Abstract
To investigate the possibility that HIV-1 replication in lymph nodes sustains the reservoir during antiretroviral therapy (ART), we looked for evidence of viral replication in 5 donors after up to 13 years of viral suppression. We characterized proviral populations in lymph nodes and peripheral blood before and during ART, evaluated the levels of viral RNA expression in single lymph node and blood cells, and characterized the proviral integration sites in paired lymph node and blood samples. Proviruses with identical sequences, identical integration sites, and similar levels of RNA expression were found in lymph nodes and blood samples collected during ART, and no single sequence with significant divergence from the pretherapy population was detected in either blood or lymph nodes. These findings show that all detectable persistent HIV-1 infection is consistent with maintenance in lymph nodes by clonal proliferation of cells infected before ART and not by ongoing viral replication during ART.
Keywords: AIDS/HIV
Keywords: Lymph, T cells
Introduction
Antiretroviral therapy (ART) effectively prevents HIV-1 disease progression. However, low-level viremia persists during ART and typically rebounds to pretherapy levels when treatment is discontinued. Rebounding virus is not genetically divergent from pre-ART HIV-1 variants (1–3), suggesting a stable viral reservoir during ART that is likely established prior to initiating treatment. Understanding the mechanisms that maintain the HIV-1 reservoir on ART is vital to developing strategies to eradicate the infection and/or to prevent viral rebound without ART.
Although there is ongoing debate about whether or not ART is fully effective (4, 5), evidence suggests that ART does effectively block HIV-1 replication in the peripheral blood of both children and adults whether initiated in acute (6–8) or in chronic infection (3, 9, 10). These results imply, and previous experimental evidence supports, that HIV-1 proviruses in peripheral blood either reside in cells infected prior to ART or in their clonal descendants (8, 11–14). Such a model suggests, assuming that there is free exchange of infected cells between the lymph nodes and blood (15, 16), that there would be a lack of viral evolution in lymph nodes during ART and that the proviral population in the lymph nodes and peripheral blood would be highly similar. However, studies have claimed to be able to detect viral evolution in tissues collected during ART (16–18), suggesting that HIV-1 replication may not be fully inhibited in all anatomical sites. Furthermore, questions have been raised about the ability of ART to effectively penetrate lymph node follicles and prevent continuing rounds of viral replication (16, 19, 20), raising the possibility that HIV-1 may persist during ART by exploiting putative sanctuary sites in lymph nodes (19, 21, 22). The hypothesis that the HIV-1 reservoir is maintained during ART by persistent viral replication in lymph nodes implies that there is only minimal trafficking of infected cells between lymph nodes and peripheral blood. If not, then proviruses in infected cells in the blood would show evidence of evolution due to accumulating mutations, especially after many years of viral suppression on ART.
To determine if the mechanisms that maintain HIV-1 proviruses in lymph nodes differ from those in peripheral blood, and whether there is ongoing exchange of infected cells between these compartments, we compared the proviral populations in lymph nodes and peripheral blood from 5 participants whose levels of viremia were suppressed on ART (<40 copies/mL) for 1.8 to 12.9 years. We addressed a number of questions related to the mechanisms that maintain the HIV-1 reservoir in lymph nodes during ART. During long-term ART, are proviral populations in lymph nodes divergent from pre-ART proviral populations in lymph nodes and peripheral blood? Are HIV-1 proviral populations divergent between paired lymph node and peripheral blood samples collected after long-term ART? Do proviral populations in lymph nodes change over time in a manner characteristic of ongoing HIV-1 replication during ART? Are the clonal populations of infected cells that persist on ART different in lymph nodes compared with peripheral blood? Do more infected cells in lymph nodes contain HIV-1 RNA compared with those in peripheral blood? Are proviruses in lymph nodes expressed at higher levels than those in peripheral blood? Are the replication-competent proviruses in lymph nodes that are induced in viral outgrowth assays divergent from pre-ART viral populations? Is there a higher number of CD4+ T cells carrying infectious, inducible proviruses in lymph nodes compared with peripheral blood during ART?
The results we obtained support the conclusion that HIV-1–infected cells persisting on long-term ART are well mixed between the peripheral blood and lymph nodes, and that the HIV-1 reservoir is sustained in the lymph nodes by proliferation of cells that were infected prior to ART and not by ongoing cycles of viral replication during ART.
Results
Donor characteristics.
Samples from 5 HIV-1–infected participants from the San Francisco SCOPE cohort (NCT00187512) (ref. 23 and Supplemental Table 1) were investigated for evidence of ongoing HIV-1 replication, as assessed by evolution of sections of the viral genome in peripheral blood and lymph nodes during ART. In these donors, viremia was well-suppressed on ART (<40 HIV-1 RNA copies/mL plasma) for 1.8 to 12.9 years. Two participants, 3720 and 2661, achieved viral suppression in early infection (<1 year and 0.3 years after the approximate date of transmission). The low pretreatment genetic diversity of the HIV-1 populations in these donors provided a sensitive probe for the detection of viral divergence in the peripheral blood or lymph nodes for periods of 1.8 years and 12.9 years, respectively. Donor 2661 had one brief ART interruption after 4.7 years, was quickly resuppressed, and subsequently maintained continuous suppression of viremia on ART for another 12.9 years. The other 3 participants initiated ART in chronic infection (2.0 to >22 years after the approximate date of transmission).
All donors had HIV-1 subtype B infection except donor 3720 who was infected by subtype C. With one exception, no participant had detectable mutations conferring resistance to current therapy in any samples studied. Donor 2669 had a history of mono- and/or dual-therapy, and, consequently, acquired reverse transcriptase inhibitor drug resistance mutations prior to achieving continuous viral suppression for 5.5 years following the addition of dolutegravir to his treatment regimen. Single-genome sequencing (SGS) of the integrase coding region in peripheral blood mononuclear cells (PBMCs) and lymph node mononuclear cells (LNMCs) did not detect mutations that would contribute to dolutegravir resistance (92 single-genome DNA sequences obtained from PBMCs and 115 from LNMCs).
To compare the evolution of HIV-1 among the different locations, pre-ART plasma and/or PBMCs and/or LNMCs were obtained from 4 of the participants. Two longitudinal on-ART LNMC samples were obtained about 1 year apart from 2 of the participants (2669 and 1079), including paired samples from contralateral inguinal lymph nodes, enabling comparison of the proviral populations in the 2 nodes. After completing this study, 1 donor interrupted ART and a plasma sample was obtained 14 days after discontinuation, allowing the rebounding plasma viral RNA population to be compared with pre-ART RNA sequence variants in plasma, to the pre-ART DNA and cell-associated RNA variants in peripheral blood, and to the on-ART DNA and cell-associated RNA variants in peripheral blood and lymph nodes. A detailed timeline of participant sampling is shown in Supplemental Table 2.
No evidence for HIV-1 evolution during ART in peripheral blood.
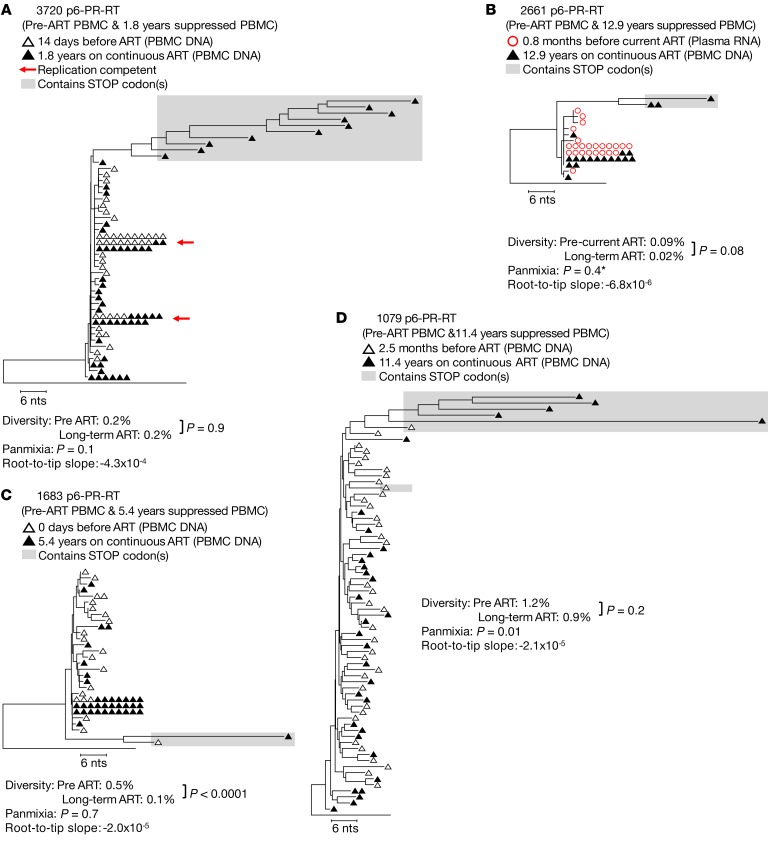
Previous studies have demonstrated the absence of detectable HIV-1 evolution in the peripheral blood during ART (3, 6, 9, 10). Because HIV-1 replication is error prone, this result shows that there is only very limited HIV-1 replication in blood. To ensure that ART fully suppressed viral replication in the peripheral blood of this set of donors, we performed SGS of the P6-PR-RT region derived from virus in the plasma or viral DNA in PBMCs collected prior to ART and after 1.8 to 12.9 years of continuous viral suppression (Figure 1). We also performed SGS on the full-length env gene in proviruses from pre- and on-ART PBMC samples from donors 1683 and 1079 (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI126714DS1). Because evolution would be a consequence of ongoing viral replication, we looked for evidence of viral evolution on ART. Evolution of the proviral sequences would cause: (i) increasing genetic diversity measured by average pairwise distance (APD), (ii) increasing genetic divergence of individual sequences from the starting population over time, as shown by a statistically significant (P < 0.001) negative test for panmixia comparing the 2 time points (24), and (iii) emergence of new viral variants, observable by increasing average and individual root-to-tip distances over time on maximum-likelihood phylogenetic trees (Figure 1 and Supplemental Figure 1).
