Abstract
Biliary atresia (BA) is a neonatal liver disease featuring cholestasis and severe liver fibrosis (LF). Despite advances in the development of surgical treatment, lacking an early diagnostic marker and intervention of LF invariably leads to death from end-stage liver disease in the early years of life. We previously reported that knockout of sphingosine 1-phosphate receptor 2 (S1PR2) protected mice from bile duct ligation (BDL)-induced cholangiocyte proliferation and LF. Our recent studies further showed that both hepatic and serum exosomal long noncoding RNA H19 (lncR-NAH19) levels are correlated with cholestatic injury in multidrug resistance 2 knockout (Mdr2−/−) mice. However, the role of lncRNAH19 in BA progression remains unclear. Here, we show that both hepatic and serum exosomal H19 levels are positively correlated with severity of fibrotic liver injuries in BA patients. H19 deficiency protects mice from BDL-induced cholangiocyte proliferation and LF by inhibiting bile-acid–induced expression and activation of S1PR2 and sphingosine kinase 2 (SphK2). Furthermore, H19 acts as a molecular sponge for members of the microRNA let-7 family, which results in up-regulation of high-mobility group AT-hook 2 (HMGA2), a known target of let-7 and enhancement of biliary proliferation. Conclusion: These results indicate that H19 plays a critical role in cholangiocyte proliferation and cholestatic liver injury in BA by regulating the S1PR2/SphK2 and let-7/HMGA2 axis. Serum exosomal H19 may represent a noninvasive diagnostic biomarker and potential therapeutic target for BA.
Biliary atresia (BA) is a severe liver disease in neonates, which is caused by obliteration of the intra-and the extrahepatic biliary duct leading to cholestasis, and progressive liver injury and fibrosis.(1-3) Early diagnosis and Kasai hepatoportoenterostomy (KHPE) helps prevent the development of cirrhosis and improve long-time survival. However, because of the lack of a reliable early diagnostic marker and limited understanding of disease pathology, BA remains the most common cause of advanced cirrhosis and the leading indication for pediatric liver transplantation.(4-6)
Sphingosine-1-phosphate is an essential bioactive lipid molecule that can directly act as an intracellular signaling molecule or function as the natural ligand of five different G protein-coupled receptors (sphingosine-1-phosphate receptors 1–5).(7-9) We recently reported that conjugated bile-acid–mediated activation of sphingosine 1-phosphate receptor 2 (S1PR2) contributes to disease progression of cholestatic liver injury (CLI) by promoting cholangiocyte proliferation and inflammatory response.(10-12) We also reported that dysregulation of several microRNAs (miRNAs) expression contributes to cholangiocyte proliferation and liver fibrosis (LF) in the cholestatic liver of BA patients.(13,14) The miRNA, let-7, belongs to a family of miRNAs required for development, growth, and metabolism regulation.(15,16) A recent study demonstrated that dysregulation of let-7 expression contributes to cholestatic liver diseases.(17,18)
Long noncoding RNAs (lncRNAs) are a group of transcripts that are longer than 200 nucleotides in length, but do not encode proteins.(19) The lncRNA, H19, is an imprinted and maternally expressed gene that is conserved between humans and the mouse and plays a vital role in regulation of cell proliferation and differentiation.(20-22) H19 is highly expressed in fetal liver, but repressed significantly after birth. Intriguingly, it has been reported that H19 is up-regulated in human liver diseases and CLI animal models, including CCl4-induced liver injury, bile duct ligation (BDL)-induced cholestatic injury, and the multidrug resistance 2 knockout (Mdr2−/−) mouse model,(23-26) implicating its important role in disease progression of CLI. Our recent studies indicate that hepatic H19 level is correlated with severity of cholestatic injury in Mdr2−/− mice.(25) Moreover, cholangiocyte-derived exosomes mediate the transfer of H19 into hepatocytes and promote cholestatic injury.(25,27) Although these studies suggest a causal link between H19 and CLI, it remains unknown whether, and to what extent, H19 is involved in regulation of cholangiocyte hyperplasia and LF in BA patients.
In the present study, we demonstrated that hepatic H19 expression was significantly up-regulated in BA patients. Consistent with our previous findings in animal CLI models, serum exosomal H19 expression level was closely correlated to severity of CLI and LF progression in BA patients.
Materials and Methods
HUMAN SPECIMENS
A total of 53 liver specimens were retrieved from biliary BA patients who underwent surgery. Eleven normal adjacent nontumor liver tissues were taken from age-matched hepatoblastoma patients and used as controls. Twenty-four serum specimens were obtained from BA patients before the surgery. Twenty-four serum samples from age-matched healthy infants were used as controls. All patients’ guardians provided written informed consent. This study was approved by the Faculty of Medicine’s Ethics Committee of Xin Hua Hospital, Shanghai Jiaotong University, China (XHEC-C-2016–063). Detailed clinical information of the patients is presented in Supporting Table S1. All methods in this study were carried out following the relevant guidelines.
ANIMALS AND BDL MOUSE MODEL
H19−/−mice (H19Δ Exon1/+) with C57/BL6 background were provided by Dr. Karl Pfeifer at the National Institutes of Health. H19−/− mice (H19 Δ Exon1–5) with C57/BL6 background were obtained from Nanjing Biomedical Research Institute of Nanjing University (Nanjing, China; Supporting Fig. S1). Mdr2−/− mice with C57BL/6J background were provided by Dr. Daniel Goldenberg at the Department of Pathology, Hadassah-Hebrew University Medical Center, Jerusalem, Israel. Mdr2−/− and H19−/− double knockout (DKO) mice were generated as described.(28) Mice were housed under a 12-hour light/dark cycle and fed standard chow and tap water ad libitum. The standard partial common BDL was performed as we described.(29) Briefly, the cystic duct and common bile duct were ligated using 7–0 nylon. Sham mice underwent similar laparotomy without BDL. In the end, the abdomen was closed by a 6–0 double-layer suture, and mice were allowed to wake up on a heating pad. After 14 days, serum and bile were collected and liver tissues were harvested for isolation of primary cholangiocytes or processed for qRT-PCR, western blotting analysis, histology, and immunohistochemistry (IHC) analysis. H19−/− mice (H19 Δ Exon1/+) BDL procedures were approved by the Virginia Commonwealth University Institutional Animal Care and Use Committee. H19−/− mice (H19Δ Exon1–5) BDL procedures were approved by Use Committee of the Xin Hua Hospital, School of Medicine, Shanghai Jiao Tong University. All animal procedures were also performed following institutional guidelines for ethical animal studies.
CELL CULTURE AND TREATMENTS
Immortalized normal large cholangiocytes (MLEs) were cultured in minimal essential medium containing 10% fetal bovine serum, penicillin G (100 U/mL), and streptomycin (100 μg/mL) at 37°C with 5% CO2 in a humidified cell-culture incubator. Mouse primary cholangiocytes were isolated by an immunoaffinity method using a rat monoclonal antibody against epithelial cell adhesion molecule (from DSHB, University of Iowa), an antigen expressed by all mouse cholangiocytes as described.(30) For H19 knockdown experiments, MLE cells were incubated with adenovirus of H19 short hairpin RNA (shRNA) for 6 hours (multiplicity of infection, 40) or control adenovirus as described.(25) The adenovirus of H19 shRNA was a gift from Dr. Li Wang (University of Connecticut). After 24 hours, cells were treated with taurocholate (TCA; 0.1 mM) for 48 hours. For the H19 overexpression study, MLE cells were plated in 60-mm plates and transfected with H19 overexpression plasmid (a gift from Dr. Jianying Wang, University of Maryland School of Medicine) or control plasmid with polyJet (SignaGen Laboratories, Rockville, MD) for 24 and 48 hours. All cells were cultured at 37°C with 5% CO2 in a humidified cell-culture incubator (Thermo scientific, Carlsbad, CA).
