Purpose of review
The ‘precision medicine’ refers to the generation of identification and classification criteria for advanced taxonomy of patients, exploiting advanced models to infer optimized clinical decisions for each disease phenotype.
Recent findings
The current article reviews new advances in the past 18 months on the microbiomics science intended as new discipline contributing to advanced ‘precision medicine’. Recently published data highlight the importance of multidimensional data in the description of deep disease phenotypes, including microbiome and immune profiling, and support the efficacy of the systems medicine to better stratify patients, hence optimizing diagnostics, clinical management and response to treatments.
Summary
The articles referenced in this review help inform the reader on new decision-support systems that can be based on multiomics patients’ data including microbiome and immune profiling. These harmonized and integrated data can be elaborated by artificial intelligence to generate optimized diagnostic pipelines and clinical interventions.
Keywords: artificial intelligence, decision-support systems, immunomics, microbiomics, personalized medicine
INTRODUCTION
The term ‘precision medicine’ refers to the new sets of methodologies and approaches able to provide optimized stratification of patients, hence generating a new ‘definition’ of disease phenotypes. Despite its inflated usage, the definition is still often generic although already linked to important scientific and clinical expectations and even to the possibility of developing new health policies and strategies. However, we can state that the ‘precision medicine’ refers more appropriately to the generation of criteria for advanced taxonomy of patients, producing models to identify and classify clinical decisions for each disease phenotype. This new perspective of patient evaluation can use both basic laboratory and clinical data (i.e. discrete variables), and omics’ data (i.e. continuous variables) produced by new-generation sequencing or NGS, proteomics, metaproteomics, metabolomics, foodomics, immunomics (proteomics and integrated serology), lipidomics, microbiomics, all of them generating large-scale or big data (Fig. 1). These data are allowing researchers and clinicians to provide a broad spectrum of clinical, genetic, immunological information on the patient, his lifestyle (e.g. feeding, family and personal relationships and habits, sports activities, stress elements) and main interactions with the environment such as exposure to pathogens, nutrients, drugs, pollution, all variables constituting the exposome. These new indicators of the patient biological or clinical state may be considered, after validation processes typical of the systems medicine, biomarkers of the phenotype of the disease, and may allow to go beyond the classic semeiotics approach, overcoming signs and symptoms to decipher the disease. These ‘omics data’ are ‘multidimensional data’, exploiting numbers of patients, genomic and metagenomic sequences, metabolites, immunological profiles as whole matrices of patients’ meaningful variables. Once the data are ‘cataloged’, that is organized in architectural systems for reading usable data, they can be harmonized and, therefore, integrated, according to univariate and multivariate statistical models, borrowed by chemometrics, which is the science of relating the measurements made on a biological system or on a chemical process with the state of the system through the precise application of statistics. By using this approach, it is possible to move from hundreds of thousands of data to a few tens through a mechanism of self-scaling (harmonization) and reduction of multidimensionality until producing a framework of both structural and functional features of the biological system. By integrating the two characteristics, it is possible to provide a complete host-exposome set of data that can provide personalized information for each patient, without introducing the bias of the a-priori selection of clinical and laboratory data (‘targeted’ approach). When metadata on human microbiome are also considered, the entire microbiota-host-exposome phenotype can be generated for each patient, including microbiome profiling as enterophenotype that can complement the classical phenotype of the disease (metadata or phenomics data). At individual level microbiota-host-exposome data may act as diagnostic and prognostic tool, while at population level they can generate baseline profiling acting as prediction factor of disease. With this approach, the description of a pathophysiological state by representative variables is performed by an ‘untargeted’ approach, without any selection a priori. This allows researchers to consider the disease as a ‘system’ represented by as many variables or ‘features’ as possible that can be collocated in a ‘precise’ or ‘personalized’ picture of the clinical history of the patient, in the context of the ‘strict’ observed disease phenotype. Moving from omics’ or big data to precision medicine, through a process of fused data down to small data, the stratification of the patients can be tremendously improved, translating this process into a potential better health care.
FIGURE 1.