Figure 1. HIV-1 P6-PR-RT proviral DNA and plasma virus RNA sequences prior to and during long-term ART.
Neighbor joining trees were constructed from single-genome P6-PR-RT proviral DNA sequences obtained from PBMCs (hollow black triangles; A, C, and D) or RNA sequences obtained from plasma virus (hollow red circles; B) prior to the most recent period of continuous viral suppression on ART and proviral DNA sequences obtained from PBMC samples taken after 1.8–12.9 years of viral suppression on ART (solid black triangles). Diversity was measured by APD, and measures were compared using an unpaired t test. Divergence was measured by a test for panmixia (to correct for multiple comparisons, populations are considered to be statistically different when the probability of panmixia is less than 0.001). Red arrows indicate sequences matching virus obtained in a viral outgrowth assay (30). Root-to-tip distances were measured using maximum likelihood trees, and the slopes of the root-to-tip distances over time were calculated by linear regression (units for slopes are substitutions/year). In cases where the slope was positive, an F-test was used to determine if the root-to-tip slopes were significantly different from zero. A is rooted on the HIV-1 subtype C consensus sequence, and B–D are rooted on the subtype B consensus sequence. Sequences containing G to A hypermutations and/or stop codons in open reading frames (indicated by shaded boxes) were excluded from all analyses. Except where unique variants were too few to test statistically (indicated by *), rakes of identical sequences were collapsed to a single variant for the test for panmixia and branch-length analysis. Results from a total of 8 samples (2 samples each from 4 patients) are represented in this figure.
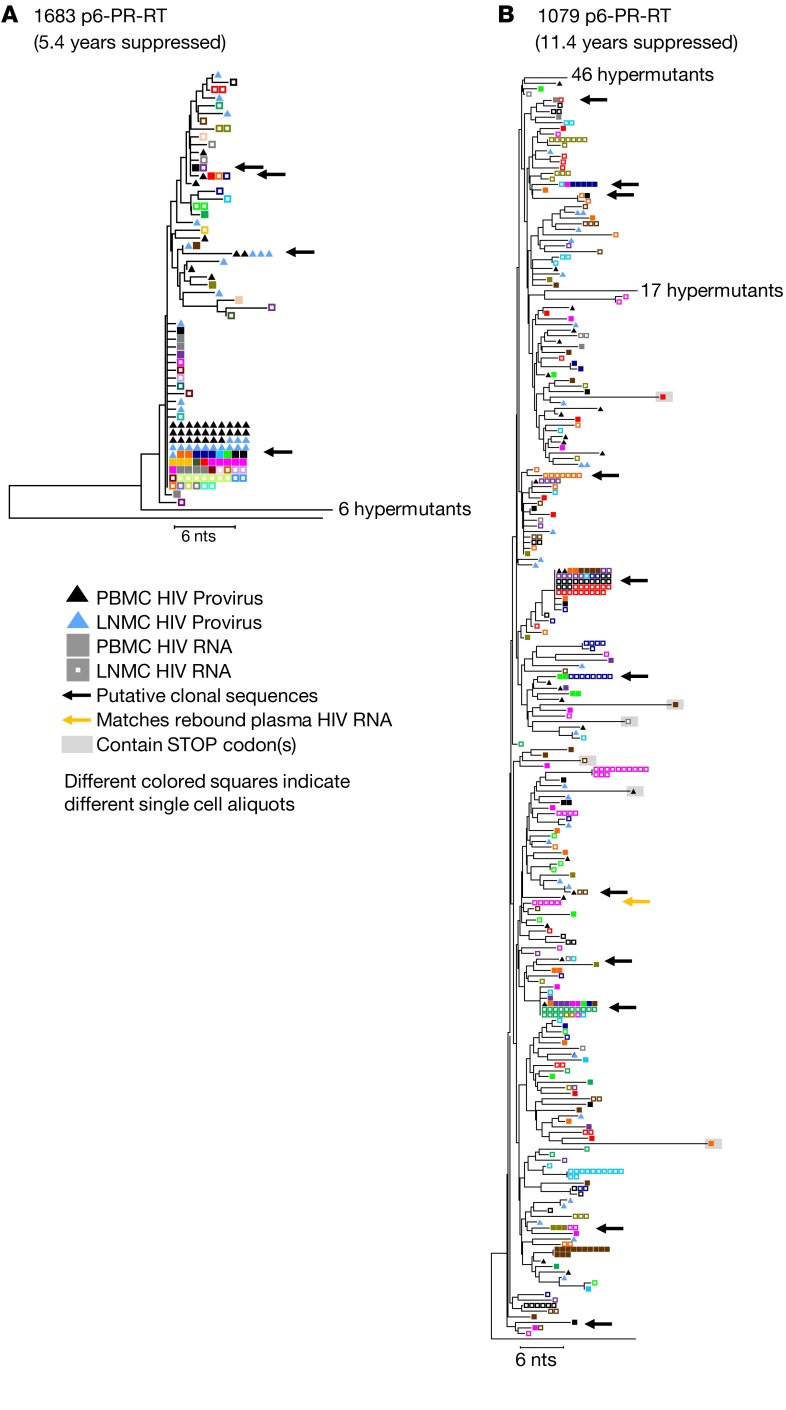
There was no evidence of HIV-1 evolution characteristic of ongoing cycles of viral replication detected (in either P6-PR-RT or env) in peripheral blood from any of the 4 individuals we studied by any of these analytical tools. Two donors (Figure 1, A and B) were treated in early infection and the HIV-1 populations did not shift from the apparent pre-ART founder viruses over 1.8 and 12.9 years of continuous treatment (probability of panmixia = 0.1 and 0.4, respectively). HIV-1 sequences with APOBEC-mediated G to A hypermutation, which are commonly seen in individuals on ART (25), as well as any other sequences containing stop codons, were excluded from these and all subsequent analyses. In donor 1683 (Figure 1C and Supplemental Figure 1A), the diversity in the proviral population decreased during ART in both P6-PR-RT and env (0.5% to 0.1%, P < 0.0001 and 1.3% to 0.6%, P < 0.0001). In this donor, the HIV-1 population became dominated by a single clonal sequence that was already present in the pre-ART population 5.4 years prior and may have been directly derived from the original founder virus. The increased proportion of this clonal variant also resulted in divergent (nonpanmictic) proviral populations prior to and during ART in P6-PR-RT (P = 5 × 10–6), a difference that disappeared when the identical sequences were collapsed to single variants (P = 0.7), as was done for all the panmixia analyses shown in the figures. This result implies that the apparent shift in the population during ART was due to clonal proliferation of infected cells rather than emergence of new variants from ongoing replication. The root-to-tip analysis also showed a potential loss of P6-PR-RT sequences on longer branches from the pre-ART population (slope = –2.0 × 10–5). Similar results were obtained from analyses of the same 2 genomic regions in peripheral blood from donor 1079 (Figure 1D and Supplemental Figure 1B), treated in chronic infection, despite the absence of prominent clonal variants in these samples after 11.4 years on ART.
As in previous studies (3, 6, 9, 10), comparative analyses of single-genome sequences obtained before therapy and after up to 13 years of suppressive ART provided no evidence for ongoing replication of HIV-1 in peripheral blood. On the contrary, all changes in the genetic structure of proviral DNA populations can be attributed to clonal proliferation of cells containing proviruses laid down prior to treatment initiation or the loss of variants during ART.
No evidence for HIV-1 evolution during ART in lymph nodes.
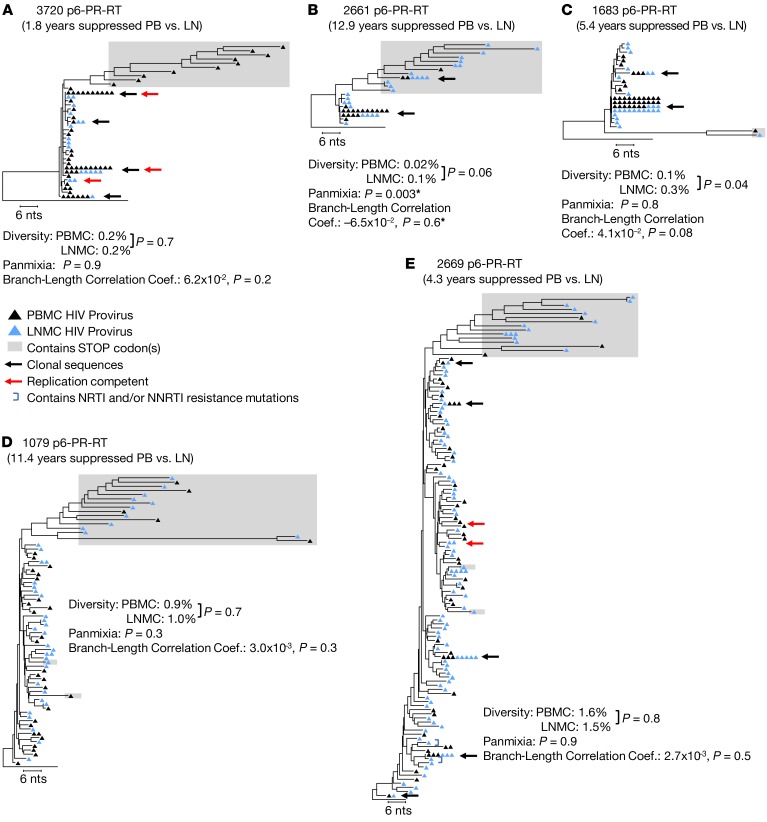
It has been suggested that the drugs used in ART do not optimally penetrate lymph nodes (19, 22), permitting persistent HIV-1 replication in lymph nodes (16, 20). If significant levels of viral replication occur in the lymph nodes during ART, then the newly emergent viral variants produced by the accumulation of reverse transcriptase errors would have to be sequestered in the lymph nodes, because they are not detected in the peripheral blood (as shown above). This argument predicts significant compartmentalization of HIV-1 DNA between the infected mononuclear cells in the lymph nodes and those in the peripheral blood. To address the possibility that HIV-1–infected cells are compartmentalized between the lymph nodes and peripheral blood, and that viral replication continues during ART at levels that sustain the HIV-1 reservoir in the lymph nodes as previously claimed (16), we compared proviral sequences of P6-PR-RT (Figure 2) and env (Supplemental Figure 2) from the lymph nodes and peripheral blood of the donors in this study.
