Abstract
The interleukin-3 receptor α subunit, CD123, is expressed in many hematologic malignancies including acute myeloid leukemia (AML) and blastic plasmacytoid dendritic cell neoplasm (BPDCN). Tagraxofusp (SL-401) is a CD123-targeted therapy consisting of interleukin-3 fused to a truncated diphtheria toxin payload. Factors influencing response to tagraxofusp other than CD123 expression are largely unknown. We interrogated tagraxofusp resistance in patients and experimental models and found that it was not associated with CD123 loss. Rather, resistant AML and BPDCN cells frequently acquired deficiencies in the diphthamide synthesis pathway, impairing tagraxofusp’s ability to ADP-ribosylate cellular targets. Expression of DPH1, encoding a diphthamide pathway enzyme, was reduced by DNA CpG methylation in resistant cells. Treatment with the DNA methyltransferase inhibitor azacitidine restored DPH1 expression and tagraxofusp sensitivity. We also developed a drug-dependent ADP-ribosylation assay in primary cells that correlated with tagraxofusp activity and may represent an additional novel biomarker. As predicted by these results and our observation that resistance also increased mitochondrial apoptotic priming, we found that the combination of tagraxofusp and azacitidine was effective in patient-derived xenografts treated in vivo. These data have important implications for clinical use of tagraxofusp and led to a phase 1 study combining tagraxofusp and azacitidine in myeloid malignancies.
Keywords: Hematology, Oncology
Keywords: Leukemias, Molecular biology, Toxins/drugs/xenobiotics
Introduction
CD123, or the interleukin-3 (IL-3) receptor (IL-3R) α chain, is a cell-surface marker associated with a variety of hematologic malignancies and is often enriched on tumor cells compared with normal stem/progenitor populations (1). CD123 expression is particularly prominent in myeloid lineage neoplasms, including acute myeloid leukemia (AML), myelodysplastic syndrome (MDS), chronic myelomonocytic leukemia (CMML), and myeloproliferative neoplasms (MPNs) (2, 3). Blastic plasmacytoid dendritic cell neoplasm (BPDCN) is an aggressive hematologic cancer with skin, lymph node, blood, and bone marrow involvement, thought to result from malignant transformation of plasmacytoid dendritic cells or their precursors (4). BPDCN may be pathologically related to myeloid neoplasms/AML and is defined by particularly high levels of CD123 cell-surface expression (5, 6). Lastly, the so-called AML leukemia stem cell (LSC) or leukemia-initiating cell (LIC) subpopulation, proposed to represent the self-renewing subset of cells that can regenerate the entire malignant hierarchy, is enriched for CD123 expression (7). For these reasons, therapeutic targeting of cells that express CD123 is an attractive strategy in blood cancers.
Tagraxofusp (SL-401, DT388IL-3) is a novel targeted therapy consisting of recombinant IL-3 fused to a truncated diphtheria toxin (DT) payload (8). Tagraxofusp delivers the cytotoxic activity of DT to cells that express CD123. After internalization, the catalytic domain of DT escapes endosomes and catalyzes ADP ribosylation of eukaryotic elongation factor 2 (eEF2), blocking protein synthesis and killing the target cell (9). In 2018, tagraxofusp was approved by the US Food and Drug Administration (FDA) for treatment of patients with BPDCN, the first approval of a drug specifically for this disease, based on the results of a phase 1/2 trial (10). Tagraxofusp is currently being evaluated in clinical trials in several additional CD123-positive hematologic malignancies, including AML, MDS, CMML, and MPNs including myelofibrosis (MF). However, the determinants of initial response and mechanisms of resistance to tagraxofusp are largely unknown.
Tagraxofusp as a single agent is effective at inducing remission, particularly in patients with BPDCN (10, 11). However, some leukemias are initially refractory to treatment and others relapse after achieving a clinical response. Therefore, the goal of this study was to characterize and target the mechanisms of resistance to tagraxofusp. We also wanted to define biomarkers of response and resistance and use our findings to develop rational combinations to enhance the efficacy of tagraxofusp-containing regimens in BPDCN and AML.
Results
CD123 is not lost after exposure to tagraxofusp.
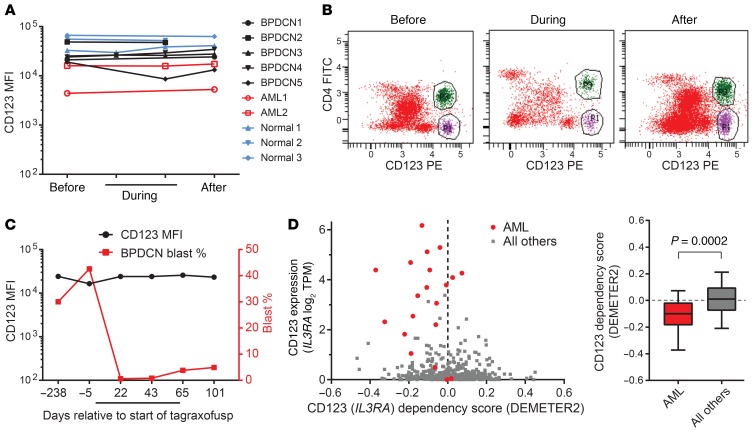
Resistance to cell-surface-targeted therapies in some hematologic malignancies is associated with loss of expression or downregulation of the target, such as CD19 after chimeric antigen receptor T (CAR-T) cells or blinatumomab, or CD22 after inotuzumab ozogamicin treatment in B cell acute lymphoblastic leukemia (B-ALL) (12, 13). Therefore, we measured the level of CD123 on the surface of AML and BPDCN cells in the bone marrow of patients before, during, and upon progression of disease during tagraxofusp treatment. We did not observe any decrease in CD123 associated with therapy, in the malignant cell population or on the surface of presumed “non-blast,” normal CD123-positive cells (Figure 1, A and B). In an illustrative patient with BPDCN, representative of all cases of BPDCN and AML analyzed, the CD123 level on the blast population was maintained even in the setting of significant changes in the relative blast frequency during initial response, ongoing treatment, and at the time of disease progression in the bone marrow (Figure 1C).
Figure 1. CD123 expression is maintained during and after tagraxofusp treatment in patients.
(A) The mean fluorescence intensity (MFI) of CD123 staining as measured by flow cytometry in bone marrow aspirates is plotted before, during, and after tagraxofusp treatment and annotated as malignant blasts of AML or BPDCN, or as putative non-blast CD123-positive cells. (B) An illustrative example of bone marrow aspirate flow cytometry from a patient with BPDCN before, during, and after tagraxofusp showing the malignant CD4+CD123+ BPDCN blasts (green) and the CD4–CD123+ non-blast cells (purple). (C) CD123 MFI and relative BPDCN blast percentage in bone marrow are plotted from another illustrative patient before, during (black bar), and after tagraxofusp treatment, showing maintenance of CD123 at a stable level despite significant changes in the relative percentage of blasts. These examples are representative of all cases in A, wherein the CD123 level was unchanged regardless of whether there was a significant change in disease burden. (D) CD123 (IL3RA) dependency score (x axis) is plotted against CD123 (IL3RA) RNA expression level in transcripts per million (TPM) for cell lines in Project Achilles. AML cell lines are labeled red (n = 20) and others are gray (n = 481). Negative dependency scores (calculated using DEMETER2) signify that cell line has a relative growth disadvantage when CD123/IL3RA is depleted in a genome-wide pooled shRNA assay. P value calculated using 2-tailed t test.
One possible explanation for this phenomenon is that CD123 (IL3RA) expression may offer a selective advantage to AML and/or BPDCN cells. We analyzed data from genome-wide RNA interference screening in 501 cell lines representing diverse cancer types from the Cancer Cell Line Encyclopedia (CCLE) and Project Achilles (14) for evidence of relative growth disadvantage upon knockdown. AML was enriched compared with cell lines derived from all other disease types for relative dependency on CD123 (P < 0.0005, Figure 1D). No BPDCN-derived cells were analyzed in the CCLE, but a prior RNA interference screen in a BPDCN cell line noted depletion of several shRNAs targeting IL3RA, consistent with dependency (15). These data suggest that myeloid lineage cancers such as AML and BPDCN may be unlikely to undergo selection for CD123-negative subclones, possibly even during therapy with a CD123-targeted agent, because loss is associated with a growth disadvantage.
Tagraxofusp-resistant cells selected in vitro maintain CD123 expression but are resistant to DT.
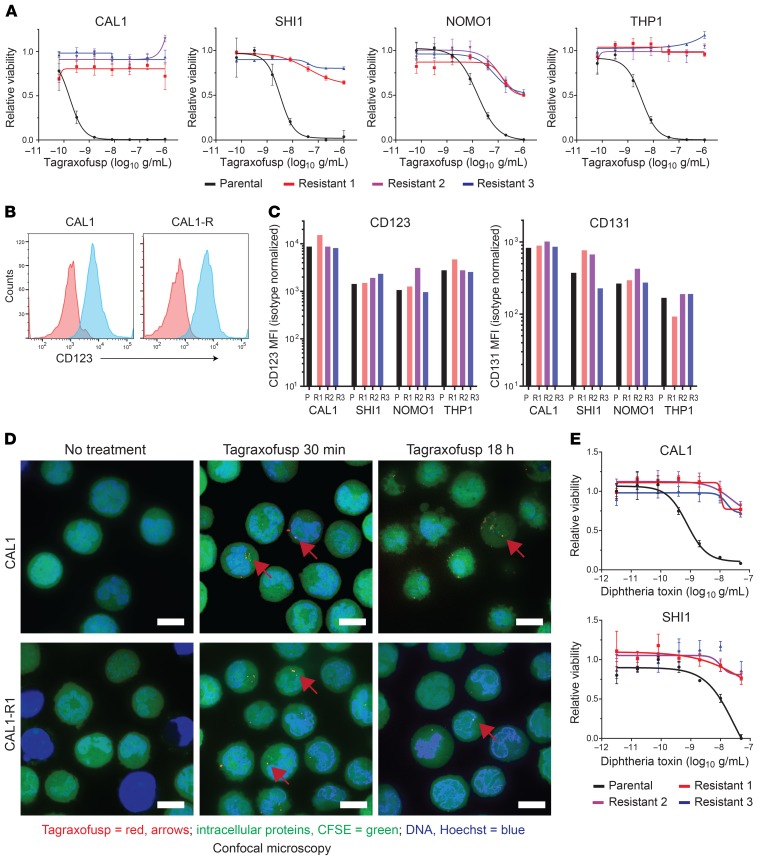
Given our findings in treated patients, we wanted to determine alternative mechanisms of resistance to tagraxofusp. We treated BPDCN (CAL1) and AML (SHI1, NOMO1, and THP1) cell lines with each lines’ approximately 95% lethal dose of tagraxofusp and re-treated repeatedly upon recovery. Within several treatment cycles, we generated biologically independent subcultures of each line that were at least 100-fold less sensitive to tagraxofusp. Some resistant cells were completely insensitive to tagraxofusp at concentrations up to 1 μg/mL, a dose that was higher than the 95% lethal dose in all parental lines tested (Figure 2A). We measured receptor subunit expression on the surface of the resistant cells by flow cytometry and found that none had a significantly altered level of CD123 or CD131, the IL-3R common β subunit, compared to their corresponding parental lines (Figure 2, B and C). Thus, although the parental lines span a 10-fold or greater range of CD123 expression and baseline tagraxofusp GI50 values (concentration that reduces growth by 50%), maintenance of CD123 expression level is a consistent feature of tagraxofusp-resistant cells.