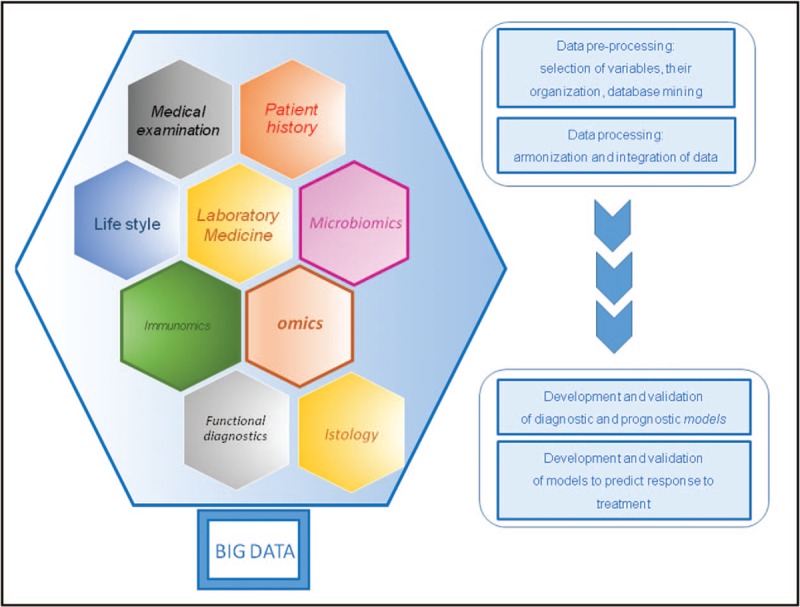
Entire set of big data that can build up a ‘precise’ physiopathological model in personalized medicine.
Box 1.
no caption available
GUT MICROBIOME, CAUSE AND DISEASE PROFILING
In a recent Nature article [1▪▪], the major modulating variables of a healthy human microbiota have been reported, showing that host genetics acts only in negligible quantity with respect to external stimuli and to modifications induced by environment (exposome). The growing availability of systems biology approaches to study intestinal microbiome has provided powerful and sophisticated analytical tools, allowing to produce multidimensional omics data (big data) able to describe without a-priori constraints a human microbiome, including variables associated with food [2▪]. The prediction of modulating effects on the symbiotic and dysbiotic state of the microbial ecosystem, through patterns of trajectories of microbial evolution, is now possible [3▪]. Therefore, the intestinal microbiota can be analysed nowadays in its ecological complexity of microbial organ within the host organism (superorganism or holobiont), characterized by continuous dynamic interactions with host, food and environment. The exhaustive description of an intestinal microbiota in the early stages of life, immediately after birth and during childhood, when the so-called physiological programming is carried out, has an important impact on neonatology and paediatrics, as it provides nutraceutical indications for the entire process of the growth [4▪▪]. Moreover, it is known that some diseases, expressed at extraintestinal level, such as allergies, are associated with perturbations of the gastrointestinal microbial ecosystem, starting since birth [5▪]. Obesity and metabolic diseases associated with obesity, such as the metabolic syndrome and type 2 diabetes, have also been linked to structural and functional characteristics of the intestinal microbiota [6▪], the last also induced by antibiotic treatments, as shown in mouse models [7▪▪]. Based on these studies, it seems plausible that the ability of the intestinal microbiota to regulate the inflammatory response is critical in the complex mechanisms related to obesity and metabolic syndrome, and even to the risk of developing some adult diseases, such as cardiovascular diseases [8▪]. In addition, for mendelian diseases, such as cystic fibrosis (CF), chronic inflammation and hence derived recurrent respiratory infections, appear to have substantial effects on the intestinal microbiota, through dynamic correlations associated with its dysbiosis [9▪]. As it is known, mutations in the Cystic Fibrosis Transmembrane conductance Regulator (CFTR) gene alter the physiology of the ion channel of the apical membranes of epithelial cells, deregulating exocrine production, with consequent malabsorption, obstruction and nutritional deficiency of the intestine. Indeed, in patients with CF the role of the intestinal bacterial proliferation and the implications of the gene defect remain controversial in endogenous inflammation and in the alteration of immunological homeostasis. A recent study produced by our group [10▪▪] has highlighted the main players of the intestinal microbiota that correlate with the gut functional alterations but also with the host's metabolism typical of CF patients. Particularly, 31 CF naïve patients were studied under clinical stability in a case–control modality, hence compared with age-matched healthy patients (1–6 years).
The profile of the intestinal microbiota obtained from the fusion of metagenomic and metabolomic data identified bacterial preponderance and deficits, with high abundances of Propionibacterium, Staphylococcus and Clostridiaceae, including Clostridium difficile, and poor presence of Eggerthella, Eubacterium, Ruminococcus, Dorea, Faecalibacterium prausnitzii, associated with over-expression of 4-aminobutyrate [gamma-aminobutyric acid (GABA)], choline, ethanol, propylbutyrate and pyridine and low levels of sarcosine, 4-methylphenol, uracil, glucose, acetate, phenol, benzaldehyde and methyl acetate [10▪▪]. These correlations represent an absolute novelty with respect to the previous acquired knowledge on intestinal microbiota profiling in paediatric disease, since induced to define as prevalent driver of intestinal microbiota dysbiosis its genetic deficit. The study demonstrated two fundamental and innovative aspects: first, the intestinal microbiota milieu is primarily modulated and induced by the alteration of the CFTR function of the host; hence the GABA and choline molecules, as they directly reflect the alterations of the intestinal transport of water and of the components regulating the intestinal osmosis, can be considered specific biomarkers of CF microbiome, while alcohols, esters cobiomarkers of the altered microbial activity, providing a new idea of disease-predicting metabolites to be investigated in the future; second, the intestinal microbiota is only secondarily modulated by patient's age, disease phenotype, colonization/infection of the pulmonary microbiota and by the chronic antibiotic treatment regime. Both evidences are extraordinary considerations if compared with what is known on intestinal microbiota/health/disease. In CF, the increase in intestinal inflammation has been repeatedly reported, possibly triggering intestinal bacterial proliferation, otherwise called blooming, favoured by the sticky material that hinders the transport of antimicrobials [11▪,12▪]. However, in naïve CF paediatric patient, our work has shown an almost independent microbiome profile from the chronic antibiotic regimen, which could be explained by the ceramide effect [10▪▪].