Figure 2. HIV-1 P6-PR-RT proviral DNA populations in lymph nodes and peripheral blood during ART.
Neighbor joining trees were constructed from single-genome P6-PR-RT proviral sequences obtained from PBMCs (black triangles) and LNMCs (blue triangles) after 1.8–12.9 years of continuous viral suppression on ART. Diversity, divergence, and root-to-tip distances were measured as described in the legend to Figure 1. Branch-length correlation coefficients were calculated from maximum likelihood trees, and significance was assessed by a permutation test as described in Methods. Black arrows indicate identical sequences that were found in both locations. The presence of identical viral sequences in both locations is likely due to clonal expansion of HIV-1–infected cells. Red arrows indicate sequences that matched virus recovered in the viral outgrowth assay. A is rooted on the HIV-1 subtype C consensus sequence; B–E are rooted on the HIV-1 subtype B consensus sequence. Sequences containing G to A hypermutation and/or stop codons in open reading frames (indicated by shaded boxes) were excluded from all analyses. Except where indicated (*), rakes of identical sequences were collapsed to a single variant for the test for panmixia and branch-length analysis. Results from a total of 10 samples (2 samples each from 5 patients) are represented in this figure.
The results of this analysis yielded no evidence of compartmentalization in any of the donors when proviral sequences from the paired peripheral blood and lymph node samples were compared, in aggregate or individually, using the same 3 analytical tools as above (Figure 2 and Supplemental Figure 2). These results (in detail below) indicate that infected cells are well-mixed between the 2 compartments, and imply the absence of significant viral replication in lymph nodes, just as in peripheral blood, during suppressive ART. Neighbor joining phylogenetic analyses of P6-PR-RT sequences from the 2 donors who were treated in early infection (Figure 2, A and B) showed, in each case, a prominent group of identical sequences in the lymph nodes that was identical to the presumed founder viruses (the most common variants observed in the peripheral blood prior to ART), suggesting that no detectable sequence divergence occurred in the lymph nodes over 1.8 or 12.9 years of viral suppression on ART in these 2 individuals (there were not enough cells available from these donors to sequence env). After sampling over half of the infected cells in an entire lymph node from donor 2661, we found only a few P6-PR-RT variants to be 1–2 nucleotides different from the sequence of the presumed founder virus and these had not diverged from the plasma virus in the pre-ART sample (APD: P = 0.08, panmixia: P = 0.2). Though the positive branch length correlation coefficient indicated a significant difference in branch lengths between lymph node and peripheral blood (r = 0.26; P = 0.0005), when identical sequences were converted into single variants this difference disappeared (r = –0.065; P = 0.6), suggesting that the difference was due to the fact that an identical variant was sampled at a disproportionately higher level in the peripheral blood than in lymph nodes or that an expanded clone carrying this variant is not equal in size across the 2 locations. As in the blood, the only noticeably divergent sequences in the lymph nodes were G to A hypermutants.
A lack of compartmentalization between peripheral blood and lymph nodes was also observed in the 3 individuals treated in chronic infection (Figure 2, C–E, and Supplemental Figure 2, A–C). Sequences obtained from paired peripheral blood and lymph nodes were well-mixed phylogenetically and by the test for panmixia, had similar measures of diversity, and had branch length correlations not significantly different from zero in both P6-PR-RT (Figure 2, C–E) and env (Supplemental Figure 2, A–C), with the exception of donor 1683. There was a slight increase in the diversity of the proviral P6-PR-RT sequences in the lymph node from this donor (from 0.1 to 0.3%, P = 0.04); however, as in donor 2661, this difference can be attributed to the disproportionate representation between the 2 locations of a major clonal variant, a difference that likely arises from the physiology of cells in those particular locations (such as expression of homing signals or responsiveness to antigen [ref. 14]) rather than a property of the provirus itself. When identical sequences were condensed into single variants, the diversity of the populations in the peripheral blood and lymph nodes was not statistically different (P = 0.7). Differences in the number of rakes of identical sequences were observed between P6-PR-RT and env, particularly in PID 1683. Such differences likely reflect the more rapid accumulation of diversity in env compared with pol and, possibly, the different selection pressures on env versus pol, including the humoral immune response and/or cell tropism. Identical sequences common to both peripheral blood and lymph nodes (indicated with black arrows in the figures) were found in the donors who initiated ART in chronic infection; these identical sequences likely resulted from clonal expansion of cells infected prior to the initiation of ART. If so, these data imply trafficking of clonally expanded cells between the 2 compartments. The presence of indistinguishable HIV-1 populations in paired peripheral blood and lymph node samples collected during ART suggests, with our level of sampling (see discussion), that ongoing HIV-1 replication in lymph nodes is likely insufficient to sustain the HIV-1 reservoir during treatment.
In addition to seeking evidence for compartmentalization between lymph nodes and peripheral blood, we also investigated the possibility of compartmentalization between 2 separate lymph nodes from the same donor at the same time point (Supplemental Figure 3). LNMCs were collected from contralateral inguinal lymph nodes of 2 donors and proviral P6-PR-RT sequences were obtained. In both donors, we found the HIV-1 populations in the paired lymph nodes to be well-mixed, showing no evidence for viral compartmentalization between nodes by any metric (for donors 1079 and 2669, respectively: APD: P = 0.8 and 0.3; panmixia: P = 0.3 and 0.7; branch length correlation: P = 0.05 and 0.7). These data imply that infected cells traffic between peripheral blood and lymph nodes and provide additional evidence that ART effectively blocks HIV-1 replication in lymph nodes as well as peripheral blood to levels that are not detectable with our level of sampling by SGS.
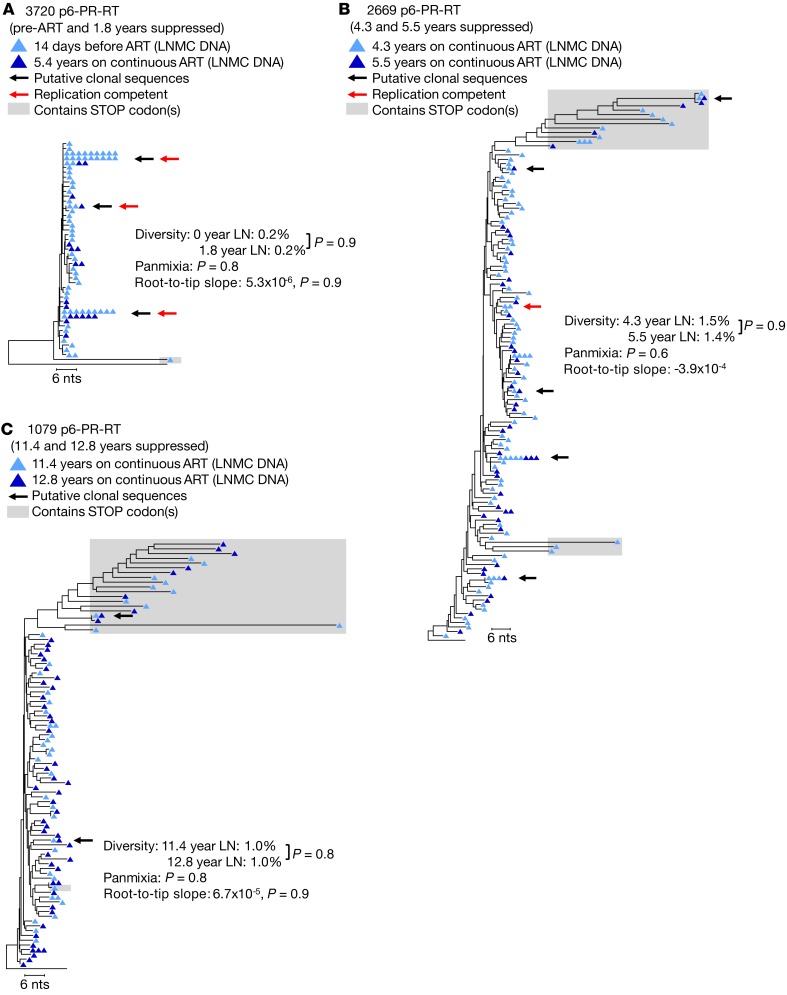
To further address the question of ongoing viral replication in lymph nodes during ART, we pooled the proviral sequences obtained from the paired lymph node samples in each donor and compared those sequences to proviral sequences obtained from a lymph node 1 year prior (during ART) or from a lymph node collected prior to ART initiation (Figure 3). Using the analytical tools described above, we observed no change in the diversity or divergence of the HIV-1 proviral populations in the longitudinal lymph node samples (for donors 3720, 1079, and 2669, respectively: APD comparison: P = 0.8, 0.9, and 0.9; panmixia: P = 0.6, 0.8, and 0.9; and branch length correlation: P = 0.9, 0.9, and 0.9). The lack of any detectable change in the proviral populations in longitudinal lymph nodes during years of therapy again shows that ART is effective at blocking viral replication in the lymph nodes.
