Abstract
Antibodies serve as critical barriers to viral infection. Humoral immunity to a virus is achieved through the dual role of antibodies in communicating the presence of invading pathogens in infected cells to effector cells and interfering with processes essential to the viral lifecycle, chiefly entry into the host cell. For individuals that successfully control infection, virus-elicited antibodies can provide lifelong surveillance and protection from future insults. One approach to understand the nature of a successful immune response has been to utilize structural biology to uncover the molecular details of the antibodies derived from vaccines or natural infection and how they interact with their cognate microbial antigens. The ability to isolate antigen specific B-cells and rapidly solve structures of functional, monoclonal antibodies in complex with viral glycoprotein surface antigens has greatly expanded our knowledge of the sites of vulnerability on viruses. In this review, we compare the adaptive humoral immune responses to HIV, influenza, and filoviruses, with a particular focus on neutralizing antibodies. The pathogenesis of each of these viruses is quite different, providing an opportunity for comparison of immune responses: HIV causes a persistent, chronic infection; influenza an acute infection with multiple exposures during a lifetime and annual vaccination; and filoviruses, a virulent, acute infection. Neutralizing antibodies that develop under these different constraints are therefore sentinels that can provide insight into the underlying humoral immune responses and important lessons to guide future development of vaccines and immunotherapeutics.
Introduction
Enveloped viruses are found across diverse viral families and cause some of the deadliest diseases known to man. Despite a spectrum of differences in their biology and pathogenesis, all enveloped viruses share two commonalities: a lipid bilayer envelope co-opted from host cells upon viral egress and the presence of surface-exposed viral glycoproteins for host cell recognition and entry. These viral glycoproteins or ‘spike’ proteins are exposed to the adaptive immune response and are the main targets of host antibodies, often being the only exposed antigen. Naturally, viruses have developed mechanisms to avoid such responses through rapid evolution of antibody-targeting epitopes, steric shielding of epitopes by glycan post-translational modifications, immune decoys such as soluble antigens that share viral spike epitopes, and immunosuppression to evade host recognition upon cellular entry. In response to the viral arms race, antibodies have in turn developed many creative solutions to overcome viral evasion, including unique structural adaptations that allow antibodies to more readily penetrate the viral armor and exploit sites of vulnerability.
The nature of the enveloped viral surface has major consequences for viral entry and, consequently, how the adaptive immune system responds. For enveloped viruses, entry hinges on the mechanics of the viral glycoprotein. Vital to the function of glycoproteins is their ability to fuse viral and host membranes together. Generally, this is achieved through a complex mechanism, where initial binding to a specific cellular host receptor usually results in drastic changes in the structure of the viral spike that enable transition to the post-fusion conformation. Successful triggering of post-fusion glycoprotein forms provides the energy necessary to fuse the viral and cellular membranes, thereby forming pores that allows the viral genomes to gain entry into the cytoplasm1,2. Once entry is achieved, viral replication takes place using a variety of strategies, both in the cytoplasm and/or nucleus, but concludes in assembly and budding of new viral progeny at the host-cell surface. For enveloped viruses, a neutralizing antibody (nAb) disrupting the viral lifecycle almost always involves binding to the viral glycoprotein, which can then disrupt entry by blocking one or more of the processes described above. Additionally, antibodies can also block viral egress by binding to glycoproteins on the infected cell surface, thus preventing viruses from budding. Thus, neutralization is achieved by preventing progeny from infecting a new cell and is a distinct mechanism from neutralization achieved from blocking entry (Figure 1). The pathogenesis of the three viruses that we describe here represent gradations of antigenic exposure to the adaptive immune system and also greatly influences the molecular nature of the antibodies formed in response to infection, which we will discuss at length later. Specific details regarding the lifecycle and pathogenesis of these viruses are summarized in Table 1. While nAbs represent the overwhelming majority of our understanding of the antibody-based immune response to enveloped viruses, especially with respect to their structure, some non-neutralizing antibodies have also been shown to provide protection in vivo; however, this review will majorly focus on nAbs.
Figure 1. Points of antibody blockade to enveloped virus entry and egress.
Like a wrench thrown into the gears of a machine, antibodies are able to neutralize or protect against viruses by blocking one or more biological processes during viral entry or exit. The mechanism of neutralization depends largely on which epitope the antibody targets, and this subsequently determines which process is slowed or inhibited completely. For HIV, antibodies may block primary CD4 receptor (1) or secondary (2) CCR5/CXCR4 co-receptor binding. Filoviruses and influenza do not enter cells at the surface, but may be blocked by preventing attachment (3) or pinocytosis (4). For filoviruses, after endocytosis, antibodies must stay attached and survive acidification and subsequently prevent endosomal cleavage (5) or block endosomal receptor binding (6). For influenza, receptor binding can be blocked at the cell surface (7). In all cases, if receptor binding occurs, antibodies may directly block fusion through binding to fusion machinery. If viruses enter the cell, antibodies can potentially block egress of progeny virus (8). Non-neutralizing pathways to antibody-based protection include tagging cells for destruction by effector cells before viruses have a chance to exit (9), for example by antibody-dependent cellular cytotoxicity (ADCC) or by complement, or through agglutination of virions which can then be destroyed by effector cells (10). Additional abbreviations for human immunodeficiency virus 1, influenza virus and filovirus are HIV, flu and filov, respectively. This image was made using BioRender.
Table 1.
Comparison of HIV, influenza and filoviral biology, taxonomy and pathogenesis.
| Glycoprotein | HIV | Influenza | Ebolaviruses |
|---|---|---|---|
| Viral Spike Name | Envelope (Env) | Hemagglutinin (HA) | Glycoprotein (GP) |
| Receptor Binding Domain | gp120 | HA1 | GP1 |
| Fusion domain | gp41 | HA2 | GP2 |
| Predidcted Glycosylation sites | 25 | ||
| Protein size | 1,195 amino acids | 522 amino acids | 667 amino acids |
| Known Receptor(s) | CD4/CCR5/CXCR5 | sialic acid | NPC1 |
| Entry strategy | cell surface receptor | endosomal acidification | endosomal |
| Classification | |||
| Group | Group VI (+ssRNA-RT) | Group V (-ssRNA) | Group V (-ssRNA) |
| Order | Unassigned | Unassigned | Mononegavirales |
| Family | Retroviridae | Orthomyxoviridae | Filoviridae |
| Subfamily | Orthoetrovirinae | n/a | n/a |
| Genera | Lentivirus | Alphainfluenzavirus | Ebolavirus |
| Betainfluenzavirus | Marburgvirus | ||
| Deltainfluenzavirus | Cuevavirus | ||
| Gammainfluenzavirus | |||
| Species | HIV1 | Influenza A | Zaire ebolavirus |
| HIV2 | Influenza B | Bundibugyo ebolavirus | |
| Influenza C | Reston ebolavirus | ||
| Influenza D | Sudan ebolavirus | ||
| Tai Forest ebolavirus | |||
| Marburg marburgvirus | |||
| Lloviu cuevavirus | |||
| Pathogenesis | |||
| Infection route | bodily fluids/blood | respitory | bodily fluids/blood |
| Latency | weeks to months | days | days |
| Duration | chronic | acute | acute |
| Yearly cases | 1–2 million | 9–50 million since 2010 | varied (1–29,000) |
| Tropism | CD4+ T-cells | epithelial cells | universal |
| FDA approved treatments | 40 | 3 | none |
The complex biology of HIV, influenza and filoviruses is difficult to categorize and each respective field has developed its own jargon to describe each virus. Here, we summarize the basic aspects of each viruses’ biology, taxonomy and pathogenesis to provide a perspective on their commonalties and divergence in a variety of categories relevant to the antibody-based immune responses. This table is not meant to convey all aspects of the viruses but rather to provide broad generalizations that allow some level of comparison.
Due to the potency, specificity, and tolerability of antibodies, they have become popular as therapeutics for viral disease and are also the intended outcome of most vaccines. It is therefore vital to understand not only where antibodies bind to viruses, but the molecular nature of the interaction. Structural biology has led the way in describing how antibodies interact with viral glycoproteins. While x-ray crystallography has long been the primary technique for solving high resolution structures, single particle electron microscopy has recently also become a powerful and increasingly used method for determining structures of viral antigen-antibody complexes, now reaching resolutions comparable to x-ray crystallography. These data are critical to structure-based design of the next generation of immunogens to elicit neutralizing and protective antibodies and in the selection and design of improved antibody therapeutics.
In this Review, we summarize recent advances in our understanding of neutralizing antibody responses to enveloped viruses, particularly from a structural perspective, with a focus on HIV, influenza, and filoviruses, for which there is a wealth of structural and immunological data. We first describe the overlapping and unique characteristics of each viral lifecycle, glycoprotein structure and pathogenesis. We highlight how structural biology has delineated the immunogenic landscape of these viruses and how antibodies have adapted to target these sites of vulnerability. Next, we discuss common and divergent themes among immune responses to these viruses and, importantly, how this information is being utilized to inform design of vaccines, therapeutics and diagnostics. Finally, we briefly comment on the role of antibodies in the recruitment of the mediators of cellular immunity, the emerging role of Fc-variants, and how lessons learned translate to other viruses and can address future outbreaks.
Neutralizing antibody and glycoprotein structures
Immunoglobulins (Ig) are produced in a wide variety in humans, each with different roles in the immune response. The Ig gamma (G) form is by far the most extensively studied class of Ig, due to its extremely important role in immunotherapeutics as well as in viral immunity. Although there are very few structures of intact IgG3–6, hundreds of structures of antibody variable domains have been described in the literature at length, usually as fragment antigen binding (Fab) alone and in complex with cognate antigen7,8. IgGs can be segregated into two identical Fab domains, which are responsible for binding and recognizing specific antigens, and a single fragment crystallizable (Fc) domain, which is responsible for communicating the presence of pathogens through binding to a limited set of cellular receptors known as Fc gamma receptors (FcγRs) (Figure 2).
Figure 2. Antibody structure and domain topology.
When discussing antibodies in the context of viral immunity, we are generally referring to the IgG isotype, and particularly IgG1, which makes up the majority of serum antibodies and is primarily responsible for protection against infection. IgGs are heterodimers with two identical heavy chains (HC) and two identical light chains (LC) linked by disulfide bonds at a flexible hinge region that separates the Fab domains and Fc domain. The HC, named for its larger size, is a single polypeptide that contains four Ig domains, including the two Fab HC domains (VH and CH1), the hinge-region, and the Fc domain (CH2 and CH3), while the LC is composed of two LC Ig domains (VL and CL). The Fc region is responsible for linking antigens to effector cells and communicating their presence to the host through binding to FcγRs on effector cells, which exist in a variety of isotypes and are expressed at varying levels and compositions depending on the particular effector cell and activation state.