When different disease phenotypes, even including those associated with a single genetic modification, are catalogued and associated with microbial enterotypes, the set of phenotypic profiles becomes pleiotropic, and instead of organism we speak of a superorganism, which implies the direct or indirect role of the microbiota on human diseases. For this reason, advanced tools of genomics are needed (i.e. NGS), capable of performing optimal studies of highly complex microbial communities. In addition to metagenomics, the microbiota is currently analysed by metabolomics. Through the production of antimicrobial compounds, volatile fatty acids and chemically modified bile acids, the intestinal microbiota creates a metabolically very reactive environment, often described as a bi-reactor. Recent studies, including those of our group, have shown that 1H-NMR, gas chromatography-mass spectrometry (GC-MS) metabolic analyses of faecal extracts can provide important clarifications on the interspecies metabolic differences of the components of the microbiota. Metabolomics may produce important diagnostic information for the main intestinal diseases, to examine metabolic cooperation host/microbiota with respect to phenotype, pathology and diet [13▪▪]. Indeed, in colon rectal cancer (CRC) patients, a GC–MS-based approach allowed to identify 17 metabolites associated with Asp metabolism, ammonia recycling, protein biosynthesis, and Trp metabolism, which are involved in tumorigenesis (i.e. Lys, heptanedioic acid, norvaline), while 42 metabolites, involved in Asp, Ala metabolism and protein biosynthesis, were not detectable in the CRC group. Targeted-metagenomics identified 76 bacteria, including Proteobacteria, Enterobacteriaceae and Fusobacteria as key discriminants of CRC, whereas Firmicutes, Clostridiales, Clostridia, Lachnospiraceae, Ruminococcaceae and Faecalibacterium were only ascribed to healthy patients considered as controls (CTRLs). Integrated analysis correlated CRC-associated microbes with metabolites, such as polyamines (cadaverine and putrescine), producing more functional insights than single datasets.
In general, mutualistic relationships in the intestinal microbiota influence metabolic health, regulate energy balance, xenobiotic metabolism, resistance to pathogen colonization, immunological maturation in children and nutritional health. On this basis, biomarker design associated with microbiota dysbiosis for specific diseases can have a direct diagnostic relapse. Applying highly performing and processive metaproteomic technologies, based on Triple-TOF mass spectrometry, Sequential Windows Acquisition of all Theoretical Precursors technology, orbitrap fusion pipelines, additional analytes from faeces or blood can produce qualitative and quantitative profiles of differentially expressed proteins/metabolites, capable of providing large in-silico reservoir of information for each individual patient [14▪▪].
With this high interdisciplinary impact and with a work at the interface between research and clinic, microbiologists and specialists in-omics sciences can now understand the role of the intestinal microbiota in physiological and pathological states, and can assign characteristics (fingerprints), able to define human endophenotypes (Fig. 2). What is already clear is that the role of the microbiota in human health is much more important than previously thought: understanding the dynamics of bacterial populations and governing them, instead of attacking them with antibiotics, could represent the correct strategy in the next future to defeat numerous diseases and the growing phenomenon of antimicrobial resistance. Microbial communities could therefore be managed in terms of their content and metabolic balance rather than in term of their removal. The classic concept of infection associated with a single organism that invades our body and reproduces itself by inducing a series of alterations is no longer acceptable. In fact, it has been discovered that some diseases appear to be caused by imbalances of microorganisms’ populations rather than by a single pathogen [15▪]. This new model can be extended to various diseases and not only be confined to infectious processes.
FIGURE 2.