Figure 3. HIV-1 P6-PR-RT proviral DNA sequences from longitudinal lymph node samples.
Neighbor joining trees were constructed from single-genome P6-PR-RT proviral sequences obtained from LNMC at 2 time points: the first prior to ART initiation or after 11.4 or 4.3 years on ART (dark blue triangles) and the second approximately 1 year later (light blue triangles). Diversity, divergence, and root-to-tip distances were measured as described in the legend to Figure 1. Black arrows indicate clonal sequences present at both time points, and red arrows indicate sequences matching virus that grew in the viral outgrowth assay. A is rooted on the HIV-1 subtype C consensus sequence, and B and C are rooted on the subtype B consensus sequence. Sequences containing G to A hypermutation and/or stop codons in open reading frames (indicated by shaded boxes) were excluded from all analyses. Results from a total of 6 samples (2 samples each from 3 patients) are represented in this figure.
HIV-1 integration site comparison of clonal populations in lymph nodes and peripheral blood.
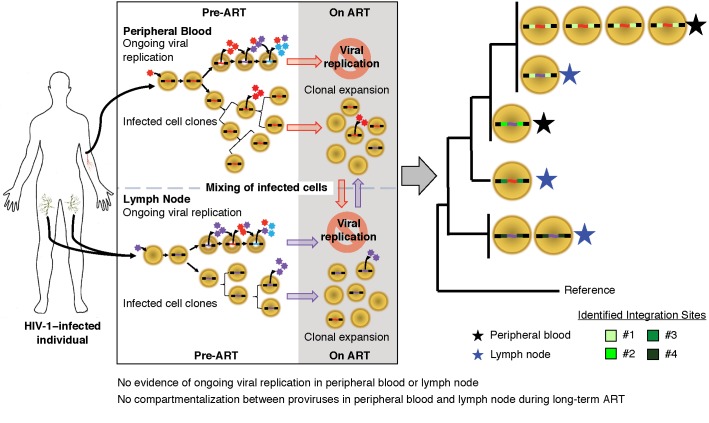
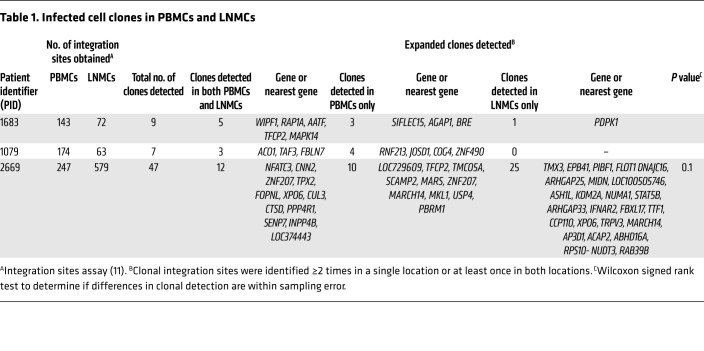
The sites at which HIV-1 proviruses are integrated can be used to monitor the clonal expansion of infected cells (11). Because HIV-1 integrates almost randomly into many sites in the human genome, finding multiple cells with exactly the same sites of integration is very strong evidence that these cells are descendants from a single infected cell. By this principle, integration site analysis can be used to monitor the trafficking and compartmentalization of populations of HIV-1–infected cells. Thus, when infected cells at 2 different locations have identical integration sites, they must be members of a clone derived from the same infected cell, and cells from that clone must have trafficked between the 2 locations. We used the integration sites assay to determine whether the same expanded clonal populations were present in lymph node and peripheral blood (Table 1). From each donor, we obtained 143–247 integration sites from the peripheral blood samples and 63–579 integration sites from the lymph nodes (full set of integration sites deposited in the Retroviral Integration Sites Database [RID], accessible by Pubmed ID at https://rid.ncifcrf.gov/). Clones were defined as proviruses with identical integration sites that were detected at least once in each of 2 different locations or samples regardless of ART status, in 2 or more different DNA fragments in the same location or sample during ART, or 3 or more times prior to ART (26). Although the presence of 2 identical integration sites is indicative of cell division, we define clones in pretherapy conservatively as requiring 3 detection events because dividing cells could die before establishing an expanded cell clone. In the 3 ART-treated donors in which the numbers of infected cells were sufficient for integration site analyses (1683, 1079, and 2669), common clones were detected between lymph nodes and peripheral blood. Of note, in the combined data set, we found evidence for selection for integrations in the BACH2 (P = 3.0 × 10–7), MKL1 (P = 0.010), and STAT5B genes (P = 3.5 × 10–4) as previously reported (11, 13). Overall, in these 3 donors, the clonal populations were not significantly different between the lymph nodes and peripheral blood (P = 0.1), confirming the absence of detectable compartmentalization of infected cells between those locations (Table 2).
Table 1. Infected cell clones in PBMCs and LNMCs.
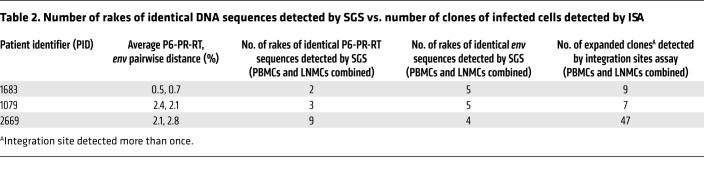
Table 2. Number of rakes of identical DNA sequences detected by SGS vs. number of clones of infected cells detected by ISA.
Expression of HIV-1 proviruses in peripheral blood and lymph nodes prior to and on ART.
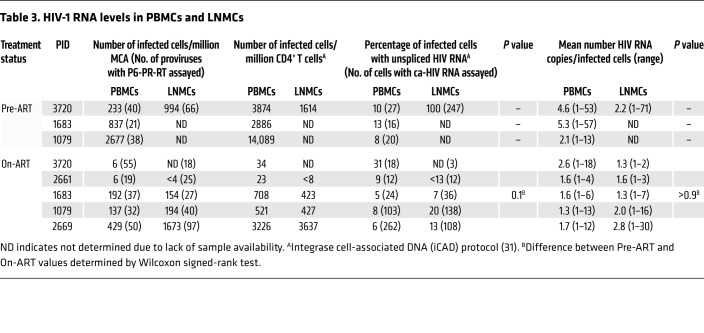
Our analysis of proviral populations showed no evidence of ongoing HIV-1 replication in, or compartmentalization between, peripheral blood and lymph nodes. In a further study, we determined the fraction of infected cells with unspliced HIV-1 RNA (monitoring P6-PR-RT) and the levels of proviral expression in single cells in each location. Using our cell-associated RNA and DNA single-genome sequencing (CARD-SGS) assay, which can detect a single HIV-1 P6-PR-RT RNA sequence in a single infected cell (27), we compared the level of unspliced viral RNA in single cells obtained from paired peripheral blood and lymph node samples (Table 3, Figures 4 and 5, Supplemental Table 3, and Supplemental Figure 4). While our previous work showed that there are single cells with high levels of HIV-1 RNA in the peripheral blood of untreated individuals, cells with similarly high levels of viral RNA were not detected in the peripheral blood of patients on ART (27). The observation of cells containing high levels of viral RNA (which may be on the viral replication pathway) in lymph nodes during ART would provide support for the proposal that there are significant levels of ongoing replication in the lymph nodes. Although cells expressing high levels of HIV-1 RNA can be seen in in situ hybridization assays on lymph nodes of HIV-infected individuals on ART (20, 28, 29), they are much more rare than in untreated individuals, and may well reflect activation of HIV-1 expression in descendants of cells infected prior to ART initiation. The low-level persistent viremia seen in most suppressed patients implies that highly expressing cells must exist, albeit at numbers averaging about 10,000-fold lower than before treatment initiation (27).
Table 3. HIV-1 RNA levels in PBMCs and LNMCs.
Figure 4. HIV-1 cell-associated RNA sequences obtained from single cells in peripheral blood and in lymph nodes during ART.
Neighbor joining trees were constructed from single-genome P6-PR-RT proviral DNA and cell-associated HIV-1 RNA sequences obtained from paired PBMCs and LNMCs sampled during continuous viral suppression on ART. Black and blue triangles indicate HIV-1 DNA sequences from PBMCs and LNMCs, respectively; solid squares and hollow squares indicate HIV-1 RNA sequences from PBMCs and LNMCs, respectively. Squares with no genetic difference of the same color indicate sequences that are assumed to be from the expression of single infected cells. Black arrows indicate sequences detected in probable clones in both the samples. The yellow arrow indicates a sequence detected in rebound plasma viremia. Trees are rooted on the subtype B consensus sequence. Results from a total of 8 samples (4 samples each from 2 patients) are represented in this figure.
