Abstract
Antisense oligonucleotides (ASOs) targeting pathologic RNAs have shown promising therapeutic corrections for many genetic diseases including myotonic dystrophy (DM1). Thus, ASO strategies for DM1 can abolish the toxic RNA gain-of-function mechanism caused by nucleus-retained mutant DMPK (DM1 protein kinase) transcripts containing CUG expansions (CUGexps). However, systemic use of ASOs for this muscular disease remains challenging due to poor drug distribution to skeletal muscle. To overcome this limitation, we test an arginine-rich Pip6a cell-penetrating peptide and show that Pip6a-conjugated morpholino phosphorodiamidate oligomer (PMO) dramatically enhanced ASO delivery into striated muscles of DM1 mice following systemic administration in comparison with unconjugated PMO and other ASO strategies. Thus, low-dose treatment with Pip6a-PMO-CAG targeting pathologic expansions is sufficient to reverse both splicing defects and myotonia in DM1 mice and normalizes the overall disease transcriptome. Moreover, treated DM1 patient–derived muscle cells showed that Pip6a-PMO-CAG specifically targets mutant CUGexp-DMPK transcripts to abrogate the detrimental sequestration of MBNL1 splicing factor by nuclear RNA foci and consequently MBNL1 functional loss, responsible for splicing defects and muscle dysfunction. Our results demonstrate that Pip6a-PMO-CAG induces long-lasting correction with high efficacy of DM1-associated phenotypes at both molecular and functional levels, and strongly support the use of advanced peptide conjugates for systemic corrective therapy in DM1.
Keywords: Therapeutics
Keywords: Genetic diseases, Muscle
Introduction
Myotonic dystrophy (DM1), one of the most common muscular dystrophies in adults (1), is an RNA-dominant disorder caused by the expression of expanded microsatellite repeats located in the 3′ untranslated region (UTR) of the DM1 protein kinase (DMPK) gene (2–4). Mutant transcripts containing an expanded CUG tract are retained within the nucleus as discrete foci (5, 6). Due to their high affinity for CUG expansions (CUGexps), RNA binding factors from the MBNL family are sequestered within the CUGexp-RNA foci, leading to their functional loss (7), which is a central pathophysiologic mechanism in DM1 (8). Thus, altered splicing regulatory activity of MBNL1 in striated muscles results in many alternative splicing misregulations, including those in CLCN1, INSR, BIN1, DMD, and SCN5A pre-mRNAs that have been associated respectively with myotonia, insulin resistance, muscle weakness, dystrophic process, and cardiac conduction defects, all symptoms of DM1 (9–14).
Given that the toxic RNA gain-of-function mechanism in DM1 is driven by a pathologic CUGexp tract within DMPK mRNAs, therapeutic approaches using modified antisense oligonucleotides (ASOs) aimed at either degrading CUGexp transcripts using RNase H–active ASOs (15–17) or releasing sequestered MBNL1 from CUGexp-containing RNAs using RNase H–inactive CAGn ASOs (1–20) have demonstrated efficient reversal of DM1-associated phenotypes. However, unlike other muscle diseases like Duchenne muscular dystrophy (DMD), the integrity of the skeletal muscle fiber membrane is not compromised in DM1, resulting in poor penetration of naked phosphorodiamidate oligomer–ASOs (PMO-ASOs) or 2′-O-methyl–ASOs (21). Backbone modifications led to the design of more potent chemistries, including 2′-O-methoxyethyl or 2′-4′–constrained ethyl–modified residues that enhanced ASO delivery and consequently therapeutic efficacy in skeletal muscles of DM1 mice (15, 17). This approach has been tested in a human clinical trial (NCT02312011); however, systemic administration of the selected naked ASO did not achieve sufficient concentrations in skeletal muscle of patients to have marked clinical benefits. Therefore, tissue uptake remains a major challenge for ASO therapies in DM1.
Among strategies to improve delivery of ASOs in DM1 skeletal muscles, cell-penetrating peptide conjugates offer an attractive avenue (22–24). Indeed, systemic administration of advanced Pip6a peptides conjugated with splice-switch oligonucleotides has shown promising results in DMD mice by restoring a truncated but functional dystrophin (25). Pip6a is a cell-penetrating peptide composed of a hydrophobic core region flanked on each side by arginine-rich domains containing β-alanine and aminohexanoyl spacers. This peptide sequence has the ability to deliver associated cargoes across the plasma and endosomal membranes and is stable to serum proteolysis (25). Here, we have investigated the potential of Pip6a conjugates to enhance RNase H–inactive CAGn-PMO delivery in skeletal muscles of DM1 mice in order to target nucleus-retained mutant-CUGexp transcripts and inhibit toxicity associated with expanded repeats.
