Abstract
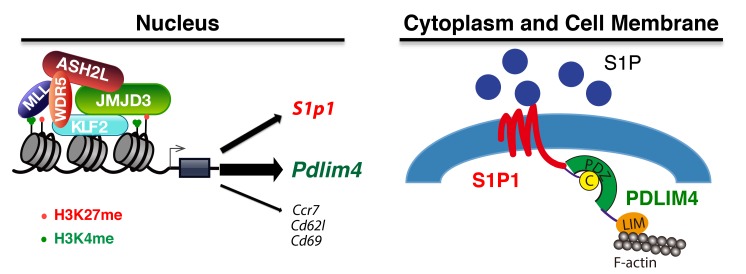
Histone H3K27 demethylase JMJD3 plays a critical role in gene expression and T cell differentiation. However, the role and mechanisms of JMJD3 in T cell trafficking remain poorly understood. Here, we show that JMJD3 deficiency in CD4+ T cells resulted in an accumulation of T cells in the thymus and reduction of T cell number in the secondary lymphoid organs. We identified PDLIM4 as a significantly downregulated target gene in JMJD3-deficient CD4+ T cells by gene profiling and ChIP-Seq analyses. We further showed that PDLIM4 functioned as an adaptor protein to interact with sphingosine-1 phosphate receptor 1 (S1P1) and filamentous actin (F-actin), thus serving as a key regulator of T cell trafficking. Mechanistically, JMJD3 bound to the promoter and gene-body regions of the Pdlim4 gene and regulated its expression by interacting with zinc finger transcription factor KLF2. Our findings have identified Pdlim4 as a JMJD3 target gene that affects T cell trafficking by cooperating with S1P1 and have provided insights into the molecular mechanisms by which JMJD3 regulates genes involved in T cell trafficking.
Keywords: Autoimmunity, Cell Biology, Immunology
Keywords: Cancer immunotherapy, Cell migration/adhesion, Cytoskeleton, Immunotherapy, T cells
Introduction
T cell development in the thymus is a multistep process. Early thymic progenitor cells (TPCs) differentiate into T cell receptor–expressing (TCR-expressing) CD4+CD8+ double-positive (DP) thymocytes in the cortex and then mature into single-positive (SP) CD4+ and CD8+ T cells in the medulla (1–3). It is known that T cell trafficking from the thymus to the periphery and then migration into the secondary lymphoid and peripheral organs are regulated by the dynamic cytoskeleton. The dynamic remodeling of the actin cytoskeleton is a key component of cell locomotion and membrane trafficking (4). Of note, cellular mobility and transmigration through the endothelium require F-actin–binding genes for complex cytoskeletal rearrangements (5). After negative and positive selection, the immature T cells express transcription factor Kruppel-like factor 2 (KLF2) and its target gene, sphingosine-1 phosphate receptor 1 (S1P1, encoded by S1pr1), which are required for T cell egress from the thymus and subsequent migration to secondary lymphoid organs (1, 3, 6–11). After thymic emigration, the trafficking of mature CD4+ or CD8+ SP T cells is a major process for allowing and regulating their immunosurveillance commitment. The motility capabilities of T cells are coupled to their ability to detect and eliminate pathogens and cancer cells. Tumors can be divided into “hot” (inflamed) and “cold” (noninflamed), according to the presence of T cells. The presence of tumor-infiltrating lymphocytes in the tumor microenvironment has been reported to correlate well with positive clinical outcomes (12, 13). Thus, the promotion of tumor-specific T cell infiltration into the tumor microenvironment would benefit T cell tumor immunotherapy. On the other hand, the abnormal motility capabilities of T cells are coupled to the development of undesirable responses against self-antigens, which is a key step in autoimmune disease progression and the failure of organ transplantation. Recent results show that S1P1 agonist FTY-720 (Gilenya) arrests T cell trafficking and prevents multiple sclerosis relapses (14). Therefore, T lymphocyte trafficking may be an important target for immunotherapy and autoimmune response. However, the molecular mechanisms responsible for T lymphocyte trafficking are still poorly understood.
The methylation state of histones is dynamically regulated by histone methyltransferases and demethylases (15–19). Of note, while trimethylation of the lysine 4 residue of histone 3 (H3K4me3) is usually associated with activation of gene expression, trimethylation of the lysine 27 residue of histone 3 (H3K27me3) is conversely associated with repression of gene expression (15–19). Methylation of H3K4 can be mediated by several histone methyltransferases (20), while demethylation of H3K4me3 is mediated by members of the lysine demethylase 2 (KDM2) and KDM5 families of lysine demethylases. Di- and trimethylation of H3K27 (H3K27me2/3) are mediated by the polycomb repressive complex 2 (PRC2), which contains the H3K27 methyltransferase enhancer of zeste homolog 2 (EZH2) protein (18, 21). On the other hand, demethylation of H3K27me2/3 is mediated by 2 H3K27 demethylases, ubiquitously transcribed tetratricopeptide repeat, X chromosome (Utx, KDM6A), and Jumonji domain containing 3 (JMJD3, KDM6B) (22–26). Genetically modified Utx-deficient mice develop severe heart defects during development, leading to embryonic lethality (27). Similarly, Jmjd3 whole-body KO mice die shortly after birth (28, 29). Although JMJD3 has been implicated in the thymic egress (30), its target genes and the regulatory mechanisms in T cell trafficking have not been reported.
Here, we show that T cell trafficking from the thymus to the spleen and lymph nodes (LNs) was markedly altered in Jmjd3-deficient T cells. Furthermore, we have identified a role for PDLIM4, a cytoskeletal protein (31, 32), in mediating T cell migration by affecting F-actin remodeling. We have also delineated mechanisms by which JMJD3 stabilizes transcription factor binding to the Pdlim4 promoter. Specifically, our results indicate that JMJD3 regulates the expression of cytoskeletal PDLIM4 by stabilizing the KLF2-ASH2L complex and thus controls T cell trafficking.
Results
Critical role of JMJD3 in T cell trafficking from the thymus to spleen and LNs.
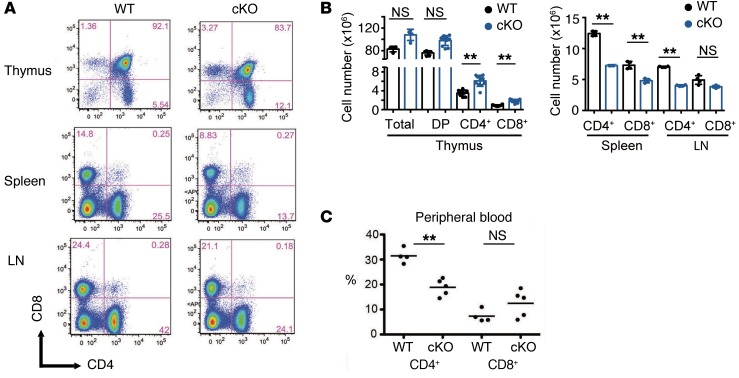
We previously generated T cell–specific deletion of Jmjd3 (Jmjd3-cKO) mice by crossing Jmjd3fl/fl mice with CD4-Cre mice and found that JMJD3 plays a critical role in T cell differentiation (28). To further define the role of JMJD3 in the homeostasis of T cell populations and trafficking, we performed flow cytometric analysis of T cells isolated from the thymus, spleen, and LNs. While thymic CD4 SP T cells and CD8 SP T cells were dramatically increased, the percentages of CD4+ T cells were markedly decreased (approximately 50%) in both spleens and LNs of Jmjd3-cKO mice as compared with WT mice (Figure 1, A and B). Similarly, CD8 SP T cells were also reduced in Jmjd3-cKO spleens, but not in the LNs (Figure 1, A and B). Furthermore, the extent of CD4+ T cells, but not CD8+ T cells, was reduced in the peripheral blood of Jmjd3-cKO mice (Figure 1C). Taken together, these results suggest that CD4-specific deletion of Jmjd3 results in marked accumulation of thymic CD4 and CD8 SP T cells, reducing their ability to migrate from the thymus to secondary lymphoid organs.
Figure 1. Analysis of T cell populations in different organs of WT and Jmjd3-cKO mice.
(A) Flow cytometry analysis of CD4 and CD8 SP T cells in the thymus, spleen, and LN from WT and Jmjd3-cKO mice. (B) Absolute cell numbers for different cell populations, including total, DP, CD4 SP (CD4+), and CD8 SP (CD8+) cells in the thymus, spleens and LNs from WT and Jmjd3-cKO mice (n = 8/group, from 2 independent experiments). Data are reported as mean + SD from 3 independent experiments. **P < 0.01, Student’s t test. (C) Percentages of CD4+ and CD8+ cells in the peripheral blood from WT and Jmjd3-cKO mice. Circles represent individual mice, and lines represent the mean. n = 4/5. **P < 0.01, Student’s t test. NS, no significant differences.
Trafficking of WT and JMJD3-deficient T cells in adoptive transfer models.