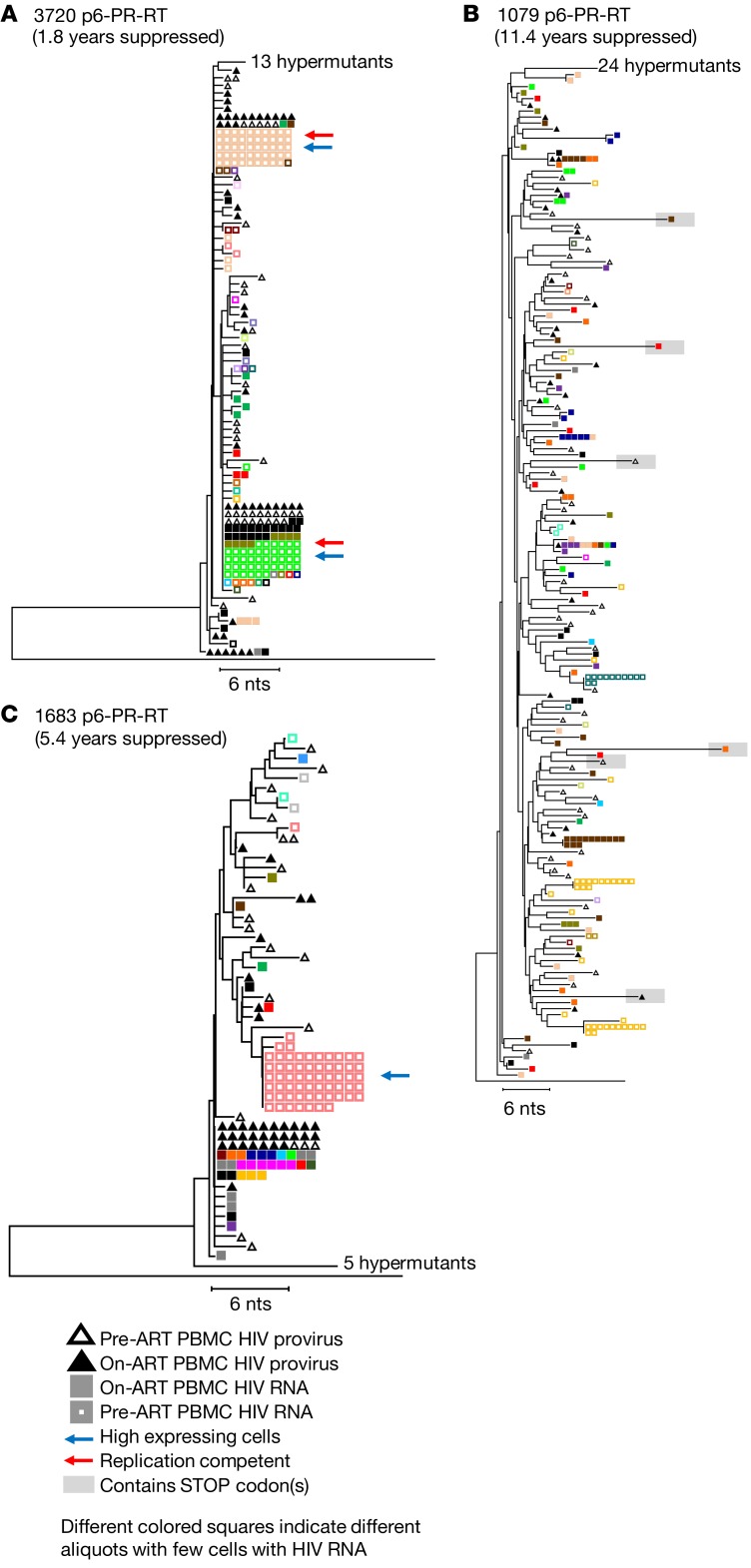
Figure 5. HIV-1 cell-associated RNA from single cells in peripheral blood prior to and during long-term ART.
Neighbor joining trees were constructed from single-genome P6-PR-RT proviral DNA and cell-associated HIV-1 RNA sequences obtained from PBMCs sampled prior to ART initiation (hollow triangles indicate proviral sequences, hollow squares indicate RNA sequences) and after 1.8–11.4 years of viral suppression on ART (solid triangles indicate proviral sequences, solid squares indicate RNA sequences). Squares of the same color indicate sequences that are assumed to be from the expression of single infected cells. The blue arrows indicate high expressing cells (>20 HIV-1 RNA copies), and red arrows indicate sequences matching virus that grew in the viral outgrowth assay. A is rooted on the HIV-1 subtype C consensus sequence, and B and C are rooted on the subtype B consensus sequence. Results from a total of 12 samples (4 samples each from 3 patients) are represented in this figure.
We found no overall difference in the fraction of infected cells that contained viral RNA in lymph nodes compared with peripheral blood during ART (median 13% in LNMCs vs. median 8% in PBMCs; P = 0.13) or in the levels of proviral expression within single cells between the lymph nodes and peripheral blood (median = 1 RNA molecule/expressing cell in both). In donor 3720, we detected a few cells in the peripheral blood and lymph node with high levels of viral RNA (containing >20 HIV-1 RNA molecules) prior to ART, while no such cells were observed in a sample taken after 1.8 years on ART (Figure 5A, cells with high viral RNA levels indicated with blue arrows; pre-ART data from lymph nodes, including 3 cells with high HIV-1 RNA levels, not shown). Similarly, in donor 1683, one cell with a high level of viral RNA was identified in the peripheral blood prior to ART, and none was found after 5.4 years on ART in either peripheral blood or lymph nodes (Figure 5B, indicated with blue arrow). In donor 1079, no cells with high levels of viral RNA were detected at either time point (Figure 5C, 353 HIV-1–infected cells assayed for HIV-1 RNA from pre-ART and 1315 on ART). Although a pre-ART sample was not available for donor 2669, we did detect one LMNC during ART with 28 molecules of HIV-1 RNA (Supplemental Figure 4C). These data suggest that cells that have high levels of viral RNA are rare and, as expected for productive infection, may be preferentially eliminated. It is also likely that latently infected cells or cells with very low levels of HIV-1 expression are not eliminated and, therefore, accumulate over the duration of infection, possibly explaining why high-expressing cells were detected before ART initiation in donors 3720 and 1683 (who were still rather early in infection) and not in donor 1079 (who had been infected for more than 4 years at the time of ART initiation). A larger study would be required to investigate this possibility.
As previously reported (27), we also found RNAs with identical P6-PR-RT sequences in different single cells, suggesting that clonally proliferating populations of infected cells include ones with proviruses that are actively expressing low levels of HIV-1 RNA. While it is possible that such cells are the source of viral rebound when ART is interrupted, it is more likely that there are always very rare highly expressing cells, as can be seen by in situ hybridization (20, 28, 29), that give rise to persistent and rebound viremia and in some cases are the source of outgrowth in viral outgrowth assays. In donor 3720, the viral outgrowth assay (30) identified multiple sequences obtained from pre-ART as replication competent (Figure 1A, Figure 2A, and Figure 3A, red arrows). Two of the viral sequences obtained in viral outgrowth assays matched RNA sequences obtained from multiple PBMCs prior to ART and from multiple PBMCs and LNMCs obtained after 1.8 years on ART (Figure 5A and Supplemental Figure 4A, red arrows). In donor 1079, a treatment interruption revealed 2 major rebound variants in the consequent viral rebound, one of which matched a set of 6 lymph node–derived, cell-associated RNA sequences obtained from a single cell after 11.4 years on ART (Figure 4B, yellow arrow). This observation suggests that rebound viremia resulted from a clone of infected cells, some of which were actively transcribing HIV-1 RNA prior to ART interruption, as previously reported (2). The other rebounding variant matched a single proviral sequence obtained from the peripheral blood during ART (data not shown but sequence submitted to GenBank, accession MK147615). Thus, persistent, rebound, and rescued virus can have the same genetic properties as the proviral DNA described above, showing no evidence of ongoing replication or evolution but strong evidence for clonal expansion of the cells responsible for it.
In estimating the fraction of infected cells that contain HIV-1 P6-PR-RT RNA, our measurements of the number of HIV-1–infected cells were based on a particular region of the genome, in the pol gene (ref. 31 and Table 3). To ensure that the region of the viral genome we analyzed did not affect our conclusions, we also used primers and probes in the LTR (R-U5) and in the gag gene (Supplemental Table 3). Though quantification by these methods resulted in varying counts of infected cells (likely due to some proviruses having large internal deletions or variants with mismatches to the primers or probes), our estimates for the fractions of infected cells with HIV-1 P6-PR-RT RNA were not significantly different between the peripheral blood and lymph nodes, regardless of the genomic region used to detect proviral sequences (P = 0.4 for LTR and P = 0.7 for gag). Because there are more CD4+ T cells in lymph nodes of some donors, and more infected cells in the lymph nodes than in the peripheral blood, the overall number of HIV-1 RNA molecules we detected could be higher in lymph nodes than in peripheral blood, as previously reported (20, 28). However, when the levels of HIV-1 RNA in single infected cells were compared, we found no difference between the cells from peripheral blood and lymph nodes during long-term ART. These findings demonstrate that lymph node cells containing high levels of HIV-1 RNA collected from HIV-infected individuals on ART, such as those detected here and previously using in situ hybridization assays (27–29), more likely reflect activation of HIV-1 expression in descendants of cells infected prior to ART initiation than from cells infected from ongoing cycles of viral replication on ART.
No single proviral sequences on ART deviate from those in pre-ART.