Results and Discussion
To test whether advanced Pip6a conjugates enhanced ASO delivery and activity in skeletal muscles of DM1 mice following systemic administration, antisense PMOs composed of a CAG7 sequence, either naked (PMO) or conjugated to Pip6a peptide (Pip6a-PMO), were injected into the tail vein of HSA-LR mice. This well-characterized DM1 mouse model expresses 250 CTG repeats in the 3′ UTR of the human skeletal muscle α actin (HSA) gene and displays molecular as well as functional DM1-associated abnormalities (26). The skeletal muscle–specific expression of CUGexp RNAs in HSA-LR mice leads to the formation of ribonuclear foci that sequester MBNL1 proteins, resulting in splicing defects and myotonia, as found in DM1 patients (11, 27).
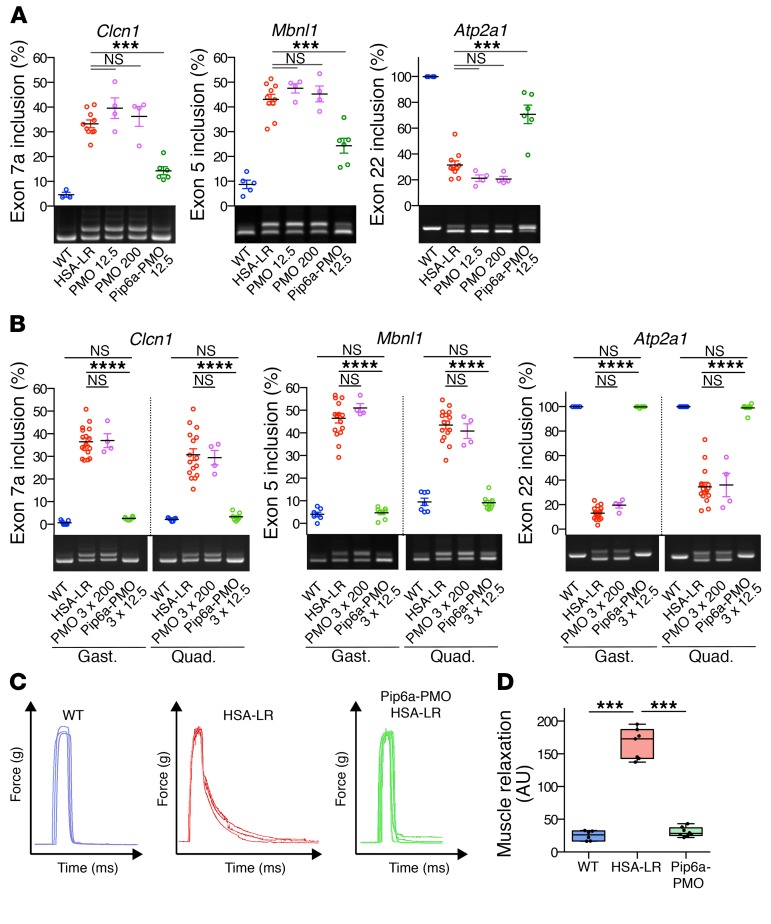
Two weeks after a single injection of a dose of 12.5 mg/kg, mice were sacrificed and the correction of DM1-splicing defects was used as a biomarker of ASO activity. Pip6a-PMO induced a partial but significant correction of several MBNL1-dependent splicing defects (e.g., Clcn1 exon 7a, Mbnl1 exon 5, and Atp2a1 exon 22) in HSA-LR gastrocnemius muscle, whereas no effect was observed with naked PMO at the same dose or even at a much higher dose of 200 mg/kg (Figure 1A). Moreover, 2 and 3 systemic injections of Pip6a-PMO (12.5 mg/kg) led to an almost complete normalization and a full correction, respectively, of DM1-splicing profiles of Clcn1, Mbnl1, and Atp2a1 genes in both HSA-LR gastrocnemius and quadriceps muscles (Supplemental Figure 1 and Figure 1B). In contrast, even 3 injections of naked PMO at a high dose of 200 mg/kg did not improve splicing defects. PMO concentration was also quantified in skeletal muscle of HSA-LR mice by a custom ELISA. Results showed PMO concentrations of greater than 1 nM 2 weeks after Pip6a-PMO treatment, whereas mice treated with naked PMO showed low picomolar concentrations, confirming the poor uptake of naked PMO in non–renal tissue (Supplemental Figure 2). Interestingly, systemic administration of Pip6a-PMO in DM1 mice also leads to therapeutic levels of PMO-CAG in heart and diaphragm tissues that are affected in DM1 disease. Despite the fact that restriction of CUGexp expression to the hACTA1 gene in HSA-LR mice does not allow the assessment of ASO therapy in cardiac tissue, our results strongly suggest that the efficient biodistribution of Pip6a-conjugated PMO into striated muscles could counteract other DM1-related symptoms, including cardiac-conduction defects and respiratory failure, which are the most common causes of death in DM1 patients.