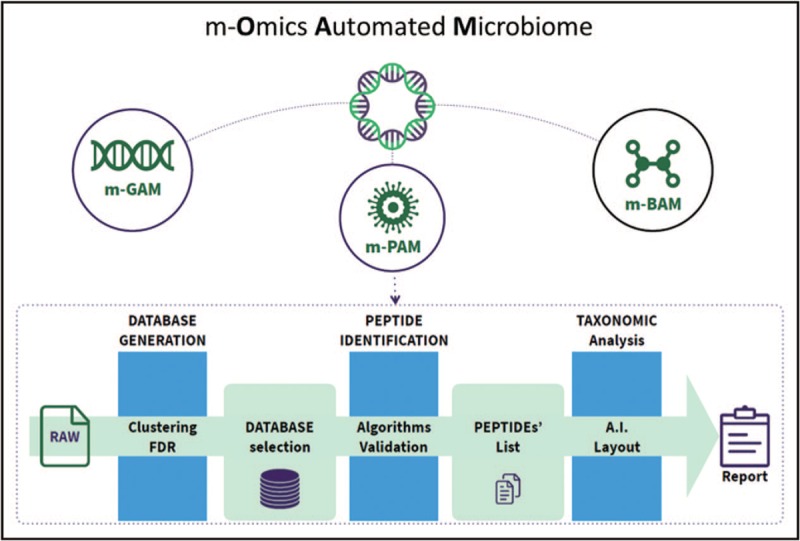
Metagenomics, metaproteomics and metabolomics platforms, interpreted by artificial intelligence, may provide automated microbiome models in health and disease. m-BAM, meta-bolomics Automated Microbiome; m-GAM, meta-Genomics Automated Microbiome; m-PAM, meta-Proteomics Automated Microbiome.
Research currently hypothesizes that gut microbiota alterations are the basis of many chronic inflammatory diseases, allergies, diabetes, obesity. The higher is the diversity, the less likely it is the possibility that external pathogens can invade and settle inside the human body. In fact, if all the niches are occupied, it becomes difficult for the invaders to invade places and become operative. However, the role of the gut microbiota in the human pathogenesis does not rely on exclusive competition: indeed, our microbiome interacts with the environment that dominates the genetic make-up [1▪▪].
PRECISION MEDICINE AND MICROBIOMICS
Researches on microbiome are increasingly exploiting omic and metaomic pipelines and algorithms that allow microbiomics researchers to characterize the gut and other districts’ human microbiomes in the most variable physiopathological conditions almost reaching the description of the gradient laying between eubiotic and dysbiotic landscapes of gut microbiome. In particular, our research group has focused its activity on the production of microbiome maps (profiles) associated with various pathological conditions such as CF, fatty liver, obesity, multidrug-resistant germ infections, autism, inflammatory bowel diseases, intestinal bowel syndromes, cancer, mental retardation, idiopathic arthritis, allergy, dermatitis, HIV, trying to address dysbiome features through dysbiosis tracking indexes (Patent ITUA20164448: Metagenomic method for in vitro diagnosis of gut dysbiosis) and integrated meta-omics data [16▪–18▪,19▪▪,20▪▪,21▪–24▪,25▪▪,26▪,27▪▪].
An interesting area of research regards microbiomics and carcinogenesis. In the study by Ponziani et al.[20▪▪], gut microbiota features associated with hepatocellular carcinoma (HCC) in patients with complex phenotypes, such as cirrhosis and nonalcoholic fatty liver disease (NAFLD), were investigated. Microbiota profile, intestinal permeability, inflammatory status and circulating mononuclear cells were considered ‘features’ to construct a correlation model in hepatocarcinogenesis. Patients with HCC showed increased levels of fecal calprotectin, while intestinal permeability was similar to patients with cirrhosis but without HCC. Plasma levels of IL8, IL13, chemokine ligand (CCL) 3, CCL4 and CCL5 were higher in the HCC group and associated with activated circulating monocytes. The fecal microbiota of patients with cirrhosis showed higher abundance of Enterobacteriaceae and Streptococcus and a reduction in Akkermansia. Bacteroides and Ruminococcaceae resulted increased in the HCC group, while Bifidobacterium was reduced. Akkermansia and Bifidobacterium were inversely correlated with calprotectin concentration, which conversely was associated with humoral and cellular inflammatory markers. This study highlighted that in patients with cirrhosis and NAFLD the gut microbiota profile and systemic inflammation are significantly correlated concurring in hepatocarcinogenesis processes [20▪▪]. Remarkably, inflammation status was found to be not correlated to microbiota profiling in juvenile idiopathic arthritis (JIA) [25▪▪]. A recent large multicenter, prospective, observational cohort study included Italian and Dutch paediatric patients affected by JIA at baseline, with inactive disease, and with persistent activity compared with healthy CTRLs [25▪▪]. Random forest models distinguished between Italian patient baseline samples and healthy CTRLs and suggested differences between Dutch patient samples and healthy CTRLs. The article showed strong evidence for dysbiosis in JIA patients. Only patient/CTRL status, age and geographic origin appear to be drivers of the microbiota profiles, regardless of disease activity stage, inflammation and markers of autoimmunity.
