Abstract
Neurotransmitters and neuromodulators are key neurochemicals that mediate cell-cell communication, maintain the body’s homeostasis, and control a wide range of biological processes. Thus, dysregulation of neurochemical signaling is associated with a range of psychiatric disorders and neurological diseases. Understanding the physiological and pathophysiological functions of neurochemicals, particularly in complex biological systems in vivo, requires tools that can probe their dynamics with high sensitivity and specificity. Recently, genetically-encoded fluorescent sensors for visualizing specific neurochemicals were developed by coupling neurochemical-sensing G protein-coupled receptors (GPCRs) with a circular-permutated fluorescent protein (cpFP). These GPCR-based sensors can monitor the dynamics of neurochemicals in behaving animals with high spatiotemporal resolution. Here, we review recent progress regarding the development and application of GPCR-based sensors for imaging neurochemicals, and we discuss future perspectives.
Graphical Abstract
Precisely probing the dynamics of specific neurochemicals in the complex biological system is the key to understand their functions. In this review, we summarized the recent progress on using GPCRs as the scaffold to engineer neurochemical-specific fluorescent sensors, which could achieve the sensitive and specific detection of neurochemicals with millisecond temporal resolution and sub-cellular spatial resolution. Taking advantage of these tools, researchers have successfully dissected the dynamics as well as function of neurochemicals in various of animal species performing distinct behaviors. With future optimizations, we anticipate that these GPCR-based sensors will be powerful tools in studying neurochemical signals.
Introduction
Since the discovery of neurotransmitters and neuromodulators in the early 1900s (Loewi 1924; Elliott 1905; Dale & Dudley 1929; Bayliss 1901), extensive studies have been performed in an attempt to understand their physiological functions. Compelling evidence now exists that communication via neurochemical transmission plays a key role in a variety of functions and behaviors ranging from the cellular level to the whole-animal level (Klinkenberg et al. 2011; Dale et al. 1936; Katz & Miledi 1961; Schultz et al. 1997; Carlson 1995). Traditional detection techniques such as microdialysis, electrochemical recording, and electrophysiology are still invasive and have limitations with respect to monitoring a population of neurons with high spatial resolution (Smith et al. 1992; Kehr 1992; Mas et al. 1995; Adams 1973). The main push forward to better understand the role of neurotransmitters from cellular to whole-animal levels greatly demands new tools with high cell type specificity, physiologically relevant affinity, high molecular specificity, millisecond temporal resolution, and subcellular spatial resolution.
Genetically encoded fluorescent sensors would be an ideal tool for detecting neurochemicals in a highly cell type-specific manner (Liang et al. 2015; Lin & Schnitzer 2016). Recently, G protein-coupled receptors (GPCRs) were used as the scaffold for generating genetically encoded sensors, due to their high ligand selectivity and physiologically relevant ligand affinity. To date, GPCR-based sensors have been developed for measuring the in vivo dynamics of several neurochemicals; these sensors include the GPCR-activation based acetylcholine (ACh) sensor (GACh sensor) for detecting acetylcholine (Jing et al. 2018), the GPCR-Activation Based dopamine (DA) sensor (GRABDA sensor) (Sun et al. 2018)and dLight (Patriarchi et al. 2018) sensors for detecting dopamine, and the GPCR-Activation Based norepinephrine (NE) sensor (GRABNE sensor) for detecting norepinephrine (Feng et al. 2019). When combined with advanced optical imaging techniques, GPCR-based sensors can provide scalable, long-term recordings of specific neurochemicals in a variety of model organisms.
Here, we review the engineering principle, properties, and in vivo applications of GPCR-based sensors. Moreover, we propose future directions for developing next-generation GPCR-based sensors with improved performance, negligible downstream signaling, an expanded color spectrum, and a wider range of ligands.
1. GPCRs have unique properties that make them highly suitable as scaffolds for engineering new sensors
Thanks to the development of optical imaging techniques, genetically-encoded fluorescent sensors have become an important tool for recording the in vivo dynamics of neurotransmitters and neuromodulators. These sensors were originally designed by combining a ligand-binding protein as the “sensing scaffold” with a circular-permutated fluorescent protein (cpFP) (Baird et al. 1999) as the “reporting module”. Previously, prokaryotic periplasmic-binding proteins (PBPs) were used as the scaffold for detecting several neurochemicals, including glutamate (iGluSnFR), GABA (iGABASnFR), and ATP (iATPSnFR), suggesting that this strategy may be applicable for in vivo use (Marvin et al. 2019; Lobas et al. 2019; Marvin et al. 2013). However, this design strategy is not feasible in cases which a suitable PBP is not available for the molecule of interest. Therefore, one approach to overcome these limitations associated with using prokaryotic PBPs as the scaffold was to search for a new group of sensing proteins (Wang et al. 2018).
1.1. The pros and cons of using GPCRs as the sensing scaffold
GPCRs are the largest family of membrane receptors, responsible for “sensing” extracellular chemicals and transducing the resulting signal into an intracellular response. Compared to prokaryotic PBPs, which bind primarily to metabolites, the broad spectrum of GPCR ligands covers most neurotransmitters and neuromodulators, thus providing an ideal natural scaffold for designing a sensor. Importantly, GPCRs have evolved to provide high ligand specificity as well as affinity suitable for detecting the in vivo dynamics of specific neurochemicals. Furthermore, although several GPCRs can sense a given neurochemical, they have unique properties; for example, in humans, five subtypes of GPCRs bind dopamine, each with a distinct combination of affinity, pharmacological selectivity and downstream signaling (Grandy et al. 1989; Dearry et al. 1990; Sunahara et al. 1991). This rich natural diversity provides a wealth of opportunities for engineering neurochemical sensors with specific properties and applications.