STATISTICAL ANALYSIS
Data statistics are presented as mean ± SD. For comparisons of different groups, one-way analysis of variance with Tukey’s post-hoc test or Student t test, using GraphPad Prism software (version 5; GraphPad Software Inc., San Diego, CA), was determined. P values <0.05 were considered statistically significant.
Results
HEPATIC H19 EXPRESSION LEVELS ARE CORRELATED TO SEVERITY OF FIBROTIC LIVER INJURY IN BA PATIENTS
A total of 53 liver specimens from BA patients and 11 control liver tissues were used in this study (Supporting Table S1). The real-time RT-PCR analysis indicated that hepatic H19 mRNA levels were significantly increased in BA patients compared to control subjects (Fig. 1A). Consistently, the fluorescence in situ hybridization (FISH) analysis also showed that H19 expression increased in livers of the BA patients when compared to control subjects (Fig. 1B and Supporting Fig. S2). To further determine whether hepatic H19 expression was correlated to disease progression in BA patients, mRNA levels of liver fibrotic marker genes, including keratin 7 (KRT7), transforming growth factor beta 1 (TGFB1), collagen type I alpha 1 (COLIA1), and actin, alpha 2, smooth muscle, aorta (ACTA2), were detected by RT-PCR. Expression of KRT7, TGFB1, COLIA1, and ACTA2 increased significantly in livers of BA patients compared to controls (Fig. 1C). Correlation analysis showed that hepatic H19 mRNA levels were positively correlated with expression levels of hepatic TGFB1 (r = 0.30; P = 0.015) and ACTA2 (r = 0.29; P = 0.017; Fig. 1D,E). Hepatocyte nuclear factor 4-alpha (HNF4α)+ hepatocytes, cytokeratin 19 (CK19)+ cholangiocytes, alpha-smooth muscle actin (α-SMA)+ stellate cells, and CD68+ macro-phages expressed H19 mRNA in livers of BA patients (Fig. 1F and Supporting Figs. S3–S6).
FIG. 1.
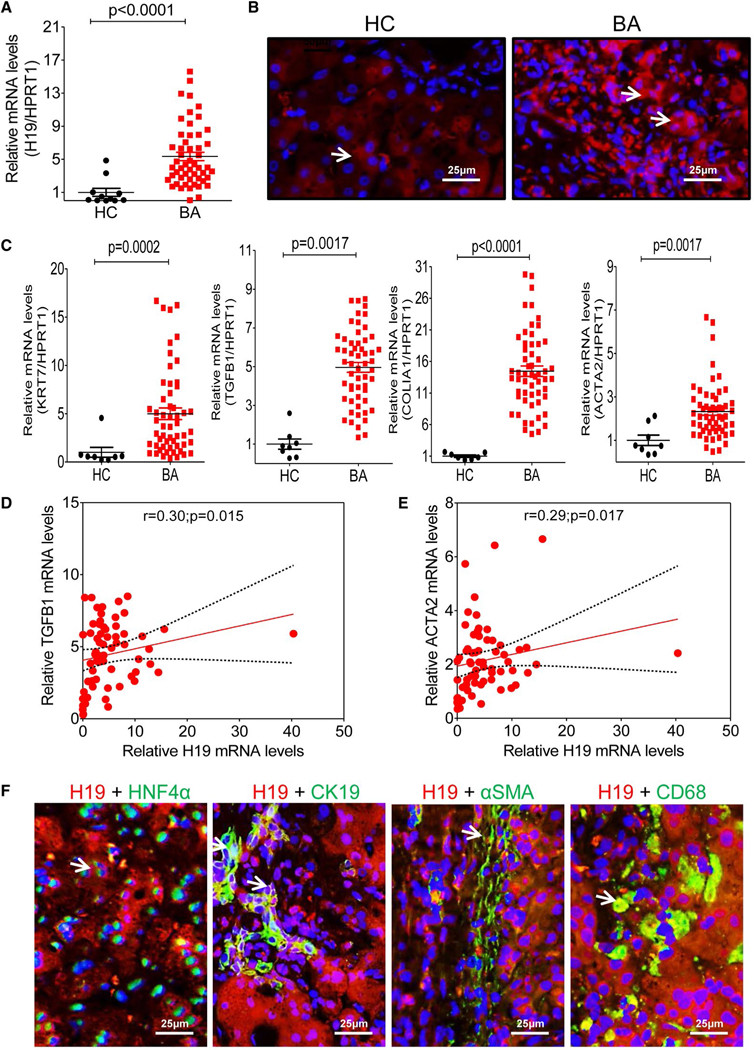
H19 increase in livers of BA patients correlated with LF progression. (A) Relative H19 mRNA levels in livers of BA patients (n = 53) and controls (n = 11) were determined by real-time RT-PCR. HPRT1 was used as an internal control. (B) Representative images of H19 FISH analysis in livers of BA (n = 8) patients and controls (n = 5). (C) Relative mRNA levels of the hepatic fibrotic marker genes, including KRT7, TGFB1, COLIA1, and ACTA2, in livers of BA patients (n = 53) and controls (n = 11). HPRT1 was used as an internal control. (D,E) Correlation analysis of H19 expression level with TGFB1 and ACTA2. (F) Representative images of costaining of H19 and HNF4α, H19 and CK19, H19 and α-SMA, and H19 and CD68 in livers of BA (n = 4–8). Arrows indicate the merged signal. Abbreviations: HC, healthy controls; HPRT1, hypoxanthine phosphoribosyltransferase 1.
Given that H19 is an imprinted and a maternally expressed gene,(31) we then compared the sex-based disparity of H19 expressed pattern and cholestatic injury in BA patients. Consistent with our previous study in Mdr2−/− mice,(25) we showed that hepatic H19 level was higher in female BA patients than in age-matched male BA patients (Supporting Fig. S7A,B). But liver functional enzyme activities were similar in male and female BA patients (Supporting Fig. S7C). Although female BA patients showed a slight increase of total bile acids—cholic acid and TCA—levels in serum and liver, they were not significantly different from male BA patients (Supporting Figs. S8–S9).
LEVELS OF SERUM EXOSOMAL H19 ARE CORRELATED WITH LF SEVERITY IN BA PATIENTS
Given that hepatic H19 was associated with liver injury and fibrosis in BA patients, H19 levels in serum may be a useful and critical marker of prognosis in liver injury and fibrosis of BA patients. To test this hypothesis, we isolated serum exosomes from BA patients with different stages of LF and age-matched controls. Transmission electron microscope (TEM) analysis showed that size of the exosomal diameter was approximately 50–150 nm (Fig. 2A). Expression of exosome marker genes, including the CD63 and CD81, was further confirmed by western blotting analysis (Fig. 2B). Similar to the findings in livers of BA patients, the RT-PCR analysis showed that H19 markedly increased in serum exosomes of BA patients compared to controls (Fig. 2C). There were 16 BA patients with different stages of LF that were classified into two groups: group 1 with mild LF (fibrosis grades I and II; 8 cases) and group 2 with severe LF (fibrosis grades III and IV; 8 cases), according to Masson’s trichrome staining (Fig. 2D and Supporting Figs. S10 and S11). The RT-PCR analysis indicated that expression of serum exosomal H19 levels in severe LF increased by around 6-fold over those with mild LF (P < 0.01; Fig. 2E).
FIG. 2.