In addition, the modulation of the intestinal microbiome following administration of pre, probiotics, nutritional modulation and resetting due to intestinal microbiota transplantation have been investigated. This massive microbiome profiling has generated more than 7000 paediatric and adult microbiota maps, generating a very advanced database of microbiome biobank (https://www.bbmri.it/nodo-nazionale/gruppi-di-lavoro/microbiota-umano-e-biobanking/) and microbiome resources (Fig. 2) that can contribute to the best definitions or taxonomy of the disease. Based on microbiomics algorithms, our group is developing the following decision-support systems (DSS) as diagnostic-clinical tools: first, dysbiosis profiles of intestinal microbiota based on targeted-metagenomics; second, identification of microbial markers and molecular biomarkers of health and disease, directly related to alterations of the intestinal microbiota; third, production of host-microbiome integrated omics data profiles; fourth, faecal microbiota transplant programs; fifth, design of computer applications, based on artificial intelligence to manage omics data and dietary profiles with the purpose to translate microbiota profiling into a score-based system for intestinal dysbiosis diagnosis and clinical-nutraceutical interventions.
CONCLUSION ON ADVANTAGES OF PRECISION MEDICINE AND ROLE OF MICROBIOMICS
The advantages of precision medicine are innumerable. First of all, we can contribute with this approach to clarify unknown etiopathogenetic mechanisms of disease; we can identify several diagnostic and prognostic biomarkers in different patients, within the framework of the same pathology, and the reason for their variability; we can understand in different individuals a differential response to the same pharmacological treatment, despite affected by the same disease.
As limitations, we have to consider the design of the study. As reported in several studies [1▪▪,6▪,18▪,25▪▪] identifying specific biomarkers for disease prediction in a global population may not be straightforward. It is extremely important to consider optimal population CTRL groups to avoid bias linked to environment, food, ethnic-specific features which can affect reproducibility of case–control studies. In addition, appropriate stratification of disease is extremely important [6▪,11▪,18▪]. Moreover, technological platforms, experimental procedures and computational algorithms need to be standardised by performing experimental settings in multiple collaborative sites including large sample sizes to avoid ‘batch effects’ and minimize biological variabilities.
However, regardless limits, precision medicine can be considered a new tool for evaluating the parameters of efficiency and effectiveness in the management of health policies. In this context, microbiomics complement deep phenotyping, providing enterophenotypes features and relationship with host genetics, metabolism, immunology, overcoming the host genetics [28▪]. As new laboratory medicine, data from microbiomics integrated with patient metadata and other omics data may provide advanced DSS. We are entering the new era of artificial intelligence that can harmonize, integrate, but especially learn from single omics models to provide new combined models of fused phenotypes/enterophenotypes associated with disease profiling. The aim is that artificial intelligence in DSSs may provide advanced taxonomy of disease phenotype to improve response to treatment through diagnostic and prognostic models of response prediction, based on score-based systems (Fig. 3). We would basically have two genomes: the human genome and the microbiome; therefore, the fluctuations in the microbial population of the microbiota would result in the manifestation of dysbiosis and, therefore, in the subsequent onset or flare-up of diseases. The ability to govern these fluctuations can represent the medicine of the future, acting on entities that can be modified, contrarily to what happens for the human genome, whose gene content cannot be modified.
FIGURE 3.
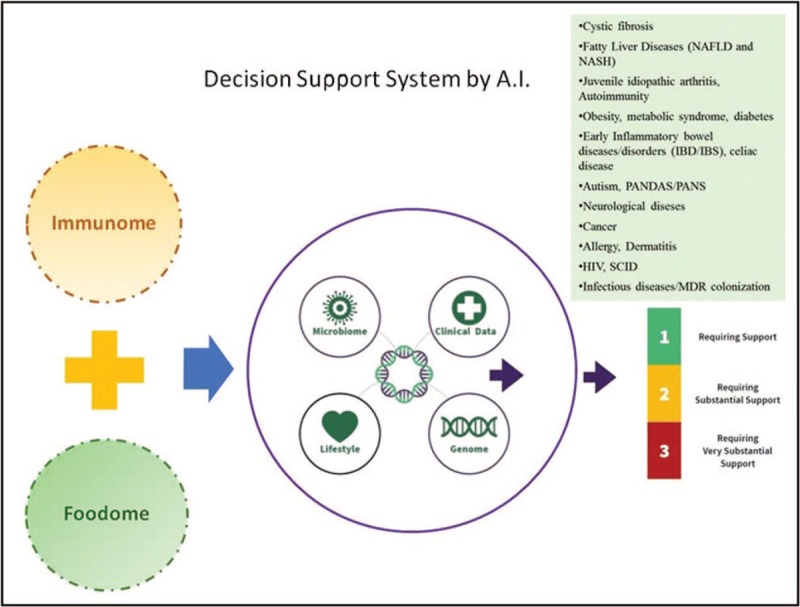
Framework of omics integration, including immunomics and foodomics, diseases and artificial intelligence to generate optimized diagnostic pipelines and clinical interventions. hiv, Human Immunodeficiency Virus; MDR, multi-drug-resistant germs; scid, severe combined immunodeficiency.
