Abstract
Mutation rate is a key determinant of the pace as well as outcome of evolution, and variability in this rate has been shown in different scenarios to play a key role in evolutionary adaptation and resistance evolution under stress caused by selective pressure. Here we investigate the dynamics of resistance fixation in a bacterial population with variable mutation rates, and we show that evolutionary outcomes are most sensitive to mutation rate variations when the population is subject to environmental and demographic conditions that suppress the evolutionary advantage of high-fitness subpopulations. By directly mapping a biophysical fitness function to the system-level dynamics of the population, we show that both low and very high, but not intermediate, levels of stress in the form of an antibiotic result in a disproportionate effect of hypermutation on resistance fixation. We demonstrate how this behavior is directly tied to the extent of genetic hitchhiking in the system, the propagation of high-mutation rate cells through association with high-fitness mutations. Our results indicate a substantial role for mutation rate flexibility in the evolution of antibiotic resistance under conditions that present a weak advantage over wildtype to resistant cells.
I. INTRODUCTION
The ability to predict the possible trajectories of a naturally evolving complex living system is key to describing and anticipating varied ecological and biomedical phenomena. Such predictability rests on an understanding of the potential for evolutionary adaptability of a given system. In asexual populations, a major mechanism responsible for evolutionary adaptation under environmental stress is the generation via genetic mutations of phenotypes able to better withstand and thrive under the stressor: resistant populations arising from within a wildtype population that may “rescue” the population from the source of stress by eventually coming to dominate the population. The rate at which such resistant mutations occur and the balance between these and more deleterious mutations are major determinants of whether the population may survive and adapt to selective evolutionary pressure [1–5], an environmental stressor that targets strain variants, or phenotypes, nonuniformly. Although the baseline mutation rate in bacteria is quite low, at about ∼10–3 per genome per generation [6,7], high prevalences of mutator strains in natural bacterial populations and clinical isolates have been observed in various studies (see [8–11] for early work and [12] for a survey), and in certain cases “hypermutability,” an increase in the mutation rate over the baseline rate, was shown to result in fitness increases and faster adaptation [5,13–18] and even be essential for survival under stress [19] by enabling genetic hitchhiking on beneficial mutations [5,20–22]. Mutation rates can increase under environmental stress [23–26], and, in particular, hypermutability may play a significant role in the rise of antibiotic resistance [27–32].
The potential for adaptability via genetic mutations is dependent on the interplay between the ensemble of phenotypes that the system can access via mutations and the rate at which such transitions may occur within this ensemble. Phenotypes are typically characterized by some intrinsic measure of evolutionary fitness, such as their growth rate or lag phase, that contributes to evolutionary success, with extrinsic conditions, such as the probability of acquisition of this trait, initial population distribution, or resource availability, held fixed. Yet evolutionary advantage is determined by an interplay of these intrinsic and extrinsic factors, and separating these dependences while considering only a subset of them is of limited utility in establishing a global picture of a system’s evolvability potential as well as specific response to selective pressure. Here, we address both with a view to investigating the extent to which mutation rate variability drives adaptation under selective pressure.
The main purpose of this paper is to demonstrate that evolution under selective pressure—an external stressor that induces a fitness gradient in a given population—may not be uniformly sensitive to mutation rate as a function of the selective pressure as well as additional fitness-determining conditions, and that this nonuniform behavior should be taken into account when deciding on an appropriate antibiotic dosing protocol. In such a situation, there is generally no information available on the mutation rate in the pathogenic bacterial population, and this rate may also change in the course of therapy, as noted above. If dosing can be restricted to ranges for which the expected evolutionary outcome is less sensitive to the mutation rate, there will be higher predictive certainty about the treatment outcome, and more reliable strategies can be developed for avoiding antibiotic resistance arising in the course of treatment.
By considering a simple deterministic model of bacterial evolution under limited resources, we show that evolutionary outcome is most sensitive to the mutation rate when there exist phenotypes in the population that have a weak advantage—expressed through either intrinsic traits or extrinsic conditions—over the phenotype that is initially dominant in the population. In Sec. II we introduce and describe our evolutionary dynamics model; in Sec. III we define and motivate our measure of mutation rate sensitivity and quantify how sensitive the evolutionary success of a population is to increases in the mutation rate. We show that the fitness advantage of the resistant mutant—as given by both intrinsic fitness and extrinsic advantage-conferring conditions—is a determining factor in the extent of this sensitivity. In Sec. IV, we focus our analysis on selective evolutionary pressure in the form of a bacterial growth inhibitor (antibiotic), and we quantify (i) the dependence of mutation rate sensitivity to this source of pressure, and (ii) the extent to which the antibiotic drives the fixation of hypermutation in the population. We conclude with a brief discussion of the ramifications of our findings in Sec. V.