Figure 1. Pip6a-PMO corrects molecular and functional defects in HSA-LR mice.
HSA-LR mice were injected with single or multiple doses of naked PMO-CAG7 (PMO) or Pip6a-PMO-CAG7 (Pip6a-PMO) by intravenous injection and analyzed 2 weeks after treatment. (A) Quantification of splicing correction induced by a single 12.5 mg/kg dose of Pip6a-PMO, or 12.5 or 200 mg/kg dose of PMO in gastrocnemius of treated mice (WT n = 4; HSA-LR n = 10; PMO n = 4; Pip6a-PMO n = 6). (B) Representative RT-PCR results and quantification of alternative splicing profiles in gastrocnemius (gast.) and quadriceps (quad.) of HSA-LR mice treated with 3 injections of PMO at 200 mg/kg or Pip6a-PMO at 12.5 mg/kg (WT n = 7; HSA-LR n = 16; PMO n = 4; Pip6a-PMO n = 8). (C) Representative maximal force/time curves obtained by in situ force measurements of Pip6a-PMO–treated mice (3 times 12.5 mg/kg) compared with WT and HSA-LR mice. (D) Correction of myotonia (measured as the area under the force/time curve during relaxation after maximal muscle contraction) in Pip6a-PMO–injected HSA-LR mice (WT n = 6; HSA-LR n = 7; Pip6a-PMO n = 8). Data are expressed as mean ± SEM, except for D (boxes, 25th–75th percentile; whiskers, min to max; black line, the mean). ***P < 0.001; ****P < 0.0001 by 1-way ANOVA with Newman-Keuls post hoc test. NS, not significant.
To determine whether Pip6a-PMO treatment can improve muscle function, we evaluated myotonia, a DM1 hallmark characterized by delayed muscle relaxation also detectable in HSA-LR mice. As shown in Figure 1, C and D, Pip6a-PMO treatment completely abolished myotonia in gastrocnemius muscles of HSA-LR mice, which is in agreement with the correction of Clcn1 exon 7a missplicing responsible for myotonia (28). In contrast, systemic injections of a control Pip6a-PMO composed of a reverse GAC7 sequence (Pip6a-Ctrl) had no effect on either DM1-splicing defects or myotonia in HSA-LR mice (Supplemental Figure 3, A and B). Altogether, these results showed that a low-dose treatment of Pip6a conjugates allows efficient systemic delivery of ASO in skeletal muscle of DM1 mice. Remarkably, optimal beneficial effects at both molecular and functional levels were achieved with a cumulative dose of Pip6a-ASO that is 5 to 10 times lower than either unconjugated or conjugated ASOs previously described and evaluated in the same DM1 mouse model (15, 17, 22, 29).
