Abstract
Accurate and reliable quantification of brain metabolites measured in vivo using 1H magnetic resonance spectroscopy (MRS) is a topic of continued interest. Aside from differences in the basic approach to quantification, the quantification of metabolite data acquired at different sites and on different platforms poses an additional methodological challenge. In this study, spectrally edited γ-aminobutyric acid (GABA) MRS data were analyzed and GABA levels were quantified relative to an internal tissue water reference. Data from 284 volunteers scanned across 25 research sites were collected using GABA+ (GABA + co-edited macromolecules) and macromolecule-suppressed GABA editing. The unsuppressed water signal from the volume of interest was acquired for concentration referencing. Whole-brain T1-weighted structural images were acquired and segmented to determine gray matter, white matter and cerebrospinal fluid voxel tissue fractions. Water-referenced GABA measurements were fully corrected for tissue-dependent signal relaxation and water visibility effects. The cohort-wide coefficient of variation was 17% for the GABA+ data and 29% for the MM-suppressed GABA data. The mean within-site coefficient of variation was 10% for the GABA+ data and 19% for the MM-suppressed GABA data. Vendor differences contributed 53% to the total variance in the GABA+ data, while the remaining variance was attributed to site- (11%) and participant-level (36%) effects. For the MM-suppressed data, 54% of the variance was attributed to site differences, while the remaining 46% was attributed to participant differences. Results from an exploratory analysis suggested that the vendor differences were related to the unsuppressed water signal acquisition. Discounting the observed vendor-specific effects, water-referenced GABA measurements exhibit similar levels of variance to creatine-referenced GABA measurements. It is concluded that quantification using internal tissue water referencing is a viable and reliable method for the quantification of in vivo GABA levels.
Keywords: Editing, GABA, MEGA-PRESS, MRS, Quantification, Tissue correction
1. Introduction
In vivo 1H magnetic resonance spectroscopy (MRS) allows noninvasive measurement of brain metabolite concentrations, but it does so only in a relative manner. Measurements usually rely on an internal reference signal and assumptions about the concentration of the reference compound. Common reference signals include the CH3 singlets of the metabolites creatine (Cr) and N-acetylaspartate (NAA), or the unsuppressed brain tissue water signal from the same volume. Opinion in the field suggests that there is no reference signal that is optimal in all applications, and discussion is ongoing about the relative merits of each (Alger, 2010; Mullins et al., 2014).
The theory and empirical feasibility of the absolute quantification of metabolites as measured by MRS is well established (Barker et al., 1993; Christiansen et al., 1993; Danielsen and Henriksen, 1994; Ernst et al., 1993; Hennig et al., 1992; Kreis et al., 1993a; Thulborn and Ackerman, 1983). Later work has further refined these approaches, particularly with respect to using brain tissue water as an internal concentration reference (Gasparovic et al., 2018, 2006; Gussew et al., 2012; Knight-Scott et al., 2003). The typical procedure for using tissue water as an internal reference is to acquire an unsuppressed water signal using the same MRS acquisition protocol as used for the water-suppressed metabolite acquisition in a voxel co-localized to the volume of interest. With proper assumptions about certain properties of the metabolite and water signals in the various tissue compartments in the volume of interest, one may infer absolute metabolite concentrations from the acquired metabolite and reference signals. This is supported by the well-characterized properties of MR-visible water in the brain and its high concentration/large signal. On the other hand, using an endogenous metabolite signal, such as Cr, as a reference to derive metabolite ratios avoids the need for a separate water acquisition and may reduce error propagation that arises during more involved signal scaling procedures, but possibly at the expense of lower signal quality. Additionally, Cr is confined to brain tissue, whereas water is in brain and cerebrospinal fluid (CSF), making accurate corrections for CSF when using water as a concentration reference particularly important. At present, while strong opinions exist on the matter, both metabolite and water referencing have advantages and disadvantages (Jansen et al., 2006), and either approach is defensible. Indeed, the reliability of each approach has been shown to be similar (Bogner et al., 2010; Saleh et al., 2016), although in relatively small studies.
It is important to note that the concentration and relaxation properties of water, Cr and NAA can change in disease (Grasso et al., 2002; Huang et al., 2001; Kantarci et al., 2000; Laule et al., 2004; Rackayova et al., 2017), aging (Marjańska et al., 2017; Neeb et al., 2006; Reyngoudt et al., 2012) and brain development (Kreis et al., 1993b; Tkáč et al., 2003). Phantom replacement, scanning a phantom of a reference compound of known concentration for comparison to in vivo measurements (Buchli and Boesiger, 1993; Duc et al., 1998; Michaelis et al., 1993; Soher et al., 1996), can – with careful attention to differences in B0/B1 inhomogeneities, amplifier transmitter/receiver gains and RF coil loading factors – also be used to determine in vivo concentrations in absolute units. This method is technically challenging, involving additional experiments before or after the scan session, and is not commonly used given the difficulties of constructing a phantom with electric conductivity similar to human tissue and the extra time that is required for scanning the phantom (Jansen et al., 2006). An alternative approach is the ERETIC (electronic reference to access in vivo concentrations) method (Barantin et al., 1997; Zoelch et al., 2017), which relies on a synthetic RF reference signal. This approach is also challenging and requires specialized hardware. For all its limitations, internal concentration referencing remains the most practicable and widely used approach in in vivo 1H MRS.
In addition to the nuances of different quantification methodologies, systematic differences in acquisition implementation and system hardware will have an impact on quantitative outcomes. This makes comparing MRS measurements collected across different sites and on different platforms non-trivial. If multi-site and multi-platform MRS studies are to be maximally useful, particularly in the era of “big data” (Bearden and Thompson, 2017; Miller et al., 2016; Van Essen et al., 2013), then the systematic effects on measurement variance must be assessed, understood and accounted for. This would then be followed by strategies for standardizing data acquisition, data processing and metabolite quantification methods.
The authors recently acquired a large multi-vendor, multi-site dataset, the purpose of which was to study the sources of variance in γ-aminobutyric acid (GABA) measurements collected by edited MRS. In the first paper describing this dataset (Mikkelsen et al., 2017), quantification was performed relative to the total Cr signal in the edit-OFF spectrum. In the current paper, GABA was quantified relative to brain tissue water, which additionally required accounting for individual differences in voxel tissue composition. In particular, this investigation aimed to determine whether quantification relative to water increases or decreases total variance (compared to Cr referencing) and discuss the impact of site- and vendor-related differences in structural image segmentation.
2. Methods
A fuller description of the acquisition and data processing methodology can be found in the original publication (Mikkelsen et al., 2017). Relevant details for this study, especially regarding quantification and tissue segmentation, are reported below.
2.1. Data collection
Data were acquired at 25 independent research sites, with each site contributing 5–12 datasets collected from consenting adult volunteers (cohort total: 284). Participants at each site were 18–35 years old, ~50% female and had no known neurological or psychiatric illnesses. Site-by-site participant demographics are provided in Table 1 in Mikkelsen et al. (2017). Scanning was conducted in accordance with ethical standards set by the institutional review board (IRB) at each site, including the sharing of anonymized data. Anonymized data files were shared securely with and analyzed by the co-authors at the Johns Hopkins University School of Medicine with local IRB approval.
Table 1.
Hardware and acquisition parameters used to collect 3D T1-weighted structural images at each site.
