Abstract
In this work we have established a collision cross section (CCS) library of primary metabolites based on analytical standards in the Mass Spectrometry Metabolite Library of Standards (MSMLS) using a commercially available ion mobility-mass spectrometer (IM-MS). From the 554 unique compounds in the MSMLS plate library, we obtained a total of 1246 CCS measurements over a wide range of biochemical classes and adduct types. Resulting data analysis demonstrated that the curated CCS library provides broad molecular coverage of metabolic pathways and highlights intrinsic mass/mobility relationships for specific metabolite super classes. The separation and characterization of isomeric metabolites were assessed, and all molecular species contained within the plate library, including isomers, were critically evaluated in order to determine the analytical separation efficiency in both the mass (m/z) and mobility (CCS/ΔCCS) dimension required for untargeted metabolomic analyses. To further demonstrate the analytical utility of CCS as an additional molecular descriptor, a well-characterized biological sample of human plasma serum (NIST SRM 1950) was examined by LC-IM-MS and used to provide a detailed isomeric analysis of carbohydrate constituents by ion mobility.
Graphical abstract
From the central dogma of molecular biology, studies of genomics, transcriptomics, and proteomics provide higher order information about gene and protein expression to better understand implicated phenotypes.1,2 However, these approaches provide limited information about real-time production of chemical species related to cellular metabolism as a function of external stimuli or phenotype of interest. To address the need for rapid characterization of cellular metabolism, metabolomics seeks to uncover molecular information on a per-molecule basis by examining expressed cellular products that can be correlated with a specific phenotype, stimuli, or other experimental conditions.3
While several analytical approaches have been utilized to study metabolism and related cellular processes (e.g. NMR, electrochemistry, etc.),4,5 mass spectrometry (MS) is gaining wide-spread adoption as a result of its high throughput, low limits of detection, and molecular specificity. Mass spectrometers can collect chemical information on the microsecond (μs) time scale,6 and with the rise of high-resolution, accurate mass techniques such as time of flight (TOF), Orbitrap, and ion cyclotron instruments, a unique chemical formula can often be generated based solely on mass measurement for a specific analyte signal.7,8 While identifying a specific chemical formula is advantageous, many metabolic pathways include isomeric molecules covering a range of biological classes, such as carbohydrates (e.g. glucose/galactose),9 nucleosides (e.g. adenosine/deoxyguanosine), and lipids (7-dehydrocholesterol/desmosterol).10 As biological function follows molecular structure, characterization of isomeric species is imperative for complete molecular identification and accurate pathway analysis. In many MS experiments, fragmentation techniques such as collision induced dissociation (CID) or electron transfer dissociation (ETD) are utilized to provide structural information about a specific analyte measured in the study.11,12 However, as many metabolite isomers are less than 300 Dalton, these compounds often possess identical fragmentation spectra at similar energy thresholds and hence molecular fingerprinting by MS/MS and high resolution precursor mass is often not specific enough to identify a unique molecular structure.13 Furthermore, as quadrupoles isolate on nominal mass, molecules with different molecular formulas but similar exact mass (i.e. nominal mass isobars) cannot be isolated, thereby complicating MS/MS analysis.14 To address these challenges, pre-separation techniques such as gas and liquid chromatography,15,16 and more recently ion mobility spectrometry,17 have been interfaced prior to mass analysis to provide enhanced structural recognition and increased analyte coverage. For un-targeted analysis, metabolomic databases (e.g. METLIN, HMDB, etc)18 include multiple descriptors of analyte information (e.g. accurate mass, ion adduct form, fragmentation pattern, and retention time) to increase confidence in molecular identification.19 With the advent of commercially-available ion mobility-mass spectrometers in 2006,20 collision cross section (CCS) has become an additional molecular descriptor for untargeted experiments. CCS measurements are being standardized across instrumental platforms using rigid experimental protocols, and as such provide a molecular descriptor independent of system settings which are transferable be-tween laboratories.17,21–23 These collected CCS measurements provide the capability to distinguish isomeric species in complex mixtures, provided enough resolution is accessible in the IM dimension.24 In order to provide additional confidence in molecular identification for untargeted metabolomic analysis, significant efforts are being made in the IM community to establish reliable CCS databases for analyzing unknown features across a range of biochemical classes, including lipids, metabolites, and xenobitics.21,22,25,26 In this work, we use uniform field IM-MS to develop a library of CCS values focused on primary metabolites established with analytical standards to facilitate chemical identification in untargeted metabolomic workflows. Furthermore, we demonstrate the utility of these measurements by analyzing a commercially available extract of human serum (NIST 1950) which has been characterized previously in traditional GC and LC-MS experiments.27,28
Experimental Methods
MSMLS Sample Preparation.
The Mass Spectrometry Metabolite Library of Standards (MSMLS, IROA technologies) is supplied as dried standards distributed across seven 96-well plates (Sigma-Aldrich; St. Louis, MO) and each well contains 5 μg of analytical standard. All solvents used to reconstitute the analytes prior to analysis, including water (H2O), metha-nol (MeOH), acetonitrile (ACN), isopropanol (IPA), and chloroform (CHCl3) were Optima LC-MS grade purchased from Fisher Scientific (Fair Lawn, NJ). Stock solutions of the hydrophilic standards were prepared by adding 100 μL 1:9 (MeOH: H2O) to each well prior to mixing on a waving rotator for 5 minutes. The stocks were then distributed in 20 μL aliquots throughout five 96-well plates (Waters part no. 186005837). Stock plates that were not immediately analyzed were capped and transferred to −80 °C for storage. Working solutions of the hydrophilic standards were prepared by adding 80 μL of water with 0.1% formic acid to the 20 μL stock solutions, sealed with plate covers (Waters part no. 186006332), and subsequently mixed on a waving rotator for 5 minutes. The hydrophobic analyte set was prepared similarly, where stock solutions were prepared with 100 μL 2:1:1:0.3 (MeOH: CHCl3: IPA: H2O), and distributed in 20 μL aliquots throughout five 96-well plates. Working solutions were prepared by adding 80 μL of 1:1 (MeOH: IPA). The concentration of the working solutions used for IM-MS analysis was 10 μg/mL.