II. THE MODEL
We consider a system under non-neutral selection in which up to two distinct phenotypes—defined by their growth rates gi—may coexist under limited resources. We denote them by “wt” for wildtype and r for resistant—designations that are intended to indicate that the r phenotype is more resistant to the evolutionary pressure considered (e.g., antibiotic, as in Sec. IV) under the non-neutral selection experienced by the population. Each of these phenotypes may be found with some baseline mutation rate μbl or with an elevated mutation rate of f × μbl, f > 1. Both forward and backward mutations are permitted with equal probability1 pwt,r; transitions between baseline-mutation phenotype and its elevated-mutation counterpart (of identical growth rate) occur with rate rμ. In addition, to account for the fitness penalty incurred due to an increased rate of deleterious mutations at higher values of f, we assume that either phenotype may experience deleterious mutations with probability Pdel. Since at non-negligible levels of selective pressure phenotypes whose resistance to the pressure is weaker than wildtype will have very low growth, we do not keep track of such low-growth populations explicitly, but they are implicitly accounted for in our model as the loss of cells from higher-growth populations via deleterious mutations. Since we assume that such loss occurs with uniform probability Pdel, when the overall genetic mutation rate of a cell is μ, the rate of deleterious mutations is given by μPdel. Note that when Pdel is high, increases in μ carry a higher penalty, implying that for hypermutation to be beneficial and counteract this penalty, resistant phenotypes would have to be significantly advantageous either by having a much higher growth rate (intrinsic advantage) or, e.g., by occurring with a high probability or being initially present in relatively high proportions (extrinsic advantage). Figure 1 shows a schematic of this model. We assume deterministic evolution under limited resources, as resources needed for growth are nearly always constrained in real biological systems, driving competition between organisms consuming the same resources. The equations governing the time evolution of this system are given by
| 2.1 |
FIG. 1.
Schematic indicating the allowed single-step transitions and their rates between phenotypes.
where and a stationary population distribution is established when the total population size reaches the resource capacity K. Note that faster-growing phenotypes will also produce exponentially more deleterious mutants as a result of their more frequent divisions, resulting in the previously noted fitness tradeoff. The four-dimensional system (Fig. 1) of Eq. (2.1) is given explicitly in Appendix A. We consider here the case in which the effect of selective pressure is limited to selecting for hypermutant variants if they are advantageous but not directly inducing hypermutability (for work on the latter, see, e.g., [35–39]). Under this assumption, hypermutation occurs independently of selective pressure, and therefore some proportion of the initial-state population would be expected to already exhibit elevated mutation rates. We assume in all that follows that cells with elevated mutation rates constitute 1% of the total initial population (distributed in proportion to the phenotype distribution) and a corresponding rate at which hypermutation-conferring mutations occur of 0.25% of cells per generation (see Appendix B for an extended discussion of these parameter choices).
III. SENSITIVITY OF EVOLUTIONARY SUCCESS TO THE MUTATION RATE
The ability of a population to survive evolutionary pressure depends on the extent to which resistant phenotypes come to dominate it and withstand potential subsequent applications of the stressor (e.g., in a serial dilutions experiment). To understand how mutation rate affects this, we consider how the stationary-state ratio of resistant cells in the population, xr/xtot, at elevated mutation rates compares with this ratio if all mutations in the system were restricted to occur at the baseline rate μbl,
| 3.1 |
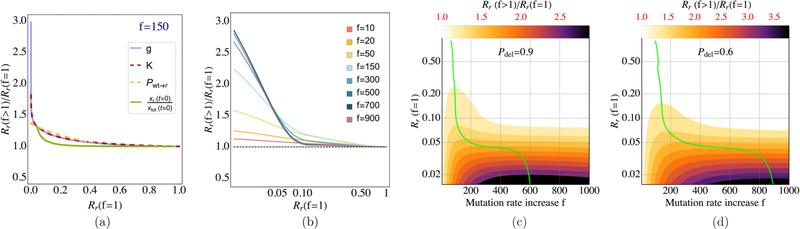
as a function of the main parameters that arise in our model, The ratio (3.1) represents the extent to which a particular (elevated) mutation rate is able to drive a successful evolutionary outcome: a larger proportion of resistant cells in the stationary-state distribution. We refer to this as the sensitivity of evolutionary success to the mutation rate. In Fig. 2(a), we show how this sensitivity correlates with our measure of baseline “evolutionary advantage” of the resistant mutant, as different components of are varied.2 For each of these model parameters, we see that at some fixed mutation rate (shown in the plot is a 150-fold increase over the baseline), sensitivity to the elevation in mutation rate is highest at low (but positive) levels of the resistant mutant’s evolutionary advantage; it decreases and eventually becomes negligible (≈1) as its evolutionary advantage increases.