The overwhelming majority of antibodies that provide sterilizing protection from infection target the viral spike. Therefore, the structure of the glycoprotein greatly influences how antibodies are selected, matured and specifically interfere with the mechanics of viral entry. Many enveloped glycoprotein structures have been solved, including those of the viruses discussed here, and have provided extensive insight into their mechanism of entry, as discussed below. Such details have led to enveloped virus membrane fusion proteins being classified into three groups based on their structure and mechanism of entry2. For the purposes of this review, we will focus on the type-I fusion glycoproteins, which compose the glycoproteins of HIV, influenza and filoviruses. However, while these three viruses are well-characterized examples, they do not represent the complete list of viruses that have contributed to our understanding of the antibody-based immune response to viruses containing type-I glycoproteins.
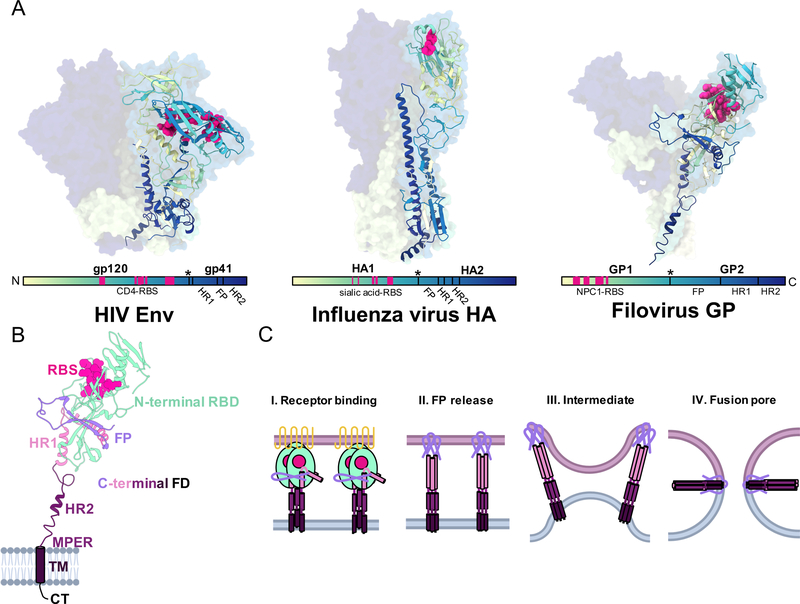
Type-I glycoproteins are expressed as single polypeptides that, after proteolytic cleavage, give rise to the pre-fusion, metastable viral spikes that are present on the surface of viruses. Type-I glycoproteins are composed of heterodimers arranged in a homotrimeric arrangement. Each protomer is identical and includes an ectodomain consisting of an N-terminal portion, the receptor binding domain (RBD) that houses the receptor binding site (RBS), and a largely C-terminal portion containing the fusion machinery, a transmembrane anchor (TM), and often some type of internal C-terminal tail. Within the fusion machinery, a hydrophobic linear sequence of amino acids, known as the ‘fusion peptide’ or ‘internal fusion loop’, is sequestered or partially buried, waiting to be released upon receptor engagement. In each of the viruses discussed here, binding to specific host-cell receptors is not sufficient to activate viral fusion. Rather, there are multiple steps required before glycoproteins become fusion competent. Once the fusion peptide is released, it is thought that it inserts itself into the host membrane and anchors the viral and host membranes together2. Helices within the fusion domain below combine to form an elongated helix and thereby ‘zip’ into their post-fusion form, creating an extended coiled-coil intermediate. The extended intermediate eventually collapses into a 6–helical bundle, bringing the inserted N-terminal fusion-peptides and C-terminal TM regions together to create the post-fusion form of the glycoprotein. This structural rearrangement provides the necessary energy to bring the viral and host membranes close together, forming a pore sufficient for the genetic material to enter into the cytoplasm. Pre-and post-fusion structures representing influenza virus9,10 HIV11–14 and Ebola virus15,16 have all been described and were instrumental in detailing the processes described above.
Covering the surface of all glycoproteins are a variety of glycans, which are typically found to be more concentrated in the N-terminal RBD domains. The extent of coverage, type of glycan and attachment, and role in the viral lifecycle varies between viruses and is not completely understood; however, one of the most commonly attributed purposes of glycans is for immune shielding17–20, as well as prevention of proteolysis10. For determination of some glycoprotein structures, glycans are trimmed back by glycosidases or occasionally deleted to facilitate crystallization and therefore are often not structurally characterized or incompletely characterized. However, in other structures, these inherently flexible glycans can be explicitly resolved in both x-ray and cryo-electron microscopy (EM) structures, particularly when bound to antibodies20. Recent efforts to model glycans in silico and by mass spectrometry21, as well as cryo-EM studies22, have also given a much greater understanding of the extent of glycosylation. Comparing structures of the type-I glycoproteins discussed here emphasizes the common structural themes among them (Figure 3). These structural motifs greatly influence the immune response and are reflected in the antibodies that are elicited, resulting in common themes among nAbs, which will be discussed in greater detail below. It should be noted that while HIV and filoviruses only have one glycoprotein on their surface, influzenza viruses have two, including hemagglutinin (HA) and neuraminidase (NA), which aids in viral egress. While neutralizaing antibodies against NA are significant and can provide protection, we will not discuss them in this review.
Figure 3. Shared structural features of type I glycoproteins.
A) The glycoproteins of HIV (PDB 5V7J), influenza (PDB 3KU3) and filoviruses (PDB 5JQ3) are widely divergent in sequence, size and shape but do share common structural features that are conserved in type I fusion mechanisms of enveloped virus entry. Here we show representative structures of glycoproteins from each of these viruses that are gradient colored from the N-terminus (light yellow) to the C-terminus (dark blue). This allows a comparison of where potential common sites of vulnerability exist on each of these structures in relation to divergent viruses, including the RBS, fusion loop, and stem. B) The sub-domain architecture of a type-I viral fusion protein is shown with this example of a GP1,2 subunit of Ebola virus GP (PDB 5JQ3). The N-terminal receptor binding domain (RBD, light green) is positioned above the C-terminal fusion domain (FD) and houses the receptor binding site (RBS, magenta). The FD contains a fusion peptide (FP, lavender), two heptad repeats (HR1 in pink and HR2 in purple), a hydrophobic membrane proximal external region (MPER, dark purple), and a transmembrane (TM, dark purple) anchor and C-terminal tail (CT, black). C) Type-I membrane fusion occurs in distinct stages. I) N-terminal RBDs bind to their cognate receptors, beginning the process of fusion. II) Receptor binding releases the FP, which pierces the host membrane and causes HR1 and HR2 to form an extended 3-helix bundle. III) During intermediate stages, it is thought that groups of viral spikes cluster to induce membrane buckling, allowing viral and host membranes to come close to each other. IV) Finally, HR2 collapses upon HR1, forming a 6-helix bundles that draws the TM domains together with the FPs, causing mixing of host and viral membranes and the formation of the fusion pore, which then permits the viral genome to exit into the host cytoplasm.
Common and divergent themes of antibody binding to enveloped glycoproteins
Antibodies can provide sterilizing protection against viral pathogens and finding antibodies that are potent against diverse strains of related viruses is highly desirable. Once such antibodies are identified and isolated, structural elucidation of their epitopes can reveal regions on the viral surface that can be targeted by vaccines and immunotherapeutics. One driver of antibody epitope mapping has been the utilization of electron microscopy to rapidly survey large number of antibodies through docking crystal structures into lower resolution EM maps23–27, and now to map polyclonal antibody responses in sera28,29. Structural studies have collectively revealed that antibodies utilize a plethora of strategies to recognize the wide range of surfaces and epitope locations on viral antigens, highlighting weak spots on the viral armor. Below we outline some of these sites of vulnerability and point out common and divergent themes.
Receptor binding site and structural mimicry
All viruses utilize a host receptor in some capacity to gain entry into cells. Therefore, it is not surprising that the receptor binding site (RBS) is a major site of vulnerability for many viruses, since this region is an obligate component of the viral lifecycle and, therefore, relatively conserved even under immune pressure (Figure 4, Row 1). Viruses have evolved unique ways to escape this pressure. For example, HIV requires both a primary receptor (CD4) that is sterically difficult for antibodies to access11,12, as well as a co-receptor (CCR5/CXCR4), whose binding site is sequestered and only formed and accessible after CD4 binds30. Nevertheless, many bnAbs have now been identified that target the CD4-binding site31,32. Influenza virus’ sialic acid binding site is easily accessed on the HA head33, and is relatively well conserved, although some mutational variability can be tolerated within this region34,35, and RBS-adjacent regions can actually vary substantially36,37. Efforts to engineer broader specificity into RBS-targeting antibodies has been met with difficulty since RBDs occupy relatively small patches on the viral glycoprotein surface compared to the size of an antibody footprint38 and, therefore, there are many places where the virus can escape neutralization by changing sequence in regions surrounding the RBS. Thus, strain specificity is more typical39, and truly broadly neutralizing antibodies (bnAbs) to the RBS are rare 40–42. The ebolaviruses on the other hand require massive proteolytic remodeling for their RBS to become exposed within the late-endosomal compartment and, therefore, the RBS is hidden from the immune system on free virions43–45. However, conservation of the RBS on all filoviruses, and the differential exposure of the RBS on Marburg virus (MARV)46,47, has shown that pan-filoviral antibodies are possible24,48 .
Figure 4. Examples of enveloped virus common and divergent sites of vulnerability targeted by neutralizing antibodies.