Collision Cross Section Measurements.
CCS measurements for the MSMLS were obtained on a commercially available drift tube ion mobility-mass spectrometer (DTIMS, Agilent 6560) operated with nitrogen gas (3.95 Torr) at room temperature (~25 °C) and using both single-field and stepped-field approaches previously established in an international in-ter-laboratory study.21 The single-field CCS values reported here were measured in triplicate, while the stepped-field values were collected in a single acquisition. Stepped-field measurements were acquired using an automated flow injection analysis (FIA) stepped-field approach described previously.29 Briefly, the FIA method was performed with a liquid chromatography system (Agilent 1290) modified with a 100 μL sample loop (Agilent part no. G4226–87303) coupled to an IM-MS (6560, Agilent). 20 μL of the working solution was injected from the 96-well plate with 1:1 (water: isopropanol) as the carrier solvent. For traditional stepped-field CCS determination by FIA, following a 0.5 s delay, an entrance potential was stepped every 0.5 min. in increments of 100 V from 1074 V to 1674 V; the first step from 1074 to 1174 occurred at 1.0 minute rather than 0.5 min. For single-field CCS determination using FIA, 4 μL of sample was injected into the carrier solvent at a flow of 800 μL/min. Data was collected for 0.5 min, followed by a 0.4 min postrun flushing cycle. A drift tube entrance voltage of 1574 V was used. DTIMS exhibits a linear relationship between drift time and CCS,6 and single-field CCS values are determined by first measuring the drift time of ions (ESI Low Concentration Tuning Mix, Agilent) with a known CCS. The calibrant ions were infused for 0.5 minutes while IM-MS spectra are collected; calibration experiments were preformed intermittently to ensure instrument stability. IM-MS Browser (Agilent, B.08) was used to plot the linear regression of the calibration ions for single field experiments, and the instrumental coefficients β and Tfix, were extracted and used to convert raw ion drift times to CCS.21 The resulting single-and stepped-field CCS library can be found in the Supporting Information.
IM-MS Source and Drift Cell Conditions.
To obtain high coverage of analytes within the MSMLS, both electrospray (Agilent Jet Stream, AJS) and chemical ionization (APCI) sources were used. The majority of the samples collected with the AJS in both ion modes were measured using the following conditions: gas temperature, 250 °C; drying gas, 8 L/min; nebulizer, 60 psig; sheath gas temperature, 300 °C; sheath gas flow, 11 L/min; capillary voltage (VCap), 3500 V; nozzle voltage, 800 V; fragmentor, 340 V; octopole 1 RF Vpp, 750 V. All metabolites were first investigated using the AJS source; those which were not observed in either ion polarity were subsequently investigated using the APCI source under the following conditions: gas temperature, 250 °C; vaporizer, 200 °C; drying gas, 7 L/min; nebulizer, 30 psig; VCap, 3800 V; corona, 5 μA; fragmentor, 350 V; octopole 1 RF Vpp 750 V. Some of the low m/z ions (typically ≤ 200 Da) exhibited metastable ion dis-sociation in the DTIMS which resulted in uncorrelated mobil-ities (Supplemental Figure S1). In these cases, we increased the fragmentor potential to > 350 V and decreased the Trap Funnel RF to ≤ 80 Vpp to culminate the ion signal into a single IM distribution. The IM-MS settings for the CCS values reported herein are as follows: 0.9 frames/s; 18 IM transients/frame; 60 ms max drift time; 600 TOF transients/IM transient; 20000 μs trap fill time; 180 μs trap release time; drift tube exit voltage, 224 V; rear funnel entrance voltage, 217.5 V; rear funnel exit voltage, 45 V.
Nonlinear Regression Analysis.
Iterative nonlinear regression modeling for the super classes was performed using GraphPad Prism 7, and 99% confidence intervals were generated for each biomolecular super class. Three fits were tested for each super class: power fit (PF), 4-parameter sigmoidal (4P), and 5-parameter sigmoidal (5P). The most parsimonious fit was chosen by a probabilistic comparison of the corrected Akaike information criterion (AICc) values.
Human Serum Preparation.
Protein precipitation was performed by adding 800 μL of ice cold MeOH to 100μL NIST 1950 serum and stored at −80 ˚C for one hour. The sample was centrifuged at 14,000 rpm for 5 minutes before collecting the supernatant. Next, 2.4 mL ice cold methyl tert-butyl ether and 800 μL ice cold water were added. The sample was vortexed then centrifuged at 10,000 rpm at 4˚C for 10 minutes. The polar and nonpolar fractions were separated and dried separately in vacuo. Samples were stored at −20 ˚C until analysis. Dried fractions were resuspended in 200 μL of the initial mobile phase solvent and analyzed via LC-IM-MS.
Liquid Chromatography.
LC-IM-MS was performed on the prepared NIST 1950 serum using HILIC chromatography for the hydrophilic layer of the liquid-liquid extraction. For this method, 4 μL of sample was injected onto a column heated to 40 °C. The Millipore SeQuant Zic-HILIC (2.1 × 100 mm, 3.5 μm) column was used with mobile phase A and B being 9:1 and 1:9 (water: acetonitrile, buffered with 5 mM ammonium formate), respectively. The mobile phase flow rate was 200 μL/min. The gradient was initially held at 98 %B from 0 to 1 minutes, decreased to 45 %B from 1 to 20 minutes, held at 45 % B from 20 to 22 minutes, increased to 98 %B from 22 to 40 minutes, and subsequently held at 98 %B from 40 to 45 minutes before the next injection.
Results and Discussion
MSMLS Plate Coverage.