GPCRs are also an ideal scaffold for engineering neurochemical sensors with high spatiotemporal resolution. For example, the genetically encoded nature of GPCR-based sensors permits their expression in specific cell types using genetic manipulation. With respect to temporal resolution, the conformational change induced by ligand binding and receptor activation occurs on the order of tens of milliseconds (Marcaggi et al. 2009; Vilardaga et al. 2003; Hoffmann et al. 2005). Thus, engineering a GPCR-based sensor based on the receptor’s conformational change will inherit millisecond temporal resolution, which is sufficient to reveal the temporal dynamics of neurotransmitter actions in vivo. Comparing with the prokaryotic PBPs as the sensor scaffold, GPCR-based sensors could in principle achieve similar sensitivity and response kinetics, but are better with affinity and selectivity in detecting corresponding neurochemicals, especially in physiological conditions.
Despite these clear advantages, several caveats should be considered when using a GPCR as the scaffold for designing a sensor. First, the assembly, folding, and trafficking of GPCR-based sensors should be considered. Because GPCRs are membrane proteins with an extracellular ligand-binding pocket, it is both essential and challenging to ensure that the GPCR-based sensors are trafficked properly to the cell surface and has the correct membrane topology. Indeed, based on our own experience when screening new sensors, the majority of recombinant GPCR-cpFP proteins have relatively poor trafficking to the plasma membrane and are therefore unable to sense extracellular neurochemicals (Feng et al. 2019; Jing et al. 2018). Second, post-translational modifications such as phosphorylation and lipid modification can potentially change the sensor’s properties, affecting its ability to accurately report extracellular neurochemicals. Lastly, expressing GPCR-based sensors may—at least in principle—alter cellular physiology, as discussed in Section 4. Taken together, these pros and cons should be weighed appropriately and—where needed—solved using rational strategies when engineering GPCR-based sensors.
1.2. The principle behind GPCR-based sensors
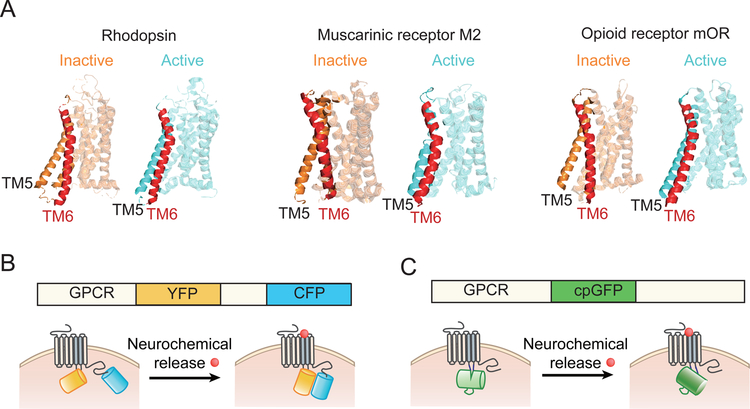
As discussed above, the most important advantage of using a GPCR as the scaffold for engineering a cpFP-based fluorescent sensor is the conformational change that occurs upon activation by a specific ligand. More than two decades ago, pioneering work by Brian Kobilka and colleagues using the conformation-sensitive dye IANBD directly revealed the ligand-induced conformational change in the β2AR (Gether et al. 1995). More recently, analyzing the crystal structure of several GPCRs, including the β2AR, M2R, μOR, and A2AR, provided further insight into the structural changes that occur when the receptor binds its ligand and interacts with its associated G protein (Haga et al. 2012; Jaakola et al. 2008; Rasmussen et al. 2007; Cherezov et al. 2007; Huang et al. 2015). Specifically, a GPCR has several conformation states, including the inactive, partially active, and full active states, with the largest difference in conformation occurring within transmembrane helices 5 and 6. Ligand binding followed by G protein interaction stabilizes the protein in the active conformation, resulting in downstream signal transduction (Venkatakrishnan et al. 2013; Rasmussen et al. 2007; Cherezov et al. 2007; Rasmussen et al. 2011; Gregorio et al. 2017; Manglik et al. 2015; Scheerer et al. 2008) (Figure 1A). Capitalizing on this conformational change, GPCR-based sensors have been engineered in which cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) were fused to the GPCR; activation of the receptor—and thus, the resulting conformational change—gives rise to a change in the FRET (fluorescence resonance energy transfer) signal, which can be detected by fluorescence imaging on CFP and YFP channels (Vilardaga et al. 2003; Hoffmann et al. 2005) (Figure 1B). Although these FRET-based GPCR sensors have a relatively low signal-to-noise ratio, which limits their use in in vivo application, they provided key proof-of-concept that the conformational change upon activation of the GPCR can be exploited for engineering new sensors.
Fig. 1.
Schematic diagram showing the conformational change induced in three GPCR upon activation, and the general structure of GPCR-based sensors. (A) The structure of rhodopsin, the muscarinic M2 receptor, and the μOR opioid receptor are shown in the inactive (left) and active (right) states. The transmembrane helix 6 (TM6) with the largest conformational change is highlighted in red. Crystal structures are from PDB database (Rhodopsin: 3C9L&3DQB; M2:3UON&4MQS; μOR:4DKL&5C1M) (B) The principle behind FRET-based GPCR sensors, with the receptor fused to the fluorophores YFP and CFP. (C) GPCR-based sensors with the GPCR fused to a single circular-permutated green fluorescent protein (cpGFP). Upon binding its ligand, the conformational change in the GPCR is sensed and reported by either FRET or cpGFP.
To develop neurochemical sensors that have a high signal-to-noise ratio and are suitable for in vivo applications, we recently combined the conformation-sensitive cpFP with GPCRs to generate the GRAB (GPCR Activation-Based) series of sensors for detecting a variety of neurochemicals, including GACh for sensing acetylcholine, GRABDA for sensing dopamine, and GRABNE for sensing norepinephrine (Jing et al. 2018; Sun et al. 2018; Feng et al. 2019). Using a similar strategy, Lin Tian’s group has developed dLight for sensing dopamine (Patriarchi et al. 2018). With GRAB sensors, the cpGFP moiety is inserted within the third intracellular loop of the GPCR; this loop connects transmembrane helices 5 and 6 and undergoes a large conformational change upon ligand binding, inducing a change in fluorescence (Figure 1C). Since most GPCRs share common activation mechanisms, this strategy could in principle be generalized to develop sensors for distinct neurochemicals. Below, we discuss the GPCR-based sensors that have been designed using this new strategy.