Figure 2. BPDCN and AML cells resistant to tagraxofusp maintain CD123 expression and internalization of tagraxofusp but are cross-resistant to full-length diphtheria toxin.
(A) BPDCN (CAL1) and AML (SHI1, NOMO1, and THP1) parental (black) and tagraxofusp-resistant (red, blue, purple) cultures were tested for sensitivity to 5-fold decreasing concentrations of tagraxofusp in an MTT assay. Each point was assessed in triplicate and plotted relative to cells growing in vehicle alone. (B) CD123 (blue) and isotype control (red) staining as measured by flow cytometry in parental CAL1 cells and in tagraxofusp-resistant CAL1 (CAL1-R) cells is shown. (C) MFI of CD123 and CD131 in the indicated BPDCN and AML parental (P) and tagraxofusp-resistant (R1–R3) cell lines is shown. (D) Confocal microscopy 30 minutes and 18 hours after exposure of parental or tagraxofusp-resistant CAL1 cells to APC-tagged tagraxofusp (red, representative foci highlighted by red arrows), costained with CFSE (intracellular proteins, green) and Hoechst 33342 (DNA, blue). Scale bars: 10 μm. (E) MTT assays for viability of CAL1 and SHI1 parental and 3 independent tagraxofusp-resistant subcultures after exposure to full-length diphtheria toxin (DT), plotted as in A.
The cytotoxic activity of full-length DT requires cell entry, escape from endosomes into the cytoplasm, and toxin-mediated ADP ribosylation of eEF2 (16). We used confocal microscopy with tagraxofusp fluorescently tagged with allophycocyanin (tagraxofusp-APC) to test if there was a difference in drug internalization into resistant cells. At early time points (30–60 minutes) after exposure to tagraxofusp-APC, we saw no difference in the pattern of intracellular APC signal in parental or resistant cells (Figure 2D). Tagraxofusp-APC killed parental cell lines with efficiency equal to that of the untagged drug but had no activity against resistant subclones. Like many hematopoietic cells (17), CAL1 and SHI1 cells express the DT ligand-binding domain cell-surface receptor, proheparin-binding EGF-like growth factor (HBEGF). This allowed us to test the cytotoxic activity of full-length DT in parental and tagraxofusp-resistant cells. Resistant sublines were similarly resistant to full-length DT and tagraxofusp (Figure 2E). Together, these data suggest that tagraxofusp resistance in AML and BPDCN cells is not due to loss of the cell-surface target or defective internalization after ligand binding.
Acquired tagraxofusp resistance is associated with loss of diphthamide synthesis pathway activity.
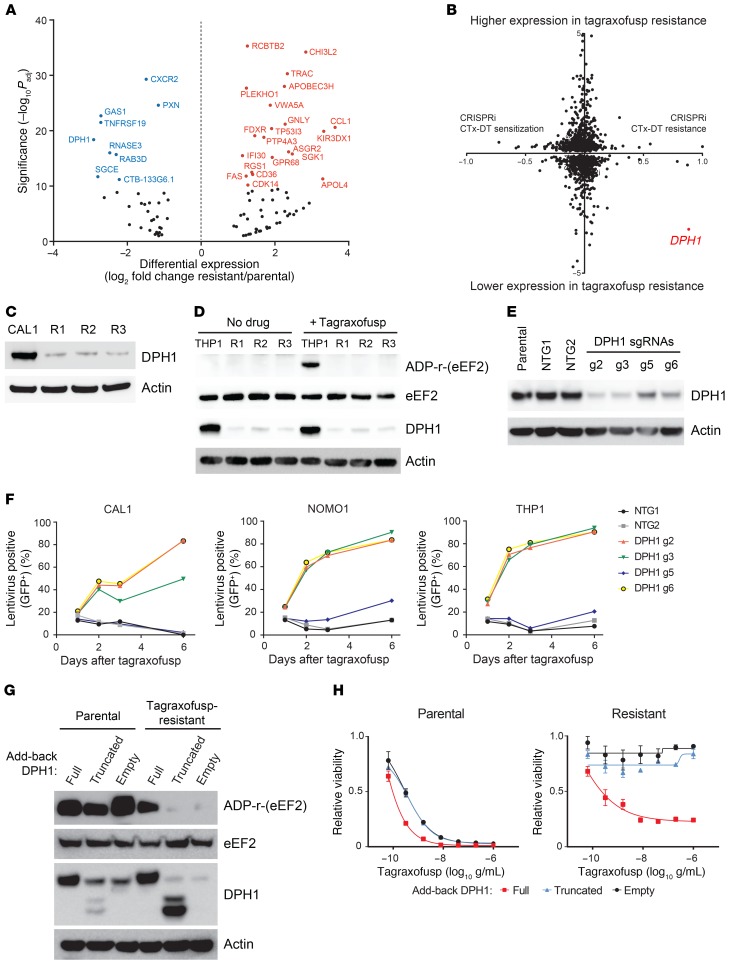
To elucidate mechanisms of tagraxofusp resistance, we performed whole-exome sequencing (WES) and whole-transcriptome RNA sequencing (RNA-seq) on parental and resistant BPDCN and AML cell lines. There were no acquired recurrently mutated genes in the exomes of 3 independent resistant CAL1 cultures by WES compared with parental cells. However, in combined RNA-seq analysis of CAL1 (BPDCN) and SHI1 (AML) tagraxofusp-resistant compared with parental cells (n = 6 independent resistant subcultures, n = 6 parental), the most downregulated gene in resistant cells was DPH1 (7.53-fold decrease, –log10 adjusted P [Padj] = 3.79 × 10–19; Figure 3A and Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI128571DS1). DPH1 encodes the first protein in an enzymatic cascade containing at least 7 members known as the diphthamide synthesis pathway or diphthamide modification pathway, which is responsible for catalyzing the conversion of histidine 715 on eEF2 to the variant amino acid diphthamide (18). Diphthamide-715 on eEF2 is the site of ADP ribosylation by DT as well as other ADP-ribosylating bacterial toxins such as Pseudomonas exotoxin A.
Figure 3. Tagraxofusp resistance is mediated by loss of the diphthamide synthesis pathway enzyme DPH1.
(A) Differentially expressed genes between CAL1 (BPDCN) and SHI1 (AML) parental cells compared to 3 independent tagraxofusp-resistant subclones each (n = 6 parental and 6 resistant total). Log2(fold change) expression in resistant compared with parental cells plotted (x axis) versus –log10 adjusted P (Padj) value (y axis). Genes names in blue (downregulated in resistant cells) or red (upregulated in resistant cells) for genes with –log10 Padj > 10. (B) Log2(fold change) in gene expression associated with tagraxofusp resistance (as from panel A), where negative values represent lower expression in resistant cells, plotted gene-by-gene versus a CRISPRi score for CTx-DTA resistance (19), where positive values represent genes that conferred CTX-DTA resistance when their expression was inhibited. DPH1 is highlighted in red. (C) Western blotting for DPH1 and actin in parental and tagraxofusp-resistant CAL1 cells. (D) In vitro ADP-ribosylation assay with or without tagraxofusp (top row) and Western blotting for eEF2, DPH1, and actin (bottom rows) for parental THP1 and tagraxofusp-resistant (R1–R3) subclones. (E) Western blotting for DPH1 and actin in parental THP1 cells, and cells transduced with independent nontargeting (NTG1-2) and DPH1-targeted (g2, g3, g5, and g6) sgRNAs. (F) Percentage lentivirus-containing (GFP+) cells plotted over time after treatment with tagraxofusp in CAL1, NOMO1, and THP1 cells transduced with the CRISPR sgRNA-containing lentiviruses as in panel E, each coexpressing GFP. (G) In vitro ADP-ribosylation in the presence of tagraxofusp (top row) and Western blotting for eEF2, DPH1, and actin (bottom rows) for parental CAL1 cells and tagraxofusp-resistant cells expressing a doxycycline-inducible full-length DPH1 cDNA, an N-terminally truncated, enzymatically inactive DPH1, or empty vector. (H) Viability after treatment with serial dilutions of tagraxofusp in parental and tagraxofusp-resistant cells expressing doxycycline-inducible DPH1 or variants as in panel G. Triplicate points plotted relative to cells in vehicle alone.
Diphthamide synthesis pathway proteins are known to be required for cytotoxicity of full-length DT, but their role in tagraxofusp sensitivity and resistance has not been studied. Given that expression of several other genes was also affected in the setting of tagraxofusp resistance, we integrated our RNA-seq data with existing data from an experiment that measured the influence of gene expression on sensitivity to a cholera toxin–DT hybrid protein (CTx-DTA) (19). In that experiment, a genome-wide CRISPR single guide RNA (sgRNA) library was introduced into human cells coexpressing a catalytically inactive Cas9 nuclease fused to either a transcriptional activator or repressor (so-called “CRISPRa” or “CRISPRi”) and relative sgRNA abundance was compared before and after exposure to CTx-DTA. When we plotted tagraxofusp-resistance-associated gene expression versus the CRISPRi score on a per-gene basis, only 1 gene, DPH1, was a “hit” in both assays — i.e., significantly lower expression in tagraxofusp-resistant cells and knockdown was advantageous in the setting of CTx-DTA exposure (Figure 3B). We confirmed loss of DPH1 protein in CAL1 tagraxofusp-resistant BPDCN cells by Western blotting (Figure 3C) and confirmed the same phenotype in an independent AML cell line, THP1 (Figure 3D). Thus, in 3 of 3 cell lines tested, tagraxofusp resistance was associated with decreased DPH1 expression.
We next performed a biochemical assay for the ability of tagraxofusp to catalyze ADP-ribosylation in cell lysates in the presence of exogenous biotin-tagged nicotine adenine dinucleotide (NAD+) as a source of ADP-ribose. In parental THP1 cells, tagraxofusp caused ADP-ribosylation of a single protein band of the expected molecular weight of eEF2 (~100 kDa), which was detectable by Western blotting with streptavidin-HRP. In contrast, in resistant cells with markedly reduced DPH1, tagraxofusp failed to catalyze detectable ADP-ribosylation of any protein in the lysate (Figure 3D). Together, these data pointed to loss of DPH1 as the source of decreased tagraxofusp-induced ADP-ribosylation activity and tagraxofusp resistance in leukemia cell lines.