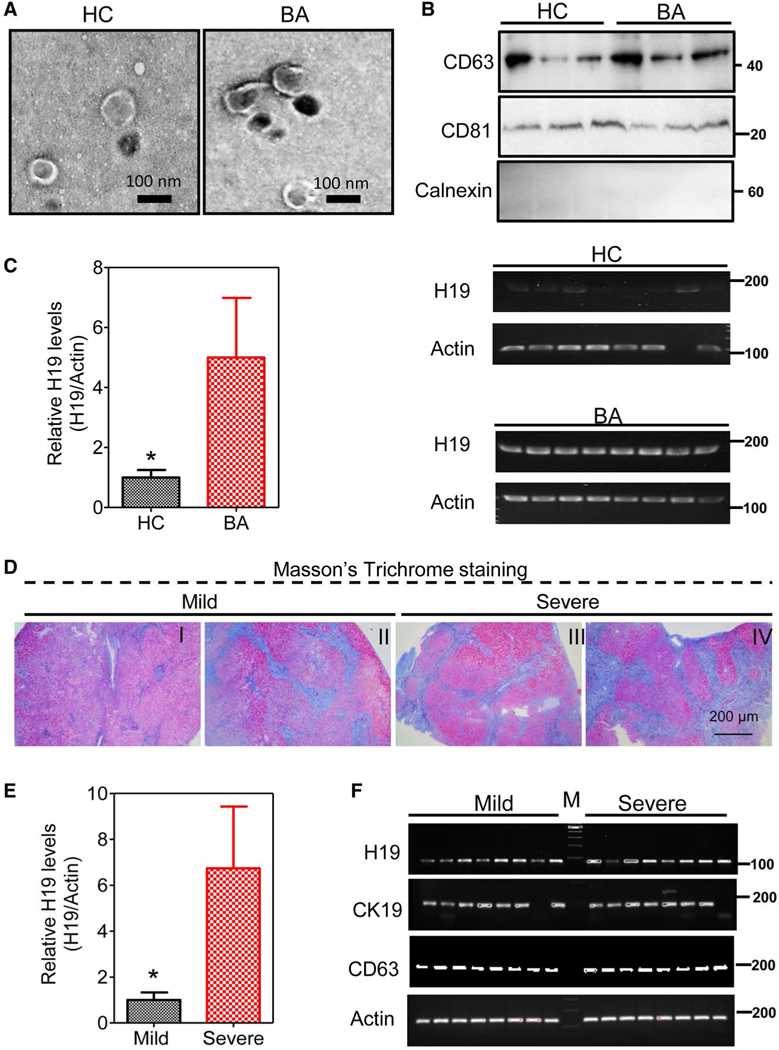
Serum exosomal H19 level is correlated with severity of LF in BA patients. (A) Representative TEM images of exosomes isolated from serum of BA patients and healthy controls. (B) Protein expression levels of CD81, CD63, and calnexin in isolated serum exosomes were determined by western blotting analysis. Representative immunoblotting images are shown. (C) Relative H19 mRNA levels in isolated exosomes from BA patients (n = 24) and healthy controls (n = 24) were determined by real-time PCR and normalized using β-actin as an internal control. Representative images of DNA agarose gels of H19 and β-actin are shown. (D) Representative images of Masson’s trichrome staining of BA patients with different grades of LF (I-IV). The group with mild fibrosis included grades I and II, and the group with severe fibrosis comprised grades III and IV. (E) Relative H19 mRNA levels in exosomes from BA patients with mild and severe LF. (F) Representative images of DNA agarose gels of H19, CK19, CD63, and β-actin in BA patients with mild and severe LF. Statistical significance relative to HC, *P < 0.05. Abbreviation: HC, healthy controls.
H19 DEFICIENCY AMELIORATES BDL-INDUCED LF AND HYPERPLASIA OF CHOLANGIOCYTES IN MICE
H19 knockout (H19−/−) mice were used to elucidate the role of H19 in BDL-induced LF and hyperplasia of cholangiocytes. After 2-week BDL, serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), and y-glutamyltransferase (GGT), alkaline phosphatase (ALP), total bilirubin (TB), and direct bilirubin (DB) levels were significantly increased in wild-type (WT) BDL mice compared to WT sham mice, but decreased in H19 / BDL mice (Fig. 3A). Hematoxylin-eosin (H&E) staining and IHC analysis showed that H19−/− BDL mice had fewer liver injuries and inflammation than WT BDL mice (Fig. 3B). Hepatic hydroxyproline levels were significantly increased in WT BDL mice, but not in H19−/− BDL mice (Fig. 4A). Masson’s Trichrome staining and Sirius Red staining further indicated that 2-week BDL significantly induced LF in WT mice, but had much less impact in H19−/− mice (Fig. 4B,C). Consistently, IHC staining and western blotting results showed that BDL significantly increased protein expression levels of α-SMA and collagen I in livers of WT BDL mice, but not in those of H19−/− BDL mice (Fig. 4D-F). To further examine the effect of BDL on cholangiocyte proliferation, intrahepatic bile duct mass was evaluated by CK19 immunofluorescence (IF) staining and western blotting analysis. BDL-induced cholangiocyte proliferation was significantly reduced in H19−/−mice (Fig. 5). In liver sections from H19−/− BDL mice, IF results showed that expression of Ck19 was significantly decreased relative to that of the WT BDL group (Fig. 5 and Supporting Fig. S12A). Additionally, the number of Ck19 and proliferating cell nuclear antigen (Pcna), Ck19 and Ki67 double-positive cells was markedly reduced in H19−/− BDL mice compared to WT BDL mice (Fig. 5C and Supporting Fig. S12B,C).
FIG. 3.
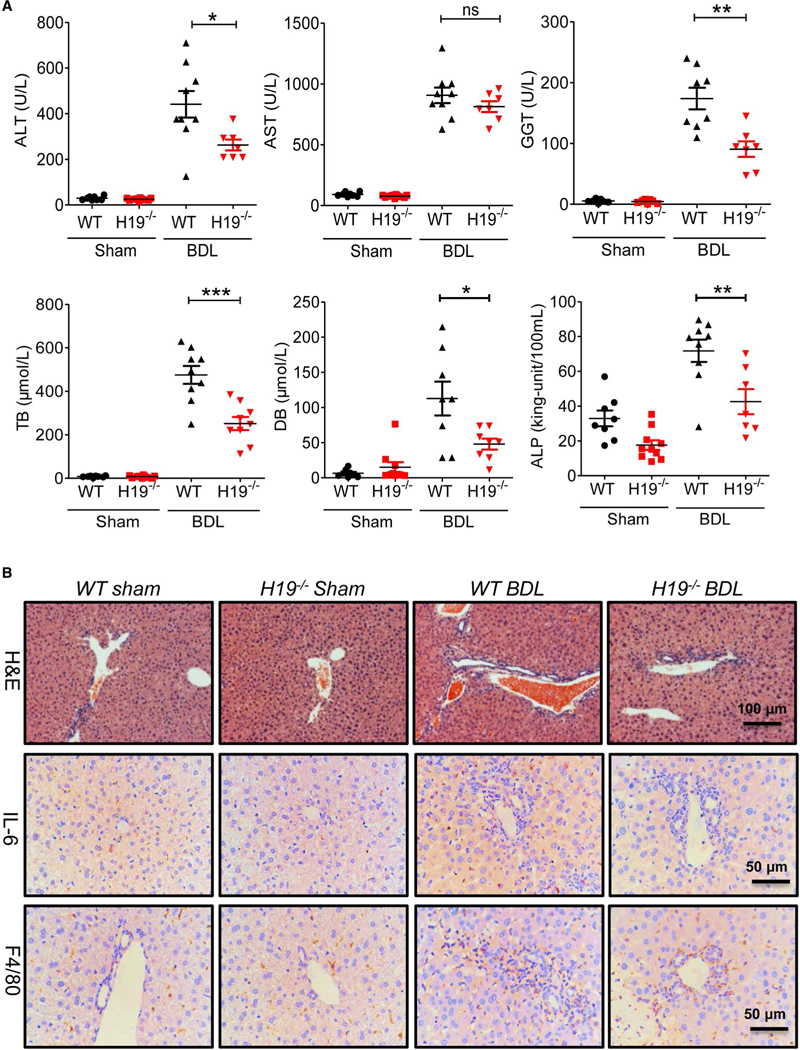
H19 knockout reduced BDL-induced liver injury and inflammation. (A) Measurement of serum AST, ALT, GGT, ALP, and bilirubin levels in WT sham, H19−/− sham, WT BDL, and H19−/− BDL. Each group, n = 6–10; *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant, WT BDL versus H19−/− BDL. (B) Representative images of H&E staining and IHC images of IL-6 and F4/80 in the livers from WT sham, H19−/− sham, WT BDL and H19−/− BDL. Abbreviation: IL-6, interleukin-6.