FIG. 2.
The sensitivity of evolution success to hypermutation [Rr(f > 1)/Rr(f = 1)] is negatively correlated with the baseline-mutation rate evolutionary advantage Rr(f = 1) of the resistant mutant and is optimized at a mutation rate increase (f ) that depends on the extent of advantage, diminishing upon further increases in this rate. (a) Rr(f > 1)/Rr(f = 1) vs Rr(f = 1) curves for individual parameters affecting the resistant mutant’s evolutionary advantage at f = 150. (b) Averages of the single-ρi curves in the appropriate ranges put through a low-pass filter for smoothness shown at multiple values of f (on a log-linear scale, for clarity of resolution between different mutation rates). The dashed black line (parity in Rr at baseline and elevated f ) indicates the point of no benefit from hypermutation; initial increases in f yield significant benefit at low-advantage conditions, which decreases and eventually becomes negligible at high-advantage conditions [Rr(f = 1) → 1]. At very high f (here ) even low-advantage mutants experience diminishing benefit from hypermutation. (c) and (d) Rr(f > 1)/Rr(f = 1) contours corresponding to plot (b) in f – Rr(f = 1) space. The green curve shows the optimal [i.e., yielding highest Rr(f > 1)/Rr(f = 1)] mutation rate increase factor f as a function of Rr(f = 1). The probability of deleterious mutations was set at Pdel = 0.9 for plots (a)–(c) and at Pdel = 0.6 in plot (d). When held constant, parameters were set at gr/gwt = 3, K/xtot(t = 0) = 102, pwt,r = 0.01, and xr(t = 0) = 0. Wildtype E. coli growth was set at gwt = 0.34 h−1 and μbl = 2 × 10−10 × Ng, with Ng the size of the E. coli genome.
Before proceeding in our analysis, we consider how the different parameters in are in fact indicative of evolutionary advantage: while it is clear how a higher growth rate gr, a larger initial proportion xr(t = 0)/xwt(t = 0), and a higher nondeleterious mutation rate lead to advantageous conditions for the resistant mutant to increase its proportions in the population, it is perhaps less obvious why a higher resource capacity produces an advantage specifically for the resistant mutant given that resource utilization is uniform in our model among the two phenotypes. The reason for this is that while the resource capacity appears a priori to be a nonselective environmental stressor, due to the exponential growth phase involved in the evolution of the system (2.1), higher resource capacity puts off the time of resource saturation, thus compounding the advantage enjoyed by phenotypes with higher growth rate gk.
By averaging over individual-ρi interpolations [Fig. 2(b)] and varying f [Figs. 2(c) and 2(d)], we observe that the largest impact of the presence of elevated mutation rates in the population is under parameter combinations that, due to any one or multiple advantage-determining parameters, result in the resistant phenotype having a weak advantage. In these circumstances, the evolutionary advantage of the resistant cells may be insufficient to establish these populations in high proportions due to competition for limited resources, and certain increases in the mutation rate may thus be critical for adaptation, even at the cost of increased deleterious mutations. When initial conditions confer a high advantage on the resistant phenotype, mutation rate increases offer negligible to negative benefit. The high growth rate of these populations and hence frequent cell divisions imply that increases in their mutation rate also drive approximately exponentially increases in deleterious mutations, and that when a strong advantage exists, the baseline-mutation phenotype will thus rise to fixation faster than its hypermutant counterpart.
