutant cells were grown at 37°C in 2×SG with vigorous shaking, and samples were taken from the cultures at various time points for RNA isolation. Total RNA was prepared as previously described [47]. The Northern blot analysis was carried out as previously described [47]. Primers used for RNA probe synthesis are shown in S2 Table.
Microscopic observation
The strains carrying gfp reporters were grown on 2×SG or LB solid medium. The expression of the GFP reporters on the colonies was analyzed with a SZX7 stereomicroscope (Olympus, Tokyo, Japan) equipped with an AdvanCam-E3Rs digital color camera (Advan Vision, Tokyo, Japan). For colony observation, the plates were tilted slightly using a small piece of cardboard (1.5 mm thickness) under the microscope to avoid detecting excitation light reflections on the surfaces of colonies and solid media. However, we could not remove reflected light completely, and some reflected light was observed as blue color signals on GFP images. Images were obtained and processed with AdvanView (Advan Vision) and Photoshop Elements (Adobe, San Jose, CA, USA). The experiments were done at least three times, and the typical examples were shown in figures.
Flow cytometry analysis
Strains harboring promoter-gfp fusions were grown on 2×SG or LB solid medium. A whole single colony was scraped from the surface of the solid medium and suspended in 1 ml of PBS buffer. Cells in the biofilms were then dispersed by repetitive pipetting and were fixed in 4% paraformaldehyde for 7 min [69]. Prior to flow cytometry analysis, the cells were subjected to mild sonication [69]. Single-cell fluorescence was measured on an Accuri C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). The number of recorded events was 50,000. The experiments were done twice, and the typical examples were shown in figures.
Competition assay
Strains were grown at 28°C overnight in LB with vigorous shaking. Cultures of the strains used for the assay contained 3.0 × 108 cells/ml on average. The cultures were diluted 100-fold in LB, and two dilutions were mixed at the indicated ratios. Aliquots (2 μl each) of the mixtures were spotted onto 2×SG solid medium, and the plates were incubated at 30°C for 48 h. GFP fluorescence on the colonies was analyzed as described above. The experiments were done three times, and the typical examples were shown in figures. The population ratios of aprE-gfp cells in these colonies were also analyzed by determining the number of cells per colonies. A whole single colony was scraped with an inoculation loop and was dissolved in 1 ml of LB in a test tube. After 10 times pipetting, the cell suspension was left for a while until the unsolved cell aggregates went down to the bottom of the tube. The dissolved cells were then serially diluted with LB, and these dilutions were plated on LB or LB plus chloramphenicol (Cm). Since aprE-gfp cells exhibited Cmr, the population ratio of aprE-gfp cells was calculated as Cmr CFUs/ total CFUs. Each population ratio was the average of 4 independent colony measurements.
Supporting information
The strains were grown at 30°C on MSgg. Scale bar, 2 mm.
(TIF)
(TIF)
ΔyitR-yitM ΔsigW and ΔnprB-yitM ΔsigW cells were added to 1.2% 2×SG agar with or without 1000 μM IPTG, and the mixtures were poured into plates. Pspac-hy-yitPOM ΔyitR-yitM and Pspac-hy-yitPOM ΔnprB-yitM cells were spotted on the lawn plates. The plates were incubated at 37°C for 24 h.
(TIF)
The genetic organization of sdpABC and sdpABC homologs in the indicated B. subtilis strains was compared with that of the corresponding locus in strains that do not have sdpABC or sdpABC homologs. Homologous genes are shown by patterned boxes of the same color. Strain names are shown to the right of the gene maps.
(TIF)
(A) The alignment of SdpC homologs. The sequences of SdpC homolog 1 (YitM), homolog 2, homolog 3, and homolog 4 are derived from B. subtilis strains NCIB3610, ATCC13952, BEST195, and OH131.1, respectively. The signal sequences and the SDP toxin sequence are shown in blue and red, respectively. Hydrophobic amino acid residues in the C-terminal regions of SdpC homologs are shown in green. Identical and similar amino acids among all of the homologs are indicated by asterisks and dots, respectively. (B) Hydropathy plots of SdpC homologs. The plots were constructed using the ExPASy website (https://web.expasy.org/protscale/) with a window size of 19.