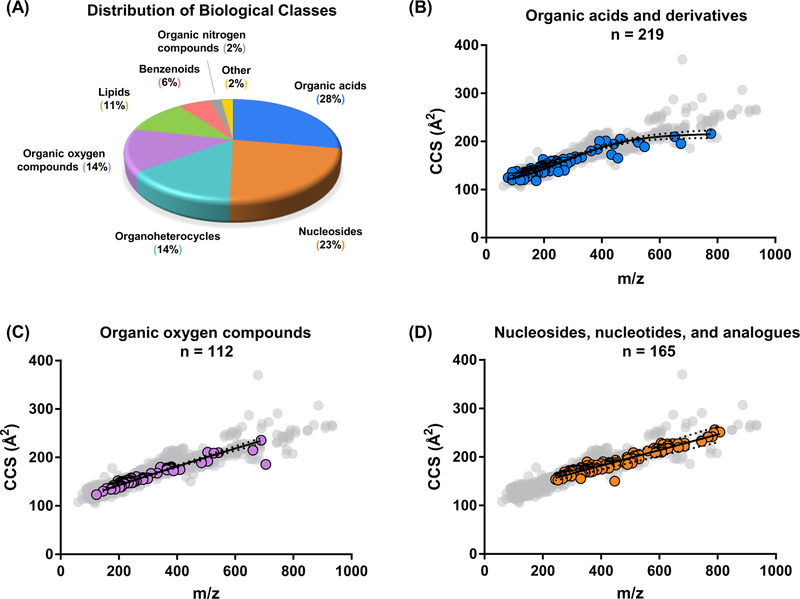
In total, the MSMLS plates analyzed in this work contained 554 unique compounds across a large breadth of biological classes found in canonical metabolite pathways (See Figure 1A). Of these 554 analytes, one or more CCS values were measured for 417, resulting in ca. 75% coverage. Of the remaining ca. 25% that did not result in an acceptable CCS measurement, approximately half of these did not yield appreciable signals, presumably because of difficulties in ionization under the conditions used here, or poor ion transmission efficiency due to the lower mass of many of these analytes and the limits of RF frequency used in ion transfer optics. For the remaining species, there is in some cases, evidence for metastable dissociation in the drift tube, resulting in uncorrelated mobility and poor peak shape (See Supplemental Figure S1). For purposes of this manuscript, we have chosen to report only values for signals demonstrating high signal intensity and reproducibility. Collectively, these 417 analytes produced 1246 CCS measurements using both positive (701 measurements) and negative ion polarities (545 measurements) across several adduct types (e.g. [M+Na]+, [M-H]−, etc., see Supplemental Figure S2). Analyte identification and relevant descriptors (chemical name, formula, KEGG ID, Metlin ID, adduct type, measured mass, CCS, and other information) have been uploaded to Metabolomics Workbench,30 and are provided in the Supporting information as two Personal Compound Database Libraries (PCDL Manager, Agilent), one corresponding to single-field measurements and the other to stepped-field CCS measurements.
Figure 1.
(A) Distribution of biological categories associated with the primary metabolites examined in the MSMLS plate library. (B, C, and D) Conformational space plots of three singly charged molecular super classes contained in the MSMLS library. Representative nonlinear regression fits (solid black lines) along with 99 % confidence intervals (black dotted lines) are shown for each. Gray dots denote all molecules CCS values obtained in the library. All CCS error bars are smaller than their respective symbols. (B) “Organic acids and derivatives” with a 4-parameter sigmoidal fit. (C) “Organic oxygen compounds” with a power fit and (D) “Nucleosides, nucleotides, and analogues” with a power fit.
Correlation Analysis.
In these data, we observed several distinct relationships between m/z and CCS for individual structural super classes represented in the MSMLS library similar to previous IM-MS literature.31–35 Mass/mobility relationships have been shown to have utility as an additional rapid identifier of biomolecular class for uncharacterized biological samples,36 making the mathematical description of these relationships by nonlinear regression modeling particularly useful. Unlike the canonical biochemical classes (nucleotides, proteins, carbohydrates, and lipids), metabolites exhibit less distinguished structural differences between chemical classes, and so several mathematical fits were investigated in order to find mass/mobility correlations which exhibit high class specificity. Fits and confidence intervals for representative super classes are shown in Figure 1 (B, C, and D), and detailed mathematical expressions are provided in the Supporting in-formation (Supplemental Equations 1-6).
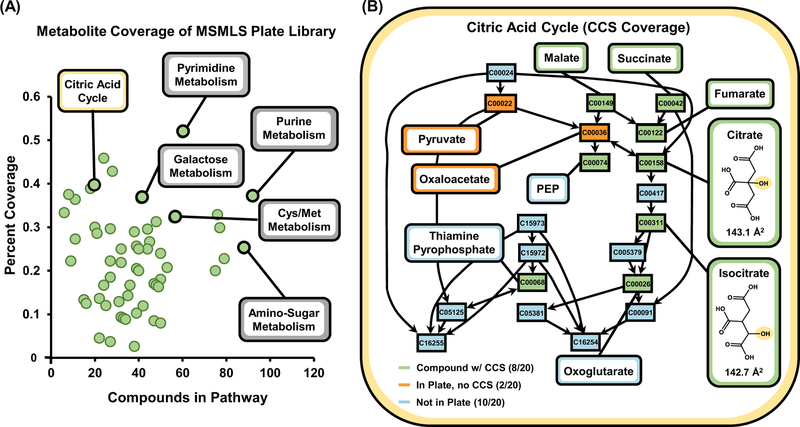
Metabolic Pathway Coverage.