A) CD4 binding site (CD4bs) antibodies, such as VRC01 (PDB 5FYJ), bind in between HIV Env protomers using their HCs (top) and mimic the immunoglobulin fold (Ig) of the actual receptor (bottom). B) CDRH3 of F045–92 (PDB 4O58), and others like it, reach into the sialic acid binding pocket (top) and closely mimic the natural ligand (bottom). C) The antibody MR78 (PDB 5UQY) uses an extended hydrophobic CDRH3 (top) to bind to the NPC1 receptor binding site (RBS). D) The fusion peptide (FP) of Env sits near the base and is contacted largely by the HC of VRC34.01 (PDB 5I8H). E) The HA FP also sits near the base on the HA stalk and similarly is contacted largely by the HC of MEDI8852 (PDB 5JW3). F) For ADI-15878 (PDB 6DZL), contacts are made across HR1 with the HC and the FP is mostly contact by the LC. G) The HIV antibody 10E8 (Env PDB 5V7J and 10E8 PDB 5T85 fit into EMDB 3312) has evolved to partially contact portions of the viral lipid membrane. H) HA stalk antibodies typically have the broadest neutralizing paratopes and CR9114 (PDB 4FQI) contacts large portions of the extended HA2 alpha-helix with its HC. I) bNab ADI-16061 (GP PDB 5JQ3 and example Fab PDB 5HJ3 fit into EMD 8698) binds far below the base of GP, contacting conserved hydropbobic residues within HR2 and the MPER. J) The long-term exposure of the immune system to HIV allows for extensive SHM and antibody evolution, producing antibodies like PGT145 (PDB 5V8L), with an extended CDRH3 that is rigidified by a beta-hairpin structure with hydrophobic residues and sulfated tyrosines. This allows the antibody to reach deep into the apex pocket of Env. K) Ebolaviruses have two glycoproteins, the viral GP trimer and the soluble GP (sGP) dimer, which is expressed in large abundance during infection and is thought to be a type of immune decoy. The first 296 amino acids of GP and sGP are shared and the protective antibody 13C6 binds to both GP (left, PDB 5KEL) and sGP (right, PDB 5KEN) near a highly conserved residue (W275).
Just as viruses have evolved complementary grooves on their protein surfaces for receptor binding, antibodies have also evolved into molecular mimics that can to some extent imitate these receptors. In such cases, binding can be quaternary in nature, involving loops that contain specific sequences or motifs similar to the actual receptor30,33,38,40,49–54. This mimicry may involve one or more complementarity determining region (CDR) loops. The structure of an antibody is thus well-suited for performing receptor mimicry, and there are examples for each of the viruses discussed here (Figure 4A–C).
HIV relies upon interaction with its receptor CD4 in a conserved binding pocket near the base of gp120 to achieve structural changes required for interaction with secondary receptors30,55. Similar to IgG, CD4 is also composed of the Ig-fold; thus, antibodies are predisposed for recognition of the CD4 epitope56, although they consist of twin Ig domains rather than one for CD4. The VRC01-class of antibodies target the CD4 binding site (Figure 4A), and are typically very broadly reactive and potent57,58. Although these antibodies mimic some aspects of CD4 binding to receptor55, they do not induce the large structural changes associated with receptor binding. The mAb b12 from a long-term survivor, although less potent and broad, more closely emulates the CD4 receptor in that it does induce some structural changes but not to the same extent as CD4 binding30,59.
Each Influenza HA monomer has a binding pocket that contacts sialic acid moieties on the cell surface as its receptor prior to entry and relies on this interaction for endocytosis. While most of the known bnAbs to influenza virus target the more conserved HA stalk, some antibodies that contact the HA head also have some breadth in their binding and neutralizing capacity33,38,40,51–53. For example, the crystal structure of F045–092 (Figure 4B), as well as other mAbs, reveals that such antibodies likely block entry by mimicking the way sialic acid binds by inserting a CDRH3 loop into the RBS on the HA head38. On this and other antibodies, a dipeptide on CDRH3 contains an Asp residue that directly mimics the carboxyl on sialic acid38,53, and a hydrophobic residue that binds in the same hydrophobic pocket as the acetamido moiety of sialic acid33,38,42,54. Occasionally CDR2 inserts into the binding site instead of CDRH352. While the strength of the monovalent interaction is often low, the added avidity of an IgG that can span HAs on the surface can subtantially boost affinity and activity52. Similarly51, many other antibodies also contact the HA RBS, but have larger footprints38 that extend into less conserved regions adjacent to the RBS and are therefore more strain-specific and less broadly neutralizing33,51,53.
A remarkable example of structural mimicry was recently demonstrated for filoviruses. Antibody MR78 (Figure 4C), which was isolated from a panel of MARV human survivor mAbs with potent neutralizing activity and targets the RBS24, was shown to bind directly to the Niemann Pick C1 (NPC1) binding site, blocking receptor binding48. The motif displayed by MR78 includes hydrophobic residues that closely mimic the loops inserted by NPC149. Ebolaviruses have evolved a more structured glycan cap15,60–62, in contrast to the one found on marburgviruses47, that occupies the RBS on the viral surface prior to cleavage by host proteases. The ebolavirus glycan cap, NPC1, and the MR78 CDR loops, all bind to GP in a similar fashion. This three-way type of mimicry demonstrates a unique way that the immune system has been able to take advantage of vulnerable sites on GP and mimic structures that ebolaviruses have evolved to avoid antibody recognition63.
Fusion peptide
Recent work has revealed the fusion peptide at the N-terminus of the membrane-proximal envelope glycoprotein as a common site of vulnerability64–66. This short, hydrophobic sequence is vital to the fusion process and must be liberated from its buried or partially sequestered location in the stalk or stem of glycoproteins to contact the host cell membrane. This makes it a difficult, but an ideal target for nAbs due to the necessary sequence conservation within a viral family. Structures of antibodies reactive with the fusion peptide have been shown for HIV66–68 (Figure 4D), influenza23,64,69,70 (Figure 4E) and ebolaviruses65,71–73 (Figure 4F). In each case, antibodies that engage the fusion peptide do so either directly or partially23 and are broadly reactive.
The glycoprotein stalk and membrane-proximal external region (MPER)
The viral stalk or stem emerges from the transmembrane anchor of glycoproteins, and contains the membrane fusion machinery. This domain is composed of the second of the two glycoproteins (HA2 for influenza virus, gp41 for HIV and GP2 for filoviruses) as well as the N and C-terminal regions of the first and membrane-distal glycoprotein (HA1 for influenza virus, gp120 for HIV and GP1 for filoviruses). This central functional role also makes it an important site of vulnerability that is often highly conserved in sequence. With the exception of HIV-1 Env, access to this region was at one time thought to be difficult or impossible, due to proximity to the hydrophobic viral membrane and spike density74,75. However, many potently neutralizing antibodies have now been discovered that can access this region for HIV (Figure 4G) and filoviruses (Figure 4I)73,76–81. The influenza stem is larger than in HIV and filoviruses and is now appreciated as a more common site of vulnerability than once thought (Figure 4H), prompting the pursuit of potential vaccine candidates that focus on this region80,82–88.
The MPER connects the heptad repeat region 2 (HR2) to the transmembrane region and is involved in the fusion machinery. Remarkably, HR2/MPER-directed mAbs for filoviruses contact epitopes nearly buried within the viral membrane (Figure 4I) and, in HIV-1, such mAbs have additionally evolved to interact with lipids in the membrane 89,90 or access transiently exposed hydrophobic residues76 (Figure 4G). There is not a well-defined MPER in influenza HA and consequently no antibodies to this specific region as for the other viruses is discussed here; however, the portion of HA that sits proximal to the membrane is referred to as the stem and does have spatially analogous regions to the MPER 91.
Virus-specific sites of vulnerability
In addition to the inherently common structural features that underlie all type-I viral glycoproteins, there are many unique features that can also serve as hotspots for eliciting potent antibodies. Determination of the structures of antibodies in complex with HIV Env, in particular, has uncovered some of the most unique examples40,92. The duration of an HIV infection allows for a long-term arms race to commence among the increasingly diverse viral population and the host immune system20,93–96. This is aided, in part, by an extensive array of glycans on the surface Env, which can help shield the underlying protein surface from immune surveillance. Strikingly though, this overabundance has given rise to antibodies that can bind directly and specifically to glycans that are highly conserved due to their high density that protects them from secondary processing 20,25,26,97–100. For example, antibody 2G12 exclusively recognizes immature high mannose glycans on HIV gp12027,101,102. While some glycan components of influenza and filoviral epitopes have been identified, they are not to the extent of HIV-directed mAbs.
HIV Env also houses many important epitopes that are buried within the central core of the viral spike, only exposed transiently through ‘conformational masking103, or exposed only after CD4 is bound30. At the apex of the HIV viral spike are several variable loops, including V1/V2 and V3, that can widely vary in sequence, but that change position after CD4 binding and participate in co-receptor binding11,12,30. These hidden epitopes are still accessible by antibodies, often through non-classical methods of antibody binding. For example, broadly neutralizing antibodies PG9 and PGT145 (Figure 4J) bind to the apex through very long CDRH3 loops (>30 residues) that can penetrate deep into recesses in the apex at the trimer axis. These Abs also bind asymmetrically where only a single Ab binds at the apex compared to most other HIV antibodies where three Fabs can be accommodated per trimer98,104. Some more recently identified antibodies can however bind to the Env apex with shorter CDRs and with a stoichiometry of up to two Fabs per trimer105,106. A common theme among such antibodies is the quaternary nature of their epitopes, requiring an intact trimeric spike and at least two protomers for binding, which is a mode of antibody binding that was not fully appreciated until the first structures of trimeric Env were determined11,12. To achieve such complicated acrobatics, these antibodies have often undergone extensive somatic hypermutation (SHM) that involves stabilizing mutations in framework regions, CDR loops that support the structure of the long inserting loops, such as CDR H3, in the paratope, and extended secondary structure within the CDR loops themselves. Conversely, some of the most potently neutralizing antibodies to influenza have far less SHM107–110 and typically much shorter CDRH3 loops, likely reflecting the much more acute nature of these infections and the possibility of first responder or “SOS-like” antibodies in the human immune repertoire that evolved as a rapid defensive against infection111,112.
On ebolaviruses, a structure referred to as the glycan cap, which covers the RBS, has been attributed to eliciting several neutralizing antibodies, despite its cleavage and removal upon entry62. The glycan cap and portions of the core GP are also shared with a second GP product known as sGP, which is secreted abundantly during infection113,114. Consequently, antibodies, such as 13C6, that react with the glycan cap are often cross-reactive to sGP115–117 (Figure 4K). Therefore, the mechanism behind how such antibodies neutralize ebolaviruses is not well understood, and marburgviruses do not produce sGP or have a defined glycan cap, rather leaving their RBS exposed on virions47.
Antibody allostery
Antibodies have also been shown to provide allosteric influence on glycoproteins, where binding in one location essentially alters a distal site. For example, HIV bnAb PGT151 binding to two sites on the HIV trimer induces asymmetry within the pliable Env structure, such that the third binding site on Env becomes inaccessible76. In a different but similar manner, antibodies have also been shown to exhibit cooperative binding, such as the binding of the Ebola virus (EBOV) antibodies FVM09 and m8C4, which do not offer effective neutralization or protection alone, but can potently neutralize EBOV in combination118. In this case, it is hypothesized that the binding of one alters the epitope of another, such that the epitope becomes more accessible.