As shown in the contour plots of Figs. 2(c) (Pdel = 0.9) and 2(d) (Pdel = 0.6), for any level of resistant mutant evolutionary advantage, there exists an optimal mutation rate (green curves) yielding the highest proportion of resistant cells. Increasing the mutation rate up to this rate provides substantial benefit for lower-advantage mutants, and further increases lead to diminishing (albeit more gradually) returns due to the tradeoff with an increased loss caused by deleterious mutations. We see [Fig. 2(c) compared with 2(d)] that the level of evolutionary advantage past which there is no gain from hypermutation is fairly robust to variations in the rate of deleterious mutations (f μblPdel), but a lower Pdel extends the range of mutation rates conferring benefit, as in that case there is little loss to deleterious mutations even at high f (see the supplemental material [40] for additional plots corresponding to different choices of Pdel and rμ).
IV. EFFECT OF SELECTIVE PRESSURE ON MUTATION RATE SENSITIVITY AND ON GENETIC HITCHHIKING
In this section, we focus on the effect of selective pressure in the form of an antibiotic that inhibits bacterial growth, and we quantify how the extent of selective pressure—different antibiotic concentrations—affects the sensitivity of evolutionary outcome to the mutation rate. The effect of antibiotic concentration on this quantity arises from the respective dependences of the phenotypes’ growth rates on this concentration. Motivated by work [34] on the response of E. coli to variations in the dosage of trimethoprim, a competitive inhibitor of dihydrofolate reductase, we assume a hyperbolic decay functional dependence for the growth rate g on the inhibitor concentration [I],
| (4.1) |
where g0 is the growth rate in the absence of an inhibitor and gI controls the extent to which the population may grow in the presence of the inhibitor. In [34] this functional dependence, with g0 and gI given explicitly as functions of various protein biophysical and cellular properties, was shown to agree with experimental measurements for several mutant phenotypes over a range of [I], and similar methods can in principle be used to derive g0 and gI from biophysical principles for a wider range of biologically relevant scenarios.
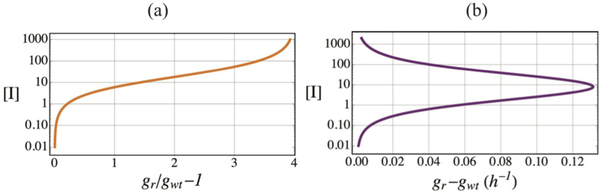
By computing the sensitivity to mutation rate, (Eq. 3.1), as a function of only the inhibitor with other parameters held fixed, we show [Fig. 3(a)] that at low levels of inhibition, where gI carries only a small fitness advantage and mutant and wildtype growth rates g([I]) are similar, there is substantial benefit to be gained from hypermutation. As inhibition is increased, the difference between mutant and wildtype growth increases, resulting in the resistant mutant easily increasing in proportions without much benefit from hypermutation; but at yet higher levels of inhibition, the role of elevated mutation rates in determining adaptation once again becomes significant. The behavior of the (intrinsic) selection coefficient gr/gwt – 1 is not revealing in this respect: it monotonically approaches a constant value at high [I]. However, the difference between gr and gwt peaks at an intermediate value of [I] and decreases at lower and higher values of [I] (Fig. 4). While the peak does not numerically coincide with the [I] concentrations yielding the lowest Rr(f > 1)/Rr(f = 1), we note that additional parameters in also affect this ratio.
FIG. 3.
(a) Hypermutation has a strong impact on resistance fixation at low and at very high levels of inhibition that is optimized at a mutation rate that depends on the inhibition level. (b) Genetic hitchhiking on resistant mutations is most pronounced in intermediate levels of inhibition. The black contour (=1) indicates no hitchhiking on resistant mutations. Parameters were set at g0,r = g0,wt = 0.34 h−1, gr,I = 5gwt,I, where gwt,I = 3.6 μg/mL, Pdel = 0.9; and K, pwt,r, and xr(t = 0) as in Fig. 2.
FIG. 4.
While the intrinsic selection coefficient gr/gwt – 1 increases monotonically as a function of inhibition (a), the difference in growth rates gr – gwt is maximized at intermediate levels of inhibition (b).
