Abstract
Intermediate filaments (IFs), in coordination with microfilaments and microtubules, form the structural framework of the cytoskeleton and nucleus, thereby providing mechanical support against cellular stresses and anchoring intracellular organelles in place. The assembly and disassembly of IFs are mainly regulated by the phosphorylation of IF proteins. These phosphorylation states can be tracked using antibodies raised against phosphopeptides in the target proteins. IFs exert their functions through interactions with not only structural proteins, but also non-structural proteins involved in cell signaling, such as stress responses, apoptosis, and cell proliferation. This review highlights findings related to how IFs regulate cell division through phosphorylation cascades and how trichoplein, a centriolar protein originally identified as a keratin-associated protein, regulates the cell cycle through primary cilium formation.
Keywords: intermediate filament, post-translational modification, site- and phosphorylation state-specific antibody, cell proliferation, cytokinesis, primary cilium
1. IFs characteristics and biological role
The cytoskeleton must be rigid to support the cell shape, resist mechanical stresses, and anchor organelles in place. There are four components of the cytoskeleton in vertebrate cells: filamentous actin (microfilaments), microtubules, septins, and intermediate filaments (IFs).1) It is widely recognized that IFs greatly contribute to the regulation of cell structure and function in coordination with microfilaments and microtubules.2–15) Microfilaments and microtubules are composed of homogeneous globular proteins named actin and tubulin, respectively. Septins are capable of self-association, polymerizing into long apolar filaments.16,17) IFs comprise a heterogeneous protein family with five major types (I–V) and one atypical type (VI).8–11) Types I and II IFs are composed of acidic keratin and neutral-basic keratin, respectively. In humans, these keratins are encoded by 28 and 26 genes, respectively, and are mainly expressed in epithelial cells. Type III IFs comprise vimentin typically expressed in mesenchymal cells, desmin expressed in muscular cells, glial fibrillary acidic protein (GFAP) expressed in astrocytes, and peripherin expressed in peripheral neurons. Type IV IFs include neurofilaments (NF-L, NF-M, and NF-H) and α-internexin expressed in neurons, and nestin expressed in neural stem cells. Type V IFs are composed of lamin A/C, B1, and B2 with ubiquitous expression. Type VI IFs include filensin and phakinin. Except for type V IFs localized on the inner aspect of the nuclear membrane, all other IFs are localized in the cytoplasm.
Types I–V IFs have similar tripartite structures, with an amino-terminal head domain, a central rod domain, and a carboxyl-terminal tail domain.2–13) The head and tail domains are variable among the types, whereas the rod domain is relatively conserved. Cytoplasmic IFs form dimers through binding of their rod domains aligned in parallel. Keratins form type I–type II heterodimers, whereas other IFs typically form homodimers. Two dimers form a tetramer in an approximately half-staggered manner through the rod domains aligned in an antiparallel orientation. Eight tetramers are aligned laterally to form a unit-length filament (ULF). The ULFs anneal end-to-end to form a non-polar structure in a nucleotide-independent manner. These processes differ from those of microfilaments and microtubules, which require ATP-bound and GTP-bound monomers, respectively, for polymerization and form polar structures wherein polymerization of F-actin and tubulin preferentially occurs at one end with depolymerization at the other end. IF precursors, composed of particles with one or a few ULFs and squiggles formed by joining of several particles, are distributed throughout the cells, and transported on either microtubules or microfilaments. The underlying mechanisms have been elucidated in some cases. For example, the movement of vimentin, peripherin, and NF proteins is mainly dependent on microtubules,18–22) whereas the movement of keratin proteins is mainly dependent on microfilaments.23) IF precursors join together to create non-polar filaments that form extensive networks throughout the cytoplasm. Typically, vimentin filaments form a mesh-like network, whereas keratin filaments form bundles designated tonofibrils.12) The typical lengths of vimentin and keratin are 0.3–0.65 µm and 0.4–2.1 µm, respectively.24) The contour lengths are about 10–20 µm in both vimentin and keratin filaments.12) Their extensibility is 280% for keratins and 300% for vimentins.12) These features of cytoplasmic IFs greatly contribute to cellular elasticity and stiffness, which support the cell shape and provide resistance against mechanical stresses.25) The cytoplasm of fibroblasts expressing vimentin is stiffer and more resistant to compressive stress than that of vimentin-null fibroblasts.26,27) Keratin works as the main component for the stiffness of keratinocytes.28,29) Type V IFs form the lamina on the inner side of the nuclear envelope and provide mechanical integrity for the nucleus.30)
Cytoplasmic IFs interact with various intracellular organelles, including the nucleus, mitochondria, endoplasmic reticulum, Golgi complex, lysosomes, ribosomes, and centrosome.13,31,32) Cytoplasmic IFs connect the nucleus to the cell cortex through Plectin 1 and Nesprin 3 and regulate nuclear size and shape, nuclear movement and position, and potentially gene expression.13,33) Vimentin associates with mitochondria through binding to Plectin 1b to regulate the mitochondrial shape.13,34) Vimentin interacts with the Golgi complex by binding to peripheral Golgi protein formiminotransferase cyclodeaminase (FTCD) and molecule interacting with CasL (MICAL), which are connected to the Golgi complex through interactions with small G protein Rab1, to regulate the Golgi organization and anterograde transport.13,31) Cytoplasmic IFs are anchored to desmosomes at sites of cell–cell interaction and hemidesmosomes at sites of cell–matrix adhesion through interactions with desmoplakin at desmosomes and Plectin 1a and BPAG1 at hemidesmosomes.13,35) K5/K14 inhibit phosphorylation of desmoplakin by protein kinase C (PKC), resulting in the stabilization of desmosomes.36) In contrast, K6/K17 stimulate phosphorylation of desmoplakin through PKC, resulting in disassembly of desmosomes.37) Cytoplasmic IFs stabilize and induce maturation of focal adhesions through binding to integrins with or without IF-associated proteins such as Plectin 1f and focal adhesion kinase.38,39) Focal adhesions also promote the assembly and organization of IFs, suggesting that cytoplasmic IFs and focal adhesions are bidirectionally regulated.13)
2. Post-translational modifications regulate IFs assembly and disassembly
The soluble cytosolic pool for IFs is much smaller than those for microfilaments and microtubules.3,4) Therefore, IFs were regarded as static cellular structures until the late 1970s. However, it was gradually revealed that vimentin existed in both phosphorylated and unphosphorylated forms in various cells40,41) and that phosphorylation was increased when vimentin underwent redistribution in cells.42,43) These findings led researchers to hypothesize that phosphorylation of IFs may stimulate disassembly of IFs.44) The first direct evidence to support this hypothesis was obtained by us2) in 1987. We found that vimentin filaments reconstituted in vitro were completely disassembled when vimentin was phosphorylated by protein kinase A (PKA) or PKC. We also demonstrated that the phosphorylation sites differed between PKA and PKC. Since then, a variety of protein kinases have been identified to regulate the assembly and disassembly of IFs.3–13,45–48) In general, the head domains of IFs, which are composed of many basic residues, are positively charged and play a key role in IF assembly. Subsequent phosphorylation at serine/threonine residues in the head domains can change the charge, resulting in disassembly of IFs by promoting IF solubility.3,4,49) This is the case for phosphorylation of vimentin by PKA, PKC, Ca2+/calmodulin-dependent protein kinase II (CaMKII), and Cdk1 kinase,50–53) phosphorylation of GFAP by PKA, PKC, and CaMKII,54,55) phosphorylation of desmin by PKA, PKC, and Cdk1 kinase,56–58) phosphorylation of K8 by PKA, p38, and JUN kinase,59–61) and phosphorylation of NF-L by PKA and PKC62,63) observed both in vitro and in cells.