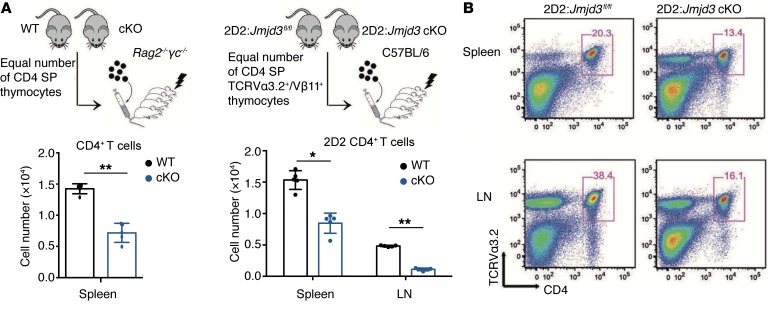
Next, we sought to determine the intrinsic trafficking properties of WT and Jmjd3-deficient T cells in Rag2–/–γc–/– recipient mice, which lack endogenous T cells. Adoptive T cell transfer of equal numbers of thymic WT and Jmjd3-deficient CD4 SP T cells demonstrated reduced numbers of splenic Jmjd3-deficient CD4+ T cells compared with splenic WT cells (Figure 2A), suggesting defective Jmjd3-deficient CD4+ T cell migration. To further substantiate these findings, we performed adoptive transfer of TCR-specific CD4+ T cells. For this purpose, we crossed TCR 2D2 transgenic mice, harboring myelin oligodendrocyte glycoprotein (MOG) peptide-specific TCRs, with Jmjd3fl/fl mice, with or without CD4-Cre to generate 2D2:Jmjd3fl/fl and 2D2:Jmjd3fl/fl:cKO mice. Next, CD4 SP thymocytes were isolated by FACS using Abs against 2D2 TCR (TCRVα3.2 and TCRVβ11) (Figure 2B). Adoptive transfer of equal numbers of these 2D2 TCR-specific T cells from 2D2:Jmjd3fl/fl (WT) and 2D2:Jmjd3-cKO mice into sublethally irradiated C57BL/6 mice demonstrated a lower percentage and a lower absolute number of 2D2:Jmjd3-cKO CD4+ T cells compared with 2D2 WT CD4+ T cells in the spleens and LNs (Figure 2B), as assessed by flow cytometry of TCRVα3.2 and CD4 cell-surface expression. Taken together, these results suggest that Jmjd3-deficient CD4+ T cells reduce the ability to migrate to the peripheral lymphoid organs compared with WT control cells.
Figure 2. Jmjd3 deficiency causes defects in T cell migration.
(A) Thymic CD4 SP cells from either WT or Jmjd3-cKO mice were purified by FACS, and equal numbers of cells were intravenously injected into sublethally irradiated Rag2–/–γc–/– mice (n = 5) (upper panel). Absolute numbers of CD4+ cells in the spleens of recipient Rag2–/–γc–/– mice were determined 24 hours after adoptive transfer (lower panel). (B) Thymic CD4 SP (TCRVα3.2+/Vβ11+) T cells from either 2D2:Jmjd3fl/fl or 2D2:Jmjd3-cKO mice were purified by FACS, and equal numbers of cells were intravenously injected into sublethally irradiated C57BL/6 mice (n = 5). Absolute numbers of CD4+ TCRVα3.2+ TCRVβ11+ cells in the spleens and LNs of recipient C57BL/6 mice were determined 24 hours after adoptive transfer (left segment) and positive cell ratio (right segment) by FACS. Data are plotted as mean + SD from 3 independent experiments. *P < 0.05, **P < 0.01, Student’s t test.
Next, we sought to determine whether the reduced trafficking ability of Jmjd3-deficient CD4+ T cells has functional consequences for the induction of experimental autoimmune encephalomyelitis (EAE), a T cell–mediated autoimmune disease of the CNS. WT and Jmjd3-cKO mice were immunized by subcutaneous injection of the antigenic peptide for MOG (MOG35–55) plus CFA and then injected with pertussis toxin (PTx) to induce EAE (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI128293DS1). WT mice developed EAE disease faster and more severely than Jmjd3-cKO mice, as evidenced by mean clinical scores (Supplemental Figure 1B).
To further study the role of JMJD3 in tumor therapy, we performed adoptive transfer of TCR-specific CD4+ T cells. For this purpose, we crossed TRP-1 TCR transgenic mice (obtained from The Jackson Laboratory, 008486), which express TCR specific for the minimal tyrosinase-related protein 1 (TRP-1) epitope, with Jmjd3fl/fl and CD4-Cre mice to generate TRP-1:Jmjd3fl/fl and TRP-1:Jmjd3fl/fl:cKO mice. Next, CD4+ T cells were isolated from splenocytes by Dynabeads Untouched Mouse CD4 Cell Kit (Invitrogen, 11415D). Equal numbers of these TRP-1 TCR-specific T cells from TRP-1:Jmjd3fl/fl (WT) and TRP-1:Jmjd3-cKO mice were adoptively transferred into B16-bearing mice (Supplemental Figure 1C). We observed smaller tumors in the mice with WT CD4+ T cells than in the mice with Jmjd3-cKO cells (Supplemental Figure 1, D and E). We next detected tumor-infiltrating T cells by flow cytometry of TCRVα3.2 and CD4 markers and demonstrated a lower percentage and absolute numbers of TRP-1:Jmjd3-cKO CD4+ T cells compared with WT TRP-1 CD4+ T cells in the tumor (Supplemental Figure 1F). Taken together, these results suggest that Jmjd3-deficient CD4+ T cells have an impaired ability to migrate to the tumor microenvironment, as compared with WT controls.
Identification of PDLIM4 as a key regulator of T cell migration.
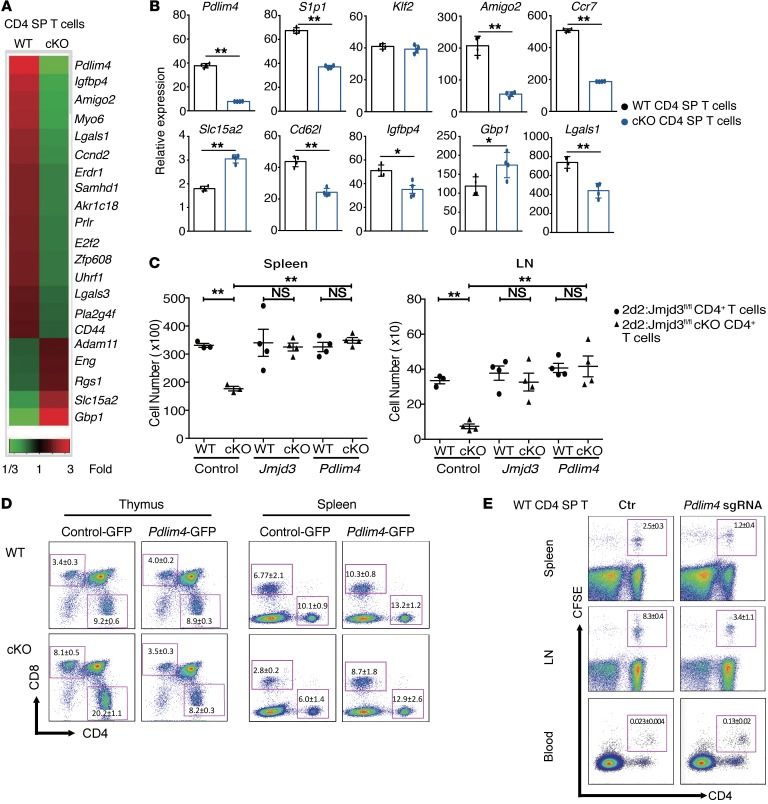
We next sought to identify JMJD3 target genes that might regulate T cell migration by performing microarray analysis of thymic CD4 SP T cells isolated from WT and Jmjd3-cKO mice. Gene expression profiling revealed that 16 genes were markedly downregulated, while 5 were upregulated in thymic Jmjd3-cKO CD4 SP T cells compared with WT cells (Figure 3A and Supplemental Table 1). Changes in genes involved in cell motility, cell death, and cell proliferation were confirmed by quantitative real-time PCR analysis (Figure 3B). Genes such as Amigo2, Igfbp4, and Lgals1 were downregulated, while Slc15a2 and Gbp1 were upregulated in thymic Jmjd3-cKO CD4 SP T cells when compared with WT CD4 SP T cells (Figure 3B). We also observed that the expression of Ccr7 and Cd62l were remarkably downregulated in Jmjd3-deficient CD4 SP T cells (Figure 3B). However, there was no significant difference in the expression levels of several other genes, such as Erdr1, Samhd1, Akr1c18, and Adam11, between WT and Jmjd3-cKO CD4 SP T cells (Supplemental Figure 2A). S1p1 and Pdlim4 (encoding the PDZ and LIM domain protein 4), but not Klf2, were significantly downregulated in thymic Jmjd3-cKO CD4 SP T cells compared with WT CD4 SP T cells (Figure 3B). We further tested their expression at the protein level. The Western blotting data showed the dramatic decrease of S1P1 in Jmjd3-cKO cells (Supplemental Figure 2B). However, we did not observe an appreciable difference in CCR7 protein between WT and Jmjd3-cKO cells. This observation was further supported by flow cytometry analysis of CCR7 on the surface of WT and Jmjd3-cKO CD4 SP T cells (Supplemental Figure 2C).
Figure 3. Identification of the JMJD3 target genes in CD4+ T cells and functional rescue of T cell defects by ectopic expression of Pdlim4.
