Abstract
T cells help regulate immunity, which makes them an important target for RNA therapies. While nanoparticles carrying RNA have been directed to T cells in vivo using protein- and aptamer-based targeting ligands, systemic delivery to T cells without targeting ligands remains challenging. Given that T cells endocytose lipoprotein particles and enveloped viruses, two natural systems with structures that can be similar to lipid nanoparticles (LNPs), we hypothesized LNPs devoid of targeting ligands could deliver RNA to T cells in vivo. To test this hypothesis, we quantified how 168 nanoparticles delivered siRNA to 9 cell types in vivo using a novel siGFP-based barcoding system and bioinformatics. We found nanomaterials containing conformationally constrained lipids formed stable LNPs, herein named constrained lipid nanoparticles (cLNPs). cLNPs delivered siRNA and sgRNA to T cells at doses as low as 0.5 mg / kg, and unlike previously reported LNPs, did not preferentially target hepatocytes. Delivery occurred via a chemical composition-dependent, size-independent mechanism. These data suggest the degree to which lipids are constrained alters nanoparticle targeting. They also suggest natural lipid trafficking pathways can promote T cell delivery, offering an alternative to active targeting approaches.
Main Text
T lymphocytes help regulate immune responses, which makes them important drug targets. For example, antibodies that block T cell CTLA-4 or PD-1 signaling can drive anti-tumor responses[1]. However, antibodies target druggable proteins, which constitute approximately 15% of total proteins. By contrast, siRNA can inhibit the translation of any gene; many ‘undruggable’ proteins were recently implicated in T cell-mediated immunity[2].
Clinically relevant RNA delivery to cells other than hepatocytes[3] remains challenging[4]. However, there have been advances in T cell siRNA delivery. For example, siRNA was delivered to T cells using a single chain antibody linked to a cationic peptide; this led to gene silencing at 5 mg / kg[5]. In a second example, nanoparticles were coated with anti-CD4 antibodies, leading to 20% target gene silencing at 1 mg / kg doses[6]. More recently, LNPs that target hepatocytes were re-targeted to T cells by coating them with CD4 antibodies, leading to 50% in vivo T cell gene silencing at 6 mg / kg doses[7]. These papers (and others)[8] achieve T cell delivery using peptide-, protein-, or aptamer-based targeting ligands, and more generally, ligand-based targeting is used throughout nanomedicine. However, ligands can make reproducible manufacturing at human scales more challenging[9].
One alternative to active targeting is to exploit endogenous lipid trafficking; notably, the only FDA approved RNA nanoparticle therapy[3] utilizes LNPs without ligands that are trafficked to hepatocytes via endogenous cholesterol transport[10]. Natural trafficking has not been exploited to promote nanoparticle delivery to T cells, yet these cells can interact with viruses and lipoprotein particles, which can have diameters similar to LNPs [11, 12]. We therefore hypothesized that LNPs could interact with T cells without targeting ligands. To test this hypothesis, we quantified how well 168 LNPs delivered siRNA to 9 cell types in vivo. Using traditional 1-by-1 in vivo approaches, this would require FACS (fluorescent activated cell sorting) analysis of hundreds of mice. Thus, to generate large scale in vivo data, we developed a siGFP / DNA barcode-based screening system. This system quantifies how over 100 nanoparticles deliver siGFP to any desired combination of on- and off-target cell types in vivo. This in vivo approach contrasts with previous LNP research, which utilizes in vitro screening to select a small number of nanoparticles for in vivo evaluation[13]. The approach is supported by evidence that in vitro nanoparticle delivery can be a poor predictor of in vivo nanoparticle delivery[14]. By combining high throughput in vivo analyses and bioinformatics, we found that a new class of materials, named conformationally constrained lipids, can form stable LNPs. We also found these ‘constrained LNPs’ (cLNPs) can deliver siRNA to T cells in vivo. These data demonstrate that the conformational state of lipids can alter LNP tropism and provide intriguing preliminary evidence that natural trafficking can promote T cell delivery, offering a potential alternative to active targeting.