In some cases, phosphorylation of IFs can promote their formation and increase their stability. Phosphorylation at Lys-Ser-Pro motifs located in the tail regions of NF-M and NF-H increases the stability of filaments in the axon.64) Phosphorylation of NF in the head region promotes the formation of filaments in the soma of neurons.64,65) A highly conserved tyrosine residue in the rod domain of K8 (Tyr267) promotes insolubility of keratin and formation of keratin filaments in cells.66)
In addition to phosphorylation, various post-translational modifications (PTMs) regulate the assembly and disassembly of IFs.5–13) Sumoylation at Lys201 in lamin A/C stabilizes the formation of lamin filaments in the inner nuclear envelope membrane.67) Mutations causing defects in sumoylation at Lys201 are associated with dilated cardiomyopathy.68) Lys207 in K18 and Lys208 in K19 are hypersumoylated by oxidative and apoptotic stresses, and consequently stimulate the formation of keratin filaments in cells and in vivo.69) Phosphorylation can also affect sumoylation. Phosphorylation at Ser74 in K8 stimulated sumoylation at Lys285 and Lys364 in K8, possibly by facilitating access of the sumoylation machinery to these regions.69) Lys207 in K18 is also a target of acetylation.70) Acetylation at Lys207 in K18 promotes the formation of keratin filaments at the perinuclear region.71)
3. Phosphorylated IFs can be detected using site- and phosphorylation state-specific antibodies
There are several ways to analyze the phosphorylation of IFs, including labeling of cells with radioactive phosphate, two-dimensional mapping of phosphorylated and unphosphorylated IFs, and mass spectrometry of IFs.5,6) However, these methods require cell lysis, making it difficult to analyze the spatiotemporal phosphorylation of IFs in cells. In 1991, we demonstrated that site- and phosphorylation state-specific antibodies could be efficiently generated by immunization with phosphorylated polypeptides (corresponding to a phosphorylated residue and its surrounding residues) as antigens.72,73) We have used these site- and phosphorylation state-specific antibodies to visualize the spatiotemporal regulation of phosphorylation in IFs, including vimentin, keratin, desmin, GFAP, α-internexin, and neurofilament IFs, through various kinases in cells.5,52,54,58,62,72–88) For example, we generated four antibodies that could selectively recognize phosphorylation at Thr7, Ser8, Ser13, and Ser38 in GFAP. Using these antibodies, we demonstrated that Ser8 was phosphorylated by Cdk1 throughout the cytoplasm from prometaphase to metaphase, whereas Thr7, Ser13, and Ser38 were phosphorylated by Rho-associated kinase (RhoK) associated with the plasma membrane and Aurora B kinase (AurB) at the spindle midzone from anaphase to telophase.72,73,77–79,84) Similarly, antibodies raised against various phosphopeptides from vimentin were successfully used to reveal the mechanisms for how phosphorylation of vimentin stimulates cell proliferation (discussed later). This method can also be applied to the generation of antibodies that recognize PTMs other than phosphorylation.5,6,86,89) Furthermore, antibodies against aberrant PTMs can be used as diagnostic and prognostic tools.7) These antibodies can also be used to assess the effects of therapeutic drugs such as kinase inhibitors that target aberrant PTMs.7)
4. IFs regulate cellular homeostasis through interactions with IF-associated proteins
IFs exert their function through interactions with not only structural proteins but also non-structural proteins involved in cell signaling, such as those associated with stress responses, apoptosis, and cell proliferation.5,6,9,11–13,90,91)
IFs are affected by various stresses, including mechanical, physical, chemical, and microbial stresses.13) These stresses stimulate phosphorylation of Ser73 in K8 by p38 and JUN kinase, thereby increasing the affinity of K8 for 14-3-3 and PKC, resulting in keratin filament remodeling and substantial increases in network elasticity.92,93) Heat shock proteins (HSPs) are also involved in IF-mediated protection against stresses.94) We identified Mrj, a DnaJ/Hsp40 family protein, as a novel binding protein for K18 using a two-hybrid system.95) We found that Mrj bound to K18 through its C-terminal region, and also bound to the constitutively expressed Hsp/c70, already identified as a binding protein for K8/K18,96) via its N-terminal region. Microinjection of an anti-Mrj antibody induced disorganization of K8/K18 filaments, but not microfilaments or microtubules, suggesting that Mrj may stabilize K8/K18 filaments by working as a chaperone with Hsp/c70.95) These interactions between IFs and HSPs play important roles in the protection of cells against various stresses.94)
Activation of caspases can lead to collapse of the IF network, because many IFs and IF-associated proteins such as desmoplakin and plectin contain caspase cleavage sites.14,97) IFs have several mechanisms to protect cells against apoptosis.13) We identified tumor necrosis factor (TNF) receptor (TNFR) 1-associated death domain protein (TRADD), an indispensable adaptor molecule for TNFR signaling, as a novel binding protein for K18 through the central rod domain.98) Overexpression of a K18 fragment containing the TRADD-binding domain rendered the cells more resistant to TNF-induced apoptosis, suggesting that resistance of epithelial cells to TNF-induced apoptosis may arise at least in part through the interaction of K18 and TRADD, which sequesters TRADD to attenuate its interaction with activated TNFR signaling.98,99) K8 also suppresses TNF-induced apoptosis through interaction with TNFR2.31) K8 and K18 suppress the delivery of Fas to the plasma membrane, which can inhibit Fas-mediated apoptosis.31) Interactions of K8/K18 with cellular FLICE inhibitory protein (cFlip) and Raf1 inhibit both TNF-mediated and Fas-mediated apoptosis.100–102)
Furthermore, IFs regulate cell proliferation through interactions with IF-associated proteins. Phosphorylation of RSX[pS/pT]XP motifs in IFs, including K17, K18, and vimentin, increases association between IFs and 14-3-3 and affect cell proliferation.7,103–106) Phosphorylation of Ser34 in K18 promotes binding to 14-3-3 and stimulates mitosis through activation of 14-3-3 signaling in the cytosol.105) Phosphorylation of Thr9 and Ser44 in K17 promotes cell growth through activation of mammalian target of rapamycin via 14-3-3 during wound healing in epithelial cells.106) Phosphorylation of Ser39 in vimentin by AKT inhibits Beclin1 through 14-3-3, and leads to inhibition of autophagy, resulting in stimulation of tumorigenesis.107) Phosphorylation of vimentin stimulates mitosis by activating signaling cascades including various kinases.2–6,80,83,108–116) Keratin-associated proteins regulate cell proliferation by regulating primary cilium formation.47,117–125) In the following sections, we will focus on these last two topics.
5. IF structure is regulated by phosphorylation cascades during mitosis
Since the pioneering work showing that phosphorylation of vimentin filaments by PKA or PKC stimulates disassembly of vimentin filaments,2) extensive studies have been performed to reveal how phosphorylation of vimentin is regulated and involved in cell proliferation. From prometaphase to metaphase, Ser41 and Ser55 in vimentin are phosphorylated by Cdk1.51,108) Phosphorylation of Ser55 by Cdk1 stimulates the activity of Polo-like kinase 1 (Plk1) through interaction with the Polo-box domain, resulting in the phosphorylation of Ser82 in vimentin and disassembly into non-filamentous vimentin particles until the end of mitosis.52,108,112) During metaphase and anaphase, PKC phosphorylates Ser33 and Ser50 in vimentin.126) From anaphase to telophase, RhoK and AurB phosphorylate Ser71 and Ser72 in vimentin, respectively.80,83,109,110) Phosphorylation of Ser71 by RhoK inhibits the formation of vimentin filaments in vitro.80) Furthermore, phosphorylation of vimentin by Cdk1, Plk1, and PKC during mitosis is observed throughout the cytoplasm.52,108,112) In contrast, phosphorylation of vimentin by RhoK and AurB is specifically observed at the cleavage furrow.5,6,83) AurB is activated by phosphorylation at Thr232. Mutation of Thr232 to Ala leads to the presence of multinucleated cells, as also observed following overexpression of vimentin with mutations at the sites phosphorylated by AurB.111)
To validate the functional importance of vimentin phosphorylation during mitosis, we generated expression vectors to express mutated vimentin proteins in which the Ser or Thr residues phosphorylated by Cdk1, Plk1, PKC, RhoK, and AurB were changed to Ala. We transfected the expression vectors into type III IF-deficient T24 cells, a urinary bladder carcinoma cell line.83,110,112,127) We found that unusual long bridge-like IF structures, designated IF-bridges, occurred during cytokinesis. The formation of IFs during interphase was not impaired. Of note, the IF-bridge phenotype was also caused by GFAP mutant proteins in which Thr7, Ser13, and Ser38, phosphorylated by both RhoK and AurB, were mutated to Ala.79,127) This was also the case for desmin mutant proteins, in which the phosphorylation sites for RhoK and/or AurB were changed to Ala or Gly.84,128) The results suggested that RhoK and AurB may regulate cleavage furrow-specific phosphorylation and segregation of type III IFs during cytokinesis.5,6)
IF-bridges are associated with two outcomes.129) In the first, IF-bridges are torn apart, presumably by cell adhesion traction forces, to allow completion of cytokinesis. In the second, cytokinesis fails to occur, resulting in tetraploidy. To analyze the effects of cytokinesis failure caused by phosphodeficient vimentin in vivo, we generated mutant mice in which vimentin could not be phosphorylated by RhoK, AurB, Cdk1, or Plk1.113) VIMSA/SA mice homozygous for the mutation exhibited microphthalmia and cataract. A significant increase in the number of mice showing tetraploidy in their vimentin-expressing cells was also observed in VIMSA/SA mice. Meanwhile, VIMWT/SA mice heterozygous for the mutation did not show any abnormal phenotype in their appearance.
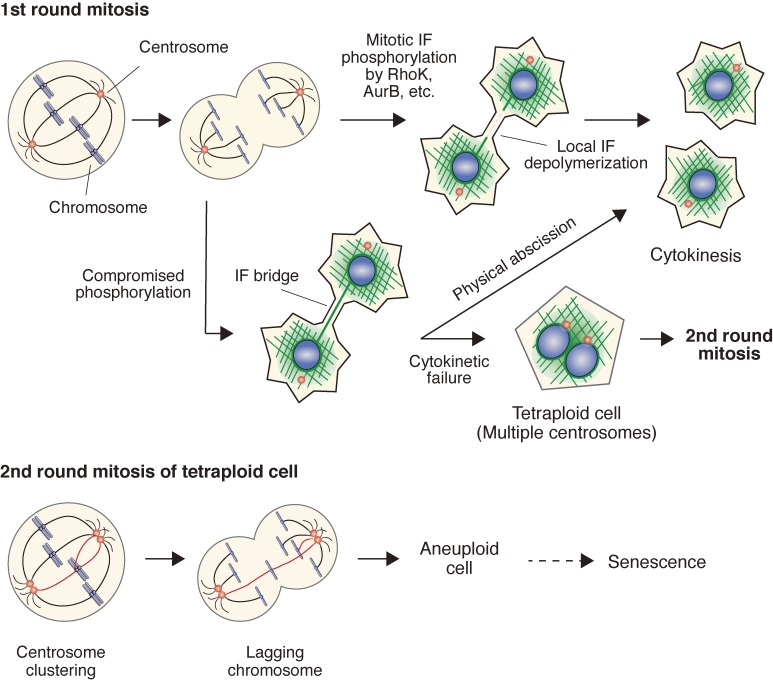
Tetraploidy can cause chromosomal instability, thus providing a route to aneuploidy and contributing to the development of aging and cancer.129,130) Loss of the tetraploidy checkpoint and subsequent catastrophic mitosis may lead to the generation of aneuploid cells. In fact, the expression of senescence-related genes was significantly increased in VIMSA/SA mice.113) Tetraploid cells continue to proliferate and generate aneuploidy, which is associated with tumorigenesis and senescence.129) Taken together, a timely succession for phosphorylation of vimentin is one of the key mechanisms involved in the regulation of cell proliferation and aging (Fig. 1).