Structures illuminate sites-of-vulnerability
The structures described here indicate that there are essentially no surfaces on viral glycoproteins that cannot be targeted by the adaptive immune response (Figure 5, Table S1). The diversity of antibody responses that occurs during infection demonstrate the extraordinary ability of the adaptive immune system to uniquely overcome viral obstacles. These studies provide valuable information that inform the development of next-generation therapeutics and vaccines that can mirror these activities.
Figure 5. The immunogenic landscape of enveloped viruses illuminated by structural biology.
Overlaying low-pass filtered structures of selected nAbs bound to trimeric glycoproteins from A, B) HIV (blues and purples), C, D) influenza (reds and oranges) and E, F) filoviruses (yellows and greens) reveals the immunogenic landscape of these viral glycoproteins and the 90 degrees of approach angles that antibodies can utilize to probe the glycoprotein surface. Abs bound to a single protomer are shown for clarity. Stripping back these antibodies clarifies how these epitopes are focused into distinct regions of vulnerability, although nearly the entire surface is susceptible to nAb binding. Several of these sites of vulnerability are equivalent across these examples, including the apex, RBS, interface of RBS and fusion domains, FP and viral stem, which includes HR1, HR2 and the MPER domains.
Implications for vaccine, therapeutic and diagnostic development
Structural data that have been amassed for enveloped viral glycoproteins in the past few decades have informed a more fundamental understanding of the complex viral lifecycle, but have also been used to directly and significantly advance efforts to generate and improve vaccines, therapeutics and diagnostics for these viruses. These efforts have been recently reviewed119; therefore, we will briefly highlight the most significant advances in each of these areas.
Efforts to generate an effective HIV-1 vaccine have been slow and challenging. Traditional routes to vaccination do not produce the bnAbs that are necessary to protect against the hugely diverse range of viral strains that are circulating in different parts of the world120–123. Failure has largely been fueled by the complex biology of HIV Env, which is highly glycosylated, is metastable, and undergoes larges structural changes during entry at the cell surface, and which can withstand large numbers of amino-acid substitutions within the so-called “hypervariable” regions124. To develop bnAbs, the adaptive response must overcome these obstacles through repeated exposure to a more diverse population of viruses, such as in long-term survivors93,125–127. Being relatively rare, it took many years to identify and isolate such bnAbs in reasonable numbers, and less than a handful were available in the 1990s128–131. Understanding how these mAbs recognize Env was further thwarted for many years by the lack of a stable and soluble form of Env that was suitable for structural studies. However, significant progress in antigen engineering resulted in the SOSIP antigen132–136 that enabled solving the first antibody-Env structures by X-ray crystallography and cryo-EM11,12.
These early results helped to shape next-generation of Env subunit vaccine designs, and have yielded two parallel efforts, exemplified mostly by the HIV and influenza fields. The first approach aims to present stable, idealized bnAb epitopes, wherein sites of vulnerability on the viral glycoprotein are presented to the immune system in an idealized way. This typically is achieved in one of two ways: 1) presentation of multiple epitopes on an intact, trimeric viral spike that is engineered for stability and that limits exposure of known non-neutralizing epitopes137–141 and 2) presentation of individual epitopes that are grafted onto protein scaffolds in order to immune-focus the antibody response to a particular site of vulnerability. Both of these approaches have advantages and disadvantages142. The epitope-focused approach has shown success as a proof-of-concept study with respiratory syncytial virus (RSV)142 and has since been expanding to show success in eliciting desired antibodies for HIV143–145, but has yet to show clinical success. However, epitope-focused vaccine designs have thus far not induced bnAbs146–148. Activity against the more complex and quaternary epitopes found on the intact, trimeric viral antigens can be lost or produce obstacles for antibodies elicited by monomeric designs. For HIV, more stabilized and engineered versions of a “native-like” trimer indeed induced stronger neutralizing antibodies titers149–151 but the responses generally lack breadth, only inducing antibodies that target the autologous immunogen. For influenza, the most broadly potent antibodies have been consistently mapped to the HA stalk. Therefore, an epitope focused approach has been much more successful, where only the HA stalk is presented to the immune system84,87,88,152–156 or presented in a chimeric form with different HA heads to focus the immune system on the conserved stem region152,153,156. In these cases, neutralizing responses can be relatively high and contain a level of breadth. One possible explanation is that the HA stalk has a larger degree of sequence conservation across the viral landscape than HIV Env, and less glycosylation.
The second approach to vaccines is to design immunogens that can stimulate specific precursor B-cells that can evolve into bnAbs. Thus, these immunogens target the non-somatic germline genes that first recombine to form the B-cell receptor (BCR) and aim to emulate the early stages of infection to initiate an immune response that has the potential to mature to breadth and potency over time143–145,149,157,158. This approach has arisen from analyzing structures of known bnAbs and engineering immunogens to bind germline versions of these bnAbs. Thus, vaccination occurs in stages, by priming bnAb precursors and then “guiding” these populations through subsequent vaccinations with immunogens that progressively drive somatic hypermutation toward bnAbs. The most advanced demonstration of these efforts has been shown by Schief and colleagues who have used state-of-the-art animal models and analytics, such as next generation sequencing (NGS), to study immune responses in great detail159,144,160,161. The germline targeting approach has two major obstacles. First, not all individuals necessarily make a high frequency, or any at all, of the naïve B-cell populations to be targeted162,163. Encouragingly though, some germline bnAb precursors have been found in naïve humans at low but sufficient frequency to initiate a response144,164,165. Indeed, as more bnAbs are found against particular epitopes, the frequency of the particular germlines in the naïve population can be taken into account in germline-targeting approaches. Additionally, longitudinal studies of the sequences and structures of bnAb evolution can provide clues as to how to better design immunogens that shepherd bnAb responses166–171. The second hurdle of this approach is that targeting human germline genes is difficult to test in animal models. Here, humanized and knock-in mice have provided a valuable tool for at least providing proof-of-concept for this approach172–174.
The B-cell origins of broadly neutralizing mAbs tend to be restricted to particular germline sequences. For example, the CD4 binding site bnAbs of HIV and the stem-directed bnAbs of influenza are often constrained to particular germline genes, namely VH1–2/VH1–4632,167,175 and VH1–69108, respectively. Interestingly, broadly potent VH1–69-derived nAbs are also found against a variety of other viral epitopes, including those found on ebolaviruses73,79,115,116 and HIV-132,167,176,177. This commonality does not have an obvious underlying reason other than the CDRH2 of VH1–69 antibodies has a hydrophobic tip that can insert into hydrophobic surfaces in viral antigens, particularly fusion domains23,81,108,109. The prevalence of VH1–69 does suggest a possible conserved immunological approach to targeting enveloped pathogens, which have evolved alongside humans for millennia111,112. Conversely, immune responses are often very diverse, particularly within a single patient, as observed in those infected by filoviruses24,115,116. In these cases, SHM tends to be low, and the polyclonal response may be more vital to overcoming more acute filoviral or seasonal influenza infections with no or limited antigenic variability. Conversely, effective responses to the hypervariable HIV reflect the longer exposure to the virus, where SHM is high within one or more predominant clonal lineages of antibodies. Influenza virus infection represents an acute infection but can occur in an individual multiple times over a lifetime through seasonal variants, and antibody responses to seasonal strains are a mixture of recall and de novo antibody responses85. Stem antibodies have greater promise for neutralization breadth due to conservation in the stem of HA, but are restricted to only a few antibody lineages, although not restricted to VH1–69. These lessons from natural infection provide important clues as to how to approach vaccine and therapeutic design, and can be tuned to the particular virus.
Vaccine and therapeutic development for filoviruses has experienced a surge in research due to a recent and unprecedented outbreak in Western Africa from 2013–2106178. Outbreaks of filovirus infection are relatively rare and most often isolated to sub-Sahara Africa; therefore, there was little information on the antibody-based response to exposure. Prior to the 2013 outbreak, there was evidence to suggest that neutralizing immune responses to infection were rare and that immunotherapy may not be possible or that vaccination may be difficult179. However, several studies that appeared during the last major outbreak showed that humanized mouse-derived mAbs, when used in combination, could provide protection. Two of these therapies, ZMab180 and MB-003181, were recombined into the tri-mAb cocktail ZMappTM,182, which demonstrated complete protection in non-human primates at late stages of infection183. Conducting trials of ZMapp™ were met with some difficulties due to the waning outbreak, and demonstration of protective efficacy in humans could not be fully substantiated184. One vaccine trial, however, did show great promise185 and there is evidence for long-term sustained protection in survivors of natural infection186. A new outbreak of EBOV in 2018 in the Democratic Republic of the Congo has seen the use of ZMapp™ as well as another tri-mAb cocktail from Regeneron 187 and as the single antibody mAb114188 and all three of these treatments are being evaluated in clinical trials in an effort to produce an FDA-approved treatment for EBOV infection.
Many new anti-Ebola Abs have been recently described24,65,71,79,115,118, including 26 new structures of antibodies/GP complexes that delineated major sites of vulnerability including the IFL, glycan cap, base, head and HR2 region of GP189. These studies demonstrate the polyclonal diversity of anti-filovirus antibodies. Additionally, several human-derived antibodies have been isolated that show broad cross-reactivity across ebolaviruses or marburgviruses and are actively being evaluated for use as immunotherapies. As for HIV and influenza, these bnAbs will likely be more restricted to particular germlines or sets of germlines. By understanding the mechanism of broadly neutralizing antibodies, it is hoped that this information will lead to pan-filoviral therapies that could replace the single species-focused ZMapp™, which would not be effective against the many other filoviral species that are pathogenic to humans.
Humanized mice offer an alternative platform to study for antibody reponses than human survivors of pandemic and epidemic viral infection, A recent study in which VelocImmune mice190 were vaccinated with either DNA-encoded or soluble EBOV GP (Makona variant) demonstrated that fully human IgGs could be produced in mice that bind to common sites of vulnerability on filoviruses that are targeted by the human immune system. Remarkably, these antibodies provide comparable protection to other therapies, including the ability to engage human Fcγ receptors, providing some level of protection through non-neutralizing means, similar to c13C6 from ZMappTM,183. This study shows that, in principal, these mice can be vaccinated against any pathogen to produce antibodies that may offer protection in humans, but on a much faster timeline, making them an attractive option to learn how to combat other emerging infectious diseases.