FIG. 4.
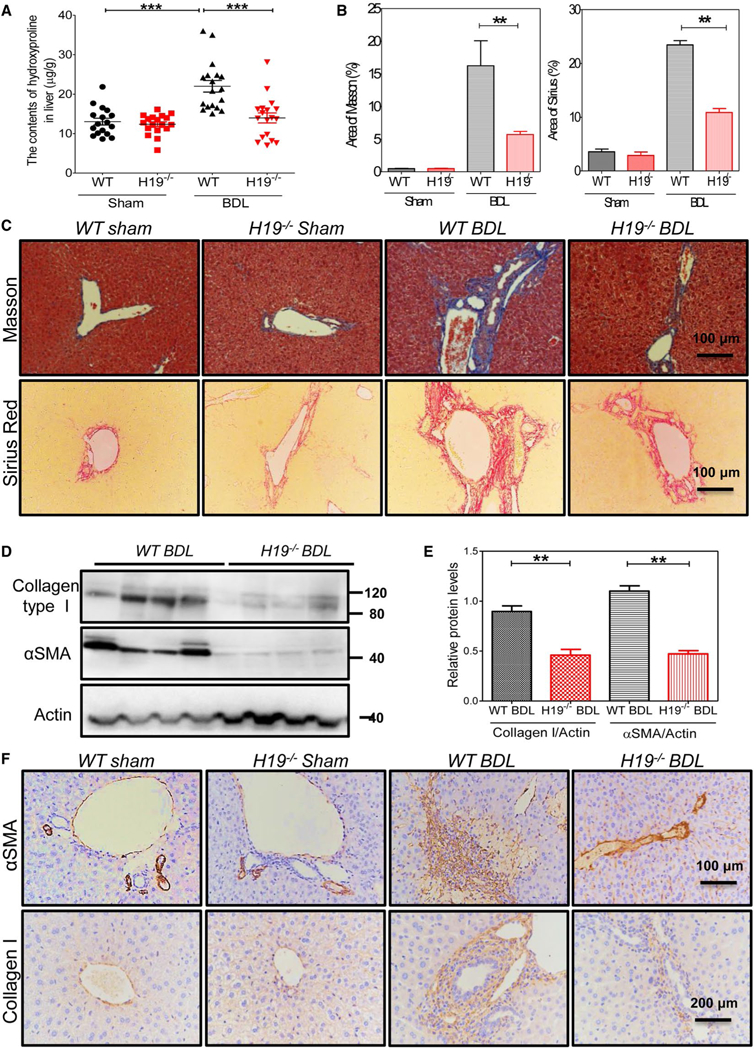
H19-deficiency impeded BDL-induced LF. (A) Quantifying collagen content with hydroxyproline assay in livers of WT sham, H19−/−sham, WT BDL, and H19−/− BDL. Each group, n = 8–16; ***P < 0.001. (B) Quantification of the fibrotic area of Masson’s trichrome images and fibrotic area of Sirius Red staining images. Sham control, WT (n = 5), H19−/− (n = 3), BDL, WT (n = 6), and H19−/− (n = 5) mice. Statistical significance relative to WT BDL group: **P < 0.01. (C) Representative images of Masson’s trichrome staining and Sirius Red staining of sham control and BDL of WT and H19−/− mice. (D) Western blotting analysis for collagen type I and α-SMA in livers of WT BDL and H19−/− BDL. (E) Quantification of panel (D). Statistical significance relative to WT BDL group: **P < 0.01. (F) Representative images of IHC for collagen type I and α-SMA in mice livers.
FIG. 5.
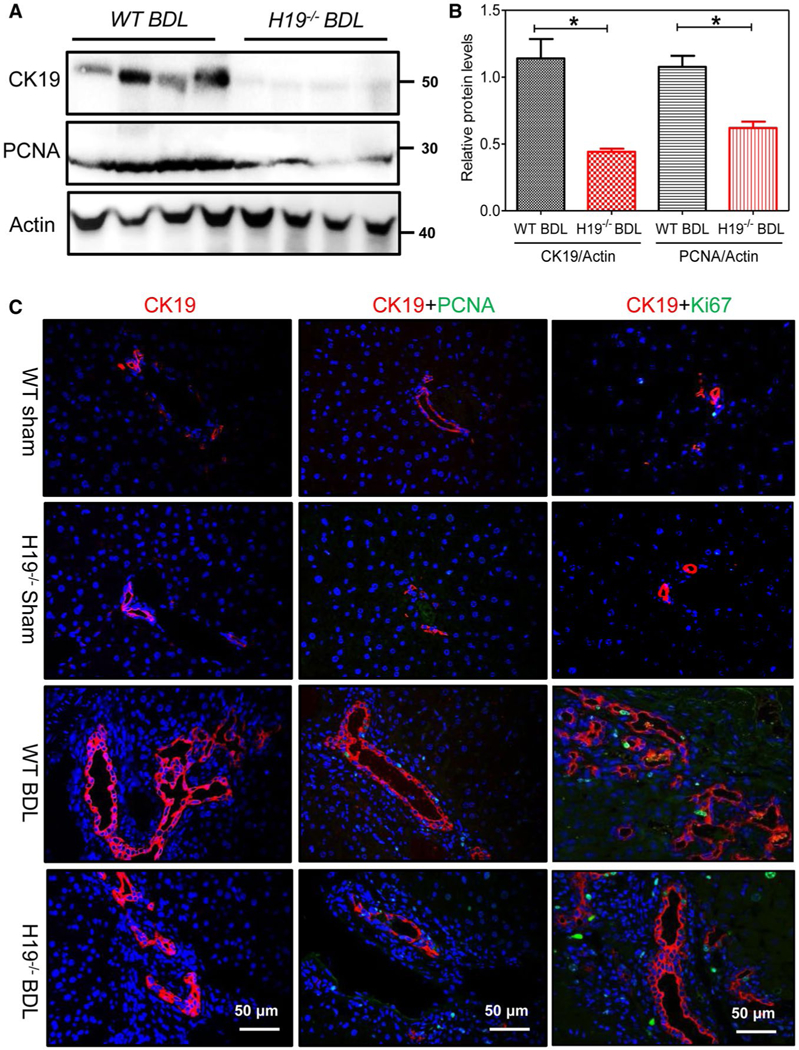
H19 deficiency inhibited BDL-induced cholangiocyte proliferation. (A) Western blotting analysis for CK19 and PCNA in WT BDL (n = 4) and H19−/−BDL (n = 4) mice. Representative images of the immune blottings are shown. (B) Relative protein expression levels of CK19 and PCNA normalized using β-actin as a loading control. (C) Representative IF images of CK19, PCNA, and Ki67 in sham controls, WT (n = 5), H19−/−(n = 3), and BDL, WT (n = 6), H19−/−(n = 5) mice. Statistical significance relative to WT: *P < 0.05.
HEPATIC H19 IS ASSOCIATED WITH UP-REGULATION OF S1PR2 AND SPHINGOSINE KINASE 2 IN BA PATIENTS
Our previous study reported that S1PR2 and sphingosine kinase 2 (SphK2) played an important role in promoting CLI. We further examined whether hepatic S1PR2 and SphK2 expression levels changed in BA patients. mRNA levels of S1PR2 and SphK2 were significantly elevated in BA patients when compared to control subjects (Fig. 6A). The IHC assay further showed that protein levels of S1PR2 and SphK2 were also increased in livers of BA patients when compared to controls (Supporting Fig. S13). Correlation analysis showed that hepatic H19 mRNA levels were positively correlated with levels of hepatic S1PR2 mRNA (r = 0.30; P = 0.02) and SphK2 (r = 0.59; P < 0.0001; Fig. 6B).
FIG. 6.