As the MSMLS was designed to provide analytical standards of primary metabolism, we also evaluated metabolite coverage using pathway analysis by inputting KEGG IDs for all of the analytes measured in our CCS database in MetaboAnalyst 4.0.37 In total, 64 pathways were covered with a wide range of biological activity including key metabolic processes such as the citric acid cycle, amino acid metabolism, and glycolysis (Figure 2A and Supplemental Table S1). Pathway coverage presented in this work is solely based on analyte coverage from the standards, and therefore provides qualitative reflection on the number of analytes in each specific pathway which are accounted for in the CCS library. MetaboAnalyst 4.0 also provides detailed infor-mation for specific pathways of interest, wherein molecular coverage can be evaluated on a per-analyte basis. For example, 10 pivotal metabolites in the citric acid cycle (see Figure 2B) are represented within the standards, and out of 20 total, 8 of these molecules exhibited a measurable CCS (green), while only 2 (orange) were observable in the mass spectrum, but did not result in a collected CCS, due to low ion intensity. Of note, many other compounds described in the KEGG pathways which are not components in the standard set (10 compounds, light blue) are protein enzymes or oxidized derivatives, and only 3 of these 10 are available for purchase as analytical standards. Hence it is unlikely that 100 % coverage of canonical pathways is obtainable with chemical standards. As an analogy, it is not necessary to have 100 % peptide coverage for a specific protein in proteomic analysis for confident identifictation.
Figure 2.
(A) KEGG pathway coverage of metabolites with measured CCS evaluated in this study. A total of 64 pathways are covered by metabolites in our CCS library based on the MSMLS. After a specific pathway is selected (B), metabolite-specific coverage can be evaluated. In many pathways, isomerization is a key intermediate in primary metabolism, noted by the callouts for citrate and isocitrate in the citric acid cycle.
Isomers in Metabolomics.
Of the more than 500 compounds in the MSMLS library, almost one-third (31%) have a chemical formula in common with another compound, forming an isomeric pair, Supplemental Figure S3. Isomeric compounds are ubiquitous in metabolomic processes across a wide range of biological classes, for example the carbohydrate rearrangements for glucose 6-phosphate isomerization to fructose 6-phosphate in glycolysis. Figure 2B highlights two key metabolic intermediates of the citric acid cycle, citrate and isocitrate, which are constitutional rearrangements of a single hydroxyl group along the central carbon chain. As these compounds have the same chemical formula, they will also possess identical masses, requiring additional separation in the chromatographic dimension for increased identification confidence in pathway analysis.17,38 In the example depicted in Figure 2B, ion mobility allows for differentiation of these two isomeric metabolites (CCS = 143.1 Å2 vs. CCS = 142.7 Å2, for citrate and isocitrate, respectively), which are indistinguishable by mass alone. Adding the ion mobility dimension to existing un-targeted workflows allows for additional separation and characterization of isomeric metabolites that interfaces within the timescale of traditional chromatographic techniques.6
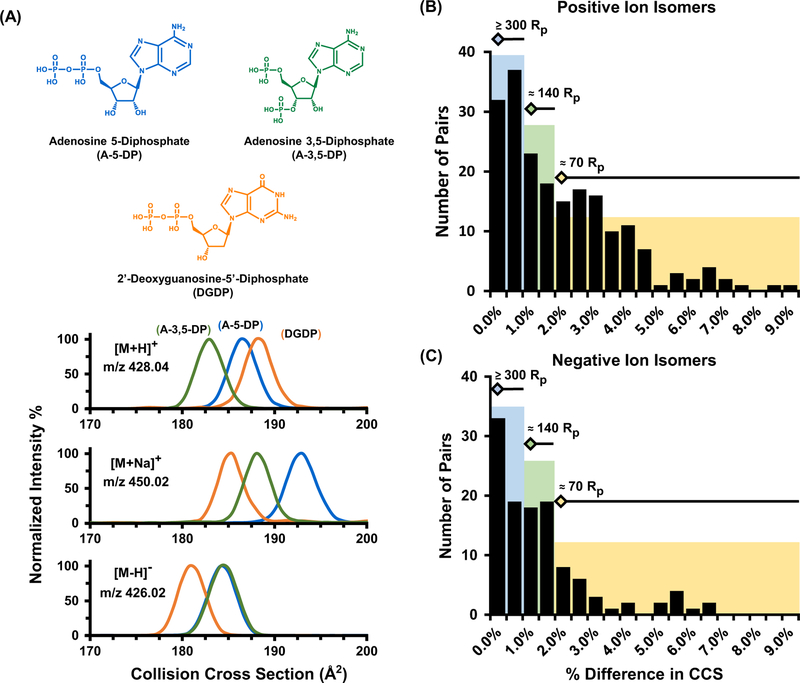
As a specific example, adenosine 5-diphosphate, adenosine 3,5-diphosphate, and 2’-deoxyguanosine-5’-diphosphate are nucleoside isomers which are key metabolites in purine metabolism and are depicted in Figure 3A. Note that the only structural difference between A-5-DP (blue) and A-3,5-DP (green) is the location of a phosphate group from the central ribose unit. These two isomers are in turn differentiated structurally from 2’-deoxyguanosine-5’-diphosphate (DGDP, orange) by molecular substitutions on the purine ring, where a hydroxyl group has been relocated from the ribose sugar to the guanine ring, as well as an amine rearrangement in the same region. Structurally, these three nucleoside compounds are also constitutional isomers, a subcategory of isomeric compounds which have been heavily characterized in previous ion mobility literature.39–41 Also noted in Figure 3A, adduct formation has a substantial effect on the overall selectivity of the IM separation. Specifically, each nucleoside isomer has a distinct cross sectional distribution which are distinguishable in the protonated [M+H]+ and sodium adducted [M+Na]+ species, however the rearrangement of the phosphate group between A-5-DP and A-3,5-DP provides no resolution for the deprotonated form observed in negative ion mode [M-H]−. Other metabolite separations in this study were more readily separated in negative ion mode such as the iso-mers L-glutamic acid and N-methyl-D-aspartic acid (see Supplemental Figure S4).
Figure 3.
(A) IM separation of nucleoside isomers (chemical structures illustrated at the top of the panel) for [M+H]+, [M+Na]+, and [M-H]− ion forms, respectively. For these particular isomers, enhanced separation is noted for the sodium adducts, while the negative mode A-5-DP [M-H]−and A-3,5-DP CCS distributions are indistinguishable. After sorting all observed isomer sets in the MSMLS dataset, pairwise matches were created and evaluated based on their percent difference in CCS. The resulting difficulty in separations is noted in panels (B) for positive and (C) negative ion forms.