| Site ID | Tx/Rx hardware | Voxel resolution (mm3) | TE/TI/TR (ms) | Scan time (m:ss) | Flip angle (deg) | Slices | FOV (mm2) | Matrix size | Acceleration (factor) |
|---|---|---|---|---|---|---|---|---|---|
| G1 | Body coil/32-ch head coil | 0.94 × 0.94 × 1 | 2.68/600/7.42 | 4:07 | 10 | 226 | 256 × 256 | 256 × 256 | ASSET (2) |
| G2 | Body coil/8-ch head coil | 0.9 × 0.9 × 1 | 2.73/650/6.24 | 2:54 | 8 | 180 | 256 × 256 | 256 × 256 | ARC (2) |
| G3 | Body coil/32-ch head coil | 1 × 1 × 1 | 2.6/500/6.4 | 4:37 | 11 | 180 | 256 × 256 | 256 × 256 | ASSET (2) |
| G4 | Body coil/8-ch head coil | 1 × 1 × 1 | 2.98/450/6.89 | 9:35 | 12 | 192 | 256 × 256 | 256 × 256 | None |
| G5 | Body coil/32-ch head coil | 0.5 × 0.5 × 0.8 | 2.1/450/7.09 | 5:39 | 12 | 232 | 256 × 256 | 512 × 512 | None |
| G6 | Body coil/8-ch head coil | 1 × 1 × 2 | 2.66/400/6.24 | 6:22 | 12 | 124 | 240 × 240 | 240 × 240 | None |
| G7 | Body coil/8-ch head coil | 1 × 1 × 1 | 3.2/450/8.2 | 4:30 | 12 | 176 | 256 × 256 | 256 × 256 | ARC (2) |
| G8 | Body coil/8-ch head coil | 1 × 1 × 1 | 4.17/450/10.19 | 5:27 | 12 | 180 | 256 × 256 | 256 × 256 | ARC (2) |
| P1 | Body coil/32-ch head coil | 1 × 1 × 1 | 3.1/865/6.9 | 7:10 | 8 | 204 | 256 × 256 | 256 × 256 | SENSE (2) |
| P2 | Body coil/32-ch head coil | 1 × 1 × 1 | 3.1/865/6.9 | 7:10 | 8 | 204 | 256 × 256 | 256 × 256 | SENSE (2) |
| P3 | Body coil/32-ch head coil | 1 × 1 × 1 | 3.1/865/6.9 | 7:10 | 8 | 204 | 256 × 256 | 256 × 256 | SENSE (2) |
| P4 | Body coil/32-ch head coil | 1 × 1 × 1 | 3.1/865/6.9 | 7:10 | 8 | 204 | 256 × 256 | 256 × 256 | SENSE (2) |
| P5 | Body coil/32-ch head coil | 1 × 1 × 1 | 3.1/865/6.9 | 7:10 | 8 | 170 | 256 × 256 | 256 × 256 | SENSE (2) |
| P6 | Body coil/8-ch head coil | 1 × 1 × 1 | 3.1/865/6.9 | 7:10 | 8 | 204 | 256 × 256 | 256 × 256 | SENSE (2) |
| P7 | Body coil/32-ch head coil | 1 × 1 × 1 | 3.1/865/6.9 | 7:10 | 8 | 204 | 256 × 256 | 256 × 256 | SENSE (2) |
| P8 | Body coil/32-ch head coil | 1 × 1 × 1 | 3.1/865/6.9 | 7:10 | 8 | 204 | 256 × 256 | 256 × 256 | SENSE (2) |
| P9 | Body coil/32-ch head coil | 1 × 1 × 1 | 3.1/865/6.9 | 7:10 | 8 | 176 | 256 × 256 | 256 × 256 | SENSE (2) |
| S1 | Body coil/32-ch head coil | 1 × 1 × 1 | 2.52/900/1900 | 4:18 | 9 | 176 | 250 × 250 | 256 × 256 | GRAPPA (2) |
| S2 | Body coil/32-ch head coil | 1 × 1 × 1 | 2.85/1050/1900 | 5:43 | 9 | 176 | 256 × 256 | 256 × 256 | GRAPPA (2) |
| S3 | Body coil/20-ch head/neck coil | 1 × 1 × 1 | 1.77/900/1900 | 4:05 | 8 | 160 | 256 × 256 | 128 × 256 | GRAPPA (2) |
| S4 | Body coil/64-ch head coil | 1 × 1 × 1 | 4.11/1000/2000 | 3:36 | 12 | 160 | 256 × 256 | 256 × 256 | GRAPPA (2) |
| S5 | Body coil/12-ch head coil | 1 × 1 × 1 | 4.6/900/1950 | 4:01 | 9 | 176 | 192 × 256 | 192 × 256 | GRAPPA (2) |
| S6 | Body coil/32-ch head coil | 1 × 1 × 1 | 2.26/900/1900 | 4:26 | 9 | 192 | 256 × 256 | 256 × 256 | GRAPPA (2) |
| S7 | Body coil/32-ch head coil | 1 × 1 × 1 | 3.03/900/2300 | 5:21 | 9 | 192 | 256 × 256 | 256 × 256 | GRAPPA (2) |
| S8 | Body coil/64-ch head coil | 1 × 1 × 1 | 3.02/900/1900 | 4:01 | 9 | 160 | 256 × 256 | 256 × 256 | GRAPPA (2) |
2.2. Data acquisition
GABA-edited MEGA-PRESS data (Mescher et al., 1998; Rothman et al., 1993) were collected at 3T at each site using a standard scan protocol. The MRI vendor breakdown was: eight GE; nine Philips; eight Siemens. Both GABA+-edited and macromolecule- (MM-) suppressed GABA-edited acquisitions were performed (Edden et al., 2012b; Henry et al., 2001). Complete details of the edited MRS acquisitions, including site-to-site idiosyncrasies, can be found in the earlier paper. Briefly, the GABA+ acquisition parameters were: TE = 68 ms; ON/OFF editing pulses = 1.9/7.46 ppm; editing pulse duration = 15 ms. The MM-suppressed GABA acquisition parameters were: TE = 68 ms (Siemens) or 80 ms (GE and Philips); ON/OFF editing pulses = 1.9/1.5 ppm; editing pulse duration = 20 ms. Common parameters were: TR = 2000 ms; 320 averages; 30 × 30 × 30 mm3 medial parietal lobe voxel (Fig. 1A). Six outer-volume saturation bands were applied in GE acquisitions (as is standard for GE PRESS-based sequences) but not in Philips or Siemens acquisitions (except for site S3, which used six saturation bands). Water suppression bandwidth scan parameters (not reported in Mikkelsen et al., 2017) were 140 Hz for Philips, 50 Hz for Siemens and 150 Hz for GE. Although some degree of control over the slice-selective gradient polarity (and the assignment of the three slice-selective pulses to the three physical dimensions of the voxel) is possible on all three vendors, no attempt was made to standardize these across vendors, nor were sites explicitly instructed to standardize gradient polarities (this was left at the discretion of scanner operators).
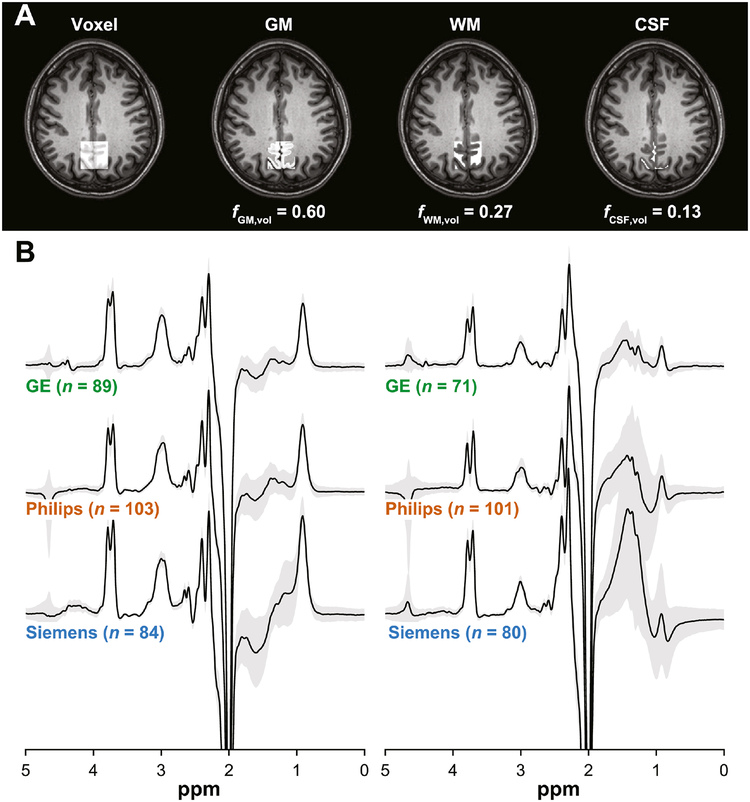
Fig. 1.
(A) Representative MRS voxel placement on a T1-weighted structural image and probabilistic partial volume voxel maps following tissue segmentation for one participant. Corresponding tissue fractions of gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF) are shown. (B) Vendor-mean GABA-edited difference spectra acquired by GABA+ and MM-suppressed GABA editing. The gray patches represent ±1 standard deviation. The associated sample sizes are shown in parentheses.
Unsuppressed water signal acquisitions were collected for internal tissue water referencing. For the GE and Philips MEGA-PRESS implementations, the water reference was automatically acquired as part of the water-suppressed metabolite scans. For GE, the reference was acquired at the end of the water-suppressed acquisitions; 16 water averages were acquired. For Philips, the reference was acquired in an interleaved manner as the water signal was also used for real-time center frequency correction (Edden et al., 2016); a single water average was acquired for every 40 water-suppressed acquisitions (8 averages in total). Acquiring a water reference on the Siemens platform requires running a separate scan with identical receiver and transmitter gains. For this, the Siemens MEGA-PRESS WIP was used, where the water suppression RF pulses were turned off but the water suppression gradients and editing pulses were left on (“Only RF off” option); 8 or 16 water averages (depending on acquisition parameters) were acquired. The TE/TR of these acquisitions were the same as the corresponding water-suppressed acquisitions. Water reference acquisitions were acquired with the transmitter frequency shifted to the water frequency (as opposed to 3.0 ppm for water-suppressed acquisitions) on Philips and Siemens. For GE, both reference and water-suppressed data were acquired with the transmitter at 2.68 ppm.
Whole-brain 3D T1-weighted structural images were acquired for accurate voxel placement and partial volume tissue correction. Sequences used were fast spoiled gradient-echo imaging (FSPGR; GE) (Low et al., 1993) and magnetization-prepared rapid gradient-echo imaging (MPRAGE; Philips/Siemens) (Mugler and Brookeman, 1990) (see Table 1 for acquisition parameters). Site-standard structural imaging protocols were used, with less effort to standardize acquisitions than the MRS protocols. Imaging data were saved in DICOM (GE and some Siemens sites) or NIfTI format (Philips and some Siemens sites). DICOM files were converted into NIfTI format for voxel segmentation and tissue segmentation purposes (see Section 2.4) using SPM12 (https://www.fil.ion.ucl.ac.uk/spm/software/spm12/).
2.3. Data processing
MRS data were processed in Gannet (Edden et al., 2014) using the pipeline described in the earlier report (Mikkelsen et al., 2017). Unsuppressed water acquisitions were processed in the same manner as the water-suppressed acquisitions and averaged. Briefly, processing steps included: frequency-and-phase correction by spectral registration (Near et al., 2015) (water-suppressed data only); 3-Hz exponential line broadening; zero-filling to yield a nominal spectral resolution of 0.061 Hz/point; and fast Fourier transformation into the frequency domain. Quality control and quality metrics were conducted and calculated as before. The linewidth of the water reference was measured as the full-width at half-maximum (FWHM) of the modeled water signal (see Section 2.5). As an independent measure of spectral linewidth, NAA FWHM linewidth was also measured from a Lorentzian-model fit of the NAA signal in the OFF spectrum.
2.4. Voxel co-registration and tissue segmentation
MRS voxels were co-registered to each volunteer’s structural image using the GannetCoRegister module in Gannet (Harris et al., 2015), which produces binary voxel masks in individual structural space. Structural images were segmented into gray matter (GM), white matter (WM) and CSF probabilistic partial volume maps using the unified tissue segmentation algorithm in SPM12 (Ashburner and Friston, 2005), executed through the GannetSegment module (Harris et al., 2015). GM, WM and CSF voxel volume fractions were calculated by multiplying the whole-brain partial volume maps for each tissue type by the corresponding binary voxel mask, summing over the partial volume estimates within each tissue-segmented voxel and then dividing these totals by the total over all three tissue-segmented voxels.