We next consider the extent to which selective pressure, the antibiotic, affects the extent of hypermutation in the population by computing the stationary-state proportion of hyper mutants in the population when an inhibitor is applied (resistant cells have a positive evolutionary advantage) relative to when no inhibition is present (neutral selection),
| 4.2 |
as a function of the inhibition [Fig. 3(a)]. Figure 3(b) shows contours of Rh([I] > 0)/Rh([I] = 0) in a two-dimensional space of f and [I]. We find [Fig. 3(b)] that genetic hitchhiking on resistant mutations as measured by Rh is most pronounced in an f – [I] phase space that up to intermediate mutation rate increases is approximately complementary to that in which hypermutation has the most pronounced beneficial effects. This effect can be explained by noting that at low inhibition, where the resistant mutant does not have significant advantage over wildtype, the acquisition of such mutations does not drastically increase the growth rate of hypermutant cells; on the other hand, when resistant mutations are highly advantageous (high inhibition), the baseline-mutation resistant mutant rises to fixation largely unaided by hypermutation, which under finite resources limits the growth potential of other subpopulations (resistant hypermutants). We note that the range of mutation rates at which we observe hitchhiking to be strongest is in keeping with experimental observations (see [12] for a review and [41] for additional recent data) of an O(101−102) increase over baseline in E. coli clinical isolates [with some data pointing to a nearly O(103) in certain cases [42]].
V. DISCUSSION
In obtaining the results presented here, we assumed deterministic dynamics. While mutations typically arise randomly and can introduce a large degree of stochasticity into the dynamics, deterministic evolution can provide important insights into processes with varying degrees of stochasticity: large populations are expected to sample a large extent of the available mutational phase space (with infinite populations sampling every possible configuration, or genotype), and experimental work [43] on evolutionary pathways in E. coli to drug resistance found that similar mutational trajectories across populations evolved in parallel. Our deterministic results, moreover, suggest that stochastic fluctuations in the mutation rate can have an outsized effect on the stationary state of the system under a broad range of conditions that suppress the evolutionary advantage of emergent resistant populations. Knowledge of the effects of these conditions in conjunction with a quantitative understanding of changes in a controllable selective pressure, such as the one we modeled here in the case of a growth inhibitor, are crucial for forming informed predictions on how variations in this main driving force of adaptation affect the dynamics of complex, high-dimensional systems and on how to best minimize the effects of stochastic fluctuations to establish a desired evolutionary outcome, such as a clinical antibiotic protocol minimizing the risk of resistance evolution.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to João Rodrigues for providing data on E. coli growth curves, and to Michael Manhart for helpful discussions. We acknowledge support from NIGMS of the National Institutes of Health under Awards No. 1R01GM124044-01 and No. 5R01GM068670-14.
APPENDIX A: EVOLUTION EQUATIONS
We consider a population subdivided into wildtype with growth rate gwt and baseline mutation rate μbl, a resistant phenotype with this baseline mutation rate and growth rate gr, and corresponding phenotypes of equal growth rate but increased mutation rate f μbl, f > 1. Population levels at time t are given by xwt,1(t), xr,1(t), xwt,f (t), and xr,f (t), respectively. Transitions (mutations) between the subpopulations are permitted in accordance with the schematic Fig. 1. We assume limited resources set by an environmental carrying capacity K, so that the subpopulation levels thus evolve in time according to the equations
| A1 |
where rμ is the rate of mutation from a baseline-mutation rate (f = 1) phenotype to an f > 1 phenotype (the rate at which mutations leading to elevated mutation rates f μbl occur) and its reverse (assumed to be equal), pr,wt ≡ pwt→r = pr→wt is the probability of mutation from wildtype to the resistant phenotype and backward, and Pdel is the probability of mutation to deleterious phenotypes, xtot = xwt,1 + xwt,1f + xr,1 + xr, f.
To compute the relative advantage or disadvantage conferred by hypermutation on the fixation of drug-resistant subpopulations [Eq. (3.1)], we numerically compute the ratio of resistant mutants (combined nonhypermutant and hypermutant types) in the total population at to the ratio that would result if no hypermutations were allowed in the system, i.e., if we set rμ = 0 and consider only phenotypes xwt,1 and xr,1. When computing these quantities for a system with an initial distribution of hypermutants of either phenotype, we assume that the rμ = 0 system has a corresponding distribution in which
with i representing either wildtype or the resistant phenotype.
In the figures shown in the main text and in the supplemental material, μbl was set at 2 × 10−10 × Ngenome per generation per cell [6], where Ngenome = 4.64 × 106 is the number of base pairs in the E. coli genome. In the results shown in the main text, when the population is not purely wildtype at t = 0, the proportion of hypermutants chosen is assumed to be distributed proportionally among the wildtype and resistant populations.