2. Overview of GPCR-based genetically encoded sensors
To track the real-time dynamics of specific neurochemicals in a complex biological system, the sensor must have high sensitivity, high specificity, rapid response kinetics, and a negligible effect on cellular physiology. Here, we summarize the various properties of GPCR-based sensors (see Table 1).
Table. 1.
The performance of currently available GPCR sensors.
| Sensor | Scaffold | Peak response (∆F/F0) | Ligand affinity | Rise-time kinetics (τON) | Decay-time kinetics (τOFF) | References |
|---|---|---|---|---|---|---|
| GACh | M3 receptor | 90% | 1 µM | 200 ms | 800 ms | Jing M, et al, Nature Biotech, 2018 |
| GRABDA1m | D2 receptor | 90% | 130 nM | 60 ms | 700 ms | Sun FM, et al, Cell, 2018 |
| GRABDA1h | D2 receptor | 90% | 10 nM | 140 ms | 2500 ms | Sun FM, et al, Cell, 2018 |
| dLight1.1 | D1 receptor | 230% | 330 nM | ND | ND | Patriarchi, T, et al, Science, 2018 |
| dLight1.2 | D1 receptor | 340% | 770 nM | 10 ms | 90 ms | Patriarchi, T, et al, Science, 2018 |
| GRABNE1m | α2A receptor | 230% | 930 nM | 70 ms | 750 ms | Feng JS, et al, Neuron, 2019 |
| GRABNE1h | α2A receptor | 130% | 83 nM | 30 ms | 2000 ms | Feng JS, et al, Neuron, 2019 |
ND, not determined. Most of these properties are from measurements in vitro in cultured cells expressing sensors, except kinetics of dLight1.1 and dLight1.2 are from slice preparation.
2.1. Sensitivity
The sensitivity of a GPCR-based sensor is based on two factors: i) the dynamic range of the fluorescence response, and ii) the receptor’s binding affinity for its ligand. After several iterative rounds of engineering and screening, and focusing primarily on the interface between the GPCR and the cpGFP, today’s GPCR-based sensors achieve a >90% change in fluorescence (∆F/F0). In addition, the signal-to-noise ratio of the fluorescence response is comparable to highly sensitive electrophysiological recording methods, indicating that the sensor has sufficient sensitivity for reporting the dynamics of neurochemical signaling (Jing et al. 2018). With respect to ligand affinity, most GPCR-based sensors retain their core GPCR’s affinity, making them suitable for detecting physiological concentrations of neurochemicals. Thus, GPCR-based sensors have sufficiently high sensitivity for monitoring neurochemical signaling at physiologically relevant dynamics; for example, the dopamine sensor GRABDA is able to report dopamine release from a single dopaminergic fiber induced by minimal electrical stimulation (Sun et al. 2018).
By introducing mutations at critical residues in the receptor—either near the ligand-binding pocket or the site of interaction with the G protein—the sensor’s affinity can be further optimized in order to increase sensitivity. For example, mutating the threonine at position 205 in the dopamine D2 receptor (T205M) increases ligand affinity (Sung et al. 2016), and introducing this mutation into the GRABDA sensor increases its affinity for dopamine by an order of magnitude (from ~130 nM to ~10 nM) without affecting the sensor’s peak response (Sun et al. 2018). Therefore, developing sensors that span a wide range of affinities will provide a panel of highly sensitive sensors for use in a range of applications.
2.2. Selectivity
GPCR-based sensors retain their receptor’s high ligand selectivity, as the reporter molecules are located primarily on the intracellular domains, largely sparing the extracellular ligand-binding pocket (see Figure 1). Thus, the GACh sensor, which is based on the muscarinic acetylcholine receptor, has a robust response to acetylcholine but is relatively insensitive to nicotine and other neurotransmitters and neuromodulators (Jing et al. 2018). GPCR-based sensors are even able to discriminate between structurally similar neurochemicals; for example, although the catecholamines dopamine and norepinephrine are structurally similar, the GPCR-based dopamine sensors GRABDA and dLight are 10-fold to 30-fold more sensitive at detecting dopamine than norepinephrine and are insensitive to other neurochemicals (Sun et al. 2018; Patriarchi et al. 2018). To discriminate ever further between these two molecules, we engineered and characterized a GRABNE sensor based on the α2A adrenergic receptor (Feng et al. 2019); this GRABNE sensor is >300-fold more sensitive at detecting norepinephrine compared to dopamine, and the sensor’s range for detecting dopamine greatly exceeds the physiological concentration of dopamine, making it an ideal sensor for reporting the dynamics of norepinephrine with high specificity. Given the high selectively for their respective ligands, we believe that the dopamine and norepinephrine sensors can be combined in order to dissect the dynamics of both molecules in vivo, with extremely high precision.
In addition to retaining selectivity for their endogenous ligand, GPCR-based sensors also retain the receptor’s sensitivity for pharmacological compounds. For example, the dopamine sensors GRABDA and dLight, which are based on the D2R and D1R receptor subtypes, respectively, respond selectively to their subtype-specific agonists and antagonists (Sun et al. 2018; Patriarchi et al. 2018).
2.3. Kinetics
Thanks to the rapid activation kinetics of GPCRs (Marcaggi et al. 2009; Vilardaga et al. 2003; Hoffmann et al. 2005), GPCR-based sensors provide high temporal resolution, with rise-time kinetics (τON) on the order of tens to hundreds of milliseconds. Although the temporal resolution of GPCR-based sensors does not reach the level of directly recording currents through ligand-gated ion channels, it is still considerably faster than other methods for measuring GPCR activation, which typically rely on measuring downstream signaling. By simultaneously recording fluorescence and electrophysiology, we found that the rise-time kinetics for the fluorescence signal in GACh and GRABNE are significantly faster than measuring the current response downstream of endogenous muscarinic and adrenergic receptors (Jing et al. 2018; Feng et al. 2019).
