Abstract
Background:
In our previous paper, we demonstrated that Connexin 43 (CX43) was highly expressed in bladder cancer (BC) tissues. But the molecular mechanism about microRNAs (miRNAs) regulation upstream of CX43 in BC has not been well elucidated and remains to be further studied. MicroRNA-139-5p (miR-139-5p) is a tumor suppressor in progression of multifarious cancers including BC. Nevertheless, the underlying mechanisms of CX43/miR-139-5p in tumorigenesis of BC are still not well illustrated. The specific objective of our study was to inquiry the effect of CX43/miR-139-5p on BC progression and its underlying mechanism.
Methods:
The bioinformatics analysis softwares were applied to predict the miRNAs in the upstream of CX43. First, the expression levels of miR-139-5p in BC tissues (tumor) and paracancer tissues (normal) were investigated using the data from The Cancer Genome Atlas database. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was used to detect the mRNA expression level of miR-139-5p in three human BC cell lines 5637, T24, ECV-304 and a human bladder epithelial immortalized cell line SV-HUC-1 (normal control). Then si-CX43, si-control, miR-139-5p mimic, and its negative control (NC) were transfected into BC cell line ECV-304. The relationship of miR-139-5p and CX43 was analyzed by dual-luciferase reporter assay. The qRT-PCR and Western blotting were used to test the mRNA and protein expression level of CX43. The proliferation of ECV-304 and T24 cells were examined by cell counting kit-8. The migration and invasion of ECV-304 cells were tested by transwell assay. To determine whether miR-139-5p would affect cell proliferation, migration and invasion by targeting CX43, we executed the rescue assay. The comparison between two groups was analyzed by Student's t test, and comparisons among multiple samples were performed by one-way analysis of variance and a Bonferroni post hoc test.
Results:
The expression of miR-139-5p was remarkably down-regulated in BC tissues (tumor vs. normal, 2.286 ± 0.017 vs. 3.211 ± 0.034, t = 11.540, P < 0.0001) and cell lines (P < 0.01 in all BC cell lines). Besides, we also indicated that over-expression of miR-139-5p reduced the proliferation of ECV-304 (P = 0.001) and T24 cells (P = 0.005). Moreover, miR-139-5p over-expression weakened the invasion (P = 0.001) and migration (P = 0.001) of ECV-304 cells. Furthermore, the relative luciferase activity of CX43-wild type construct was distinctly lessened by up-regulation of miR-139-5p (miR-139-5p mimic NC vs. miR-139-5p mimic, 0.916 ± 0.063 vs. 0.356 ± 0.048, t = 7.085, P = 0.002), nevertheless the activity of CX43-mutant type construct was untouched (miR-139-5p mimic NC vs. miR-139-5p mimic, 0.918 ± 0.057 vs. 0.878 ± 0.039, t = 0.577, P = 0.595). Finally, the rescue assay revealed that CX43 deletion enhanced the depressor effect of miR-139-5p on ECV-304 cell proliferation (P < 0.01), invasion (P = 0.028), and migration (P = 0.014).
Conclusion:
MiR-139-5p, as a tumor-suppressor, repressed cell proliferation, invasion, and migration in BC, which might be achieved by regulating CX43.
Keywords: Bladder cancer, Connexin 43, MicroRNAs
Introduction
Bladder cancer (BC), with 549,393 new cases in 2018, is the most familiar malignant tumor in the urinary system of men and women.[1,2] Because of the introduction of novel diagnostic approaches, the development of imaging and surgical techniques, the hoisting of chemotherapy, as well as the improvement of urinary tumor markers, BC diagnosis and treatment have made some progress in recent decades.[3] However, these treatments have some limitations in clinic and the 5-year survival rate has improved very little. Therefore, the underlying mechanism of BC needs further probe to find more effective treatments for BC.
Connexins (CX), which are found to be highly expressed in various cancers, are composed by 21 proteins, of which Connexin 43 (CX43) is the most rifely expressed and diffusely researched protein.[4] In previous article, the high expression of CX43 was shown in BC tissues.[5] MicroRNAs (miRNAs), short non-coding RNAs, display momentous modulatory functions in genetic expression.[6] Besides, many miRNAs are engaged in cell survival of cancers.[7,8] Lin et al[9] reported that miRNA-206 (miR-206) restrained the metastasis of breast cancer cells via modulating CX43. The regulation mechanism of miRNA on CX43 in BC is still unknown. According to Hu et al,[10] up-regulation of miR-139-5p could suppress the effects on BC cells. There were a large body of literatures which were concerned with miR-139-5p acting as a remedial molecule for patients with BC.[11,12] However, CX43 and miR-139-5p acting together on cancers have not been reported.
This study aimed to explore the biological effect of CX43/miR-139-5p on BC and their potential molecular mechanism.