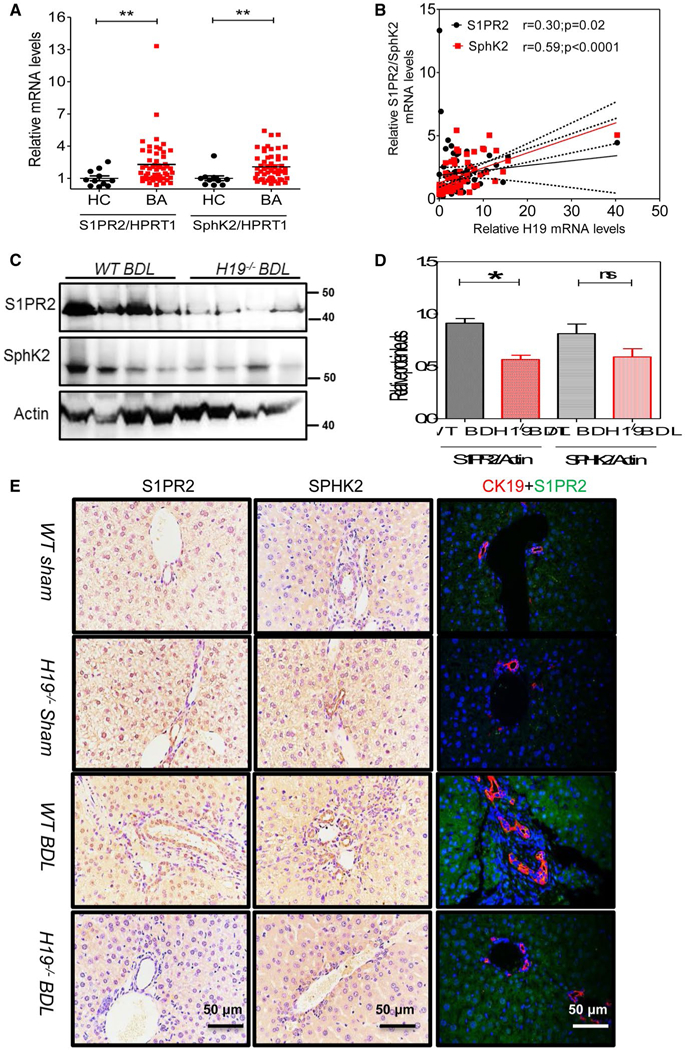
H19 deficiency restrained hepatic S1PR2/SphK2 signaling in BDL mice. (A) Relative mRNA levels of S1PR2, SphK2, and HMGA2 in livers of BA patients (n = 53) and controls (n = 11). (B) Correlation analysis of H19 mRNA levels with S1PR2 or SphK2 mRNA levels. (C) Representative western blotting images of S1PR2 and SphK2 in BDL WT and H19−/− mice (n = 4) are shown. (D) Relative protein expression levels of S1PR2 and SphK2 were normalized using β-actin as a loading control. (E) Representative IHC images of S1PR2 and SphK2 and IF images of S1PR2 and CK19 in sham controls, WT (n = 5) and H19−/− (n = 3), and BDL mice, WT (n = 6), and H19−/− (n = 5) mice. Statistical significance relative to WT BDL group: *P < 0.05; **P < 0.01; ns, not significant. Abbreviation: HC, healthy controls.
To further determine whether the key genes involved in bile acid synthesis were changed in BA patients, we measured mRNA levels of farnesoid X receptor, small heterodimer partner (SHP), cytochrome P450 family 7 subfamily A member 1, and cytochrome P450 family 27 subfamily A member 1. Results showed that there was no significant alteration between BA patients and controls (Supporting Fig. S14). However, mRNA and protein expression levels of an RNA-binding protein, human antigen R (HuR),(32) were increased in livers of BA patients compared to controls (Supporting Fig. S15). Hepatic H19 mRNA level was positively correlated with hepatic HuR mRNA level (r = 0.58; P < 0.0001; Supporting Fig. S15B).
H19 DEFICIENCY ATTENUATES S1PR2/SphK2 SIGNALING IN CHOLESTATIC LIVER
We previously reported that S1PR2 deficiency significantly reduced BDL-induced cholangiocyte proliferation and CLI.(33) To determine whether S1PR2 plays a role in H19-mediated LF and cholangiocyte proliferation in the cholestatic liver, we first measured the protein levels of S1PR2 and SphK2. Western blotting results indicated that hepatic protein expression level of S1PR2 was significantly decreased in H19−/− BDL mice relative to that of WT BDL mice (Fig. 6C,D). SphK2 protein level was also decreased, but not statistically significantly. Furthermore, we also showed that BDL increased S1PR2 expression in CK19-positive cells. The number of CK19 and S1PR2 double-positive cholangiocytes was reduced in H19−/−BDL mice compared to WT BDL mice (Fig. 6E and Supporting Fig. S16). Moreover, S1PR2 expression level was increased in livers of Mdr2−/− mice, but was significantly reduced in livers of Mdr2−/−/H19−/− DKO mice (Supporting Fig. S17). In addition, we also showed that BDL-induced HuR expression was markedly inhibited in H19−/− mice (Supporting Fig. S18). Recently, we reported that TCA activates S1PR2 in both hepatocytes and cholangiocytes.(11,34) To determine the effects of H19 on expression of S1PR2 in cholangiocytes, primary mouse cholangiocytes from WT and H19−/− mice were isolated and mRNA expression levels of S1PR2 were measured. Results showed that H19 deficiency reduced S1PR2 expression in mice primary cholangiocytes (Supporting Fig. S19A). To further determine the effect of H19 on bile-acid-induced up-regulation of S1PR2, an H19 shRNA was used to knock down expression of H19 in cultured mouse large cholangiocytes. It showed that TCA-induced expression of S1PR2 in mouse cholangiocytes was significantly inhibited by H19 shRNA (Supporting Fig. S19B).
H19 ACTS AS A MOLECULAR SPONGE TO PROMOTE THE let-7/HMGA2 AXIS IN CHOLESTATIC LIVER
Consistent with a previous study,(35) the bioinformatic analysis (RNAhybrid) demonstrated putative complementary sequences for let-7 in human H19 (Supporting Fig. S20). Given that HEK293 cells can express appreciable levels of let-7 miR-NAs, but no endogenous H19,(35) we transfected psi-CHECK2-let-7 4× (Sensor) together with WT H19 plasmid (pH19 WT, Sponge) or let-7-binding site mutated H19 plasmid (pH19 MUT) into HEK293 cells to examine whether H19 acts as a molecular sponge to sequester let-7. Luciferase activity of let-7 sensor was increased in response to pH 19 WT in a dose-dependent manner, but remained unchanged with the pH19 MUT, suggesting that ectopically expressed WT H19 specifically sequestered endogenous let-7 and impeded endogenous let-7 from inhibiting let-7 sensor activity (Fig. 7A). Sequence complementarity and conservation analyses with TargetScan revealed that high-mobility group AT-hook 2 (HMGA2) is a potential target of let-7 miRNAs (Supporting Fig. S21). To verify that HMGA2 is a true target of let-7 miRNAs, we generated luciferase reporter plasmids containing WT or mutated let-7 miRNA-binding sites in the 3′ untranslated regions (3′-UTRs) of human HMGA2. Luciferase reporter assay revealed that let-7 miRNAs significantly repressed the activity of WT HMGA2–3′UTR, but not mutated HMGA2–3′UTR (Fig. 7B). Consistent with the results of the luciferase reporter gene assay, overexpression of let-7 miRNAs significantly inhibited HMGA2 protein expression in human HuCCT1 cells (Fig. 7C,D). We further examined expression levels of let-7 family members in human BA patients. Only let-7b was significantly down-regulated in BA patients (Fig. 7E).
FIG. 7.