(TIF)
(DOCX)
(DOCX)
(PDF)
Acknowledgments
We would like to thank Prof. Hisaji Maki and Associate Prof. Masahiro Akiyama for their helpful advice and support.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
KK was supported by JSPS KAKENHI [Grant Number JP17K07721] (https://www.jsps.go.jp/j-grantsinaid/index.html). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010. January;8(1):15–25. 10.1038/nrmicro2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raaijmakers JM, Mazzola M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu Rev Phytopathol. 2012;50:403–24. 10.1146/annurev-phyto-081211-172908 [DOI] [PubMed] [Google Scholar]
- 3.Liu G, Chater KF, Chandra G, Niu G, Tan H. Molecular regulation of antibiotic biosynthesis in streptomyces. Microbiol Mol Biol Rev. 2013. March;77(1):112–43. 10.1128/MMBR.00054-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein T. Bacillus subtilis antibiotics: structures, syntheses and specific functions. Mol Microbiol. 2005. May;56(4):845–57. 10.1111/j.1365-2958.2005.04587.x [DOI] [PubMed] [Google Scholar]
- 5.Davies D. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov. 2003. February;2(2):114–22. 10.1038/nrd1008 [DOI] [PubMed] [Google Scholar]
- 6.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010. April;35(4):322–32. 10.1016/j.ijantimicag.2009.12.011 [DOI] [PubMed] [Google Scholar]
- 7.Ratcliff WC, Denison RF. Alternative actions for antibiotics. Science. 2011. April 29;332(6029):547–8. 10.1126/science.1205970 [DOI] [PubMed] [Google Scholar]
- 8.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005. January;13(1):20–6. [DOI] [PubMed] [Google Scholar]
- 9.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010. September;8(9):623–33. 10.1038/nrmicro2415 [DOI] [PubMed] [Google Scholar]
- 10.Rendueles O, Ghigo JM. Multi-species biofilms: how to avoid unfriendly neighbors. FEMS Microbiol Rev. 2012. September;36(5):972–89. 10.1111/j.1574-6976.2012.00328.x [DOI] [PubMed] [Google Scholar]
- 11.Mulcahy H, Charron-Mazenod L, Lewenza S. Extracellular DNA chelates cations and induces antibiotic resistance in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2008. November;4(11):e1000213 10.1371/journal.ppat.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Billings N, Millan M, Caldara M, Rusconi R, Tarasova Y, Stocker R, Ribbeck K. The extracellular matrix Component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2013;9(8):e1003526 10.1371/journal.ppat.1003526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng BS, Zhang W, Harrison JJ, Quach TP, Song JL, Penterman J, Singh PK, Chopp DL, Packman AI, Parsek MR. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol. 2013. October;15(10):2865–78. 10.1111/1462-2920.12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toska J, Ho BT, Mekalanos JJ. Exopolysaccharide protects Vibrio cholerae from exogenous attacks by the type 6 secretion system. Proc Natl Acad Sci U S A. 2018. July 31;115(31):7997–8002. 10.1073/pnas.1808469115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doroshenko N, Tseng BS, Howlin RP, Deacon J, Wharton JA, Thurner PJ, Gilmore BF, Parsek MR, Stoodley P. Extracellular DNA impedes the transport of vancomycin in Staphylococcus epidermidis biofilms preexposed to subinhibitory concentrations of vancomycin. Antimicrob Agents Chemother. 2014. December;58(12):7273–82. 10.1128/AAC.03132-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh R, Sahore S, Kaur P, Rani A, Ray P. Penetration barrier contributes to bacterial biofilm-associated resistance against only select antibiotics, and exhibits genus-, strain- and antibiotic-specific differences. Pathog Dis. 2016. August;74(6). pii: ftw056 10.1093/femspd/ftw056 [DOI] [PubMed] [Google Scholar]
- 17.Yan L, Boyd KG, Adams DR, Burgess JG. Biofilm-specific cross-species induction of antimicrobial compounds in bacilli. Appl Environ Microbiol. 2003. July;69(7):3719–27. 10.1128/AEM.69.7.3719-3727.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kreth J, Merritt J, Bordador C, Shi W, Qi F. Transcriptional analysis of mutacin I (mutA) gene expression in planktonic and biofilm cells of Streptococcus mutans using fluorescent protein and glucuronidase reporters. Oral Microbiol Immunol. 2004. August;19(4):252–6. [DOI] [PubMed] [Google Scholar]