On the other hand, the fluorescence decay kinetics (τOFF) of GPCR-based sensors range from approximately 100 ms to 2000 ms. Taken together, the kinetics of GPCR-based sensors are similar to—or in some cases, faster than—the kinetics of native metabotropic GPCRs, making these sensors an ideal tool for reliably measuring the dynamics of neurochemical signals.
2.4. Influence on cellular physiology
As mentioned above, GPCRs play an important role in cellular physiology by transducing extracellular signals (i.e., ligand binding) to intracellular downstream pathways. Therefore, expressing GPCR-based sensors should ideally have little effect on normal cellular physiology. In principle, GPCR-based sensors could affect cellular physiology by activating downstream signaling processes or by competing with endogenous receptors for ligand binding. With respect to downstream signaling, we systematically tested two major pathways downstream of GPCRs, the G protein-dependent pathway and the arrestin-dependent pathway. Generally speaking, all GPCR-based sensors have significantly reduced—or even negligible—coupling with these downstream pathways, possibly due to steric hindrance from the bulky cpGFP that occupies the region where the G protein and arrestin proteins bind the receptor. Moreover, the third intracellular loop, which is important for downstream signaling, was truncated during the engineering process, possibly contributing to the decreased signal coupling in GPCR-based sensors.
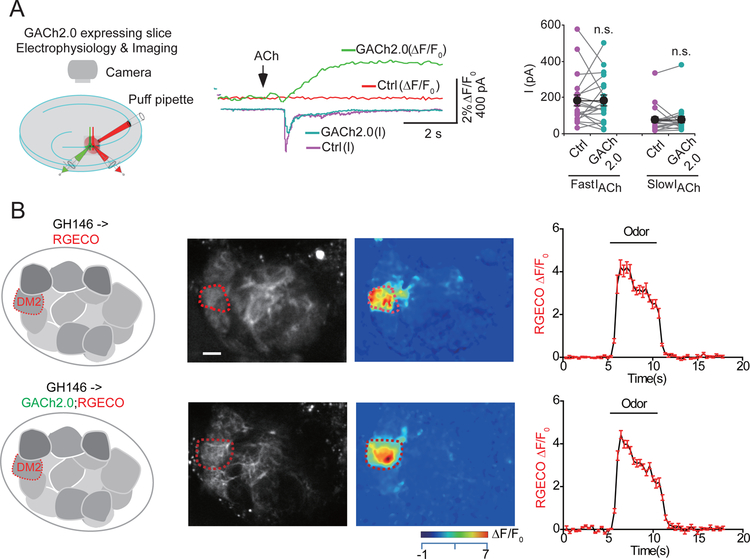
Whether a GPCR-based sensor competes with endogenous GPCRs for ligand binding depends largely on the sensor’s ligand affinity and expression level. As mentioned above, most GPCR-based sensors have a similar ligand affinity as their corresponding endogenous receptor, and the sensor’s affinity can be further tuned by mutagenesis. Using electrophysiology and calcium imaging of neuronal activity, we found that expressing GPCR-based sensors does not affect the physiological properties of endogenous receptors with respect to sensing neurochemicals or mediating synaptic transmission (Jing et al. 2018; Sun et al. 2018; Feng et al. 2019) (Figure 2). We have also shown that chronic expression of the GPCR-based sensors did not cause observable changes on cellular properties including electrophysiological signals and calcium responses (Jing et al. 2018; Sun et al. 2018; Feng et al. 2019). Ideally, by increasing the sensor’s fluorescence response and optimizing the sensor’s ligand affinity, one can then optimize the sensor’s expression level in order to obtain a high signal-to-noise recording of neurochemicals while minimizing the effect on cellular physiology.
Fig. 2.
GPCR-based sensors do not alter cellular physiology. (A) Cells expressing the acetylcholine sensor GACh2.0 have similar ACh-induced currents compared to control (Ctrl) neurons in cultured brain slices. (B) Expressing GACh2.0 in the Drosophila antennal lobe does not affect the odorant-evoked calcium response measured using the genetically encoded calcium indicator RGECO. Scale bar, 10μm. Figures are modified from original research paper(Jing et al. 2018).
3. In vivo applications for GPCR-based sensors
Given the highly complex morphological and physiological properties of neurons, the ability to monitor the dynamics of specific neurochemicals at the single-cell level, in real time, would provide valuable insights in the function of these chemicals with respect to regulating neuronal activity and behavior. Because they are genetically encoded, GPCR-based sensors such as GRAB and dLight can be expressed in specific cell types in specific brain regions, and they have the sensitivity and ligand specificity needed to report the endogenous dynamics of specific neurochemicals.
