Abstract
Background:
The phenotypic switching of Candida spp. plays an important role in the development of vulvovaginal candidiasis (VVC). Farnesol, as a quorum-sensing molecule in Candida albicans, has the ability to prevent yeast-to-hyphal conversion in vitro. However, the mechanism underlying this ability is unclear. This study aimed to investigate changes in protein levels to better understand how farnesol impacts processes contributing to VVC.
Methods:
The isobaric tag for relative and absolute quantitation technique was used to detect protein expression in C. albicans strain SC5314 (ATCC MYA-2876) with or without farnesol exposure. Proteins with a threshold fold change greater than 1.5 were screened and considered differentially expressed proteins. All the altered proteins were analyzed using Gene Ontology annotation, Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation, and metabolic pathway annotation.
Results:
Between the farnesol-exposed group and the farnesol-unexposd group, we detected 297 altered proteins among all 2047 tested proteins based on a threshold fold change of more than 1.5 (P < 0.05). Eighty-seven of the 297 altered proteins exhibited metabolic enzyme activity and participated in 85 metabolic pathways according to KEGG pathway analysis. Most of these metabolic pathways were associated with central carbon metabolism processes. In the sterol synthesis pathway, which involves the synthesis of farnesol, ERG25 (methylsterol monooxygenase) and ERG4 (delta 24(24(1))-sterol reductase) were both down-regulated in the farnesol-exposed group. All six altered proteases associated with the oxidative phosphorylation process were down-regulated in the farnesol-exposed group relative to the farnesol-unexposed group.
Conclusions:
The mechanisms underlying farnesol-induced phenotype switching involves the adjustment of metabolic activities and epigenetic modification. Exogenous farnesol had an evident, but non-deterministic effect on the synthesis of ergosterol. The potential drug activity of farnesol warrants further investigation.
Keywords: Farnesol, Candida albicans, Proteomic analysis, Morphogenesis
Introduction
Candida albicans (C. albicans) is a polymorphic fungus that exists as either a commensal organism or an opportunistic pathogen. It is capable of causing superficial to life-threatening infection.[1] Although vulvovaginal candidiasis (VVC) is considered a superficial fungal infection, it is known for its characteristic of high recurrence.[2] As a result, it can cause long-term suffering.[3]C. albicans, as an opportunistic pathogen, exhibits several virulence traits, such as degradation enzyme production,[4] biofilm formation, and phenotype switching.[1] Penotypic switching was critical for cells adaptation in response to changes environment. During the development of VVC, filamentous growth was key cytotoxicity changing in C. albicans adaptation. Such growth is correlated with altered metabolism activities and increased pathogenic activities, such as adhesion, invasion, and hydrolase secretion.[1,5] All of these activities are important for the success of this organism as an opportunistic pathogen.[6]
Farnesol is an intermediate product in the fungal ergosterol biosynthetic process. Farnesol, as a quorum-sensing molecule (QSM), is able to prevent the hyphal formation in C. albicans.[7,8] Hence, it is thought to play a critical role in the physiological adaptation and pathogenic processes in C. albicans.[9] Studies have discovered that farnesol is involved in regulating cell density, controlling the growth of hyphae,[8,10] blocking the formation of biofilm,[11,12] and inducing cell apoptosis.[13] These characteristics suggest the potential of farnesol as a novel tool to combat fungal infections or as a potential drug treatment for candidiasis. However, the mechanism by which farnesol prevents C. albicans from invading its host is unknown.[14]
In our previous study, we found that in addition to inhibiting germination in C. albicans at low concentrations, farnesol did not disturb the growth, metabolism, or cytomorphology of vaginal Lactobacillus spp., even at high concentrations.[15] This characteristic of farnesol is very important for restoring and maintaining a healthy vaginal environment during vaginitis treatment.[16] To elucidate the mechanisms underlying farnesol's cytotoxicity to C. albicans, we used an isobaric tag for relative and absolute quantitation (iTRAQ) analysis of protein expression in C. albicans strain SC5314 to investigate altered proteins by comparing farnesol-treated C. albicans cells to normal, control C. albicans cells. We systematically analyzed protein expression in C. albicans exposed 100 μmol farnesol environment; this concentration was selected based on previous work demonstrating that this concentration could markedly inhibit yeast-to-hypha conversion in C. albicans. Interestingly, a number of altered proteins were found by comparing these two groups. The systematic protein analysis in this study provides insight into the mechanisms responsible for farnesol-related C. albicans morphogenesis and virulence.