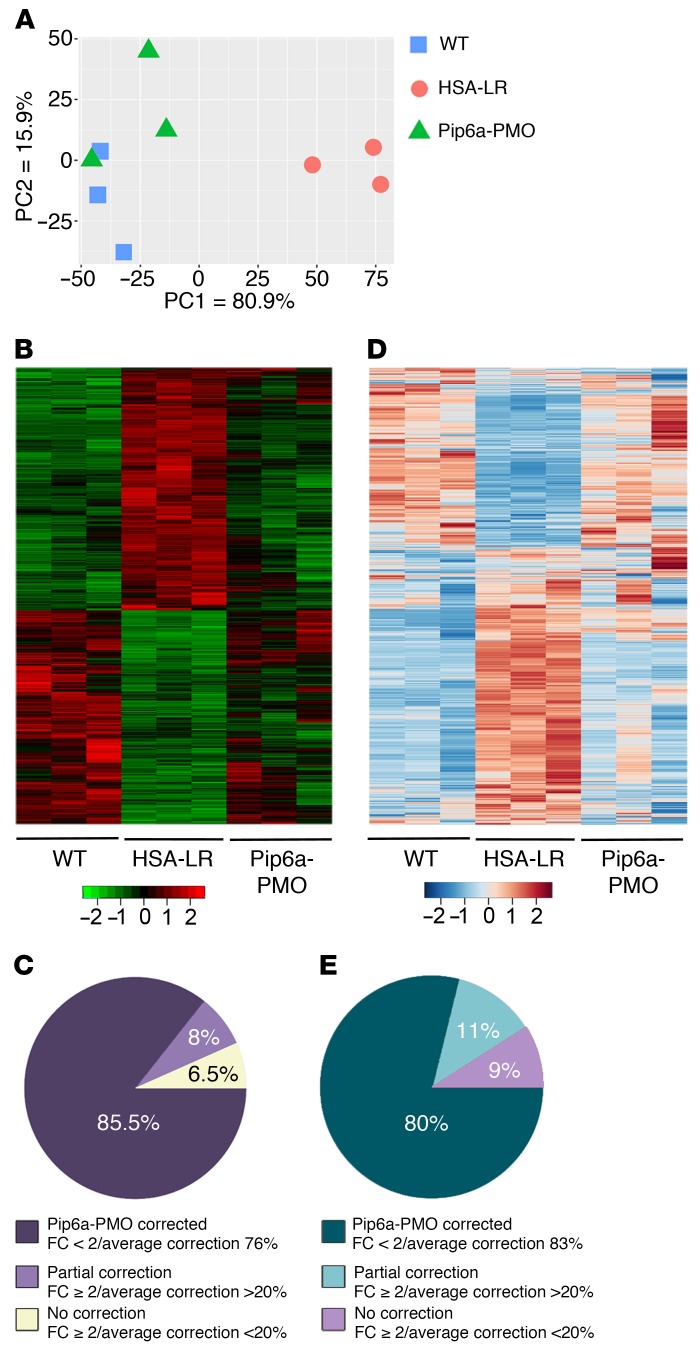
To examine the overall effect of Pip6a-PMO on the skeletal muscle transcriptome of DM1 mice, we performed deep, paired-end RNA sequencing. Principal component analysis (PCA) showed that gene expression in the gastrocnemius muscle of HSA-LR mice treated with Pip6a-PMO was shifted toward that of WT mice (Figure 2A). Moreover, the heatmap generated with all the transcripts having a significant change (n = 3176; adj. P < 0.1; no threshold in fold change [FC]) confirmed the global gene expression correction in treated HSA-LR mice (Figure 2B). Strikingly, the vast majority (85%) of the most significant changes observed at the gene expression level in HSA-LR mice (FC ≥ 2 and adj. P < 0.1; 376 transcripts) were corrected in treated mice with an average correction index of 76% (Figure 2C and Supplemental Table 1). In addition, most of the deregulated genes containing 7 or more CTG repeats in skeletal muscles were also corrected in treated HSA-LR mice (Supplemental Table 2). Transcriptomic analysis also confirmed that Pip6a-PMO treatment induced a global correction of alternative splicing misregulation measured in HSA-LR mice (adj. P < 0.1; no threshold in FC; Figure 2D). Thus, 80% of the most deregulated splicing events in HSA-LR mice (FC ≥ 2 and adj. P < 0.1; 339 events) were normalized in treated mice an average correction index of 83% (Figure 2E), also leading to correction of the deregulated biological pathways (Supplemental Figure 4 and Supplemental Table 3). Similar results were obtained from the quadriceps muscle of the same treated mice (Supplemental Figure 5), indicating that the muscle transcriptome of DM1 mice could be almost normalized by Pip6a-PMO treatment targeting pathologic CUGexps. In addition, PCA and heatmap analysis (FC ≥ 1.5; P < 0.05; n = 118) of the proteome assessed by label-free mass spectrometry revealed that protein expression in the quadriceps muscle of treated HSA-LR mice also tends to shift from a disease toward a WT profile, supporting the correction of the DM1 muscle phenotype by Pip6a-PMO treatment (Supplemental Figure 6 and Supplemental Table 4).
Figure 2. Treatment with Pip6a-PMO normalizes global transcriptome at both expression and splicing levels.