2.5. Quantification
The 3.0 ppm edited GABA signal was modeled as described previously (Mikkelsen et al., 2017). The water spectrum was modeled between 3.8 and 5.6 ppm with a Gaussian-Lorentzian function with phase and linear baseline parameters using nonlinear least-squares fitting. GABA measurements were quantified in pseudo-absolute molality units (approximating moles of GABA per kg of solute water) and corrected for partial volume effects (Gasparovic et al., 2006) based on the following equation:
| (1) |
where CG is the GABA concentration in institutional units (i.u.) and CW is the assumed molal concentration of MR-visible solute water (55.51 mol/kg); IG and IW are the GABA and water signal integrals, respectively; HW and HG are the number of 1H protons that give rise to the water and 3.0 ppm GABA signals (both 2), respectively; MM is a correction factor for the contribution of the co-edited macromolecule signal in the GABA+ signal, assumed to be 0.45 for GABA+ acquisitions and 1 for the MM-suppressed acquisitions; and κ is the editing efficiency, assumed to be 0.5. TEG, TEW, TRG and TRW are the echo and repetition times of the GABA-edited and water acquisitions, respectively. T1W,i is the longitudinal relaxation time of water in GM (assumed to be 1331 ms), WM (assumed to be 832 ms) (Wansapura et al., 1999) or CSF (assumed to be 3817 ms) (Lu et al., 2005); T2W,i is the transverse relaxation time of water in GM (assumed to be 110 ms), WM (assumed to be 79.2 ms) (Wansapura et al., 1999) or CSF (assumed to be 503 ms) (Piechnik et al., 2009). T1G and T2G are the longitudinal and transverse relaxation times of GABA, assumed to be 1310 and 88 ms, respectively (Edden et al., 2012a; Puts et al., 2013). The tissue-dependent water contents βi (as defined in Ernst et al., 1993) are assumed to be 0.78, 0.65 and 0.97 for grey matter, white matter and CSF, respectively. fi,vol is the volume fraction of GM, WM or CSF in the MRS voxel. Note that this equation equates to the molality equation in Gasparovic et al. (2006), with additional terms to account for MM fraction and editing efficiency that are particular to edited MRS of GABA.
Fit quality for the water peak model (the fit error) was assessed by normalizing the standard deviation (SD) of the model fit residuals to the amplitude of the modeled signal (Edden et al., 2014). This metric, the degree to which the measured signal cannot accurately be modeled as a Gaussian-Lorentzian, captures eddy current artifacts and some aspects of sub-optimal shimming.
To examine whether systematic effects on the variance of the GABA+ and MM-suppressed GABA data were attributed to the water acquisition, water-referenced Cr measurements were also quantified. The 3.0 ppm Cr signal in the OFF spectrum was modeled as described in the original publication. The longitudinal and transverse relaxation times of Cr were assumed to be 1350 and 154 ms, respectively (Mlynárik et al., 2001). MM and κ were not applied. Finally, the degree of association between participants’ water-referenced, tissue-corrected GABA values and their previously quantified GABA/Cr values as reported in Mikkelsen et al. (2017) was examined. It should be noted that these measurements are not independent (the GABA integral being a common factor) and, therefore, a strong correlation was expected.
2.6. Exploratory analysis
The results revealed systematically higher water-referenced GABA+ measurements from the Siemens sites as compared to the GE and Philips measurements (see Section 3). This level of variation was not apparent in the Cr-referenced GABA+ measurements that were previously reported (Mikkelsen et al., 2017). To reconcile this, an unplanned exploratory analysis was conducted in which the Siemens GABA measurements were referenced to a water signal acquired by a separate unsuppressed short-TE PRESS acquisition. These separate data were collected alongside the MEGA-PRESS data for the purpose of studying conventional MRS data acquired across the sites. The water reference data from this dataset enabled the unplanned exploratory analysis reported here. This acquisition was acquired at TE/TR = 35/2000 ms from a voxel in the same location as the MEGA-PRESS acquisition. Concentrations were quantified according to Eq. (1) without additional correction for any amplifier gain differences as it was assumed that the gain would not have changed between the PRESS and MEGA-PRESS acquisitions.
2.7. Statistical analysis
Linear mixed-effects models were fit to the water-referenced GABA data in R (version 3.5.2; R Core Team, 2018) using the lme4 package (Bates et al., 2015) and maximum likelihood for parameter estimation. An unconditional model (Eq. (1) in Mikkelsen et al., 2017) was fit to calculate variance partition coefficients (VPCs) to estimate the proportion of total variance attributed to vendor-, site- and participant-related effects. Secondary, conditional linear mixed-effects models (Eq. (5) in Mikkelsen et al., 2017) were also fit to the data to assess the impact of NAA linewidth, fGM,vol, age and sex, and to test the association with GABA/Cr measurements. Goodness-of-fit was calculated as a log-likelihood statistic. Significance testing was performed using chi-square likelihood ratio tests, which were bootstrapped 2,000 times using parametric bootstrapping (Halekoh and Højsgaard, 2014). Effects were tested in the following order: vendor and site; NAA linewidth and fGM,vol; age and sex. If an effect was significant, the relevant variable was retained in the next model; if not, it was removed. Unconditional linear mixed-effects models were also fit to the voxel tissue fractions to test for site and vendor effects. Post-hoc pairwise comparisons were corrected for multiple comparisons using the Holm-Bonferroni method (Holm, 1979). A p-value less than 0.05 was considered significant.
3. Results
GABA+ data from seven volunteers and MM-suppressed GABA data from 19 volunteers were removed from further analysis following quality control of the MRS data (largely due to excessive lipid contamination). All MM-suppressed GABA data from site G3 were excluded as consistent, excessive center frequency offsets (approximately –0.1 ppm on average) resulted in extremely small or absent GABA signals. One further dataset was removed because the unusually small water reference signal indicated an acquisition error. Vendor-mean GABA-edited difference spectra are shown in Fig. 1B.
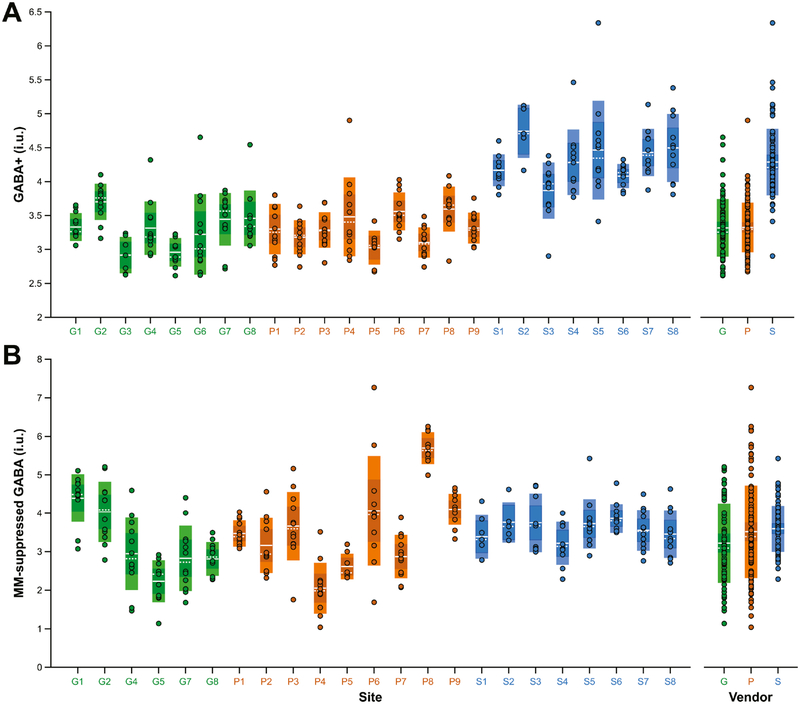
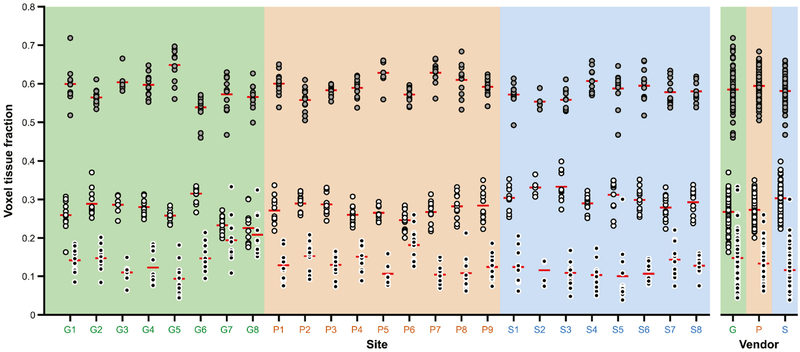
Fig. 2A shows the GABA+ values arranged by site and by vendor. Mean ± 1 SD (standard error of the mean, SEM) GABA+ measurements were 3.32 ± 0.42 (0.04) i.u. for GE, 3.32 ± 0.36 (0.04) i.u. for Philips and 4.29 ± 0.49 (0.05) i.u. for Siemens. Siemens values were on average 29% higher than the GE (pholm < 0.001) and Philips (pholm < 0.001) values. The cohort-wide average was 3.61 ± 0.61 (0.04) i.u. Coefficients of variation (CVs) were 12.7%, 10.8% and 11.3% for GE, Philips and Siemens, and 16.9% across all sites and vendors. The mean within-site CV was 9.5%. Fig. 2B shows the MM-suppressed GABA values arranged by site and by vendor. Mean ± 1 SD MM-suppressed GABA measurements were 3.22 ± 1.02 (0.12) i.u. for GE, 3.52 ± 1.20 (0.12) i.u. for Philips and 3.60 ± 0.59 (0.07) i.u. for Siemens. Siemens MM-suppressed values were on average 12% higher than the GE (pholm > 0.9) and only 2% higher than Philips (pholm > 0.9) values. The cohort-wide average was 3.46 ± 1.00 (0.06) i.u. CVs were 31.7%, 34.0% and 16.4% for GE, Philips and Siemens, and 28.8% across all sites and vendors. The mean within-site CV was 18.7%. GM, WM and CSF fractions are displayed in Fig. 3. Across the cohort, the average (and CV of) fGM,vol, fWM,vol and fCSF,vol was 0.59 ± 0.04 (6.9%), 0.28 ± 0.04 (14.0%) and 0.13 ± 0.05 (36.7%), respectively. Values of GABA+, fGM,vol, fWM,vol and fCSF,vol for each site are listed in Table 2.