Figure 1.
Compromised IF phosphorylation leads to defective cytokinesis, tetraploidy, and aneuploidy. From anaphase to telophase, Rho kinase and Aurora B kinase phosphorylate type III IFs, leading to local IF depolymerization and cytokinesis. When this phosphorylation is compromised, unusual long bridge-like IF structures are formed, leading to cytokinetic failure and tetraploidy, a condition in which cells have four homologous sets of chromosomes. Mitosis of tetraploid cells frequently causes lagging chromosomes through merotelic attachment, resulting in aneuploidy, a deviation from a multiple of the haploid chromosome number in which some of the chromosomes are missing or present in excess. Aneuploidy is frequently observed in cancer cells and senescent cells.
6. Cell proliferation is regulated by trichoplein, a keratin-associated protein, through the primary cilium
Using two-hybrid systems, we identified trichoplein (TCHP) as a novel binding protein for various keratins, including K18, K16, K14, K8, K6a, and K5.117) TCHP contains a domain with a low degree of sequence similarity to trichohyalin and plectin, designated the trichohyalin/plectin homology domain (TPHD). In polarized epithelial cells, TCHP colocalizes with K8/K18 filaments in the apical region and is also concentrated at desmosomes, suggesting its involvement in the organization of the apical network of keratin filaments and desmosomes in simple epithelial cells.117)
Keratin filaments not only associate with desmosomes and hemidesmosomes at the plasma membrane but also extend into the cytoplasm to provide a scaffold for other cytoskeletal elements including the centrosome and microtubules.131–134) Plectin can associate with IFs, microfilaments, and microtubules.135–137) Thus, TCHP may localize with cytoskeletal elements such as the centrosome through the TPHD in certain conditions, because the centrosome is associated with IFs, microfilaments, and microtubules.138–140) We found that expression of TCHP is concentrated in desmosomes and centrosomes in polarized and mitotic cells, respectively.117,118) In proliferating cells, TCHP is localized at the distal to subdistal ends of the mother centriole through binding to the centrosomal proteins Odf2 and ninein.118)
In most non-dividing cells, the centrosome moves to the cell surface where the mother centriole differentiates into a basal body to nucleate a cilium.47,120,123,125) A primary cilium is found in most types of quiescent vertebrate cells. Many proteins are involved in assembly and maintenance of the primary cilium.125) We found that TCHP was expressed in the mother centriole, but not in the basal body as the nucleation site for the primary cilium.119) Based on these findings, we hypothesized that TCHP may function as a suppressor of primary cilium assembly. We subsequently demonstrated that exogenous expression of TCHP inhibited primary cilium assembly in serum-starved cells, whereas knockdown of TCHP induced primary cilium assembly in serum-fed cells.119) We further found that TCHP bound to and activated Aurora A kinase (AurA) at the centrosome and that binding to AurA was required for suppression of primary cilium assembly by TCHP.119) The downstream targets of AurA that regulate primary cilium assembly remain largely unknown.125)
Knockdown of TCHP or AurA induced primary cilium assembly and G0/G1 arrest in serum-fed cells, whereas these effects were prevented when primary cilium formation was blocked by simultaneous knockdown of IFT20, suggesting that the primary cilium works as a brake for cell cycle progression. In fact, primary cilia are lost in a wide range of cancers.47,120,123,125) Therefore, it is very important to elucidate the mechanisms that regulate the expression of TCHP, which functions as a suppressor of primary cilium assembly, to clarify the underlying molecular mechanism for tumor cell proliferation and facilitate the development of novel treatment strategies against cancer.47,120,123,125)
Given that TCHP was stabilized by proteasome inhibitors in serum-starved RPE cells, we performed global screening of E3 ligases and identified potassium channel tetramerization domain-containing 17 (KCTD17), which is associated with the scaffold protein Cullin 3 (Cul3) and RBX1 in an E3 ligase complex called CRL3KCTD17, as an E3 ligase that can polyubiquitinate TCHP.121) However, the activity of CRL3KCTD17 remained constant regardless of the presence or absence of serum, leading us to hypothesize the existence of deubiquitinases (DUBs) that can deubiquitinate TCHP in a serum-dependent manner. Through global screening of DUBs, we identified ubiquitin-specific peptidase 8 (USP8) as a DUB that can stabilize TCHP. We also demonstrated that USP8 was activated by EGFR tyrosine kinase through phosphorylation of Tyr717 and Tyr810.124) Knockdown of EGFR or USP8 suppressed the TCHP-AurA pathway, induced unscheduled ciliogenesis, and caused cell cycle arrest of RPE1 cells, even in the presence of serum.124) These effects were inhibited when ciliogenesis was abrogated by depletion of IFT20 or CEP164.124) The findings suggested that EGF signaling regulates not only the well-known kinase cascades, such as the mitogen-activation protein kinase and PI3K-AKT cascades, but also primary cilium dynamics. RTK signaling is frequently activated in cancer cells.141,142) EGFR family receptors are amplified and/or mutated in a variety of human tumors, including glioma, non-small-cell lung carcinoma, breast cancer, gastric cancer, and ovarian cancer.143,144) Amplification or mutation of EGFR family receptors can lead to overexpression and/or constitutive activation of EGFR signaling in these tumor tissues.143–145) The finding that activation of EGFR suppresses ciliogenesis through activation of the USP8-TCHP-AurA pathway provides a novel explanation for the loss of primary cilia frequently observed in cancer cells with aberrant RTK signaling121,124) (Fig. 2). In 1979, Tucker et al. found that primary cilia were assembled when cultured mouse 3T3 fibroblasts exited the cell cycle under serum deprivation (i.e., G0 phase).146,147) They also reported that when the quiescent fibroblasts were stimulated with serum, primary cilia were disassembled after the serum stimulation.146,147) Deciliation after serum stimulation corresponded to the G0/G1 transition.148) Our findings provide a molecular basis for the regulation of cell cycle through assembly and disassembly of primary cilia in response to growth factors.
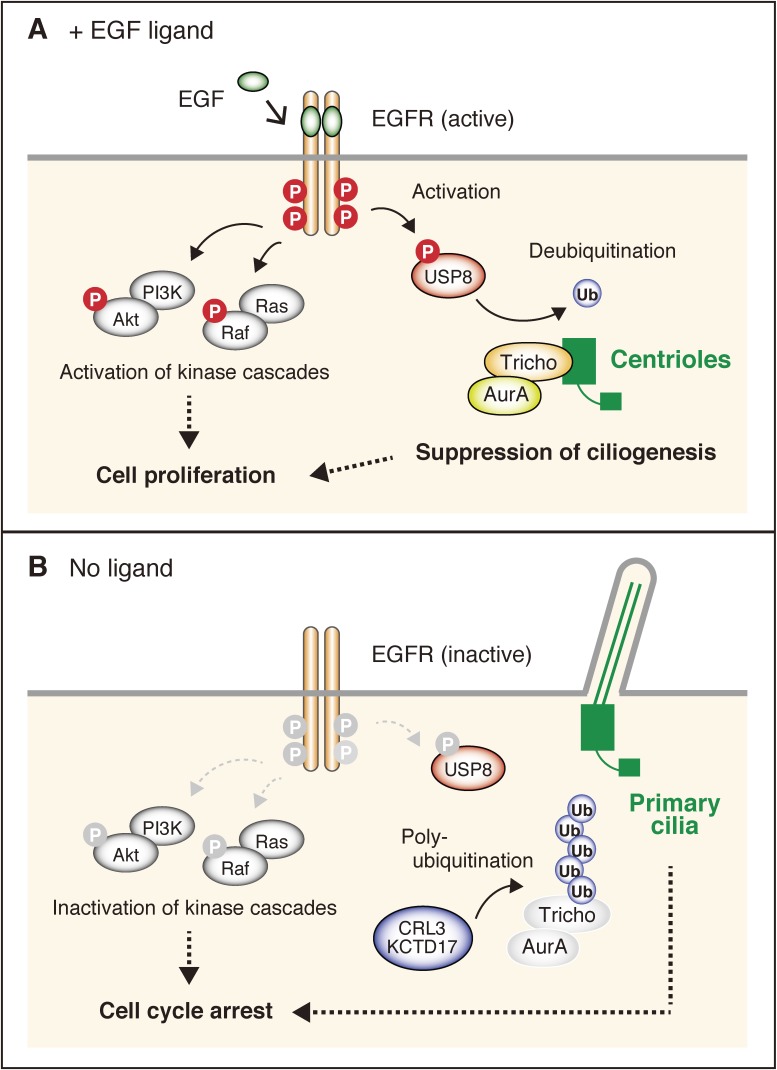
Figure 2.
Trichoplein promotes cell proliferation through suppression of ciliogenesis in the presence of EGF. A) EGF activates USP8 through phosphorylation. Activated USP8 deubiquitinates and stabilizes trichoplein at the mother centriole. Trichoplein suppresses ciliogenesis through activation of Aurora A kinase, which stimulates cell proliferation. B) In the absence of EGF signaling, USP8 is not activated. Unphosphorylated USP8 (inactive USP8) allows for the polyubiquitination of TCHP by CRL3-KCTD17, which culminates in TCHP degradation and primary cilium formation. Cell cycle arrest occurs after formation of the primary cilium.