Methods
Prediction of the miRNAs on upstream of CX43
The miRNA target gene prediction sites miRanda (http://miranda.org.uk/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/), TargetScan (http://www.targetscan.org/), miRDB (http://mirdb.org/) were used to predict the miRNAs on the upstream of CX43. The miRNAs were obtained from the intersection of the above four sites and the lowly expressed miRNAs in BC analyzed in The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/) database. In this study, we utilized the gene expression profile dataset from the public database TCGA to study the differential expression of miR-139-5p in BC tissues (tumor) and paracancer tissues (normal). The dataset has 437 samples, including 418 BC tissues and 19 normal samples.
Cell culture and transfection
Human BC cell lines 5637, T24, ECV-304 and human bladder epithelial immortalized cell line SV-HUC-1 (normal control) were acquired from American Type Culture Collection (USA). Routine culture was performed using Roswell Park Memorial Institute-1640 medium in a routine culture with penicillin (100 U/mL), streptomycin (0.1 mg/mL), and fetal bovine serum (10%).
si-CX43-1 (Forward: 5’-GCCCAAACTGATGGTGTCAAT-3’), si-CX43-2 (Forward: 5’-CAATTCTTCTTGCCGCAATTA-3’), si-CX43-3 (Forward: 5’-GCCCAAACTGATGGTGTCAAT-3’), and si-con (Forward: 5’-AATTCTCCGAACGTGTCACGT-3’) were synthesized by Shenzhen Huada Gene Research Institute (Guangdong, China). MiR-139-5p mimic/mimic negative control (NC) was bought from RuiboBio (Guangzhou, China). According to the instructions of Lipofectamine 2000 (Invitrogen, Shanghai, China), transfection was performed. After 24 h, the efficiency of transfection can be observed by quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
qRT-PCR
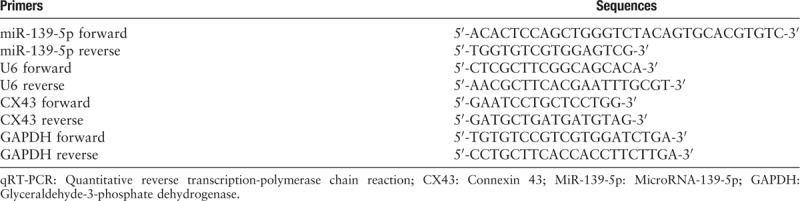
We extracted total RNA from ECV-304 cells using TRIzol reagent (Invitrogen). For reverse transcription of miRNAs, miScript II RT Kit (QIAGEN, Germany) was used. To determine miR-139-5p expression level by qPCR, MiScript SYBR® Green PCR Kit (QIAGEN) was used. PrimeScript RT Reagent Kit (Takara, Dalian, China) was used to form complementary DNA for detecting the mRNA expression level of CX43. SYBR® Premix Ex Taq II (TaKara) was applied to qPCR. U6 expression was used to normalize miR-139-5p expression. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was used to normalize CX43 expression. 2−ΔΔCt method was used to calculate miR-139-5p and CX43 relative expression levels. All the primers used for qPCR were illustrated in Table 1.
Table 1.
The primers used in qRT-PCR.
Western blotting
First, cells were gathered and lysed using radioimmunoprecipitation assay buffer (Beyotime, Jiangsu, China) based on the manufacturer’ protocol. Then the membrane was sealed by 5% skim milk powder for 1 h. Following that, the membrane was incubated with the primary antibodies overnight at 4°C. Then, Tris-buffered saline containing 0.1% Tween-20 (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) was used to wash the membrane three times for 5 min each. The membrane was probed with the secondary antibody for 1 h at room temperature and washed and added enhanced chemi-luminescence (ProteinTech Group, Inc., Chicago, IL, USA). GAPDH was used as the internal control. The QUANTITY ONE software (version 4.6.9; Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to scan the gray value.
Cell counting kit-8
After 24-h transfection, the cells were digested and counted, and a cell suspension was prepared. 100-μL cell suspension was taken into 96-well plates at the standard of 1000 cells per well. Then the cells were incubated in a 37°C incubator with 5% CO2 for 1.5 h. The 10-μL cell counting kit-8 (CCK-8) reagent (CWBio, Beijing, China) was added to each well. Finally, the optical density at 450 nm excitation light (A450nm) was detected every 24 h using the microplate reader.
Transwell migration and invasion experiments
The invasive or migratory capacity of ECV-304 cells was detected using transwell chambers (Merck KGaA, Germany) with or without matrigel (BD Bioscience, CA, USA) in the upper chamber, respectively. After 24 h transfection, cell suspension was prepared and added to the upper chamber (1 × 104 cells for invasion assay and 5 × 103 cells for migration assay). In the lower chamber, 500 μL of complete medium was added. After being kept at 37°C overnight, remanent cells in the upper chamber were wiped off using cotton swab. The invaded or migrated cells on the lower surface were fixed with 4% paraformaldehyde for half an hour and stained with 0.1% crystal violet for 20 min. The invaded and migrated cells were captured under a microscope (Olympus, Tokyo, Japan) and cell numbers were counted in five random fields.