To confirm that loss of DPH1 was both necessary and sufficient to create tagraxofusp resistance, we performed a series of knockout and add-back experiments. First, we generated cell lines stably expressing the Cas9 nuclease and transduced them with lentiviruses expressing 1 of 4 independent sgRNAs targeting the DPH1 locus or nontargeting controls, each linked to GFP. At high multiplicity of infection (MOI) resulting in greater than 80% GFP positivity, we confirmed decreased expression of DPH1 protein in several independent DPH1 sgRNA–transduced cultures (Figure 3E). In cultures infected at lower MOI (resulting in ~20% GFP positivity), we observed a growth advantage after treatment with tagraxofusp only in the DPH1 sgRNA–transduced cells but not in the control-transduced cells, particularly in those harboring sgRNAs associated with the greatest loss of DPH1 protein (Figure 3F). Therefore, we concluded that loss of DPH1 is sufficient to confer relative tagraxofusp resistance in AML cells.
Next, we cloned a full-length DPH1 cDNA or an N-terminally truncated cDNA, deleting a domain known to be required for DPH1 catalytic activity (20), into a doxycycline-inducible lentiviral expression vector. We transduced full-length or truncated dox-on DPH1, or empty vector–containing viruses into parental or tagraxofusp-resistant CAL1 BPDCN cells and selected with puromycin. We then performed Western blotting and the in vitro ADP-ribosylation biochemical assay in cell lysates after doxycycline induction. We found that expression of the full-length DPH1, but not the enzymatic activity–deficient DPH1 or empty vector, restored the ADP-ribosylation activity of tagraxofusp in resistant cells (Figure 3G). Re-expression of full-length DPH1 also restored the cytotoxic activity of tagraxofusp in resistant cells to GI50 values comparable to those of parental cells (Figure 3H). Of interest, overexpression of DPH1 in parental cells was also associated with modestly increased sensitivity to tagraxofusp, suggesting that DPH1 level and/or diphthamide synthesis pathway activity is rate limiting for the pathway and could serve as a biomarker of sensitivity to tagraxofusp.
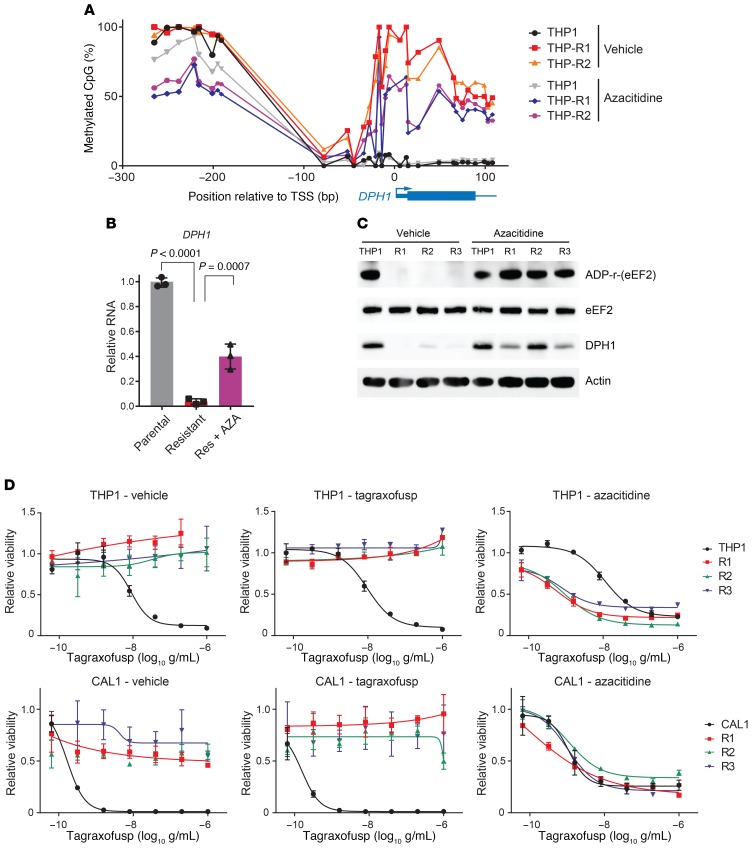
DPH1 locus DNA methylation correlates with tagraxofusp resistance and both are reversible by azacitidine.
DPH1 expression is known to be regulated by DNA methylation, and sensitivity to a CD22–Pseudomonas exotoxin A fusion in a B-ALL cell line was previously shown to be modulated by CpG methylation at the DPH1 locus (21). Therefore, we quantified CpG methylation in parental and tagraxofusp-resistant THP1 cells using bisulfite DNA sequencing. We found that tagraxofusp-resistant cells had acquired hypermethylation of CpG motifs in the promoter region and first exon of DPH1 (Figure 4A). CpGs further upstream, between –300 and –80 bases from the transcription start site (TSS), showed no significant change in methylation, suggesting that increased DPH1-promoter methylation associated with tagraxofusp resistance may confer a specific advantage. Given this finding, we hypothesized that azacitidine, a DNA methyltransferase inhibitor or DNA hypomethylating agent (HMA) might reverse resistance-associated DPH1 hypermethylation and restore DPH1 expression. We treated tagraxofusp-resistant or parental THP1 cells with a noncytotoxic dosing regimen of azacitidine (300 nM 2 days on/2 days off, for 2 weeks; Supplemental Figure 1A). Azacitidine treatment reduced DPH1 CpG DNA methylation and partially restored mRNA expression as measured by quantitative reverse transcription PCR (RT-PCR) (Figure 4, A and B). We observed similar findings in tagraxofusp-resistant CAL1 BPDCN cells (Supplemental Figure 1, A–C).
Figure 4. Tagraxofusp resistance is associated with hypermethylation of DPH1 locus CpGs, and azacitidine restores diphthamide pathway activity and tagraxofusp sensitivity.
(A) Percentage of methylated CpGs in the DPH1 locus are shown for the indicated genomic positions in parental THP1 cells and 2 independent tagraxofusp-resistant subclones, before and after 2 weeks of pulsatile treatment with noncytotoxic doses of azacitidine. (B) Quantitative RT-PCR for DPH1 expression in parental THP1 cells and a tagraxofusp-resistant subclone treated with vehicle or azacitidine (n = 3 replicates each). Dots represent relative expression, bars are ±SD. Conditions compared by 1-way ANOVA with Dunnett’s multiple-comparisons correction, adjusted P values shown. Data are representative of 2 independent resistant subclones with similar results. (C) In vitro ADP-ribosylation assay in the presence of tagraxofusp (top row) and Western blotting for eEF2, DPH1, and actin (bottom rows) are shown for parental THP1 and 3 independent tagraxofusp-resistant subclones (R1–R3) after 2 weeks of pulsatile treatment with noncytotoxic doses of azacitidine or vehicle. (D) Tagraxofusp cytotoxicity assays in parental and tagraxofusp-resistant AML (THP1) and BPDCN (CAL1) cells after 2 weeks of pulsatile treatment with noncytotoxic doses of azacitidine or vehicle, or with weekly exposure to 1 μg/mL tagraxofusp. Each point was assessed in triplicate and plotted relative to cells growing in vehicle alone.
Next, we tested if azacitidine restored sensitivity to tagraxofusp in AML and BPDCN cells. First, we asked if the partial restoration of DPH1 mRNA we observed after long-term noncytotoxic exposure to azacitidine (Figure 4B) was sufficient to reverse the functional defect in ADP-ribosylation activity by tagraxofusp. Using the in vitro biochemical activity assay described above, we found that 2-week azacitidine-treated THP1 cells showed complete restoration of tagraxofusp-mediated ADP-ribosylation activity (Figure 4C). In agreement with those results, azacitidine treatment also re-sensitized resistant AML and BPDCN cells to the cytotoxic activity of tagraxofusp (Figure 4D). Controls, including resistant cells cultured for 2 weeks with repeated dosing of tagraxofusp or maintained in vehicle alone, showed no change in tagraxofusp sensitivity. Given that azacitidine likely affects expression of many genes, not only DPH1, we also tested whether cells in which DPH1 is targeted by CRISPR (Figure 3E) were resistant to the sensitizing effect of azacitidine. DPH1-knockdown cells did not have increased tagraxofusp sensitivity after 2-week azacitidine treatment, whereas cells resistant to tagraxofusp and harboring a control nontargeting sgRNA were relatively re-sensitized (Supplemental Figure 2D). These data support the conclusion that DPH1 is at least among the dominant targets of azacitidine’s ability to restore tagraxofusp sensitivity in cells with acquired resistance.
Increased apoptotic priming in tagraxofusp-resistant cells associated with increased sensitivity to and synergy with chemotherapy.
To address the possibility that resistant cells have lost the ability to undergo apoptosis, which is important in considering how to sequence or combine therapies with tagraxofusp, we performed BH3 profiling. This assay quantifies the apoptotic priming, or propensity to undergo cell death via mitochondrial apoptosis, after stimulation of permeabilized cells with BH3 domain–containing peptides derived from proapoptotic BCL-2 family proteins (22). We have used BH3 profiling to predict BCL-2 family dependence and therapeutic sensitivity in several types of cancer (23, 24).
We performed BH3 profiling on parental cells and those with acquired resistance to tagraxofusp. The most striking finding was that across multiple resistant subclones of both AML and BPDCN cells, tagraxofusp resistance was associated with an increase in overall apoptotic priming (Supplemental Figure 2A). This finding was unexpected, as, in most cases, resistance to one cancer therapy is associated with decreased overall apoptotic priming and decreased sensitivity to subsequent alternative therapies (25, 26). In addition, selective peptide treatments showed that resistant SHI1 cells were more dependent on BCL-2 and/or BCL-XL (based on increased sensitivity to BAD and HRK peptides), which suggests there could be specific changes in BCL-2 family dependencies in the setting of resistance.
Consistent with the prediction that tagraxofusp-resistant cells would be sensitized to treatments that act via mitochondrial apoptosis, we found that tagraxofusp-resistant cells were more sensitive than parental cells to treatment with several chemotherapeutic agents (Supplemental Figure 2B). These results led us to ask if there was also up-front synergy between tagraxofusp and conventional chemotherapy in parental cells. Using isobologram analysis and the method of Chou-Talalay, we determined that tagraxofusp and chemotherapy were synergistic (combination index [CI] < 1) across diverse cytotoxic compounds and cell lines (Supplemental Figure 2C). Together, these data suggest that the combination of tagraxofusp and chemotherapy, particularly azacitidine due to its additional ability to reverse tagraxofusp resistance, might be effective in AML, BPDCN, and possibly other CD123-expressing malignancies.
Tagraxofusp and azacitidine are effective in combination in vivo.