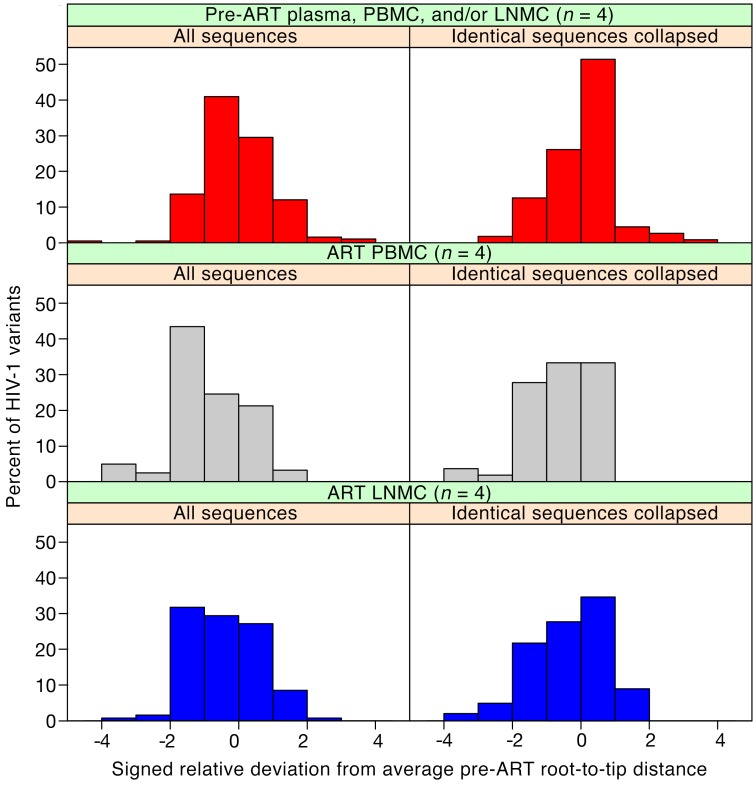
Our results clearly show that there is no detectable evolution of the bulk population in either peripheral blood or lymph nodes for almost 13 years of suppressive ART. However, it remains possible that a fraction of the proviral DNA population may have arisen by low-level ongoing replication and that these sequences may not have been visible in analyses of the population as a whole. Such variants would only be visible in the populations when analyzed individually, as distinct outliers in the overall distribution of a diversity metric. To test for the presence of such variants in the combined patient data, we normalized the root-to-tip distances of all HIV-1 RNA or DNA sequences from pre-ART in each patient, when available, and divided the results into bins, yielding a histogram centered around 0, as shown in Figure 6 (red bar graph). We then did the same analysis with the P6-PR-RT proviral sequences from the on-ART peripheral blood and lymph node samples, normalizing individual distances to the respective donor’s pre-ART data, as shown in Figure 6 (grey and blue graphs, respectively). Thus normalized, all patient data could be aggregated, allowing visualization of evolution of the peripheral blood and lymph node populations overall, as well as sensitive detection of small subpopulations that might be evolving separately from the bulk population. Analyses were performed both on total sequences (left panels) and with groups of identical sequences collapsed to remove the effects of clonal expansion (right panels).
Figure 6. HIV-1 root-to-tip distances normalized to pre-ART average.
Signed relative standard deviations were calculated for each sequence (including either all sequences or only each distinct variant) from each patient, as described in Methods. These data were separated into categories of pre-ART, on-ART PBMCs, and on-ART LNMCs, and the values from all patients were aggregated and plotted separately for each group in red, grey, and blue for pre-ART, on-ART PBMCs, and on-ART LNMCs, respectively, in bin sizes of 1 SD. A negative relative standard deviation indicates a regression from the mean and a positive value indicates an increase from the mean. Patient 2669 was excluded from this analysis due to lack of pre-ART sampling. Sequences from patient 2661 included pre-ART plasma sequence data, and sequences from patient 3720 included both pre-ART PBMC and LNMC data. All sequences containing G to A hypermutation and/or stop codons in open reading frames were excluded from the analysis. The number of samples for each panel was 4.
Consistent with our previous analyses, the pre-ART and on-ART data sets yielded very similar histograms, despite the samples being drawn from different anatomical locations and separated by very long times. In the plots, any single sequence with a genetic distance of more than 2 standard deviations from the mean of the pre-ART sequence distribution would be revealed as a significant outlier. Since we examined a total of about 125 sequences in each set, we expected about 5% of the 122 PBMC or 129 LNMC sequences to be between 2 and 3 standard deviations from the mean by chance, and none to be greater than 3. As the figure shows, the number of such outlier sequences is actually less than the 6 sequences expected (0/122 total sequences in the on-ART PBMCs and 1/129 sequences in the on-ART LNMCs) and very similar to the frequency of such outliers in the pre-ART data. Given that HIV-1 genomes accumulate mutations at about 1% per year in untreated patients (32) and 1 standard deviation in this analysis corresponds to a genetic distance of between 0.08% and 0.9% from the root in these donors, 3 standard deviations represents a very low bar based on the time between ART initiation and the on-ART sampling, yet not a single provirus in 251 studied in the 2 tissues was able to cross it. Thus, if there is ongoing replication, it can only involve, at most, 0.4% (upper bound of 95% CI = 1.2%) of the virus population in either blood or lymph nodes in ART-suppressed patients.
Discussion
The question of whether ongoing viral replication sustains the HIV-1 reservoir during ART has important implications for the design of curative interventions. Some have concluded that viral replication in lymph nodes continues at levels that maintain the HIV-1 reservoir (16), and others have claimed that viral replication may persist during ART, but in only a small subset of cells (20, 33). If HIV-1 replication persists on ART to the extent necessary to maintain the viral reservoir, then the accumulation of new proviral mutations would be apparent after long-term ART. The acquisition of new mutations would be especially evident in patients who initiated treatment in early infection with a homogeneous virus population, such as donor 2661. In this donor, the number of HIV-1–infected cells that persisted on ART was small compared with the individuals who initiated ART in chronic infection, consistent with previous reports (34–37). As a result, all 150 million LNMCs obtained from an entire lymph node were investigated for HIV-1 DNA or RNA. We found that fewer than 600 infected cells persisted in the lymph node from this individual after almost 13 years on ART and less than 13% had unspliced HIV-1 RNA. Extrapolating this finding to the approximately 500–700 lymph nodes in the body, we can estimate that about 300,000–420,000 HIV-1–infected LNMCs persisted in this individual and about 39,000–55,000 had unspliced HIV-1 RNA. In contrast, the donors who initiated ART in chronic infection and had been sustained on ART for a shorter duration (about 1–5 years), according to our calculations, had as many as 175 million infected LMNCs in the entire body and as many as 22.8 million with unspliced HIV-1 RNA, 456,000 of which may be high expressers. In total, we examined 251 lymph node and peripheral blood proviral-derived sequences from the donors who had pre-ART sampling and we failed to detect a single provirus with a significant number of new mutations in either location in any of the donors. Thus, if there is a fraction of the proviral population in either lymph node or peripheral blood representing virus generated by ongoing replication, it must be very small. With our level of sampling, we can report with 95% confidence that, if there is a replicating population, it is less than 1.2% of the total HIV-1–infected cells that persist on ART. Additionally, as viral RNA+ cells do not necessarily indicate replication competency, less than 1.2% is a highly conservative estimate of the upper bound of a potential replicating pool (25, 38).
By contrast, the clonal proliferation of infected cells that contain replication-competent proviruses can generate large pools of infected cells, some of which persist during long-term ART (14, 39–41). Our integration site assay data show that clonal populations of infected cells that are detectable after long-term ART in the peripheral blood and lymph nodes are not significantly different and provides support for the conclusion that cell proliferation is likely to be the most important mechanism that maintains the persistent viral DNA in blood and lymph nodes. These data, taken together with the finding that the HIV-1 populations in peripheral blood and lymph nodes are not genetically divergent and that infected cells in those locations contain similar levels of viral RNA, show that this process is not different in these compartments. Importantly, these data also show that infected cells are trafficked between the blood and the lymph nodes, as well as between lymph nodes. This lack of compartmentalization further implies that ongoing cycles of viral replication during ART do not persist in lymph nodes at a level that sustains the HIV-1 reservoir. Our findings support the conclusion that the viral DNA reservoir is maintained largely or entirely by clonal proliferation of cells that were infected prior to initiating ART and that this mechanism is important in both the peripheral blood and the lymph nodes. In sum, our results refute the idea that development of new antiretroviral drugs (with improved targeting to lymph nodes, for example) will be of value toward achieving a cure of HIV-1 infection. The very low frequency of virus-producing cells detectable by in situ PCR assays (20, 28, 29) may be important biologically for other reasons not addressed in this study, but our results imply that they are more likely to be due to expression of rare proviruses in descendants of cells infected prior to therapy than from ongoing viral replication. Likewise, the low number of high-expressing cells, though likely to be the source of persistent viremia on ART and rebound viremia after stopping ART, is unlikely to result in ongoing cycles of viral replication in the presence of effective therapy. These results make it very clear that survival and expansion of cells infected prior to therapy constitute the major, if not the only, barrier to a cure of HIV-1 infection. Future strategies for curative interventions must focus on methods to target the populations of clonally expanded HIV-1–infected cells harboring intact HIV-1 proviruses.
Methods
Participant cohort and sample collection.
Samples from the 5 HIV-1 infected participants were obtained from the San Francisco SCOPE cohort (NCT00187512) (ref. 23 and Supplemental Table 1). PBMCs were separated with Ficoll and resuspended in FBS with 10% DMSO. To disaggregate LNMCs from lymph node biopsies, excess fat was trimmed and the node was minced and strained on 70-μm and 40-μm nylon cell strainers. Samples were shipped on dry ice and stored in liquid nitrogen until processed.
Sequence data availability.
All sequence data are available on GenBank at accession numbers MK145079-MK148700.
HIV-1 quantification.