- 19.Nandi M, Berry C, Brassinga AK, Belmonte MF, Fernando WG, Loewen PC, de Kievit TR. Pseudomonas brassicacearum strain DF41 kills Caenorhabditis elegans through biofilm-dependent and biofilm-independent mechanisms. Appl Environ Microbiol. 2016. December;82(23):6889–6898. 10.1128/AEM.02199-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rendueles O, Beloin C, Latour-Lambert P, Ghigo JM. A new biofilm-associated colicin with increased efficiency against biofilm bacteria. ISME J. 2014. June;8(6):1275–88. 10.1038/ismej.2013.238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waite RD, Curtis MA. Pseudomonas aeruginosa PAO1 pyocin production affects population dynamics within mixed-culture biofilms. J Bacteriol. 2009. February;191(4):1349–54. 10.1128/JB.01458-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oluyombo O, Penfold CN, Diggle SP. Competition in biofilms between cystic fibrosis isolates of Pseudomonas aeruginosa is shaped by R-pyocins. MBio. 2019. January 29;10(1). pii: e01828–18. 10.1128/mBio.01828-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson MS, Garcia EC, Cotter PA. Kind discrimination and competitive exclusion mediated by contact-dependent growth inhibition systems shape biofilm community structure. PLoS Pathog. 2014. April 17;10(4):e1004076 10.1371/journal.ppat.1004076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Branda SS, González-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A. 2001. September 25;98(20):11621–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Branda SS, González-Pastor JE, Dervyn E, Ehrlich SD, Losick R, Kolter R. 2004. Genes involved in formation of structured multicellular communities by Bacillus subtilis. J Bacteriol. 2004. June;186(12):3970–9. 10.1128/JB.186.12.3970-3979.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol. 2006. February;59(4):1229–38. 10.1111/j.1365-2958.2005.05020.x [DOI] [PubMed] [Google Scholar]
- 27.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010. February 2;107(5):2230–4 10.1073/pnas.0910560107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi K, Iwano M. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol. 2012. July;85(1):51–66 10.1111/j.1365-2958.2012.08094.x [DOI] [PubMed] [Google Scholar]
- 29.Hobley L, Ostrowski A, Rao FV, Bromley KM, Porter M, Prescott AR, MacPhee CE, van Aalten DM, Stanley-Wall NR. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci U S A. 2013. August 13;110(33):13600–5. 10.1073/pnas.1306390110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamon MA, Stanley NR, Britton RA, Grossman AD, Lazazzera BA. Identification of AbrB-regulated genes involved in biofilm formation by Bacillus subtilis. Mol Microbiol. 2004. May;52(3):847–60. 10.1111/j.1365-2958.2004.04023.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol Microbiol. 2005. February;55(3):739–49. 10.1111/j.1365-2958.2004.04440.x [DOI] [PubMed] [Google Scholar]
- 32.Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol Microbiol. 2006. February;59(4):1216–28. 10.1111/j.1365-2958.2005.05019.x [DOI] [PubMed] [Google Scholar]
- 33.Verhamme DT, Murray EJ, Stanley-Wall NR. DegU and Spo0A jointly control transcription of two loci required for complex colony development by Bacillus subtilis. J Bacteriol. 2009. January;191(1):100–8. 10.1128/JB.01236-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.López D, Fischbach MA, Chu F, Losick R, Kolter R. Structurally diverse natural products that cause potassium leakage trigger multicellularity in Bacillus subtilis. Proc Natl Acad Sci U S A. 2009. January 6;106(1):280–5. 10.1073/pnas.0810940106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mielich-Süss B, Lopez D. Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ Microbiol. 2015. March;17(3):555–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Straight PD, Fischbach MA, Walsh CT, Rudner DZ, Kolter R. A singular enzymatic megacomplex from Bacillus subtilis. Proc Natl Acad Sci U S A. 2007. January 2;104(1):305–10. 10.1073/pnas.0609073103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abriouel H, Franz CM, Ben Omar N, Gálvez A. Diversity and applications of Bacillus bacteriocins. FEMS Microbiol Rev. 2011. January;35(1):201–32. 10.1111/j.1574-6976.2010.00244.x [DOI] [PubMed] [Google Scholar]