Dual-luciferase vector construction experiment
The transfected cells were planted in 96-well plates (the density was 1 × 104/well) and cultured for 48 h. MiR-139-5p mimic and its NC, and PGL3-CX43-3’UTR-wild type (wt) (Promega, Madison, Wisconsin, USA) or PGL3-CX43-3’UTR-mutant type (mut) (Promega) were co-transfected into cells using Lipofectamine 2000 (Invitrogen). After 24 h transfection, the luciferase activity was assayed using the Dual-Luciferase Reporter Assay System (Promega). The ratio of the firefly luciferin intensity value to the Renilla fluorescein intensity value was calculated.
Statistical analysis
All experiments were done in duplicate and repeated three times. All data conformed to the normal distribution and the homogeneity of the variance, and were expressed as mean ± standard deviation. The experimental data were analyzed by SPSS 22.0 statistical analysis software (SPSS Inc., Chicago, IL, USA). The comparison between two groups was analyzed by Student's t test. The comparisons among multiple samples were performed by one-way analysis of variance and a Bonferroni post hoc test. P < 0.05 was considered statistically significant.
Results
Low expression of miR-139-5p in BC tissues
To identify potential upstream regulatory miRNAs for CX43, we first used miRanda, miRWalk, TargetScan, and miRDB to predict miRNAs on the upstream of CX43 and 120 miRNAs were obtained from these databases. Then we used the OncomiR website to analyze miRNAs with low expression in BC in the TCGA database and 22 miRNAs were obtained. After that, two common miRNAs (hsa-miR-1 and hsa-miR-139-5p) were obtained from the intersection of 120 miRNAs from the above four software predictions and 22 miRNAs from TCGA database [Figure 1A]. Based on comprehensive literature analysis, miR-139-5p was selected as the regulatory miRNA of CX43 for the following study. By analyzing the data about miR-139-5p in OncomiR website, we concluded that miR-139-5p was less expressed in BC tissues (n = 418) than that in paracancer tissues (n = 19) (tumor vs. normal, 2.286 ± 0.017 vs. 3.211 ± 0.034, t = 11.540, P < 0.0001) [Figure 1B].
Figure 1.
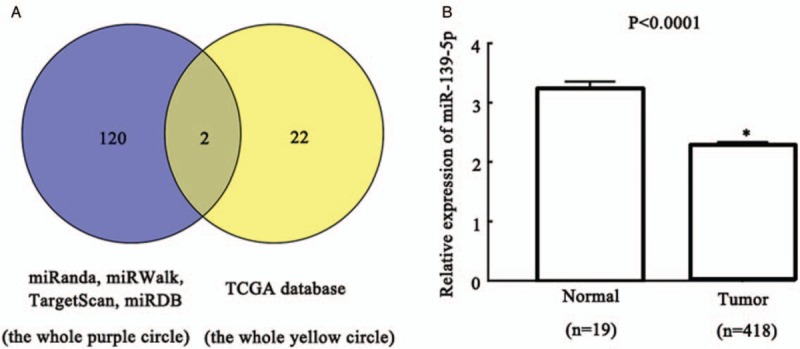
MiR-139-5p was preliminary predicted as a candidate upstream miRNA of CX43 and it is lowly expressed in BC tissues. (A) Venn diagram for common miRs in miRs predict websites miRanda, miRWalk, TargetScan, miRDB (the purple round on the left) and TCGA database (the yellow round on the right). (B) The mRNA expression level of miR-139-5p in BC tissues (tumor) and paracancer tissues (normal). ∗P < 0.0001 compared with Normal group. BC: Bladder cancer; CX43: Connexin 43; MiR-139-5p: MicroRNA-139-5p; TCGA: The Cancer Genome Atlas.
Low expression of miR-139-5p in BC cell lines
We used the qRT-PCR assay to investigate the expression level of miR-139-5p in BC. The results revealed that miR-139-5p was less expressed in BC cell lines than in normal cell lines (P < 0.01 in all BC cell lines), especially the lowest expression of miR-139-5p in ECV-304 cell line. So, ECV-304 cell line was used in the subsequent experiments [Figure 2A]. In order to investigate the effect of miR-139-5p on BC, miR-139-5p mimic/mimic NC were transfected into ECV-304 cells. As presented in Figure 2B, miR-139-5p mimic treatment significantly increased miR-139-5p expression, compared with miR-139-5p mimic NC group (P < 0.01).
Figure 2.