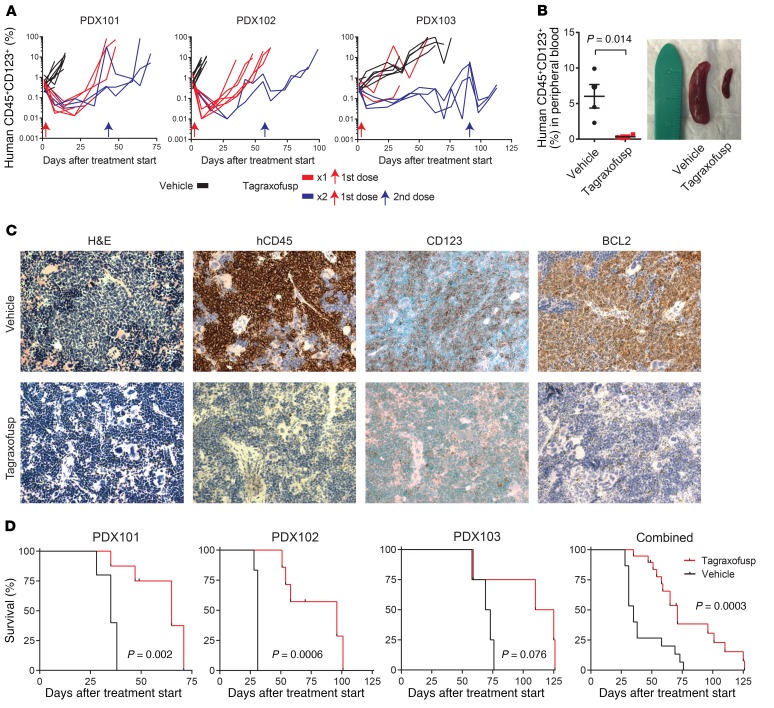
To test these predictions in primary human leukemia cells in vivo, we established tagraxofusp therapeutic models using BPDCN patient–derived xenografts (PDXs) (27). First, we tested the activity of single-agent tagraxofusp in 3 independent BPDCN PDXs. Each PDX was injected into a cohort of recipients as a secondary transplant of cryopreserved splenocytes from the initial PDX generation. When the average peripheral blood disease burden reached 0.2%, we randomized animals to receive vehicle or tagraxofusp, given daily for 5 days to mimic the dosing schedule used in patients. In a subset, we gave one additional cycle of therapy at the time of overt disease progression. We followed the peripheral blood leukemia burden by weekly flow cytometry and observed a significant pharmacodynamic effect of tagraxofusp (Figure 5A). To confirm disease response in tissue, we sacrificed a subset of vehicle or tagraxofusp-treated animals after 7 days and observed reduction in splenomegaly, associated with decreased splenic infiltration of human BPDCN cells (human CD45, CD123, BCL-2 positive) and increases in normal hematopoietic elements (Figure 5, B and C). Tagraxofusp treatment resulted in prolonged median overall survival compared with vehicle in each PDX individually and in combined analysis (71 vs. 35 days, P = 0.0003; Figure 5D).
Figure 5. Tagraxofusp is active in BPDCN patient–derived xenografts (PDXs) in vivo.
(A) Human CD45+CD123+ cells as a percentage of the peripheral blood mononuclear cells in NSG mice engrafted with 1 of 3 BPDCN PDXs, treated at day 0 (red arrow) with 5 days of tagraxofusp (red line) or vehicle (black line). A subset of animals in each group was re-treated with another cycle at the time when greater than 50% of animals showed progression (>5% in peripheral blood; blue lines represent animals that received 2 treatments and blue arrows are the time of the second treatment). (B) Peripheral blood disease burden measured by CD45+CD123+ flow cytometry and representative spleen size reduction in animals treated with tagraxofusp as compared to vehicle harvested 7 days after treatment (n = 4 each). (C) Sections from mouse spleens harvested on day 7 after treatment with tagraxofusp or vehicle and stained with hematoxylin & eosin (H&E) or the indicated antibodies by immunohistochemistry. Original magnification, ×40. (D) Kaplan-Meier overall survival curves for recipients of each PDX (left) and for all PDXs combined (right) that received tagraxofusp (red, n = 19) or vehicle (black, n = 15). Curves compared by log-rank test.
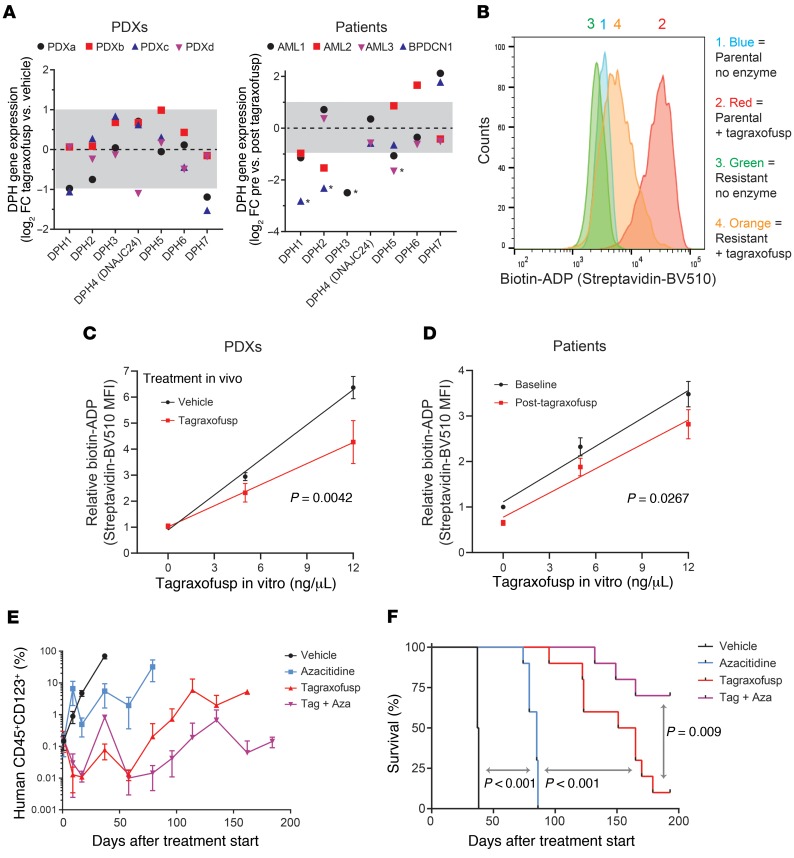
Next, we asked if the adaptations we observed after tagraxofusp exposure in cell lines were observed in PDXs and primary samples. While the second cycle of PDX treatment was effective (Supplemental Figure 3A), the time to progression was shorter, suggesting acquisition of relative resistance in vivo (Figure 5A). We noted that, as in patients treated with tagraxofusp and in resistant cell lines, CD123 expression on the surface of PDX blasts was not decreased at the time of progression after tagraxofusp (Supplemental Figure 3B). Therefore, we performed RNA-seq on CD45+CD123+ sorted cells from 4 relapsed PDXs and from 4 patients (3 AML and 1 BPDCN) before and after treatment with tagraxofusp. We observed downregulation of DPH1 (2-fold or more) in 2 of 4 PDXs and in 3 of 3 evaluable patients (Figure 6A). One patient sample, AML3, had baseline DPH1 expression below the level of detection, which is of unclear significance. In addition, we saw decreased expression of several other diphthamide synthesis pathway genes in some samples after tagraxofusp exposure, and there were also a few instances of increased expression (Figure 6A). While we were encouraged that changes in DPH1 expression were largely consistent with our cell line data, we reasoned that a functional assay might more clearly define the overall contribution of the entire diphthamide pathway to tagraxofusp resistance in primary cells.
Figure 6. Impaired ADP-ribosylation activity in resistant cells and effectiveness of tagraxofusp plus azacitidine in vivo.
(A) Log2(fold change) (log2FC) in RNA expression for diphthamide genes in individual PDXs (left) and patients (right) after exposure to tagraxofusp in vivo compared to vehicle (PDXs) or baseline (patients). Dotted line represents no change (log2FC = 0) and gray area is less than 2-fold change in expression (log2FC –1 to 1). *Genes below 0.1 fragments per kilobase of transcript per million mapped reads (FPKM) after tagraxofusp, in which case an FPKM of 0.1 was used to calculate fold change. Genes that had FPKM less than 0.1 before treatment are not shown. (B) Flow cytometry–based in vitro enzymatic labeling assay for tagraxofusp-induced ADP-ribosylation activity in parental and tagraxofusp-resistant CAL1 cells, showing decreased labeling with biotin-NAD+ in the setting of tagraxofusp resistance. Staining detected with streptavidin-BV510 fluorescence. (C) Flow cytometry–based enzymatic labeling assay with increasing doses of tagraxofusp measuring ADP-ribosylation activity as in panel B for BPDCN PDXs harvested from spleen or bone marrow at the time of disease progression after in vivo treatment with vehicle (n = 8) or tagraxofusp (n = 10). Curves compared by regression modeling of least-squares fit. (D) Flow cytometry–based ADP-ribosylation activity over increasing concentrations of tagraxofusp added in vitro measured in single CD45+CD123+ cells collected from the bone marrow of 5 patients before and after in vivo treatment with tagraxofusp (curves compared as in panel D). (E) Disease burden measured by peripheral blood human CD45+CD123+ flow cytometry in animals engrafted with BPDCN PDX cells after 2 cycles of treatment (at days 0 and 30) with vehicle, azacitidine, tagraxofusp, or the combination of tagraxofusp and azacitidine (Tag + Aza). (F) Kaplan-Meier overall survival curves from the time of treatment start for animals after treatments described in panel E (n = 10 mice per arm for panels E and F). Curves compared pairwise by log-rank test.
Given that loss of any diphthamide pathway member can inhibit diphthamide generation and DT cytotoxicity (19, 20, 28), we designed a series of biomarker assays to measure the ability of tagraxofusp to ADP-ribosylate substrates in permeabilized cells, independent of toxin internalization (see Methods for details). We reasoned that this type of assay would be agnostic to specific upstream alteration(s) in the diphthamide pathway and represent an integrated functional readout of tagraxofusp ADP-ribosylation activity in relevant cells. Specifically, we developed tests to measure biotinylated-NAD incorporation either by microscopy in cells fixed to glass slides or by flow cytometry of permeabilized cells in solution. Using these assays, we found impairment in drug-dependent biotin labeling in tagraxofusp-resistant cell lines (Figure 6B and Supplemental Figure 3C).