Fig. 2.
(A) Water-referenced GABA+ and (B) MM-suppressed GABA measurements fully corrected for partial volume effects, displayed by site and by vendor. The boxes shaded with lighter colors represent ±1 standard deviation and the darker boxes represent the 95% confidence interval. The solid white lines denote the mean, while the dashed white lines denote the median. Sites are colored by vendor (GE sites in green, Philips sites in orange, Siemens sites in blue).
Fig. 3.
Gray matter (GM), white matter (WM) and cerebrospinal fluid (CSF) voxel tissue fractions, displayed by site and by vendor. GM = gray fill; WM = white fill; CSF = black fill. The red lines denote the mean. Sites are colored by vendor (GE sites with a green background, Philips sites with an orange background, Siemens sites with a blue background).
Table 2.
Quantification, tissue fractions and (TE = 68 ms) water data quality metrics, displayed by site and by vendor (shown as mean ± 1 standard deviation (CV)).
| Site ID | GABA+ (i.u.) | MM-s GABA (i.u.) | Cr (i.u.) | fGM,vol | fWM,vol | fCSF,vol | Water fit error (%) | Water linewidth (Hz) |
|---|---|---|---|---|---|---|---|---|
| G1 | 3.33 ± 0.20 (6.1%) | 4.40 ± 0.61 (13.9%) | 11.69 ± 1.00 | 0.60 ± 0.05 | 0.26 ± 0.04 | 0.14 ± 0.03 | 0.95 ± 0.09 | 9.43 ± 0.48 |
| G2 | 3.70 ± 0.26 (7.1%) | 4.04 ± 0.78 (19.3%) | 11.19 ± 0.61 | 0.56 ± 0.02 | 0.29 ± 0.03 | 0.15 ± 0.03 | 0.57 ± 0.11 | 9.98 ± 0.73 |
| G3 | 2.92 ± 0.26 (9.0%) | DE | 10.16 ± 0.69 | 0.60 ± 0.03 | 0.29 ± 0.02 | 0.11 ± 0.03 | 0.43 ± 0.09 | 9.46 ± 0.76 |
| G4 | 3.31 ± 0.39 (11.7%) | 2.95 ± 0.93 (31.6%) | 10.54 ± 0.52 | 0.60 ± 0.03 | 0.28 ± 0.02 | 0.12 ± 0.04 | 0.42 ± 0.06 | 9.34 ± 0.40 |
| G5 | 2.96 ± 0.20 (6.9%) | 2.23 ± 0.54 (24.1%) | 10.33 ± 0.41 | 0.65 ± 0.05 | 0.26 ± 0.01 | 0.09 ± 0.04 | 0.55 ± 0.11 | 9.79 ± 0.63 |
| G6 | 3.22 ± 0.59 (18.2%) | DNA | 9.65 ± 0.76 | 0.54 ± 0.04 | 0.32 ± 0.02 | 0.15 ± 0.04 | 0.65 ± 0.07 | 9.80 ± 0.92 |
| G7 | 3.44 ± 0.38 (11.1%) | 2.83 ± 0.84 (29.7%) | 11.28 ± 0.55 | 0.57 ± 0.05 | 0.23 ± 0.02 | 0.19 ± 0.06 | 0.47 ± 0.11 | 8.72 ± 0.88 |
| G8 | 3.46 ± 0.41 (11.7%) | 2.81 ± 0.43 (15.2%) | 11.10 ± 0.49 | 0.57 ± 0.03 | 0.23 ± 0.05 | 0.21 ± 0.05 | 0.41 ± 0.09 | 8.40 ± 0.42 |
| AUGE | 3.32 ± 0.42 (12.7%) | 3.22 ± 1.02 (31.7%) | 10.78 ± 0.90 | 0.58 ± 0.05 | 0.27 ± 0.04 | 0.15 ± 0.05 | 0.56 ± 0.19 | 9.37 ± 0.84 |
| P1 | 3.30 ± 0.37 (11.1%) | 3.47 ± 0.33 (9.6%) | 11.46 ± 0.78 | 0.60 ± 0.03 | 0.27 ± 0.04 | 0.13 ± 0.04 | 0.47 ± 0.07 | 8.78 ± 0.53 |
| P2 | 3.18 ± 0.25 (7.8%) | 3.16 ± 0.71 (22.4%) | 11.55 ± 0.71 | 0.56 ± 0.03 | 0.29 ± 0.02 | 0.15 ± 0.04 | 0.43 ± 0.07 | 8.74 ± 0.40 |
| P3 | 3.29 ± 0.26 (7.8%) | 3.66 ± 0.88 (24.1%) | 11.24 ± 0.94 | 0.58 ± 0.02 | 0.29 ± 0.03 | 0.13 ± 0.03 | 0.84 ± 0.11 | 9.02 ± 0.43 |
| P4 | 3.48 ± 0.58 (16.5%) | 2.05 ± 0.65 (31.7%) | 11.86 ± 0.56 | 0.59 ± 0.02 | 0.26 ± 0.02 | 0.15 ± 0.03 | 0.32 ± 0.06 | 8.78 ± 0.38 |
| P5 | 3.03 ± 0.25 (8.1%) | 2.61 ± 0.32 (12.3%) | 11.32 ± 0.75 | 0.63 ± 0.03 | 0.27 ± 0.02 | 0.11 ± 0.03 | 0.75 ± 0.08 | 9.06 ± 0.24 |
| P6 | 3.56 ± 0.27 (7.7%) | 4.07 ± 1.42 (34.9%) | 13.19 ± 1.19 | 0.57 ± 0.02 | 0.25 ± 0.03 | 0.18 ± 0.05 | 0.67 ± 0.17 | 8.71 ± 0.53 |
| P7 | 3.10 ± 0.22 (7.0%) | 2.87 ± 0.55 (19.3%) | 11.88 ± 0.47 | 0.63 ± 0.03 | 0.27 ± 0.03 | 0.10 ± 0.03 | 0.69 ± 0.11 | 10.21 ± 0.62 |
| P8 | 3.59 ± 0.33 (9.1%) | 5.70 ± 0.40 (7.1%) | 13.84 ± 0.54 | 0.61 ± 0.04 | 0.28 ± 0.03 | 0.11 ± 0.04 | 0.40 ± 0.04 | 9.03 ± 0.29 |
| P9 | 3.31 ± 0.22 (6.7%) | 4.10 ± 0.40 (9.6%) | 11.13 ± 0.93 | 0.59 ± 0.02 | 0.28 ± 0.04 | 0.12 ± 0.03 | 0.50 ± 0.06 | 8.77 ± 0.31 |
| All Philips | 3.32 ± 0.36 (10.8%) | 3.52 ± 1.20 (34.0%) | 11.95 ± 1.17 | 0.59 ± 0.04 | 0.27 ± 0.03 | 0.13 ± 0.04 | 0.56 ± 0.19 | 9.01 ± 0.62 |
| S1 | 4.17 ± 0.23 (5.5%) | 3.39 ± 0.56 (16.5%) | 14.76 ± 0.84 | 0.57 ± 0.03 | 0.30 ± 0.03 | 0.12 ± 0.05 | 0.39 ± 0.08 | 9.40 ± 0.72 |
| S2 | 4.74 ± 0.39 (8.1%) | 3.76 ± 0.52 (13.7%) | 17.24 ± 0.28 | 0.55 ± 0.02 | 0.33 ± 0.02 | 0.12 ± 0.03 | 0.38 ± 0.03 | 9.22 ± 0.36 |
| S3 | 3.87 ± 0.41 (10.6%) | 3.75 ± 0.76 (20.1%) | 14.86 ± 0.98 | 0.56 ± 0.03 | 0.33 ± 0.04 | 0.11 ± 0.04 | 0.30 ± 0.06 | 8.63 ± 0.39 |
| S4 | 4.28 ± 0.48 (11.1%) | 3.22 ± 0.55 (17.1%) | 14.05 ± 0.57 | 0.61 ± 0.03 | 0.29 ± 0.02 | 0.10 ± 0.04 | 0.33 ± 0.05 | 8.81± 0.31 |
| S5 | 4.46 ± 0.72 (16.2%) | 3.73 ± 0.63 (17.0%) | 14.45 ± 1.60 | 0.59 ± 0.05 | 0.31± 0.04 | 0.10 ± 0.07 | 0.42 ± 0.11 | 9.45 ± 0.92 |
| S6 | 4.08 ± 0.17 (4.2%) | 3.86 ± 0.37 (9.5%) | 14.57 ± 0.42 | 0.59 ± 0.04 | 0.30 ± 0.03 | 0.11± 0.03 | 0.44 ± 0.08 | 9.12 ± 0.28 |
| S7 | 4.43 ± 0.34 (7.8%) | 3.55 ± 0.52 (14.7%) | 13.88 ± 0.96 | 0.58 ± 0.03 | 0.28 ± 0.03 | 0.14 ± 0.04 | 0.46 ± 0.08 | 8.98 ± 0.39 |
| S8 | 4.50 ± 0.50 (11.0%) | 3.46 ± 0.61 (17.6%) | 15.52 ± 0.61 | 0.58 ± 0.03 | 0.29 ± 0.03 | 0.13 ± 0.02 | 0.33 ± 0.07 | 8.84 ± 0.36 |
| All Siemens | 4.29 ± 0.49 (11.3%) | 3.60 ± 0.59 (16.4%) | 14.73 ± 1.19 | 0.58 ± 0.04 | 0.30 ± 0.04 | 0.12 ± 0.04 | 0.38 ± 0.10 | 9.04 ± 0.57 |
| Overall | 3.61 ± 0.61 (16.9%) | 3.46 ± 1.00 (28.8%) | 12.42 ± 1.94 | 0.59 ± 0.04 | 0.28 ± 0.04 | 0.13 ± 0.05 | 0.51 ± 0.19 | 9.14 ± 0.70 |
CV, coefficient of variation; DE, data excluded; DNA, data not acquired; MM-s, MM-suppressed.