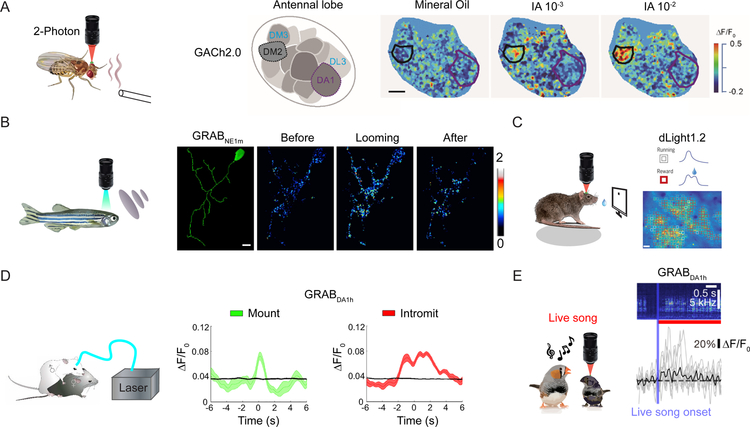
By combining genetic manipulation with optical recording, it is now possible to use GPCR-based sensors to measure neurochemical release in vivo in a wide range of awake, freely behaving animals. For example, transgenic Drosophila expressing GRABDA sensors have used to probe the compartment-specific release of dopamine in response to various physiologically relevant stimuli in the olfactory center mushroom body. Similarly, the GACh sensor has been used to report odorant-evoked acetylcholine release in specific glomeruli within the Drosophila antennal lobe, providing evidence that these sensors can be used to monitor spatially restricted neurotransmitter release (Figure 3A). In transgenic zebrafish, both GRABDA and GRABNE were used to measure neurotransmitter release evoked by a visual looming stimulation, even down to the single-cell level (Figure 3B). In freely behaving mice, GPCR-based sensors have been used to report sensory stimulation‒evoked acetylcholine release and behavior-related dopamine and norepinephrine release using fiber photometry recording and in vivo two-photon imaging (Figure 3C and D) (Jing et al. 2018; Sun et al. 2018; Patriarchi et al. 2018; Dong et al. 2019; de Jong et al. 2019; Corre et al. 2018; Feng et al. 2019; Mohebi et al. 2019). In principle, the GPCR-based sensors could also be used together with mini-scopes to achieve real-time imaging of neurochemicals in free-behaving animals. In addition, GRABDA sensors have been used successfully in zebra finches (songbirds) in order to measure the contribution of dopaminergic modulation in vocal learning (Figure 3E) (Tanaka et al. 2018).
Fig. 3.
In vivo applications of GPCR-based sensors for studying behavior in a variety of animal models and preparations. (A) Two-photon imaging was used to measure acetylcholine release evoked by odorant application in transgenic Drosophila (left), with high spatial resolution in the antennal lobe area of GH146-Gal4:UAS-GACh2.0 flies. The images at the right show the fluorescence response following application of mineral oil (as a control) and two concentrations of the odorant isoamyl acetate (IA). (B) In vivo single-cell confocal imaging of a transgenic (HuC:GRABNE1m) zebrafish was used to measure noradrenergic activity before, during, and after a visual looming stimulus. (C) Imaging of task-related dopaminergic activity was measured at the single-cell level using dLight1.2 expressed in layer 2/3 of the M1 cortex in mice. (D) Fiber photometry recordings of the GRABDA sensor was used to monitor dopamine release in the nucleus accumbens (NAc) of a freely moving male mouse during mounting and intromission. (E) The GRABDA1h sensor was used to record an increase in dopamine levels in juvenile male birds in response to a live tutor song. Scale bars, 10μm in (A) and (B), 50μm in (C). Figures are modified from original research paper(Jing et al. 2018; Sun et al. 2018; Tanaka et al. 2018; Feng et al. 2019; Patriarchi et al. 2018) with permission from the publisher.
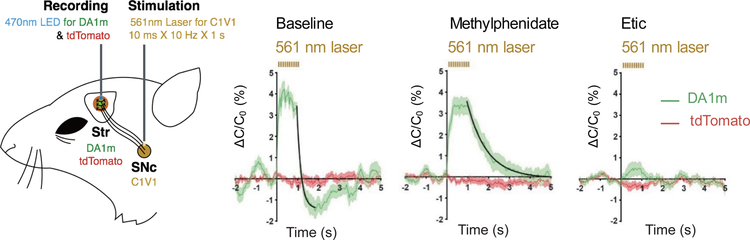
To further investigate the relationship between neuromodulation and physiological function, monitoring of neuromodulators must be combined with methods to control activity, for example optogenetics and chemogenetics. In this respect, it is interesting to note that the dLight sensor was recently combined with optogenetics or chemogenetics in order to study dopaminergic modulation in mice (Augustine et al. 2019; Patriarchi et al. 2018). Similarly, we successfully combined optogenetics with real-time monitoring of dopaminergic activity using GRABDA sensors (Figure 4)(Sun et al. 2018). Thus, combining these robust methods can provide a powerful new set of tools for studying neuromodulation. Using several GPCR-based sensors that specifically report different neurochemicals, it will likely be possible to dissect in detail whether the same group of neurons or different groups of neurons receive distinct forms of neuromodulation, as well as how this functional heterogeneity in terms of receiving neuromodulation affects their function within the neural circuit.
Fig. 4.
GRABDA can detect both the release and modulation of dopamine evoked by optogenetic stimulation. The red-activated excitatory opsin C1V1 was expressed in the substantia nigra pars compacta (SNc) and used to optically activate dopaminergic neurons, while GRABDA1m was expressed in the striatum (Str) and used to measure dopamine. The GRABDA1m fluorescence signal was prolonged by application of the dopamine transporter blocker methylphenidate and blocked by application of the D2R-specific antagonist Etic. tdTomato was co-expressed with GRABDA1m and imaged simultaneously in order to rule out any possible artifacts. Figures are modified from original research paper(Sun et al. 2018) with permission from the publisher.
4. Future directions and perspectives for GPCR-based sensors
The broad applicability of GPCR-based sensors makes them well suited for monitoring in vivo neurochemical activity with millisecond temporal resolution. Importantly, these sensors can be optimized further, and the variety of GPCR-based sensors can be expanded in order to sense a wider range of physiologically relevant molecules with negligible effects on the cell’s normal physiological processes.