The flow cytometry–based ADP-ribosylation assay is particularly attractive because it allows costaining with cell-surface markers, which facilitates measurement of tagraxofusp activity in defined populations of single cells isolated from blood or more complex tissues such as spleen or bone marrow (Supplemental Figure 3D). Thus, we applied this assay to human leukemias before and after exposure to tagraxofusp treatment in vivo. Among primary BPDCN PDXs harvested from animals with progressive disease after treatment with tagraxofusp, we observed decreased tagraxofusp-dependent ADP-ribosylation activity in CD45+CD123+ cells compared with leukemias progressing after treatment with vehicle alone (Figure 6C). Similarly, in 5 patients (2 AML and 3 BPDCN) that had persistent or recurrent bone marrow blasts after treatment with 1 or 2 cycles of tagraxofusp, we observed decreased tagraxofusp-dependent ADP-ribosylation activity in CD45+CD123+ cells (Figure 6D). These data suggest that one mechanism of resistance in primary leukemias treated in vivo is associated with decreased ADP-ribosylation of cellular substrates by tagraxofusp, which may be caused by downregulation of diphthamide pathway members or other upstream alterations. The ADP-ribosylation biochemical assays described here are therefore potential biomarker tests for tagraxofusp activity that could be explored prospectively and in larger patient cohorts.
Finally, we tested if tagraxofusp and azacitidine were an effective combination in primary human leukemias in vivo. We injected recipient mice with BPDCN PDX cells and when the average peripheral blood leukemia burden was greater than 0.2%, we randomized animals to 1 of 4 groups for treatment with 2 cycles of vehicle, azacitidine alone, tagraxofusp alone, or azacitidine/tagraxofusp in combination. Both azacitidine and tagraxofusp alone prolonged the time to leukemia progression, but the combination of azacitidine and tagraxofusp was more effective than either single agent (Figure 6E). This correlated with a prolongation of overall survival after treatment with tagraxofusp plus azacitidine that was greater than either agent alone (Figure 6F). At 200 days, we sacrificed the remaining 1 tagraxofusp-treated mouse and 7 tagraxofusp-plus-azacitidine–treated mice and measured CD45+CD123+ human leukemia cell burden in peripheral blood, spleen, and bone marrow. Whereas the 1 remaining tagraxofusp-treated animal had detectable leukemia cells in spleen and bone marrow, only 1 of 7 combination-treated mice had measurable residual disease above a detection threshold of 0.1% in the bone marrow (Supplemental Figure 3E).
Discussion
Understanding the mechanisms of response and resistance to tagraxofusp are important in optimizing its use in BPDCN, AML, and other malignancies that express CD123. A deeper understanding of the factors associated with tagraxofusp sensitivity might lead to new biomarkers of disease- or patient-specific drug activity. For example, drug-induced ADP-ribosylation activity in tumor cell lysates or permeabilized tumor cells, as we described here, is a potential predictive biomarker that merits further evaluation in prospective studies.
We observed no evidence of CD123 loss as a mechanism of resistance in cell lines, PDXs, or in leukemias from patients treated with tagraxofusp. Rather, resistance was associated with loss of diphthamide synthesis pathway activity and insensitivity to DT, and at least in some cells this was via DNA-methylation-mediated downregulation of DPH1. Tagraxofusp-resistant cells also had increased apoptotic priming and were hypersensitive to cytotoxic agents, which provides one strategy for targeting resistance by combination with chemotherapy. We also found that azacitidine restored DPH1 expression and reversed tagraxofusp resistance, and that tagraxofusp and azacitidine prolonged survival of in vivo leukemia models compared with either agent alone. Based on these data, we have initiated a multicenter phase 1 trial of the combination of tagraxofusp and azacitidine in patients with AML or high-risk MDS (clinicaltrials.gov: NCT03113643). Our data also suggest that tagraxofusp with azacitidine, or with conventional chemotherapy, could be synergistic in other CD123-positive diseases and support development of additional clinical studies to evaluate combination treatment.
Other bacterial toxin conjugates are in various stages of development for cancer therapy, including moxetumomab pasudotox, an anti-CD22 monoclonal antibody linked to pseudomonas exotoxin (PT) that has been tested in several hematologic malignancies and was recently approved for treatment of relapsed/refractory hairy cell leukemia (29). Both DT and PT catalyze ADP-ribosylation of eEF2 to inhibit protein synthesis, and deletion or loss of diphthamide synthesis genes promotes resistance to both toxins. Therefore, it is possible that resistance mechanisms may be shared across this class of toxin immunoconjugates. However, while in vitro resistance to moxetumomab pasudotox in cell lines was associated with downregulation of diphthamide synthesis pathway genes (21), this did not seem to be the dominant mechanism of resistance in B-ALL treated in vivo (30). Rather, moxetumomab resistance was associated with a developmental state change, karyotypic abnormalities, and loss of surface CD22 expression. Yet, despite this difference, azacitidine and moxetumomab were also an effective combination in B-ALL cell lines treated in vivo (30). Thus, mechanisms of in vivo resistance to diphthamide-targeting toxins may be disease or cell-type dependent. Given that CD123 loss was specifically associated with myeloid lineage growth disadvantage in genetic screens, it is possible that malignancies such as AML, MDS, and BPDCN are forced to exploit alternative tagraxofusp-resistance mechanisms other than loss of the target.
Acquisition of non–CD123-associated mechanisms of resistance is particularly important given the current clinical development of various distinct agents targeting CD123. In addition to tagraxofusp, there are anti-CD123 antibody-drug conjugates, CD123-CD3 bispecific antibodies, and CD123-targeted CAR-T cells in development for treatment of AML, MDS, BPDCN, and other hematologic malignancies. If resistance to CD123-targeted agents in myeloid cells is more likely to be related to the payload rather than loss of the target, then there might not be cross-resistance to different drug classes. This should be considered when designing entry criteria for clinical trials, specifically that prior therapy with another CD123-targeted agent should not necessarily exclude eligibility.
In this study, tagraxofusp resistance was associated with increased apoptotic priming and sensitivity to conventional chemotherapy, an observation consistent with data showing that engineered single gene knockouts of DPH1-4 result in hypersensitivity to cell death induced by tumor necrosis factor α (31). Diphthamide-eEF2 is thought to regulate protein translation by preventing ribosomal slippage and –1 frameshifting (32). This is in line with studies showing that loss of DPH3 results in decreased levels of the antiapoptotic protein XIAP via impairment of translation fidelity (33) and leads to enhanced sensitivity to cytotoxic agents (34). Regardless of the upstream cause of the increased priming we observe in BPDCN and AML cells, these data suggest that combinations of tagraxofusp with agents targeting BCL-2 or BCL-2 family members may be particularly active. Given that we previously found BPDCN was markedly dependent on BCL-2 and sensitive to venetoclax (24), understanding potential synergy between tagraxofusp and BCL-2 inhibition is of significant interest for future studies.
Relatively little is understood about the diphthamide pathway in normal biology, outside of its role in toxin-mediated cytotoxicity. For example, DPH1 is frequently codeleted with TP53 on chromosome 17 in several human cancers (35), and haploinsufficiency of Dph1 promoted spontaneous tumor development in mice, alone or in concert with Tp53 deletion (36). However, the mechanism of DPH1’s tumor suppressor activity and whether it involves regulation of translation is largely unknown. Cells resistant to tagraxofusp might provide another model to understand the normal function of diphthamide-eEF2. It is intriguing to speculate that alterations in gene expression, protein abundance, or translational fidelity associated with loss of diphthamide pathway activity may create additional novel targets for therapy. Given that these changes could be lineage and/or patient specific, the consequences of tagraxofusp resistance and diphthamide synthesis alterations likely need to be assessed in each unique situation.
Methods
Cell lines
Cell lines were from ATCC or DSMZ, except CAL1 cells that were provided by T. Maeda (Nagasaki University, Nagasaki, Japan) (37). Cell line identity was verified by short tandem repeat (STR) profiling in the DFCI Molecular Diagnostics Core and cells were verified to be mycoplasma-free by regular testing at least every 6 months. Tagraxofusp-resistant cultures were generated by first determining the lethal dose for approximately 95% (LD95) of parental cells using an MTT assay as outlined below. Cultures were treated with the LD95 and allowed to recover to confluence. The LD95 treatment was repeated up to 3 times. All cell lines developed resistance within 2–4 treatments. Resistant cultures were re-treated with tagraxofusp at 1 μg/mL every approximately 2 weeks to maintain the resistant state.
Flow cytometry
Cells were washed with phosphate-buffered saline (PBS) with 2% fetal bovine serum (FBS) and then stained with 1:100 dilutions of CD123 conjugated to phycoerythrin (CD123-PE) (BD Pharmingen, 555644), CD131-PE (eBioscience, 12-1319-41), CD4-APC (BD Pharmingen, 561840), or PE–mouse IgG kappa isotype (BD Pharmingen, 550617), for 20 minutes at 4°C. Samples were then washed twice with PBS plus 2% FBS and analyzed. Flow cytometry was performed on patient samples using a BD FACSCanto II system and analyzed using BD FACSDIVA software (BD Biosciences). Cell-line flow was performed using a Cytoflex flow cytometer (Beckman Coulter, B53012) and analyzed using FlowJo software, version 10. Antigen levels were normalized by subtracting the mean fluorescence intensity (MFI) of the isotype control. MFI accuracy and reproducibility were verified using CD-Chex Plus reagents, positive controls manufactured from normal human peripheral blood for flow cytometry immunophenotyping (Streck Labs).
Cancer Dependency Map analysis
Data for IL3RA (CD123) RNA interference from the Cancer Dependency Map (14) of Project Achilles and the CCLE were downloaded from the publicly available DepMap data portal (https://depmap.org/portal/) on September 6, 2018.
Confocal microscopy
Tagraxofusp was conjugated to allophycocyanin (APC) by OneWorld Biotech. Briefly, tagraxofusp was dialyzed against PBS containing 4 mM EDTA and then treated with 2-iminothiolane to introduce a terminal sulfhydryl group on primary amines, APC was added for the conjugation reaction, and then unreacted sulfhydryl groups were blocked by N-ethylmaleimide. Cells were treated with 1 μg/mL of tagraxofusp-APC for the indicated times and washed twice with PBS. Cell were stained in PBS with a mixture of 5 μg/mL Hoechst 33342 (Life Technologies, H1399) and 5 μM CFSE (Thermo Fisher Scientific, C34554) for 20 minutes at room temperature in the dark, and then washed 3 times in PBS. Cells were adhered to glass slides using a Cytospin centrifuge (300 g for 3 minutes) and imaged used a Yokogawa Spinning Disk Confocal Microscope at ×100 magnification.