(A) Heatmap from microarray analysis of upregulated and downregulated genes in WT and Jmjd3-cKO thymic CD4 SP T cells. (B) Real-time PCR analysis of a panel of genes between WT and Jmjd3-cKO thymic CD4 SP T cells. Expression levels are given as the ratio of the target gene to the control gene to correct for variations in the starting amount of mRNA (gene/Gapdh ×1000). n = 4. *P < 0.05; **P < 0.01, Student’s t test. (C) CD4+ T cells from 2D2:Jmjd3fl/fl (WT) mice or 2D2:Jmjd3-cKO mice were activated with MOG35–55 peptide in vitro before transduction with GFP-expressing retroviral vectors containing Jmjd3 or Pdlim4. Equal numbers of GFP+CD4+ T cells were intravenously injected into sublethally irradiated C57BL/6 mice (n = 4). Absolute numbers of TCRVα3.2+/Vβ11+GFP+CD4+ T cells in spleens and LNs were determined by flow cytometry 48 hours after adoptive transfer. Data are presented as mean + SD from 3 independent experiments. **P < 0.01, 1-way ANOVA with Tukey’s multiple comparisons test. (D) WT and Jmjd3-cKO bone marrow cells overexpressing control GFP or Pdlim4-GFP were transplanted into lethally irradiated C57BL/6 (WT) mice to generate chimeric mice. Flow cytometric analysis of CD4+ and CD8+ T cells from the thymus and the spleens of chimeric mice. n = 3/group; 1 experiment. (E) Thymic CD4 SP T cells were isolated from WT mice. Pdlim4 KO was generated using the CRISPR-Cas9 system. Cells were labeled with CFSE and then intravenously injected into sublethally irradiated C57BL/6 mice. After 48 hours, spleens, LNs, and peripheral blood were analyzed by flow cytometry for CD4+ and CFSE-stained cells. Experiments were repeated 3 independent times. n = 3/group; 1 experiment.
Since PDLIM4 has been identified as a modifier of actin filament dynamics through its interaction with α-actinin and F-actin (33), we postulated that PDLIM4 might contribute to the migration defects in Jmjd3-cKO CD4+ T cells. To test this possibility, we constructed GFP-expressing retroviral vectors containing Jmjd3 and Pdlim4. Splenic T cells were collected from 2D2:Jmjd3fl/fl (WT) and 2D2:Jmjd3-cKO mice and activated in vitro before transduction with GFP-containing viruses. GFP+CD4+ T cells were sorted, and equal numbers of GFP+ cells were adoptively transferred into irradiated C57BL/6 recipient mice. After 48 hours, single-cell suspensions were prepared from the isolated spleens and LNs of the recipient mice. MOG35–55 peptide–specific donor cells were analyzed by flow cytometry using 2D2 TCR–specific Abs (TCRVα3.2 and TCRVβ11). Ectopic expression of Jmjd3 or Pdlim4, but not the empty vector control, restored the number of 2D2:Jmjd3-cKO CD4+ T cells in the spleens and LNs similarly to 2D2:Jmjd3fl/fl (WT) CD4+ cells (Figure 3C), suggesting that Jmjd3 or Pdlim4 may rescue the defects in T cell migration.
To further determine whether PDLIM4 could rescue the CD4+ T cell migration defect due to Jmjd3 deficiency, we isolated bone marrow cells from either WT or Jmjd3-cKO mice and overexpressed control-GPF or Pdlim4-GFP in these bone marrow cells (Figure 3D). We generated chimeric mice, in which these bone marrow cells were transplanted into sublethally irradiated C57BL/6 mice. Flow cytometric analysis of splenocytes and thymocytes from these chimeric mice showed that significantly higher percentages of Jmjd3-cKO CD4+ T cells transduced with control-GFP in thymus, but reduced percentages of T cells accumulated in the spleen when compared with WT CD4+ T cells. However, thymic and splenic accumulation of Jmjd3-deficient CD4+ T cells was restored upon ectopic expression of Pdlim4 (Figure 3D).
In order to further demonstrate the physiological role of PDLIM4 in CD4+ T cell migration, we isolated thymic CD4 SP T cells from WT mice, generated Pdlim4-KO CD4+ T cells using the CRISPR-Cas9 system (Supplemental Figure 3A), labeled the cells with CFSE, and then adoptively transferred them to irradiated C57BL/6 mice. After 48 hours, we determined the number of T cells by FACS analysis and found an increased number of Pdlim4-KO CD4+ T cells in the peripheral blood of mice, but reduced numbers in spleen and LNs, suggesting a decreased ability of these cells to migrate into spleen and LNs (Figure 3E). To further determine whether Pdlim4 KO inhibits CD4+ T cell migration, we isolated bone marrow cells from WT mice and knocked out Pdlim4 using the CRISPR-Cas9 system to generate chimeric mice by transferring Pdlim4-KO bone marrow cells into irradiated recipient mice. FACS analysis of cells isolated from these chimeric mice showed higher percentages of Pdlim4-KO CD4+ T cells accumulated in the thymus, but reduced percentages of Pdlim4-KO CD4+ T cells accumulated in the spleen and LNs (Supplemental Figure 3B). Taken together, these results suggest that PDLIM4, the target of JMJD3, plays a critical role in T cell trafficking.
PDLIM4 regulates T cell migration through interaction with S1P1 and modulation of F-actin organization.
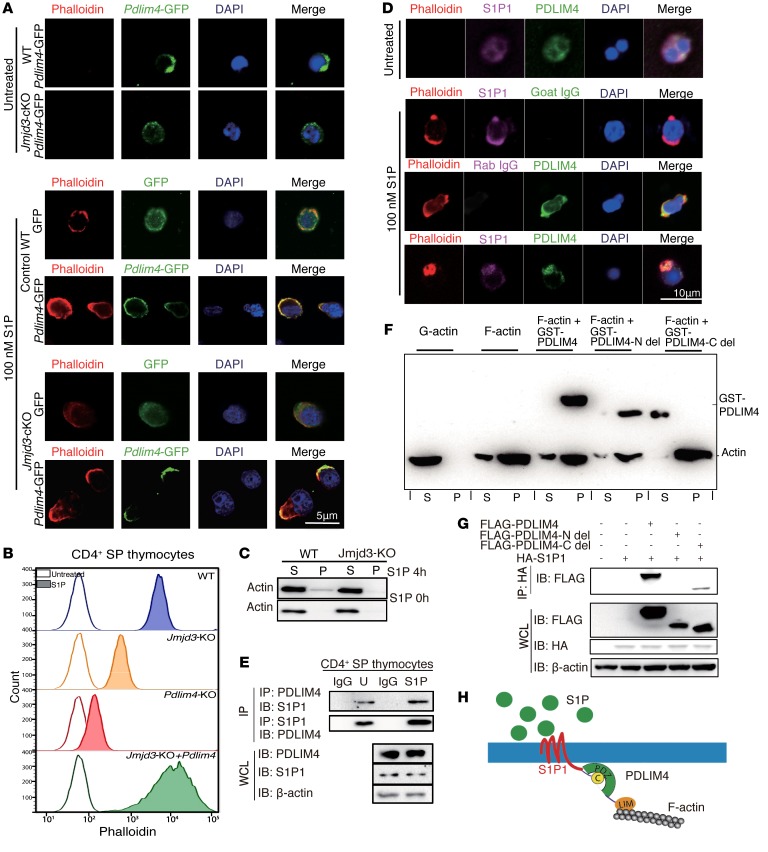
PDLIM4 has been identified as a modifier of actin filament dynamics through its interaction with α-actinin and F-actin in muscle cells and nonmuscle epithelial cells (33). Furthermore, PDLIM4 cytoskeleton protein contains 1 PDZ domain, which has been reported to play a key role in anchoring receptor proteins in the membrane to cytoskeletal components (34). Since S1P treatment can cause actin cytoskeleton remodeling, we next sought to determine whether S1P treatment regulates PDLIM4-mediated actin remodeling in T cells, which may be required for T cell trafficking. To this end, we isolated WT and Jmjd3-deficient CD4 SP thymocytes, treated them with S1P after 12 hours of starvation with serum-free medium, and stained F-actin with phalloidin after T cell fixation. As shown in Figure 4A, in the WT and Jmjd3-deficient CD4+ T cells transfected with Pdlim4-GFP, we observed specific localization of PDLIM4 in the lamellipodium structure of the F-actin remodeling area. In contrast, we did not observe the specific localization of GFP in the lamellipodium structure of the F-actin remodeling area in the WT and Jmjd3-deficient CD4+ T cells transfected with GFP. We next determined whether ectopic expression of PDLIM4 promoted F-actin formation using WT thymic CD4 SP T cells, Jmjd3-cKO CD4 SP T cells, Pdlim4-KO CD4 SP T cells, and Pdlim4-expressing Jmjd3-cKO T cells, followed by phalloidin staining and FACS analysis. We found that F-actin organization was defective in Pdlim4-KO and Jmjd3-cKO CD4 SP T cells compared with WT and PDLIM4-expressing cells based on low phalloidin staining after 30 minutes of 50 nM S1P treatment (Figure 4B). Notably, PDLIM4-expressing cells exhibited the highest phalloidin staining for F-actin (Figure 4B), suggesting that PDLIM4 enhances F-actin formation. To further determine the distribution of insoluble F-actin (pellet) and the soluble G-actin (supernatant), we treated WT and Jmjd3-cKO CD4 SP T cells with S1P and lysed the treated cells. After removal of cellular debris by low centrifugation at 350 g for 5 minutes, the supernatants were further centrifuged at 150,000 g for 90 minutes to separate F-actin (in pellet) from soluble G-actin (in supernatant). The relative distributions of insoluble F-actin (pellet) and the soluble G-actin (supernatant) were analyzed by Western blot. We found that polymerized F-actin in the Jmjd3-cKO CD4 SP T cells was dramatically decreased when compared with that in WT cells (Figure 4C). Collectively, our results suggest that JMJD3 or PDLIM4 deficiency markedly reduces F-actin after S1P treatment, which can be rescued by ectopic expression of PDLIM4.