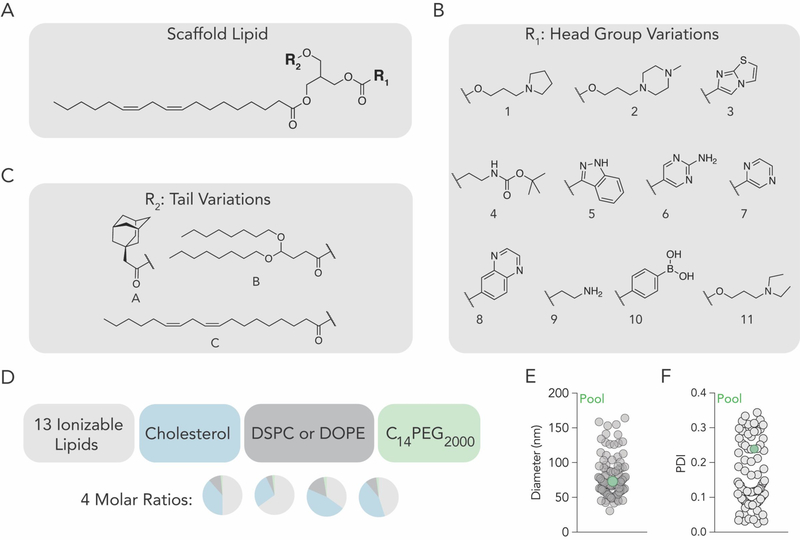
We synthesized 13 chemically diverse lipids containing amines or boronic acid. The library was constructed to investigate whether the structure of the (i) head groups and (ii) lipid tail affected delivery. We purified a ‘scaffold’ containing the unsaturated lipid linoleate and two ester bonds (Figure 1A, Figure S1A, Supporting Information). This scaffold did not have any ionizable components; we attached head group variants to the reactive sites, in order to create chemical diversity. At reactive site 1, we added 11 head groups (labeled 1–11) via esterification, resulting in head groups linked by ester or carbonate linkages, respectively (Fig 1B). At reactive site 2, we added 3 lipid tails (labeled A-C) with diverse structures (Figure 1C) via esterification. Tail A contained adamantane, a constrained lipid with a defined ‘armchair’ structure. Tail B contained 2 lipid tails, bringing the total number of tails to 3. The control tail, C, was linoleate; this created a construct with two identical lipid tails. After synthesis, we confirmed the chemical structure of all 13 lipids using high resolution mass spectroscopy or 1H-NMR (Figure S1B–D, Supporting Information). We named each lipid with the nomenclature ‘head group number – tail letter’ (e.g. 11-A).
Figure 1.
Lipid nanoparticles containing a constrained lipid can form stable LNPs. (A) Ionizable lipid scaffold to which (B) tail variants and (C) head group variants were added. (D) Using 4 molar rations, each of the 13 ionizable lipids were formulated with cholesterol, lipid-PEG, and either DSPC or DOPE, to create 104 distinct LNPs. (E) Hydrodynamic diameter and (F) polydispersity index of all formulated LNPs, measured individually.
We then investigated whether the 13 ionizable lipids formed stable LNPs. We measured the hydrodynamic diameter of LNPs carrying a siRNA targeting GFP[12] (siGFP) as well as a DNA barcode[15]; the LNPs were formulated using microfluidics[16]. The siGFP was chemically modified to reduce immunostimulation and enhance on-target silencing via preferential antisense RISC loading (Figure S2A, Supporting Information). To minimize the chance our results were affected by other constituents added to the LNP, we added previously validated constituents: C14PEG2000, unmodified cholesterol, and either 1–2-distearoyl-sn-glycero-3-phosphocholine (DSPC) or 1–2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) (Figure 1D). As a control to ensure our results were not affected by the molar ratio of the 4 components, we formulated each of the 13 lipids with 2 phospholipids and 4 molar ratios, producing a total of 104 chemically distinct LNPs (Figure 1D). Encouragingly, 100 of the 104 LNPs formed small, monodisperse populations, as evidenced by hydrodynamic diameter and polydispersity index; these 100 LNPs were pooled together. The diameter for individual LNPs varied between 30 and 170 nm. As a control, we also measured the diameter and polydispersity of the pooled LNPs. We found them to be 76 nm (Figure 1E) and 0.23 (Figure 1F), respectively, which was within the range of the individuals comprising the pool. We then analyzed the hydrodynamic diameter as a function of ionizable lipid (Figure S2B, Supporting Information), molar ratio of the four constituents (Figure S2C, Supporting Information), and the type of phospholipid (DSPC / DOPE) added to the formulation (Figure S2D, Supporting Information). In all cases, the average diameter varied between 50 and 100 nm. These data led us to conclude these lipids could form LNPs with hydrodynamic diameters similar to lipoproteins and viruses.