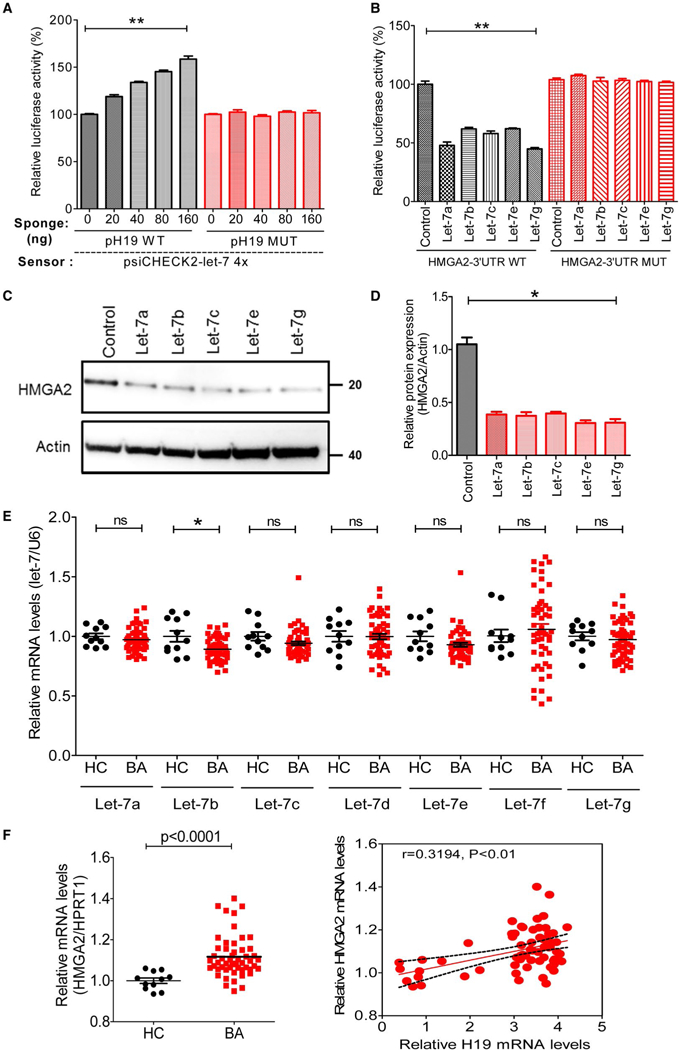
H19 altered the let-7/HMGA2 pathway by sponging the let-7 miRNAs. (A) let-7 sensor (psiCHECK2-let-7 4×) was transfected into HEK293 cells, together with 0, 20, 40, 80, or 160 ng of sponge plasmid pH19 WT or pH19 MUT. Renilla luciferase activity was normalized to firefly luciferase. Statistical significance relative to the 0-ng group: **P < 0.01. (B) The psiCHECK2 plasmid that contained HMGA2–3′UTR WT or HMGA2–3′UTR MUT were transfected into HEK293 cells, together with let-7a, let-7b, let-7c, let-7e, or let-7g. Statistical significance relative to control plasmid group: **P < 0.01. (C) The mimics for let-7a, let-7b, let-7c, let-7e, or let-7 g transfected into human biliary cell line HuccT1 for 48 hours, and the protein, HMGA2, was determined by western blotting. (D) Relative protein expression levels of HMGA2 were normalized using β-actin as a loading control. (E) Relative levels of let-7 miRNA family members, including let-7a, let-7b, let-7c, let-7d, let-7e, let-7f, and let-7g, in livers of BA patients (n = 53) and controls (n = 11). RNU6B was used as an internal control. (F) Relative mRNA levels of HMGA2 in livers of BA patients (n = 53) and controls (n = 11). Correlation analysis of H19 mRNA levels with HMGA2 mRNA levels. *P < 0.05; **P < 0.01; ns, not significant. Abbreviations: HC, healthy controls; MUT, mutant; RNU6B, U6 small noncoding RNA.
Consistently, mRNA expression level of HMGA2 was markedly increased in BA patients, which was correlated with H19 expression level (r = 0.3194; P < 0.01; Fig. 7F). In addition, mRNA levels of S1PR2 mRNA and HMGA2 were positively correlated with progression of liver injury in BA livers (Supporting Fig. S22).
Discussion
Abnormal cholangiocyte proliferation and LF are hallmarks of BA,(5,36) but the underlying cause(s) and pathogenesis are still unclear. We have investigated an lncRNA H19 regulatory mechanism involved in cholestasis-induced cholangiocyte proliferation andLF. Our study suggests that H19 promotes cholangiocyte proliferation and LF by regulating the S1PR2/SphK2 and let-7/HMGA2 signaling pathway.
BA is a pediatric liver disease that is primarily as a result of extrahepatic biliary tree obstruction. Presently, there level. One is to develop an early diagnosis, which is critical to increasing the percentage of successful KHPE. The other one is to prevent progressive liver injury and fibrosis.(5) lncRNAs have essential biological functions, and H19 is one of the few well-characterized lncRNAs, which is an imprinted and maternally expressed transcript.(37,38) Aberrant expression of H19 has been associated with numerous disease conditions.(39-43) The current study showed that BA patients had significantly higher levels of H19 mRNA in livers when compared to control subjects, which indicates that H19 may play a role in the pathogenesis of BA. Indeed, the correlation analysis showed that hepatic H19 mRNA levels were positively correlated with hepatic TGFB1 mRNA and ACTA2, suggesting that H19 mRNA levels could be used to monitor LF progression in BA patients. Similar to the findings in mouse CLI models,(24) the current study showed that H19 levels were markedly increased in serum exosomes isolated from BA patients compared to those in controls. Given that exosomal and hepatic H19 levels are associated with LF in BA patients, serum exosomal H19 levels may be utilized as a useful marker for monitoring LF progression of BA patients. The BDL mouse model further showed that H19 deficiency significantly blocked BDL-induced LF progression and reduced cholangiocyte proliferation. Recent studies indicated that S1PR2 plays an important role in regulating hepatic metabolism through activation of SphK2.(10) We also reported that S1PR2 null mice were protected from BDL-induced bile duct proliferation and LF.(33) Furthermore, we also demonstrated that up-regulation of hepatic S1PR2 was correlated to the increased SphK2 protein level in female Mdr2−/− mice with severe LF.(25) Similar to our previous findings, we have demonstrated that mRNA and protein levels of S1PR2 and SphK2 were markedly increased in liver and cholangiocytes under cholestasis conditions in BDL WT mice, whereas they were significantly reduced in H19−/− BDL mice. These results indicated that H19 might contribute to BDL-induced LF and cholangiocyte proliferation by enhancing activation of S1PR2/SphK2 signaling pathways. Our previous in vitro studies demonstrated that TCA dose dependently increased S1PR2 expression and promoted cholangiocyte proliferation.(33) In the present study, we identified that TCA-induced up-regulation of S1PR2 in cholangiocytes was blocked by H19 knockdown.
In the current study, we also showed that levels of the RNA-binding protein, HuR, increased significantly in both human livers of BA patients and BDL mouse livers. Moreover, H19 mRNA levels were positively correlated with HuR mRNA expressions in livers of BA patients. It is reported that an increase of the levels of HuR prevented the processing of miR-675 from H19, given that a precursor of miR-675 is embedded within the first exon of H19. Increased miR-675 expression resulted in reduced cell proliferation in the face of pathological stress.(44,45) It also has been identified that H19 can function as a molecular sponge for the miRNAs, such as let-7.(35) Recently, several studies have shown that let-7 miRNAs are involved in many biliary and cholestatic diseases. Glaser et al. reported that inhibition of let-7a in BDL mice increases biliary proliferation(46) Let-7b/7d/7i decreased primary biliary cirrhosis and engaged in inflammation processes in cholangiocytes.(47,48) In the study, we found that let-7 family members did not change significantly in livers of BA patients compared to controls. Intriguingly, let-7 miRNAs overexpression reduced HMGA2 by targeting 3′UTR of HMGA2 miRNA. Moreover, H19 acted as a “sponge” to restrain the activity of let-7 miRNAs. This may explain why H19 does not have much impact on let-7 miRNA expression, but decreases their bioavailability.