Figure 4. PDLIM4 regulates T cell migration by interaction with S1P1 and modulation of F-actin reorganization.
(A) Immunofluorescence microscopy images of WT and Jmjd3-cKO CD4+ T cells infected with lentivirus containing GFP or PDLIM4-GFP plasmids. Cells were starved 12 hours and treated with 100 nM S1P for 3 hours at 37°C. GFP or PDLIM4-GFP was detected with green fluorescence. Actin filaments were labeled with rhodamine-conjugated phalloidin (red). Nuclei were stained with DAPI (blue). (B) FACS analysis of phalloidin-labeled F-actin in untreated and S1P-treated WT, Jmjd3-cKO, Pdlim4-KO, and Pdlim4-expressing Jmjd3-cKO CD4 SP thymocytes. (C) F-actin (pellet [P]) and G-actin (supernatant [S]) from untreated and treated WT, Jmjd3-cKO CD4 SP thymocytes were detected by Western blot. (D) Immunofluorescence microscopy of untreated and S1P-treated WT CD4+ SP cells stained with FITC-conjugated Abs to detect PDLIM4, Cy5-conjugated Abs to detect S1P1, and rhodamine-conjugated phalloidin to detect actin filaments. Goat IgG and rabbit IgG were used as isotype controls. Merged images indicate colocalization of proteins. (E) Co-IP analysis of endogenous interaction of PDLIM4 with S1P1 in untreated and S1P-treated CD4 SP thymocytes. (F) Cosedimentation assay was performed using GST-PDLIM4, GST-PDLIM4 N-del, or GST-PDLIM4 C-del with F-actin, and subsequent analysis of supernatants and pellets by Western blot analysis. (G) 293T cells were cotransfected with FLAG-Pdlim4-N-del, C-del, or full-length Pdlim4 along with HA-S1p1. WCLs were immunoprecipitated with anti-HA beads and immunoblotted with anti-FLAG Ab. Three independent experiments were repeated with similar results. (H) A schematic diagram of the proposed model showing that PDLIM4 interacts with the S1P1 protein at the N-terminal PDZ domain and binds F-actin by the C-terminal LIM domain.
S1P1 is a G protein–coupled receptor that binds to the bioactive signaling molecule S1P and activates intracellular signaling pathways that lead to cytoskeleton remodeling and T cell egress from the thymus (35). We next used different immunofluorescences to label PDLIM4, S1P1, and phalloidin to stain F-actin in untreated and S1P-treated WT SP CD4+ T cells and observed colocalization of PDLIM4 with S1P1 on lamellipodium structure in the F-actin remodeling area (Figure 4D). Colocalization of PDLIM4 in S1P1 was quantified by using NIS-Elements Analysis and reached 95% upon S1P stimulation, whereas only 40% of colocalization was observed in the untreated cells (Supplemental Figure 4A). Consistently, co-IP analysis revealed that PDLIM4 was associated with S1P1 in CD4 SP thymocytes (Figure 4E). To determine the specific domains of PDLIM4 involved in the interaction with F-actin, we used purified recombinant GST-PDLIM4 in an in vitro cosedimentation assay to investigate the interaction of PDLIM4 and F-actin. The polymerized actin was incubated with GST-PDLIM4, GST-PDLIM4-N-del (PDZ domain deletion), or GST PDLIM4-C-del (LIM domain deletion), followed by high-speed centrifugation at 150,000 g for 90 minutes. The pellets and supernatants were subjected to SDS-PAGE separation after fractionation. We found that GST-PDLIM4 or GST-PDLIM4-N-del, but not GST-PDLIM4-C-del, was in the pellet fraction (Figure 4F), suggesting that the C terminus of PDLIM4 interacts with F-actin. Since PDLIM4 interacts with S1P1 (Figure 4E), we next determined which domain of PDLIM4 was required for interacting with S1P1. We constructed FLAG-tagged Pdlim4-N-del (PDZ domain deletion), C-del (LIM domain deletion), full-length Pdlim4, and HA-tagged S1P1, and cotransfected into HEK293T cells. Co-IP analysis revealed that the PDZ domain of PDLIM4 was required for interacting with S1P1 (Figure 4G). Additionally, we mutated 1 cysteine amino acid to proline amino acid (C330P ) in the C-terminal helix-8 region of S1P1. The co-IP results showed that S1P1 could not interact with PDLIM4 containing a mutation in the helix-8 region (Supplemental Figure 4B). Based on these findings, we propose a model showing that the PDLIM4 interacts with the S1P1 protein at the N-terminal PDZ domain and binds to the actin cytoskeleton through its C-terminal LIM domain (Figure 4H).
Expression of Pdlim4 is coregulated by JMJD3 and KLF2.
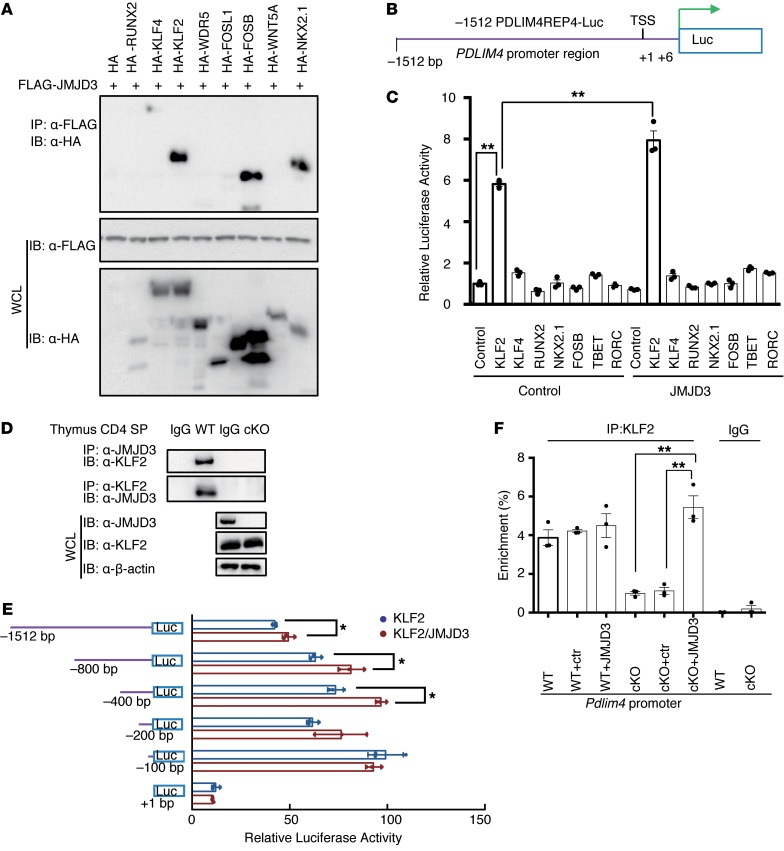
During T cell differentiation, JMJD3 interacts with key differentiation regulatory transcription factors, such as T-bet (28, 36, 37). Hence, we postulated that JMJD3 might also interact with factors involved in T cell migration, such as KLF2 and S1P1, which are master regulators of T cell emigration from the thymus (6–11). To test this possibility, we cotransfected 293T cells with FLAG-Jmjd3 and HA-tagged transcription factor genes, including Runx2, Klf4, Klf2, Wdr5, Fosl1, FosB, Wnt5a, and Nkx2.1 (Figure 5A). Co-IP and immunoblot analysis revealed that JMJD3 interacted with KLF2, FosB, and Nkx2.1 in vitro (Figure 5A). A dual-luciferase assay was performed in 293T cells cotransfected with Klf2, Klf4, Runx2, Nkx2.1, FosB, T-bet, and Rorc with or without Jmjd3 and –1512 bp upstream of mouse Pdlim4 promoter–linked episomal luciferase vector (Figure 5B). Only KLF2 harbored transcriptional activity in regulating Pdlim4 expression, and JMJD3 significantly enhanced the ability of KLF2 to induce Pdlim4 promoter–driven luciferase activity (Figure 5C). JMJD3 did not enhance Pdlim4 promoter–driven luciferase activity without KLF2, suggesting coregulation of Pdlim4 by JMJD3 and KLF2. To determine whether endogenous interactions occur between KLF2 and JMJD3, we collected thymic WT and Jmjd3-cKO CD4+ SP T cell lysates and co-IP with anti-JMJD3 or anti-KLF2 Abs and then immunoblotted with anti-KLF2 or anti-JMJD3 Abs. We observed that KLF2 interacted with JMJD3 in CD4+ T cells (Figure 5D). Next, we dissected the precise Pdlim4 promoter sequences that were responsible for KLF2 and JMJD3 binding. We constructed a series of Pdlim4 promoter fragment–linked episomal luciferase vectors and cotransfected 293T cells with KLF2 alone or KLF2 and JMJD3 together (Figure 5E). The dual-luciferase assay indicated that sequences –100 bp upstream of the Pdlim4 transcriptional start site (TSS) were required for KLF2 binding and sequences –800 bp upstream of the Pdlim4 TSS were required for JMJD3 and KLF2 binding (Figure 5E); the luciferase reporter assay using CD4 SP T cells showed consistent results (Supplemental Figure 5A). To address whether the interaction between JMJD3 and KLF2 is required for KLF2 binding to the Pdlim4 promoter, we performed a ChIP–quantitative PCR (ChIP-qPCR) assay on thymic CD4 SP T cells from WT or Jmjd3-cKO mice. We found that the binding of KLF2 to the Pdlim4 promoter was defective in Jmjd3-deficient CD4 SP T cells, suggesting that JMJD3 is required for KLF2 binding to the Pdlim4 promoter (Supplemental Figure 5B). This defective binding of KLF2 to the Pdlim4 promoter could be rescued by expression of JMJD3 in Jmjd3-cKO CD4 SP T cells (Figure 5F). Taken together, our results indicate that both JMJD3 and KLF2 are required for binding to the promoter region of Pdlim4 and coregulate the expression of Pdlim4.