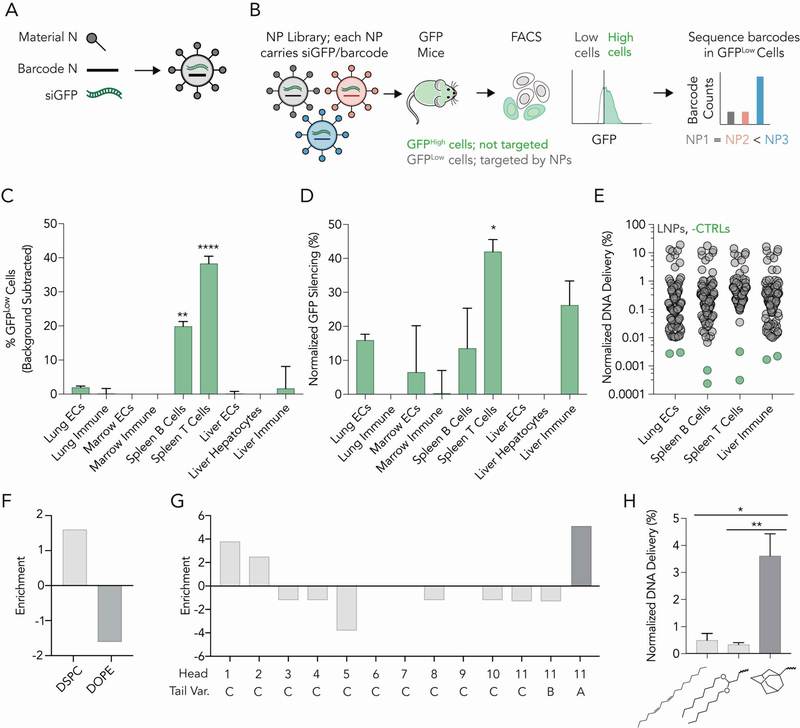
We evaluated how each LNP delivered siRNA to target cells (in this case, T lymphocytes) as well as 8 off-target cell types in vivo (Fig. S3, Supporting Information). Our approach utilizes DNA barcodes and siGFP, to evaluate how many distinct LNPs functionally delivered siGFP, in any combination of target cells, in a single mouse. We formulated LNP-1, with chemical structure 1, to carry siGFP and DNA barcode 1; we separately formulated LNP-N, with chemical structure N, to carry siGFP and DNA barcode N (Figure 2A). We included naked barcodes as a negative control[14], since DNA does not readily enter cells. We pooled the LNPs together, and intravenously injected them into mice that constitutively express GFP under a CAG promoter (Figure 2B). The GFP acted as the functional delivery readout; LNPs which functionally delivered siGFP into the cytoplasm would have lower GFP protein expression. Thus, 3 days after injecting mice, we isolated GFPLow cells using fluorescence activated cell sorting (FACS), and deep sequenced the DNA barcodes in GFPLow cells. In this way, we isolated barcodes co-localized with cells in which GFP protein silencing occurred. After sequencing the barcodes, we calculated normalized delivery, i.e., the number of barcodes for each individual barcode, divided by the total number of barcodes within that sample. Normalized delivery is analogous to counts per million in RNAseq experiments[17]. Since GFP is expressed in all cell types, this assay allows us to (i) compare GFP knockdown in any combination of on- / off-target cells and (ii) identify LNPs that co-localized in GFPLow cells.
Figure 2.
A high-throughput siRNA screen for in vivo activity reveals LNPs with constrained lipids have biological activity in T cells. (A) Nanoparticles were formulated to carry a distinct DNA barcode and siGFP. (B) Of the 104 LNPs formulated, we pooled 100 stable LNPs together, and administered them to mice expressing GFP. After 3 days, we isolated GFPLow cells and sequenced the DNA barcodes within that population. (C) Percent GFPLow cells in 9 cell types. Two-way ANOVA, **P<0.01, ****P<0.0001. (D) Normalized decrease in GFP MFI in 9 cell types. Two-way ANOVA, *P<0.05. (E) Normalized DNA delivery in lung endothelial cells, splenic B and T cells, as well as liver immune cells. (F) Enrichment of DSPC-containing LNPs in splenic T cells. (G) Enrichment for each of the 13 ionizable lipids. (H) Normalized DNA delivery of LNPs formulated with head group 11 and tail L, S, or A. One-way ANOVA, *P<0.05, **P<0.01.
Three days after injecting a total dose of 1.5 mg / kg into mice (100 distinct LNPs, 0.015 mg / kg / particle on average), we quantified GFP silencing in 9 cell types. Compared to PBS treated mice, there was an increased number of GFPLow splenic B cells and splenic T cells (Figure 2C). The average GFP protein silencing, quantified by mean fluorescent intensity, was greatest in splenic T cells, followed by liver immune cells, splenic B cells, and lung endothelial cells (Figure 2D). Surprisingly, we found no evidence of silencing in hepatocytes (Figure 2C,D), which are preferentially targeted by many LNPs[4]. We then monitored the controls included in our data; we sequenced the GFPLow splenic T cells as well as lung endothelial cells, splenic B cells, and liver immune cells. In all 4 cell types, the normalized delivery of both negative controls (naked barcodes) was lower than barcodes delivered by LNPs, as expected (Figure 2E).