4.1. Optimizing performance
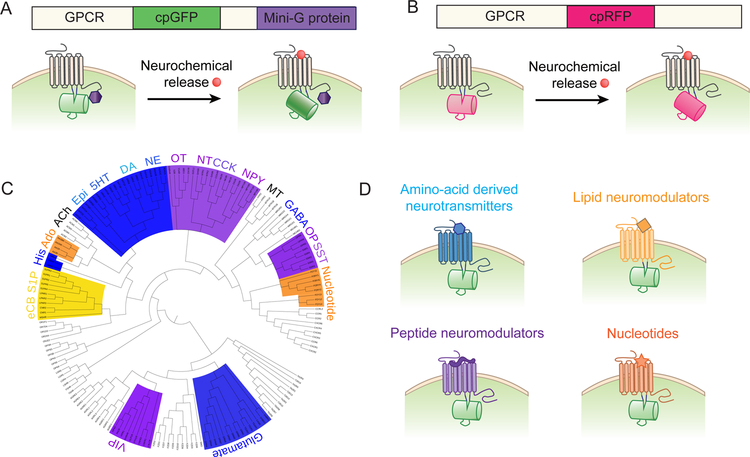
In principle, existing GPCR-based sensors can be improved by increasing their dynamic range, tuning their ligand affinity and response kinetics, and further reducing downstream coupling. The availability of the crystal structure of GPCRs provides important information regarding the residues that are critical for ligand binding and conformational changes, which can help facilitate the rational design and optimization of GPCR-based sensors. Inserting mutations near the domain responsible for maintaining the receptor in the inactive state could alter the ligand binding‒induced conformational change and/or ligand affinity (Ballesteros et al. 2001; Shapiro et al. 2002), possibly affecting the performance of the resulting GPCR-based sensor. Moreover, obtaining the crystal structure of GPCR-based sensors would be helpful for optimizing their performance using systematic and targeted mutagenesis. To minimize potential downstream coupling, we developed a strategy to prevent binding of the endogenous G protein to the GPCR-based sensor while retaining the full fluorescence signal (Figure 5A). This strategy involves attaching a G protein-derived “mini-G protein” or peptide (Carpenter & Tate 2016; Hamm et al. 1988) to the C-terminus of the sensor, thereby suppressing activation of endogenous G proteins through competitive binding (Wu et al. 2019).
Fig. 5.
Overview of future directions for developing GPCR-based sensors. (A) Downstream signaling via GPCR-based sensors can be prevented by fusing a mini-G protein or G protein‒derived peptide to the C-terminal domain, serving as a competitive inhibitor of endogenous G proteins. (B) The color spectrum of GPCR-based sensors can be expanded by replacing cpGFP with another fluorescent protein, for example, cpRFP. (C) Phylogenetic tree covering the family of neurochemical-activated GPCRs, including their respective ligands. (D) In principle, GPCR-based sensors can be developed and optimized for detecting a wide range of neurochemicals. Abbreviations: 5HT, 5-hydroxytryptamine (serotonin); ACh, acetylcholine; Ado, adenosine; CCK, cholecystokinin; DA, dopamine; eCB, endocannabinoid; Epi; epinephrine; GABA, γ-aminobutyric acid; His, histamine; MT, Melatonin; NE, norepinephrine; NPY, neuropeptide Y; NT, neurotensin; OP, opioid; OT, oxytocin; S1P, sphingosine-1-phosphate; SST, somatostatin (somatotropin release inhibiting factor); VIP, vasoactive intestinal peptide.
4.2. Expanding the color spectrum
Current GPCR-based sensors include a single fluorescent protein, which prevents the simultaneous measurement of multiple cellular events. Moreover, the excitation and emission spectra of GFP-based proteins have relatively shallow penetration, making them less suitable for in vivo imaging of deeper structures. By replacing the cpGFP moiety with cpRFP, red-wavelength calcium and voltage sensors have been developed (Dana et al. 2016; Zhao et al. 2011; Akerboom et al. 2013; Abdelfattah et al. 2016). In principle, a GPCR-based sensor containing cpRFP could be engineered (Figure 5B). In the future, the color spectrum of GPCR sensors can be expanded, which will not only allow the simultaneous measurement of neurochemical signals and calcium/voltage signals, but also the sensors to be used simultaneously with other optics-based actuators such as channelrhodopsins and other optically controlled molecules such as photoswitchable kinases (Zhou et al. 2017), thus providing a comprehensive functional map of neurochemical activity.
4.3. Subcellular targeting
It has been suggested that subcellular application of neurochemicals might activate localized receptor pools that function as separate computation units. In principle, genetically-encoded GPCR-based sensors could be targeted to specific subcellular compartments within the neuron, for example, the axon or postsynaptic spines. Additional engineering steps would be required to target the sensor to a specific subcellular structure, for example by fusing the sensor with a subcellular targeting motif similar to the strategy used for calcium sensors (Broussard et al. 2018). Comparing the dynamics of neurochemicals in different compartments within a single neuron—ideally in combination with a calcium and/or voltage indicator in order to monitor their downstream effects—will likely reveal new insights in neuromodulation.
4.4. Expanding the detection range
An exciting new direction for designing future GPCR-based sensors is to expand the repertoire of sensors to include additional neurochemicals (Figure 5C). This will open new avenues in the field of neuroscience, as most neurochemicals—particularly lipids and neuropeptides—currently lack a suitable method for studying their physiological properties distribution in vivo. From a mechanistic perspective, the currently known GPCR structures suggest that most GPCRs—particularly those in the rhodopsin family—have a highly conserved activation mechanism, with the largest conformational change occurring in transmembrane helices 5 and 6 (see Figure 1A). Indeed, Lin Tian’s group successfully used the strategy for designing dLight sensors using other GPCRs, including the β1, β2, and α2 adrenergic receptors, the κ- and μ-type opioid receptors, the 5-hydroxytryptamine (serotonin) receptor 2A, and the melatonin type 2 receptor (Patriarchi et al. 2018). In addition, we developed a series of GRAB sensors selective for acetylcholine, dopamine, and norepinephrine, supporting the notion that this strategy can also be expanded to develop a wide range of new GPCR-based sensors. Of course, each new sensor will require extensive protein engineering to optimize its performance in vivo. Nevertheless, we believe that the introduction of a wide family of GPCR-based sensors will greatly benefit the entire neuroscience community (Figure 5D).