We also hypothesized that proteins with a TPHD may have similar functions to TCHP. By searching a public database, we found that Albatross (also known as Fas-binding factor 1) and nudE-like 1 (Ndel1) may possess a TPHD.122,149) Both Albatross and Ndel1 can bind to IFs, actin, and/or microtubules, are concentrated in desmosomes and the centrosome in polarized and mitotic cells, respectively, and regulate primary cilium assembly.122,149–157) Of note, Ndel1 suppresses ciliogenesis,122) whereas Albatross induces ciliogenesis.153) Ndel1 acts as an upstream regulator of the TCHP-AurA pathway for suppression of ciliogenesis.122) Ndel1 indirectly inhibits the ubiquitination of trichoplein by CRL3KCTD17 and suppresses ciliogenesis in the presence of serum, while Ndel1 is degraded via the ubiquitin-proteasome system in the absence of serum, resulting in the disappearance of TCHP at the mother centriole and the promotion of ciliogenesis.122) The molecules involved in ubiquitin-proteasome system-mediated Ndel1 degradation remain to be identified. Albatross acts as a downstream target of CEP83 required for docking of the centrosome to the plasma membrane.153) Taken together, TCHP, Albatross, and Ndel1, all of which have a TPHD, may localize at either desmosomes/hemidesmosomes or the centrosome through interactions with IFs, microfilaments, and/or microtubules dependent on the context and regulate cell proliferation by regulating primary cilium formation.
7. Future directions
With the rapid advance of scientific knowledge and technologies, we can begin to reveal these novel IF-associated proteins. For example, Lin et al.158) discovered that IFs, including vimentin, peripherin, and NF, interact with toxic proline-arginine poly-dipeptides, which are encoded by the C9orf72 hexanucleotide expansions frequently observed in amyotrophic lateral sclerosis (ALS), through a low complexity domain (LCD) in the head domain of IFs. Aggregation of NF is associated with various neurodegenerative diseases, including ALS.159) The toxic proline-arginine poly-dipeptides may cause aggregation of NF through interaction with the LCD.
Mutations in genes coding IFs are associated with more than 80 human diseases.160–162) There is an urgent and growing need for the development of therapeutic drugs that target these diseases caused by dysfunction of IFs. Several drugs can ameliorate the abnormal phenotypes arising from dysfunction of IFs. For example, sulforaphane ameliorated skin blistering in K14-knockout mice and abnormal thickening of the palms and soles in K16-knockout mice.163–165) Sulforaphane suppresses Keap1, a negative regulator of Nrf2, resulting in activation of Nrf2-dependent K16 and K17 expression.163–168) PKC412, a pan-kinase inhibitor, reverted the disrupted keratin filament network caused by a R90C mutation in K18 by promoting dephosphorylation of non-muscle myosin heavy chain IIA (NMHC-IIA) and the association between NMHC-IIA and K8/K18, resulting in stabilization of the keratin filament network.169) FiVe1 selectively and irreversibly inhibited the proliferation of mesenchymally transformed breast cancer cells and soft tissue sarcoma cells by causing mitotic catastrophe through its promotion of vimentin filament disorganization.170) Image-based high-throughput screening revealed that simvastatin, one of the most commonly used drugs for hypercholesterolemia, induced apoptosis of vimentin-positive cancer cells by promoting disorganization of vimentin filaments.171)
Zebrafish is widely recognized as a useful tool for examining the associations between genes and drugs and assessing the therapeutic and toxicological effects of drugs in vivo.124,172–175) It was demonstrated that knock-in of a desmin mutation into zebrafish caused disorganization of their cardiac and skeletal muscles and that doxycycline could ameliorate these abnormal phenotypes.176) Combined with genome-editing technology such as CRISPR/Cas9, zebrafish has also been successfully used to examine the complex phenotypes caused by impaired primary cilia.124,177)
Further studies are necessary to completely elucidate the regulation and function of IFs and IF-associated proteins. For example, TCHP binds to and activates AurA at the centrosome, resulting in the suppression of primary cilium assembly and cell cycle progression.119,121,124) The downstream targets of AurA, however, remain largely elusive.125) It has also been shown that IFs and IF-associated proteins can regulate cell differentiation.8,13,15,178) The complete picture of how IFs and IF-associated proteins regulate cell differentiation remains to be fully elucidated. Clarification of these molecular mechanisms facilitates the development of novel treatment strategies against various diseases caused by the impairment of IFs and IF-associated proteins.
Acknowledgments
We sincerely apologize to all researchers whose important works are not cited in this review. This work was supported in part by the Japan Society for the Promotion of Science KAKENHI (19K07318 to YN, 17K08269 to KK, and 15H02398 to MI), Takeda Science Foundation (YN, KK, and MI), and the Naito Foundation (MI). The authors thank Alison Sherwin, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Profile
Masaki Inagaki was born in Aichi Prefecture in 1956 and graduated from Mie University School of Medicine in 1982. He received his PhD degree in 1986. After working as a principal investigator at Aichi Cancer Center Research Institute, he moved to Tokyo Metropolitan Institute of Gerontology as a chief at 1992. He then moved to Aichi Cancer Center Research Institute as the chief of the Division of Biochemistry at 1997. He has been Professor of Physiology at Mie University since 2016. Using site- and phosphorylation state-specific antibodies that he developed, he has revealed novel mechanisms of cell cycle regulation through the phosphorylation of various proteins including intermediate filaments (IF) and IF-associated proteins. He has also revealed that the ubiquitin-proteasome system regulates the dynamics of primary cilia formation and cell proliferation induced by growth factors.
References
- 1).Fischer R.S., Fowler V.M. (2015) Thematic Minireview Series: The state of the cytoskeleton in 2015. J. Biol. Chem. 290, 17133–17136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2).Inagaki M., Nishi Y., Nishizawa K., Matsuyama M., Sato C. (1987) Site-specific phosphorylation induces disassembly of vimentin filaments in vitro. Nature 328, 649–652. [DOI] [PubMed] [Google Scholar]
- 3).Inagaki N., Ito M., Nakano T., Inagaki M. (1994) Spatiotemporal distribution of protein kinase and phosphatase activities. Trends Biochem. Sci. 19, 448–452. [DOI] [PubMed] [Google Scholar]
- 4).Inagaki M., Matsuoka Y., Tsujimura K., Ando S., Tokui T., Takahashi T., et al. (1996) Dynamic property of intermediate filaments: Regulation by phosphorylation. BioEssays 18, 481–487. [Google Scholar]
- 5).Izawa I., Inagaki M. (2006) Regulatory mechanisms and functions of intermediate filaments: A study using site- and phosphorylation state-specific antibodies. Cancer Sci. 97, 167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6).Goto H., Inagaki M. (2014) New insights into roles of intermediate filament phosphorylation and progeria pathogenesis. IUBMB Life 66, 195–200. [DOI] [PubMed] [Google Scholar]
- 7).Snider N.T., Omary M.B. (2014) Post-translational modifications of intermediate filament proteins: Mechanisms and functions. Nat. Rev. Mol. Cell Biol. 15, 163–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8).Lowery J., Kuczmarski E.R., Herrmann H., Goldman R.D. (2015) Intermediate filaments play a pivotal role in regulating cell architecture and function. J. Biol. Chem. 290, 17145–17153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9).Loschke F., Seltmann K., Bouameur J.E., Magin T.M. (2015) Regulation of keratin network organization. Curr. Opin. Cell Biol. 32, 56–64. [DOI] [PubMed] [Google Scholar]
- 10).Herrmann H., Aebi U. (2016) Intermediate filaments: Structure and assembly. Cold Spring Harb. Perspect. Biol. 8, a018242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11).Robert A., Hookway C., Gelfand V.I. (2016) Intermediate filament dynamics: What we can see now and why it matters. BioEssays 38, 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12).Sanghvi-Shah R., Weber G.F. (2017) Intermediate filaments at the junction of mechanotransduction, migration, and development. Front. Cell Dev. Biol. 5, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13).Etienne-Manneville S. (2018) Cytoplasmic intermediate filaments in cell biology. Annu. Rev. Cell Dev. Biol. 34, 1–28. [DOI] [PubMed] [Google Scholar]
- 14).Omary M.B., Coulombe P.A., McLean W.H. (2004) Intermediate filament proteins and their associated diseases. N. Engl. J. Med. 351, 2087–2100. [DOI] [PubMed] [Google Scholar]
- 15).Jacob J.T., Coulombe P.A., Kwan R., Omary M.B. (2018) Types I and II keratin intermediate filaments. Cold Spring Harb. Perspect. Biol. 10, a018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16).Kinoshita M. (2003) Assembly of mammalian septins. J. Biochem. 134, 491–496. [DOI] [PubMed] [Google Scholar]