Transcriptomic analysis by RNA sequencing was performed on total RNA isolated from gastrocnemius muscles of treated HSA-LR mice compared with HSA-LR and WT mice (n = 3). (A and B) Principal component analysis (A) and heatmap graphic (B) of all significantly expressed transcripts (adj. P < 0.1) reveal a global correction of the gene expression profile with Pip6a-PMO treatment. (C) Global correction of gene expression by Pip6a-PMO treatment (n = 376; FC ≥ 2, adj. P < 0.1): 85.5% of transcripts return to FC < 2 with an average correction index of 76%; 8% of transcripts remain at FC ≥ 2 but with correction index > 20%; 6.5% of transcripts are not corrected. (D) Heatmap graphic of all significant deregulated exon_bin (normalized counts) reveals a global correction of missplicing events with Pip6a-PMO treatment. (E) Overall correction of alternative splicing profiles by Pip6a-PMO treatment (n = 339 splicing events; FC ≥ 2, adj. P < 0.1): 80% of events return to FC < 2 with an average correction index of 83%; 11% remain at FC ≥ 2 but with correction index > 20%; 8% are not corrected.
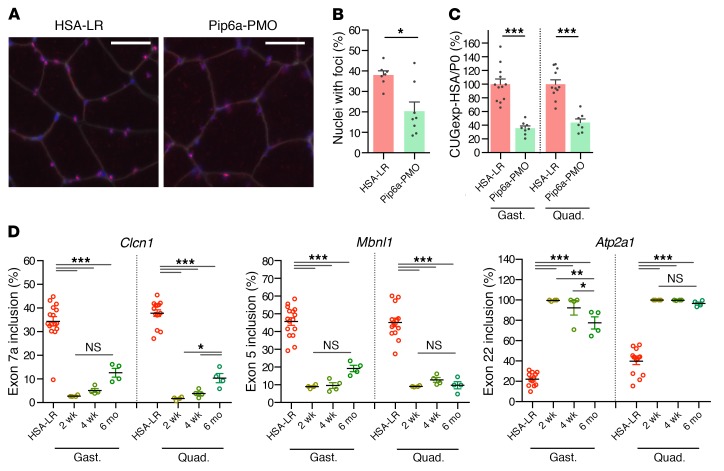
To further determine the consequences of Pip6a-PMO for CUGexp-RNA behavior, we examined nuclear RNA foci number and CUGexp-RNA levels since it has been shown previously that both features were reduced by RNase H–inactive CAGn ASOs (18, 19). Likewise, a similar mechanism of action, which is not yet fully explained, was also observed in Pip6a-PMO–treated HSA-LR mice, as (i) missplicing reversal due to the release of functional MBNL1 from CUGexp-RNA foci was associated with a 50% reduction in the number of myonuclei containing RNA foci in treated muscles (Figure 3, A and B, and Supplemental Figure 7); and (ii) a 60% decrease of CUGexp-RNA steady-state levels was detected following Pip6a-PMO treatment, whereas Pip6a-Ctrl treatment had no effect on CUGexp-RNA expression (Figure 3C and Supplemental Figure 3C).
Figure 3. Treatment with Pip6a-PMO normalizes DM1-specific phenotype.
HSA-LR mice were injected in the tail vein with three 12.5 mg/kg doses of Pip6a-PMO and analyzed 2 weeks after treatment. (A) Representative images of gastrocnemius muscle section stained for CUGexp foci (FISH, red), fiber membranes (WGA, gray), and nuclei (Hoechst, blue). Scale bars: 50 μm. (B) Quantification of the number of nuclei with foci in gastrocnemius muscle sections (HSA-LR n = 7; Pip6a-PMO n = 8). (C) Quantification of HSA transcripts levels normalized to murine Rlp0 (P0) by qRT-PCR (HSA-LR n = 12; Pip6a-PMO n = 8). (D) Quantification of splicing correction in gastrocnemius (gast.) and quadriceps (quad.) at 2 weeks, 4 weeks, and 6 months after treatment (HSA-LR n = 16; n = 4 for each time point of Pip6a-PMO). Data are expressed as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001 by Mann-Whitney test (B) or 1-way ANOVA with Newman-Keuls post hoc test (C and D). NS, not significant.
In addition, the duration of Pip6a-PMO action in HSA-LR skeletal muscles was evaluated because the activity and beneficial effect of nuclease-resistant ASOs could be maintained for several weeks in vivo (15, 30–32). Correction of splicing changes was used as a molecular biomarker and the Pip6a-PMO effect was assessed up to 6 months after treatment. As observed in Figure 3D, splicing misregulation of Clcn1 exon 7a, Mbnl1 exon 5, and Atp2a1 exon 22 pre-mRNAs was completely normalized in both HSA-LR gastrocnemius and quadriceps muscles until 4 weeks after systemic administration. Moreover, a significant 80% to 100% of correction of these splicing defects was still measured 26 weeks after treatment, supporting a long-lasting activity of Pip6a-PMO, which remained nearly complete for a 6-month period. Furthermore, mild histological changes observed in liver or kidney 2 weeks after Pip6a-PMO injections were mostly reversed 6 months later (Supplemental Table 5), supporting transient and reversible side effects of Pip6a-PMO treatment.