Methods
Organisms and growth conditions
Farnesol (Sigma-Aldrich Chemical, St. Louis, MO, USA) was obtained as a 3 mol/L stock solution and diluted to a 30 mmol/L solution in 100% methanol. For each of three independent experiments, an aliquot of glycerol stock of C. albicans strain was resuscitated and grown on Sabouraud Dextrose Agar (Gibco-Thermo, Waltham, USA). Cultures were grown overnight in yeast extract peptone dextrose medium (Gibco-Thermo) in an orbital shaker (38×g) at 35°C under aerobic conditions. Cultures were harvested and washed twice in sterile phosphate-buffered saline (PBS). Next, the cultures were re-suspended to an optical density of 0.5 McFarland, which were used as seed cultures for all experiments. A seed culture (1 mL) was inoculated into 100 mL RPM1640 (Gibco-Thermo) medium and shaken (72×g) at 35°C for 6 h. Cells treated with 100 μmol/L farnesol were termed “group F.” As a negative control, cells treated without farnesol were termed “group O.” Three pairs of group F and group O samples were subjected to proteomic profiling by iTRAQ and liquid chromatography-tandem mass spectrometry (LC-MS/MS).
Sample preparation
For crude protein extraction, the harvested cells were re-suspended in PBS. Then, protein lysis buffer (4% sodium dodecyl sulfate [SDS], 0.1% phenylmethylsulfonyl fluoride, 1× cocktail) was added to the fungal mass at a buffer-to-thallus volume ratio of 1:50. A few glass beads were added. The mixture was vibrated at high frequency with a tissue grind mill for 2 min, followed by 2 min cooling on ice; this process was repeated five times. The lysate was sonicated for 5 min (with a cycle of sonicate 5 s, rest 10 s) and then subjected to 10 min of boiling. The lysate was then cooled and centrifuged at 15,575×g for 30 min at 4°C. The supernatant was collected and stored at − 80°C. The protein concentration was measured by a commercially available kit, the bicinchoninic acid (BCA) Protein Assay Kit (Biovision, Francisco, USA). The protein concentration was calculated according to a standard curve. Based on the manufacturer's instructions, equal amounts of protein were boiled with loading buffer and subjected to SDS polyacrylamide gel electrophoresis to evaluate the quality of protein and precision of quantification.
Trypsin digestion and iTRAQ labeling
The reagents used were provided with the iTRAQ kit. 100 μg protein was taken from each sample and was subjected to the following processes. Each pair repeated three times. The protein samples reduced by dithiothreitol at 60°C for 1.0 h, alkylated with iodoacetamide at 37°C for 0.5 h in the dark, added five times volume pre-cooled acetone into the tube and incubated at −20°C for 1 h. After centrifugation at 13,350×g for 20 min at 4°C, discarded the supernatant, and added 20 μL dissolution buffer to dissolve the precipitated pellet, the protein then digested with trypsin (Sigma-Aldrich Chemical) at an enzyme-to-substrate ratio of 1:20 at 37°C. The resulting peptides were de-salted and vacuum dried, then labeled according to the manufacturer's protocol for iTRAQ kit. Finally, labeled peptide mixtures pooled, de-salted and dried by vacuum centrifugation.
Reverse phase high-performance liquid chromatograph analysis
The pooled peptide mixtures were fractionated into 40 fractions by ultra-performance liquid chromatography (WATERS, Milford, USA) using a C18 column (WATERS beh C18 2.1 × 50 mm, 1.7 μm). The fractions were then combined into ten fractions to lower the complexity of the peptide mixture and enhance the accuracy and throughput of protein identification. Chromatography was performed with solvent A (Milli-Q, Billerica, USA) (water with 2% acetonitrile [ACN] and 20 mmol/L ammonium formate, pH = 10) and solvent B (90% ACN). Peptides were eluted at 600 nL/min for 5% to 35% solvent B over 16 min. The ultraviolet detection wavelength was 214 nm.