- 17).Valadares N.F., d’Muniz Pereira H., Ulian Araujo A.P., Garratt R.C. (2017) Septin structure and filament assembly. Biophys. Rev. 9, 481–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18).Yoon M., Moir R.D., Prahlad V., Goldman R.D. (1998) Motile properties of vimentin intermediate filament networks in living cells. J. Cell Biol. 143, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19).Ho C.L., Martys J.L., Mikhailov A., Gundersen G.G., Liem R.K. (1998) Novel features of intermediate filament dynamics revealed by green fluorescent protein chimeras. J. Cell Sci. 111, 1767–1778. [DOI] [PubMed] [Google Scholar]
- 20).Prahlad V., Yoon M., Moir R.D., Vale R.D., Goldman R.D. (1998) Rapid movements of vimentin on microtubule tracks: Kinesin-dependent assembly of intermediate filament networks. J. Cell Biol. 143, 159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21).Prahlad V., Helfand B.T., Langford G.M., Vale R.D., Goldman R.D. (2000) Fast transport of neurofilament protein along microtubules in squid axoplasm. J. Cell Sci. 113, 3939–3946. [DOI] [PubMed] [Google Scholar]
- 22).Helfand B.T., Loomis P., Yoon M., Goldman R.D. (2003) Rapid transport of neural intermediate filament protein. J. Cell Sci. 116, 2345–2359. [DOI] [PubMed] [Google Scholar]
- 23).Kolsch A., Windoffer R., Leube R.E. (2009) Actin-dependent dynamics of keratin filament precursors. Cell Motil. Cytoskeleton 66, 976–985. [DOI] [PubMed] [Google Scholar]
- 24).Block J., Schroeder V., Pawelzyk P., Willenbacher N., Koster S. (2015) Physical properties of cytoplasmic intermediate filaments. Biochim. Biophys. Acta 1853, 3053–3064. [DOI] [PubMed] [Google Scholar]
- 25).Charrier E.E., Janmey P.A. (2016) Mechanical properties of intermediate filament proteins. Methods Enzymol. 568, 35–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26).Guo M., Ehrlicher A.J., Mahammad S., Fabich H., Jensen M.H., Moore J.R., et al. (2013) The role of vimentin intermediate filaments in cortical and cytoplasmic mechanics. Biophys. J. 105, 1562–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27).Mendez M.G., Restle D., Janmey P.A. (2014) Vimentin enhances cell elastic behavior and protects against compressive stress. Biophys. J. 107, 314–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28).Ramms L., Fabris G., Windoffer R., Schwarz N., Springer R., Zhou C., et al. (2013) Keratins as the main component for the mechanical integrity of keratinocytes. Proc. Natl. Acad. Sci. U.S.A. 110, 18513–18518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29).Seltmann K., Fritsch A.W., Kas J.A., Magin T.M. (2013) Keratins significantly contribute to cell stiffness and impact invasive behavior. Proc. Natl. Acad. Sci. U.S.A. 110, 18507–18512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30).Turgay Y., Medalia O. (2017) The structure of lamin filaments in somatic cells as revealed by cryo-electron tomography. Nucleus (Austin, Tex.) 8, 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31).Toivola D.M., Tao G.Z., Habtezion A., Liao J., Omary M.B. (2005) Cellular integrity plus: Organelle-related and protein-targeting functions of intermediate filaments. Trends Cell Biol. 15, 608–617. [DOI] [PubMed] [Google Scholar]
- 32).Schwarz N., Leube R.E. (2016) Intermediate filaments as organizers of cellular space: How they affect mitochondrial structure and function. Cells 5, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33).Wilhelmsen K., Litjens S.H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., et al. (2005) Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 171, 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34).Wiche G., Osmanagic-Myers S., Castanon M.J. (2015) Networking and anchoring through plectin: A key to IF functionality and mechanotransduction. Curr. Opin. Cell Biol. 32, 21–29. [DOI] [PubMed] [Google Scholar]
- 35).Jones J.C., Kam C.Y., Harmon R.M., Woychek A.V., Hopkinson S.B., Green K.J. (2017) Intermediate filaments and the plasma membrane. Cold Spring Harb. Perspect. Biol. 9, a025866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36).Kroger C., Loschke F., Schwarz N., Windoffer R., Leube R.E., Magin T.M. (2013) Keratins control intercellular adhesion involving PKC-α-mediated desmoplakin phosphorylation. J. Cell Biol. 201, 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37).Loschke F., Homberg M., Magin T.M. (2015) Keratin isotypes control desmosome stability and dynamics through PKCα. J. Invest. Dermatol. 136, 202–213. [DOI] [PubMed] [Google Scholar]
- 38).Leube R.E., Moch M., Windoffer R. (2015) Intermediate filaments and the regulation of focal adhesion. Curr. Opin. Cell Biol. 32, 13–20. [DOI] [PubMed] [Google Scholar]
- 39).Gregor M., Osmanagic-Myers S., Burgstaller G., Wolfram M., Fischer I., Walko G., et al. (2014) Mechanosensing through focal adhesion-anchored intermediate filaments. FASEB J. 28, 715–729. [DOI] [PubMed] [Google Scholar]
- 40).Lazarides E. (1982) Intermediate filaments: A chemically heterogeneous, developmentally regulated class of proteins. Annu. Rev. Biochem. 51, 219–250. [DOI] [PubMed] [Google Scholar]
- 41).Cabral F., Gottesman M.M. (1979) Phosphorylation of the 10-nm filament protein from Chinese hamster ovary cells. J. Biol. Chem. 254, 6203–6206. [PubMed] [Google Scholar]
- 42).Evans R.M., Fink L.M. (1982) An alteration in the phosphorylation of vimentin-type intermediate filaments is associated with mitosis in cultured mammalian cells. Cell 29, 43–52. [DOI] [PubMed] [Google Scholar]
- 43).Gard D.L., Lazarides E. (1982) Cyclic AMP-modulated phosphorylation of intermediate filament proteins in cultured avian myogenic cells. Mol. Cell. Biol. 2, 1104–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44).Celis J.E., Larsen P.M., Fey S.J., Celis A. (1983) Phosphorylation of keratin and vimentin polypeptides in normal and transformed mitotic human epithelial amnion cells: Behavior of keratin and vimentin filaments during mitosis. J. Cell Biol. 97, 1429–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45).Inagaki M., Nakamura Y., Takeda M., Nishimura T., Inagaki N. (1994) Glial fibrillary acidic protein: Dynamic property and regulation by phosphorylation. Brain Pathol. 4, 239–243. [DOI] [PubMed] [Google Scholar]
- 46).Inagaki M., Inagaki N., Takahashi T., Takai Y. (1997) Phosphorylation-dependent control of structures of intermediate filaments: A novel approach using site- and phosphorylation state-specific antibodies. J. Biochem. 121, 407–414. [DOI] [PubMed] [Google Scholar]
- 47).Goto H., Inoko A., Inagaki M. (2013) Cell cycle progression by the repression of primary cilia formation in proliferating cells. Cell. Mol. Life Sci. 70, 3893–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48).Inada H., Togashi H., Nakamura Y., Kaibuchi K., Nagata K., Inagaki M. (1999) Balance between activities of Rho kinase and type 1 protein phosphatase modulates turnover of phosphorylation and dynamics of desmin/vimentin filaments. J. Biol. Chem. 274, 34932–34939. [DOI] [PubMed] [Google Scholar]
- 49).Inagaki M., Takahara H., Nishi Y., Sugawara K., Sato C. (1989) Ca2+-dependent deimination-induced disassembly of intermediate filaments involves specific modification of the amino-terminal head domain. J. Biol. Chem. 264, 18119–18127. [PubMed] [Google Scholar]
- 50).Ando S., Tanabe K., Gonda Y., Sato C., Inagaki M. (1989) Domain- and sequence-specific phosphorylation of vimentin induces disassembly of the filament structure. Biochemistry 28, 2974–2979. [DOI] [PubMed] [Google Scholar]
- 51).Chou Y.H., Ngai K.L., Goldman R. (1991) The regulation of intermediate filament reorganization in mitosis. p34cdc2 phosphorylates vimentin at a unique N-terminal site. J. Biol. Chem. 266, 7325–7328. [PubMed] [Google Scholar]
- 52).Kusubata M., Tokui T., Matsuoka Y., Okumura E., Tachibana K., Hisanaga S., et al. (1992) p13suc1 suppresses the catalytic function of p34cdc2 kinase for intermediate filament proteins, in vitro. J. Biol. Chem. 267, 20937–20942. [PubMed] [Google Scholar]
- 53).Inagaki N., Goto H., Ogawara M., Nishi Y., Ando S., Inagaki M. (1997) Spatial patterns of Ca2+ signals define intracellular distribution of a signaling by Ca2+/Calmodulin-dependent protein kinase II. J. Biol. Chem. 272, 25195–25199. [DOI] [PubMed] [Google Scholar]
- 54).Inagaki M., Gonda Y., Nishizawa K., Kitamura S., Sato C., Ando S., et al. (1990) Phosphorylation sites linked to glial filament disassembly in vitro locate in a non-α-helical head domain. J. Biol. Chem. 265, 4722–4729. [PubMed] [Google Scholar]
- 55).Nakamura Y., Takeda M., Aimoto S., Hojo H., Takao T., Shimonishi Y., et al. (1992) Assembly regulatory domain of glial fibrillary acidic protein. A single phosphorylation diminishes its assembly-accelerating property. J. Biol. Chem. 267, 23269–23274. [PubMed] [Google Scholar]
- 56).Geisler N., Weber K. (1988) Phosphorylation of desmin in vitro inhibits formation of intermediate filaments; identification of three kinase A sites in the aminoterminal head domain. EMBO J. 7, 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57).Kitamura S., Ando S., Shibata M., Tanabe K., Sato C., Inagaki M. (1989) Protein kinase C phosphorylation of desmin at four serine residues within the non-α-helical head domain. J. Biol. Chem. 264, 5674–5678. [PubMed] [Google Scholar]