HIV-1 DNA levels were determined using the integrase cell-associated DNA assay (31) or using a droplet digital PCR (ddPCR) assay (42). For ddPCR, HIV-1 DNA copies per million cells were measured in triplicate with a duplexed ddPCR assay measuring the amount of DNA corresponding to HIV-1 gag (43), pol, and the RU5 region of the HIV-1 LTR. The amount of cellular DNA was measured by CCR5 quantitation. The RU5 region of the HIV-1 LTR was amplified using the following primers: RU5-F, 5′-CTTAAGCCTCAATAAAGCTTGCC-3′; RU5-R, 5′-GGATCTCTAGTTACCAGAGTC-3′; and RU5-probe (Hex Zen), 5′-AGTAGTGTGTGCCCGTCTG-3′. Prior to amplification, DNA was extracted as in ref. 31 and randomly sheared with a Branson ultrasonic cup horn sonifier (Emerson) at 60% amplification in pulse mode for 5 seconds. This step was repeated 3 times. Samples were then heated to 100°C for 15 minutes and snap-cooled on ice. A 20 μL PCR mastermix was made which gave a final concentration of 1x ddPCR Supermix for Probes (BioRad), 750 nM forward and reverse primers, 250 μM probe, and 5 μL DNA template. End-point PCR was performed with the following conditions: 95°C for 10 minutes, then 40 cycles of 94°C for 30 seconds, 55°C for 1 minute with a ramp rate of 2°C/second, followed by a final 98°C for 10 minutes. After the reactions were completed, the samples were held at 12°C. Reactions were then read on the QX200 Droplet Reader (Bio-Rad) and analyzed using the Quantasoft software version 1.7.4 (Bio-Rad) with a user-defined threshold. Averaged triplicate HIV DNA measurements were normalized to one million PBMCs using concurrent triplicate CCR5 measurements.
Cell-associated RNA and DNA single-genome sequencing assay.
CARD-SGS was performed on the P6-PR-RT region of HIV-1 as previously described (27). In brief, CARD-SGS is performed by diluting cells to near the endpoint for those with HIV-1 unspliced RNA and performing SGS on reverse-transcribed RNA. Using single ACH2 cells, we previously showed that our method can detect a single HIV-1 P6-PR-RT RNA molecule in a single cell (26). Because the reverse transcription step is known to introduce errors at a rate of about 10–4 per sequenced nucleotide of viral cDNA sequences (44), single RT-PCR variants that differed by a single nucleotide from a group of 5 or more identical sequences within the same aliquot were counted with the majority. HIV-1 proviral SGS was performed on full-length env as previously described (45).
SGS of the full-length integrase coding region was carried out on PBMC cell-associated DNA using the same PCR parameters as those described for P6-PR-RT (46) but using different PCR primers: IntF1, 5′-CATCTAGCTTTGCAGGATTCG-3′ and IntR1, 5′- CTGACCCAAATGCCAGTCTC-3′ and for nested PCR: IntF2, 5′-GGAAAAGGTCTACCTGGCATG-3′ and IntR2, 5′-TCTCCTGTATGCAGACCCCA-3′. Sequences were aligned using ClustalW and all sequences that would clearly render the virus defective for replication (stop codons within the coding region) were omitted from downstream analyses. Population genetic diversity was calculated as average pairwise P distance (APD) using MEGA (www.megasoftware.net). Neighbor joining trees were constructed using MEGA and rooted on consensus B or C as appropriate.
Integration sites assay (ISA).
ISA was performed as previously described (11) using patient-specific primers to the 5′ and 3′ LTRs. The full set of integration sites obtained were submitted to the Retroviral Integration Sites Database (https://rid.ncifcrf.gov/) and can be accessed using the Pubmed ID for this paper. The primer sequences are as follows: 3′LTR TGTGGACTCTGGTAACTAGAGATCCCTC, 5′LTR TCAGGGAAGTAGCCTTGTGTGTGGT (3720 PCR 1); 3′LTR CCCTTTGTGGTAAGTGTGGAAAATC, 5′LTR TCTTGGCTCTTCTTGGAGTAAACTA (3720 nested PCR); 3′LTR TGTGACTCTGGTACTAGAGATCCCTC, 5′LTR TCAGGGAAGTAGCCTTGTGTGTGGT (1683 PCR 1); 3′LTR CCCTTTTAGTCAGTGTGAAAATC, 5′LTR TCTTGCCTTTGCTGGGAGTAAATTA (1683 nested PCR); 3′LTR TGTGACTCTGGTACTAGAGATCCCTC, 5′LTR TCAGGGAAGTAGCCTTGTGTGTGGT (1079 PCR 1); 3′LTR TAAAAAGTGGCTAAGATCCAGAGC, 5′LTR CCTATTTAGTCAGTGTGGAAAATC (1079 nested PCR); 3′LTR TGTGACTCTGGTAACTAGAGATCCCTC, 5′LTR TCAGGGAAGTAGCCTTGTGTGTGGT (2669 PCR 1); 3′LTR CTTGTCTTTTCTGGGAGTGAATTA, 5′LTR CCCTTTTAGTCAGTGTGGAAAATC (2669 nested PCR).
Signed relative deviation analysis of single proviruses.
Root-to-tip distances from patient ML trees containing all pre-ART sequences, pre-ART sequences with identical sequences collapsed to a single variant, all on-ART proviral sequences from PBMCs and LNMCs, and on-ART proviral sequences from PBMCs and LNMCs with identical sequences collapsed to a single variant were generated by phyML as described above. Normalization was done by taking the average and standard deviation of root-to-tip distances from the pre-ART sequence data. Signed relative standard deviations (sRSD) were then calculated as:
(Equation 1)
for each sequence i in each patient. The sRSD were then plotted as histograms, as shown in Figure 6. The upper limit of the confidence interval was determined by the “rule of 3” for an unseen event (47).
Statistics.
Implementation of the Hudson test for geographic subdivision — a test for panmixia — was run using an in-house program and a significance cutoff of less than 10–3 was applied to account for multiple comparisons (24, 48). Maximum likelihood (ML) trees were generated using the best fit model from the Smart model selection and estimated in PhyML (49, 50). Trees were rooted against consensus HIV-1 subtype B or C sequences as appropriate. Root-to-tip distances were plotted against the time of sample collection and evaluated via F-test on the resulting linear regression (units for root-to-tip are substitutions/year). Branch-length correlation coefficients for compartmentalization were calculated as described in Critchlow et al. using an in-house program (51). Differences in APD were tested as previously described (6). Other standard statistical analyses and summary statistics were done in R version 3.3.1. and are noted in figure legends. In-house programs for the test for panmixia (java), root-to-tip analysis (python3), and branch-length tree correlation coefficient (python3) are available online at www.github.com/michaelbale
Study approval.
Samples from the 5 HIV-1–infected participants were obtained from the San Francisco SCOPE cohort (NCT00187512) (23) (Supplemental Table 1) and studies were approved by the internal review board of the University of California San Francisco. Written informed consent was received from the participants prior to inclusion in the study.
Author contributions
WRM performed SGS, analyzed data, and wrote the paper. MJB analyzed data, performed statistical analyses, and wrote the paper. JS and VKM performed SGS and analyzed data. AW and BFK performed SGS. AM performed SGS, performed ddPCR, and analyzed data. SCP wrote the paper. MDS performed VOA. EMA developed the ddPCR assay. JCC performed iCARD assay and performed FACS analyses. EKH performed iCARD assays. WS and XW analyzed data. DW performed the integration sites assay (ISA). JMM processed patient samples and performed FACS analyses. RH enrolled patients, analyzed data, and wrote the paper. JWM wrote the paper and contributed data. SHH analyzed data and wrote the paper. SGD conceived of idea, enrolled patients, and wrote the paper. JMC and MFK conceived of the idea, analyzed data, and wrote the paper.
Supplementary Material
Acknowledgments
We thank Connie Kinna, Anne Arthur, Valerie Turnquist, and Sue Toms for administrative support, Valerie Boltz and Frank Maldarelli for useful discussions, and Adam Capoferri and Joe Meyer for help with graphics. We also thank the Viral Evolution Core in the AIDS and Cancer Virus Program, Christine Fennessey, Leslie Lipkey, Laura Newman, and Carolyn Reid for sequencing support. We thank the following funding sources: NCI Intramural funding (to MFK); NIH Intramural AIDS Targeted Antiviral Program (IATAP) Award (to 2015–2016 Principal Investigator, MFK; “Characterizing HIV Proviruses in Clonally Expanded Cells in Vivo”); NIH IATAP Award (to 2017–2018 Principal Investigator, MFK; “Single Cell Analysis of Proviral Expression in Expanded Clones During ART”); Leidos Biomedical Research, Inc. subcontracts 12XS547 (to JWM) and 13SX110 (to JMC); and the NCI, federal funds under contract no. HHSN261200800001E. JMC was a Research Professor of the American Cancer Society.
Version 1. 07/30/2019
In-Press Preview
Version 2. 09/23/2019
Electronic publication
Version 3. 11/01/2019
Print issue publication
Funding Statement
NCI Intramural funding to MFKNIH Intramural AIDS Targeted Antiviral Program (IATAP) Award, 2015-2016 Principal Investigator: MFK Title: Characterizing HIV Proviruses in Clonally Expanded Cells in Vivo.NIH IATAP Award, 2017-2018 Principal Investigator: MFK Title: Single Cell Analysis of Proviral Expression in Expanded Clones During ART. Leidos Biomedical Research, Inc. Subcontracts 12XS547 (to J.W.M.) and 13SX110 (to J.M.C.). Federal funds from the NCI under Contract No. HHSN261200800001E. J.M.C. was a Research Professor of the American Cancer Society
Footnotes
Conflict of interest: JWM is a consultant for and has received grants from Gilead Sciences and owns shares of Cocrystal Pharma Inc.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(11):4629–4642.https://doi.org/10.1172/JCI126714.
Contributor Information
William R. McManus, Email: wrmcmanus01@gmail.com.
Michael J. Bale, Email: Michael.Bale@nih.gov.
Jonathan Spindler, Email: jspindler@mail.nih.gov.
Ann Wiegand, Email: wieganda@mail.nih.gov.
Andrew Musick, Email: andrew.musick@nih.gov.
Sean C. Patro, Email: sean.patro@nih.gov.
Michele D. Sobolewski, Email: mis135@pitt.edu.