Acknowledgments
We thank Jianzhi Zeng and Ao Dong for valuable suggestions regarding this review. This work was supported by the National Basic Research Program of China (973 Program; grant 2015CB856402), the General Program of National Natural Science Foundation of China (project 31671118), the NIH BRAIN Initiative grant (U01NS103558), the Beijing Brian Initiative of Beijing Municipal Science & Technology Commission (Z181100001518004), the Junior Thousand Talents Program of China, grants from the Peking-Tsinghua Center for Life Sciences, and the State Key Laboratory of Membrane Biology at Peking University School of Life Sciences (to Y.L.).
List of abbreviations
- ACh
acetylcholine
- ATP
adenosine triphosphate
- A2AR
adenosine receptor A2a
- β2AR
β2 adrenergic receptor
- cpFP
circular-permutated fluorescent protein
- CFP
cyan fluorescent protein
- DA
dopamine
- FRET
Foster resonance energy transfer
- GPCR
G-Protein-Coupled receptor
- GACh
GPCR-activation based acetylcholine sensor
- GRABDA
GPCR-activation based dopamine sensor
- GRABNE
GPCR-activation based norepinephrine sensor
- GABA
γ-aminobutyric acid
- M2R
muscarinic acetylcholine receptor 2
- NE
norepinephrine
- PBP
periplasmic binding protein
- TM
transmembrane helix
- μOR
μ opioid receptor
- YFP
yellow fluorescent protein
Footnotes
Competing interests
M.J. and Y.L. have filed patent applications, the value of which could be affected by this publication.
References
- Abdelfattah AS, Farhi SL, Zhao Y et al. (2016) A Bright and Fast Red Fluorescent Protein Voltage Indicator That Reports Neuronal Activity in Organotypic Brain Slices. J Neurosci 36, 2458–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PTKJBHRN (1973) Voltammetry in brain tissue - a new neurophysiological measurement. Brain Research 55, 209–213. [DOI] [PubMed] [Google Scholar]
- Akerboom J, Carreras Calderon N, Tian L et al. (2013) Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustine V, Ebisu H, Zhao Y, Lee S, Ho B, Mizuno GO, Tian L and Oka Y (2019) Temporally and Spatially Distinct Thirst Satiation Signals. Neuron [DOI] [PMC free article] [PubMed]
- Baird GS, Zacharias DA and Tsien RY (1999) Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci U S A 96, 11241–11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros JA, Jensen AD, Liapakis G, Rasmussen SG, Shi L, Gether U and Javitch JA (2001) Activation of the beta 2-adrenergic receptor involves disruption of an ionic lock between the cytoplasmic ends of transmembrane segments 3 and 6. J Biol Chem 276, 29171–29177. [DOI] [PubMed] [Google Scholar]
- Bayliss BYWM (1901) The mechanism of pancreatic secretion. J Physiol [DOI] [PMC free article] [PubMed]
- Broussard GJ, Liang Y, Fridman M, Unger EK, Meng G, Xiao X, Ji N, Petreanu L and Tian L (2018) In vivo measurement of afferent activity with axon-specific calcium imaging. Nature neuroscience 21, 1272–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson ASMLCA (1995) Crucial role of the accumbens nucleus in the neurotransmitter interactions regulating motor control in mice. J Neural Transm [DOI] [PubMed]
- Carpenter B and Tate CG (2016) Engineering a minimal G protein to facilitate crystallisation of G protein-coupled receptors in their active conformation. Protein engineering, design & selection : PEDS 29, 583–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA et al. (2007) High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science 318, 1258–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corre J, van Zessen R, Loureiro M, Patriarchi T, Tian L, Pascoli V and Luscher C (2018) Dopamine neurons projecting to medial shell of the nucleus accumbens drive heroin reinforcement. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale HH and Dudley HW (1929) The presence of histamine and acetylcholine in the spleen of the ox and the horse. The Journal of Physiology 68, 97–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale HH, Feldberg W and Vogt M (1936) Release of acetylcholine at voluntary motor nerve endings. The Journal of Physiology 86, 353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H, Mohar B, Sun Y et al. (2016) Sensitive red protein calcium indicators for imaging neural activity. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong JW, Afjei SA, Pollak Dorocic I, Peck JR, Liu C, Kim CK, Tian L, Deisseroth K and Lammel S (2019) A Neural Circuit Mechanism for Encoding Aversive Stimuli in the Mesolimbic Dopamine System. Neuron 101, 133–151 e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearry A, Gingrich JA, Falardeau P, Fremeau RT, Bates MD and Caron MG (1990) Molecular cloning and expression of the gene for a human D1 dopamine receptor. Nature 347, 72–76. [DOI] [PubMed] [Google Scholar]
- Dong H, Wang J, Yang YF, Shen Y, Qu WM and Huang ZL (2019) Dorsal Striatum Dopamine Levels Fluctuate Across the Sleep-Wake Cycle and Respond to Salient Stimuli in Mice. Front Neurosci 13, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott TR (1905) Report xciii. the action of adrenalin. British Medical Journal 2, 127–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhang C, Lischinsky JE et al. (2019) A Genetically Encoded Fluorescent Sensor for Rapid and Specific In Vivo Detection of Norepinephrine. Neuron 102, 745–761 e748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gether U, Lin S and Kobilka BK (1995) Fluorescent labeling of purified beta 2 adrenergic receptor. Evidence for ligand-specific conformational changes. J Biol Chem 270, 28268–28275. [DOI] [PubMed] [Google Scholar]
- Grandy DK, Marchionni MA, Makam H et al. (1989) Cloning of the cDNA and gene for a human D2 dopamine receptor. Proc Natl Acad Sci U S A 86, 9762–9766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorio GG, Masureel M, Hilger D et al. (2017) Single-molecule analysis of ligand efficacy in beta(2)AR-G-protein activation. Nature 547, 68-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm H, Deretic D, Arendt A, Hargrave P, Koenig B and Hofmann K (1988) Site of G protein binding to rhodopsin mapped with synthetic peptides from the alpha subunit. Science 241, 832–835. [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Gaietta G, Bunemann M et al. (2005) A FlAsH-based FRET approach to determine G protein-coupled receptor activation in living cells. Nat Methods 2, 171–176. [DOI] [PubMed] [Google Scholar]