Drug toxicity and apoptosis assays
Cell lines were cultured in RPMI-1640 with 10% FBS (except SHI1 was in IMDM + 20% FBS) and were plated in a 96-well plate at a concentration of 5,000 cells per 120 μL media per well. Compounds were added to the cells in serial 5-fold dilutions. After a 72-hour incubation at 37°C, viability was determined by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. In brief, 20 μL of 5 mg/mL MTT (EMD Millipore, 475989) in PBS was added to each well and incubated for 2 hours 37°C, and then 100 μL MTT lysis buffer (100 g SDS dissolved in 50% DMF/water solution containing 12.5 mL glacial acetic acid and 2.56 mL 1N HCl) was added to each well followed by additional 4-hour incubation 37°C. Absorbance values were measured using a SpectraMax M3 plate reader (Molecular Devices) at 570 and 630 nm. Viability curve values were generated using the nonlinear regression (curve fit) function in Prism (GraphPad Software). For drug synergy experiments, tagraxofusp was diluted serially 5-fold from 1 μg/mL to 64 pg/mL in rows of a 96-well plate. Drug no. 2 was added and diluted serially 5-fold in columns. The setup yielded 96 data points (1 control [no drug], 7 doses of tagraxofusp only, 11 doses of drug no. 2 only, and 77 different drug concentration combinations). Each plate was prepared in triplicate and after 72 hours, an MTT assay was performed as above. The data were analyzed for synergy using the Compusyn software (http://www.combosyn.com/index.html). BH3 profiling was performed as previously described (22).
WES
Exome sequencing was performed using the Agilent SureSelect All Exon V2 probe–based capture using an in-solution hybrid selection method to generate Illumina exome sequencing libraries (38). Pooled libraries were normalized to 2 nM and denatured using 0.2N NaOH prior to sequencing. Flowcell cluster amplification and sequencing were performed according to the manufacturer’s protocols using the HiSeq 2500. Each run was a 76-bp paired-end with a dual 8-base index barcode read. Data were analyzed using the Broad Picard Pipeline, which includes de-multiplexing and data aggregation. Sequencing data were aligned and processed including mutation calling as previously described (39).
RNA-seq and analysis
Total RNA was quantified using the Quant-iT RiboGreen RNA Assay Kit and normalized to 5 ng/μL. Two hundred nanograms of each sample was transferred into library preparation, which was an automated variant of the Illumina TruSeq Stranded mRNA Sample Preparation Kit. This method preserves strand orientation of the RNA transcript. It uses oligo(dT) beads to select mRNA from the total RNA sample. It was followed by heat fragmentation and cDNA synthesis from the RNA template. The resultant cDNA then underwent library preparation (end repair, base ‘A’ addition, adapter ligation, and enrichment) using indexed adapters for multiplexing. After enrichment the libraries were quantified by qPCR using the KAPA Library Quantification Kit for Illumina Sequencing Platforms and then pooled equimolarly. The entire process was in 96-well format and all pipetting was done by either Agilent Bravo or Hamilton Starlet. Pooled libraries were normalized to 2 nM and denatured using 0.1N NaOH prior to sequencing. Flowcell cluster amplification and sequencing were performed according to the manufacturer’s protocols using the HiSeq 2500. Each run was a 101-bp paired-end with an 8-base index barcode read. Data were analyzed using the Broad Picard Pipeline, which includes de-multiplexing and data aggregation. RNA-seq reads were aligned to the B37 version of human genome using TopHat version 1.4. Gene level read count values and fragments per kilobase of transcript per million mapped reads (FPKM) were determined using the RNA-SeQC pipeline (40). Differential gene expression analysis was performed using DESeq2 (41) with parameter minReplicatesForReplace=Inf to keep outlier genes. For unbiased genome-wide differential expression analysis, we excluded genes with low statistical significance (Padj. > 0.1), effect size (absolute mean log2[fold change] < 1), or expression level (maximum log2[FPKM] < 1) across experimental conditions. Data are deposited in the NCBI’s Gene Expression Omnibus (GEO GSE131147).
Western blotting
Lysates were prepared from cell lines in RIPA buffer (Boston BioProducts, BP-115) with protease inhibitor cocktail (Thermo Fisher Scientific, 862209) and sonicated before quantification by BCA assay (Thermo Fisher Scientific, 23225). Samples were prepared with 15 μg of total protein and SDS-Sample Buffer (Boston BioProducts, BP-110R), boiled for 5 minutes at 98°C, and recovered by spinning at 12,000 g for 5 minutes at 4°C before loading onto the gel. The gel was run for 80 minutes at 120 V in SDS-Running Buffer (Boston BioProducts, BP-177) before being transferred to a PVDF membrane (Bio-Rad, 1620177) for 80 minutes at 100 V. Blots were blocked in 5% dried milk (AppliChem, A0830) in Tris-buffered saline (TBS) with 0.1% Tween for 1 hour before being incubated overnight in antibodies recognizing DPH1 (Abcam, ab185960, 1:1000), eEF2 (Abcam, 40812, 1:1000), or β-actin (Sigma-Aldrich, A5441, 1:10,000). The blots were washed 3 times in TBS with 0.1% Tween before being incubated with either rabbit (Santa Cruz Biotechnology, SC-2004) or mouse (Santa Cruz Biotechnology, SC-2005) secondary HRP-conjugated antibodies before being imaged using ECL substrate (Bio-Rad, 170-5061) on an ImageQuant LAS-4000 (GE Healthcare, 28-9607-59AB).
In vitro ADP-ribosylation assay for detection on protein blot
Lysates were prepared from cell lines in RIPA buffer (Boston BioProducts, BP-115) with protease inhibitor (Thermo Fisher Scientific, 1862209) and sonicated before quantification with BCA assay (Thermo Fisher Scientific, 23225). Samples were prepared using 50 μg total protein, 100 ng tagraxofusp, and 5 μM biotinylated NAD+ (Trevigen, 460-500-01) in ADP-Ribosylation Buffer (final concentrations: 20 mM Tris-HCl [pH 7.4], 1 mM EDTA, 50 mM DTT), to 30 μL total volume and incubated at room temperature for 30 minutes. SDS-Sample Buffer (Boston BioProducts, BP-110R) was then added to each sample and then boiled at 98°C for 5 minutes, recovered by spinning at 12,000 g for 5 minutes at 4°C, and the entire sample was loaded onto an SDS-PAGE gel. Gels were run, transferred, and blocked as per above in the Western blot methods. For biotinylated ADP detection, blots were incubated with streptavidin-HRP (Abcam, ab7403) at 1:1000 dilution, with chemiluminescence detection by ECL as above.
CRISPR-Cas9 gene targeting and treatment with tagraxofusp
Cell lines stably expressing the Cas9 nuclease were generated by infection with the empty lentiCRISPRv2 lentivirus (not containing an sgRNA guide) using standard methods (https://www.addgene.org/viral-vectors/lentivirus/lenti-guide/). Cells were selected in puromycin and FLAG-Cas9 expression was confirmed by Western blot. Next, sgRNAs (see target sequences below) were subcloned into a lentiviral expression vector that coexpresses GFP (pLKO5.sgRNA.EFS.GFP, Addgene) via the BsmBI restriction site per standard methods as above. Cas9-expressing cell lines were infected at a density of 250,000 cells in 1.5 mL media in the presence of 4 μg/mL polybrene (Santa Cruz Biotechnology, SC-134220). Forty-eight hours after infection, cell lines were analyzed via a CytoFlex flow cytometer (Beckman Coulter, B53012) for infection efficiency. CRISPR-Cas9 cell lines were then analyzed by Western blotting or treated with tagraxofusp at 1 μg/mL and analyzed by flow cytometry every 24 hours for 6 days.
sgRNA sequences.
hDPH1_sgRNA_2_For, CACCGTTCACGGAGGCCGAAGTGA; hDPH1_sgRNA_2_Rev, AAACTCACTTCGGCCTCCGTGAAC; hDPH1_sgRNA_3_For, CACCGTGATGGGTGACGTGACCTAC; hDPH1_sgRNA_3_Rev, AAACGTAGGTCACGTCACCCATCAC; hDPH1_sgRNA_5_For, CACCGCCTCTTTGCCTGTACCATTG; hDPH1_sgRNA_5_Rev, AAACCAATGGTACAGGCAAAGAGGC; hDPH1_sgRNA_6_For, CACCGCCTTTCCAAGATATCCACAA; hDPH1_sgRNA_6_Rev, AAACTTGTGGATATCTTGGAAAGGC.
Control guides.
NTG1_For, CACCGACGGAGGCTAAGCGTCGCAA; NTG1_Rev, AAACTTGCGACGCTTAGCCTCCGTC; NTG2_For, CACCGCGCTTCCGCGGCCCGTTCAA; NTG2_Rev, AAACTTGAACGGGCCGCGGAAGCGC.
DPH1 re-expression experiments
A codon-optimized HA-tagged human DPH1 DNA fragment was synthesized by Twist Bioscience designed using their Codon Optimization for Synthesis software. The codon-optimized sequence was ATGTACCCGTACGATGTGCCGGACTACGCAGCCGCTGCAGGAGGGCGCAGACAGGTCATGGCGGCGCTTGTAGTCTCTGGAGCGGCCGAACAGGGCGGGCGCGATGGCCCGGGTAGGGGGCGAGCACCAAGGGGGAGAGTGGCTAACCAGATCCCCCCAGAAATACTCAAGAATCCACAGCTTCAGGCCGCTATTCGAGTTCTTCCTTCTAATTACAATTTTGAAATTCCCAAAACAATTTGGAGAATTCAGCAAGCCCAAGCCAAAAAAGTGGCCCTGCAGATGCCGGAGGGCCTTTTGCTCTTCGCGTGTACAATTGTGGATATTTTGGAGCGGTTTACAGAAGCGGAGGTGATGGTGATGGGGGACGTAACATATGGCGCGTGTTGCGTAGACGATTTTACGGCCAGAGCGTTGGGCGCGGATTTCCTGGTCCATTATGGTCACAGCTGCCTGATTCCAATGGATACGAGTGCTCAAGACTTCCGAGTCTTGTACGTGTTTGTCGATATTCGGATTGATACAACACACCTGCTCGATAGCCTCCGCCTTACGTTTCCACCGGCCACCGCCCTGGCTTTGGTTTCTACCATACAATTCGTTTCAACTCTCCAGGCGGCAGCGCAGGAACTTAAAGCAGAATATAGAGTAAGTGTTCCGCAGTGCAAACCCCTTTCTCCCGGTGAAATACTCGGCTGTACGTCCCCTCGACTCTCTAAAGAGGTTGAGGCTGTCGTCTATCTTGGTGATGGGAGATTCCATCTGGAGAGTGTCATGATAGCAAATCCCAATGTTCCAGCTTATAGGTACGACCCATACTCAAAAGTTCTTTCCCGAGAGCATTACGACCACCAGAGGATGCAAGCGGCGCGGCAGGAAGCAATTGCTACTGCACGCTCAGCCAAATCTTGGGGGCTGATATTGGGTACATTGGGGCGGCAGGGTTCCCCAAAAATTCTCGAACACCTTGAGAGTCGCCTTCGCGCTCTCGGCCTGTCATTCGTCAGACTTTTGTTGAGTGAGATTTTTCCATCCAAACTTTCATTGCTGCCGGAAGTGGACGTATGGGTACAGGTTGCGTGCCCCAGACTCTCCATTGATTGGGGGACGGCTTTCCCTAAACCGCTCCTCACACCCTATGAAGCCGCGGTCGCACTCCGAGACATCTCATGGCAACAACCATACCCCATGGATTTCTATGCCGGGAGTAGTTTGGGTCCCTGGACTGTCAACCATGGTCAAGATAGGAGGCCGCATGCGCCTGGTAGACCCGCACGGGGCAAAGTCCAAGAGGGTAGTGCCCGACCTCCCAGTGCAGTAGCCTGTGAAGATTGCTCTTGTAGGGATGAGAAAGTCGCACCTCTCGCCCCTTGA.