The broad range of chemical diversity present in small molecules presents unique advantages in the range of ion types that can be utilized. Collectively, these results demonstrate the advantage of utilizing both ion polarities in untargeted analysis wherein various charge adducts formed during the ionization process can be exploited in order to substantially enhance the selectivity in IM-MS analysis, by increasing the absolute CCS difference between isomers. This allows a significant improvement in separation without instrumental upgrades that would otherwise be necessary to achieve improved separation via increased resolving power. This enhanced separation, in turn, provides additional confidence in identification through CCS library matches and enhanced ion mobility resolution. A potential future direction in the field will utilize molecular modeling and machine learning approaches for prediction of adduct specific CCS values and optimal separation conditions.
IM-MS Separation in Primary Metabolites.
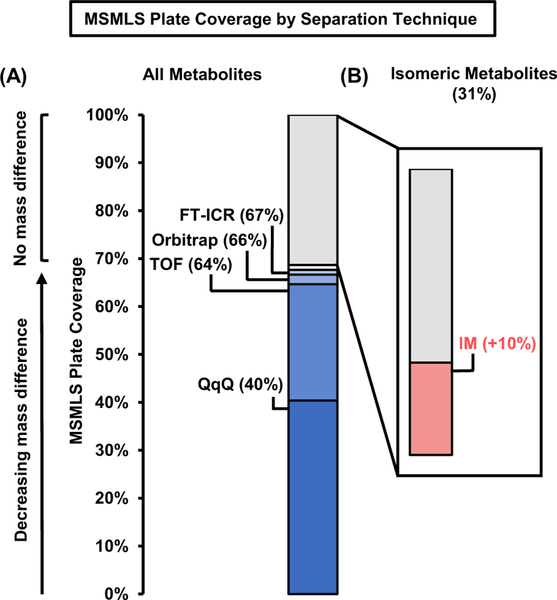
In addition to enhanced separation through charged, adduct formation, recent advances in ion mobility resolving power have provided increased separation coverage of isomeric species.42,43 In order to determine the resolving power in the IM dimension needed for untargeted metabolomic experiments, we analyzed pair-wise matches of all isomers which provided a usable CCS and binned the resulting pairs by percent difference in cross section (%ΔCCS). In brief, analytes with identical chemical formulas were grouped into isomeric sets and were subsequently matched in a pairwise comparison. Each pairwise match was generated using an enumeration strategy wherein a percent difference in CCS was calculated for each possible combination of isomers. Most isomeric sets consist of 2–3 compounds, whereas the largest isomeric set was comprised of 9 unique analytes (see Supplemental Figure S3). In one example, there are 5 sugar compounds which share the same chemical formula (C6H13O9PNa+, exact m/z 283.0195), which results in a total of 10 pairwise isomer matches in this analysis. The percent difference in CCS for all isomer matches were calculated, and the compiled results for all isomer pairings are displayed in Figure 3B (positive ion mode) and Figure 3C (negative ion mode). Approximately half of the isomer pairs generated are ≥ 2.0% different in CCS and require ca. 70 resolving power (CCS/ΔCCS) to separate at half height.43,44 In order to separate additional isomers, more resolving power would be required (ca. 140 for ~1.0–2.0% difference in CCS, and ca. >300 for ~0.5% CCS difference). Currently, only two commercially available IM-MS platforms provide this level of resolving power (i.e. atmospheric pressure DTIMS and trapped IMS),39,44 although several research instrument prototypes have been developed which are capable of accessing resolving powers in excess of 300 (CCS/ΔCCS).6,45
While IM instruments are continually increasing in resolving power capabilities, current untargeted metabolomic work-flows identification is based first on primary mass measurement and subsequently supported with retention time, isotope ratios, and fragmentation matching. From this view-point, it is also imperative to describe how much resolving power in the mass dimension is necessary for metabolomic studies. By sorting the entire MSMLS library based on primary mass alone, our analysis shows that most analytes (64%) are resolvable based only on the mass dimension utilizing 40,000 mass resolving power (e.g. high resolution TOF, see Figure 4). Increasing levels of mass resolving power (300,000 for Orbitrap and up to 40,000,000 for FT-ICR, respectively)46,47 provides minimal increases in resolution of these metabolites (ca. 3% more). As ca. 30% of the compounds in the MSMLS library are isomers, essentially no level of increased mass spectrometry efficiency (short of exited state isomer resolution with MS resolving power of ca. 10 billion as theorized by Marshall and coworkers.48) will be able to resolve these compounds, and hence orthogonal separation techniques are still required (i.e. GC, LC, or IM). Modest resolving power for commercially available IM instrumentation (ca. 70 CCS/ΔCCS) resolves an additional 10% of compounds in the library, which outweighs the benefits of additional mass resolving power beyond 40,000 (e.g. TOF MS). We note, however, that this analysis does not consider mass measurement accuracy, which is typically higher for FTMS instruments (Orbitrap and FT-ICR). Nevertheless, in order to obtain the widest scope of molecular coverage in untargeted workflows, possessing sequential separa-tion dimensions based on chemical affinity, gas-phase area, and m/z (LC-IM-MS) would strengthen analyte identification strategies
Figure 4.
MSMLS plate coverage using different separation strategies. (A) Many analytes contained in the library can be resolved in the mass dimension at modest resolving power (TOF Rp = 40,000), with only incremental increases in coverage resulting from the use of an instrument with significantly higher resolving power (FT-ICR Rp = 40,000,000). (B) The addition of IM prior to mass analysis allows for isomeric separation and thus increases plate coverage by 10%.