We then performed a large in vivo structure-function analysis, using the DNA sequencing data to evaluate whether any nanoparticle material properties promoted delivery to splenic T cells. First, we calculated the enrichment for different nanoparticle properties (Figure S4, Supporting Information). Briefly, we calculated the odds a nanoparticle with a particular property would show up by chance in particles that (i) performed in the top 10%, and separately, (ii) particles that performed in the bottom 10%. Nanoparticles formulated with DSPC were enriched in effective particles (i.e., particles with high normalized delivery), whereas nanoparticles formulated with DOPE were enriched in particles that performed poorly (Figure 2F). To validate these results, we then compared the normalized delivery for all LNPs formulated with DSPC and DOPE, respectively, and found that DSPC-containing LNPs outperformed DOPE-containing LNPs (Figure S5A, Supporting Information). As an additional validation, we calculated the normalized delivery of ‘paired’ LNPs, i.e., LNPs that had the same molar ratios and ionizable lipids (but different phospholipids). We found that DSPC LNPs outperformed their paired DOPE containing LNP (Figure S5B, Supporting Information). Based on these data, we concluded that the phospholipid contained within the LNP affected splenic T cell delivery. We therefore limited future chemical analysis to DSPC containing formulations.
We then analyzed enrichment for the 13 ionizable lipids, in order to evaluate the effect of the lipid tail and head group, and found 3 ionizable lipids were enriched (Figure 2G). As a control, we plotted enrichment of each headgroup versus headgroup molecular weight, hydrophobicity (LogP), and polar surface area
(Figure S6A–D, Supporting Information); we did not observe correlations between these traits and enrichment. The lipid that was most enriched, 11-A, contained a conformationally constrained adamantane tail. Notably, the enrichment (Figure 2G) and normalized DNA delivery (Figure 2H) of 11-A was higher than 11-B and 11-C, the two compounds with identical head groups, but unconstrained lipid tails. We then performed a paired analysis using compounds with the same molar ratio and found that adamantane containing tails outperformed other tail structures (Figure S6E, Supporting Information). Finally, as a control, we analyzed how LNP size affected delivery. We plotted normalized T cell delivery against LNP size and found no relationship (Figure S6F, Supporting Information). Taken together, these data provided initial evidence that adamantane-containing LNPs could deliver nucleic acids to T cells in vivo, without the use of a targeting ligand.
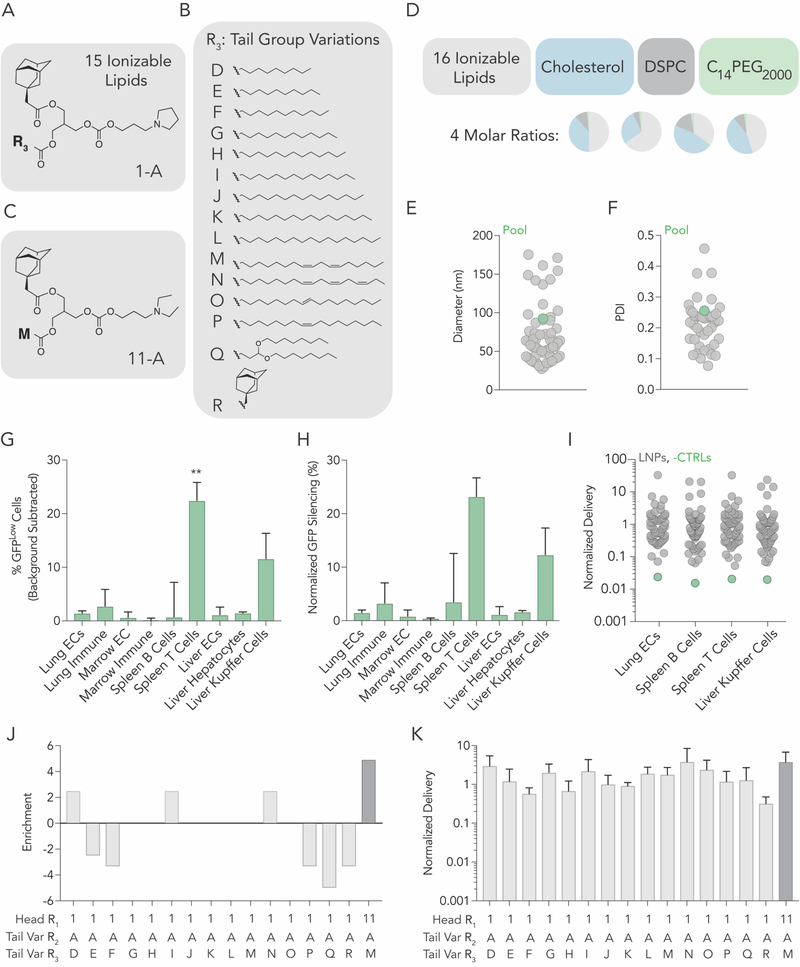
To further investigate the relationship between adamantane and siRNA delivery, we performed a second in vivo screen with adamantane-containing LNPs. The second library was designed based on the results of the first screen. To do this, we first synthesized new ionizable lipids, using the most enriched components from the first screen, head group 1 (Figure 1B) and adamantane tail (tail A, Figure 1C). More specifically, at reactive site 3, we added 15 lipids tails (labeled D-S) with diverse