In summary, based on previously published studies and the current study, we propose that H19 plays a vital role in regulating cholangiocyte proliferation and LF by regulating S1PR2/SphK2– and let-7/HMGA2–mediated pathways under cholestatic conditions in BA patients (Fig. 8).
FIG. 8.
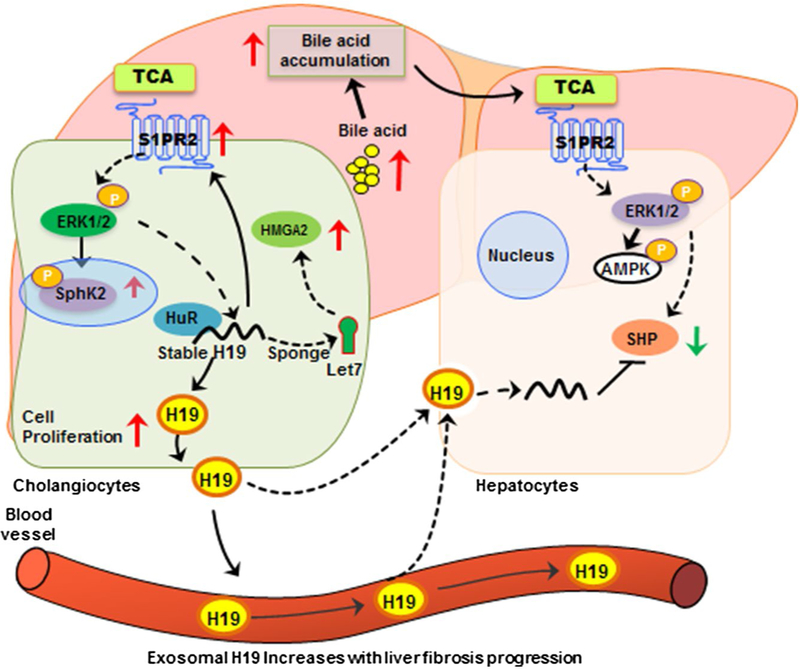
Schematic diagram of potential mechanisms involved in H19-mediated cholangiocyte proliferation and LF in BA patients. H19 mRNA increased in livers of BA patients and released into circulation by exosomal package. On the one hand, hepatic H19 enhanced S1PR2 expression, an important bile acid receptor, leading to activation of the downstream mediator, SphK2, and promoted cholestatic injury. On the other hand, H19 acted as a molecular sponge to restrain the bioactivity of let-7 miRNAs, resulting in increased let-7 miRNAs targeting HMGA2 expression, leading to enhancement of cholangiocyte growth. Abbreviations: AMPK, 5′ adenosine monophosphate–activated protein kinase; ERK1/2, extracellular signal regulated kinase 1 and 2.
Supplementary Material
Acknowledgments
Supported by the National Natural Science Foundation of China (81770517, 81630039) and Shanghai Key Laboratory of Pediatric Gastroenterology and Nutrition (17DZ2272000). This work was partially supported by National Institutes of Health Grant R01 DK104893 and R01DK-057543; VA Merit Award I01BX004033 and 1I01BX001390; and Research Career Scientist Award IK6BX004477.
Abbreviations:
- ACTA2
actin, alpha 2, smooth muscle, aorta
- ALP
alkaline phosphatase
- ALT
alanine aminotransferase
- AST
aspartate aminotransferase
- BA
biliary atresia
- BDL
bile duct ligation
- CK19
cytokeratin 19
- CLI
cholestatic liver injury
- COLIA1
collagen type I alpha 1
- DB
direct bilirubin
- GGT
γ-glutamyltransferase
- H&E
hematoxylin-eosin
- HMGA2
high-mobility group AT-hook 2
- HNF4-α
hepatocyte nuclear factor 4-alpha
- HuR
human antigen R
- IF
immunofluorescence
- IHC
immunohistochemistry
- KRT7
keratin 7
- LF
liver fibrosis
- lncRNA
long non-coding RNA
- miRNA
microRNA
- MLEs
mouse large cholangiocytes
- PCNA
proliferating cell nuclear antigen
- S1PR2
sphingosine 1-phosphate receptor 2
- shRNA
short hairpin RNA
- SphK2
sphingosine kinase 2
- α-SMA
alpha-smooth muscle actin
- TB
total bilirubin
- TCA
taurocholate
- TGFB1
transforming growth factor beta 1
- 3’-UTRs
3’untranslated regions
- WT
wild type
Footnotes
Supporting Information
Additional Supporting Information may be found at onlinelibrary.wiley.com/doi/10.1002/hep.30698/suppinfo.
Potential conflict of interest: Nothing to report.
references
- 1 ).Hartley JL, Davenport M, Kelly DA. Biliary atresia. Lancet 2009;374:1704–1713. [DOI] [PubMed] [Google Scholar]
- 2 ).Muraji T, Ohtani H, Ieiri S. Unique manifestations of biliary atresia provide new immunological insight into its etiopathogenesis. Pediatr Surg Int 2017;33:1249–1253. [DOI] [PubMed] [Google Scholar]
- 3 ).Sokol RJ, Mack C. Etiopathogenesis of biliary atresia. Semin Liver Dis 2001;21:517–524. [DOI] [PubMed] [Google Scholar]
- 4 ).Lampela H, Kosola S, Heikkila P, Lohi J, Jalanko H, Pakarinen MP. Native liver histology after successful portoenterostomy in biliary atresia. J Clin Gastroenterol 2014;48:721–728. [DOI] [PubMed] [Google Scholar]
- 5 ).Bezerra JA, Wells RG, Mack CL, Karpen SJ, Hoofnagle JH, Doo E, et al. Biliary atresia: clinical and research challenges for the 21(st) century. Hepatology 2018. March 31 10.1002/hep.29905. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6 ).Verkade HJ, Bezerra JA, Davenport M, Schreiber RA, Mieli-Vergani G, Hulscher JB, et al. Biliary atresia and other cholestatic childhood diseases: advances and future challenges. J Hepatol 2016;65:631–642. [DOI] [PubMed] [Google Scholar]
- 7 ).Tsai HC, Han MH. Sphingosine-1-phosphate (S1P) and S1P signaling pathway: therapeutic targets in autoimmunity and inflammation. Drugs 2016;76:1067–1079. [DOI] [PubMed] [Google Scholar]
- 8 ).Takabe K, Paugh SW, Milstien S, Spiegel S. “Inside-out” signaling of sphingosine-1-phosphate: therapeutic targets. Pharmacol Rev 2008;60:181–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9 ).Li C, Zheng S, You H, Liu X, Lin M, Yang L, Li L. Sphingosine 1-phosphate (S1P)/S1P receptors are involved in human liver fibrosis by action on hepatic myofibroblasts motility. J Hepatol 2011;54:1205–1213. [DOI] [PubMed] [Google Scholar]
- 10 ).Nagahashi M, Takabe K, Liu R, Peng K, Wang X, Wang Y, et al. Conjugated bile acid-activated S1P receptor 2 is a key regulator of sphingosine kinase 2 and hepatic gene expression. Hepatology 2015;61:1216–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11 ).Liu R, Zhao R, Zhou X, Liang X, Campbell DJ, Zhang X, et al. Conjugated bile acids promote cholangiocarcinoma cell invasive growth through activation of sphingosine 1-phosphate receptor 2. Hepatology 2014;60:908–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12 ).Liu R, Li X, Qiang X, Luo L, Hylemon PB, Jiang Z, et al. Taurocholate induces cyclooxygenase-2 expression via the sphin-gosine 1-phosphate receptor 2 in a human cholangiocarcinoma cell line. J Biol Chem 2015;290:30988–31002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13 ).Xiao Y, Wang J, Yan W, Zhou Y, Chen Y, Zhou K, et al. Dysregulated miR-124 and miR-200 expression contribute to cholangiocyte proliferation in the cholestatic liver by targeting IL-6/STAT3 signalling. J Hepatol 2015;62:889–896. [DOI] [PubMed] [Google Scholar]
- 14 ).Xiao Y, Wang J, Chen Y, Zhou K, Wen J, Wang Y, et al. Up-regulation of miR-200b in biliary atresia patients accelerates proliferation and migration of hepatic stallate cells by activating PI3K/Akt signaling. Cell Signal 2014;26:925–932. [DOI] [PubMed] [Google Scholar]
- 15 ).Jun-Hao ET, Gupta RR, Shyh-Chang N. Lin28 and let-7 in the metabolic physiology of aging. Trends Endocrinol Metab 2016;27:132–141. [DOI] [PubMed] [Google Scholar]
- 16 ).Su JL, Chen PS, Johansson G, Kuo ML. Function and regulation of let-7 family microRNAs. Microrna 2012;1:34–39. [DOI] [PubMed] [Google Scholar]