LC-MS/MS analysis
The fractionated samples were analyzed by QTrap 6500 mass spectrometer (AB Sciex, Beijing, China) interfaced with nanoLC 400-3 (Eksigent, Washington D.C., USA). The column parameters were C18, 5 μm, and 150 mm × 75 μm. Peptides were re-suspended in buffer A (0.1% formic acid [FA], 2% ACN) and centrifuged at 20,000×g for 2 min. The supernatant was transferred into the column and eluted with a linear gradient of 5% buffer B (0.1% FA in 98% ACN) for 5 min, 5% to 40% solvent B for 50 min and 40% to 80% for 1 min at a constant flow rate of 300 nL/min on a nanoLC 400-3 (Eksigent, SiliconVstreet) system. The mass spectrometric analysis parameters were as follows: spray voltage 2500 v; full scam m/z range 350 to 1250. Up to 30 of the most intense precursors were selected for fragmentation per cycle, with a dynamic exclusion time of 25 s.
Statistical analysis
To ensure highly robust data, four successive processes performed: database searching, data quality control, data quantification, and quantitative quality control. For protein identification, MS/MS spectra were searched using Mascot (version 2.3.0, Matrix Science, London, UK) against the UniProt_TREMBL_Candida albicans database. All the identified proteins were subjected to protein quantification analysis. Statistical analyses were performed using SPSS software version 21.0 (IBM SPSS Inc., Chicago, IL, USA). To compare the differences between the two groups, independent-samples t tests were performed. The differentially expressed proteins (DEPs) were screened according to a threshold fold change of more than 1.5. Differences were considered statistically significant at P < 0.05. All the altered proteins were subjected to Gene Ontology (GO) analysis via Blast2GO software (https://www.blast2go.com/blast2go-pro).
Results
Statistical analysis of differentially expressed proteins
Clustering analysis was performed with Multiple Array Viewer software, as shown in Figure 1A. By constructing a scatter diagram and performing median analysis of the protein abundance values in each sample, we found that the interclass distributions of group O and group F were similar. A volcano plot was constructed to show the quantitative distributions of the DEPs in group F vs. group O (F/O) (with the median value of sample O used as internal reference). Proteomic analysis of farnesol-exposed cells compared with unexposed cells revealed a significant number of DEPs, as shown in Figure 1B. In total, 2047 proteins were identified, and 1958 of these proteins were quantified. A total of 297 DEPs (F/O) were screened according to the threshold fold change of >1.5 and P < 0.05. Among these altered proteins, 238 proteins were up-regulated, and 59 proteins were down-regulated. All these 297 altered proteins were subjected to GO analysis performed by Blast2GO software (https://www.blast2go.com/blast2go-pro) and classified among the GO categories cellular component, molecular function, and biological process, as shown in Figure 2.
Figure 1.
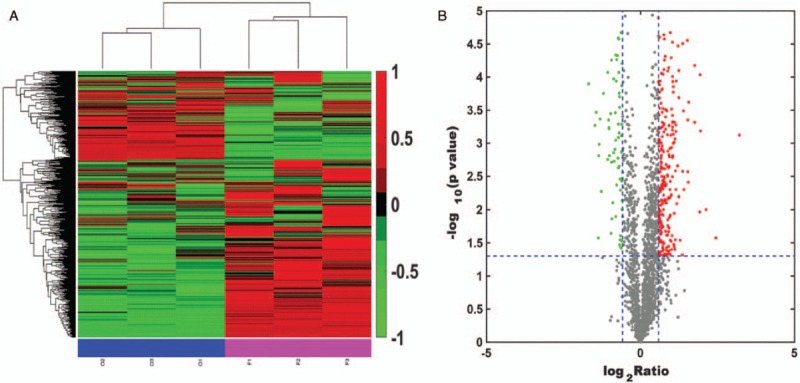
Identification of differentially expressed proteins in a farnesol-exposed group (sample F) relative to a farnesol-unexposed group (sample O). (A) O1, O2, and O3 represent three replicates of sample O; F1, F2, and F3 represent three replicates of sample F. Each column represents a sample, and each line represents a gene. Proteins with similar expression patterns in different samples are clustered together. Red represents up-regulation, and green represents down-regulation. (B) Volcano plot representing protein abundance changes (F [farnesol-exposed group] vs. O [farnesol-unexposed group]). The X-axis presents the protein quantification ratio (F/O); the Y-axis presents the P-values from replicate experiments. Each point represents a protein. Red points are up-regulated proteins, and green points are down-regulated proteins.