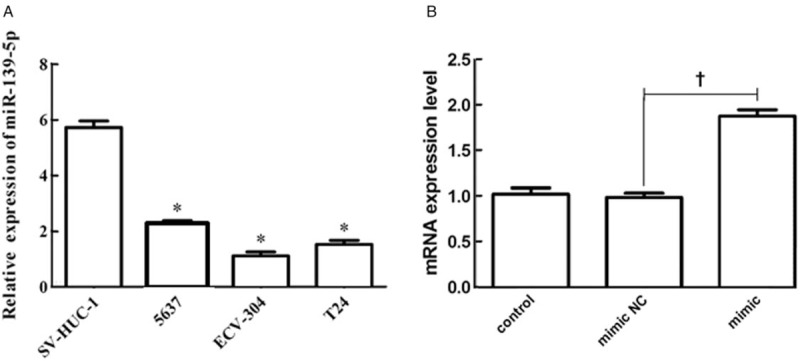
MiR-139-5p was lowly expressed in BC cell lines. (A) The relative expressions of miR-139-5p in normal cell line SV-HUC-1 and BC cell lines 5637, T24, and ECV-304 were detected by qRT-PCR. (B) The expressions of miR-139-5p in BC cells under miR-139-5p mimic transfection were tested by qRT-PCR. ∗P < 0.01 compared with SV-HUC-1 group. †P < 0.05 compared with mimic NC group. BC: Bladder cancer; MiR-139-5p: MicroRNA-139-5p; NC: Negative control; qRT-PCR: Quantitative reverse transcription-polymerase chain reaction.
Effect of over-expressed miR-139-5p on BC cell properties
The over-expression of miR-139-5p in ECV-304 cells was used to further investigate the possible biological roles of miR-139-5p in BC. The results of the CCK8 assay demonstrated that the miR-139-5p mimic group was significantly lower than the other two groups, indicating that over-expression of miR-139-5p restrained the proliferation of ECV-304 (P = 0.001) [Figure 3A] and T24 cells (P = 0.005) [Figure 3B]. The results revealed that the proliferation of ECV-304 and T24 cells were similar. Therefore, in the following experiments, we chose one cell line of them. As shown in Figure 3C and 3D, the number of crystal violet-stained cells in the invasion/migration assay was lessened in miR-139-5p mimic groups than in miR-139-5p mimic NC groups (P = 0.001, P = 0.001), presenting that over-expression of miR-139-5p suppressed the invasion and migration of ECV-304 cells. The results in this chapter indicated that over-expression of miR-139-5p restrained the properties of BC cells.
Figure 3.
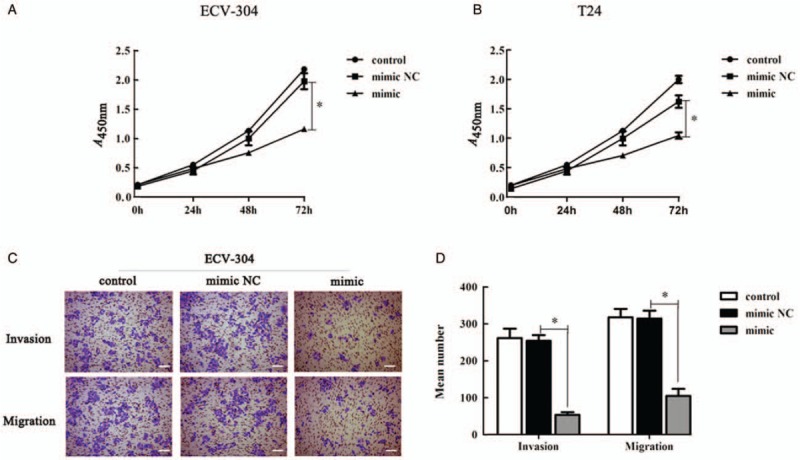
Over-expression of miR-139-5p restrained the proliferation, invasion, and migration potentials of BC cells. (A) CCK8 assay determined the role of miR-139-5p on the proliferation of ECV-304 cells. (B) CCK8 assay determined the role of miR-139-5p on the proliferation of T24 cells. (C) Transwell assay (bar = 200 μm) was used to detect ECV-304 cell migration and invasion. Representative images of crystal violet staining were shown on the upper panel (original magnification ×200). (D) Quantification of (C). ∗P < 0.01 compared with mimic NC group. BC: Bladder cancer; CCK-8: Cell counting kit-8; MiR-139-5p: MicroRNA-139-5p; NC: Negative control.
Possible binding sites of CX43 for miR-139-5p
To identify the potential miRNAs targeting CX43 mRNA, we utilized the miRanda, miRWalk, TargetScan, and miRDB algorithm to detect the sequence of CX43 mRNA. MiR-139-5p was authenticated as an altitudinal reliance candidate [Figure 4A]. To detect the targeting between miR-139-5p and CX43, CX43-wt and CX43-mut, targeting sequences were cloned to the luciferase on-limits reading frame [Figure 4A]. The relative luciferase activity of CX43-wt construct was distinctly lessened by up-regulation of miR-139-5p (miR-139-5p mimic NC vs. miR-139-5p mimic, 0.916 ± 0.063 vs. 0.356 ± 0.048, t = 7.085, P = 0.002), nevertheless in the same conditions, the activity of CX43-mut construct was untouched (miR-139-5p mimic NC vs. miR-139-5p mimic, 0.918 ± 0.057 vs. 0.878 ± 0.039, t = 0.577, P = 0.595) [Figure 4B]. By qRT-PCR and Western blotting, we discovered that the mRNA (P = 0.005) and protein (P = 0.002) expression of CX43 in ECV-304 cells declined dramatically in miR-139-5p mimic group compared to the miR-139-5p mimic NC group [Figure 4C–E]. These results verified that CX43 was a direct target of miR-139-5p, which was negatively regulated by miR-139-5p.