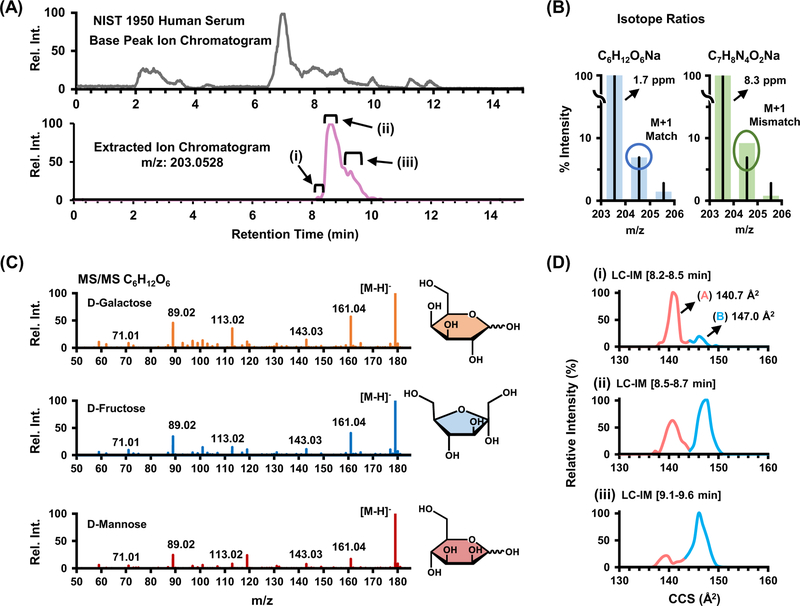
LC-IM-MS Characterization of NIST 1950 Serum
The NIST 1950 human serum standard has been previously characterized in the literature,27,28,49 and is analyzed in this work to underscore the importance of isomeric characterization in un-targeted experiments. Separation and characterization of isomeric species in biological extracts often requires multiple steps of chemical separation in order to gain increased confidence in assigning molecular structure. For example, the base peak chromatogram in Figure 5A shows the molecular complexity of the NIST 1950 human serum and the extracted ion chromatogram (lower trace) details a specific molecular feature at m/z 203.0528 that elutes into an unresolved broad peak over a ca. 2 minute chromatographic window. This broad distribution in the elution profile indicates the potential presence of multiple isomeric forms with similar, yet not identical, retention times. Although TOF MS has high resolving power (ca. 40,000), potentially two chemical formula are within 10 ppm of the measured m/z (C6H12O6Na and C7H8N4O2Na, at 1.7 ppm and 8.3 ppm respectively; see Figure 5C). While assignment of this feature to chemical formula C6H12O6Na is more probable due to lower observed mass error, isotope ratios were used to confirm this molecular formula assignment, wherein the relative abundance of the M+1 peak in the serum more closely aligns with the isotope model for C6H12O6Na as opposed to C7H8N4O2Na (Figure 5B). However, even after a specific molecular formula is determined, 9 potential isomers (including both constitutional rearrangements and stereochemistry for this chemical formula) exist within the MSMLS standards, all carbohydrates. These isomers possess almost identical fragmentation profiles (see [M-H]− ion, Figure 5C), and sophisticated algorithms for identification by MS/MS are needed, an observation which has been previously noted in other carbohydrate studies.50 Note that Figure 5C utilizes the deprotonated ion of C6H12O6, as the [M+Na]+ species noted in the other panels provides no fragmentation spectra due to ejection of the sodium charge carrier during collisional activation. Although previous studies utilized relative abundance ratios of fragment ions to determine molecular structure, this technique is time intensive and currently is not readily amended to rapid structural determination in untargeted workflows.50 Similar to the chromatographic profile, ion mobility distributions obtained at three separate time points in the chromatogram (roman numerals) also indicate two separate chemical species present in the serum (Figure 5D). The collision cross sections measured for these two distributions helps narrow potential chemical structures from 9 potential isomeric forms down to 4 tentative identifications based on a CCS match within 1%. The smaller distribution at 140.7 Å2 (light red, A) closely aligns with 3 isomers of C6H12O6 in the standards (fructose, galactose and mannose at ca. 141.5 Å2. The larger distribution at 147.0 Å2 (light blue, B) closely aligns with α-D-glucose, which is noted at 146.3 Å2 in the database. Alt-hough in this example ion mobility does not provide definitive identification of the compounds observed in the NIST serum, it does significantly reduce the possible candidates from the 9 potential structures noted in the database. By using collision cross section as additional metric for tentative identification, additional confidence in identifying molecular signatures can be gained in untargeted metabolomics.
Figure 5.
(A) HILIC base peak chromatogram for NIST 1950 human serum sample and (lower trace) the extracted ion chromatogram of m/z 203.0528, which consists of two distributions of interest that were further examined by isotope ratio pattern and ion mobility for structural char-acterization. (B) Expected and measured isotope ratio abundances for two possible chemical formulas corresponding to m/z 203 within 10 ppm. The chemical formula C6H12O6 [M+Na]+ more closely aligns with experimental measurements from the NIST serum both on basis of mass accuracy (2 ppm) and isotope ratio pattern (M+1). (C) Fragmentation spectra for isomers of with shared chemical formula C6H12O6 [M-H]−. (D) Selected ion mobility distributions for m/z 203 extracted over three time points in the chromatographic dimension.
Conclusions.
In this work we have developed a CCS library based on primary metabolites obtained from the MSMLS library of analyte standards. As many key intermediates across metabolic pathways are formed through isomerization processes, utilizing orthogonal dimensions of separation in addition to mass analysis is imperative to fully characterize metabolic pathways. The intrinsic mass/mobility relationships for metabolites noted in this work, and others, illustrates a reproducible method of characterization for biochemical classes which interfaces seamlessly into the timescale of traditional LC/GC-MS workflows. Furthermore, we demonstrated that while additional resolving power in the m/z dimension is always advantageous, the diminishing returns of these efforts may not offset the additional analysis time required for ultrahigh resolution mass acquisition (i.e. FT processes). However, orthogonal separation techniques such as LC and IM can often resolve many isomeric forms, facilitating their identification for a more comprehensive understanding of the biochemical implications of experimental samples. Finally, we have demonstrated the advantages of adding CCS as a molecular descriptor in untargeted metabolomic analyses through characterization of a well-studied human serum extract (NIST 1950) by LC-IM-MS.