Figure 5. JMJD3 regulates Pdlim4 expression by interacting with KLF2.
(A) Screening of transcription factors interacting with JMJD3. 293T cells were cotransfected with HA-tagged Runx2, Klf4, Klf2, Wdr5, Fosl1, FosB, Wnt5a, Nkx2.1, and FLAG-Jmjd3. WCLs were immunoprecipitated with anti-FLAG Abs and immunoblotted with anti-HA Abs. (B) Schematic presentation of the Pdlim4 promoter–driven luciferase construct. The promoter region –1512 bp upstream was cloned into an episomal luciferase vector. (C) The transcriptional activity of proteins interacting with JMJD3 in regulating Pdlim4 was evaluated by dual-luciferase assay. Data are presented as mean ± SD from 3 independent experiments (n = 3). **P < 0.01, 1-way ANOVA with Tukey’s multiple comparisons test. (D) Thymic CD4 SP T cells were isolated from WT and Jmjd3-cKO mice and immunoprecipitated with anti-JMJD3 or anti-KLF2 Abs and protein (A+G) beads. The immunoprecipitated product was immunoblotted with anti-KLF2 or anti-JMJD3 Abs. (E) Mapping the KLF2 and JMJD3 binding regions of the Pdlim4 promoter using a dual-luciferase assay. Different regions of the Pdlim4 promoter were cloned into the episomal luciferase vector and then were cotransfected with Klf2 and Jmjd3 into 293T cells. Data are presented as mean ± SD from 3 independent experiments (n = 3). *P < 0.05, Student’s t test. (F) ChIP-qPCR analysis of percentage of enrichment of KLF2 at the Pdlim4 promoter in WT and Jmjd3-cKO thymic CD4 SP T cells after ectopic expression. Jmjd3 IgG was used as isotype control. n = 3. **P < 0.01, 1-way ANOVA with Tukey’s multiple comparisons test.
JMJD3 deficiency alters the methylation state of H3K27 and H3K4 on the Pdlim4 promoter.
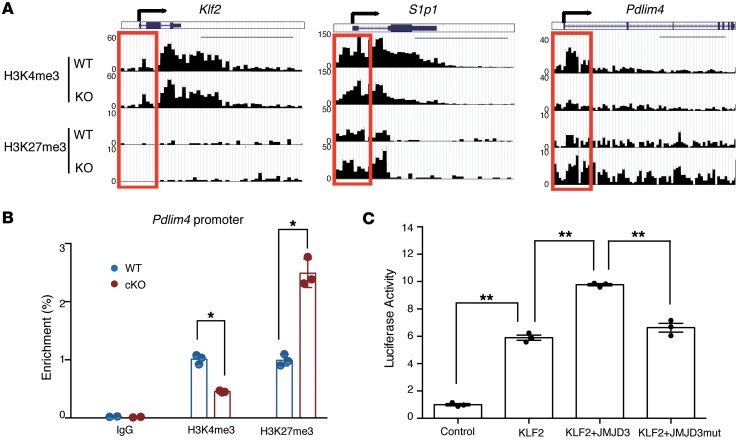
We previously reported that Jmjd3 deletion specifically increased H3K27 di- and trimethylation in CD4+ T cells and also affected H3K4 methylation (28). By dissecting the ChIP-Seq data for individual genes of interest, we found that Pdlim4 and S1p1, but not Klf2, were highly bivalently marked in Jmjd3-cKO CD4 SP T cells (Figure 6A). Both Klf2 and S1p1 genes contained high levels of H3K4me3, but little or low levels of H3K27me3. Conversely, the upstream promoter regions of the TSS and even gene-body regions of Pdlim4 harbored high levels of H3K27me3 in Jmjd3-cKO CD4 SP T cells, which were higher than in WT CD4 SP T cells (Figure 6A). For genes such as Ccr7, Cd62l, Cd69, and Klf4, we did not observe appreciable changes in H3K4me3 and H3K27me3 levels (Supplemental Figure 6).
Figure 6. H3K27me3 and H3K4me3 levels in the Pdlim4 promoter in WT and Jmjd3-deficient T cells.
(A) ChIP-Seq analysis of H3K27me3 and H3K4me3 levels in the promoter and gene-body regions of Klf2, S1p1, and Pdlim4 in thymic CD4 SP T cells isolated from WT and Jmjd3-cKO mice. Red boxes indicate 2 kb regions around the TSS. Scale bars: 5 kb. (B) Validation of methylation changes on the Pdlim4 gene in WT and Jmjd3-cKO CD4 SP T cells by ChIP-qPCR. Data represent mean ± SD from 3 independent experiments. n = 3. *P < 0.05, Student’s t test. (C) Luciferase assay was performed on 293T cells cotransfected with Pdlim4 promoter–linked episomal luciferase vector and with Klf2 in the presence of WT or mutant Jmjd3 (a loss of demethylase function mutation). Data are presented as mean ± SD from 3 independent experiments. n = 3. **P < 0.01, 1-way ANOVA with Tukey’s multiple comparisons test.
To validate our ChIP-Seq data on the methylation status of Pdlim4, we performed ChIP-qPCR using WT CD4 SP T and Jmjd3-deficient CD4 SP T cells and Abs that were used to target H3K4me3 and H3K27me3. Interestingly, we observed a marked decrease in H3K4me3 and a marked increase in H3K27me3 levels on the Pdlim4 gene promoter in Jmjd3-deficient CD4+ T cells as compared with WT cells (Figure 6B), suggesting that downregulation of Pdlim4 in Jmjd3-cKO CD4 SP T cells may be a result of changes in these epigenetic markers. To determine whether the demethylase activity of JMJD3 is required for regulating Pdlim4 expression, a luciferase assay was performed on 293T cells cotransfected with Pdlim4 promoter–linked episomal luciferase vector and with Klf2 in the presence of WT or mutant Jmjd3 (a loss of demethylase function mutation). WT Jmjd3 significantly enhanced KLF2-mediated Pdlim4 promoter activity, whereas mutant JMJD3 failed to enhance KLF2-mediated Pdlim4 promoter activity (Figure 6C). Together, these results suggest that Pdlim4 gene expression in CD4 SP T cells is dependent on JMJD3-dependent methylation of H3K27/H3K4 on Pdlim4, which may regulate T cell trafficking.
JMJD3 stabilizes the interaction between KLF2 and WDR5 and regulates Pdlim4 expression.