- 58).Kusubata M., Matsuoka Y., Tsujimura K., Ito H., Ando S., Kamijo M., et al. (1993) cdc2 kinase phosphorylation of desmin at three serine/threonine residues in the amino-terminal head domain. Biochem. Biophys. Res. Commun. 190, 927–934. [DOI] [PubMed] [Google Scholar]
- 59).Ando S., Tokui T., Yano T., Inagaki M. (1996) Keratin 8 phosphorylation in vitro by cAMP-dependent protein kinase occurs within the amino- and carboxyl-terminal end domains. Biochem. Biophys. Res. Commun. 221, 67–71. [DOI] [PubMed] [Google Scholar]
- 60).Woll S., Windoffer R., Leube R.E. (2007) p38 MAPK-dependent shaping of the keratin cytoskeleton in cultured cells. J. Cell Biol. 177, 795–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61).He T., Stepulak A., Holmstrom T.H., Omary M.B., Eriksson J.E. (2002) The intermediate filament protein keratin 8 is a novel cytoplasmic substrate for c-Jun N-terminal kinase. J. Biol. Chem. 277, 10767–10774. [DOI] [PubMed] [Google Scholar]
- 62).Gonda Y., Nishizawa K., Ando S., Kitamura S., Minoura Y., Nishi Y., et al. (1990) Involvement of protein kinase C in the regulation of assembly-disassembly of neurofilaments in vitro. Biochem. Biophys. Res. Commun. 167, 1316–1325. [DOI] [PubMed] [Google Scholar]
- 63).Sihag R.K., Nixon R.A. (1991) Identification of Ser-55 as a major protein kinase A phosphorylation site on the 70-kDa subunit of neurofilaments. Early turnover during axonal transport. J. Biol. Chem. 266, 18861–18867. [PubMed] [Google Scholar]
- 64).Kirkcaldie M.T.K., Dwyer S.T. (2017) The third wave: Intermediate filaments in the maturing nervous system. Mol. Cell. Neurosci. 84, 68–76. [DOI] [PubMed] [Google Scholar]
- 65).Mukhopadhyay R., Kumar S., Hoh J.H. (2004) Molecular mechanisms for organizing the neuronal cytoskeleton. BioEssays 26, 1017–1025. [DOI] [PubMed] [Google Scholar]
- 66).Snider N.T., Park H., Omary M.B. (2013) A conserved rod domain phosphotyrosine that is targeted by the phosphatase PTP1B promotes keratin 8 protein insolubility and filament organization. J. Biol. Chem. 288, 31329–31337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67).Zhang Y.Q., Sarge K.D. (2008) Sumoylation regulates lamin A function and is lost in lamin A mutants associated with familial cardiomyopathies. J. Cell Biol. 182, 35–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68).Boudreau É., Labib S., Bertrand A.T., Decostre V., Bolongo P.M., Sylvius N., et al. (2012) Lamin A/C mutants disturb sumo1 localization and sumoylation in vitro and in vivo. PLoS One 7, e45918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69).Snider N.T., Weerasinghe S.V., Iniguez-Lluhi J.A., Herrmann H., Omary M.B. (2011) Keratin hypersumoylation alters filament dynamics and is a marker for human liver disease and keratin mutation. J. Biol. Chem. 286, 2273–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70).Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., et al. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840. [DOI] [PubMed] [Google Scholar]
- 71).Snider N.T., Leonard J.M., Kwan R., Griggs N.W., Rui L., Omary M.B. (2013) Glucose and SIRT2 reciprocally mediate the regulation of keratin 8 by lysine acetylation. J. Cell Biol. 200, 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72).Nishizawa K., Yano T., Shibata M., Ando S., Saga S., Takahashi T., et al. (1991) Specific localization of phosphointermediate filament protein in the constricted area of dividing cells. J. Biol. Chem. 266, 3074–3079. [PubMed] [Google Scholar]
- 73).Yano T., Taura C., Shibata M., Hirono Y., Ando S., Kusubata M., et al. (1991) A monoclonal antibody to the phosphorylated form of glial fibrillary acidic protein: Application to a non-radioactive method for measuring protein kinase activities. Biochem. Biophys. Res. Commun. 175, 1144–1151. [DOI] [PubMed] [Google Scholar]
- 74).Inagaki M., Gonda Y., Matsuyama M., Nishizawa K., Nishi Y., Sato C. (1988) Intermediate filament reconstitution in vitro. The role of phosphorylation on the assembly-disassembly of desmin. J. Biol. Chem. 263, 5970–5978. [PubMed] [Google Scholar]
- 75).Yano T., Tokui T., Nishi Y., Nishizawa K., Shibata M., Kikuchi K., et al. (1991) Phosphorylation of keratin intermediate filaments by protein kinase C, by calmodulin-dependent protein kinase and by cAMP-dependent protein kinase. Eur. J. Biochem. 197, 281–290. [DOI] [PubMed] [Google Scholar]
- 76).Tanaka J., Ogawara M., Ando S., Shibata M., Yatani R., Kusagawa M., et al. (1993) Phosphorylation of a 62 kd porcine α-internexin, a newly identified intermediate filament protein. Biochem. Biophys. Res. Commun. 196, 115–123. [DOI] [PubMed] [Google Scholar]
- 77).Matsuoka Y., Nishizawa K., Yano T., Shibata M., Ando S., Takahashi T., et al. (1992) Two different protein kinases act on a different time schedule as glial filament kinases during mitosis. EMBO J. 11, 2895–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78).Sekimata M., Tsujimura K., Tanaka J., Takeuchi Y., Inagaki N., Inagaki M. (1996) Detection of protein kinase activity specifically activated at metaphase-anaphase transition. J. Cell Biol. 132, 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79).Kosako H., Amano M., Yanagida M., Tanabe K., Nishi Y., Kaibuchi K., et al. (1997) Phosphorylation of glial fibrillary acidic protein at the same sites by cleavage furrow kinase and Rho-associated kinase. J. Biol. Chem. 272, 10333–10336. [DOI] [PubMed] [Google Scholar]
- 80).Goto H., Kosako H., Tanabe K., Yanagida M., Sakurai M., Amano M., et al. (1998) Phosphorylation of vimentin by Rho-associated kinase at a unique amino-terminal site that is specifically phosphorylated during cytokinesis. J. Biol. Chem. 273, 11728–11736. [DOI] [PubMed] [Google Scholar]
- 81).Nagata K., Izawa I., Inagaki M. (2001) A decade of site- and phosphorylation state-specific antibodies: Recent advances in studies of spatiotemporal protein phosphorylation. Genes Cells 6, 653–664. [DOI] [PubMed] [Google Scholar]
- 82).Goto H., Tanabe K., Manser E., Lim L., Yasui Y., Inagaki M. (2002) Phosphorylation and reorganization of vimentin by p21-activated kinase (PAK). Genes Cells. 7, 91–97. [DOI] [PubMed] [Google Scholar]
- 83).Goto H., Yasui Y., Kawajiri A., Nigg E.A., Terada Y., Tatsuka M., et al. (2003) Aurora-B regulates the cleavage furrow-specific vimentin phosphorylation in the cytokinetic process. J. Biol. Chem. 278, 8526–8530. [DOI] [PubMed] [Google Scholar]
- 84).Kawajiri A., Yasui Y., Goto H., Tatsuka M., Takahashi M., Nagata K., et al. (2003) Functional significance of the specific sites phosphorylated in desmin at cleavage furrow: Aurora-B may phosphorylate and regulate type III intermediate filaments during cytokinesis coordinatedly with Rho-kinase. Mol. Biol. Cell 14, 1489–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85).Goto H., Kiyono T., Tomono Y., Kawajiri A., Urano T., Furukawa K., et al. (2006) Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat. Cell Biol. 8, 180–187. [DOI] [PubMed] [Google Scholar]
- 86).Goto H., Inagaki M. (2007) Production of a site- and phosphorylation state-specific antibody. Nat. Protoc. 2, 2574–2581. [DOI] [PubMed] [Google Scholar]
- 87).Makihara H., Inaba H., Enomoto A., Tanaka H., Tomono Y., Ushida K., et al. (2016) Desmin phosphorylation by Cdk1 is required for efficient separation of desmin intermediate filaments in mitosis and detected in murine embryonic/newborn muscle and human rhabdomyosarcoma tissues. Biochem. Biophys. Res. Commun. 478, 1323–1329. [DOI] [PubMed] [Google Scholar]
- 88).Inaba H., Yamakawa D., Tomono Y., Enomoto A., Mii S., Kasahara K., et al. (2018) Regulation of keratin 5/14 intermediate filaments by CDK1, Aurora-B, and Rho-kinase. Biochem. Biophys. Res. Commun. 498, 544–550. [DOI] [PubMed] [Google Scholar]
- 89).Goto H., Tanaka H., Kasahara K., Inagaki M. (2016) Phospho-specific antibody probes of intermediate filament proteins. Methods Enzymol. 568, 85–111. [DOI] [PubMed] [Google Scholar]
- 90).Herrmann H., Hesse M., Reichenzeller M., Aebi U., Magin T.M. (2003) Functional complexity of intermediate filament cytoskeletons: From structure to assembly to gene ablation. Int. Rev. Cytol. 223, 83–175. [DOI] [PubMed] [Google Scholar]
- 91).Coulombe P.A., Wong P. (2004) Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat. Cell Biol. 6, 699–706. [DOI] [PubMed] [Google Scholar]
- 92).Flitney E.W., Kuczmarski E.R., Adam S.A., Goldman R.D. (2009) Insights into the mechanical properties of epithelial cells: The effects of shear stress on the assembly and remodeling of keratin intermediate filaments. FASEB J. 23, 2110–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93).Sivaramakrishnan S., Schneider J.L., Sitikov A., Goldman R.D., Ridge K.M. (2009) Shear stress induced reorganization of the keratin intermediate filament network requires phosphorylation by protein kinase C ζ. Mol. Biol. Cell 20, 2755–2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94).Toivola D.M., Strnad P., Habtezion A., Omary M.B. (2010) Intermediate filaments take the heat as stress proteins. Trends Cell Biol. 20, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95).Izawa I., Nishizawa M., Ohtakara K., Ohtsuka K., Inada H., Inagaki M. (2000) Identification of Mrj, a DnaJ/Hsp40 family protein, as a keratin 8/18 filament regulatory protein. J. Biol. Chem. 275, 34521–34527. [DOI] [PubMed] [Google Scholar]