Supplementary Material
ACKNOWLEDGMENT
Financial support for aspects of this research was provided by The National Institutes of Health (NIH NIGMS R01GM092218 and NCI R03CA222–452-01) and under Assistance Agreement No. 83573601 awarded by the U. S. Environmental Protection Agency. This work has not been formally reviewed by EPA. The views expressed in this document are solely those of the authors and do not necessarily reflect those of the funding agencies and organizations. EPA does not endorse any products or commercial services mentioned in this publication.
Footnotes
ASSOCIATED CONTENT
Supporting Information.
Experimental IM-MS spectra illustrating metastable ion fragmentation during mobility analysis for fragile ion clusters, MetaboAnalyst Pathway analysis results, mass/mobility fitting equations, ion adduct types, and isomer results are included in the Supporting Information in addition to the CCS library and PCDL information generated from this study.
The Supporting information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.8b04322.
The authors declare no competing financial interest.
REFERENCES
- (1).CRICK F. Nature 1970, 227 (5258), 561–563. [DOI] [PubMed] [Google Scholar]
- (2).Joyce AR; Palsson BØ Nat. Rev. Mol. Cell Biol 2006, 7 (3), 198–210. [DOI] [PubMed] [Google Scholar]
- (3).Goodacre R; Broadhurst D; Smilde AK; Kristal BS; Baker JD; Beger R; Bessant C; Connor S; Capuani G; Craig A; Ebbels T; Kell DB; Manetti C; Newton J; Paternostro G; Somorjai R; Sjöström M; Trygg J; Wulfert F. Metabolomics 2007, 3 (3), 231–241. [Google Scholar]
- (4).Sumner S; Snyder R; Burgess J; Myers C; Tyl R; Sloan C; Fennell T. J. Appl. Toxicol 2009, 29 (8), 703–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Kimmel DW; Dole WP; Cliffel DE J. Lipids 2017, 2017, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).May JC; McLean JA Analytical Chemistry. 2015, pp 1422–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Kind, T.; Tsugawa, H.; Cajka, T.; Ma, Y.; Lai, Z.; Mehta, S. S.; Wohlgemuth, G.; Kumar, D.; Showalter, M. R.; Arita, M.; Fiehn, O. 2018, No. March 2017, 513–532. [DOI] [PMC free article] [PubMed]
- (8).Kim S; Rodgers RP; Marshall AG Int. J. Mass Spectrom 2006, 251 (2–3), 260–265. [Google Scholar]
- (9).Hofmann J; Hahm HS; Seeberger PH; Pagel K. Nature 2015, 526 (7572), 241–244. [DOI] [PubMed] [Google Scholar]
- (10).Kyle JE; Zhang X; Weitz KK; Monroe ME; Ibrahim YM; Moore RJ; Cha J; Sun X; Lovelace ES; Wagoner J; Polyak SJ; Metz TO; Dey SK; Smith RD; Burnum-Johnson KE; Baker ES Analyst 2016, 141 (5), 1649–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Yost RA; Enke CG; McGilvery DC; Smith D; Morrison JD Int. J. Mass Spectrom. Ion Phys 1979, 30 (2), 127–136. [Google Scholar]
- (12).Lareau NM; May JC; McLean JA Analyst 2015, 140 (10), 3335–3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dodds JN; May JC; McLean JA Anal. Chem 2017, 89 (1), 952–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Ekroos K; Chernushevich IV; Simons K; Shevchenko A. Anal. Chem 2002, 74 (5), 941–949. [DOI] [PubMed] [Google Scholar]
- (15).Kanani H; Chrysanthopoulos PK; Klapa MI J. Chromatogr. B Anal. Technol. Biomed. Life Sci 2008, 871 (2), 191–201. [DOI] [PubMed] [Google Scholar]
- (16).Lu W; Bennett BD; Rabinowitz JD J. Chromatogr. B Anal. Technol. Biomed. Life Sci 2008, 871 (2), 236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Mairinger T; Causon TJ; Hann S. Curr. Opin. Chem. Biol 2018, 42, 9–15. [DOI] [PubMed] [Google Scholar]
- (18).Smith C. a; O’Maille G; Want EJ; Qin C; Trauger SA; Brandon TR; Custodio DE; Abagyan R; Siuzdak G. Ther. Drug Monit 2005, 27 (6), 747–751. [DOI] [PubMed] [Google Scholar]
- (19).Schrimpe-Rutledge AC; Codreanu SG; Sherrod SD; McLean JA J. Am. Soc. Mass Spectrom 2016, 27 (12), 1897–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Pringle SD; Giles K; Wildgoose JL; Williams JP; Slade SE; Thalassinos K; Bateman RH; Bowers MT; Scrivens JH Int. J. Mass Spectrom 2007, 261 (1), 1–12. [Google Scholar]
- (21).Stow SM; Causon TJ; Zheng X; Kurulugama RT; Mairinger T; May JC; Rennie EE; Baker ES; Smith RD; McLean JA; Hann S; Fjeldsted JC Anal. Chem 2017, 89 (17), 9048–9055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Paglia G; Williams JP; Menikarachchi L; Thompson JW; Tyldesley-Worster R; Halldórsson S; Rolfsson O; Moseley A; Grant D; Langridge J; Palsson BO; Astarita G. Anal. Chem 2014, 86 (8), 3985–3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Paglia G; Astarita G. Nat. Protoc 2017, 12 (4), 797–813. [DOI] [PubMed] [Google Scholar]