structures (Fig 3A,B) via esterification. We also included the top-ranked LNP from our first screen (Figure 3C). Thus, we synthesized 16 ionizable lipids in total. We named each lipid in the following manner: ‘head group number (R1) – tail letter (R2) – tail letter (R3)’ (e.g. 11-A-M). We formulated each LNP with C14PEG2000, cholesterol, and DSPC. Once again, to control against effects driven by the molar ratio of the components, we formulated all 16 ionizable lipids at 4 molar ratios, for a total of 64 LNPs (Figure 3D). Each LNP carried siGFP and a unique barcode. 55 of the LNPs formed small, monodisperse populations, based on the hydrodynamic diameter and polydispersity index, and were therefore pooled together. The diameter for individual LNPs varied between 20 and 200 nm, with an average of 92 nm (Figure 3E) and an average PDI of 0.20 (Figure 3F). We plotted hydrodynamic diameter for each ionizable lipid (Figure S7A, Supporting Information) and each molar ratio (Figure S7B, Supporting Information), and found that all the compositions formed LNPs within the 20–200 nm range.
Figure 3.
A second high-throughput siRNA screen suggests that adamantanes can deliver siRNA to T cells. (A) 15 cLNPs can be formed with the top performing head group and (B) 15 tail variations. (C) The top performing constrained lipid from screen 1 was included. (D) Using 4 molar rations, each of the 16 ionizable lipids were formulated with cholesterol, lipid-PEG, and DSPC to create 64 distinct LNPs. (E) Hydrodynamic diameter and (F) polydispersity index of all formulated LNPs, measured individually. (G) Percent GFPLow cells in 9 cell types. Two-way ANOVA, *P<0.05. (I) Normalized decrease in GFP MFI in 9 cell types. (E) Normalized DNA delivery in lung endothelial cells, splenic B and T cells, as well as liver Kupffer cells. (J) Enrichment for each of the 16 ionizable lipids. (K) Normalized DNA delivery of each LNP formulated.
We then administered all 55 LNPs intravenously to mice at a total dose of 1.5 mg / kg. Three days later, we quantified GFP silencing in 9 cell types. Once again, we (i) found GFPLow splenic T cells (Figure 3G), (ii) measured GFP protein silencing in splenic T cells (Figure 3H), (iii) found no evidence of silencing in hepatocytes (Figure 3G,H), and (iv) found no relationship between nanoparticle size and delivery (Figure S7C, Supporting Information). As a control, we quantified the normalized delivery of both negative controls (naked barcodes), and found they were lower than barcodes delivered by LNPs (Figure 3I). We then evaluated whether the molecule added to reactive site 3 (Figure 3B) altered delivery when adamantane was present. Using enrichment (Figure 3J), we found reactive group D, I, and M were enriched. We were unable to identify why these variants performed well. However, compared to the top performing cLNP from screen 1, we found delivery was not greatly improved with any alteration to the reactive site 3 molecule (Figure 3K). It is interesting to note that the top performing cLNP from screen 1 (11-A-M) was enriched more than any other cLNP (Figure 3J). These data provided additional evidence to support the hypothesis that LNPs can deliver siRNA to T cells without targeting ligands.