Figure 4.
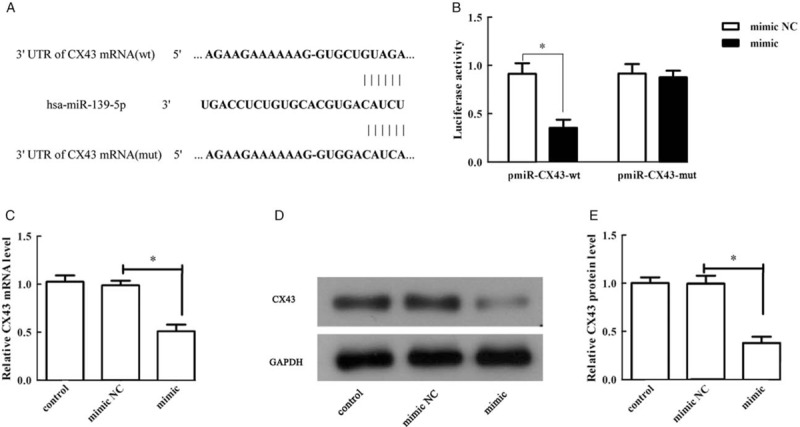
CX43 was a direct target of miR-139-5p, and was negatively modulated by miR-139-5p. (A) The binding sites between CX43 and miR-139-5p were predicted by Targetscan. (B) The relative luciferase activity of CX43-wt and CX43-mut. (C) The relative mRNA expression level of CX43 in ECV-304 cells. (D) The protein expression of CX43 in ECV-304 cells. (E) Quantification of (D). ∗P < 0.01 compared with mimic NC group. CX43: Connexin 43; MiR-139-5p: MicroRNA-139-5p; mut: Mutant type; NC: Negative control; wt: Wild type.
CX43 deletion strengthened inhibitory effect of miR-139-5p in BC cells
To further manifest whether miR-139-5p displayed its function via targeting CX43, we designed co-transfection of CX43 knockdown combined with miR-139-5p mimic. CCK8 results indicated that over-expression of miR-139-5p suppressed the ECV-304 cell proliferation (P < 0.01), and CX43 silence accelerated the elevated role of miR-139-5p on the proliferation of ECV-304 cells (P < 0.01) [Figure 5A]. Transwell assay results revealed that CX43 silence stressed the inhibitory influence of miR-139-5p on the invasion (P = 0.028) and migration (P = 0.014) of BC cells [Figure 5B and 5C]. Together these results provide important insights into that CX43 ablation enhanced the inhibitory effect of miR-139-5p on cell proliferation, invasion, and migration in BC cells.
Figure 5.
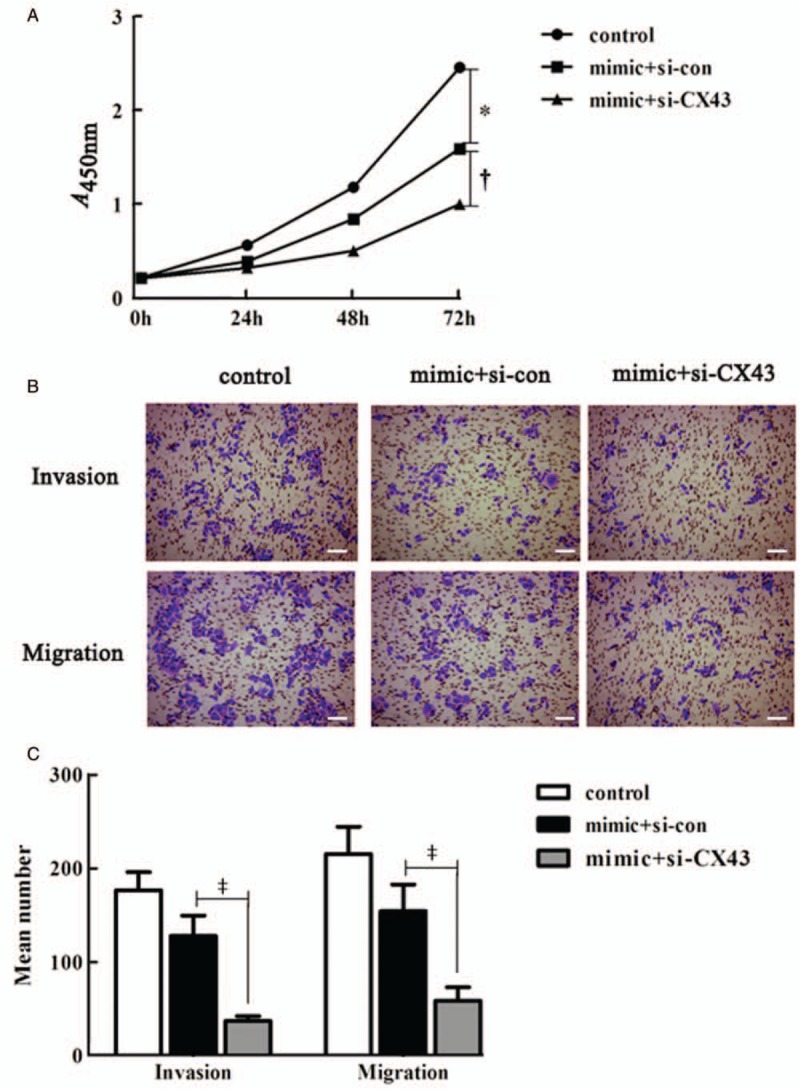
CX43 deletion strengthened the inhibitory effect of miR-139-5p in bladder cancer cells. (A) The proliferation ability of ECV-304 cells. (B) The numbers of invaded and migrated ECV-304 cells (bar = 200 μm). Representative images of crystal violet staining were shown on the upper panel (original magnification ×200). (C) Quantification of (B). ∗P < 0.01 compared with control group. †P < 0.01 compared with mimic + si-con group. ‡P < 0.05 compared with mimic + si-con group. CX43: Connexin 43; MiR-139-5p: MicroRNA-139-5p; Si-con: Si-control.
Discussion
In our present paper, we observed that miR-139-5p was lowly expressed in BC tissues. Besides, miR-139-5p was lowly expressed in BC cell lines and over-expression of miR-139-5p could alleviate the property of BC cells. Moreover, CX43 was a direct target of miR-139-5p and CX43 deletion could strengthen the inhibitory effect of miR-139-5p in BC cells.