The future of microbiota is already present in current diagnostics and clinical practice and, therefore, it is already precision medicine.
Acknowledgements
We wish to thank Simone Gardini of Genomeup for graphical assistance. The article is dedicated to the lovely memory of Prof Valerio Nobili.
Financial support and sponsorship
The work was supported by the Ministry of Health, Italian, Ricerca Corrente 201802G004314 assigned to LP, Children's Hospital Bambino Gesù, IRCCS.
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
REFERENCES
- 1▪▪.Rothschild D, Weissbrod O, Barkan E, et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018; 555:210–215. [DOI] [PubMed] [Google Scholar]; An outstanding review on genotype and microbiome data from 1046 healthy individuals with distinct origins and common environment that demonstrates how gut microbiome is not significantly associated with host genetics.
- 2▪.Braconi D, Bernardini G, Millucci L, Santucci A. Foodomics for human health: current status and perspectives. Expert Rev Proteomics 2018; 15:153–164. [DOI] [PubMed] [Google Scholar]; In this review, authors discuss the complex relationships connecting food, nutrition and human health to develop functional foods and nutraceuticals, personalized nutrition based on interplay among gut microbiota, diet and diseases.
- 3▪.Angulo MT, Moog CH, Liu YY. A theoretical framework for controlling complex microbial communities. Nat Commun 2019; 10:1045. [DOI] [PMC free article] [PubMed] [Google Scholar]; A key review on control (CTRL) frameworks of microbial communities based on the new notion of structural accessibility and ecology driver species, which microbiologists can potentially manipulate to CTRL whole communities towards desired states.
- 4▪▪.Cerdó T, Diéguez E, Campoy C. Early nutrition and gut microbiome: interrelationship between bacterial metabolism, immune system, brain structure and neurodevelopment. Am J Physiol Endocrinol Metab 2019; [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]; The nonsystematic review explores disturbances of diet during pregnancy and early postnatal life, which may affect colonization of gut microbiota during early life, influencing infant health and leading to potential long-lasting consequences later in life.
- 5▪.Fyhrquist N. The human microbiota and its relationship with allergies. Gastroenterol Clin North Am 2019; 48:377–387. [DOI] [PubMed] [Google Scholar]; Authors focus on the understanding of mechanisms by which the human microbiota may be regulating the immune system, and changes in the composition of the microbiota may produce effects, linked with an increased risk of developing chronic inflammatory disorders, including allergies.
- 6▪.Thingholm LB, Rühlemann MC, Koch M, et al. Obese individuals with and without type 2 diabetes show different gut microbial functional capacity and composition. Cell Host Microbe 2019; 26:252–264.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]; Co-occurrence of microbial features for obesity and type 2 diabetes poses challenges in disentangling unique to each condition. This article differentiates microbes linked to these interrelated metabolic diseases and identifies dietary and medication exposures to consider in future studies.
- 7▪▪.Sun L, Zhang X, Zhang Y, et al. Antibiotic-induced disruption of gut microbiota alters local metabolomes and immune responses. Front Cell Infect Microbiol 2019; 9:99. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study suggests that antibiotic-induced changes in gut microbiota might contribute to the inflammation responses through the alternation of metabolic status, providing novel insights on different interactions between gut microbiota, metabolic functions and immune host responses.
- 8▪.Schiattarella GG, Sannino A, Esposito G, et al. Diagnostics and therapeutic implications of gut microbiota alterations in cardiometabolic diseases. Trends Cardiovasc Med 2019; 29:141–147. [DOI] [PubMed] [Google Scholar]; The article summarizes the role of gut microbiota in pathophysiology of the most common cardiometabolic disorders, discussing the potential therapeutic opportunities offered by interventions aimed at modifying microbiome composition and activity.
- 9▪.Héry-Arnaud G, Boutin S, Cuthbertson L, et al. The lung and gut microbiome: what has to be taken into consideration for cystic fibrosis? J Cyst Fibros 2019; 18:13–21. [DOI] [PubMed] [Google Scholar]; The review focuses on main points raised during the 15th European Cystic Fibrosis Society Basic Science preconference symposium focused on microbiome, asking the question ‘The lung and gut microbiome: what has to be considered for cystic fibrosis (CF)?’, which dealt with technical considerations, pathophysiology and clinical implications of the microbiome in CF.