- 96).Liao J., Lowthert L.A., Ghori N., Omary M.B. (1995) The 70-kDa heat shock proteins associate with glandular intermediate filaments in an ATP-dependent manner. J. Biol. Chem. 270, 915–922. [DOI] [PubMed] [Google Scholar]
- 97).Marceau N., Schutte B., Gilbert S., Loranger A., Henfling M.E., Broers J.L., et al. (2007) Dual roles of intermediate filaments in apoptosis. Exp. Cell Res. 313, 2265–2281. [DOI] [PubMed] [Google Scholar]
- 98).Inada H., Izawa I., Nishizawa M., Fujita E., Kiyono T., Takahashi T., et al. (2001) Keratin attenuates tumor necrosis factor-induced cytotoxicity through association with TRADD. J. Cell Biol. 155, 415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99).Caulin C., Ware C.F., Magin T.M., Oshima R.G. (2000) Keratin-dependent, epithelial resistance to tumor necrosis factor-induced apoptosis. J. Cell Biol. 149, 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100).Lee J.C., Schickling O., Stegh A.H., Oshima R.G., Dinsdale D., Cohen G.M., et al. (2002) DEDD regulates degradation of intermediate filaments during apoptosis. J. Cell Biol. 158, 1051–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101).Dinsdale D., Lee J.C., Dewson G., Cohen G.M., Peter M.E. (2004) Intermediate filaments control the intracellular distribution of caspases during apoptosis. Am. J. Pathol. 164, 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102).Gilbert S., Loranger A., Marceau N. (2004) Keratins modulate c-Flip/extracellular signal-regulated kinase 1 and 2 antiapoptotic signaling in simple epithelial cells. Mol. Cell. Biol. 24, 7072–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103).Green K.J., Bohringer M., Gocken T., Jones J.C. (2005) Intermediate filament associated proteins. Adv. Protein Chem. 70, 143–202. [DOI] [PubMed] [Google Scholar]
- 104).Freeman A.K., Morrison D.K. (2011) 14-3-3 Proteins: Diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 22, 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105).Ku N.O., Michie S., Resurreccion E.Z., Broome R.L., Omary M.B. (2002) Keratin binding to 14-3-3 proteins modulates keratin filaments and hepatocyte mitotic progression. Proc. Natl. Acad. Sci. U.S.A. 99, 4373–4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106).Kim S., Wong P., Coulombe P.A. (2006) A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature 441, 362–365. [DOI] [PubMed] [Google Scholar]
- 107).Wang R.C., Wei Y., An Z., Zou Z., Xiao G., Bhagat G., et al. (2012) Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108).Tsujimura K., Ogawara M., Takeuchi Y., Imajoh-Ohmi S., Ha M.H., Inagaki M. (1994) Visualization and function of vimentin phosphorylation by cdc2 kinase during mitosis. J. Biol. Chem. 269, 31097–31106. [PubMed] [Google Scholar]
- 109).Kosako H., Goto H., Yanagida M., Matsuzawa K., Fujita M., Tomono Y., et al. (1999) Specific accumulation of Rho-associated kinase at the cleavage furrow during cytokinesis: Cleavage furrow-specific phosphorylation of intermediate filaments. Oncogene 18, 2783–2788. [DOI] [PubMed] [Google Scholar]
- 110).Yasui Y., Goto H., Matsui S., Manser E., Lim L., Nagata K., et al. (2001) Protein kinases required for segregation of vimentin filaments in mitotic process. Oncogene 20, 2868–2876. [DOI] [PubMed] [Google Scholar]
- 111).Yasui Y., Urano T., Kawajiri A., Nagata K., Tatsuka M., Saya H., et al. (2004) Autophosphorylation of a newly identified site of Aurora-B is indispensable for cytokinesis. J. Biol. Chem. 279, 12997–13003. [DOI] [PubMed] [Google Scholar]
- 112).Yamaguchi T., Goto H., Yokoyama T., Sillje H., Hanisch A., Uldschmid A., et al. (2005) Phosphorylation by Cdk1 induces Plk1-mediated vimentin phosphorylation during mitosis. J. Cell Biol. 171, 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113).Matsuyama M., Tanaka H., Inoko A., Goto H., Yonemura S., Kobori K., et al. (2013) Defect of mitotic vimentin phosphorylation causes microophthalmia and cataract via aneuploidy and senescence in lens epithelial cells. J. Biol. Chem. 288, 35626–35635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114).Kasahara K., Goto H., Izawa I., Kiyono T., Watanabe N., Elowe S., et al. (2013) PI 3-kinase-dependent phosphorylation of Plk1-Ser99 promotes association with 14-3-3γ and is required for metaphase-anaphase transition. Nat. Commun. 4, 1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115).Ikawa K., Satou A., Fukuhara M., Matsumura S., Sugiyama N., Goto H., et al. (2014) Inhibition of endocytic vesicle fusion by Plk1-mediated phosphorylation of vimentin during mitosis. Cell Cycle 13, 126–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116).Tanaka H., Goto H., Inoko A., Makihara H., Enomoto A., Horimoto K., et al. (2015) Cytokinetic failure-induced tetraploidy develops into aneuploidy, triggering skin aging in phosphovimentin-deficient mice. J. Biol. Chem. 290, 12984–12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117).Nishizawa M., Izawa I., Inoko A., Hayashi Y., Nagata K., Yokoyama T., et al. (2005) Identification of trichoplein, a novel keratin filament-binding protein. J. Cell Sci. 118, 1081–1090. [DOI] [PubMed] [Google Scholar]
- 118).Ibi M., Zou P., Inoko A., Shiromizu T., Matsuyama M., Hayashi Y., et al. (2011) Trichoplein controls microtubule anchoring at the centrosome by binding to Odf2 and ninein. J. Cell Sci. 124, 857–864. [DOI] [PubMed] [Google Scholar]
- 119).Inoko A., Matsuyama M., Goto H., Ohmuro-Matsuyama Y., Hayashi Y., Enomoto M., et al. (2012) Trichoplein and Aurora A block aberrant primary cilia assembly in proliferating cells. J. Cell Biol. 197, 391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120).Izawa I., Goto H., Kasahara K., Inagaki M. (2015) Current topics of functional links between primary cilia and cell cycle. Cilia 4, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121).Kasahara K., Kawakami Y., Kiyono T., Yonemura S., Kawamura Y., Era S., et al. (2014) Ubiquitin-proteasome system controls ciliogenesis at the initial step of axoneme extension. Nat. Commun. 5, 5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122).Inaba H., Goto H., Kasahara K., Kumamoto K., Yonemura S., Inoko A., et al. (2016) Ndel1 suppresses ciliogenesis in proliferating cells by regulating the trichoplein-Aurora A pathway. J. Cell Biol. 212, 409–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123).Goto H., Inaba H., Inagaki M. (2017) Mechanisms of ciliogenesis suppression in dividing cells. Cell. Mol. Life Sci. 74, 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124).Kasahara K., Aoki H., Kiyono T., Wang S., Kagiwada H., Yuge M., et al. (2018) EGF receptor kinase suppresses ciliogenesis through activation of USP8 deubiquitinase. Nat. Commun. 9, 758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125).Nishimura Y., Kasahara K., Shiromizu T., Watanabe M., Inagaki M. (2019) Primary cilia as signaling hubs in health and disease. Adv. Sci. (Weinh) 6, 1801138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126).Takai Y., Ogawara M., Tomono Y., Moritoh C., Imajoh-Ohmi S., Tsutsumi O., et al. (1996) Mitosis-specific phosphorylation of vimentin by protein kinase C coupled with reorganization of intracellular membranes. J. Cell Biol. 133, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127).Yasui Y., Amano M., Nagata K., Inagaki N., Nakamura H., Saya H., et al. (1998) Roles of Rho-associated kinase in cytokinesis; Mutations in Rho-associated kinase phosphorylation sites impair cytokinetic segregation of glial filaments. J. Cell Biol. 143, 1249–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128).Kawajiri A., Inagaki M. (2004) Approaches to study phosphorylation of intermediate filament proteins using site-specific and phosphorylation state-specific antibodies. Methods Cell Biol. 78, 353–371. [DOI] [PubMed] [Google Scholar]
- 129).Tanaka K., Goto H., Nishimura Y., Kasahara K., Mizoguchi A., Inagaki M. (2018) Tetraploidy in cancer and its possible link to aging. Cancer Sci. 109, 2632–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130).Brown A., Geiger H. (2018) Chromosome integrity checkpoints in stem and progenitor cells: Transitions upon differentiation, pathogenesis, and aging. Cell. Mol. Life Sci. 75, 3771–3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131).Eriksson J.E., Dechat T., Grin B., Helfand B., Mendez M., Pallari H.M., et al. (2009) Introducing intermediate filaments: From discovery to disease. J. Clin. Invest. 119, 1763–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132).Eriksson J.E., Opal P., Goldman R.D. (1992) Intermediate filament dynamics. Curr. Opin. Cell Biol. 4, 99–104. [DOI] [PubMed] [Google Scholar]
- 133).Steinert P.M. (1993) Structure, function, and dynamics of keratin intermediate filaments. J. Invest. Dermatol. 100, 729–734. [DOI] [PubMed] [Google Scholar]
- 134).Fuchs E., Weber K. (1994) Intermediate filaments: Structure, dynamics, function, and disease. Annu. Rev. Biochem. 63, 345–382. [DOI] [PubMed] [Google Scholar]
- 135).Foisner R., Bohn W., Mannweiler K., Wiche G. (1995) Distribution and ultrastructure of plectin arrays in subclones of rat glioma C6 cells differing in intermediate filament protein (vimentin) expression. J. Struct. Biol. 115, 304–317. [DOI] [PubMed] [Google Scholar]