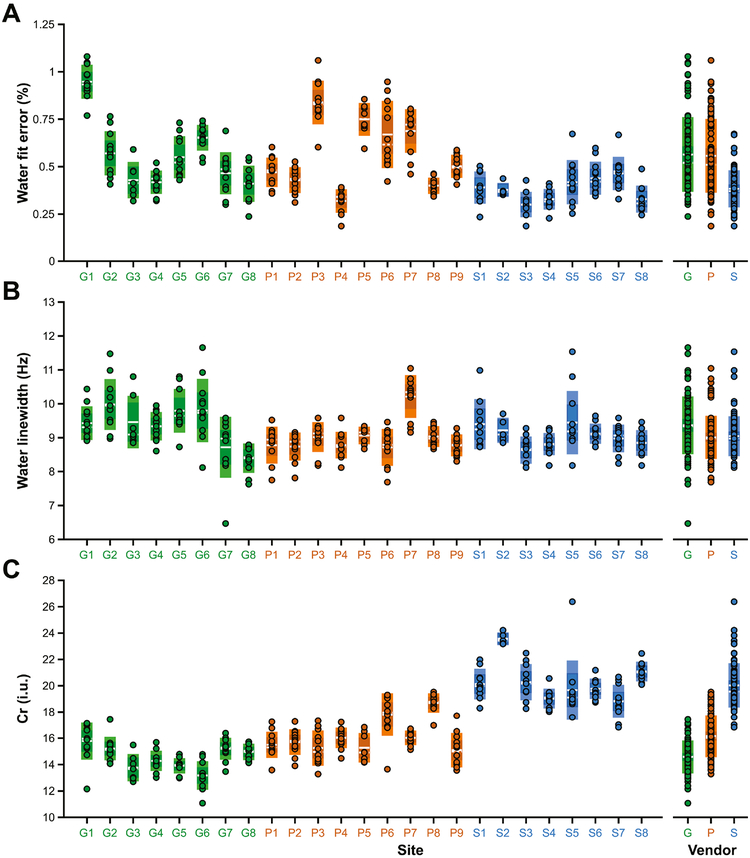
Water fit errors and linewidths and Cr measurements (for TE = 68 ms data) are displayed in Fig. 4, with site- and vendor-averaged values given in Table 2. On average, fit errors were small, and lower for Siemens (0.38%) compared to GE (0.56%) and Philips (0.56%). Water linewidths were similar across the vendors, with Philips (9.01 ± 0.62 Hz) and Siemens (9.04 ± 0.57 Hz) showing slightly smaller linewidths compared to GE (9.37 ± 0.84 Hz). Average water-referenced Cr measurements were 14.6 ± 1.2 i.u. for GE, 16.2 ± 1.6 i.u. for Philips and 20.0 ± 1.6 i.u. for Siemens, based on TE = 68 ms data. The Siemens MM-suppressed acquisition, also acquired at TE = 68 ms, gave an average Cr measurement of 21.7 ± 2.1 i.u. Cr measurements from MM-suppressed acquisitions acquired at TE = 80 ms were 16.9 ± 1.2 i.u. for GE and 18.1 ± 1.7 i.u. for Philips, 14% higher on average.
Fig. 4.
Quality metrics and water-referenced Cr measurements (from the TE = 68 ms data), displayed by site and by vendor. (A) water fit error; (B) water linewidth; (C) Cr measurements fully corrected for partial volume effects. The boxes shaded with lighter colors represent ±1 standard deviation and the darker boxes represent the 95% confidence interval. The solid white lines denote the mean, while the dashed white lines denote the median. Sites are colored by vendor (GE sites in green, Philips sites in orange, Siemens sites in blue).
3.1. Linear mixed-effects analyses
A summary of the linear mixed-effects analyses on the GABA+ measurements is given in Table S1. The unconditional linear mixed-effects model showed that vendor and site effects contributed significantly to the total amount of variance in the data: χ2(1) = 28.36, pboot = 0.001 and χ2(1) = 28.89, pboot < 0.001, respectively. Based on the calculated VPCs, 53.6% of the variance was accounted for by vendor-level differences, while 10.7% was accounted for by site-level differences. The remaining proportion of variance (35.7%) was attributed to individual differences in participants (see Table 3). The same model applied to MM-suppressed GABA showed that site effects contributed significantly to the total variance: χ2(1) = 131.53, pboot = 0.001, but vendor did not: χ2(1) < 0.1, pboot = 0.65 (see Table S2). The corresponding VPCs were 53.6% (site level) and 46.4% (participant level) (see Table 3).
Table 3.
Summary of variance partition analyses. Cr-referenced values are reproduced from Mikkelsen et al., 2017.
| GABA+ (i.u.) | MM-suppressed GABA (i.u.) | GABA+/Cr | MM-suppressed GABA/Cr | |
|---|---|---|---|---|
| Vendor | 53.6% | - | 8.2% | - |
| Site | 10.7% | 53.6% | 19.7% | 50.4% |
| Participant | 35.7% | 46.4% | 72.1% | 49.6% |
The voxel tissue fractions exhibited significant site-related effects for fGM,vol [χ2(1) = 56.33, pboot = 0.001], fWM,vol [χ2(1) = 46.77, pboot < 0.001] and fCSF,vol [χ (1) = 47.22, pboot < 0.001], but only fWM,vol showed an additional vendor-related effect [χ2(1) = 4.08, pboot = 0.01]. Corresponding VPCs for fGM,vol were: vendor < 0.1%; site = 31.4%; participant = 68.6%. For fWM,vol these were: vendor = 12.8%; site = 24.9%; participant = 62.3%. For fCSF,vol these were: vendor = 1.8%; site = 28.5%; participant = 69.7%. Pairwise comparisons showed that, at the vendor level, the Siemens fWM,vol values were significantly higher than the GE (pholm = 0.001) and Philips (pholm = 0.003) values.
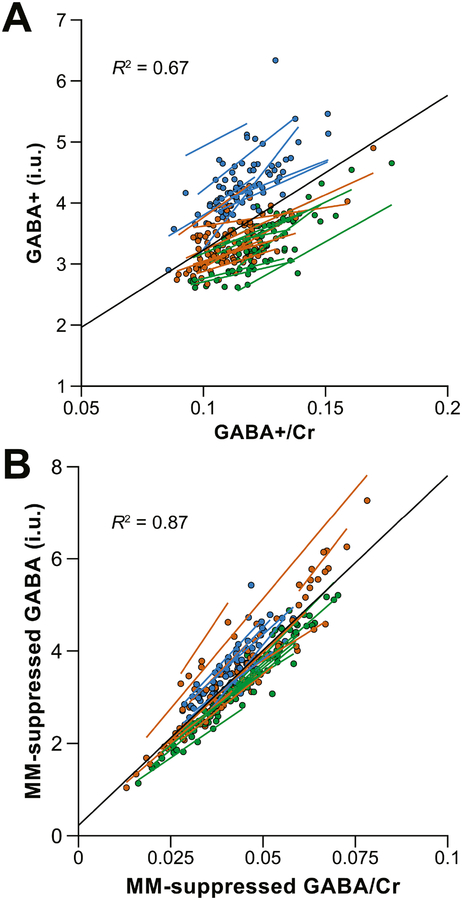
Based on the conditional linear mixed-effects analyses, neither GABA+ nor MM-suppressed GABA levels were significantly impacted by the effects of NAA linewidth [χ2(5) = 4.65, pboot = 0.21; χ2(3) = 1.19, pboot = 0.61]. GABA+ levels did, however, show a relationship with fGM,vol [χ2(5) = 10.28, pboot = 0.02], whereas MM-suppressed GABA levels did not [χ2(3) = 5.50, pboot = 0.08]. There were no significant effects of age [χ2(5) = 1.19, pboot = 0.75; χ2(3) = 0.85, pboot = 0.69] or sex [χ2(5) = 1.56, pboot = 0.67; χ2(3) = 2.34, pboot = 0.35] on the GABA+ or MM-suppressed data, respectively. As shown in Fig. 5, the water-referenced and Cr-referenced measurements were strongly related, for both GABA+ and MM-suppressed acquisitions [χ2(5) = 257.2, pboot < 0.001; χ2(3) = 495.7, pboot < 0.001] as expected.
Fig. 5.
Scatterplots illustrating the relationship between (A) water-referenced GABA+ measurements and GABA+/Cr ratios and (B) water-referenced MM-suppressed GABA measurements and MM-suppressed GABA/Cr ratios. Individual measurements are color-coded by vendor (GE in green, Philips in orange, Siemens in blue). The black regression line shows the relationship between GABA+/Cr and water-referenced GABA+ over the entire dataset. Additional color-coded regression lines are shown for each site. R2 values (i.e., the effect sizes) are also displayed.