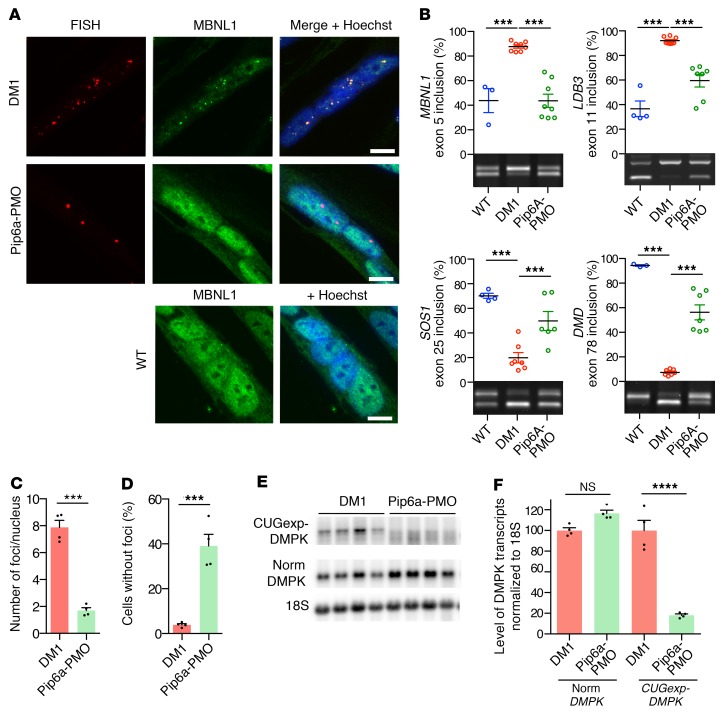
Lastly, molecular effects of Pip6a-PMO were assessed in human DM1 muscle cells carrying a large expansion (2600 repeats) and expressing within its natural context both mutant CUGexp-DMPK and nonmutated normal DMPK transcripts (33). Pip6a-PMO treatment induced the displacement and relocation of MBNL1 from RNA foci toward the nucleoplasm as observed in WT muscle cells (Figure 4A). Moreover, MBNL1-dependent splicing defects of LDB3, MBNL1, SOS1, and DMD pre-mRNAs that are present in DM1 differentiated muscle cells were significantly corrected by Pip6a-PMO, confirming the functional restoration of MBNL1 (Figure 4B). Similar results were obtained in another DM1 cell line carrying 1300 CTG and as described previously (33) (Supplemental Figure 8). Importantly, no splicing changes were observed in DM1 muscle cells treated with Pip6a-Ctrl or naked PMO nor in WT muscle cells treated with Pip6a-PMO (Supplemental Figure 9). As a consequence of MBNL1 displacement from CUGexp-RNA foci, Pip6a-PMO treatment induced a 79% decrease in the number of foci per nucleus and was associated with a 40% increase in the number of nuclei without foci (Figure 4, C and D), which was consistent with a 77% decrease of CUGexp-DMPK transcript levels in treated DM1 cells (Figure 4, E and F). A direct measurement of CUGexp-DMPK transcripts using single-molecule RNA FISH also confirmed the reduced level of CUGexp-DMPK mRNA by Pip6a-PMO treatment in muscle cells expressing the human DMPK gene with 1500 CTG (Supplemental Figure 10). Remarkably, products of nonmutated DMPK alleles were not affected in DM1-treated cells, supporting a specific targeting of Pip6a-PMO composed of a CAG7 sequence to mutant DMPK transcripts containing a CUGexp tract (Figure 4, E and F).
Figure 4. Pip6a-PMO corrects DM1-specific molecular symptoms in DM1 muscle cells.
Four-day-differentiated immortalized DM1 myoblasts (2600 CTG repeats) were treated with 1 μM Pip6a-PMO and analyzed after 24 hours. (A) Combined FISH (Cy3-CAG, red) and immunofluorescence (MBNL1, green) on DM1 or WT differentiated myoblasts. Scale bars: 10 μm. (B) Quantification of splicing corrections by RT-PCR (WT n = 4; DM1 and Pip6a-PMO n = 7). (C) Quantification of mean number of foci per nucleus in treated DM1 differentiated myoblasts (n = 4; >500 nuclei per n). (D) Quantification of the number of nuclei without foci in treated DM1 differentiated myoblasts (n = 4; >500 nuclei per n). (E and F) Levels of mutant DMPK and normal DMPK transcripts analyzed by Northern blot using a DMPK probe (n = 4). Data are expressed as mean ± SEM. ***P < 0.001; ****P < 0.0001 by 1-way ANOVA with Newman-Keuls post hoc test (B and F) or Mann-Whitney test (C and D). NS, not significant.