Future perspectives
The plethora of structural information that has been generated in the past decade has opened many doors for understanding how the adaptive immune system recognizes and neutralizes enveloped and other viruses, resulting in exciting new vaccine and antibody therapeutic development opportunities. One trait that is shared amongst nearly all neutralizing antibodies examined thus far and reviewed here is the restricted range of angles (90°) of approach to viral spikes (with respect to the viral membrane). The majority of neutralizing antibodies approach at a steeper angle nearly perpendicular to the membrane (PG9, PGT145, CAP256, CH01–4, VRC38, C05, 13C6), while others approach at a nearly parallel angle to the membrane (VRC01, FI6, CR8781, 10E8, KZ52, 4G7, ADI-16061). These allowable angles then likely reflect the window in which naïve B-cells receptors can successfully engage the viral membrane-bound antigen for the prolonged periods required for activation, with the viral membrane a providing some constraint and limiting angles beyond 90°. Upward angles are unfavorable for soluble IgGs as the membrane provides a steric constraint for approaching glycoproteins for binding141, although strain-specific neutralizing antibodies that approach GP at such an angle are not unheard of for filoviruses61,182,189. Additionally, the density of viral spikes on the surface of a virus can facilitate bivalent binding, which is possible for GP191 and HA38,51,52,192, but perhaps less so for HIV, where the spike density can be quite low75,193–195. In this case, engineering bivalent binding within a single antigen may be an effective approach to overcoming lower monovalent binding affinity to broadly reactive epitopes195,196.
The examples above demonstrate the near infinite capacity of the adaptive immune system to evolve in response to diverse antigenic insults that humans and animals encounter. The diverse epitopes targeted by acute ebolavirus infection115 demonstrate how a polyclonal antibody response with low SHM can be very effective, while the incredible breadth and potency of monoclonal bnAbs isolated from chronically infected HIV patients reveal the extremes of SHM that antibodies can accommodate to overcome huge antigenic diversity197–200. Despite this adaptive potential, the immune system still has a difficult time to keep pace with antigenically variable viruses like influenza and HIV that have high mutations rates94. Superficially, one would expect influenza to be an easier target for antibodies, particularly with yearly boosts via seasonal infection or vaccination. Yet, bnAbs are rare and typically do not persist and means that the world-wide human population is under constant threat of a new influenza pandemic. On the other hand, ebolaviruses may well be a relatively easy target for the adaptive immune system, but its spectacularly rapid pathogenesis normally results in mortality before effective antibodies can made. We sometimes take for granted the wonderful arsenal of vaccines that have already been developed, largely by empirical methods, and which result in lasting immunity with impressive potency. Of course, most of the viruses for which these vaccines are aimed at have little variability. Ironically, we have a relatively limited understanding of the sequence and molecular of antibody responses to historical vaccines and a much greater understanding of the antibody responses to pathogens that continue to outpace current vaccines. Even then, the monoclonal antibodies that have been successfully isolated and structurally characterized are almost certainly underrepresent the true diversity of immune responses. Thus, the pursuit of new antibodies, and therefore new pathways to bnAbs, remains highly valuable. Understanding polyclonal responses to large antigenic surfaces during infection/vaccination 29 also holds future promise for improving vaccines that increase on-and decrease off-target responses.
As noted above, structure-based design for HIV has already generated some encouraging results in animal models that demonstrate it is possible to drive the path of antibody evolution towards a neutralizing but not a broadly neutralizing response at present by vaccination with candidate immunogens144,145,159,164,165,173. Here, vaccine design increasingly benefits from a deeper understanding of the basic biological processes that happen in B-cell germinal centers165,201,202, as well as antigen display and uptake.
There is also renewed interest in the role of the innate immune pathways in antibody-based protection, as well as the role of non-neutralizing antibodies118,203–206. Fc-mediated protection has been shown to play at least some part in providing protection from all viruses discussed here. Even neutralizing antibodies have often been found to rely on some level of Fc-mediated function to realize their full potency207,208. While there is a basic understanding of the antibody-based innate immune response209–212, there is still much to be learned about the subtleties of the molecular nature of effector cell activation. Future studies to address the role of Fc-mediated protection in individuals that effectively control HIV replication, as well as in those that survive filovirus infection, will enhance vaccine and therapeutic research. A more detailed molecular understanding of the immune activation complex, and what type of antibody-antigen interaction results in a potent innate immune response, could help to improve antibody therapeutic selection and engineering. Further, these types of studies may provide information that will guide antibody designs that can specifically recruit effector cell subsets and immune responses, such as NK cells and ADCC, which have shown great promise in augmenting antibody neutralization213.
Based on the incredible advances described above, it is conceivable that, in the not so distant future, we will be able to rationally design vaccines that elicit antibodies with epitope specificity and broad antigen reactivity. An exciting new challenge will be the design of vaccines that also elicit antibodies that can also potently and specifically recruit desired effector functions. Integrating lessons from different viruses, including those described here and others, will continue to provide insights to arm researchers in their quest to vanquish the most formidable of pathogens.
Supplementary Material
Acknowledgements.
This work was supported by NIAID UM1AI100663 (Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery), U19 AI109762 (Viral Hemorrhagic Fever Immunotherapeutic Consortium), NIH grant R56 AI127371, and the Bill & Melinda Gates Collaboration for AIDS Vaccine Discovery (OPP1084519) and the International AIDS Vaccine Initiative. The authors declare no competing interests.
References
- 1.White JM & Whittaker GR Fusion of Enveloped Viruses in Endosomes. Traffic 17, 593–614, doi: 10.1111/tra.12389 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison SC Viral membrane fusion. Virology 479–480, 498–507, doi: 10.1016/j.virol.2015.03.043 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris LJ, Larson SB, Hasel KW & McPherson A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry 36, 1581–1597, doi: 10.1021/bi962514+ (1997). [DOI] [PubMed] [Google Scholar]
- 4.Harris LJ, Skaletsky E. & McPherson A. Crystallographic structure of an intact IgG1 monoclonal antibody. J Mol Biol 275, 861–872, doi: 10.1006/jmbi.1997.1508 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Saphire EO et al. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293, 1155–1159, doi: 10.1126/science.1061692 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Scapin G. et al. Structure of full-length human anti-PD1 therapeutic IgG4 antibody pembrolizumab. Nat Struct Mol Biol 22, 953–958, doi: 10.1038/nsmb.3129 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Stanfield RL & Wilson IA Antibody Structure. Microbiol Spectr 2, doi: 10.1128/microbiolspec.AID-0012-2013 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Wilson IA & Stanfield RL Antibody-antigen interactions: new structures and new conformational changes. Curr Opin Struct Biol 4, 857–867 (1994). [DOI] [PubMed] [Google Scholar]
- 9.Bullough PA, Hughson FM, Skehel JJ & Wiley DC Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43, doi: 10.1038/371037a0 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Wilson IA, Skehel JJ & Wiley DC Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289, 366–373 (1981). [DOI] [PubMed] [Google Scholar]
- 11.Julien JP et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science 342, 1477–1483, doi: 10.1126/science.1245625 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lyumkis D. et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science 342, 1484–1490, doi: 10.1126/science.1245627 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan DC, Fass D, Berger JM & Kim PS Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 (1997). [DOI] [PubMed] [Google Scholar]
- 14.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ & Wiley DC Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430, doi: 10.1038/387426a0 (1997). [DOI] [PubMed] [Google Scholar]
- 15.Lee JE et al. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 454, 177–182, doi: 10.1038/nature07082 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weissenhorn W, Carfi A, Lee KH, Skehel JJ & Wiley DC Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell 2, 605–616 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Cook JD & Lee JE The secret life of viral entry glycoproteins: moonlighting in immune evasion. PLoS Pathog 9, e1003258, doi: 10.1371/journal.ppat.1003258 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tate MD et al. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 6, 1294–1316, doi: 10.3390/v6031294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu NC & Wilson IA A Perspective on the Structural and Functional Constraints for Immune Evasion: Insights from Influenza Virus. J Mol Biol 429, 2694–2709, doi: 10.1016/j.jmb.2017.06.015 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crispin M, Ward AB & Wilson IA Structure and Immune Recognition of the HIV Glycan Shield. Annu Rev Biophys, doi: 10.1146/annurev-biophys-060414-034156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao L. et al. Global site-specific N-glycosylation analysis of HIV envelope glycoprotein. Nat Commun 8, 14954, doi: 10.1038/ncomms14954 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, de Val N, Lyumkis D. & Ward AB Model Building and Refinement of a Natively Glycosylated HIV-1 Env Protein by High-Resolution Cryoelectron Microscopy. Structure 23, 1943–1951, doi: 10.1016/j.str.2015.07.020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ekiert DC et al. Antibody recognition of a highly conserved influenza virus epitope. Science 324, 246–251, doi: 10.1126/science.1171491 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flyak AI et al. Mechanism of human antibody-mediated neutralization of Marburg virus. Cell 160, 893–903, doi: 10.1016/j.cell.2015.01.031 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Julien JP et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A 110, 4351–4356, doi: 10.1073/pnas.1217537110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong L. et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol 20, 796–803, doi: 10.1038/nsmb.2594 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murin CD et al. Structure of 2G12 Fab2 in complex with soluble and fully glycosylated HIV-1 Env by negative-stain single-particle electron microscopy. J Virol 88, 10177–10188, doi: 10.1128/JVI.01229-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams KL et al. Superinfection Drives HIV Neutralizing Antibody Responses from Several B Cell Lineages that Contribute to a Polyclonal Repertoire. Cell Rep 23, 682–691, doi: 10.1016/j.celrep.2018.03.082 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bianchi M. et al. Electron-Microscopy-Based Epitope Mapping Defines Specificities of Polyclonal Antibodies Elicited during HIV-1 BG505 Envelope Trimer Immunization. Immunity 49, 288–300 e288, doi: 10.1016/j.immuni.2018.07.009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ozorowski G. et al. Open and closed structures reveal allostery and pliability in the HIV-1 envelope spike. Nature 547, 360–363, doi: 10.1038/nature23010 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Georgiev IS, Gordon Joyce M, Zhou T. & Kwong PD Elicitation of HIV-1-neutralizing antibodies against the CD4-binding site. Curr Opin HIV AIDS 8, 382–392, doi: 10.1097/COH.0b013e328363a90e (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou T. et al. Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors. Cell 161, 1280–1292, doi: 10.1016/j.cell.2015.05.007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu R. et al. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol 20, 363–370, doi: 10.1038/nsmb.2500 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Sastre A. Influenza virus receptor specificity: disease and transmission. Am J Pathol 176, 1584–1585, doi: 10.2353/ajpath.2010.100066 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens J. et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312, 404–410, doi: 10.1126/science.1124513 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Wu NC et al. A complex epistatic network limits the mutational reversibility in the influenza hemagglutinin receptor-binding site. Nat Commun 9, 1264, doi: 10.1038/s41467-018-03663-5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu NC et al. Diversity of Functionally Permissive Sequences in the Receptor-Binding Site of Influenza Hemagglutinin. Cell Host Microbe 22, 247–248, doi: 10.1016/j.chom.2017.07.001 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee PS et al. Receptor mimicry by antibody F045–092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 5, 3614, doi: 10.1038/ncomms4614 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu NC et al. In vitro evolution of an influenza broadly neutralizing antibody is modulated by hemagglutinin receptor specificity. Nat Commun 8, 15371, doi: 10.1038/ncomms15371 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Julien JP, Lee PS & Wilson IA Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA. Immunol Rev 250, 180–198, doi: 10.1111/imr.12005 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laursen NS & Wilson IA Broadly neutralizing antibodies against influenza viruses. Antiviral Res 98, 476–483, doi: 10.1016/j.antiviral.2013.03.021 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee PS & Wilson IA Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr Top Microbiol Immunol 386, 323–341, doi: 10.1007/82_2014_413 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bornholdt ZA et al. Host-Primed Ebola Virus GP Exposes a Hydrophobic NPC1 Receptor-Binding Pocket, Revealing a Target for Broadly Neutralizing Antibodies. MBio 7, e02154–02115, doi: 10.1128/mBio.02154-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandran K, Sullivan NJ, Felbor U, Whelan SP & Cunningham JM Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 308, 1643–1645, doi: 10.1126/science.1110656 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miller EH et al. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J 31, 1947–1960, doi: 10.1038/emboj.2012.53 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gnirss K. et al. Cathepsins B and L activate Ebola but not Marburg virus glycoproteins for efficient entry into cell lines and macrophages independent of TMPRSS2 expression. Virology 424, 3–10, doi: 10.1016/j.virol.2011.11.031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.King LB et al. The Marburgvirus-Neutralizing Human Monoclonal Antibody MR191 Targets a Conserved Site to Block Virus Receptor Binding. Cell Host Microbe 23, 101–109 e104, doi: 10.1016/j.chom.2017.12.003 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hashiguchi T. et al. Structural basis for Marburg virus neutralization by a cross-reactive human antibody. Cell 160, 904–912, doi: 10.1016/j.cell.2015.01.041 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang H. et al. Ebola Viral Glycoprotein Bound to Its Endosomal Receptor Niemann-Pick C1. Cell 164, 258–268, doi: 10.1016/j.cell.2015.12.044 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin YP et al. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci U S A 109, 21474–21479, doi: 10.1073/pnas.1218841110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ekiert DC et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489, 526–532, doi: 10.1038/nature11414 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee PS et al. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci U S A 109, 17040–17045, doi: 10.1073/pnas.1212371109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whittle JR et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci U S A 108, 14216–14221, doi: 10.1073/pnas.1111497108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt AG et al. Viral receptor-binding site antibodies with diverse germline origins. Cell 161, 1026–1034, doi: 10.1016/j.cell.2015.04.028 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kwong PD et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393, 648–659, doi: 10.1038/31405 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garrett TP, Wang J, Yan Y, Liu J. & Harrison SC Refinement and analysis of the structure of the first two domains of human CD4. J Mol Biol 234, 763–778, doi: 10.1006/jmbi.1993.1625 (1993). [DOI] [PubMed] [Google Scholar]
- 57.Wu X. et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science 329, 856–861, doi: 10.1126/science.1187659 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou T. et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science 329, 811–817, doi: 10.1126/science.1192819 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou T. et al. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445, 732–737, doi: 10.1038/nature05580 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bale S. et al. Structural basis for differential neutralization of ebolaviruses. Viruses 4, 447–470, doi: 10.3390/v4040447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dias JM et al. A shared structural solution for neutralizing ebolaviruses. Nat Struct Mol Biol 18, 1424–1427, doi: 10.1038/nsmb.2150 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JE & Saphire EO Ebolavirus glycoprotein structure and mechanism of entry. Future Virol 4, 621–635, doi: 10.2217/fvl.09.56 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crowe JE Jr. Principles of Broad and Potent Antiviral Human Antibodies: Insights for Vaccine Design. Cell Host Microbe 22, 193–206, doi: 10.1016/j.chom.2017.07.013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hashem AM et al. Universal antibodies against the highly conserved influenza fusion peptide cross-neutralize several subtypes of influenza A virus. Biochem Biophys Res Commun 403, 247–251, doi: 10.1016/j.bbrc.2010.11.030 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Murin CD, Bruhn JF, Bornholdt ZA, Copps J, Stanfield R, Ward AB Structural basis of pan-ebolavirus neutralization by an antibody targeting the glycoprotein fusion loop. Cell Rep 24, 2723–2732, doi: 10.1016/j.celrep.2018.08.009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Gils MJ et al. An HIV-1 antibody from an elite neutralizer implicates the fusion peptide as a site of vulnerability. Nat Microbiol 2, 16199, doi: 10.1038/nmicrobiol.2016.199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kong R. et al. Fusion peptide of HIV-1 as a site of vulnerability to neutralizing antibody. Science 352, 828–833, doi: 10.1126/science.aae0474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu K. et al. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat Med 24, 857–867, doi: 10.1038/s41591-018-0042-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kallewaard NL et al. Structure and Function Analysis of an Antibody Recognizing All Influenza A Subtypes. Cell 166, 596–608, doi: 10.1016/j.cell.2016.05.073 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prabhu N. et al. Monoclonal antibodies against the fusion peptide of hemagglutinin protect mice from lethal influenza A virus H5N1 infection. J Virol 83, 2553–2562, doi: 10.1128/JVI.02165-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao X. et al. Immunization-Elicited Broadly Protective Antibody Reveals Ebolavirus Fusion Loop as a Site of Vulnerability. Cell 169, 891–904 e815, doi: 10.1016/j.cell.2017.04.038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Furuyama W. et al. Discovery of an antibody for pan-ebolavirus therapy. Sci Rep 6, 20514, doi: 10.1038/srep20514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wec AZ et al. Antibodies from a Human Survivor Define Sites of Vulnerability for Broad Protection against Ebolaviruses. Cell 169, 878–890 e815, doi: 10.1016/j.cell.2017.04.037 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu P. et al. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci U S A 100, 15812–15817, doi: 10.1073/pnas.2634931100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu P. et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852, doi: 10.1038/nature04817 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Lee JH, Ozorowski G. & Ward AB Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351, 1043–1048, doi: 10.1126/science.aad2450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang J. et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature 491, 406–412, doi: 10.1038/nature11544 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cardoso RM et al. Broadly neutralizing anti-HIV antibody 4E10 recognizes a helical conformation of a highly conserved fusion-associated motif in gp41. Immunity 22, 163–173, doi: 10.1016/j.immuni.2004.12.011 (2005). [DOI] [PubMed] [Google Scholar]
- 79.Flyak AI et al. Broadly neutralizing antibodies from human survivors target a conserved site in the Ebola virus glycoprotein HR2-MPER region. Nat Microbiol, doi: 10.1038/s41564-018-0157-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krammer F. & Palese P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 3, 521–530, doi: 10.1016/j.coviro.2013.07.007 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dreyfus C, Ekiert DC & Wilson IA Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J Virol 87, 7149–7154, doi: 10.1128/JVI.02975-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khanna M, Sharma S, Kumar B. & Rajput R. Protective immunity based on the conserved hemagglutinin stalk domain and its prospects for universal influenza vaccine development. Biomed Res Int 2014, 546274, doi: 10.1155/2014/546274 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Erbelding EJ et al. A Universal Influenza Vaccine: The Strategic Plan for the National Institute of Allergy and Infectious Diseases. J Infect Dis, doi: 10.1093/infdis/jiy103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Impagliazzo A. et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349, 1301–1306, doi: 10.1126/science.aac7263 (2015). [DOI] [PubMed] [Google Scholar]
- 85.Krammer F, Garcia-Sastre A. & Palese P. Is It Possible to Develop a “Universal” Influenza Virus Vaccine? Potential Target Antigens and Critical Aspects for a Universal Influenza Vaccine. Cold Spring Harb Perspect Biol 10, doi: 10.1101/cshperspect.a028845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krammer F, Pica N, Hai R, Margine I. & Palese P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 87, 6542–6550, doi: 10.1128/JVI.00641-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nachbagauer R. et al. A universal influenza virus vaccine candidate confers protection against pandemic H1N1 infection in preclinical ferret studies. NPJ Vaccines 2, 26, doi: 10.1038/s41541-017-0026-4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yassine HM et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21, 1065–1070, doi: 10.1038/nm.3927 (2015). [DOI] [PubMed] [Google Scholar]
- 89.Irimia A. et al. Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design. PLoS Pathog 13, e1006212, doi: 10.1371/journal.ppat.1006212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Irimia A, Sarkar A, Stanfield RL & Wilson IA Crystallographic Identification of Lipid as an Integral Component of the Epitope of HIV Broadly Neutralizing Antibody 4E10. Immunity 44, 21–31, doi: 10.1016/j.immuni.2015.12.001 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Friesen RH et al. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci U S A 111, 445–450, doi: 10.1073/pnas.1319058110 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burton DR Antibodies, viruses and vaccines. Nat Rev Immunol 2, 706–713, doi: 10.1038/nri891 (2002). [DOI] [PubMed] [Google Scholar]