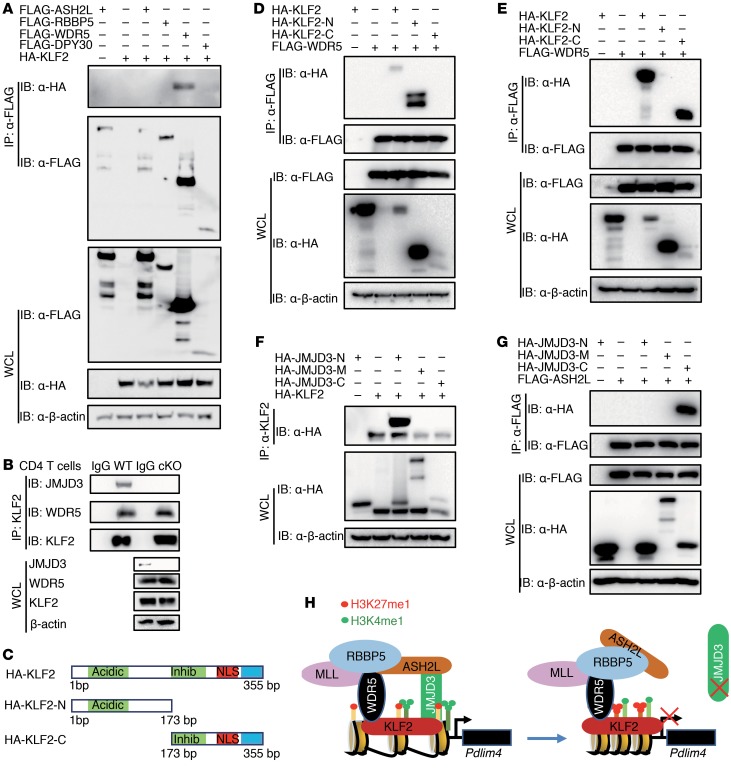
JMJD3 also coregulates H3K4 methylation levels by interacting with the H3K4 methyltransferase complex and transcription factors (28). To determine whether KLF2 also interacts with the H3K4 methyltransferase complex and JMJD3, we performed co-IP and immunoblot analyses on 293T cells cotransfected with HA-Klf2 and FLAG-tagged Ash2l, Rbbp5, Wdr5, and Dpy30. KLF2 specifically interacted with WDR5, but not ASH2L, RBBP5, and DPY30 (Figure 7A). To confirm the endogenous interaction of KLF2 with WDR5, we isolated CD4 SP T cell lysates, co-IP with anti-KLF2 Abs, and immunoblotted with anti-JMJD3, anti-WDR5, and anti-KLF2 Abs, respectively. Our results showed that KLF2 interacted with WDR5 and that JMJD3 was required for stabilizing this interaction (Figure 7B and Supplemental Figure 5C). To dissect the regions responsible for this interaction, we generated HA-tagged N- and C-terminal regions of KLF2 (Figure 7C). 293T cells were cotransfected with HA-Klf2, HA-Klf2-N, or HA-Klf2-C along with either FLAG-Wdr5 or FLAG-Jmjd3. We collected whole cell lysates (WCLs), immunoprecipitated with anti-FLAG beads, and immunoblotted with anti-HA Ab. Our results revealed that the N-terminal region of KLF2 interacts with FLAG-WDR5 and that the C-terminal region of KLF2 interacts with FLAG-JMJD3 (Figure 7, D and E). To address which regions of JMJD3 are involved in the interaction with either KLF2 or ASH2L, we cotransfected 293T cells with HA-KLF2 and HA-tagged N-terminal, C-terminal, or M-middle regions of Jmjd3. WCLs were immunoprecipitated with anti-KLF2 Ab and immunoblotted with anti-HA Ab. We found that the N-terminal region of JMJD3 interacts with KLF2 (Figure 7F). We also performed co-IP and immunoblot analysis using 293T cells, which were cotransfected with FLAG-ASH2L and HA N-terminal, C-terminal, and M-middle regions of JMJD3 and found that the C-terminal region of JMJD3 interacts with ASH2L (Figure 7G). Taken together, our results demonstrate that JMJD3 regulates Pdlim4 expression by interacting with the transcription factor KLF2 and the H3K4 methyltransferase complex protein ASH2L (Figure 7H).
Figure 7. JMJD3 regulates PDLIM4 by facilitating the interaction between KLF2 and WDR5.
(A) 293T cells were cotransfected with HA-Klf2 and FLAG-tagged Ash2L, RbBP5, Wdr5, or Dpy30. WCLs were immunoprecipitated with anti-FLAG beads and immunoblotted with anti-HA Abs. (B) WCLs from WT and Jmjd3-cKO CD4 SP T cells were immunoprecipitated with anti-KLF2 Abs and immunoblotted with anti-JMJD3, WDR5, and KLF2 Abs, respectively. (C) Schematic presentation of cloned HA-tagged N- and C-terminal regions of KLF2. (D) 293T cells were cotransfected with HA–N-terminal, HA–C-terminal, or full-length Klf2 along with FLAG-Wdr5. WCLs were immunoprecipitated with anti-FLAG beads and immunoblotted with anti-HA Abs. (E) 293T cells were cotransfected with HA–N-terminal, HA–C-terminal, or full-length Klf2 along with FLAG-Jmjd3. WCLs were immunoprecipitated with anti-FLAG beads and immunoblotted with anti-HA Abs. (F) 293T cells were cotransfected with HA–N-terminal, HA–C-terminal, or the middle region of Jmjd3 along with HA-Klf2. WCLs were immunoprecipitated with anti-Klf2 Abs and immunoblotted with anti-HA Abs. (G) 293T cells were cotransfected with HA–N-terminal, HA–C-terminal, or the middle region of JMJD3 along with FLAG-Ash2L. WCLs were immunoprecipitated with anti-FLAG Abs and immunoblotted with anti-HA Abs. (H) Schematic diagram of proposed mechanism of how JMJD3 and KLF2 regulate Pdlim4 expression. JMJD3 interacts with the MLL4 protein ASH2L and forms a stable complex with KLF2, which is capable of binding to the Pdlim4 promoter. This permits JMJD3 to alter the H3K27 methylation state of Pdlim4 and permits ASH2L to alter H3K4 methylation to control Pdlim4 expression.
Discussion
During development, early TPCs differentiate into TCR-expressing CD4+CD8+ DP thymocytes in the cortex of the thymus and then mature into SP CD4+ and CD8+ T cells in the medulla (1–3). After negative and positive selection, CD4 SP T cells emigrate from the thymus to the periphery, e.g., peripheral blood, spleen, and LNs. The epigenetic modifier JMJD3 has been shown to play a critical role in macrophage and T cell differentiation, but its role and mechanism in T cell migration are not clear. In this study, we demonstrate that Jmjd3 ablation results in diminished emigration of mature CD4 SP T cells from the thymus and decreased numbers of CD4+ T cells in secondary lymphoid organs. T cell migration from the thymus to secondary lymphoid organs is regulated by S1P1, whereas T cell entry into resting LNs typically requires CD62L and CCR7 (1, 38). It is known that KLF2 regulates expression of S1p1, Cd62l, and Ccr7 (1). S1P1 is highly expressed in mature SP T cells and mediates SP T cell migration along S1P concentration gradients from the thymus to the periphery. Deficiency in KLF2 or S1P1 leads to impaired thymic egress (3, 8, 39). T cell migration phenotypes in T cell–specific Jmjd3-deficient mice are similar to those in KLF2-deficient mice (6), leading to the accumulation of mature SP T cells in the thymus, which is associated with lower numbers of SP T cells in the spleen and LNs as well as downregulation of S1p1, Cd62l, and Ccr7. To further understand how JMJD3 affects T cell migration, we identified a new JMJD3 target gene, Pdlim4, which plays an important role in T cell trafficking. Although Pdlim4 is the most downregulated gene, other genes, including S1p1, Cd62l, and Ccr7, but not Klf2, are also downregulated at the RNA level in Jmjd3-deficient mature SP T cells. Ectopic expression of Pdlim4 in Jmjd3-deficient CD4+ T cells restores their ability to migrate in both the spleen and LNs. Using chimeric mice and adoptive T cell transfer experiments, we demonstrated that ablation of Pdlim4 in CD4+ T cells reduces splenic T cell accumulation. Furthermore, we showed that removal of H3K27me3 by JMJD3 is a critical event in regulating Pdlim4 and S1p1 expression, but not in the promoter regions of Cd62l and Ccr7. Thus, our study identified a role for PDLIM4 in T cell migration.
PDLIM4 has been identified as a modifier of actin filament dynamics in muscle cells and nonmuscle epithelial cells (31, 32), but how PDLIM4 regulates T cell migration remains unknown. Our findings presented in this study indicate that PDLIM4 overexpression could rescue the F-actin assembly and remodeling deficiency in Jmjd3-deficient CD4+ T cells, thus demonstrating the critical role of PDLIM4 for F-actin organization in thymic CD4 SP T cells. Furthermore, S1P/S1P1 signaling is also known to induce cytoskeleton remodeling by increasing polymerization of actin filaments in T cells (40), but how S1P/S1P1 signaling is linked to cytoskeleton remodeling is not clear. To understand the molecular mechanisms of how PDLIM4 regulates T cell migration, we found that PDLIM4 interacts with S1P1 through the N-terminal PDZ domain and anchors to F-actin through the C-terminal LIM domain. S1P1 has been reported as interacting with other proteins, such as P-Rex1, through the PDZ domain (41). However, the triple-serine sequence on the C-terminus of S1P1 does not represent a typical PDZ motif, thus indicating that the interaction between S1P1 and the PDZ domain may require an internal PDZ motif that still locates at the intracellular C terminus of S1P1. Another previous report illustrates that the conserved helix-8 motif in class-A GPCR is critical for PDZ domain recognition (42). We found helix-8 motif mutation abolishes the interaction between S1P1 and PDLIM4, suggesting that S1P1 C-terminal helix structure is critical for PDZ domain recognition. Thus, our mechanistic studies suggest that PDLIM4 acts as an adaptor bridging between S1P1 signaling and F-actin remodeling during T cell migration.
Previous studies have shown that, due to hypermethylation, significant inhibition of Pdlim4 expression in prostate cancer is observed (43, 44), but the epigenetic factors responsible for this hypermethylation at the Pdlim4 promoter remain to be identified. Consistent with these observations, our ChIP-PCR and ChIP-Seq analyses showed a substantial increase in H3K27me3 and a decrease in H3K4me3 in the promoter and gene-body regions of Pdlim4 as well as S1pr1 (but to lesser extent) in Jmjd3-deficient CD4 SP T cells. However, JMJD3 alone did not enhance Pdlim4 expression without KLF2, suggesting coregulation of Pdlim4 by JMJD3 and KLF2. In contrast, we observed a marked reduction in H3K4me3 (with a small increase in H3K27me3) in the promoter and gene-body regions of S1p1. Not only did JMJD3 harbor H3K27 demethylase activity, it also associated with H3K4 methyltransferase complexes and affected H3K4me3 levels at gene promoters. Indeed, we found that JMJD3 directly interacted with ASH2L, a core component of the H3K4 methyltransferase complex (28, 36, 37). We further showed that KLF2 interacted with WDR5, another component of the H3K4 methyltransferase complex. However, JMJD3 was required for stabilizing the interaction between KLF2 and WDR5. Based on these findings, we propose that JMJD3 regulates Pdlim4 and known KLF2-regulated genes (S1p1, Cd62l, and Ccr7) by interacting with KLF2, WDR5, and ASH2L in the H3K4 methyltransferase complex. JMJD3 ablation disrupts the KLF2-JMJD3-ASH2L complex, leading to an increase in H3K27me3 and/or a reduction in H3K4me3 in the promoter and gene body of Pdlim4, thus downregulating target gene expression. The changes in H3K4me3 and/or H3K27me3 in the gene body are supported by findings that JMJD3 is involved in the protein complex engaged in transcriptional elongation (45). Thus, our study provides the molecular insights by which JMJD3 regulates Pdlim4 gene expression through interaction with KLF2, WDR5, and ASH2L as a transcription complex.