- 10▪▪.Vernocchi P, Del Chierico F, Russo A, et al. Gut microbiota signatures in cystic fibrosis: loss of host CFTR function drives the microbiota enterophenotype. PLoS One 2018; 13:e0208171. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study provides a predictive and functional model of the gut microbiota enterophenotype of paediatric patients affected by CF under clinical stability, intrinsically linked to disease, regardless of age and only partially related to oral antibiotic administration or lung colonization/infection.
- 11▪.Antosca KM, Chernikova DA, Price CE, et al. Altered stool microbiota of infants with cystic fibrosis shows a reduction in genera associated with immune programming from birth. J Bacteriol 2019; 201: pii: e00274-19. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study reports that faecal microbiota of CF infants show an altered but largely unchanging within-individual bacterial diversity (alpha diversity) over the first year of life, in contrast to the healthy infants. The data clarify that current therapeutics do not establish in CF infants a gut microbiota like that in healthy infants and further studies are necessary.
- 12▪.Enaud R, Hooks KB, Barre A, et al. Intestinal inflammation in children with cystic fibrosis is associated with Crohn's-like microbiota disturbances. J Clin Med 2019; 8: pii: E645. [DOI] [PMC free article] [PubMed] [Google Scholar]; The main objective of this study is to assess gut microbiota composition in CF children depending on their intestinal inflammation. These results aim to link gut microbiota and intestinal inflammation in CF, suggesting the key role of the gut-lung axis in the CF evolution.
- 13▪▪.Yang Y, Misra BB, Liang L, et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics 2019; 9:4101–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]; Driven by the host's genetic makeup and environmental exposures, the gut microbiome and its metabolites are herein discussed as the causes and regulators of colon rectal cancer (CRC) pathogenesis. The article provides substantial evidence of a novel interplay between the gut microbiome and metabolome, which is drastically perturbed in CRC.
- 14▪▪.Zhang B, Kuster B. Proteomics is not an island: multiomics integration is the key to understanding biological systems. Mol Cell Proteomics 2019; 18 8 Suppl 1:S1–S4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Proteomic profiles reflect cellular responses to genomic, epigenomic and environmental alterations. Integrating proteomics data with data from genomics, epigenomics, transcriptomics, metabolomics, image-omics and phenomics holds great potential for revealing novel biology and transforming clinical practice.
- 15▪.Del Chierico F, Cardile S, Pietrobattista A, et al. Liver transplantation and gut microbiota profiling in a child colonized by a multi-drug resistant Klebsiella pneumoniae: a new approach to move from antibiotic to ‘eubiotic’ control of microbial resistance. Int J Mol Sci 2018; 19: pii: E1280. [DOI] [PMC free article] [PubMed] [Google Scholar]; In chronic liver disease with indications for liver transplantation, MDR colonization can affect the liver transplantation outcome. The perioperative antibiotic regimen can select Klebsiella pneumoniae MDR. Mechanisms modulating the gut ecosystem should be taken into account in MDR colonization clinical management.
- 16▪.Altomare A, Putignani L, Del Chierico F, et al. Gut mucosal-associated microbiota better discloses inflammatory bowel disease differential patterns than faecal microbiota. Dig Liver Dis 2019; 51:648–656. [DOI] [PubMed] [Google Scholar]; The aim of the study is to evaluate the mucosal and faecal microbiota composition in a cohort of inflammatory bowel diseases (IBD) patients compared with healthy CTRLs. The profiles of faecal microbiota, more similar to CTRLs, suggest that microbiota adhering to the gut mucosa better discriminates patients from CTRLs, with the identification of some interesting biomarkers.
- 17▪.Ponziani FR, Putignani L, Paroni Sterbini F, et al. Influence of hepatitis C virus eradication with direct-acting antivirals on the gut microbiota in patients with cirrhosis. Aliment Pharmacol Ther 2018; 48:1301–1311. [DOI] [PubMed] [Google Scholar]; The article investigates the correlation between direct-acting antivirals (DAAs) in HCV eradication and gut microbiota composition, intestinal and systemic inflammatory. The DAAs does not improve intestinal barrier function.
- 18▪.Del Chierico F, Abbatini F, Russo A, et al. Gut microbiota markers in obese adolescent and adult patients: age-dependent differential patterns. Front Microbiol 2018; 9:1210. [DOI] [PMC free article] [PubMed] [Google Scholar]; The article focuses on gut microbiota composition in obese adolescents and adults compared with age-matched normal weight volunteers to assemble age-related and obesity-related microbiota profiles. The study reveals unique features of gut microbiota in terms of ecological patterns, microbial composition and metabolism in obese patients, assigning novel age-related obesity bacterial markers.
- 19▪▪.Quagliariello A, Del Chierico F, Russo A, et al. Gut microbiota profiling and gut-brain crosstalk in children affected by pediatric acute-onset neuropsychiatric syndrome and pediatric autoimmune neuropsychiatric disorders associated with streptococcal infections. Front Microbiol 2018; 9:675. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study investigates the relationship between paediatric acute-onset neuropsychiatric syndrome and paediatric autoimmune neuropsychiatric disorders and gut microbiota. Streptococcus infections alter gut microbiota leading to a proinflammatory status through the selection of specific bacteria associated with gut inflammation and immune response activation.