- (24).Pu Y; Ridgeway ME; Glaskin RS; Park MA; Costello CE; Lin C. Anal. Chem 2016, 88 (7), 3440–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Paglia G; Angel P; Williams JP; Richardson K; Olivos HJ; Thompson JW; Menikarachchi L; Lai S; Walsh C; Moseley A; Plumb RS; Grant DF; Palsson BO; Langridge J; Geromanos S; Astarita G. Anal. Chem 2015, 87 (2), 1137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Zheng X; Aly NA; Zhou Y; Dupuis KT; Bilbao A; Paurus VL; Orton DJ; Wilson R; Payne SH; Smith RD; Baker ES Chem. Sci 2017, 8, 7724–7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Simón-Manso Y; Lowenthal MS; Kilpatrick LE; Sampson ML; Telu KH; Rudnick PA; Mallard WG; Bearden DW; Schock TB; Tchekhovskoi DV; Blonder N; Yan X; Liang Y; Zheng Y; Wallace WE; Neta P; Phinney KW; Remaley AT; Stein SE Anal. Chem 2013, 85 (24), 11725–11731. [DOI] [PubMed] [Google Scholar]
- (28).Telu KH; Yan X; Wallace WE; Stein SE; Simón-Manso Y. Rapid Commun. Mass Spectrom 2016, 30 (5), 581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Nichols CM; May JC; Sherrod SD; McLean JA Analyst 2018, 143 (7), 1556–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Sud M; Fahy E; Cotter D; Azam K; Vadivelu I; Burant C; Edison A; Fiehn O; Higashi R; Nair KS; Sumner S; Subramaniam S. Nucleic Acids Res 2016, 44 (D1), D463–D470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).May JC; Goodwin CR; Lareau NM; Leaptrot KL; Morris CB; Kurulugama RT; Mordehai A; Klein C; Barry W; Darland E; Overney G; Imatani K; Stafford GC; Fjeldsted JC; McLean JA Anal. Chem 2014, 86 (4), 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Hines KM; Ashfaq S; Davidson JM; Opalenik SR; Wikswo JP; McLean JA Anal. Chem 2013, 85 (7), 3651–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hines KM; Ross DH; Davidson KL; Bush MF; Xu L. Anal. Chem 2017, 89 (17), 9023–9030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Dwivedi P; Wu P; Klopsch SJ; Puzon GJ; Xun L; Hill HH Metabolomics 2008, 4 (1), 63–80. [Google Scholar]
- (35).McLean JA J. Am. Soc. Mass Spectrom 2009, 20 (10), 1775–1781. [DOI] [PubMed] [Google Scholar]
- (36).Goodwin CR; Fenn LS; Derewacz DK; Bachmann BO; McLean JA J. Nat. Prod 2012, 75 (1), 48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Chong J; Soufan O; Li C; Caraus I; Li S; Bourque G; Wishart DS; Xia J. Nucleic Acids Res 2018, 46 (W1), W486–W494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).May JC; McLean JA Annu. Rev. Anal. Chem 2016, 9 (1), 387–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Groessl M; Graf S; Knochenmuss R. Analyst 2015, 140 (20), 6904–6911. [DOI] [PubMed] [Google Scholar]
- (40).Li H; Giles K; Bendiak B; Kaplan K; Siems WF; Hill HH Anal. Chem 2012, 84 (7), 3231–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Giles K; Williams JP; Campuzano I. Rapid Commun. Mass Spectrom 2011, 25 (11), 1559–1566. [DOI] [PubMed] [Google Scholar]
- (42).D’Atri V; Causon T; Hernandez-Alba O; Mutabazi A; Veuthey JL; Cianferani S; Guillarme DJ Sep. Sci 2018, 41 (1), 20–67. [DOI] [PubMed] [Google Scholar]
- (43).Dodds JN; May JC; McLean JA Anal. Chem 2017, 89 (22), 12176–12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Fernandez-Lima FA; Kaplan DA; Park MA Rev. Sci. Instrum 2011, 82 (12). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Deng L; Ibrahim YM; Baker ES; Aly NA; Hamid AM; Zhang X; Zheng X; Garimella SVB; Webb IK; Prost SA; Sandoval JA; Norheim RV; Anderson GA; Tolmachev AV; Smith RD ChemistrySelect 2016, 1 (10), 2396–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).N. Nikolaev E; N. Vladimirov G; Jertz R; Baykut G. Mass Spectrom 2013, 2, S0010–S0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Zubarev RA; Makarov A. Anal. Chem 2013, 85 (11), 5288–5296. [DOI] [PubMed] [Google Scholar]
- (48).Marshall AG; Hendrickson CL; Shi SD-H Anal. Chem 2002, 74 (9), 252 A–259 A. [DOI] [PubMed] [Google Scholar]
- (49).Bowden JA; Heckert A; Ulmer CZ; Jones CM; Koelmel JP; Abdullah L; Ahonen L; Alnouti Y; Armando AM; Asara JM; Bamba T; Barr JR; Bergquist J; Borchers CH; Brandsma J; Breitkopf SB; Cajka T; Cazenave-Gassiot A; Checa A; Cinel MA; Colas RA; Cremers S; Dennis EA; Evans JE; Fauland A; Fiehn O; Gardner MS; Garrett TJ; Gotlinger KH; Han J; Huang Y; Neo AH; Hyötyläinen T; Izumi Y; Jiang H; Jiang H; Jiang J; Kachman M; Kiyonami R; Klavins K; Klose C; Köfeler HC; Kolmert J; Koal T; Koster G; Kuklenyik Z; Kurland IJ; Leadley M; Lin K; Maddipati KR; McDougall D; Meikle PJ; Mellett NA; Monnin C; Moseley MA; Nandakumar R; Oresic M; Patterson R; Peake D; Pierce JS; Post M; Postle AD; Pugh R; Qiu Y; Quehenberger O; Ramrup P; Rees J; Rembiesa B; Reynaud D; Roth MR; Sales S; Schuhmann K; Schwartzman ML; Serhan CN; Shevchenko A; Somerville SE; St. John-Williams L; Surma MA; Takeda H; Thakare R; Thompson JW; Torta F; Triebl A; Trötzmüller M; Ubhayasekera SJK; Vuckovic D; Weir JM; Welti R; Wenk MR; Wheelock CE; Yao L; Yuan M; Zhao XH; Zhou S. J. Lipid Res 2017, 58 (12), 2275–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Fang TT; Bendiak B. J. Am. Chem. Soc 2007, 129 (31), 9721–9736. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.