3.2. Exploratory analysis
Using the Siemens short-TE water reference brought down the Siemens GABA+ values to an average of 3.65 ± 0.44 i.u. (an 18% reduction), reducing the discrepancy with the other vendors to 10% (boxplots plotted in Fig. S1). Corresponding VPCs were: vendor = 12.0%; site = 24.2%; participant = 63.8%, with vendor and site effects remaining significant: χ2(1) = 3.92, pboot = 0.01 and χ2(1) = 32.77, pboot < 0.001, respectively. The short-TE-referenced Siemens Cr measurements were also reduced on average (by 18%, to 17.1 ± 2.1 i.u.) compared to the initial analysis, again closer to the GE and Philips Cr measurements (Fig. S2).
4. Discussion
In this second paper describing a large multi-vendor, multi-site GABA-edited MRS dataset, it has been shown that water-referenced GABA measurements, including tissue correction based on variously acquired T1-weighted structural images, can be applied across sites and vendors with relatively low levels of variance. Water-referenced quantification shows very similar levels of performance to Cr referencing, as reported previously (Mikkelsen et al., 2017), with the notable exception of an additional vendor-related effect.
4.1. Water vs. Cr referencing
One objective of this study was to compare quantitative outcomes of water and Cr referencing. Within site, water- and Cr-referenced GABA+ measurements both show variance of 9.5%. Levels of site-related variance are also similar (mean within-vendor CV: 11.6% vs. 11.3%, respectively). The major difference between the water- and Cr-referenced results was the systematic effect of vendor in the water-referenced data.
It is not clear why the water-referenced GABA+ values from the Siemens data were larger than the estimates from the other two vendors. This substantial vendor-related difference was not observed in the Cr-referenced data reported previously (Mikkelsen et al., 2017). Higher GABA+ values suggest a lower-than-expected water signal. Referencing to a short-TE PRESS water acquisition attenuated the discrepancy somewhat, suggesting that the Siemens MEGA-PRESS WIP water reference signal is most likely at issue. It is notable that the water fit errors were lower in the Siemens data, suggesting that the water signal is closer to a Gaussian-Lorentzian lineshape than the other vendors, and that the data undergo differing degrees of preprocessing (e.g., downsampling from the analog-to-digital converter sampling rate to the specified acquisition rate), with potentially different dynamic range performance – the water reference signal is ~10,000 times larger than the GABA signal and acquired with the same receiver gains. MM-suppressed data on Siemens were acquired with TE = 68 ms, compared to TE = 80 ms on the other vendors, a decision made in response to the TE-independent timing of editing pulses in that sequence. This clear difference in acquisition approach (which can only really be avoided by further sequence standardization; see, e.g., Saleh et al., 2019) means the MM-suppressed data offer little insight on the vendor water signal scaling effect. At this stage, it has not been possible to isolate the cause of this discrepancy, and communication with Siemens experts has not yielded a conclusive explanation.
Aside from this vendor effect, it was clear that the variation in the water-referenced GABA+ measurements was similar to the GABA+/Cr measurements. This suggests that the reliability of the two referencing strategies is comparable, consistent with previous smaller studies (Bogner et al., 2010; Saleh et al., 2016). This is perhaps surprising as several additional corrections were performed to obtain the water-referenced values, which introduces more sources of error into the quantification.
4.2. Water referencing approach
Quantification in this study used a best-practice approach, whereby the water reference signal was corrected for partial volume effects and relaxation. It has been shown that failing to account for these effects will lead to quantification errors (Gasparovic et al., 2006, 2018). These errors can be particularly pronounced at longer TEs (Gasparovic et al., 2006) or when there is large variability in tissue compartment fractions across cohorts (Harris et al., 2015; Mato Abad et al., 2014; Mikkelsen et al., 2016; Tal et al., 2012). The relatively low level of variance in the present dataset suggests that incorporating image-based voxel segmentation into the quantification routine did not add significant variance into the data.
Nonetheless, subtle differences in quantification and tissue correction methodologies can have important consequences on reported results. For instance, the linear relationship between GABA+ levels and age, seen when applying a simple CSF tissue correction, can largely be attributed to the dependency of GABA+ levels on tissue composition (Maes et al., 2018; Porges et al., 2017). Additionally, the units of measurement of water-referenced metabolite concentrations, and the information content of the values, will depend on the quantification approach used. Metabolite concentrations have been reported in molar, molal and institutional units (Gasparovic et al., 2018; Jansen et al., 2006; Knight-Scott et al., 2003; Kreis et al., 1993a). Interpretation of stated concentrations, and particularly comparisons across studies, can be challenging. In this study, measurements were reported in institutional units of molality.
Not every source of variance was captured in the statistical analysis. In the statistical model used here, participant-level variance not only accounts for true biological differences between individuals but also measurement error. One systematic site-level factor was the diversity of the T1-weighted structural imaging protocols, leading to heterogeneity in T1-weighted contrast and image signal-to-noise ratio, which lead to small but significant site-to-site differences in voxel segmentation. There is an extensive literature on the successes and limitations of image segmentation (Clark et al., 2006; Eggert et al., 2012; Klauschen et al., 2009), and while segmentation algorithms aim to be robust against the effects of imaging parameters, segmentation remains a challenging undertaking. The substantial tissue differences in water T2, particularly given the medium TE of the water acquisitions (68 ms), and in GABA and MR-visible water concentrations, suggest that accurate segmentation is important for reproducible water-referenced quantification. It should also be noted that the MRS voxel masks that are generated for tissue segmentation purposes correspond to the nominal excited volume, which, because of acquisition parameters, voxel placement and chemical shift displacement errors, will not necessarily be centered on the excited signals of interest. In this study, the Philips and Siemens voxel masks were centered on the GABA and water reference signals, whereas the GE voxel masks centered on the 2.68 ppm signals (see Section 2.2). This variation will have introduced a degree of error in the water-referenced GABA measurements.
4.3. Macromolecular contamination
In quantifying the 3.0 ppm GABA+ signal in this work, a fixed signal fraction of co-edited macromolecule contribution to the peak was assumed, an assumption that may not be valid for studies in neurological and psychiatric disorders. As presented previously (Mikkelsen et al., 2017) for Cr-referenced values, MM-suppressed measures showed greater relative variance than GABA+ measures. The degree of similarity between Cr-referenced and water-referenced measurements was similar for the macromolecule-suppressed and GABA+ datasets.
4.4. Towards further standardization
As highlighted in (Mikkelsen et al., 2017), it is likely that differences in acquisition sequence contribute to variance at the level of both vendor (e.g., factors inherited from vendor PRESS sequences) and site (e.g., differences in implementation of MEGA editing), which might be removed/attenuated by further standardization. In a parallel piece of work, we have implemented a cross-vendor standardized MEGA-PRESS sequence on GE, Philips, Siemens and Canon systems (Saleh et al., 2019), which addresses some of these issues.
4.5. Conclusion
In summary, GABA+ levels were quantified using brain tissue water as an internal concentration reference across 25 sites and low levels of within-site variance were observed. This level of variability is similar to that seen for GABA+ measurements quantified relative to Cr. Given the concern that observed effects might be driven by changes in the reference signal, it is often helpful to quantify both water- and metabolite-referenced measurements. Study-specific expectations of reference signal stability (e.g., between-group differences in clinical populations or known changes in water content and signal relaxation) might suggest one concentration reference a priori. That said, the present results do not show a clear reason to prefer one reference signal for MRS quantification, and it can be concluded that water-referenced measurements of GABA+ are sufficiently reliable to be applied in multi-site studies.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R01 EB016089, R01 EB023963 and P41 EB015909. Data collection was supported by the Shandong Provincial Key Research and Development Plan of China (2016ZDJS07A16) and the National Natural Science Foundation of China for Young Scholars (no. 81601479). AJW was supported by NIA grants K01 AG050707 and R01 AG054077 and the University of Florida (UF), the Center for Cognitive Aging and Memory (CAM) and the McKnight Brain Research Foundation (MBRF). DAE was supported by NIH grant F31 ES028081. ECP was supported by NIAAA grant K01 AA025306 and UF, CAM and MBRF. HJZ was supported by DFG grant SFB 974 TP B07. IDW and NH thank The Wellcome Trust, the NIHR-Sheffield Biomedical Research Centre and Mrs. J. Bigley of the University of Sheffield MRI Unit for her assistance with data acquisition. JJP was supported by NIAAA grant K23 AA020842. KMC was supported by NIH grants R01 MH095014 and R01 NS096207. MPS was supported by NIH grant F32 EY025121. NAJP receives salary support from NIH grant R00 MH107719. SPS was supported by the Research Foundation - Flanders (G089818N), the Excellence of Science (EOS, 30446199, MEMODYN) and the KU Leuven Research Fund (C16/15/070). The authors acknowledge implementation contributions from a number of employees of Siemens Medical Solutions, including Dr. Keith Heberlein and Dr. Sinyeob Ahn, to the Siemens WIP sequences, which are shared with several research sites under sequence-specific agreements.