Full-length and an N-terminally truncated isoform missing the first 75 amino acids of DPH1 (amplified via PCR from the above sequence to insert a start site followed by the CCG codon [in bold text] through the stop codon) were subcloned into the pDONR221 vector (Thermo Fisher Scientific). N-terminal truncation of DPH1 is known to abolish its activity and cannot rescue DT sensitivity in DPH1-knockout yeast cells (20). cDNAs were then transferred to a lentiviral doxycycline-inducible expression vector (pCW57.1, Addgene) by gateway cloning. Lentivirus was produced in 293T cells using standard procedures and parental or tagraxofusp-resistant cells were spinfected in the presence of polybrene. After 48 hours, puromycin was added to the cultures at a final concentration of 1 μg/mL. After 5 days, puromycin was removed and doxycycline was added to the cultures at 200 ng/mL to induce expression of the transduced cDNAs. After 24 hours, lysates were harvested for Western blotting and in vitro ADP-ribosylation activity assay, and for tagraxofusp cytotoxicity assays.
DNA methylation analysis
DNA was prepared from each cell line using an All Prep DNA/RNA Mini kit (Qiagen, 80204). Bisulfite modification, PCR amplification, and pyrosequencing to quantitate the CpG methylation fraction was performed by EpigenDx using the following assays: AD2737-FS, spanning –265 to –191 from the TSS, ADS2738-FS2 (–78 to +26 from the TSS), and ADS2738-FS3 (+49 to +108 from the TSS). Data are expressed as percentage of methylated cytosine per total cytosine at each CpG locus.
qRT-PCR
RNA was extracted from cell lines using TRIzol (Life Technologies, 15596018) and quantified via NanoDrop. The High-Capacity cDNA Reverse Transcriptase Kit (Thermo Fisher Scientific, 4368814) was used to generate cDNA from each cell line. qRT-PCR was then performed for human DPH1, using GAPDH as the internal control, using the primers below, and SYBR Green PCR master mix per the manufacturer’s instructions (Thermo Fisher Scientific, 4367659). Relative quantitation was calculated using the ΔΔCt method. DPH1 For, CTGGTCGTATCCGGGGC; and DPH1 Rev, CAGCTGAGGGTTCTTCAGGA; GAPDH For, CCTGCACCACCAACTGCTT; GAPDH Rev, CCATCACGCCACAGTTTCC.
Long-term azacitidine treatment
Parental and tagraxofusp-resistant cells were set up in triplicate for 3 cohorts of treatment: no treatment, tagraxofusp (resistant clones only), and azacitidine (all clones). The cells were treated for 2 days with either 1 μg/mL tagraxofusp or 300 nM azacitidine (Selleckchem, 320-67-2), respectively. After 2 days, the media were removed and replaced with fresh media for 2 days. This cycle was repeated 4 times (2 days on/2 days off, total of 16 days), for a total of 4 treatments. After the final drug treatments, fresh media were added, and cells were allowed to recover for 1 week before performing assays.
PDX experiments
BPDCN PDXs were generated as previously described (27). For PDX experiments in this study, 8-week-old female NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; Jackson Laboratories, 005557) mice were injected intravenously with 1 × 106 leukemic splenocytes in “passage 1” (i.e., PDXs that had already grown in a first round of NSG animals). Three to 5 recipients of each PDX were sampled every 1–2 weeks via tail vein peripheral blood flow cytometry to quantitate human CD45+CD123+ cells. When the average peripheral blood disease burden was greater than 0.2%, all animals were randomly assigned to vehicle or treatment arms. In the single-agent experiments, tagraxofusp was diluted in PBS to 10 μg/mL and was administered intraperitoneally (i.p.) at 0.1 mg/kg/d for 5 days. A subset of animals was sacrificed 7 days after treatment for pharmacodynamic studies and pathological analysis. Remaining mice were monitored every 1–2 weeks by peripheral blood sampling. In a subset of animals in the tagraxofusp treatment group, when average disease burden exceeded 5%, they were randomly assigned to re-treatment with vehicle or tagraxofusp as above. In the combination experiment, animals were transplanted with PDX cells and monitored for disease as above. When average peripheral blood disease burden was greater than 0.2%, all animals were randomly assigned to vehicle or treatment arms. Here, tagraxofusp was given i.p. at 0.1 mg/kg/d for 5 days starting on day 1 and azacitidine was given i.p. at 2.5 mg/kg/d for 7 days starting on day 1. On day 30, all mice received a second cycle of treatment at a reduced dose by treatment arm: tagraxofusp at 0.05 mg/kg/d for 4 days and azacitidine at 1.25 mg/kg/d for 4 days.
Histochemical ADP-ribosylation assay on slides
Cells were adhered to glass slides using a Cytospin centrifuge (300 g for 3 minutes), fixed in 3% paraformaldehyde in PBS for 15 minutes at room temperature, and then rinsed 5 times in PBS. Next, slides were incubated in cold methanol at –20°C for 1 minute and then rinsed 5 times in PBS. Next, slides were incubated in 3% hydrogen peroxide in water for 10 minutes at room temperature, washed 4 times for 5 minutes in PBS with 0.1% Tween-20 (PBS-T), incubated with protein block (Leica, RE7102) for 30 minutes at room temperature, and then washed 4 times for 5 minutes in PBS-T. For enzymatic labeling, cells were incubated in a buffer containing 20 mM Tris-Cl pH 7.4, 50 mM DTT, 50 μM biotinylated NAD+ (Trevigen, 4670), with or without 20 ng/μL tagraxofusp at 37°C for 1 hour in a humid box. After incubation, slides were washed 4 times for 5 minutes each in PBS-T, and then incubated with streptavidin-poly HRP (Thermo Fisher Scientific, 21140) in PBS containing 1% BSA for 30 minutes at room temperature in a humid box. Slides were washed 4 times for 5 minutes each in PBS-T and treated with DAB chromogen (Vector Labs, SK-4105) diluted in DAB diluent per the manufacturer’s instructions. Cells were washed 4 times for 5 minutes each in PBS-T, counterstained with hematoxylin (Leica, RE7107) for 3 minutes, washed in water once, and then dehydrated (washed in tap water 3 times, 100% ethanol 4 times, and xylene 4 times) and air dried for at least 15 minutes. Slides were mounted in Cytoseal mounting media (Thermo Fisher Scientific, 8312-4) under a glass coverslip and visualized.
Flow cytometry–based single-cell ADP-ribosylation assay
Cells were washed with PBS and stained with Zombie NIR fixable viability dye (Biolegend, 423105) diluted 1:250 in PBS for 15 minutes at room temperature. Cells were washed in FACS buffer (PBS with 2% FBS) for 5 minutes, followed by staining with fluorescent antibodies, anti–human CD45-FITC (Biolegend, 368508) and anti–human CD123-APC (BD Biosciences, 658172), diluted 1:100 in FACS buffer for 30 minutes at 4°C, followed by washing in FACS buffer for 5 minutes. Cells were fixed in 3% paraformaldehyde diluted in PBS for 20 minutes at room temperature, followed by washing with PBS for 5 minutes. Next, cells were permeabilized with 1× perm/wash buffer containing saponin (Thermo Fisher Scientific, BDB554723) per the manufacturer’s instructions for 20 minutes at 4°C, washed in perm/wash buffer, blocked with streptavidin block from the endogenous biotin-blocking kit (Thermo Fisher Scientific, E21390) diluted 1:1 in perm/wash buffer for 20 minutes at room temperature, washed once in perm/wash, and then blocked with biotin block (Thermo Fisher Scientific, E21390) diluted 1:1 in perm/wash buffer for 20 minutes at room temperature. Cells were washed with perm/wash and then incubated with protein block (Leica, RE7102) for 30 minutes at room temperature diluted 1:1 in perm/wash buffer. For the enzymatic reaction, cells were incubated in the tagraxofusp reaction cocktail (0–12 ng/μL tagraxofusp, as indicated; 1 μM biotinylated NAD+ [Trevigen, 4670]; 50 mM DTT; in perm/wash buffer containing 20 mM Tris-HCl, pH 7.4, and 1 mM EDTA) for 30 minutes at room temperature. Cells were washed twice with perm/wash buffer, incubated with streptavidin-BV510 diluted 1:100 in perm/wash buffer for 30 minutes at 4°C, washed in FACS buffer once, and then detected on a Cytoflex flow cytometer (Beckman Coulter, B53012).
Statistics
Statistical analyses were performed using Prism software (GraphPad) or DEseq2 for RNA-seq, as indicated. Data are plotted as mean ± SD, except as where indicated in legends. Box-and-whisker plots show a horizontal bar at the median, a box at the 25th–75th percentile bounds, and whiskers at the 5%-95% percentiles. Statistical significance calculations on pairwise comparisons were performed using 2-tailed t tests. When more than one sample was compared to the same control, Dunnett’s test was used to correct for multiple comparisons. Survival analyses were performed on curves generated using the Kaplan-Meier method and groups were compared using the log-rank test. A P value less than 0.05 was considered significant.
Study approval
All experiments were approved by the Dana-Farber Institutional Animal Care and Use Committee (IACUC). All human studies were approved by the appropriate Institutional Review Board (IRB) and written informed consent was obtained from all participants prior to inclusion in the study.
Author contributions
KT, TP, JS, ALC, KLJ, CAJ, JM, JCA, CMJ, and AAL designed and conducted experiments. KT, TP, JS, MG, AL, JWC, OP, DMW, JM, CMJ, and AAL analyzed data. RWL and CLB provided reagents. KT, TP, DMW, JM, JCA, CMJ, and AAL wrote the manuscript. For co–first authorship order assignment, KT performed and also led TP and JS in methods optimization and execution of the majority of experiments.
Supplementary Material
Acknowledgments
This work was supported by Ludwig Cancer Research (to TP, JS, AL, JCA, and AAL), National Cancer Institute grant CA066996 (to AL), Stand Up to Cancer and The V Foundation (TVF) SU2C-TVF Convergence Scholar Awards grant D2015-037 and Ramon y Cajal Programme, Ministerio de Economia y Competitividad grant RYC-2015-18357 (to JM), the Gerstner Family Foundation (to CMJ), Stemline Therapeutics (to AAL), National Cancer Institute grant CA225191-01 (to AAL), and the Doris Duke Charitable Foundation (DDCF) grant 2017065 (to AAL). No DDCF funds were used in animal experiments.