- 93.Burton DR & Mascola JR Antibody responses to envelope glycoproteins in HIV-1 infection. Nat Immunol 16, 571–576, doi: 10.1038/ni.3158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Burton DR, Poignard P, Stanfield RL & Wilson IA Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science 337, 183–186, doi: 10.1126/science.1225416 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Burton DR, Stanfield RL & Wilson IA Antibody vs. HIV in a clash of evolutionary titans. Proc Natl Acad Sci U S A 102, 14943–14948, doi: 10.1073/pnas.0505126102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Caskey M, Klein F. & Nussenzweig MC Broadly Neutralizing Antibodies for HIV-1 Prevention or Immunotherapy. N Engl J Med 375, 2019–2021, doi: 10.1056/NEJMp1613362 (2016). [DOI] [PubMed] [Google Scholar]
- 97.Mouquet H. et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proc Natl Acad Sci U S A 109, E3268–3277, doi: 10.1073/pnas.1217207109 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.McLellan JS et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480, 336–343, doi: 10.1038/nature10696 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Julien JP et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS Pathog 9, e1003342, doi: 10.1371/journal.ppat.1003342 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pejchal R. et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science 334, 1097–1103, doi: 10.1126/science.1213256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Calarese DA et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300, 2065–2071, doi: 10.1126/science.1083182 (2003). [DOI] [PubMed] [Google Scholar]
- 102.Stanfield RL, De Castro C, Marzaioli AM, Wilson IA & Pantophlet R. Crystal structure of the HIV neutralizing antibody 2G12 in complex with a bacterial oligosaccharide analog of mammalian oligomannose. Glycobiology 25, 412–419, doi: 10.1093/glycob/cwu123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kwong PD et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420, 678–682, doi: 10.1038/nature01188 (2002). [DOI] [PubMed] [Google Scholar]
- 104.Lee JH et al. A Broadly Neutralizing Antibody Targets the Dynamic HIV Envelope Trimer Apex via a Long, Rigidified, and Anionic beta-Hairpin Structure. Immunity 46, 690–702, doi: 10.1016/j.immuni.2017.03.017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cale EM et al. Virus-like Particles Identify an HIV V1V2 Apex-Binding Neutralizing Antibody that Lacks a Protruding Loop. Immunity 46, 777–791 e710, doi: 10.1016/j.immuni.2017.04.011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang H. et al. Asymmetric recognition of HIV-1 Envelope trimer by V1V2 loop-targeting antibodies. Elife 6, doi: 10.7554/eLife.27389 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kashyap AK et al. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci U S A 105, 5986–5991, doi: 10.1073/pnas.0801367105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lang S. et al. Antibody 27F3 Broadly Targets Influenza A Group 1 and 2 Hemagglutinins through a Further Variation in VH1–69 Antibody Orientation on the HA Stem. Cell Rep 20, 2935–2943, doi: 10.1016/j.celrep.2017.08.084 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sui J. et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16, 265–273, doi: 10.1038/nsmb.1566 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Throsby M. et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS One 3, e3942, doi: 10.1371/journal.pone.0003942 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lerner RA Rare antibodies from combinatorial libraries suggests an S.O.S. component of the human immunological repertoire. Mol Biosyst 7, 1004–1012, doi: 10.1039/c0mb00310g (2011). [DOI] [PubMed] [Google Scholar]
- 112.Lerner RA Combinatorial antibody libraries: new advances, new immunological insights. Nat Rev Immunol 16, 498–508, doi: 10.1038/nri.2016.67 (2016). [DOI] [PubMed] [Google Scholar]
- 113.Pallesen J. et al. Structures of Ebola virus GP and sGP in complex with therapeutic antibodies. Nat Microbiol 1, 16128, doi: 10.1038/nmicrobiol.2016.128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.de La Vega MA, Wong G, Kobinger GP & Qiu X. The multiple roles of sGP in Ebola pathogenesis. Viral Immunol 28, 3–9, doi: 10.1089/vim.2014.0068 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bornholdt ZA et al. Isolation of potent neutralizing antibodies from a survivor of the 2014 Ebola virus outbreak. Science 351, 1078–1083, doi: 10.1126/science.aad5788 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Flyak AI et al. Cross-Reactive and Potent Neutralizing Antibody Responses in Human Survivors of Natural Ebolavirus Infection. Cell 164, 392–405, doi: 10.1016/j.cell.2015.12.022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wilson JA et al. Epitopes involved in antibody-mediated protection from Ebola virus. Science 287, 1664–1666 (2000). [DOI] [PubMed] [Google Scholar]
- 118.Howell KA et al. Cooperativity Enables Non-neutralizing Antibodies to Neutralize Ebolavirus. Cell Rep 19, 413–424, doi: 10.1016/j.celrep.2017.03.049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kwong PD & Mascola JR HIV-1 Vaccines Based on Antibody Identification, B Cell Ontogeny, and Epitope Structure. Immunity 48, 855–871, doi: 10.1016/j.immuni.2018.04.029 (2018). [DOI] [PubMed] [Google Scholar]
- 120.Flynn NM et al. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis 191, 654–665, doi: 10.1086/428404 (2005). [DOI] [PubMed] [Google Scholar]
- 121.Pitisuttithum P. et al. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis 194, 1661–1671, doi: 10.1086/508748 (2006). [DOI] [PubMed] [Google Scholar]
- 122.Dolin R. et al. The safety and immunogenicity of a human immunodeficiency virus type 1 (HIV-1) recombinant gp160 candidate vaccine in humans. NIAID AIDS Vaccine Clinical Trials Network. Ann Intern Med 114, 119–127 (1991). [DOI] [PubMed] [Google Scholar]
- 123.Cooney EL et al. Safety of and immunological response to a recombinant vaccinia virus vaccine expressing HIV envelope glycoprotein. Lancet 337, 567–572 (1991). [DOI] [PubMed] [Google Scholar]
- 124.Burton DR et al. HIV vaccine design and the neutralizing antibody problem. Nat Immunol 5, 233–236, doi: 10.1038/ni0304-233 (2004). [DOI] [PubMed] [Google Scholar]
- 125.Gonzalez N. et al. Characterization of broadly neutralizing antibody responses to HIV-1 in a cohort of long term non-progressors. PLoS One 13, e0193773, doi: 10.1371/journal.pone.0193773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gray ES et al. The neutralization breadth of HIV-1 develops incrementally over four years and is associated with CD4+ T cell decline and high viral load during acute infection. J Virol 85, 4828–4840, doi: 10.1128/JVI.00198-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kelsoe G. & Haynes BF Host controls of HIV broadly neutralizing antibody development. Immunol Rev 275, 79–88, doi: 10.1111/imr.12508 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Burton DR et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266, 1024–1027 (1994). [DOI] [PubMed] [Google Scholar]
- 129.Muster T. et al. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J Virol 67, 6642–6647 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Trkola A. et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J Virol 70, 1100–1108 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Buchacher A. et al. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res Hum Retroviruses 10, 359–369, doi: 10.1089/aid.1994.10.359 (1994). [DOI] [PubMed] [Google Scholar]
- 132.Binley JM et al. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol 74, 627–643 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sanders RW et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol 76, 8875–8889 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sanders RW et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog 9, e1003618, doi: 10.1371/journal.ppat.1003618 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sanders RW & Moore JP Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol Rev 275, 161–182, doi: 10.1111/imr.12481 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sanders RW, Wilson IA & Moore JP HIV’s Achilles’ Heel. Sci Am 315, 50–55, doi: 10.1038/scientificamerican1216-50 (2016). [DOI] [PubMed] [Google Scholar]
- 137.Torrents de la Pena A. et al. Improving the Immunogenicity of Native-like HIV-1 Envelope Trimers by Hyperstabilization. Cell Rep 20, 1805–1817, doi: 10.1016/j.celrep.2017.07.077 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Voss JE et al. Elicitation of Neutralizing Antibodies Targeting the V2 Apex of the HIV Envelope Trimer in a Wild-Type Animal Model. Cell Rep 21, 222–235, doi: 10.1016/j.celrep.2017.09.024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.de Taeye SW et al. Immunogenicity of Stabilized HIV-1 Envelope Trimers with Reduced Exposure of Non-neutralizing Epitopes. Cell 163, 1702–1715, doi: 10.1016/j.cell.2015.11.056 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kulp DW & Schief WR Advances in structure-based vaccine design. Curr Opin Virol 3, 322–331, doi: 10.1016/j.coviro.2013.05.010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kulp DW et al. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat Commun 8, 1655, doi: 10.1038/s41467-017-01549-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Correia BE et al. Proof of principle for epitope-focused vaccine design. Nature 507, 201–206, doi: 10.1038/nature12966 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jardine J. et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716, doi: 10.1126/science.1234150 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jardine JG et al. HIV-1 broadly neutralizing antibody precursor B cells revealed by germline-targeting immunogen. Science 351, 1458–1463, doi: 10.1126/science.aad9195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Jardine JG et al. HIV-1 VACCINES. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349, 156–161, doi: 10.1126/science.aac5894 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Correia BE et al. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure 18, 1116–1126, doi: 10.1016/j.str.2010.06.010 (2010). [DOI] [PubMed] [Google Scholar]
- 147.McLellan JS et al. Design and characterization of epitope-scaffold immunogens that present the motavizumab epitope from respiratory syncytial virus. J Mol Biol 409, 853–866, doi: 10.1016/j.jmb.2011.04.044 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ofek G. et al. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci U S A 107, 17880–17887, doi: 10.1073/pnas.1004728107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.de Taeye SW, Moore JP & Sanders RW HIV-1 Envelope Trimer Design and Immunization Strategies To Induce Broadly Neutralizing Antibodies. Trends Immunol 37, 221–232, doi: 10.1016/j.it.2016.01.007 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sanders RW et al. HIV-1 VACCINES. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349, aac4223, doi: 10.1126/science.aac4223 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pauthner M. et al. Elicitation of Robust Tier 2 Neutralizing Antibody Responses in Nonhuman Primates by HIV Envelope Trimer Immunization Using Optimized Approaches. Immunity 46, 1073–1088 e1076, doi: 10.1016/j.immuni.2017.05.007 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Krammer F. et al. Assessment of influenza virus hemagglutinin stalk-based immunity in ferrets. J Virol 88, 3432–3442, doi: 10.1128/JVI.03004-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Krammer F. et al. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J Virol 88, 2340–2343, doi: 10.1128/JVI.03183-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mallajosyula VV et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci U S A 111, E2514–2523, doi: 10.1073/pnas.1402766111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wohlbold TJ et al. Vaccination with soluble headless hemagglutinin protects mice from challenge with divergent influenza viruses. Vaccine 33, 3314–3321, doi: 10.1016/j.vaccine.2015.05.038 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hai R. et al. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol 86, 5774–5781, doi: 10.1128/JVI.00137-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Goo L, Chohan V, Nduati R. & Overbaugh J. Early development of broadly neutralizing antibodies in HIV-1-infected infants. Nat Med 20, 655–658, doi: 10.1038/nm.3565 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.van den Kerkhof TL et al. Early development of broadly reactive HIV-1 neutralizing activity in elite neutralizers. AIDS 28, 1237–1240, doi: 10.1097/QAD.0000000000000228 (2014). [DOI] [PubMed] [Google Scholar]