APPENDIX B: RATE OF ACQUISITION rμ OF INCREASED MUTATION RATE AND INITIAL PROPORTION OF HYPERMUTANTS
To estimate a biologically reasonable rμ, we consider a simple system consisting of a wildtype f = 1 phenotype and a wildtype f > 1, both with fitness gwt, which can mutate into
each other with rate rμ:
| (A2) |
The steady-state (stationary distribution) proportion of hypermutants in the total population will be given by
| 3 |
at time τ after resources have been saturated.3 Hypermutation can be caused by various mechanisms; studies that focused on pathogenic E. coli have found comparatively high (>1%) proportions of mutators in bacterial isolates (3.6% in [44] and 1.9% in [9]); a separate study that looked specifically for MMR deactivation in E. coli found a much lower proportion (0.24%) when both commensal and pathogenic E. coli were included [8]. A later study [10] found, however, that when other sources of hypermutation were included besides MMR, E. coli cells exhibiting increased mutation rates—of up to two orders of magnitude from the baseline mutation rate—constituted as much as 14% of the total population, most being mild mutators, with both commensal and pathogenic strains included in the study. The highest mutation rates were found to correspond to MMR deficiencies, with lower increases due to other mechanisms. Note that since rμ is a neutral-selection rate, studies of mutator proportions that were conducted under conditions of adaptive evolution will likely overestimate this parameter, and we therefore restrict our data to studies of natural isolates, noting that even in those cases adaptive evolution in the recent past may have taken place.
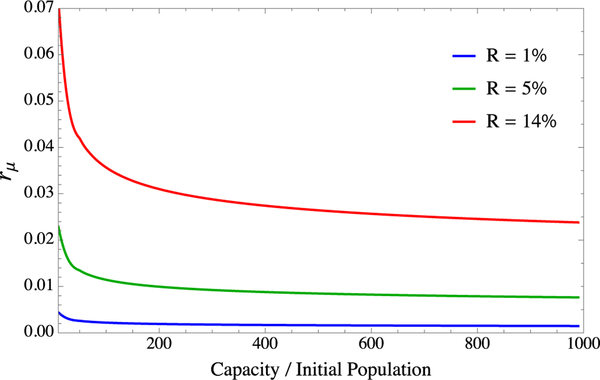
We compute which rμ values yield the stationary distribution ratio (3) for different carrying capacities by taking gwt as in the main text to be 0.34 h−1 under no inhibition. The results for different values of R are shown in Fig. 5. Since we consider a uniform distribution4 of mutation rate increase factors f and the 14% figure is heavily tipped toward mild mutators, using this figure will likely overestimate the mutation rate rμ in our model for higher values of f. For the purpose of the plots in the main text, we set on the lower end at rμ = 0.25% and an initial proportion of 1% hypermutating cells in the population.
FIG. 5.
Computed values of rμ from the system (2) for different expected steady-state proportions of resistant cells as the availability of resources is varied. Across a wide range of carrying capacities, rμ only varies from ∼0.5% to ∼1.5%. Plots in the main text employ rμ = 0.25%, corresponding to an initial hypermutant population of 1% of the total population.
Footnotes
While resistance can result from the accumulation of a series of mutations, first-generation mutants—for which the mutation is reversible by a single-point mutation—can already exhibit discernibly increased resistance [33,34], with further substantial increase in resistance found in some second-generation mutants, for which the equal probability assumption can be though of as a “first-order” approximation.
Since we consider here determine dynamics, the ratio directly projects to the stationary state should therefore be viewed as both a final outcome (at stationary state) as well as an indicator of the evolutionary advantage conferred by the system’s intrinsic and extrinsic conditions
Since our goal in the analysis of this Appendix is to obtain a first-order estimate for rμ that is independent of the precise mutation rate, we omit the potential effect of deleterious mutations here via Pdel, as the magnitude of this effect will depend on the actual mutation frequency f μbl of the hypermutators.
A nonuniform distribution can be incorporated by multiplying rμ by a probability distribution that depends on f —as this adds additional degrees of freedom to the model, we avoid doing so here.