In summary, our studies provide genetic evidence that T cell–specific deletion of Jmjd3 results in multiple defects in T cell migration. Gene profiling and ChIP-Seq analysis identified a JMJD3 target gene, Pdlim4. Importantly, PDLIM4 bridges S1P1-mediated extracellular signaling and F-actin formation, which is critical for T cell migration and trafficking through its PDZ domain in the N-terminus and LIM domain in the C-terminus. Mechanistically, JMJD3 regulates Pdlim4 expression through interaction with KLF2, WDR5, and ASH2L in the promoter and gene-body regions of Pdlim4. Thus, our results provide insights into the molecular mechanisms by which JMJD3 and its target PDLIM4 regulate T cell migration.
Methods
Mice.
Jmjd3fl/fl:CD4-Cre mice were generated as previously described (28). C57BL/6 and 2D2 mice were obtained from The Jackson Laboratory. Rag2–/–γc–/– mice were purchased from Taconic. RAG1–BwTRP-1 mice were purchased from the Jackson Laboratory (catalog 008684) and crossed with Jmjd3fl/fl:CD4-Cre mice. Mouse strains are listed in Supplemental Table 3. All mice were rederived using standard embryo transfer and maintained in pathogen-free animal facilities at the Houston Methodist Research Institute.
Preparation of mouse CD4+ T cells from thymus and spleen.
Thymus and spleen were removed from mice, and single-cell suspensions were prepared by mincing and passing through stainless mesh. After removing RBCs by lysis with Tris-NH4Cl solution, the cells were suspended in RPMI 1640 medium with 1% fatty acid–free BSA. The spleen and thymus cells were purified to greater than 90% of CD4+ T cells using a mouse CD4 Enrichment Kit (Thermo Fisher).
IP and Western blot analysis.
For IP, cells were lysed in an ice-cold lysis buffer (40 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 1% Triton X-100) with proteinase inhibitor. Supernatants were incubated overnight with primary Ab (2 μg), and immunocomplexes were allowed to bind to the protein A/G beads for 90 minutes at 4°C. Immunoprecipitates were washed 3 times with the lysis buffer. Western blotting was performed under conventional conditions after extracting the samples in SDS sample buffer. Protein extracts were separated by SDS-PAGE and electrotransferred onto PVDF membrane (Millipore). The membranes were exposed to primary Abs, washed, and incubated with secondary Abs, and the proteins were visualized by using Pierce Western Blotting Substrate Plus (Thermo Fisher). See complete unedited blots in Supplemental Figure 7. All Ab information is listed in Supplemental Table 3.
Flow cytometry.
To detect the expression of surface molecules, T cells were first incubated with an anti-Fc receptor Ab (24G2) to reduce nonspecific binding of mAbs, and then they were labeled with the appropriate fluorescent mAbs. Appropriate fluorescein-conjugated, isotype-matched control mAbs were used as negative controls. Cells were analyzed with BD FACS Aria II.
Real-time RT-PCR.
Total RNA was extracted from cultured cells with TRIzol (Invitrogen) according to the manufacturer’s instructions. Oligo (dT) primers and Superscript III reverse transcriptase (Invitrogen) were used for the generation of cDNA from mRNA. Gene expression was determined by quantitative PCR with SYBR Green MasterMix (ABI), and the reactions were run on an ABI PRISM 7900HT Sequence Detection System (Life Science). Primers used for real-time PCR analysis are shown in Supplemental Table 2. Expression levels are given as the ratio of the target gene to the control gene to correct for variations in the initial amount of mRNA (gene/Gapdh ×1000).
Chemotaxis, phalloidin staining, immunofluorescence, and confocal image analysis.
To analyze actin cytoskeleton remodeling induced by S1P and other stimulation, CD4 SP T cells were starved 12 hours in RPMI 1640 medium with 1% fatty acid–free BSA at 37°C. Starved T cells were mixed with an equal volume of Matrigel and coated on a confocal dish. After 30 minutes at 37°C, the diluted S1P was added to outside Matrigel for 3 hours at 37°C in 5% CO2. Cells were washed and fixed with 4% formaldehyde in PBS at room temperature for 20 minutes. Whenever required, cells were first labeled with Abs against extracellular markers, such as CD4, and washed, blocked, and permeabilized in PBS containing 3% FBS and 0.5% Triton X-100 for phalloidin staining, which is specific staining for F-actin. CD4+ T cells in Matrigel- or poly-lysine–coated dish were processed for immunofluorescence, as described (33). Slides were imaged using the Nikon A1Si Confocal Imaging System. Image analysis was performed using ImageJ software (NIH).
Bone marrow chimeras.
Bone marrow cells were isolated from female C57BL6/J mice (8 to 10 weeks of age). The cells were transfected with either retrovirus vector expressing Pdlim4 or lentivirus vector containing Pdlim4 CRISPR/Cas9 genome-editing elements. These bone marrow cells were injected into lethally irradiated (9.5 Gy) recipient mice (female C57BL6/J mice at 8 to 10 weeks old).
Retrovirus production and transduction.
Jmjd3 and Pdlim4 genes were cloned into retrovirus expression vector with a GFP-expressing cassette (pCLIG-IRES-GFP). Retroviral particles were produced in 293T cells transfected with retroviral vectors plus pCL-ECO retroviral packaging plasmid using Lipofectamine 2000 (Invitrogen). Sorted naive CD4+ T cells were plated and activated with anti-CD3 and anti-CD28 for 1 day, transduced with retroviral supernatants, and spun at 1000 g for 1.5 hours at 32°C. After spin infection, cells were cultured in T cell culture medium and harvested on day 5 for subsequent T cell transfer experiments.
Pdlim4 KO by CRISPR-Cas9 and T cell migration analysis.
Naive thymic CD4 SP T cells collected from Cas9 knockin mice (The Jackson Laboratory, 024858) were transduced with lentivirus containing Pdlim4 sgRNA or control sgRNA plasmids (LentiGuide-puro; Addgene, 52963). An equal number of cells (10 million) were labeled by CFSE and intravenously injected into sublethally irradiated C57BL/6 mice, and 48 hours later, peripheral blood was collected from the recipient mice. Single-cell suspensions were prepared from the isolated spleens and LNs of the recipient mice. The CFSE-labeled CD4 SP T cells isolated from the spleens, LNs, and peripheral blood were analyzed using FACS assay.
F-actin binding assay.
F-actin cosedimentation assay was performed as described previously (44) using the Actin Binding Protein Spin Down Assay Kit (Cytoskeleton Inc., BK001) and recombinant GST-PDLIM4 fusion protein. GST-PDLIM4 was mixed with F-actin in F-actin buffer at room temperature for 30 minutes and centrifuged at 150,000 g for 1.5 hours at 24°C. The supernatants were then removed, and the pellets were suspended in the same buffer. Aliquots of pellets and supernatants were mixed with the loading dye and run on an SDS-PAGE gel, followed by Western blot analysis.
Microarray, ChIP-Seq, and Chip-PCR analyses.
RNA was extracted from 3 sorted biological replicates of CD4 SP thymocytes from WT and Jmjd3-cKO mice. Gene expression profiling was conducted at the Genomics and Microarray Core Facility of the UT Southwestern Medical Center (Dallas, Texas, USA) with Illumina whole-genome gene expression arrays. Gene transcripts with greater than 1.5-fold difference in expression were analyzed with Ingenuity pathway analysis software. CD4+ T cells from WT and Jmjd3-cKO mice were purified, and a total of 200 ng of DNA was used for the ChIP-Seq library construction. Illumina sequencing was performed according to previously described protocols (46–48). Sequencing reads from H3K4me3 and H3K27me3 ChIP-Seq libraries were aligned to the mouse mm8 genome using ELAND software (omicX). In order to reduce PCR amplification bias, only one uniquely mapped read per genomic position was retained. The histone modification changes upon JMJD3 KO were assessed as follows. The H3K27me3 sequencing reads in WT and cKO samples were counted in 4 kb windows centered at TSS or p300 ChIP-Seq peaks. The resulting read counts were used as input to the DEGseq algorithm56 to identify differentially methylated genes and enhancers. The DEGseq q value cutoff of 0.05 and the fold-change cutoff of 2 were used to identify statistically significant methylation changes. The raw and analyzed ChIP-Seq were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE58775). ChIP-PCR experiments were performed using the Imprint Chromatin Immunoprecipitation Kit (MilliporeSigma) according to the manufacturer’s instructions. Samples in triplicate were used for all experiments. All primers are listed in Supplemental Table 2.
Statistics.
Data were analyzed with GraphPad Prism 4.0 software. Results represent the mean ± SD where applicable. Statistical analyses between 2 groups were assessed using 2-tailed Student’s t test. Statistical analyses among 3 or more groups were assessed by 1-way ANOVA with Tukey’s multiple comparisons test. For all tests, a P value of less than 0.05 was considered statistically significant.