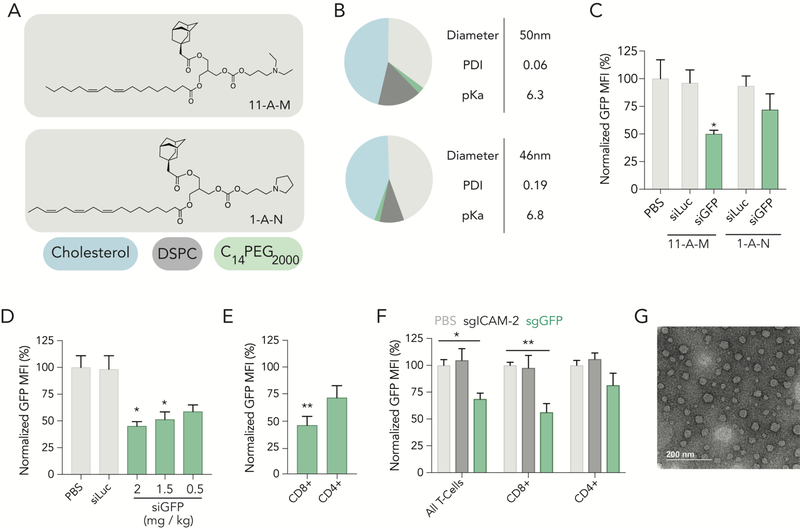
Like all high throughput screening systems, the value of this siGFP / DNA barcode assay is related to its ability to make predictions. We therefore evaluated whether the top-ranked LNPs from our first (11-A-M) and second (1-A-N) screens delivered siRNA in vivo (Figure 4A). We formulated each LNP with siGFP and analyzed physical traits; each LNP had similar hydrodynamic diameter, polydispersity and pKa (Figure 4B). We then intravenously injected mice with 1.5 mg / kg; 3 days later, we isolated cells and measured GFP protein expression. When compared to mice treated with a non-targeting, chemically modified siRNA (siLuc), we found 11-A-M silenced GFP more than 1-A-N (Figure 4C). To validate the activity of 1-A-N, we formulated it with siCD45, and quantified CD45 silencing three days after injecting mice with 1.5 mg / kg; we found statistically significant silencing in T cells (Figure S7D,E). Based on these head-to-head data, we focused on compound 11-A-M. We performed a siRNA gene silencing dose response in vivo, and found that 11-A-M silenced GFP as doses as low as 0.5 mg / kg (Figure 4D). We quantified silencing in subsets of T cells, focusing on CD4+ and CD8+ cells, respectively, and observed more potent protein silencing in CD8+ T cells (Figure 4E). During this experiment, we also evaluated whether 11-A-M delivered siGFP to other common ‘off-target’ cell types by quantifying GFP silencing. Recapitulating observations made in both screens, we observed no significant silencing at doses as high as 1.5 mg / kg in other cell types, including hepatic T cells, bone marrow T cells, and splenic macrophages (Figure S8A-H, Supporting Information). We then quantified on- and off-target biodistribution using QUANT, a highly sensitive digital droplet PCR-based method we recently reported[15]. We formulated 11-A-M to carry the chemically modified QUANT barcode and injected 1.0 mg / kg intravenously. 24 hours later, we isolated cell types using FACS, and measured biodistribution in 8 cell types. We found that 11-A-M distribution was highest in splenic CD8+ T cells, CD4+ T cells, and B cells. Distribution was lower in splenic macrophages as well as endothelial cells, Kupffer cells, hepatocytes, and T cells isolated from the liver (Figure S9A-B, Supporting Information). We also quantified biodistribution mediated by 1-A-N in all 8 cell types, and found that splenic macrophages acted as the primary ‘sink’ for this LNP (Figure S9C–D, Supporting Information). These data suggest that the 11-A-M cLNP may preferentially silence genes in splenic CD8+ T cells.
Figure 4.
cLNPs deliver small RNAs that change gene expression in CD8+ T cells. (A) Structure of ionizable lipid 11-A-M, top performing cLNP from screen 1 and structure of ionizable lipid 1-A-N, top performing cLNP from screen 2. B) Molar composition and characteristics of 11-A-M and 1-A-N. (C) Normalized GFP expression in splenic CD3+ T cells 72 hours after treatment of 2 cLNP carrying siLuc at a dose of 1.5 mg / kg or siGFP at doses of 1.5 mg / kg. (D) Normalized GFP MFI in splenic CD3+ T cells 72 hours after treatment of cLNP carrying either siLuc or siGFP at various doses. One-way ANOVA, *P<0.05. (E) Normalized GFP MFI in splenic CD8+ and CD4+ T cells 72 hours after treatment of cLNP carrying siGFP at a dose of 2.0 mg / kg. unpaired 2-tail t-test, **P<0.01. (F) Normalized GFP MFI in splenic CD3+ T cells as well as CD8+ and CD4+ T cells after treatment of cLNPs carrying sgRNA at a dose of 2.0 mg / kg. Two-way ANOVA, *P<0.05, **P<0.01. (G) Transmission electron microscopy image of 11-A-M formulated to carry siGFP.
Finally, we utilized 11-A-M to facilitate in vivo gene editing in T cells. We formulated it to carry a chemically modified sgRNA targeting GFP (Figure S10, Supporting Information) into mice[18] constitutively expressing Cas9 and GFP. Five days after administration, we quantified GFP expression in CD3+ T cells as well as in CD4+ and CD8+ T Cells. We observed a similar tropism; GFP protein was silenced more robustly in CD8+ than in CD4+ T cells (Figure 4F). We did not observe changes in GFP expression when a control sgRNA targeting the gene ICAM-2 was administered to the mice (Figure 4F). Notably, cLNPs did not lead to weight loss 24 hours after administration in any experiment (Figure S10, Supporting Information). Taken together, these data led us to conclude that cLNPs without targeting ligands can deliver siRNA and sgRNA to splenic T cells. Finally, we analyzed additional traits of 11-A-M cLNPs formulated with siGFP using transmission electron microscopy. We found that the cLNPs formed small, monodisperse spherical structures (Figure 4G) and that cLNPs encapsulated approximately 75% of the formulated RNA (Figure S11, Supporting Information).