Appendix
A subset of the data presented in this work has been made available on the NITRC portal in the “Big GABA” project repository (https://www.nitrc.org/projects/biggaba/) and is distributed freely under a non-commercial Creative Commons license. Community members are encouraged to make use of this resource for developing and optimizing new MRS methods. This data resource can also serve as a normative dataset against which clinical data may be compared or for quality assurance purposes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alger JR, 2010. Quantitative Proton Magnetic Resonance Spectroscopy and Spectroscopic Imaging of the Brain. Top. Magn. Reson. Imaging 21, 115–128. doi: 10.1097/RMR.0b013e31821e568f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ, 2005. Unified segmentation. Neuroimage 26, 839–851. doi: 10.1016/j.neuroimage.2005.02.018 [DOI] [PubMed] [Google Scholar]
- Barantin L, Le Pape A, Akoka S, 1997. A new method for absolute quantitation MRS metabolites. Magn. Reson. Med 38, 179–182. doi: 10.1002/mrm.1910380203 [DOI] [PubMed] [Google Scholar]
- Barker PB, Soher BJ, Blackband SJ, Chatham JC, Mathews VP, Bryan RN, 1993. Quantitation of proton NMR spectra of the human brain using tissue water as an internal concentration reference. NMR Biomed 6, 89–94. doi: 10.1002/nbm.1940060114 [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker BM, Walker SC, 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw 67. doi: 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Bearden CE, Thompson PM, 2017. Emerging Global Initiatives in Neurogenetics: The Enhancing Neuroimaging Genetics through Meta-analysis (ENIGMA) Consortium. Neuron 94, 232–236. doi: 10.1016/j.neuron.2017.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogner W, Gruber S, Doelken M, Stadlbauer A, Ganslandt O, Boettcher U, Trattnig S, Doerfler A, Stefan H, Hammen T, 2010. In vivo quantification of intracerebral GABA by single-voxel 1H-MRS—How reproducible are the results? Eur. J. Radiol 73, 526–531. doi: 10.1016/j.ejrad.2009.01.014 [DOI] [PubMed] [Google Scholar]
- Buchli R, Boesiger P, 1993. Comparison of methods for the determination of absolute metabolite concentrations in human muscles by 31P MRS. Magn. Reson. Med 30, 552–558. doi: 10.1002/mrm.1910300505 [DOI] [PubMed] [Google Scholar]
- Christiansen P, Henriksen O, Stubgaard M, Gideon P, Larsson HBW, 1993. In vivo quantification of brain metabolites by 1H-MRS using water as an internal standard. Magn. Reson. Imaging 11, 107–118. doi: 10.1016/0730-725X(93)90418-D [DOI] [PubMed] [Google Scholar]
- Clark KA, Woods RP, Rottenberg DA, Toga AW, Mazziotta JC, 2006. Impact of acquisition protocols and processing streams on tissue segmentation of T1 weighted MR images. Neuroimage 29, 185–202. doi: 10.1016/j.neuroimage.2005.07.035 [DOI] [PubMed] [Google Scholar]
- Danielsen ER, Henriksen O, 1994. Absolute quantitative proton NMR spectroscopy based on the amplitude of the local water suppression pulse. Quantification of brain water and metabolites. NMR Biomed 7, 311–318. doi: 10.1002/nbm.1940070704 [DOI] [PubMed] [Google Scholar]
- Duc CO, Weber OM, Trabesinger AH, Meier D, Boesiger P, 1998. Quantitative 1H MRS of the human brain in vivo based on the stimulation phantom calibration strategy. Magn. Reson. Med 39, 491–6. doi: 10.1002/mrm.1910390320 [DOI] [PubMed] [Google Scholar]
- Edden RAE, Intrapiromkul J, Zhu H, Cheng Y, Barker PB, 2012a. Measuring T2 in vivo with J-difference editing: Application to GABA at 3 Tesla. J. Magn. Reson. Imaging 35, 229–234. doi: 10.1002/jmri.22865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RAE, Oeltzschner G, Harris AD, Puts NAJ, Chan KL, Boer VO, Schär M, Barker PB, 2016. Prospective frequency correction for macromolecule-suppressed GABA editing at 3T. J. Magn. Reson. Imaging 44, 1474–1482. doi: 10.1002/jmri.25304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RAE, Puts NAJ, Barker PB, 2012b. Macromolecule-suppressed GABA-edited magnetic resonance spectroscopy at 3T. Magn. Reson. Med 68, 657–661. doi: 10.1002/mrm.24391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edden RAE, Puts NAJ, Harris AD, Barker PB, Evans CJ, 2014. Gannet: A batch-processing tool for the quantitative analysis of gamma-aminobutyric acid-edited MR spectroscopy spectra. J. Magn. Reson. Imaging 40, 1445–1452. doi: 10.1002/jmri.24478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert LD, Sommer J, Jansen A, Kircher T, Konrad C, 2012. Accuracy and Reliability of Automated Gray Matter Segmentation Pathways on Real and Simulated Structural Magnetic Resonance Images of the Human Brain. PLoS One 7, e45081. doi: 10.1371/journal.pone.0045081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Kreis R, Ross BD, 1993. Absolute quantitation of water and metabolites in the human brain. I. Compartments and water. J. Magn. Reson. Ser. B 102, 1–8. doi: 10.1006/jmrb.1993.1055 [DOI] [Google Scholar]
- Gasparovic C, Chen H, Mullins PG, 2018. Errors in 1H-MRS estimates of brain metabolite concentrations caused by failing to take into account tissue-specific signal relaxation. NMR Biomed 31, e3914. doi: 10.1002/nbm.3914 [DOI] [PubMed] [Google Scholar]
- Gasparovic C, Song T, Devier D, Bockholt HJ, Caprihan A, Mullins PG, Posse S, Jung RE, Morrison LA, 2006. Use of tissue water as a concentration reference for proton spectroscopic imaging. Magn. Reson. Med 55, 1219–1226. doi: 10.1002/mrm.20901 [DOI] [PubMed] [Google Scholar]
- Grasso G, Alafaci C, Passalacqua M, Morabito A, Buemi M, Salpietro FM, Tomasello F, 2002. Assessment of Human Brain Water Content by Cerebral Bioelectrical Impedance Analysis: A New Technique and Its Application to Cerebral Pathological Conditions. Neurosurgery 50, 1064–1074. doi: 10.1097/00006123-200205000-00023 [DOI] [PubMed] [Google Scholar]
- Gussew A, Erdtel M, Hiepe P, Rzanny R, Reichenbach JR, 2012. Absolute quantitation of brain metabolites with respect to heterogeneous tissue compositions in 1H-MR spectroscopic volumes. Magn. Reson. Mater. Physics, Biol. Med 25, 321–333. doi: 10.1007/s10334-012-0305-z [DOI] [PubMed] [Google Scholar]
- Halekoh U, Højsgaard S, 2014. A Kenward-Roger approximation and parametric bootstrap methods for tests in linear mixed models - The R package pbkrtest. J. Stat. Softw 59, 1–32. doi: 10.18637/jss.v059.i0926917999 [DOI] [Google Scholar]
- Harris AD, Puts NAJ, Edden RAE, 2015. Tissue correction for GABA-edited MRS: Considerations of voxel composition, tissue segmentation, and tissue relaxations. J. Magn. Reson. Imaging 42, 1431–1440. doi: 10.1002/jmri.24903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig J, Pfister H, Ernst T, Ott D, 1992. Direct absolute quantification of metabolites in the human brain within vivo localized proton spectroscopy. NMR Biomed 5, 193–199. doi: 10.1002/nbm.1940050406 [DOI] [PubMed] [Google Scholar]
- Henry P-G, Dautry C, Hantraye P, Bloch G, 2001. Brain GABA editing without macromolecule contamination. Magn. Reson. Med 45, 517–520. doi: [DOI] [PubMed] [Google Scholar]
- Holm S, 1979. A simple sequentially rejective multiple test procedure. Scand. J. Stat 6, 65–70. [Google Scholar]
- Huang W, Alexander GE, Chang L, Shetty HU, Krasuski JS, Rapoport SI, Schapiro MB, 2001. Brain metabolite concentration and dementia severity in Alzheimer’s disease: A 1H MRS study. Neurology 57, 626–632. doi: 10.1212/WNL.57.4.626 [DOI] [PubMed] [Google Scholar]
- Jansen JFA, Backes WH, Nicolay K, Kooi ME, 2006. 1H MR Spectroscopy of the Brain: Absolute Quantification of Metabolites. Radiology 240, 318–332. doi: 10.1148/radiol.2402050314 [DOI] [PubMed] [Google Scholar]
- Kantarci K, Jack CR, Xu YC, Campeau NG, O’Brien PC, Smith GE, Ivnik RJ, Boeve BF, Kokmen E, Tangalos EG, Petersen RC, 2000. Regional metabolic patterns in mild cognitive impairment and Alzheimer’s disease: A 1H MRS study. Neurology 55, 210–217. doi: 10.1212/WNL.55.2.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauschen F, Goldman A, Barra V, Meyer-Lindenberg A, Lundervold A, 2009. Evaluation of automated brain MR image segmentation and volumetry methods. Hum. Brain Mapp 30, 1310–1327. doi: 10.1002/hbm.20599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight-Scott J, Haley AP, Rossmiller SR, Farace E, Mai VM, Christopher JM, Manning C. a., Simnad VI, Siragy HM, 2003. Molality as a unit of measure for expressing 1H MRS brain metabolite concentrations in vivo. Magn. Reson. Imaging 21, 787–797. doi: 10.1016/S0730-725X(03)00179-6 [DOI] [PubMed] [Google Scholar]
- Kreis R, Ernst T, Ross BD, 1993a. Absolute quantitation of water and metabolites in the human brain. II. Metabolite concentrations. J. Magn. Reson. Ser. B 102, 9–19. doi: 10.1006/jmrb.1993.1056 [DOI] [Google Scholar]
- Kreis R, Ernst T, Ross BD, 1993b. Development of the human brain: In vivo quantification of metabolite and water content with proton magnetic resonance spectroscopy. Magn. Reson. Med 30, 424–437. doi: 10.1002/mrm.1910300405 [DOI] [PubMed] [Google Scholar]