- Jing M, Zhang P, Wang G et al. (2018) A genetically encoded fluorescent acetylcholine indicator for in vitro and in vivo studies. Nature biotechnology 36, 726–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz B and Miledi R (1961) The localized action of ‘end-plate drugs’ in the twitch fibres of the frog. The Journal of Physiology 155, 399–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehr J (1992) Microdialysis/HPLC On-Line Dopamine and Metabolites Vol. 12, pp. 1–4. CMA/Microdialysis AB. [Google Scholar]
- Klinkenberg I, Sambeth A and Blokland A (2011) Acetylcholine and attention Vol. 221, pp. 430–442. [DOI] [PubMed] [Google Scholar]
- Liang R, Broussard GJ and Tian L (2015) Imaging chemical neurotransmission with genetically encoded fluorescent sensors. ACS chemical neuroscience 6, 84–93. [DOI] [PubMed] [Google Scholar]
- Lin MZ and Schnitzer MJ (2016) Genetically encoded indicators of neuronal activity. Nature neuroscience 19, 1142–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobas MA, Tao R, Nagai J, Kronschlager MT, Borden PM, Marvin JS, Looger LL and Khakh BS (2019) A genetically encoded single-wavelength sensor for imaging cytosolic and cell surface ATP. Nat Commun 10, 711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewi O (1924) Weiteres ??ber Humorale ??bertragbarkeit der Herznervenwirkung. Klinische Wochenschrift 3, 680–681. [Google Scholar]
- Manglik A, Kim TH, Masureel M et al. (2015) Structural insights into the dynamic process of β<inf>2</inf>-adrenergic receptor signaling. Cell 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcaggi P, Mutoh H, Dimitrov D, Beato M and Knopfel T (2009) Optical measurement of mGluR1 conformational changes reveals fast activation, slow deactivation, and sensitization. Proc Natl Acad Sci U S A 106, 11388–11393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Borghuis BG, Tian L et al. (2013) An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat Methods 10, 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marvin JS, Shimoda Y, Magloire V et al. (2019) A genetically encoded fluorescent sensor for in vivo imaging of GABA. Nature Methods 16, 763–770. [DOI] [PubMed] [Google Scholar]
- Mas M, Fumero B and González-Mora J (1995) Voltammetric and microdialysis monitoring of brain monoamine neurotransmitter release during sociosexual interactions. Behavioural Brain Research 71. [DOI] [PubMed] [Google Scholar]
- Mohebi A, Pettibone JR, Hamid AA, Wong JT, Vinson LT, Patriarchi T, Tian L, Kennedy RT and Berke JD (2019) Dissociable dopamine dynamics for learning and motivation. Nature 570, 65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarchi T, Cho JR, Merten K et al. (2018) Ultrafast neuronal imaging of dopamine dynamics with designed genetically encoded sensors. Science 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Fung JJ et al. (2011) Structure of a nanobody-stabilized active state of the beta(2) adrenoceptor. Nature 469, 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Choi HJ, Rosenbaum DM et al. (2007) Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature 450, 383–387. [DOI] [PubMed] [Google Scholar]
- Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe HW, Hofmann KP and Ernst OP (2008) Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502. [DOI] [PubMed] [Google Scholar]
- Schultz W, Dayan P and Montague PR (1997) A neural substrate of prediction and reward. Science 275, 1593–1599. [DOI] [PubMed] [Google Scholar]
- Shapiro DA, Kristiansen K, Weiner DM, Kroeze WK and Roth BL (2002) Evidence for a model of agonist-induced activation of 5-hydroxytryptamine 2A serotonin receptors that involves the disruption of a strong ionic interaction between helices 3 and 6. J Biol Chem 277, 11441–11449. [DOI] [PubMed] [Google Scholar]
- Smith AD, Olson RJ and Justice JB (1992) Quantitative microdialysis of dopamine in the striatum: effect of circadian variation. Journal of Neuroscience Methods 44, 33–41. [DOI] [PubMed] [Google Scholar]
- Sun F, Zeng J, Jing M et al. (2018) A Genetically Encoded Fluorescent Sensor Enables Rapid and Specific Detection of Dopamine in Flies, Fish, and Mice. Cell 174, 481–496 e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunahara RK, Guan H-C, O’Dowd BF et al. (1991) Cloning of the gene for a human dopamine D5 receptor with higher affinity for dopamine than D1. Nature 350, 614–619. [DOI] [PubMed] [Google Scholar]
- Sung YM, Wilkins AD, Rodriguez GJ, Wensel TG and Lichtarge O (2016) Intramolecular allosteric communication in dopamine D2 receptor revealed by evolutionary amino acid covariation. Proc Natl Acad Sci U S A 113, 3539–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Sun FM, Li YL and Mooney R (2018) A mesocortical dopamine circuit enables the cultural transmission of vocal behaviour. Nature 563, 117-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF and Babu MM (2013) Molecular signatures of G-protein-coupled receptors. Nature 494, 185–194. [DOI] [PubMed] [Google Scholar]
- Vilardaga JP, Bunemann M, Krasel C, Castro M and Lohse MJ (2003) Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nature biotechnology 21, 807–812. [DOI] [PubMed] [Google Scholar]
- Wang H, Jing M and Li Y (2018) Lighting up the brain: genetically encoded fluorescent sensors for imaging neurotransmitters and neuromodulators. Curr Opin Neurobiol 50, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Feng J, Jing M, Li Y, Luo Q, Ding J and Fu L (2019) G protein-assisted optimization of GPCR-activation based (GRAB) sensors 24. [Google Scholar]
- Zhao Y, Araki S, Wu J et al. (2011) An expanded palette of genetically encoded Ca(2)(+) indicators. Science 333, 1888–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XX, Fan LZ, Li P, Shen K and Lin MZ (2017) Optical control of cell signaling by single-chain photoswitchable kinases. Science 355, 836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]