The proteins of CX family constitute the gap junction channel. The channel admits diminutive molecules to pass first-hand among neighboring cells and they are metabolically and electrically coupled.[13,14] In human, CX are constituted of 21 proteins, among which ten CX genes mutations are associated with 28 different genetic diseases. Six of these diseases are ascribed to CX43 mutations.[14,15] Recent evidence suggested that after pernicious transformation of cancers, the aberrant regulation of CX and its role in cellular communication might reinforce or restrain tumorigenesis and metastasis.[16]CX43, as a member of CX family, was alleviated in a few of solid tumors, including breast cancer,[17] ovarian cancer,[18] and lung cancer.[19] In our previous study, we found that CX43 was highly expressed in BC tissues.[5,20] Once again, we verified that CX43 might be a potential target for BC treatment.
To predict potential upstream regulatory miRNAs for CX43, we used four prediction softwares miRanda, miRWalk, TargetScan, and miRDB and found that miR-139-5p is one of the miRNAs on the upstream of CX43. Increasing evidences have illustrated that miRs could administer the expression of many genes by replenishing the target mRNAs 3′UTR. We then verified that miR-139-5p could direct bind to the 3′UTR of CX43 by luciferase reporter assay. MiRNAs, about 22 nucleotides in length, are the same category of diminutive non-coding cellular RNAs and can suppress their target genes via dehiscing mRNA molecule or restraining their translation to disturb the post-transcriptional pathways.[21] Growing evidences have revealed that many miRs have been deemed as cancer regulators.[22,23] It has previously been observed that CX43 and/or miR-125b may be remedial targets for human breast cancer.[24] It was indicated that miR-1 targeted CX43 expression to regulate the vascular tutelar effect of angiotensin II.[25] However, there are few reports on the role of miR-139-5p in CX43 expression. Our study demonstrated that miR-139-5p was a direct target of CX43 and CX43 deletion could strengthen the inhibitory effect of miR-139-5p in BC cells.
Our results also indicated that miR-139-5p over-expression restrained the proliferation, invasion, and migration of BC cells. Increasing number of articles evidenced that many miRs may act as tumor-promoters or tumor-inhibitors,[26] such as miR-146b[27] and miR-373[28] function as tumor-promoters, conversely miR-27a[29] and miR-34b-3p[30] function as tumor-inhibitors in BC. Some researches hold the views that miR-139-5p, as a tumor-inhibitor and a latent modulatory miRNA, is normally down-graded in pernicious cancers, such as non-small cell lung cancer,[31] endometrial cancer,[32] colorectal cancer,[33] and BC.[11] Consistent with previous researches, in this paper, we revealed that over-expression of miR-139-5p was dramatically lessened in BC tissues and cell lines. Moreover, restraining of miR-139-5p could recover the inhibitory influences on proliferation, invasion, and migration of BC cell. These suggested that miR-139-5p played as a tumor-inhibitor in BC progression and metastasis.