Figure 2.
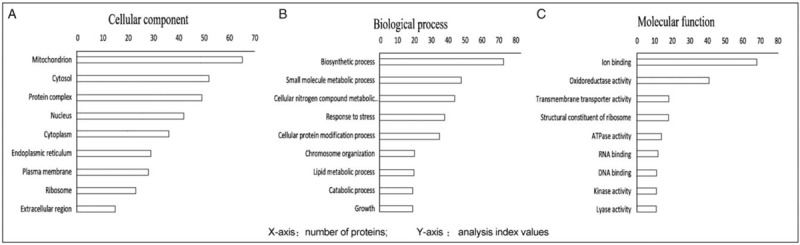
Gene Ontology analysis of differentially expressed proteins. (A) Cellular component category, (B) biological process category, (C) molecular function category.
Metabolic enzyme function analysis of DEPs
According to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database classification criteria, we identified 87 DEPs annotated as metabolic enzymes, including Hydrolases > Oxidoreductases > Transferases > Lyases > Isomerases > Ligases. To analyze the information in molecular interaction networks, such as pathways and complexes (the “Pathway” database), information about genes and proteins generated by genome projects (including gene databases) and information about biochemical compounds and reactions (including compound and reaction databases), we searched all 87 DEPs in the KEGG database and obtained 85 KEGG matches. The category biosynthetic process included the greatest number of altered proteins, followed by small-molecule metabolic process, cellular nitrogen compounds metabolic process, response to stress, and cellular protein modification process. Among the altered proteins in farnesol-exposed cells, the majority were involved in central carbon metabolism and oxidative phosphorylation, as shown in Table 1.
Table 1.
The identified differentially expressed proteins involved in metabolic pathways.
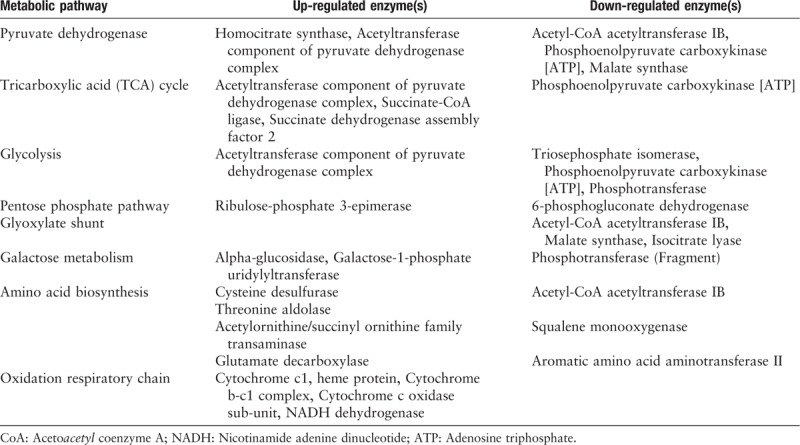
Regarding the glycolysis process, in C. albicans exposed to farnesol, several altered proteins that exhibit glycolytic enzyme activity, such as triosephosphate isomerase, phosphoenolpyruvate carboxykinase, and phosphotransferase, were found to be down-regulated, whereas the acetyltransferase component of pyruvate dehydrogenase complex was up-regulated. Moreover, we found five altered enzymes, including up-regulated enzymes (homocitrate synthase, acetyltransferase component of pyruvate dehydrogenase complex) and down-regulated enzymes (acetyl-CoA acetyltransferase IB, phosphoenolpyruvate carboxykinase. and malate synthase), that may be involved in the pyruvate metabolic pathway. Furthermore, we found several altered proteins that participate in the tricarboxylic acid cycle process. For example, acetyltransferase component of pyruvate dehydrogenase complex, succinate-CoA ligase, and succinate dehydrogenase assembly factor 2 were up-regulated while phosphoenolpyruvate carboxykinase was down-regulated in the farnesol-exposed C. albicans. In this study, four altered enzymes involved in the oxidation respiratory chain were all up-regulated in cells exposed to 100 μmol farnesol, as shown in Table 1. In the ergosterol biosynthesis pathway, we found only two enzymes, ERG25 (delta24(24(1))-sterol reductase) and ERG4 (methylsterol monooxygenase), that were down-regulated; this down-regulation may affect the ergosterol biosynthesis pathway in fungi, as shown in Figure 3.