Nanoparticles that deliver RNA systemically to non-hepatocytes are difficult to design[4], in large part because (i) there is no high throughput method to study nanoparticle siRNA delivery in vivo and (ii) natural trafficking mechanisms to non-liver cells remain elusive. This universal problem in nanomedicine slows the development of all RNA therapies; currently, scientists perform high throughput nanoparticle assays in vitro, even though cell culture does not[14] recapitulate all the factors that affect delivery in vivo. Notably, the results from our first siGFP screen predicted that preferential T cell delivery would occur; these data were confirmed by the second siGFP screen, and by several in vivo experiments with cLNPs selected from the library. These data suggest that high throughput in vivo siRNA screens can identify nanoparticles with novel tropism. The screening data (Figure 2C,D and Figure 3G,H) suggested that other immune cell subsets may be targeted with LNPs. Although our current data do not allow us to predict how endosomal escape varies between T cells and other immune cells, we believe future studies utilizing traditional cell signaling techniques may elucidate genes and pathways that govern (and differentiate) endosomal escape in subsets of immune cells. Notably, evidence suggests a given immune cell ‘type’ actually encompasses a spectrum of transcriptionally and phenotypically distinct cells[19]. We therefore hypothesize that the relationship between uptake and cytoplasmic delivery will vary along this spectrum.
It is important to acknowledge the limitations of our work. First, the siGFP system will not work with unstable nanoparticles. It is critical to (i) analyze the size and polydispersity of each individual nanoparticle before pooling, (ii) include the naked DNA barcode control, (iii) use untreated GFP mice to gate during FACS, and (iv) individually confirm any lead candidates identified by the screen. Second, although we observed protein silencing in T cells at 0.5 mg / kg doses, we will need to reduce this dose more than 30-fold before it matches the potency of a FDA-approved siRNA delivery vehicle in mice[20]. We anticipate iterative in vivo approaches we recently reported[21] may further improve T cell LNP potency. Lastly, we did not identify the natural trafficking pathways that promoted delivery to T cells.
However, we believe that identifying the genes or pathways that promote LNP delivery to T cells without ligands constitutes an exciting scientific opportunity. In this way, we hope future work on cLNPs will lead to more effective, scalable RNA immunotherapies[22], as well as fundamental advances in our understanding of T cell lipid trafficking. More generally, these data may inspire other efforts to identify natural mechanisms to target cells which – to date – have only been targeted with active ligands.
Experimental Methods
Lipids were attached to the scaffold via esterification. Nucleic acids were diluted in citrate buffer; nanoparticle components were diluted in 100% ethanol. The phases were mixed together via microfluidics[16]. LNP hydrodynamic diameter was measured using dynamic light scattering. pKa was calculated using the TNS assay, as we previously reported[21]. Mice were purchased from Jackson Laboratory; all were 5–8 weeks old. N = 4–5 mice per group were injected intravenously via the lateral tail vein. All animal experiments were performed in accordance with the Georgia Tech IACUC. Sequencing was performed on MiniSeq™ using Illumina protocols.