In general, we demonstrated that miR-139-5p was down-regulated in BC tissues and cell lines. In addition, miR-139-5p may function as a miRNA on the upstream of CX43. Besides, over-expression of miR-139-5p alleviated cell proliferation and metastasis of BC. Furthermore, CX43 mediated the effects of miR-139-5p on cell proliferation, invasion, and migration in BC cells. Therefore, these findings hinted that miR-139-5p and CX43 hold significant potential to serve as therapeutic molecules for BC. However, there are some limitations in our study. This is just a preliminary experiment in the role of CX43 and miR-139-5p on BC. It may need more experiments in other cell lines to support our findings. Moreover, the in-vivo experimental studies are needed to further confirm our results. These experiments are in progress, which will be reported in future articles.
Conflicts of interest
None.
Footnotes
How to cite this article: Chi Q, Wang ZY, Li HY, Song DB, Xu H, Ma G, Wang ZM, Li XM. Tumor-suppressor microRNA-139-5p restrains bladder cancer cell line ECV-304 properties via targeting Connexin 43. Chin Med J 2019;132:2354–2361. doi: 10.1097/CM9.0000000000000455
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Bausch K, Roth E, Heinz S, Horst D, Mathia S, Vlajnic T, et al. Urinary Calprotectin loses specificity as tumour marker due to sterile leukocyturia associated with bladder cancer. PLoS One 2019; 14:e0213549.doi: 10.1371/journal.pone.0213549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powles T, Al E. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature 2014; 194:558–562. doi: 10.1016/j.juro.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Wu Y-T, Chang H-M, Huang H-F, Sheng J-Z, Leung PCK. Bone morphogenetic protein 2 regulates cell–cell communication by down-regulating connexin43 expression in luteinized human granulosa cells. Mol Hum Reprod 2016; 23:155–165. doi: 10.1093/molehr/gaw078. [DOI] [PubMed] [Google Scholar]
- 5.Ai XL, Chi Q, Qiu Y, Li HY, Li DJ, Wang JX, et al. Gap junction protein connexin43 deregulation contributes to bladder carcinogenesis via targeting MAPK pathway. Mol Cell Biochem 2017; 428:109–118. doi: 10.1007/s11010-016-2921-9. [DOI] [PubMed] [Google Scholar]
- 6.Jin Y, Zhou TY, Cao JN, Feng QT, Fu YJ, Xu X, et al. MicroRNA-206 downregulates Connexin 43 in cardiomyocytes to induce cardiac arrhythmias in a transgenic mouse model. Heart Lung Circ 2018; 10:31916–31917. doi: 10.1016/j.hlc.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Lin-Hong J, He-da Z, Jin-hai T. MiR-30a: a novel biomarker and potential therapeutic target for cancer. J Oncol 2018; 8:1–9. doi: 10.1155/2018/5167829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu R, Li H, Wu S, Qu J, Yuan H, Zhou Y, et al. MicroRNA-1246 regulates the radio-sensitizing effect of curcumin in bladder cancer cells via activating P53. Int Urol Nephrol 2019; 5:24.doi: 10.1007/s11255-019-02210-5. [DOI] [PubMed] [Google Scholar]
- 9.Lin ZJ, Ming J, Yang L, Du JZ, Wang N, Luo HJ. Mechanism of regulatory effect of microRNA-206 on Connexin 43 in distant metastasis of breast cancer. Chin Med J 2016; 129:424–434. doi: 10.4103/0366-6999.176071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Y, Deng C, Zhang H, Zhang J, Peng B, Hu C. Long non-coding RNA XIST promotes cell growth and metastasis through regulating miR-139-5p mediated Wnt/β-catenin signaling pathway in bladder cancer. Oncotarget 2017; 8:94554–94568. doi: 10.18632/oncotarget.21791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Luo H, Yang R, Li C, Tong Y, Fan L, Liu X, et al. MicroRNA-139-5p inhibits bladder cancer proliferation and self-renewal by targeting the Bmi1 oncogene. Tumour Biol 2017; 39:1010428317718414.doi: 10.1177/1010428317718414. [DOI] [PubMed] [Google Scholar]
- 12.Yonemori M, Seki N, Yoshino H, Matsushita R, Miyamoto K, Nakagawa M, et al. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci 2016; 107:1233–1242. doi: 10.1111/cas.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathieu V. Introduction: connexins, pannexins and their channels as gatekeepers of organ physiology. Cell Mol Life Sci 2015; 72:2775.doi: 10.1007/s00018-015-1958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srinivas M, Verselis VK, White TW. Human diseases associated with connexin mutations. BBA-Biomembranes 2018; 1860:192–201. doi: 10.1016/j.bbamem.2017.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laird DW, Naus CC, Lampe PD. SnapShot: connexins and disease. Cell 2017; 170:1260–1260.e1. doi: 10.1016/j.cell.2017.08.034. [DOI] [PubMed] [Google Scholar]