References
- [1].Lynch M, Ackerman MS, Gout J-F, Long H, Sung W, Thomas WK, and Foster PL, Genetic drift, selection and the evolution of the mutation rate, Nat. Rev. Genet. 17, 704 (2016). [DOI] [PubMed] [Google Scholar]
- [2].Denamur E and Matic I, Evolution of mutation rates in bacteria, Mol. Microbiol. 60, 820 (2006). [DOI] [PubMed] [Google Scholar]
- [3].de Visser JAG, The fate of microbial mutators, Microbiology 148, 1247 (2002). [DOI] [PubMed] [Google Scholar]
- [4].Sniegowski PD, Gerrish PJ, Johnson T, Shaver A et al. , The evolution of mutation rates: separating causes from consequences, Bioessays 22, 1057 (2000). [DOI] [PubMed] [Google Scholar]
- [5].Taddei F, Radman M, Maynard-Smith J, Toupance B, Gouyon P-H, and Godelle B, Role of mutator alleles in adaptive evolution, Nature (London) 387, 700 (1997). [DOI] [PubMed] [Google Scholar]
- [6].Lee H, Popodi E, Tang H, and Foster PL, Rate and molecular spectrum of spontaneous mutations in the bacterium escherichia coli as determined by whole-genome sequencing, Proc. Natl. Acad. Sci. (USA) 109, E2774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Drake JW, Charlesworth B, Charlesworth D, and Crow JF, Rates of spontaneous mutation, Genetics 148, 1667 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gross MD and Siegel EC, Incidence of mutator strains in escherichia coli and coliforms in nature, Mutat. Res. Lett. 91, 107 (1981). [DOI] [PubMed] [Google Scholar]
- [9].LeClerc JE, Li B, Payne WL, Cebula TA et al. , High mutation frequencies among escherichia coli and salmonella pathogens, Science 274, 1208 (1996). [DOI] [PubMed] [Google Scholar]
- [10].Matic I, Radman M, Taddei F, Picard B, Doit C, Bingen E, Denamur E, and Elion J, Highly variable mutation rates in commensal and pathogenic escherichia coli, Science 277, 1833 (1997). [DOI] [PubMed] [Google Scholar]
- [11].Denamur E, Bonacorsi S, Giraud A, Duriez P, Hilali F, Amorin C, Bingen E, Andremont A, Picard B, Taddei F et al. , High frequency of mutator strains among human uropathogenic escherichia coli isolates, J. Bacteriol. 184, 605 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hall LM and Henderson-Begg SK, Hypermutable bacteria isolated from humans—a critical analysis, Microbiology 152, 2505 (2006). [DOI] [PubMed] [Google Scholar]
- [13].Chao L and Cox EC, Competition between high and low mutating strains of escherichia coli, Evolution 37, 125 (1983). [DOI] [PubMed] [Google Scholar]
- [14].Tenaillon O, Toupance B, Le Nagard H, Taddei F, and Godelle B, Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria, Genetics 152, 485 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Heo M and Shakhnovich EI, Interplay between pleiotropy and secondary selection determines rise and fall of mutators in stress response, PLoS Comput. Biol. 6, e1000710 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ram Y and Hadany L, The evolution of stress-induced hypermutation in asexual populations, Evolution 66, 2315 (2012). [DOI] [PubMed] [Google Scholar]
- [17].Ram Y and Hadany L, Stress-induced mutagenesis and complex adaptation, Proc. R. Soc. London, Ser. B 281, 20141025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lukačišinová M, Novak S, and Paixão T, Stress-induced mutagenesis: Stress diversity facilitates the persistence of mutator genes, PLoS Comput. Biol. 13, e1005609 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Swings T, Van den Bergh B, Wuyts S, Oeyen E, Voordeckers K, Verstrepen KJ, Fauvart M, Verstraeten N, and Michiels J, Adaptive tuning of mutation rates allows fast response to lethal stress in Escherichia coli, eLife 6, e22939 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Smith JM and Haigh J, The hitch-hiking effect of a favourable gene, Genet. Res. 23, 23 (1974). [PubMed] [Google Scholar]
- [21].Gentile CF, Yu S-C, Serrano SA, Gerrish PJ, and Sniegowski PD, Competition between high- and higher-mutating strains of escherichia coli, Biol. Lett. 7, 422 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, and Taddei F, Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut, Science 291, 2606 (2001). [DOI] [PubMed] [Google Scholar]
- [23].D. M. Fitzgerald, P. Hastings, and S. M. Rosenberg (unpublished).