Figure 3.
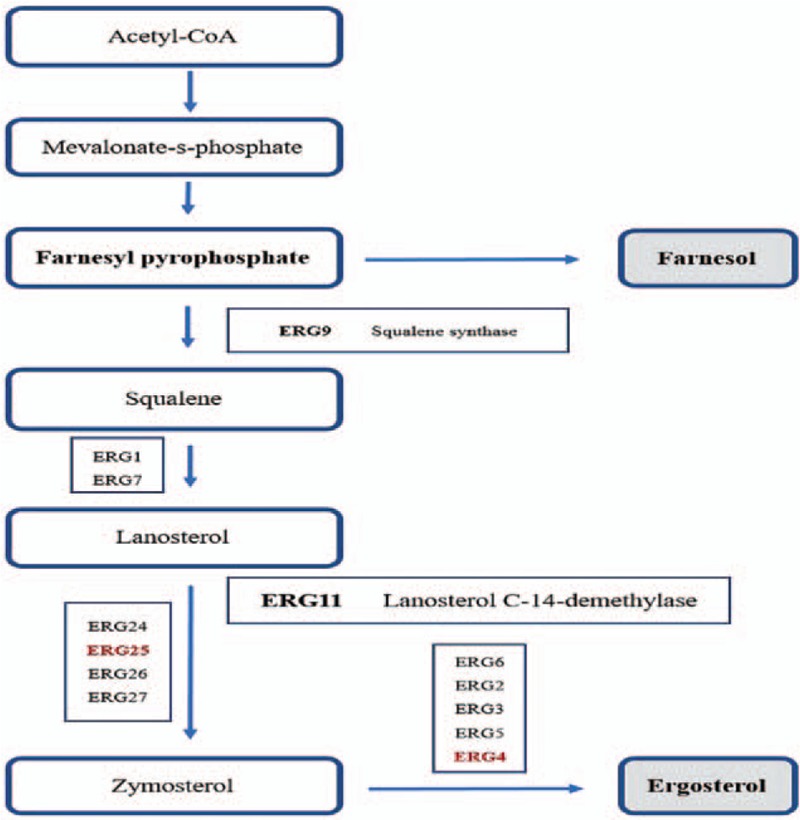
The ergosterol biosynthesis pathway. Proteases presented in red text were methylsterol monooxygenase (ERG25) and delta 24(24(1))-sterol reductase (ERG4), which were both down-regulated in farnesol-exposed cells. The detailed biosynthesis pathway is available at https://www.genome.jp/kegg/pathway.htm. CoA: Acetoacetyl coenzyme A.
Discussion
Farnesol is a QSM produced by C. albicans.[7] It can opportunistically regulate the growth of C. albicans cells in response to cell density to optimize growth under different environmental conditions.[8,9,17] In addition, it has been investigated for its anti-microbial potential and proposed as a potential novel drug for candidiasis treatment due to its complex activities, such as blocking the yeast-to-hyphal switch,[10] blocking biofilm formation,[12] affecting interspecies communication,[8] and mediating cell death.[18] In the present study, we systematically analyzed protein levels in a farnesol environment to elucidate the mechanisms underlying farnesol activities and cytotoxicity in C. albicans. As a result, we found that 87 of the 297 identified altered proteins exhibited metabolic enzyme activity when cells were exposed to farnesol. The metabolic enzymes tended to be associated with central carbon metabolism processes, such as glycometabolism, amino acid metabolism, and lipid metabolic process. Previous studies have found that primary metabolic pathways in C. albicans, such as central carbon metabolism and sterol biosynthesis, are closely associated with dimorphic transition and virulence. For example, pyruvate dehydrogenase multi-complex enzyme is associated with filamentous growth; acetyl-CoA is required for the morphogenesis of C. albicans.[19] In this study, we found two up-regulated enzymes and three down-regulated enzymes in farnesol-exposed C. albicans that are potentially involved in the pyruvate metabolic pathway. The protein analysis implied that phenotype switching induced by farnesol involve the adjustment of metabolic activities and cytotoxicity induction; however, the relationships between farnesol activities and primary metabolism adjustment are stronger than we thought.