- 20▪▪.Ponziani FR, Bhoori S, Castelli C, et al. Hepatocellular carcinoma is associated with gut microbiota profile and inflammation in nonalcoholic fatty liver disease. Hepatology 2019; 69:107–120. [DOI] [PubMed] [Google Scholar]; The study explores gut microbiota features associated with hepatocellular carcinoma (HCC) in patients with cirrhosis and nonalcoholic fatty liver disease (NAFLD), constructing a correlation model on HCC features, that suggest that gut microbiota profile and systemic inflammation are significantly correlated.
- 21▪.Nobili V, Putignani L, Mosca A, et al. Bifidobacteria and lactobacilli in the gut microbiome of children with nonalcoholic fatty liver disease: which strains act as health players? Arch Med Sci 2018; 14:81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]; The article discusses that Bifidobacteria seem to have a protective role against the development of NAFLD and obesity, highlighting their possible use in developing novel, targeted and effective probiotics.
- 22▪.Lo Presti A, Zorzi F, Del Chierico F, et al. Fecal and mucosal microbiota profiling in irritable bowel syndrome and inflammatory bowel disease. Front Microbiol 2019; 10:1655. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study provides an overview of the alterations of gut microbiota in patients affected by IBD and irritable bowel syndrome (IBS) and may aid in identifying potential mucosal biomarkers for IBD and IBS.
- 23▪.Sessa L, Reddel S, Manno E, et al. Distinct gut microbiota profile in antiretroviral therapy-treated perinatally HIV-infected patients associated with cardiac and inflammatory biomarkers. AIDS 2019; 33:1001–1011. [DOI] [PubMed] [Google Scholar]; The article investigates correlation between gut microbiota profiles, markers of inflammation, vascular endothelial activation (VEA) and microbial translocation in perinatally HIV-infected patients under antiretroviral therapy, showing distinctive gut microbiota profiles associated with inflammation, microbial translocation and VEA.
- 24▪.Reddel S, Del Chierico F, Quagliariello A, et al. Gut microbiota profile in children affected by atopic dermatitis and evaluation of intestinal persistence of a probiotic mixture. Sci Rep 2019; 9:4996. [DOI] [PMC free article] [PubMed] [Google Scholar]; The study shows an alteration in microbiota composition of children affected by atopic dermatitis, with the depletion or absence of some species, opening the way to new probiotic interventions.
- 25▪▪.van Dijkhuizen EHP, Del Chierico F, Malattia C, et al. Model driven paediatric European digital repository consortium. Microbiome analytics of the gut microbiota in patients with juvenile idiopathic arthritis: a longitudinal observational cohort study. Arthritis Rheumatol 2019; 71:1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]; The article describes a multicentre, prospective, observational cohort study to assess the composition of gut microbiota in Italian and Dutch patients with juvenile idiopathic arthritis at baseline, with inactive disease and with persistent activity compared with healthy CTRLs. Only patient/CTRL status, age and geographic origin appear to be drivers of the microbiota dysbiotic profiles, regardless of disease activity stage, inflammation and markers of autoimmunity.
- 26▪.Nobili V, Mosca A, Alterio T, et al. Fighting fatty liver diseases with nutritional interventions, probiotics, symbiotics, and fecal microbiota transplantation (FMT). Adv Exp Med Biol 2019; 1125:85–100. [DOI] [PubMed] [Google Scholar]; The review describes the current state of knowledge on the ways fatty liver diseases can be addressed with nutritional interventions, probiotics, symbiotics and fecal microbiota transplantation (FMT).
- 27▪▪.Gargiullo L, Del Chierico F, D’Argenio P, et al. Gut microbiota modulation for multidrug-resistant organism decolonization: present and future perspectives. Front Microbiol 2019; 10:1704. [DOI] [PMC free article] [PubMed] [Google Scholar]; The narrative review discusses current knowledge of microbiota-mediated mechanisms of antimicrobial resistance and strategies for MDR colonization CTRL. It focuses on FMT for MDR intestinal decolonization and report updated literature on its current clinical use.
- 28▪.Cammarota G, Putignani L, Gasbarrini A. Gut microbiome beats two to zero host genome. Hepatobiliary Surg Nutr 2019; 8:378–380. [DOI] [PMC free article] [PubMed] [Google Scholar]; Microbiome data can better predict many disease-related human traits than models that use only host genetic. The gut microbiome ‘beats two to zero’ host genome to interpret host phenotype; exposome definitely dominates over host genetics in modulating gut microbiota.