Study approval.
These studies were reviewed and approved by the Institutional Animal Care and Use Committee at the Houston Methodist Research Institute.
Author contributions
CF, QL, and JZ contributed equally to this work. Authorship order among co–first authors is alphabetical by surname. CF, QL, JZ, and RFW conceived and designed the experiments. CF, QL, JZ, ML, CX, BY, JC, JY, and XL performed the experiments and developed the reagents. CF, QL, JZ, HYW, and RFW analyzed and interpreted the data. CF, QL, JZ, and RFW wrote the paper.
Supplementary Material
Acknowledgments
We would like to thank Olga A. Guryanova (Lerner Research Institute, Cleveland Clinic, Cleveland, Ohio, USA) for the LCMV-Pdlim4-eGFP plasmids and Jana S. Burchfield, Joshua C. Korb, and Kalpana Mujoo for critical reading and editing of the manuscript. This work was supported by grants from the National Cancer Institute and the National Institute on Drug Abuse, NIH (R01CA101795 and U54CA210181), the Cancer Prevention and Research Institute of Texas (CPRIT) (DP150099, RP150611, and RP170537), and the Department of Defense (DoD) Congressionally Directed Medical Research Programs Breast Cancer Research Program (CDMRP BCRP) (BC151081 to RFW).
Version 1. 08/08/2019
In-Press Preview
Version 2. 09/30/2019
Electronic publication
Version 3. 11/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(11):4745–4757.https://doi.org/10.1172/JCI128293.
Contributor Information
Chuntang Fu, Email: cfu2@houstonmethodist.org.
Qingtian Li, Email: Qingtian.Li@bcm.edu.
Jia Zou, Email: Jia.Zou.GR@dartmouth.edu.
Changsheng Xing, Email: cxing@houstonmethodist.org.
Mei Luo, Email: mluo@houstonmethodist.org.
Bingnan Yin, Email: byin@houstonmethodist.org.
Junjun Chu, Email: jchu@houstonmethodist.org.
Jiaming Yu, Email: jiamingyuxxm@hotmail.com.
Xin Liu, Email: xliu3@houstonmethodist.org.
Helen Y. Wang, Email: ywang4@houstonmethodist.org.
Rong-Fu Wang, Email: rwang3@houstonmethodist.org.
References
- 1.Love PE, Bhandoola A. Signal integration and crosstalk during thymocyte migration and emigration. Nat Rev Immunol. 2011;11(7):469–477. doi: 10.1038/nri2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrie HT. Cell migration and the control of post-natal T-cell lymphopoiesis in the thymus. Nat Rev Immunol. 2003;3(11):859–866. doi: 10.1038/nri1223. [DOI] [PubMed] [Google Scholar]
- 3.Spiegel S, Milstien S. The outs and the ins of sphingosine-1-phosphate in immunity. Nat Rev Immunol. 2011;11(6):403–415. doi: 10.1038/nri2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee SH, Dominguez R. Regulation of actin cytoskeleton dynamics in cells. Mol Cells. 2010;29(4):311–325. doi: 10.1007/s10059-010-0053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samstag Y, Eibert SM, Klemke M, Wabnitz GH. Actin cytoskeletal dynamics in T lymphocyte activation and migration. J Leukoc Biol. 2003;73(1):30–48. doi: 10.1189/jlb.0602272. [DOI] [PubMed] [Google Scholar]
- 6.Carlson CM, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442(7100):299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 7.Lee K, et al. Molecular mechanism of Jmjd3-mediated interleukin-6 gene regulation in endothelial cells underlying spinal cord injury. J Neurochem. 2012;122(2):272–282. doi: 10.1111/j.1471-4159.2012.07786.x. [DOI] [PubMed] [Google Scholar]
- 8.Matloubian M, et al. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427(6972):355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 9.Sebzda E, Zou Z, Lee JS, Wang T, Kahn ML. Transcription factor KLF2 regulates the migration of naive T cells by restricting chemokine receptor expression patterns. Nat Immunol. 2008;9(3):292–300. doi: 10.1038/ni1565. [DOI] [PubMed] [Google Scholar]
- 10.Weinreich MA, Takada K, Skon C, Reiner SL, Jameson SC, Hogquist KA. KLF2 transcription-factor deficiency in T cells results in unrestrained cytokine production and upregulation of bystander chemokine receptors. Immunity. 2009;31(1):122–130. doi: 10.1016/j.immuni.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamada T, Park CS, Mamonkin M, Lacorazza HD. Transcription factor ELF4 controls the proliferation and homing of CD8+ T cells via the Krüppel-like factors KLF4 and KLF2. Nat Immunol. 2009;10(6):618–626. doi: 10.1038/ni.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Woude LL, Gorris MAJ, Halilovic A, Figdor CG, de Vries IJM. Migrating into the tumor: a roadmap for T cells. Trends Cancer. 2017;3(11):797–808. doi: 10.1016/j.trecan.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Trujillo JA, Sweis RF, Bao R, Luke JJ. T cell-inflamed versus non-T cell-inflamed tumors: a conceptual framework for cancer immunotherapy drug development and combination therapy selection. Cancer Immunol Res. 2018;6(9):990–1000. doi: 10.1158/2326-6066.CIR-18-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garris CS, et al. Defective sphingosine 1-phosphate receptor 1 (S1P1) phosphorylation exacerbates TH17-mediated autoimmune neuroinflammation. Nat Immunol. 2013;14(11):1166–1172. doi: 10.1038/ni.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10(7):457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22(9):1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8(4):307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 18.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–179. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 20.Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48(4):491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 22.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 23.Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104(47):18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jepsen K, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450(7168):415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- 25.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 26.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449(7163):689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 27.Jin C, et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 2011;14(2):161–172. doi: 10.1016/j.cmet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Li Q, et al. Critical role of histone demethylase Jmjd3 in the regulation of CD4+ T-cell differentiation. Nat Commun. 2014;5:5780. doi: 10.1038/ncomms6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satoh T, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11(10):936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 30.Manna S, et al. Histone H3 Lysine 27 demethylases Jmjd3 and Utx are required for T-cell differentiation. Nat Commun. 2015;6:8152. doi: 10.1038/ncomms9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boumber YA, et al. RIL, a LIM gene on 5q31, is silenced by methylation in cancer and sensitizes cancer cells to apoptosis. Cancer Res. 2007;67(5):1997–2005. doi: 10.1158/0008-5472.CAN-06-3093. [DOI] [PubMed] [Google Scholar]
- 32.Guryanova OA, Drazba JA, Frolova EI, Chumakov PM. Actin cytoskeleton remodeling by the alternatively spliced isoform of PDLIM4/RIL protein. J Biol Chem. 2011;286(30):26849–26859. doi: 10.1074/jbc.M111.241554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vallenius T, Scharm B, Vesikansa A, Luukko K, Schäfer R, Mäkelä TP. The PDZ-LIM protein RIL modulates actin stress fiber turnover and enhances the association of alpha-actinin with F-actin. Exp Cell Res. 2004;293(1):117–128. doi: 10.1016/j.yexcr.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Lee HJ, Zheng JJ. PDZ domains and their binding partners: structure, specificity, and modification. Cell Commun Signal. 2010;8:8. doi: 10.1186/1478-811X-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zachariah MA, Cyster JG. Thymic egress: S1P of 1000. F1000 Biol Rep. 2009;1:60. doi: 10.3410/B1-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22(21):2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2010;40(4):594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masopust D, Soerens AG. Tissue-resident T cells and other resident leukocytes. Annu Rev Immunol. 2019;37:521–546. doi: 10.1146/annurev-immunol-042617-053214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cyster JG, Schwab SR. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu Rev Immunol. 2012;30:69–94. doi: 10.1146/annurev-immunol-020711-075011. [DOI] [PubMed] [Google Scholar]
- 40.Mudd JC, et al. Impaired T-cell responses to sphingosine-1-phosphate in HIV-1 infected lymph nodes. Blood. 2013;121(15):2914–2922. doi: 10.1182/blood-2012-07-445783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Paik JH, Paik JH, Wang Z, Hla T, Wu D. Role of guanine nucleotide exchange factor P-Rex-2b in sphingosine 1-phosphate-induced Rac1 activation and cell migration in endothelial cells. Prostaglandins Other Lipid Mediat. 2005;76(1-4):95–104. doi: 10.1016/j.prostaglandins.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 42.Sensoy O, Weinstein H. A mechanistic role of Helix 8 in GPCRs: Computational modeling of the dopamine D2 receptor interaction with the GIPC1-PDZ-domain. Biochim Biophys Acta. 2015;1848(4):976–983. doi: 10.1016/j.bbamem.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vanaja DK, et al. PDLIM4 repression by hypermethylation as a potential biomarker for prostate cancer. Clin Cancer Res. 2006;12(4):1128–1136. doi: 10.1158/1078-0432.CCR-05-2072. [DOI] [PubMed] [Google Scholar]
- 44.Vanaja DK, et al. PDLIM4, an actin binding protein, suppresses prostate cancer cell growth. Cancer Invest. 2009;27(3):264–272. doi: 10.1080/07357900802406319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen S, et al. The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev. 2012;26(12):1364–1375. doi: 10.1101/gad.186056.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 47.Qi HH, et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466(7305):503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.