- 38.Koskiniemi S, Lamoureux JG, Nikolakakis KC, t'Kint de Roodenbeke C, Kaplan MD, Low DA, Hayes CS. Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A. 2013. April 23;110(17):7032–7. 10.1073/pnas.1300627110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu WT, Yang YL, Xu Y, Lamsa A, Haste NM, Yang JY, Ng J, Gonzalez D, Ellermeier CD, Straight PD, Pevzner PA, Pogliano J, Nizet V, Pogliano K, Dorrestein PC. Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc Natl Acad Sci U S A. 2010. September 14;107(37):16286–90. 10.1073/pnas.1008368107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.González-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003. July 25;301(5632):510–3 10.1126/science.1086462 [DOI] [PubMed] [Google Scholar]
- 41.Pérez Morales TG, Ho TD, Liu WT, Dorrestein PC, Ellermeier CD. Production of the cannibalism toxin SDP is a multistep process that requires SdpA and SdpB. J Bacteriol. 2013. July;195(14):3244–51. 10.1128/JB.00407-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lamsa A, Liu WT, Dorrestein PC, Pogliano K. The Bacillus subtilis cannibalism toxin SDP collapses the proton motive force and induces autolysis. Mol Microbiol. 2012. May;84(3):486–500. 10.1111/j.1365-2958.2012.08038.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006. February 10;124(3):549–59. 10.1016/j.cell.2005.11.041 [DOI] [PubMed] [Google Scholar]
- 44.Fujita M, Losick R. Evidence that entry into sporulation in Bacillus subtilis is governed by a gradual increase in the level and activity of the master regulator Spo0A. Genes Dev. 2005. September 15;19(18):2236–44. 10.1101/gad.1335705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.López D, Vlamakis H, Losick R, Kolter R. Cannibalism enhances biofilm development in Bacillus subtilis. Mol Microbiol. 2009. November;74(3):609–18. 10.1111/j.1365-2958.2009.06882.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lyons NA, Kraigher B, Stefanic P, Mandic-Mulec I, Kolter R. A Combinatorial Kin Discrimination System in Bacillus subtilis. Curr Biol. 2016. March 21;26(6):733–42. 10.1016/j.cub.2016.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi K. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol Microbiol. 2007. October;66(2):395–409. 10.1111/j.1365-2958.2007.05923.x [DOI] [PubMed] [Google Scholar]
- 48.Verhamme DT, Kiley TB, Stanley-Wall NR. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol Microbiol. 2007. July;65(2):554–68. 10.1111/j.1365-2958.2007.05810.x [DOI] [PubMed] [Google Scholar]
- 49.Quisel JD, Burkholder WF, Grossman AD. In vivo effects of sporulation kinases on mutant Spo0A proteins in Bacillus subtilis. J Bacteriol. 2001. November;183(22):6573–8. 10.1128/JB.183.22.6573-6578.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leighton TJ, Doi RH. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem. 1971. May 25;246(10):3189–95. [PubMed] [Google Scholar]
- 51.Butcher BG, Helmann JD. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol Microbiol. 2006. May;60(3):765–82. 10.1111/j.1365-2958.2006.05131.x [DOI] [PubMed] [Google Scholar]
- 52.Cao M, Bernat BA, Wang Z, Armstrong RN, Helmann JD. FosB, a cysteine-dependent fosfomycin resistance protein under the control of sigma(W), an extracytoplasmic-function sigma factor in Bacillus subtilis. J Bacteriol. 2001. April;183(7):2380–3. 10.1128/JB.183.7.2380-2383.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamada Y, Tikhonova EB, Zgurskaya HI. YknWXYZ is an unusual four-component transporter with a role in protection against sporulation-delaying-protein-induced killing of Bacillus subtilis. J Bacteriol. 2012. August;194(16):4386–94. 10.1128/JB.00223-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang X, Gaballa A, Cao M, Helmann JD. Identification of target promoters for the Bacillus subtilis extracytoplasmic function sigma factor, sigma W. Mol Microbiol. 1999. January;31(1):361–71. 10.1046/j.1365-2958.1999.01180.x [DOI] [PubMed] [Google Scholar]
- 55.Marlow VL, Cianfanelli FR, Porter M, Cairns LS, Dale JK, Stanley-Wall NR. The prevalence and origin of exoprotease-producing cells in the Bacillus subtilis biofilm. Microbiology. 2014. January;160(Pt 1):56–66. 10.1099/mic.0.072389-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spizizen J. Transformation of Biochemically Deficient Strains of Bacillus subtilis by Deoxyribonucleate. Proc Natl Acad Sci U S A. 1958. October 15;44(10):1072–8. 10.1073/pnas.44.10.1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.West SA, Diggle SP, Buckling A, Gardner A, Griffin AS. The Social lives of microbes. Ann Rev Ecol Evol Syst. 2007;38:53–77. [Google Scholar]