- 159.Sok D. et al. Priming HIV-1 broadly neutralizing antibody precursors in human Ig loci transgenic mice. Science 353, 1557–1560, doi: 10.1126/science.aah3945 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Havenar-Daughton C. et al. The human naive B cell repertoire contains distinct subclasses for a germline-targeting HIV-1 vaccine immunogen. Sci Transl Med 10, doi: 10.1126/scitranslmed.aat0381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Havenar-Daughton C, Abbott RK, Schief WR & Crotty S. When designing vaccines, consider the starting material: the human B cell repertoire. Curr Opin Immunol 53, 209–216, doi: 10.1016/j.coi.2018.08.002 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Arnaout R. et al. High-resolution description of antibody heavy-chain repertoires in humans. PLoS One 6, e22365, doi: 10.1371/journal.pone.0022365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.DeKosky BJ et al. In-depth determination and analysis of the human paired heavy-and light-chain antibody repertoire. Nat Med 21, 86–91, doi: 10.1038/nm.3743 (2015). [DOI] [PubMed] [Google Scholar]
- 164.Abbott RK et al. Precursor Frequency and Affinity Determine B Cell Competitive Fitness in Germinal Centers, Tested with Germline-Targeting HIV Vaccine Immunogens. Immunity 48, 133–146 e136, doi: 10.1016/j.immuni.2017.11.023 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Havenar-Daughton C. et al. Direct Probing of Germinal Center Responses Reveals Immunological Features and Bottlenecks for Neutralizing Antibody Responses to HIV Env Trimer. Cell Rep 17, 2195–2209, doi: 10.1016/j.celrep.2016.10.085 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Doria-Rose NA et al. Developmental pathway for potent V1V2-directed HIV-neutralizing antibodies. Nature 509, 55–62, doi: 10.1038/nature13036 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bonsignori M. et al. Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody. Cell 165, 449–463, doi: 10.1016/j.cell.2016.02.022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rantalainen K. et al. Co-evolution of HIV Envelope and Apex-Targeting Neutralizing Antibody Lineage Provides Benchmarks for Vaccine Design. Cell Rep 23, 3249–3261, doi: 10.1016/j.celrep.2018.05.046 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Landais E. et al. Broadly Neutralizing Antibody Responses in a Large Longitudinal Sub-Saharan HIV Primary Infection Cohort. PLoS Pathog 12, e1005369, doi: 10.1371/journal.ppat.1005369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Landais E. et al. HIV Envelope Glycoform Heterogeneity and Localized Diversity Govern the Initiation and Maturation of a V2 Apex Broadly Neutralizing Antibody Lineage. Immunity 47, 990–1003 e1009, doi: 10.1016/j.immuni.2017.11.002 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Doria-Rose NA et al. New Member of the V1V2-Directed CAP256-VRC26 Lineage That Shows Increased Breadth and Exceptional Potency. J Virol 90, 76–91, doi: 10.1128/JVI.01791-15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Dosenovic P. et al. Immunization for HIV-1 Broadly Neutralizing Antibodies in Human Ig Knockin Mice. Cell 161, 1505–1515, doi: 10.1016/j.cell.2015.06.003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Escolano A. et al. Sequential Immunization Elicits Broadly Neutralizing Anti-HIV-1 Antibodies in Ig Knockin Mice. Cell 166, 1445–1458 e1412, doi: 10.1016/j.cell.2016.07.030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lee EC et al. Complete humanization of the mouse immunoglobulin loci enables efficient therapeutic antibody discovery. Nature Biotechnology 32, 356–U219, doi: 10.1038/nbt.2825 (2014). [DOI] [PubMed] [Google Scholar]
- 175.Acharya P. et al. CD4-binding-Site Recognition by VH1–46 Germline-derived HIV-1 Neutralizers. Aids Res Hum Retrov 30, A120–A121, doi:DOI 10.1089/aid.2014.5231.abstract (2014). [DOI] [Google Scholar]
- 176.Gorny MK et al. Preferential use of the VH5–51 gene segment by the human immune response to code for antibodies against the V3 domain of HIV-1. Mol Immunol 46, 917–926, doi: 10.1016/j.molimm.2008.09.005 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Huang CC et al. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc Natl Acad Sci U S A 101, 2706–2711, doi: 10.1073/pnas.0308527100 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Baize S. et al. Emergence of Zaire Ebola virus disease in Guinea. N Engl J Med 371, 1418–1425, doi: 10.1056/NEJMoa1404505 (2014). [DOI] [PubMed] [Google Scholar]
- 179.Oswald WB et al. Neutralizing antibody fails to impact the course of Ebola virus infection in monkeys. PLoS Pathog 3, e9, doi: 10.1371/journal.ppat.0030009 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Qiu X. et al. Sustained protection against Ebola virus infection following treatment of infected nonhuman primates with ZMAb. Sci Rep 3, 3365, doi: 10.1038/srep03365 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Pettitt J. et al. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Sci Transl Med 5, 199ra113, doi: 10.1126/scitranslmed.3006608 (2013). [DOI] [PubMed] [Google Scholar]
- 182.Murin CD et al. Structures of protective antibodies reveal sites of vulnerability on Ebola virus. Proc Natl Acad Sci U S A 111, 17182–17187, doi: 10.1073/pnas.1414164111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Qiu X. et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 514, 47–53, doi: 10.1038/nature13777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Group PIW et al. A Randomized, Controlled Trial of ZMapp for Ebola Virus Infection. N Engl J Med 375, 1448–1456, doi: 10.1056/NEJMoa1604330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Henao-Restrepo AM et al. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ca Suffit!). Lancet 389, 505–518, doi: 10.1016/S0140-6736(16)32621-6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rimoin AW et al. Ebola Virus Neutralizing Antibodies Detectable in Survivors of theYambuku, Zaire Outbreak 40 Years after Infection. J Infect Dis 217, 223–231, doi: 10.1093/infdis/jix584 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pascal KE et al. Development of clinical-stage human monoclonal antibodies that treat advanced Ebola virus disease in non-human primates. J Infect Dis, doi: 10.1093/infdis/jiy285 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Misasi J. et al. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J Virol 86, 3284–3292, doi: 10.1128/JVI.06346-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Saphire EO et al. Systematic Analysis of Monoclonal Antibodies against Ebola Virus GP Defines Features that Contribute to Protection. Cell 174, 938–952 e913, doi: 10.1016/j.cell.2018.07.033 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Murphy AJ et al. Mice with megabase humanization of their immunoglobulin genes generate antibodies as efficiently as normal mice. P Natl Acad Sci USA 111, 5153–5158, doi: 10.1073/pnas.1324022111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Beniac DR et al. The organisation of Ebola virus reveals a capacity for extensive, modular polyploidy. PLoS One 7, e29608, doi: 10.1371/journal.pone.0029608 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Harris A. et al. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci U S A 103, 19123–19127, doi: 10.1073/pnas.0607614103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Liu J, Bartesaghi A, Borgnia MJ, Sapiro G. & Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113, doi: 10.1038/nature07159 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Liu J, Wright ER & Winkler H. 3D visualization of HIV virions by cryoelectron tomography. Methods Enzymol 483, 267–290, doi: 10.1016/S0076-6879(10)83014-9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Klein JS & Bjorkman PJ Few and Far Between: How HIV May Be Evading Antibody Avidity. Plos Pathogens 6, doi:ARTN e1000908 10.1371/journal.ppat.1000908 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Galimidi RP et al. Intra-spike crosslinking overcomes antibody evasion by HIV-1. Cell 160, 433–446, doi: 10.1016/j.cell.2015.01.016 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Walker LM et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326, 285–289, doi: 10.1126/science.1178746 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Scheid JF et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458, 636–640, doi: 10.1038/nature07930 (2009). [DOI] [PubMed] [Google Scholar]
- 199.Simek MD et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J Virol 83, 7337–7348, doi: 10.1128/JVI.00110-09 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Walker LM et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477, 466–470, doi: 10.1038/nature10373 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Cirelli KM & Crotty S. Germinal center enhancement by extended antigen availability. Curr Opin Immunol 47, 64–69, doi: 10.1016/j.coi.2017.06.008 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Crotty S. Raging evolution of a B cell response to a viral infection. Nat Rev Immunol 18, 79, doi: 10.1038/nri.2017.219 (2018). [DOI] [PubMed] [Google Scholar]
- 203.Horwitz JA et al. Non-neutralizing Antibodies Alter the Course of HIV-1 Infection In Vivo. Cell 170, 637–648 e610, doi: 10.1016/j.cell.2017.06.048 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Mayr L, Su B. & Moog C. Role of nonneutralizing antibodies in vaccines and/or HIV infected individuals. Curr Opin HIV AIDS 12, 209–215, doi: 10.1097/COH.0000000000000357 (2017). [DOI] [PubMed] [Google Scholar]
- 205.Forthal D, Hope TJ & Alter G. New paradigms for functional HIV-specific nonneutralizing antibodies. Curr Opin HIV AIDS 8, 393–401, doi: 10.1097/COH.0b013e328363d486 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kajihara M. et al. Inhibition of Marburg virus budding by nonneutralizing antibodies to the envelope glycoprotein. J Virol 86, 13467–13474, doi: 10.1128/JVI.01896-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Bournazos S. et al. Broadly neutralizing anti-HIV-1 antibodies require Fc effector functions for in vivo activity. Cell 158, 1243–1253, doi: 10.1016/j.cell.2014.08.023 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bournazos S. & Ravetch JV Anti-retroviral antibody FcgammaR-mediated effector functions. Immunol Rev 275, 285–295, doi: 10.1111/imr.12482 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Bournazos S. & Ravetch JV Fcgamma Receptor Function and the Design of Vaccination Strategies. Immunity 47, 224–233, doi: 10.1016/j.immuni.2017.07.009 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Nimmerjahn F. & Ravetch JV Fcgamma receptors: old friends and new family members. Immunity 24, 19–28, doi: 10.1016/j.immuni.2005.11.010 (2006). [DOI] [PubMed] [Google Scholar]
- 211.Nimmerjahn F. & Ravetch JV Fc-receptors as regulators of immunity. Adv Immunol 96, 179–204, doi: 10.1016/S0065-2776(07)96005-8 (2007). [DOI] [PubMed] [Google Scholar]
- 212.Nimmerjahn F. & Ravetch JV Fcgamma receptors as regulators of immune responses. Nat Rev Immunol 8, 34–47, doi: 10.1038/nri2206 (2008). [DOI] [PubMed] [Google Scholar]
- 213.Pelegrin M, Naranjo-Gomez M & Piechaczyk M. Antiviral Monoclonal Antibodies: Can They Be More Than Simple Neutralizing Agents? Trends Microbiol 23, 653–665, doi: 10.1016/j.tim.2015.07.005 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.