The compounds generated in the ergosterol biosynthesis pathway, which are essential for membrane integrity and permeability, are necessary for the maintenance of cellular structure in fungi. The sterol biosynthesis pathway is targeted by several anti-fungal agents, such as azole drugs.[20,21] Farnesol, as the most well-characterized QSM involved in C. albicans morphogenesis, is generated in the ergosterol biosynthesis pathway.[13] In the present study, we found that both ERG25 (methylsterol monooxygenase) and ERG4 (delta 24(24(1))-sterol reductase) that participate in the ergosterol biosynthesis process were down-regulated in C. albicans exposed to exogenous farnesol. However, neither of these two proteins are critical enzymes in this pathway. Thus, the impact of farnesol on the biosynthesis of ergosterol may be nondeterministic.
Mitochondrial release of reactive oxygen species (ROS), which can damage cellular components, is an early event in apoptotic cell death. C. albicans cells must use specific defense mechanisms to neutralize ROS, such as regulating oxidation respiratory chain activity.[9,14,22] In this study, we consistently found that four altered enzymes that participate in the oxidation respiratory chain were up-regulated in cells exposed to 100 μmol farnesol. Additionally, the expression of phospholipase and mannosyltransferase, which are associated with cytotoxicity and adhesion, was down-regulated. These responses warrant further study.
This study was a preliminary study. We found many altered enzymes involved in many central carbon metabolism pathways, which provide a basis for future studies. Studies focusing on a specific signaling pathway are needed. Such work will be valuable for enhancing our understanding of the pathogenesis of C. albicans.
Our previous study showed that farnesol significantly affects the morphogenetic process in C. albicans in a concentration-dependent manner. Farnesol (100 μmol) can markedly inhibit the yeast-to-hypha conversion in C. albicans.[15] Based on previous work, we systematically analyzed protein expression in C. albicans in a 100 μmol farnesol environment and found a number of altered proteins. The processes central carbon metabolism, response to stress and cellular protein modification process were highly affected by the presence of farnesol. The systematic analysis of protein expression in this study provided insight into the metabolic mechanisms responsible for farnesol-related C. albicans morphogenesis and virulence.
Funding
This work was supported by a grant from the National Natural Science Foundation of China (No. 81571394).
Conflicts of interest
None.
Footnotes
How to cite this article: Wang FJ, Liu ZH. Systematic analysis of protein expression in Candida albicans exposed to farnesol. Chin Med J 2019;132:2348–2353. doi: 10.1097/CM9.0000000000000420.
References
- 1.Noble SM, Gianetti BA, Witchley JN. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol 2017; 15:96–108. doi: 10.1038/nrmicro.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denning DW, Kneale M, Sobel JD, Rautemaa-Richardson R. Global burden of recurrent vulvovaginal candidiasis: a systematic review. Lancet Infect Dis 2018; 18:e339–e347. doi: 10.1016/S1473-3099(18)30103-8. [DOI] [PubMed] [Google Scholar]
- 3.Leusink P, van de Pasch S, Teunissen D, Laan ET, Lagro-Janssen AL. The relationship between vulvovaginal candidiasis and provoked vulvodynia: a systematic review. J Sex Med 2018; 15:1310–1321. doi: 10.1016/j.jsxm.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Cassone A. Vulvovaginal Candida albicans infections: pathogenesis, immunity and vaccine prospects. BJOG 2015; 122:785–794. doi: 10.1111/1471-0528.12994. [DOI] [PubMed] [Google Scholar]