- Laule C, Vavasour IM, Moore GRW, Oger J, Li DKB, Paty DW, MacKay AL, 2004. Water content and myelin water fraction in multiple sclerosis. A T2 relaxation study. J. Neurol 251, 284–93. doi: 10.1007/s00415-004-0306-6 [DOI] [PubMed] [Google Scholar]
- Low RN, Francis IR, Herfkens RJ, Jeffrey RB, Glazer GM, Foo TK, Shimakawa A, Pelc NJ, 1993. Fast multiplanar spoiled gradient-recalled imaging of the liver: pulse sequence optimization and comparison with spin-echo MR imaging. Am. J. Roentgenol 160, 501–509. doi: 10.2214/ajr.160.3.8381572 [DOI] [PubMed] [Google Scholar]
- Lu H, Nagae-Poetscher LM, Golay X, Lin D, Pomper M, van Zijl PCM, 2005. Routine clinical brain MRI sequences for use at 3.0 Tesla. J. Magn. Reson. Imaging 22, 13–22. doi: 10.1002/jmri.20356 [DOI] [PubMed] [Google Scholar]
- Maes C, Hermans L, Pauwels L, Chalavi S, Leunissen I, Levin O, Cuypers K, Peeters R, Sunaert S, Mantini D, Puts NAJ, Edden RAE, Swinnen SP, 2018. Age-related differences in GABA levels are driven by bulk tissue changes. Hum. Brain Mapp 39, 3652–3662. doi: 10.1002/hbm.24201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjańska M, McCarten JR, Hodges J, Hemmy LS, Grant A, Deelchand DK, Terpstra M, 2017. Region-specific aging of the human brain as evidenced by neurochemical profiles measured noninvasively in the posterior cingulate cortex and the occipital lobe using 1 H magnetic resonance spectroscopy at 7 T. Neuroscience 354, 168–177. doi: 10.1016/j.neuroscience.2017.04.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato Abad V, Quirós A, García-Álvarez R, Loureiro JP, Álvarez-Linera J, Frank A, Hernández-Tamames JA, 2014. The Partial Volume Effect in the Quantification of 1H Magnetic Resonance Spectroscopy in Alzheimer’s Disease and Aging. J. Alzheimer’s Dis 42, 801–811. doi: 10.3233/JAD-140582 [DOI] [PubMed] [Google Scholar]
- Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R, 1998. Simultaneous in vivo spectral editing and water suppression. NMR Biomed 11, 266–272. doi: [DOI] [PubMed] [Google Scholar]
- Michaelis T, Merboldt KD, Bruhn H, Hänicke W, Frahm J, 1993. Absolute concentrations of metabolites in the adult human brain in vivo: quantification of localized proton MR spectra. Radiology 187, 219–227. doi: 10.1148/radiology.187.1.8451417 [DOI] [PubMed] [Google Scholar]
- Mikkelsen M, Barker PB, Bhattacharyya PK, Brix MK, Buur PF, Cecil KM, Chan KL, Chen DY-T, Craven AR, Cuypers K, Dacko M, Duncan NW, Dydak U, Edmondson DA, Ende G, Ersland L, Gao F, Greenhouse I, Harris AD, He N, Heba S, Hoggard N, Hsu T, Jansen JFA, Kangarlu A, Lange T, Lebel RM, Li Y, Lin CE, Liou J, Lirng J-F, Liu F, Ma R, Maes C, Moreno-Ortega M, Murray SO, Noah S, Noeske R, Noseworthy MD, Oeltzschner G, Prisciandaro JJ, Puts NAJ, Roberts TPL, Sack M, Sailasuta N, Saleh MG, Schallmo M, Simard N, Swinnen SP, Tegenthoff M, Truong P, Wang G, Wilkinson ID, Wittsack H-J, Xu H, Yan F, Zhang C, Zipunnikov V, Zöllner HJ, Edden RAE, 2017. Big GABA: Edited MR spectroscopy at 24 research sites. Neuroimage 159, 32–45. doi: 10.1016/j.neuroimage.2017.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen M, Singh KD, Brealy JA, Linden DEJ, Evans CJ, 2016. Quantification of γ-aminobutyric acid (GABA) in 1H MRS volumes composed heterogeneously of grey and white matter. NMR Biomed 29, 1644–1655. doi: 10.1002/nbm.3622 [DOI] [PubMed] [Google Scholar]
- Miller KL, Alfaro-Almagro F, Bangerter NK, Thomas DL, Yacoub E, Xu J, Bartsch AJ, Jbabdi S, Sotiropoulos SN, Andersson JLR, Griffanti L, Douaud G, Okell TW, Weale P, Dragonu I, Garratt S, Hudson S, Collins R, Jenkinson M, Matthews PM, Smith SM, 2016. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat. Neurosci 19, 1523–1536. doi: 10.1038/nn.4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynárik V, Gruber S, Moser E, 2001. Proton T1 and T2 relaxation times of human brain metabolites at 3 Tesla. NMR Biomed 14, 325–331. doi: 10.1002/nbm.713 [DOI] [PubMed] [Google Scholar]
- Mugler JP, Brookeman JR, 1990. Three-dimensional magnetization-prepared rapid gradient-echo imaging (3D MP RAGE). Magn. Reson. Med 15, 152–157. doi: 10.1002/mrm.1910150117 [DOI] [PubMed] [Google Scholar]
- Mullins PG, McGonigle DJ, O’Gorman RL, Puts NAJ, Vidyasagar R, Evans CJ, Cardiff Symposium on MRS of GABA, Edden RAE, 2014. Current practice in the use of MEGA-PRESS spectroscopy for the detection of GABA. Neuroimage 86, 43–52. doi: 10.1016/j.neuroimage.2012.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Near J, Edden R, Evans CJ, Paquin R, Harris A, Jezzard P, 2015. Frequency and phase drift correction of magnetic resonance spectroscopy data by spectral registration in the time domain. Magn. Reson. Med 73, 44–50. doi: 10.1002/mrm.25094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeb H, Zilles K, Shah NJ, 2006. Fully-automated detection of cerebral water content changes: Study of age- and gender-related H2O patterns with quantitative MRI. Neuroimage 29, 910–922. doi: 10.1016/j.neuroimage.2005.08.062 [DOI] [PubMed] [Google Scholar]
- Piechnik SK, Evans J, Bary LH, Wise RG, Jezzard P, 2009. Functional changes in CSF volume estimated using measurement of water T2 relaxation. Magn. Reson. Med 61, 579–586. doi: 10.1002/mrm.21897 [DOI] [PubMed] [Google Scholar]
- Porges EC, Woods AJ, Lamb DG, Williamson JB, Cohen RA, Edden RAE, Harris AD, 2017. Impact of tissue correction strategy on GABA-edited MRS findings. Neuroimage 162, 249–256. doi: 10.1016/j.neuroimage.2017.08.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puts NAJ, Barker PB, Edden RAE, 2013. Measuring the longitudinal relaxation time of GABA in vivo at 3 Tesla. J. Magn. Reson. Imaging 37, 999–1003. doi: 10.1002/jmri.23817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackayova V, Cudalbu C, Pouwels PJW, Braissant O, 2017. Creatine in the central nervous system: From magnetic resonance spectroscopy to creatine deficiencies. Anal. Biochem 529, 144–157. doi: 10.1016/j.ab.2016.11.007 [DOI] [PubMed] [Google Scholar]
- Reyngoudt H, Claeys T, Vlerick L, Verleden S, Acou M, Deblaere K, De Deene Y, Audenaert K, Goethals I, Achten E, 2012. Age-related differences in metabolites in the posterior cingulate cortex and hippocampus of normal ageing brain: A 1H-MRS study. Eur. J. Radiol 81, e223–e231. doi: 10.1016/j.ejrad.2011.01.106 [DOI] [PubMed] [Google Scholar]
- Rothman DL, Petroff OA, Behar KL, Mattson RH, 1993. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proc. Natl. Acad. Sci. U. S. A 90, 5662–5666. doi: 10.1073/pnas.90.12.5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleh MG, Near J, Alhamud A, Robertson F, van der Kouwe AJW, Meintjes EM, 2016. Reproducibility of macromolecule suppressed GABA measurement using motion and shim navigated MEGA-SPECIAL with LCModel, jMRUI and GANNET. Magn. Reson. Mater. Physics, Biol. Med 29, 863–874. doi: 10.1007/s10334-016-0578-8 [DOI] [PubMed] [Google Scholar]
- Saleh MG, Rimbault D, Mikkelsen M, Oeltzschner G, Wang AM, Jiang D, Alhamud A, Near J, Schär M, Noeske R, Murdoch JB, Ersland L, Craven AR, Dwyer GE, Grüner ER, Pan L, Ahn S, Edden RAE, 2019. Multi-vendor standardized sequence for edited magnetic resonance spectroscopy. Neuroimage 189, 425–431. doi: 10.1016/j.neuroimage.2019.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soher BJ, van Zijl PCM, Duyn JH, Barker PB, 1996. Quantitative proton MR spectroscopic imaging of the human brain. Magn. Reson. Med 35, 356–363. doi: 10.1002/mrm.1910350313 [DOI] [PubMed] [Google Scholar]
- Tal A, Kirov II, Grossman RI, Gonen O, 2012. The role of gray and white matter segmentation in quantitative proton MR spectroscopic imaging. NMR Biomed 25, 1392–1400. doi: 10.1002/nbm.2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulborn KR, Ackerman JJH, 1983. Absolute molar concentrations by NMR in inhomogeneous B1. A scheme for analysis of in vivo metabolites. J. Magn. Reson 55, 357–371. doi: 10.1016/0022-2364(83)90118-X [DOI] [Google Scholar]
- Tkáč I, Rao R, Georgieff MK, Gruetter R, 2003. Developmental and regional changes in the neurochemical profile of the rat brain determined by in vivo 1H NMR spectroscopy. Magn. Reson. Med 50, 24–32. doi: 10.1002/mrm.10497 [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K, 2013. The WU-Minn Human Connectome Project: An overview. Neuroimage 80, 62–79. doi: 10.1016/j.neuroimage.2013.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansapura JP, Holland SK, Dunn RS, Ball WS, 1999. NMR relaxation times in the human brain at 3.0 Tesla. J. Magn. Reson. Imaging 9, 531–538. doi: [DOI] [PubMed] [Google Scholar]
- Zoelch N, Hock A, Heinzer-Schweizer S, Avdievitch N, Henning A, 2017. Accurate determination of brain metabolite concentrations using ERETIC as external reference. NMR Biomed 30, e3731. doi: 10.1002/nbm.3731 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.