Supplementary Material
References
- [1].Khalil DN, Smith EL, Brentjens RJ, Wolchok JD, Nat Rev Clin Oncol 2016, 13, 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shifrut E, Carnevale J, Tobin V, Roth TL, Woo JM, Bui CT, Li PJ, Diolaiti ME, Ashworth A, Marson A, Cell 2018, 175, 1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rizk M, Tuzmen S, Pharmacogenomics and personalized medicine 2017, 10, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Lorenzer C, Dirin M, Winkler AM, Baumann V, Winkler J, J Control Release 2015, 203, 1. [DOI] [PubMed] [Google Scholar]
- [5].Kumar P, Ban HS, Kim SS, Wu H, Pearson T, Greiner DL, Laouar A, Yao J, Haridas V, Habiro K, Yang YG, Jeong JH, Lee KY, Kim YH, Kim SW, Peipp M, Fey GH, Manjunath N, Shultz LD, Lee SK, Shankar P, Cell 2008, 134, 577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ramishetti S, Kedmi R, Goldsmith M, Leonard F, Sprague AG, Godin B, Gozin M, Cullis PR, Dykxhoorn DM, Peer D, ACS nano 2015, 9, 6706. [DOI] [PubMed] [Google Scholar]
- [7].Kedmi R, Veiga N, Ramishetti S, Goldsmith M, Rosenblum D, Dammes N, Hazan-Halevy I, Nahary L, Leviatan-Ben-Arye S, Harlev M, Behlke M, Benhar I, Lieberman J, Peer D, Nat Nanotechnol 2018, 13, 214. [DOI] [PubMed] [Google Scholar]
- [8].Smith TT, Stephan SB, Moffett HF, McKnight LE, Ji W, Reiman D, Bonagofski E, Wohlfahrt ME, Pillai SPS, Stephan MT, Nat Nanotechnol 2017, 12, 813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Cheng Z, Al Zaki A, Hui JZ, Muzykantov VR, Tsourkas A, Science 2012, 338, 903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Akinc A, Querbes W, De S, Qin J, Frank-Kamenetsky M, Jayaprakash KN, Jayaraman M, Rajeev KG, Cantley WL, Dorkin JR, Butler JS, Qin L, Racie T, Sprague A, Fava E, Zeigerer A, Hope MJ, Zerial M, Sah DW, Fitzgerald K, Tracy MA, Manoharan M, Koteliansky V, Fougerolles A, Maier MA, Mol Ther 2010, 18, 1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Majorovits E, Nejmeddine M, Tanaka Y, Taylor GP, Fuller SD, Bangham CR, PLoS One 2008, 3, e2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Paunovska K, Gil CJ, Lokugamage MP, Sago CD, Sato M, Lando GN, Gamboa Castro M, Bryksin AV, Dahlman JE, ACS nano 2018, 12, 8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dahlman JE, Barnes C, Khan OF, Thiriot A, Jhunjunwala S, Shaw TE, Xing Y, Sager HB, Sahay G, Speciner L, Bader A, Bogorad RL, Yin H, Racie T, Dong Y, Jiang S, Seedorf D, Dave A, Singh Sandhu K, Webber MJ, Novobrantseva T, Ruda VM, Lytton-JeanAbigail KR, Levins CG, Kalish B, Mudge DK, Perez M, Abezgauz L, Dutta P, Smith L, Charisse K, Kieran MW, Fitzgerald K, Nahrendorf M, Danino D, Tuder RM, von Andrian UH, Akinc A, Panigrahy D, Schroeder A, Koteliansky V, Langer R, Anderson DG, Nat Nano 2014, 9, 648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Paunovska K, Sago CD, Monaco CM, Hudson WH, Castro MG, Rudoltz TG, Kalathoor S, Vanover DA, Santangelo PJ, Ahmed R, Bryksin AV, Dahlman JE, Nano Lett 2018, 18, 2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sago CD, Lokugamage MP, Lando GN, Djeddar N, Shah NN, Syed C, Bryksin AV, Dahlman JE, Nano Lett 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen D, Love KT, Chen Y, Eltoukhy AA, Kastrup C, Sahay G, Jeon A, Dong Y, Whitehead KA, Anderson DG, J Am Chem Soc 2012, 134, 6948. [DOI] [PubMed] [Google Scholar]
- [17].Lokugamage MP, Sago CD, Dahlman JE, Current Opinion in Biomedical Engineering 2018, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Platt RJ, Chen S, Zhou Y, Yim MJ, Swiech L, Kempton HR, Dahlman JE, Parnas O, Eisenhaure TM, Jovanovic M, Graham DB, Jhunjhunwala S, Heidenreich M, Xavier RJ, Langer R, Anderson DG, Hacohen N, Regev A, Feng G, Sharp PA, Zhang F, Cell 2014, 159, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, Jardine L, Dixon D, Stephenson E, Nilsson E, Grundberg I, McDonald D, Filby A, Li W, De Jager PL, Rozenblatt-Rosen O, Lane AA, Haniffa M, Regev A, Hacohen N, Science 2017, 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, Sah DW, Stebbing D, Crosley EJ, Yaworski E, Hafez IM, Dorkin JR, Qin J, Lam K, Rajeev KG, Wong KF, Jeffs LB, Nechev L, Eisenhardt ML, Jayaraman M, Kazem M, Maier MA, Srinivasulu M, Weinstein MJ, Chen Q, Alvarez R, Barros SA, De S, Klimuk SK, Borland T, Kosovrasti V, Cantley WL, Tam YK, Manoharan M, Ciufolini MA, Tracy MA, de Fougerolles A, MacLachlan I, Cullis PR, Madden TD, Hope MJ, Nat Biotechnol 2010, 28, 172. [DOI] [PubMed] [Google Scholar]
- [21].Sago CD, Lokugamage MP, Islam FZ, Krupczak BR, Sato M, Dahlman JE, Journal of the American Chemical Society 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Wang C, Ye Y, Hu Q, Bellotti A, Gu Z, Adv Mater 2017, 29. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.