- 5.Tong Y, Tang J. Candida albicans infection and intestinal immunity. Microbiol Res 2017; 198:27–35. doi: 10.1016/j.micres.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Jabra-Rizk MA, Kong EF, Tsui C, Nguyen MH, Clancy CJ, Fidel PJ, et al. Candida albicans pathogenesis: fitting within the host-microbe damage response framework. Infect Immun 2016; 84:2724–2739. doi: 10.1128/IAI.00469-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langford ML, Atkin AL, Nickerson KW. Cellular interactions of farnesol, a quorum-sensing molecule produced by Candida albicans. Future Microbiol 2009; 4:1353–1362. doi: 10.2217/fmb.09.98. [DOI] [PubMed] [Google Scholar]
- 8.Egbe NE, Dornelles TO, Paget CM, Castelli LM, Ashe MP. Farnesol inhibits translation to limit growth and filamentation in C. albicans and S. cerevisiae. Microb Cell 2017; 4:294–304. doi: 10.15698/mic2017.09.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wongsuk T, Pumeesat P, Luplertlop N. Fungal quorum sensing molecules: role in fungal morphogenesis and pathogenicity. J Basic Microbiol 2016; 56:440–447. doi: 10.1002/jobm.201500759. [DOI] [PubMed] [Google Scholar]
- 10.Lindsay AK, Deveau A, Piispanen AE, Hogan DA. Farnesol and cyclic AMP signaling effects on the hypha-to-yeast transition in Candida albicans. Eukaryot Cell 2012; 11:1219–1225. doi: 10.1128/EC.00144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monteiro DR, Arias LS, Fernandes RA, Deszo DSL, de Castilho M, Da RT, et al. Antifungal activity of tyrosol and farnesol used in combination against Candida species in the planktonic state or forming biofilms. J Appl Microbiol 2017; 123:392–400. doi: 10.1111/jam.13513. [DOI] [PubMed] [Google Scholar]
- 12.Sebaa S, Boucherit-Otmani Z, Courtois P. Effects of tyrosol and farnesol on Candida albicans biofilm. Mol Med Rep 2019; 19:3201–3209. doi: 10.3892/mmr.2019.9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta P, Sharma M, Arora N, Pruthi V, Poluri KM. Chemistry and biology of farnesol and its derivatives: quorum sensing molecules with immense therapeutic potential. Curr Top Med Chem 2018; 18:1937–1954. doi: 10.2174/1568026619666181210124159. [DOI] [PubMed] [Google Scholar]
- 14.Polke M, Leonhardt I, Kurzai O, Jacobsen ID. Farnesol signalling in Candida albicans – more than just communication. Crit Rev Microbiol 2018; 44:230–243. doi: 10.1080/1040841X.2017.1337711. [DOI] [PubMed] [Google Scholar]
- 15.Wang F, Liu Z, Zhang D, Niu X. In vitro activity of farnesol against vaginal Lactobacillus spp. Eur J Obstet Gynecol Reprod Biol 2017; 212:25–29. doi: 10.1016/j.ejogrb.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 16.Bradford LL, Ravel J. The vaginal mycobiome: a contemporary perspective on fungi in women's health and diseases. Virulence 2017; 8:342–351. doi: 10.1080/21505594.2016.1237332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polke M, Jacobsen ID. Quorum sensing by farnesol revisited. Curr Genet 2017; 63:791–797. doi: 10.1007/s00294-017-0683-x. [DOI] [PubMed] [Google Scholar]
- 18.Xia J, Qian F, Xu W, Zhang Z, Wei X. In vitro inhibitory effects of farnesol and interactions between farnesol and antifungals against biofilms of Candida albicans resistant strains. Biofouling 2017; 33:283–293. doi: 10.1080/08927014.2017.1295304. [DOI] [PubMed] [Google Scholar]
- 19.Han TL, Cannon RD, Villas-Boas SG. The metabolic basis of Candida albicans morphogenesis and quorum sensing. Fungal Genet Biol 2011; 48:747–463. doi: 10.1016/j.fgb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Lv QZ, Yan L, Jiang YY. The synthesis, regulation, and functions of sterols in Candida albicans: well-known but still lots to learn. Virulence 2016; 7:649–659. doi: 10.1080/21505594.2016.1188236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wiederhold NP. The antifungal arsenal: alternative drugs and future targets. Int J Antimicrob Agents 2018; 51:333–339. doi: 10.1016/j.ijantimicag.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Jung YY, Hwang ST, Sethi G, Fan L, Arfuso F, Ahn KS. Potential anti-inflammatory and anti-cancer properties of farnesol. Molecules 2018; 23:E2827.doi: 10.3390/molecules23112827. [DOI] [PMC free article] [PubMed] [Google Scholar]