- 58.Lyons NA, Kraigher B, Stefanic P, Mandic-Mulec I, Kolter R. A Combinatorial Kin Discrimination System in Bacillus subtilis. Curr Biol. 2016. March 21;26(6):733–42. 10.1016/j.cub.2016.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsuge K, Ano T, Hirai M, Nakamura Y, Shoda M. The genes degQ, pps, and lpa-8 (sfp) are responsible for conversion of Bacillus subtilis 168 to plipastatin production. Antimicrob Agents Chemother. 1999. September;43(9):2183–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koumoutsi A, Chen XH, Vater J, Borriss R. DegU and YczE positively regulate the synthesis of bacillomycin D by Bacillus amyloliquefaciens strain FZB42. Appl Environ Microbiol. 2007. November;73(21):6953–64. 10.1128/AEM.00565-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen XH, Scholz R, Borriss M, Junge H, Mögel G, Kunz S, Borriss R. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J Biotechnol. 2009. March 10;140(1–2):38–44. 10.1016/j.jbiotec.2008.10.015 [DOI] [PubMed] [Google Scholar]
- 62.Kobayashi K. Plant methyl salicylate induces defense responses in the rhizobacterium Bacillus subtilis. Environ Microbiol. 2015. April;17(4):1365–76. 10.1111/1462-2920.12613 [DOI] [PubMed] [Google Scholar]
- 63.Iwase T, Uehara Y, Shinji H, Tajima A, Seo H, Takada K, Agata T, Mizunoe Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010. May 20;465(7296):346–9. 10.1038/nature09074 [DOI] [PubMed] [Google Scholar]
- 64.Baidamshina DR, Trizna EY, Holyavka MG, Bogachev MI, Artyukhov VG, Akhatova FS, Rozhina EV, Fakhrullin RF, Kayumov AR. Targeting microbial biofilms using Ficin, a nonspecific plant protease. Sci Rep. 2017. April 7;7:46068 10.1038/srep46068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitrofanova O, Mardanova A, Evtugyn V, Bogomolnaya L, Sharipova M. Effects of Bacillus serine proteases on the bacterial biofilms. Biomed Res Int. 2017;2017:8525912 10.1155/2017/8525912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumar L, Cox CR, Sarkar SK. Matrix metalloprotease-1 inhibits and disrupts Enterococcus faecalis biofilms. PLoS One. 2019. January 11;14(1):e0210218 10.1371/journal.pone.0210218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kobayashi K. Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J Bacteriol. 2007. July;189(13):4920–31. 10.1128/JB.00157-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uchiyama I, Mihara M, Nishide H, Chiba H. MBGD update 2015: microbial genome database for flexible ortholog analysis utilizing a diverse set of genomic data. Nucleic Acids Res. 2015. January;43(Database issue):D270–6. 10.1093/nar/gku1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vlamakis H, Aguilar C, Losick R, Kolter R. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev. 2008. April 1;22(7):945–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The strains were grown at 30°C on MSgg. Scale bar, 2 mm.
(TIF)
(TIF)
ΔyitR-yitM ΔsigW and ΔnprB-yitM ΔsigW cells were added to 1.2% 2×SG agar with or without 1000 μM IPTG, and the mixtures were poured into plates. Pspac-hy-yitPOM ΔyitR-yitM and Pspac-hy-yitPOM ΔnprB-yitM cells were spotted on the lawn plates. The plates were incubated at 37°C for 24 h.
(TIF)
The genetic organization of sdpABC and sdpABC homologs in the indicated B. subtilis strains was compared with that of the corresponding locus in strains that do not have sdpABC or sdpABC homologs. Homologous genes are shown by patterned boxes of the same color. Strain names are shown to the right of the gene maps.
(TIF)
(A) The alignment of SdpC homologs. The sequences of SdpC homolog 1 (YitM), homolog 2, homolog 3, and homolog 4 are derived from B. subtilis strains NCIB3610, ATCC13952, BEST195, and OH131.1, respectively. The signal sequences and the SDP toxin sequence are shown in blue and red, respectively. Hydrophobic amino acid residues in the C-terminal regions of SdpC homologs are shown in green. Identical and similar amino acids among all of the homologs are indicated by asterisks and dots, respectively. (B) Hydropathy plots of SdpC homologs. The plots were constructed using the ExPASy website (https://web.expasy.org/protscale/) with a window size of 19.
(TIF)
(DOCX)
(DOCX)
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.