Version 1. 08/22/2019
In-Press Preview
Version 2. 10/14/2019
Electronic publication
Version 3. 11/01/2019
Print issue publication
Footnotes
JWC’s present address is: Department of Pathology and Centre for Lymphoid Cancer, BC Cancer, Vancouver, Canada.
JM’s present address is: Institute for Bioengineering of Catalonia, Barcelona, Spain.
Conflict of interest: RWL and CLB are full-time paid employees of Stemline Therapeutics. TP is currently a full-time paid employee of AbbVie but was not during the time this work was done. CMJ is currently a full-time paid employee of Novartis but was not during the time this work was done. AL receives research support from AbbVie, AstraZeneca, and Novartis; and is a cofounder or advisory board member of Zeno Pharmaceuticals, Vivid Bioscience, Flash Therapeutics, and Dialectic Therapeutics. JM is a consultant for Oncoheroes Biosciences and Vivid Biosciences. AAL receives research support from AbbVie and Stemline Therapeutics and is a consultant for N-of-One. JCA and AAL have applied for a patent regarding biomarkers and methods to overcome resistance to bacterial toxin-based therapeutics (PCT/US2018/058222).
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(11):5005–5019.https://doi.org/10.1172/JCI128571.
See the related Commentary at Hitting the bullseye with a nonlethal payload: resistance in CD123-positive malignancies.
Contributor Information
Katsuhiro Togami, Email: Katsuhiro_Togami@DFCI.HARVARD.EDU.
Timothy Pastika, Email: timpastika@gmail.com.
Jason Stephansky, Email: jason.stephansky@gmail.com.
Mahmoud Ghandi, Email: mghandi@gmail.com.
Amanda L. Christie, Email: alchristie84@gmail.com.
Kristen L. Jones, Email: KristenL_Jones@DFCI.HARVARD.EDU.
Carl A. Johnson, Email: carljohnson930@gmail.com.
Ross W. Lindsay, Email: RLindsay@stemline.com.
Christopher L. Brooks, Email: cbrooks@stemline.com.
Anthony Letai, Email: anthony_letai@dfci.harvard.edu.
Jeffrey W. Craig, Email: jeffrey.craig@bccancer.bc.ca.
Olga Pozdnyakova, Email: opozdnyakova@partners.org.
David M. Weinstock, Email: dweinstock@partners.org.
Joan Montero, Email: jmontero@ibecbarcelona.eu.
Jon C. Aster, Email: jaster@partners.org.
Cory M. Johannessen, Email: cory.johannessen@gmail.com.
Andrew A. Lane, Email: andrew_lane@dfci.harvard.edu.
References
- 1.Jordan CT. Unique molecular and cellular features of acute myelogenous leukemia stem cells. Leukemia. 2002;16(4):559–562. doi: 10.1038/sj.leu.2402446. [DOI] [PubMed] [Google Scholar]
- 2.Testa U, Pelosi E, Frankel A. CD 123 is a membrane biomarker and a therapeutic target in hematologic malignancies. Biomark Res. 2014;2(1):4. doi: 10.1186/2050-7771-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehninger A, et al. Distribution and levels of cell surface expression of CD33 and CD123 in acute myeloid leukemia. Blood Cancer J. 2014;4:e218. doi: 10.1038/bcj.2014.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khoury JD. Blastic plasmacytoid dendritic cell neoplasm. Curr Hematol Malig Rep. 2018;13(6):477–483. doi: 10.1007/s11899-018-0489-z. [DOI] [PubMed] [Google Scholar]
- 5.Garnache-Ottou F, et al. Extended diagnostic criteria for plasmacytoid dendritic cell leukaemia. Br J Haematol. 2009;145(5):624–636. doi: 10.1111/j.1365-2141.2009.07679.x. [DOI] [PubMed] [Google Scholar]
- 6.Sullivan JM, Rizzieri DA. Treatment of blastic plasmacytoid dendritic cell neoplasm. Hematology Am Soc Hematol Educ Program. 2016;2016(1):16–23. doi: 10.1182/asheducation-2016.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordan CT, et al. The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia. 2000;14(10):1777–1784. doi: 10.1038/sj.leu.2401903. [DOI] [PubMed] [Google Scholar]
- 8.Alkharabsheh O, Frankel AE. Clinical activity and tolerability of SL-401 (tagraxofusp): recombinant diphtheria toxin and interleukin-3 in hematologic malignancies. Biomedicines. 2019;7(1):E6. doi: 10.3390/biomedicines7010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.FitzGerald DJ. Targeted diphtheria toxin to treat BPDCN. Blood. 2014;124(3):310–312. doi: 10.1182/blood-2014-06-578633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pemmaraju N, et al. Tagraxofusp in blastic plasmacytoid dendritic-cell neoplasm. N Engl J Med. 2019;380(17):1628–1637. doi: 10.1056/NEJMoa1815105. [DOI] [PubMed] [Google Scholar]
- 11.Frankel AE, et al. Activity of SL-401, a targeted therapy directed to interleukin-3 receptor, in blastic plasmacytoid dendritic cell neoplasm patients. Blood. 2014;124(3):385–392. doi: 10.1182/blood-2014-04-566737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhojwani D, et al. Inotuzumab ozogamicin in pediatric patients with relapsed/refractory acute lymphoblastic leukemia. Leukemia. 2019;33(4):884–892. doi: 10.1038/s41375-018-0265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruella M, Maus MV. Catch me if you can: leukemia escape after CD19-directed T cell immunotherapies. Comput Struct Biotechnol J. 2016;14:357–362. doi: 10.1016/j.csbj.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsherniak A, et al. Defining a cancer dependency map. Cell. 2017;170(3):564–576.e16. doi: 10.1016/j.cell.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceribelli M, et al. A druggable TCF4- and BRD4-dependent transcriptional network sustains malignancy in blastic plasmacytoid dendritic cell neoplasm. Cancer Cell. 2016;30(5):764–778. doi: 10.1016/j.ccell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collier RJ. Understanding the mode of action of diphtheria toxin: a perspective on progress during the 20th century. Toxicon. 2001;39(11):1793–1803. doi: 10.1016/S0041-0101(01)00165-9. [DOI] [PubMed] [Google Scholar]
- 17.Vinante F, Rigo A. Heparin-binding epidermal growth factor-like growth factor/diphtheria toxin receptor in normal and neoplastic hematopoiesis. Toxins (Basel) 2013;5(6):1180–1201. doi: 10.3390/toxins5061180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Su X, Lin Z, Lin H. The biosynthesis and biological function of diphthamide. Crit Rev Biochem Mol Biol. 2013;48(6):515–521. doi: 10.3109/10409238.2013.831023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert LA, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159(3):647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdel-Fattah W, Scheidt V, Uthman S, Stark MJ, Schaffrath R. Insights into diphthamide, key diphtheria toxin effector. Toxins (Basel) 2013;5(5):958–968. doi: 10.3390/toxins5050958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu X, et al. Methylation of the DPH1 promoter causes immunotoxin resistance in acute lymphoblastic leukemia cell line KOPN-8. Leuk Res. 2013;37(11):1551–1556. doi: 10.1016/j.leukres.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryan JA, Brunelle JK, Letai A. Heightened mitochondrial priming is the basis for apoptotic hypersensitivity of CD4+ CD8+ thymocytes. Proc Natl Acad Sci U S A. 2010;107(29):12895–12900. doi: 10.1073/pnas.0914878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montero J, et al. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell. 2015;160(5):977–989. doi: 10.1016/j.cell.2015.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montero J, et al. Blastic plasmacytoid dendritic cell neoplasm is dependent on BCL2 and sensitive to venetoclax. Cancer Discov. 2017;7(2):156–164. doi: 10.1158/2159-8290.CD-16-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ni Chonghaile T, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 2011;334(6059):1129–1133. doi: 10.1126/science.1206727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vo TT, et al. Relative mitochondrial priming of myeloblasts and normal HSCs determines chemotherapeutic success in AML. Cell. 2012;151(2):344–355. doi: 10.1016/j.cell.2012.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Townsend EC, et al. The public repository of xenografts enables discovery and randomized phase II-like trials in mice. Cancer Cell. 2016;29(4):574–586. doi: 10.1016/j.ccell.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasetto M, et al. Whole-genome RNAi screen highlights components of the endoplasmic reticulum/Golgi as a source of resistance to immunotoxin-mediated cytotoxicity. Proc Natl Acad Sci U S A. 2015;112(10):E1135–E1142. doi: 10.1073/pnas.1501958112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreitman RJ, et al. Moxetumomab pasudotox in relapsed/refractory hairy cell leukemia. Leukemia. 2018;32(8):1768–1777. doi: 10.1038/s41375-018-0210-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller F, et al. 5-Azacytidine prevents relapse and produces long-term complete remissions in leukemia xenografts treated with Moxetumomab pasudotox. Proc Natl Acad Sci U S A. 2018;115(8):E1867–E1875. doi: 10.1073/pnas.1714512115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahl S, et al. Loss of diphthamide pre-activates NF-κB and death receptor pathways and renders MCF7 cells hypersensitive to tumor necrosis factor. Proc Natl Acad Sci U S A. 2015;112(34):10732–10737. doi: 10.1073/pnas.1512863112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, et al. Diphthamide modification on eukaryotic elongation factor 2 is needed to assure fidelity of mRNA translation and mouse development. Proc Natl Acad Sci U S A. 2012;109(34):13817–13822. doi: 10.1073/pnas.1206933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Argüelles S, Camandola S, Cutler RG, Ayala A, Mattson MP. Elongation factor 2 diphthamide is critical for translation of two IRES-dependent protein targets, XIAP and FGF2, under oxidative stress conditions. Free Radic Biol Med. 2014;67:131–138. doi: 10.1016/j.freeradbiomed.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Villahermosa D, Knapp K, Fleck O. A mutated dph3 gene causes sensitivity of Schizosaccharomyces pombe cells to cytotoxic agents. Curr Genet. 2017;63(6):1081–1091. doi: 10.1007/s00294-017-0711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen CM, Behringer RR. OVCA1: tumor suppressor gene. Curr Opin Genet Dev. 2005;15(1):49–54. doi: 10.1016/j.gde.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 36.Chen CM, Behringer RR. Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev. 2004;18(3):320–332. doi: 10.1101/gad.1162204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeda T, et al. A novel plasmacytoid dendritic cell line, CAL-1, established from a patient with blastic natural killer cell lymphoma. Int J Hematol. 2005;81(2):148–154. doi: 10.1532/IJH97.04116. [DOI] [PubMed] [Google Scholar]
- 38.Fisher S, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12(1):R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng SY, et al. Targetable vulnerabilities in T- and NK-cell lymphomas identified through preclinical models. Nat Commun. 2018;9(1):2024. doi: 10.1038/s41467-018-04356-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeLuca DS, et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012;28(11):1530–1532. doi: 10.1093/bioinformatics/bts196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.