In conclusion, our study demonstrates that Pip6a cell-penetrating peptide improves the penetration of ASOs in striated muscles of DM1 mice after systemic delivery. Thus, low-dose treatment with a Pip6a-conjugated PMO directed against pathogenic CUGexp repeats is sufficient to achieve an effective concentration of ASOs in muscle fibers and induces an efficient and long-lasting correction of molecular and functional phenotypes in DM1 mice. Altogether, these results strongly support the development of ASO-conjugate peptides for further clinical trial in DM1 as well as other microsatellite expansion disorders.
Methods
See the supplemental methods for a full description of all experimental procedures as well as complete unedited blots.
Statistics.
All group data are expressed as mean ± SEM. Comparisons were performed by Mann-Whitney test or 1-way ANOVA followed by Newman-Keuls or Tukey’s post hoc test using Prism 6 software (GraphPad Software, Inc.). Differences between groups were considered significant when P ≤ 0.05.
Study approval.
Experiments on HSA-LR mice were carried out according to French legislation and Ethics committee approval (number 1760-2015091512001083v6).
Data availability.
Complete raw data generated from RNA sequencing were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE134926).
Author contributions
Experiments were performed by AFK, MAV, LA, AH, DS, AF, LR, and DJ. AA produced the Pip6a-PMO. Transcriptomic analysis was performed by NN. Proteomics data were generated and analyzed by AH. Data were analyzed by AFK, MAV, GB, GG, JP, MJG, MJAW, and DF. The study was designed and written by MJAW and DF. As part of a joint collaborative work, co–first and co–last authors order was decided to ensure both teams have equitable representation.
Supplementary Material
Acknowledgments
This work was supported by the Association Francaise contre les Myopathies/AFM-Telethon (grant 15758), the Association Institut de Myologie, Muscular Dystrophy UK (16GRO-PG36-0091), the Medical Research Council (MR/P01741X/1), and the John Fell Fund (152/058). We would like to thank Mégane Lemaitre from the Muscle Function Evaluation platform (UMS28), and C. Thornton for the MBNL1 polyclonal antibody and the HSALR mouse model.
Version 1. 09/03/2019
In-Press Preview
Version 2. 09/30/2019
Electronic publication
Version 3. 11/01/2019
Print issue publication
Footnotes
Conflict of interest: MJG and MJAW are cofounders of PepGen, a company committed to the enhanced peptide delivery of oligonucleotide therapeutics for treatment of neuromuscular and other diseases.
Copyright: © 2019 Klein et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: J Clin Invest. 2019;129(11):4739–4744.https://doi.org/10.1172/JCI128205.
See the related Commentary at Better living through peptide-conjugated chemistry: next-generation antisense oligonucleotides.
Contributor Information
Arnaud F. Klein, Email: arnaud.klein@sorbonne-universite.fr.
Miguel A. Varela, Email: miguel.varela@paediatrics.ox.ac.uk.
Ludovic Arandel, Email: ludovic.arandel@upmc.fr.
Ashling Holland, Email: aholland@pepgen.net.
Naira Naouar, Email: naira.naouar@upmc.fr.
Andrey Arzumanov, Email: arzuman@waitrose.com.
David Seoane, Email: david.seoanemiraz@dpag.ox.ac.uk.
Lucile Revillod, Email: revillod.lucile@gmail.com.
Guillaume Bassez, Email: guillaume.bassez@aphp.fr.
Arnaud Ferry, Email: arnaud.ferry@upmc.fr.
Dominic Jauvin, Email: Dominic.Jauvin@crchudequebec.ulaval.ca.
Jack Puymirat, Email: jack.puymirat@crchul.ulaval.ca.
Michael J. Gait, Email: mgait@mrc-lmb.cam.ac.uk.
Denis Furling, Email: denis.furling@upmc.fr.