Victoria K. Musick, Email: vkm2@hood.edu.
Elizabeth M. Anderson, Email: Elizabeth.Anderson@pennmedicine.upenn.edu.
Joshua C. Cyktor, Email: jcc114@pitt.edu.
Elias K. Halvas, Email: ekh2@pitt.edu.
Wei Shao, Email: shaow@mail.nih.gov.
Daria Wells, Email: wellsd2@mail.nih.gov.
Xiaolin Wu, Email: forestwu@mail.nih.gov.
Brandon F. Keele, Email: keelebf@mail.nih.gov.
Jeffrey M. Milush, Email: jeffrey.milush@ucsf.edu.
Rebecca Hoh, Email: rhoh@php.ucsf.edu.
John W. Mellors, Email: jwm1@pitt.edu.
Stephen H. Hughes, Email: hughesst@mail.nih.gov.
Steven G. Deeks, Email: Steven.Deeks@ucsf.edu.
John M. Coffin, Email: john.coffin@tufts.edu.
Mary F. Kearney, Email: kearneym@mail.nih.gov.
References
- 1.Joos B, et al. HIV rebounds from latently infected cells, rather than from continuing low-level replication. Proc Natl Acad Sci USA. 2008;105(43):16725–16730. doi: 10.1073/pnas.0804192105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kearney MF, et al. Origin of rebound plasma HIV includes cells with identical proviruses that are transcriptionally active before stopping of antiretroviral therapy. J Virol. 2016;90(3):1369–1376. doi: 10.1128/JVI.02139-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kearney MF, et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog. 2014;10(3):e1004010. doi: 10.1371/journal.ppat.1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbloom DIS, Hill AL, Laskey SB, Siliciano RF. Re-evaluating evolution in the HIV reservoir. Nature. 2017;551(7681):E6–E9. doi: 10.1038/nature24634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kearney MF, et al. Ongoing HIV replication during ART reconsidered. Open Forum Infect Dis. 2017;4(3):ofx173. doi: 10.1093/ofid/ofx173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Zyl GU, et al. No evidence of HIV replication in children on antiretroviral therapy. J Clin Invest. 2017;127(10):3827–3834. doi: 10.1172/JCI94582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Josefsson L, et al. The HIV-1 reservoir in eight patients on long-term suppressive antiretroviral therapy is stable with few genetic changes over time. Proc Natl Acad Sci USA. 2013;110(51):E4987–E4996. doi: 10.1073/pnas.1308313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Stockenstrom S, et al. Longitudinal genetic characterization reveals that cell proliferation maintains a persistent HIV type 1 DNA pool during effective HIV therapy. J Infect Dis. 2015;212(4):596–607. doi: 10.1093/infdis/jiv092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vancoillie L, et al. Longitudinal sequencing of HIV-1 infected patients with low-level viremia for years while on ART shows no indications for genetic evolution of the virus. Virology. 2017;510:185–193. doi: 10.1016/j.virol.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Brodin J, et al. Establishment and stability of the latent HIV-1 DNA reservoir. Elife. 2016;5:e18889. doi: 10.7554/eLife.18889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maldarelli F, et al. HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science. 2014;345(6193):179–183. doi: 10.1126/science.1254194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey JR, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80(13):6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner TA, et al. HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science. 2014;345(6196):570–573. doi: 10.1126/science.1256304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simonetti FR, et al. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci USA. 2016;113(7):1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.von Andrian UH, Mempel TR. Homing and cellular traffic in lymph nodes. Nat Rev Immunol. 2003;3(11):867–878. doi: 10.1038/nri1222. [DOI] [PubMed] [Google Scholar]
- 16.Lorenzo-Redondo R, et al. Persistent HIV-1 replication maintains the tissue reservoir during therapy. Nature. 2016;530(7588):51–56. doi: 10.1038/nature16933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnell G, Joseph S, Spudich S, Price RW, Swanstrom R. HIV-1 replication in the central nervous system occurs in two distinct cell types. PLoS Pathog. 2011;7(10):e1002286. doi: 10.1371/journal.ppat.1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Marle G, Gill MJ, Kolodka D, McManus L, Grant T, Church DL. Compartmentalization of the gut viral reservoir in HIV-1 infected patients. Retrovirology. 2007;4:87. doi: 10.1186/1742-4690-4-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fletcher CV, et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci USA. 2014;111(6):2307–2312. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Estes JD, et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med. 2017;23(11):1271–1276. doi: 10.1038/nm.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cory TJ, Schacker TW, Stevenson M, Fletcher CV. Overcoming pharmacologic sanctuaries. Curr Opin HIV AIDS. 2013;8(3):190–195. doi: 10.1097/COH.0b013e32835fc68a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Solas C, Lafeuillade A, Halfon P, Chadapaud S, Hittinger G, Lacarelle B. Discrepancies between protease inhibitor concentrations and viral load in reservoirs and sanctuary sites in human immunodeficiency virus-infected patients. Antimicrob Agents Chemother. 2003;47(1):238–243. doi: 10.1128/AAC.47.1.238-243.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deeks SG, et al. Immune activation set point during early HIV infection predicts subsequent CD4+ T-cell changes independent of viral load. Blood. 2004;104(4):942–947. doi: 10.1182/blood-2003-09-3333. [DOI] [PubMed] [Google Scholar]
- 24.Achaz G, et al. A robust measure of HIV-1 population turnover within chronically infected individuals. Mol Biol Evol. 2004;21(10):1902–1912. doi: 10.1093/molbev/msh196. [DOI] [PubMed] [Google Scholar]
- 25.Bruner KM, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 2016;22(9):1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coffin JM, et al. Clones of infected cells arise early in HIV-infected individuals. JCI Insight. 2019;4(12):128432. doi: 10.1172/jci.insight.128432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wiegand A, et al. Single-cell analysis of HIV-1 transcriptional activity reveals expression of proviruses in expanded clones during ART. Proc Natl Acad Sci USA. 2017;114(18):E3659–E3668. doi: 10.1073/pnas.1617961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baxter AE, et al. Single-cell characterization of viral translation-competent reservoirs in HIV-infected individuals. Cell Host Microbe. 2016;20(3):368–380. doi: 10.1016/j.chom.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deleage C, et al. Defining HIV and SIV reservoirs in lymphoid tissues. Pathog Immun. 2016;1(1):68–106. doi: 10.20411/pai.v1i1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laird GM, Rosenbloom DI, Lai J, Siliciano RF, Siliciano JD. Measuring the frequency of latent HIV-1 in resting CD4+ T cells using a limiting dilution coculture assay. Methods Mol Biol. 2016;1354:239–253. doi: 10.1007/978-1-4939-3046-3_16. [DOI] [PubMed] [Google Scholar]
- 31.Hong F, et al. Novel assays for measurement of total cell-associated HIV-1 DNA and RNA. J Clin Microbiol. 2016;54(4):902–911. doi: 10.1128/JCM.02904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kearney M, et al. Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J Virol. 2009;83(6):2715–2727. doi: 10.1128/JVI.01960-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boritz EA, Douek DC. Perspectives on human immunodeficiency virus (HIV) cure: HIV persistence in tissue. J Infect Dis. 2017;215(suppl_3):S128–S133. doi: 10.1093/infdis/jix005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rainwater-Lovett K, et al. Paucity of intact non-induced provirus with early, long-term antiretroviral therapy of perinatal HIV infection. PLoS ONE. 2017;12(2):e0170548. doi: 10.1371/journal.pone.0170548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ananworanich J, et al. Markers of HIV reservoir size and immune activation after treatment in acute HIV infection with and without raltegravir and maraviroc intensification. J Virus Erad. 2015;1(2):116–122. doi: 10.1016/S2055-6640(20)30482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ananworanich J, et al. Reduced markers of HIV persistence and restricted HIV-specific immune responses after early antiretroviral therapy in children. AIDS. 2014;28(7):1015–1020. doi: 10.1097/QAD.0000000000000178. [DOI] [PubMed] [Google Scholar]
- 37.Crowell TA, et al. Initiation of antiretroviral therapy before detection of colonic infiltration by HIV reduces viral reservoirs, inflammation and immune activation. J Int AIDS Soc. 2016;19(1):21163. doi: 10.7448/IAS.19.1.21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho YC, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hosmane NN, et al. Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: potential role in latent reservoir dynamics. J Exp Med. 2017;214(4):959–972. doi: 10.1084/jem.20170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bui JK, et al. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 2017;13(3):e1006283. doi: 10.1371/journal.ppat.1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorenzi JC, et al. Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA. Proc Natl Acad Sci USA. 2016;113(49):E7908–E7916. doi: 10.1073/pnas.1617789113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson EM, Maldarelli F. Quantification of HIV DNA using droplet digital PCR techniques. Curr Protoc Microbiol. 2018;51(1):e62. doi: 10.1002/cpmc.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer S, et al. New real-time reverse transcriptase-initiated PCR assay with single-copy sensitivity for human immunodeficiency virus type 1 RNA in plasma. J Clin Microbiol. 2003;41(10):4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mansky LM, Temin HM. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69(8):5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Palmer S, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol. 2005;43(1):406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eypasch E, Lefering R, Kum CK, Troidl H. Probability of adverse events that have not yet occurred: a statistical reminder. BMJ. 1995;311(7005):619–620. doi: 10.1136/bmj.311.7005.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol Biol Evol. 1992;9(1):138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- 49.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 50.Lefort V, Longueville JE, Gascuel O. SMS: Smart model selection in PhyML. Mol Biol Evol. 2017;34(9):2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Critchlow DE, Li S, Nourijelyani K, Pearl DK. Some statistical methods for phylogenetic trees with application to HIV disease. Math Comput Model. 2000;32(1–2):69–81. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.