- 17 ).McDaniel K, Wu N, Zhou T, Huang L, Sato K, Venter J, et al. Amelioration of ductular reaction by stem cell derived extracellular vesicles in MDR2 knockout mice via let-7 microRNA. Hepatology 2019. February 5 10.1002/hep.30542. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18 ).McDaniel K, Hall C, Sato K, Lairmore T, Marzioni M, Glaser S, et al. Lin28 and let-7: roles and regulation in liver diseases. Am J Physiol Gastrointest Liver Physiol 2016;310:G757–G765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19 ).Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013;154:26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20 ).Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell 2013;152:743–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21 ).Liang WC, Fu WM, Wang YB, Sun YX, Xu LL, Wong CW, et al. H19 activates Wnt signaling and promotes osteoblast differentiation by functioning as a competing endogenous RNA. Sci Rep 2016;6:20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22 ).Giovarelli M, Bucci G, Ramos A, Bordo D, Wilusz CJ, Chen CY, et al. H19 long noncoding RNA controls the mRNA decay promoting function of KSRP. Proc Natl Acad Sci U S A 2014;111:E5023–E5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23 ).Zhang Y, Liu C, Barbier O, Smalling R, Tsuchiya H, Lee S, et al. Bcl2 is a critical regulator of bile acid homeostasis by dictating Shp and lncRNA H19 function. Sci Rep 2016;6:20559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24 ).Wan XY, Xu LY, Li B, Sun QH, Ji QL, Huang DD, et al. Chemical conversion of human lung fibroblasts into neuronal cells. Int J Mol Med 2018;41:1463–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25 ).Li X, Liu R, Yang J, Sun L, Zhang L, Jiang Z, et al. The role of long noncoding RNA H19 in gender disparity of cholestatic liver injury in multidrug resistance 2 gene knockout mice. Hepatology 2017;66:869–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26 ).Liu C, Yang Z, Wu J, Zhang L, Lee S, Shin DJ, et al. Long noncoding RNA H19 interacts with polypyrimidine tract-binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology 2018;67:1768–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27 ).Li X, Liu R, Huang Z, Gurley EC, Wang X, Wang J, et al. Cholangiocyte-derived exosomal long noncoding RNA H19 promotes cholestatic liver injury in mouse and humans. Hepatology 2018;68:599–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28 ).Liu R, Li X, Zhu W, Wang Y, Zhao D, Wang X, et al. Cholangiocyte-derived exosomal LncRNA H19 promotes hepatic stellate cell activation and cholestatic liver fibrosis. Hepatology 2019. April 15 10.1002/hep.30662. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29 ).Aoki H, Aoki M, Yang J, Katsuta E, Mukhopadhyay P, Ramanathan R, et al. Murine model of long-term obstructive jaundice. J Surg Res 2016;206:118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30 ).Glaser SS, Gaudio E, Rao A, Pierce LM, Onori P, Franchitto A, et al. Morphological and functional heterogeneity of the mouse intrahepatic biliary epithelium. Lab Invest 2009;89:456–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31 ).Tremblay KD, Saam JR, Ingram RS, Tilghman SM, Bartolomei MS. A paternal-specific methylation imprint marks the alleles of the mouse H19 gene. Nat Genet 1995;9:407–413. [DOI] [PubMed] [Google Scholar]
- 32 ).Zou T, Jaladanki SK, Liu L, Xiao L, Chung HK, Wang JY, et al. H19 Long noncoding RNA regulates intestinal epithelial barrier function via microRNA 675 by interacting with RNA-binding protein HuR. Mol Cell Biol 2016;36:1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33 ).Wang Y, Aoki H, Yang J, Peng K, Liu R, Li X, et al. The role of sphingosine 1-phosphate receptor 2 in bile-acid-induced cholangiocyte proliferation and cholestasis-induced liver injury in mice. Hepatology 2017;65:2005–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34 ).Studer E, Zhou X, Zhao R, Wang Y, Takabe K, Nagahashi M, et al. Conjugated bile acids activate the sphingosine-1-phosphate receptor 2 in primary rodent hepatocytes. Hepatology 2012;55:267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35 ).Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol Cell 2013;52:101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36 ).Cheung AC, Lorenzo Pisarello MJ, LaRusso NF. Pathobiology of biliary epithelia. Biochim Biophys Acta Mol Basis Dis 2018;1864:1220–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37 ).Khosla S, Aitchison A, Gregory R, Allen ND, Feil R. Parental allele-specific chromatin configuration in a boundary-imprinting-control element upstream of the mouse H19 gene. Mol Cell Biol 1999;19:2556–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38 ).Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature 1991;351:153–155. [DOI] [PubMed] [Google Scholar]
- 39 ).Stuhlmuller B, Kunisch E, Franz J, Martinez-Gamboa L, Hernandez MM, Pruss A, et al. Detection of oncofetal h19 RNA in rheumatoid arthritis synovial tissue. Am J Pathol 2003;163:901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40 ).Iempridee T Long non-coding RNA H19 enhances cell proliferation and anchorage-independent growth of cervical cancer cell lines. Exp Biol Med (Maywood) 2017;242:184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41 ).Geng H, Bu HF, Liu F, Wu L, Pfeifer K, Chou PM, et al. In inflamed intestinal tissues and epithelial cells, interleukin 22 signaling increases expression of H19 long noncoding RNA, which promotes mucosal regeneration. Gastroenterology 2018;155:144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42 ).Yoruker EE, Keskin M, Kulle CB, Holdenrieder S, Gezer U. Diagnostic and prognostic value of circulating lncRNA H19 in gastric cancer. Biomed Rep 2018;9:181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43 ).Zhang K, Luo Z, Zhang Y, Zhang L, Wu L, Liu L, et al. Circulating lncRNA H19 in plasma as a novel biomarker for breast cancer. Cancer Biomark 2016;17:187–194. [DOI] [PubMed] [Google Scholar]
- 44 ).Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 2012;14:659–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45 ).Zou T, Jaladanki SK, Liu L, Xiao L, Chung HK, Wang JY, et al. H19 long noncoding RNA regulates intestinal epithelial barrier function via microRNA 675 by interacting with RNA-binding protein HuR. Mol Cell Biol 2016;36:1332–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46 ).Glaser S, Meng F, Han Y, Onori P, Chow BK, Francis H, et al. Secretin stimulates biliary cell proliferation by regulating expression of microRNA 125b and microRNA let7a in mice. Gastroenterology 2014; 146:1795–1808.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47 ).Zhang M, Liu F, Jia H, Zhang Q, Yin L, Liu W, et al. Inhibition of microRNA let-7i depresses maturation and functional state of dendritic cells in response to lipopolysaccharide stimulation via targeting suppressor of cytokine signaling 1. J Immunol 2011;187:1674–1683. [DOI] [PubMed] [Google Scholar]
- 48 ).Padgett KA, Lan RY, Leung PC, Lleo A, Dawson K, Pfeiff J, et al. Primary biliary cirrhosis is associated with altered hepatic microRNA expression. J Autoimmun 2009;32:246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