- 16.Jiang JX, Penuela S. Connexin and pannexin channels in cancer. BMC Cell Biol 2016; 17:12.doi: 10.1186/s12860-016-0094-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grek CL, Rhett JM, Bruce JS, Ghatnekar GS, Yeh ES. Connexin 43, breast cancer tumor suppressor: missed connections? Cancer Lett 2016; 374:117–126. doi: 10.1016/j.canlet.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Qiu X, Cheng JC, Klausen C, Chang HM, Fan Q, Leung PC. EGF-induced Connexin43 negatively regulates cell proliferation in human ovarian cancer. J Cell Physiol 2015; 231:111–119. doi: 10.1002/jcp.25058. [DOI] [PubMed] [Google Scholar]
- 19.Sun Q, Wang D, Ai G, Tian L, Zhao L, Chen R, et al. Epac1 is involved in cell cycle progression in lung cancer through PKC and Cx43 regulation. Folia Histochem Cyto 2018; 56:21–26. doi: 10.5603/FHC.a2018.0004. [DOI] [PubMed] [Google Scholar]
- 20.Poyet C, Buser L, Roudnicky F, Detmar M, Hermanns T, Mannhard D, et al. Connexin 43 expression predicts poor progression-free survival in patients with non-muscle invasive urothelial bladder cancer. J Clin Pathol 2015; 68:819–824. doi: 10.1136/jclinpath-2015-202898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 22.Majem B, Parrilla A, Jimenez C, Suarez-Cabrera L, Barber M, Marin A, et al. MicroRNA-654-5p suppresses ovarian cancer development impacting on MYC, WNT and AKT pathways. Oncogene 2019; 7:5.doi: 10.1038/s41388-019-0860-0. [DOI] [PubMed] [Google Scholar]
- 23.Nam RK, Benatar T, Wallis CJD, Kobylecky E, Amemiya Y, Sherman C, et al. MicroRNA-139 is a predictor of prostate cancer recurrence and inhibits growth and migration of prostate cancer cells through cell cycle arrest and targeting IGF1R and AXL. Prostate 2019; 79:1422–1438. doi: 10.1002/pros.23871. [DOI] [PubMed] [Google Scholar]
- 24.Maqbool R, Rashid R, Ismail R, Niaz S, Chowdri NA, Hussain MU. The carboxy-terminal domain of connexin 43 (CT-Cx43) modulates the expression of p53 by altering miR-125b expression in low-grade human breast cancers. Cell Oncol (Dordr) 2015; 38:443–451. doi: 10.1007/s13402-015-0240-x. [DOI] [PubMed] [Google Scholar]
- 25.Lei Y, Peng X, Li T, Liu L, Yang G. ERK and miRNA-1 target Cx43 expression and phosphorylation to modulate the vascular protective effect of angiotensin II. Life Sci 2019; 216:59–66. doi: 10.1016/j.lfs.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 26.Fu Y, Shao ZM, He QZ, Jiang BQ, Wu Y, Zhuang ZG. Hsa-miR-206 represses the proliferation and invasion of breast cancer cells by targeting Cx43. Eur Rev Med Pharmacol Sci 2015; 19:2091–2104. [PubMed] [Google Scholar]
- 27.Zhang X, Liu X, Jing Z, Bi J, Li Z, Liu X, et al. The circINTS4/miR-146b/CARMA3 axis promotes tumorigenesis in bladder cancer. Cancer Gene Ther 2019; 2:6.doi: 10.1038/s41417-019-0085-y. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Xu Z, Wang X. miRNA-373 promotes urinary bladder cancer cell proliferation, migration and invasion through upregulating epidermal growth factor receptor. Exp Ther Med 2019; 17:1190–1195. doi: 10.3892/etm.2018.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang C, Liao X, Jin H, Xie F, Zheng F, Li J, et al. MEG3, as a competing endogenous RNA, binds with miR-27a to promote PHLPP2 protein translation and impairs bladder cancer invasion. Mol Ther Nucleic Acids 2019; 16:51–62. doi: 10.1016/j.omtn.2019.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan Y, Zhang T, Zhou L, Liu S, Liang C. MiR-34b-3p represses the multidrug-chemoresistance of bladder cancer cells by regulating the CCND2 and P2RY1 genes. Med Sci Monit 2019; 25:1323–1335. doi: 10.12659/MSM.913746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yong-Hao Y, Xian-Guo W, Ming X, Jin-Ping Z. Expression and clinical significance of miR-139-5p in non-small cell lung cancer. J Int Med Res 2019; 47:867–874. doi: 10.1177/0300060518815379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Li C, Jiang Y, Wan Y, Zhou S, Cheng W. Tumor-suppressor role of miR-139-5p in endometrial cancer. Cancer Cell Int 2018; 18:51.doi: 10.1186/s12935-018-0545-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyoshi J, Toden S, Yoshida K, Toiyama Y, Alberts SR, Kusunoki M, et al. MiR-139-5p as a novel serum biomarker for recurrence and metastasis in colorectal cancer. Sci Rep 2017; 7:43393.doi: 10.1038/srep43393. [DOI] [PMC free article] [PubMed] [Google Scholar]