- [24].MacLean RC, Torres-Barceló C, and Moxon R, Evaluating evolutionary models of stress-induced mutagenesis in bacteria, Nat. Rev. Genet. 14, 221 (2013). [DOI] [PubMed] [Google Scholar]
- [25].Galhardo RS, Hastings PJ, and Rosenberg SM, Mutation as a stress response and the regulation of evolvability, CRC Crit. Rev. Biochem. Mol. Biol. 42, 399 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Foster PL, Stress-induced mutagenesis in bacteria, CRC Crit. Rev. Biochem. Mol. Biol. 42, 373 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hammerstrom TG, Beabout K, Clements TP, Saxer G, and Shamoo Y, Acinetobacter baumannii repeatedly evolves a hypermutator phenotype in response to tigecycline that effectively surveys evolutionary trajectories to resistance, PloS One 10, e0140489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jolivet-Gougeon A, Kovacs B, Le Gall-David S, Le Bars H, Bousarghin L, Bonnaure-Mallet M, Lobel B, Guillé F, Soussy C-J, and Tenke P, Bacterial hypermutation: clinical implications, J. Med. Microbiol. 60, 563 (2011). [DOI] [PubMed] [Google Scholar]
- [29].Eliopoulos GM and Blázquez J, Hypermutation as a factor contributing to the acquisition of antimicrobial resistance, Clin. Infect. Dis. 37, 1201 (2003). [DOI] [PubMed] [Google Scholar]
- [30].Chopra I, O’Neill AJ, and Miller K, The role of mutators in the emergence of antibiotic-resistant bacteria, Drug Resist. Updates 6, 137 (2003). [DOI] [PubMed] [Google Scholar]
- [31].Giraud A, Matic I, Radman M, Fons M, and Taddei F, Mutator bacteria as a risk factor in treatment of infectious diseases, Antimicrob. Agents Chemother. 46, 863 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Martinez J and Baquero F, Mutation frequencies and antibiotic resistance, Antimicrob. Agents Chemother. 44, 1771 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Palmer AC, Toprak E, Baym M, Kim S, Veres A, Bershtein S, and Kishony R, Delayed commitment to evolutionary fate in antibiotic resistance fitness landscapes, Nat. Commun. 6, 7385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rodrigues JV, Bershtein S, Li A, Lozovsky ER, Hartl DL, and Shakhnovich EI, Biophysical principles predict fitness landscapes of drug resistance, Proc. Natl. Acad. Sci. (USA) 113, E1470 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Phillips I, Culebras E, Moreno F, and Baquero F, Induction of the sos response by new 4-quinolones, J. Antimicrob. Chemother. 20, 631 (1987). [DOI] [PubMed] [Google Scholar]
- [36].Piddock LJ and Wise R, Induction of the sos response in escherichia coli by 4-quinolone antimicrobial agents, FEMS Microbiol. Lett. 41, 289 (1987). [Google Scholar]
- [37].Ysern P, Clerch B, Castańo M, Gibert I, Barbé J, and1/2 M. Llagostera, Induction of sos genes in escherichia coli and mutagenesis in salmonella typhimurium by fluoroquinolones, Mutagenesis 5, 63 (1990). [DOI] [PubMed] [Google Scholar]
- [38].Ren L, Rahman MS, and Humayun MZ, Escherichia coli cells exposed to streptomycin display a mutator phenotype, J. Bacteriol. 181, 1043 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pérez-Capilla T, Baquero M-R, Gómez-Gómez J-M, Ionel A, Martín S, and Blázquez J, Sos-independent induction of dinb transcription by β-lactam-mediated inhibition of cell wall synthesis in escherichia coli, J. Bacteriol. 187, 1515 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].See Supplemental Material at http://link.aps.org/supplemental/ 10.1103/PhysRevE.99.022424 for discussions of the mutation rate choice in the main text plots, the effects of variations in the rate of deleterious mutations, and the time evolution growth curves (fixation dynamics) for the different subpopulations in the model.
- [41].Robert L, Ollion J, Robert J, Song X, Matic I, and Elez M, Mutation dynamics and fitness effects followed in single cells, Science 359, 1283 (2018). [DOI] [PubMed] [Google Scholar]
- [42].Pang P, Lundberg A, and Walker G, Identification and characterization of the mutl and muts gene products of salmonella typhimurium lt2., J. Bacteriol. 163, 1007 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Toprak E, Veres A, Michel J-B, Chait R, Hartl DL, and Kishony R, Evolutionary paths to antibiotic resistance under dynamically sustained drug selection, Nat. Genet. 44, 101 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Jyssum K, Observation of two types of genetic instability in Escherichia coli, Acta Pathol. Microbiol. Immunol. Scand. 48, 113 (1960). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.