- 136).Svitkina T.M., Verkhovsky A.B., Borisy G.G. (1996) Plectin sidearms mediate interaction of intermediate filaments with microtubules and other components of the cytoskeleton. J. Cell Biol. 135, 991–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137).Wiche G. (1998) Role of plectin in cytoskeleton organization and dynamics. J. Cell Sci. 111, 2477–2486. [DOI] [PubMed] [Google Scholar]
- 138).Scliwa M., Honer B. (1993) Microtubules, centrosomes and intermediate filaments in directed cell movement. Trends Cell Biol. 3, 377–380. [DOI] [PubMed] [Google Scholar]
- 139).Petry S., Vale R.D. (2015) Microtubule nucleation at the centrosome and beyond. Nat. Cell Biol. 17, 1089–1093. [DOI] [PubMed] [Google Scholar]
- 140).Farina F., Gaillard J., Guerin C., Coute Y., Sillibourne J., Blanchoin L., et al. (2016) The centrosome is an actin-organizing centre. Nat. Cell Biol. 18, 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141).Lemmon M.A., Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142).Zañudo J., Steinway S., Albert R. (2018) Discrete dynamic network modeling of oncogenic signaling: Mechanistic insights for personalized treatment of cancer. Curr. Opin. Syst. Biol. 9, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143).Appert-Collin A., Hubert P., Cremel G., Bennasroune A. (2015) Role of ErbB receptors in cancer cell migration and invasion. Front. Pharmacol. 6, 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144).Mishra R., Hanker A.B., Garrett J.T. (2017) Genomic alterations of ERBB receptors in cancer: Clinical implications. Oncotarget 8, 114371–114392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145).Witsch E., Sela M., Yarden Y. (2010) Roles for growth factors in cancer progression. Physiology (Bethesda) 25, 85–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146).Tucker R.W., Pardee A.B., Fujiwara K. (1979) Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell 17, 527–535. [DOI] [PubMed] [Google Scholar]
- 147).Tucker R.W., Scher C.D., Stiles C.D. (1979) Centriole deciliation associated with the early response of 3T3 cells to growth factors but not to SV40. Cell 18, 1065–1072. [DOI] [PubMed] [Google Scholar]
- 148).Pugacheva E.N., Jablonski S.A., Hartman T.R., Henske E.P., Golemis E.A. (2007) HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell 129, 1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149).Sugimoto M., Inoko A., Shiromizu T., Nakayama M., Zou P., Yonemura S., et al. (2008) The keratin-binding protein Albatross regulates polarization of epithelial cells. J. Cell Biol. 183, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150).Sasaki S., Shionoya A., Ishida M., Gambello M.J., Yingling J., Wynshaw-Boris A., et al. (2000) A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron 28, 681–696. [DOI] [PubMed] [Google Scholar]
- 151).Niethammer M., Smith D.S., Ayala R., Peng J., Ko J., Lee M.S., et al. (2000) NUDEL is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron 28, 697–711. [DOI] [PubMed] [Google Scholar]
- 152).Bradshaw N.J., Hayashi M.A. (2017) NDE1 and NDEL1 from genes to (mal)functions: Parallel but distinct roles impacting on neurodevelopmental disorders and psychiatric illness. Cell. Mol. Life Sci. 74, 1191–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153).Tanos B.E., Yang H.J., Soni R., Wang W.J., Macaluso F.P., Asara J.M., et al. (2013) Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 27, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154).Sumigray K.D., Chen H., Lechler T. (2011) Lis1 is essential for cortical microtubule organization and desmosome stability in the epidermis. J. Cell Biol. 194, 631–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155).Sumigray K.D., Lechler T. (2011) Control of cortical microtubule organization and desmosome stability by centrosomal proteins. Bioarchitecture 1, 221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156).Liang Y., Yu W., Li Y., Yang Z., Yan X., Huang Q., et al. (2004) Nudel functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein. J. Cell Biol. 164, 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157).Kholmanskikh S.S., Dobrin J.S., Wynshaw-Boris A., Letourneau P.C., Ross M.E. (2003) Disregulated RhoGTPases and actin cytoskeleton contribute to the migration defect in Lis1-deficient neurons. J. Neurosci. 23, 8673–8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158).Lin Y., Mori E., Kato M., Xiang S., Wu L., Kwon I., et al. (2016) Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167, 789–802.E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159).Didonna A., Opal P. (2019) The role of neurofilament aggregation in neurodegeneration: Lessons from rare inherited neurological disorders. Mol. Neurodegener. 14, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160).Sun J., Groppi V.E., Gui H., Chen L., Xie Q., Liu L., et al. (2016) High-throughput screening for drugs that modulate intermediate filament proteins. Methods Enzymol. 568, 163–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161).Danielsson F., Peterson M.K., Caldeira Araujo H., Lautenschlager F., Gad A.K.B. (2018) Vimentin diversity in health and disease. Cells 7, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162).Tsikitis M., Galata Z., Mavroidis M., Psarras S., Capetanaki Y. (2018) Intermediate filaments in cardiomyopathy. Biophys. Rev. 10, 1007–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163).Kerns M., DePianto D., Yamamoto M., Coulombe P.A. (2010) Differential modulation of keratin expression by sulforaphane occurs via Nrf2-dependent and -independent pathways in skin epithelia. Mol. Biol. Cell 21, 4068–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164).Kerns M.L., DePianto D., Dinkova-Kostova A.T., Talalay P., Coulombe P.A. (2007) Reprogramming of keratin biosynthesis by sulforaphane restores skin integrity in epidermolysis bullosa simplex. Proc. Natl. Acad. Sci. U.S.A. 104, 14460–14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165).Kerns M.L., Hakim J.M., Lu R.G., Guo Y., Berroth A., Kaspar R.L., et al. (2016) Oxidative stress and dysfunctional NRF2 underlie pachyonychia congenita phenotypes. J. Clin. Invest. 126, 2356–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166).Morimitsu Y., Nakagawa Y., Hayashi K., Fujii H., Kumagai T., Nakamura Y., et al. (2002) A sulforaphane analogue that potently activates the Nrf2-dependent detoxification pathway. J. Biol. Chem. 277, 3456–3463. [DOI] [PubMed] [Google Scholar]
- 167).Thimmulappa R.K., Mai K.H., Srisuma S., Kensler T.W., Yamamoto M., Biswal S. (2002) Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 62, 5196–5203. [PubMed] [Google Scholar]
- 168).Zhang D.D., Hannink M. (2003) Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol. Cell. Biol. 23, 8137–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169).Kwan R., Chen L., Looi K., Tao G.Z., Weerasinghe S.V., Snider N.T., et al. (2015) PKC412 normalizes mutation-related keratin filament disruption and hepatic injury in mice by promoting keratin-myosin binding. Hepatology (Baltimore, Md.) 62, 1858–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170).Bollong M.J., Pietila M., Pearson A.D., Sarkar T.R., Ahmad I., Soundararajan R., et al. (2017) A vimentin binding small molecule leads to mitotic disruption in mesenchymal cancers. Proc. Natl. Acad. Sci. U.S.A. 114, E9903–E9912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171).Trogden K.P., Battaglia R.A., Kabiraj P., Madden V.J., Herrmann H., Snider N.T. (2018) An image-based small-molecule screen identifies vimentin as a pharmacologically relevant target of simvastatin in cancer cells. FASEB J. 32, 2841–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172).MacRae C.A., Peterson R.T. (2015) Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 14, 721–731. [DOI] [PubMed] [Google Scholar]
- 173).Nishimura Y., Murakami S., Ashikawa Y., Sasagawa S., Umemoto N., Shimada Y., et al. (2015) Zebrafish as a systems toxicology model for developmental neurotoxicity testing. Congenit. Anom. (Kyoto) 55, 1–16. [DOI] [PubMed] [Google Scholar]
- 174).Nishimura Y., Inoue A., Sasagawa S., Koiwa J., Kawaguchi K., Kawase R., et al. (2016) Using zebrafish in systems toxicology for developmental toxicity testing. Congenit. Anom. (Kyoto) 56, 18–27. [DOI] [PubMed] [Google Scholar]
- 175).Naert T., Vleminckx K. (2018) CRISPR/Cas9 disease models in zebrafish and Xenopus: The genetic renaissance of fish and frogs. Drug Discov. Today. Technol. 28, 41–52. [DOI] [PubMed] [Google Scholar]
- 176).Ramspacher C., Steed E., Boselli F., Ferreira R., Faggianelli N., Roth S., et al. (2015) Developmental alterations in heart biomechanics and skeletal muscle function in desmin mutants suggest an early pathological root for desminopathies. Cell Reports 11, 1564–1576. [DOI] [PubMed] [Google Scholar]
- 177).May-Simera H.L., Wan Q., Jha B.S., Hartford J., Khristov V., Dejene R., et al. (2018) Primary cilium-mediated retinal pigment epithelium maturation is disrupted in ciliopathy patient cells. Cell Reports 22, 189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178).Omary M.B. (2017) Intermediate filament proteins of digestive organs: Physiology and pathophysiology. Am. J. Physiol. Gastrointest. Liver Physiol. 312, G628–G634. [DOI] [PMC free article] [PubMed] [Google Scholar]