References
- 1. Harper PS. Myotonic Dystrophy. London, UK: Oxford University Press; 2001. [Google Scholar]
- 2.Mahadevan M, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3’ untranslated region of the gene. Science. 1992;255(5049):1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 3.Brook JD, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell. 1992;68(4):799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 4.Fu YH, et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255(5049):1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- 5.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128(6):995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3’ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci U S A. 1997;94(14):7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller JW, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19(17):4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee KY, et al. Compound loss of muscleblind-like function in myotonic dystrophy. EMBO Mol Med. 2013;5(12):1887–1900. doi: 10.1002/emmm.201303275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29(1):40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 10.Charlet-BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10(1):45–53. doi: 10.1016/S1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 11.Mankodi A, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10(1):35–44. doi: 10.1016/S1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 12.Rau F, et al. Abnormal splicing switch of DMD’s penultimate exon compromises muscle fibre maintenance in myotonic dystrophy. Nat Commun. 2015;6:7205. doi: 10.1038/ncomms8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freyermuth F, et al. Splicing misregulation of SCN5A contributes to cardiac-conduction delay and heart arrhythmia in myotonic dystrophy. Nat Commun. 2016;7:11067. doi: 10.1038/ncomms11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fugier C, et al. Misregulated alternative splicing of BIN1 is associated with T tubule alterations and muscle weakness in myotonic dystrophy. Nat Med. 2011;17(6):720–725. doi: 10.1038/nm.2374. [DOI] [PubMed] [Google Scholar]
- 15.Wheeler TM, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488(7409):111–115. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JE, Bennett CF, Cooper TA. RNase H-mediated degradation of toxic RNA in myotonic dystrophy type 1. Proc Natl Acad Sci U S A. 2012;109(11):4221–4226. doi: 10.1073/pnas.1117019109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jauvin D, et al. Targeting DMPK with antisense oligonucleotide improves muscle strength in myotonic dystrophy type 1 mice. Mol Ther Nucleic Acids. 2017;7:465–474. doi: 10.1016/j.omtn.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wheeler TM, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325(5938):336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulders SA, et al. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci U S A. 2009;106(33):13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wojtkowiak-Szlachcic A, et al. Short antisense-locked nucleic acids (all-LNAs) correct alternative splicing abnormalities in myotonic dystrophy. Nucleic Acids Res. 2015;43(6):3318–3331. doi: 10.1093/nar/gkv163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.González-Barriga A, et al. Cell membrane integrity in myotonic dystrophy type 1: implications for therapy. PLoS ONE. 2015;10(3):e0121556. doi: 10.1371/journal.pone.0121556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leger AJ, et al. Systemic delivery of a peptide-linked morpholino oligonucleotide neutralizes mutant RNA toxicity in a mouse model of myotonic dystrophy. Nucleic Acid Ther. 2013;23(2):109–117. doi: 10.1089/nat.2012.0404. [DOI] [PubMed] [Google Scholar]
- 23.Lehto T, Ezzat K, Wood MJA, El Andaloussi S. Peptides for nucleic acid delivery. Adv Drug Deliv Rev. 2016;106(Pt A):172–182. doi: 10.1016/j.addr.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 24.McClorey G, Banerjee S. Cell-penetrating peptides to enhance delivery of oligonucleotide-based therapeutics. Biomedicines. 2018;6(2):E51. doi: 10.3390/biomedicines6020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Betts C, et al. Pip6-PMO, a new generation of peptide-oligonucleotide conjugates with improved cardiac exon skipping activity for DMD treatment. Mol Ther Nucleic Acids. 2012;1(8):e38. doi: 10.1038/mtna.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mankodi A, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289(5485):1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 27.Lin X, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15(13):2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 28.Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117(12):3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sobczak K, Wheeler TM, Wang W, Thornton CA. RNA interference targeting CUG repeats in a mouse model of myotonic dystrophy. Mol Ther. 2013;21(2):380–387. doi: 10.1038/mt.2012.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu B, et al. Long-term rescue of dystrophin expression and improvement in muscle pathology and function in dystrophic mdx mice by peptide-conjugated morpholino. Am J Pathol. 2012;181(2):392–400. doi: 10.1016/j.ajpath.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller CM, Harris EN. Antisense oligonucleotides: treatment strategies and cellular internalization. RNA Dis. 2016;3(4):e1393. doi: 10.14800/rd.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rinaldi C, Wood MJA. Antisense oligonucleotides: the next frontier for treatment of neurological disorders. Nat Rev Neurol. 2018;14(1):9–21. doi: 10.1038/nrneurol.2017.148. [DOI] [PubMed] [Google Scholar]
- 33.Arandel L, et al. Immortalized human myotonic dystrophy muscle cell lines to assess therapeutic compounds. Dis Model Mech. 2017;10(4):487–497. doi: 10.1242/dmm.027367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complete raw data generated from RNA sequencing were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE134926).