Abstract
Decoding brain functional states underlying cognitive processes from functional MRI (fMRI) data using multivariate pattern analysis (MVPA) techniques has achieved promising performance for characterizing brain activation patterns and providing neurofeedback signals. However, it remains challenging to decode subtly distinct brain states for individual fMRI data points due to varying temporal durations and dependency among different cognitive processes. In this study, we develop a deep learning based framework for brain decoding by leveraging recent advances in intrinsic functional network modeling and sequence modeling using long short-term memory (LSTM) recurrent neural networks (RNNs). Particularly, subject-specific intrinsic functional networks (FNs) are computed from resting state fMRI data and are used to characterize functional signals of task fMRI data with a compact representation for building brain decoding models, and LSTM RNNs are adopted to learn brain decoding mappings between functional profiles and brain states. Validation results on fMRI data from the HCP dataset have demonstrated that brain decoding models built on training data using the proposed method could learn discriminative latent feature representations and effectively distinguish subtly distinct working memory tasks of different subjects with significantly higher accuracy than conventional decoding models. Informative FNs of the brain decoding models identified as brain activation patterns of working memory tasks were largely consistent with the literature. The method also obtained promising decoding performance on motor and social cognition tasks. Our results suggest that LSTM RNNs in conjunction with FNs could build interpretable, highly accurate brain decoding models.
Keywords: brain decoding, working memory, intrinsic functional networks, recurrent neural networks, long short-term memory
Introduction
Multivariate pattern analysis (MVPA) of functional MRI (fMRI) data has been a successful technique for characterizing brain activation patterns and providing neurofeedback signals (Gonzalez-Castillo et al., 2015; Haxby et al., 2001; LaConte, 2011; Rose et al., 2016; Watanabe et al., 2017), and a variety of MVPA methods have been proposed to improve the brain decoding of fMRI data (Davatzikos et al., 2005; Fan et al., 2006; Huth et al., 2016; Jang et al., 2017; Loula et al., 2017; Shirer et al., 2012; Wang et al., 2018).
Most existing fMRI based brain decoding studies focus on identification of functional signatures that are informative for distinguishing different brain states. Particularly, the informative functional signatures are commonly identified as brain activations evoked by task stimuli under a general linear model (GLM) framework (Mumford et al., 2012). Such a procedure of identifying brain activations is equivalent to a supervised feature selection procedure in machine learning, which may improve the sensitivity of the brain decoding. In addition to feature selection under the GLM framework, several studies select regions of interests (ROIs) related to the brain decoding tasks based on a priori anatomical/functional knowledge (Huth et al., 2016). A two-step strategy (Loula et al., 2017) that swaps the functional signature identification from spatial domain to temporal domain has recently been proposed to decode brain activities of fMRI data in the time domain, aiming to overcome the curse of dimensionality problem caused by a large number of spatial functional signatures used for the brain decoding. The aforementioned methods build brain decoding models based on task-specific functional signatures, which may limit their general applications to decoding of brain states associated with other tasks.
Other than task-specific functional signatures identified in a supervised manner, several methods have proposed to build brain decoding models based on whole-brain functional measures. In particular, brain decoding models could be built on whole-brain functional connectivity patterns based on resting-state brain networks identified using independent component analysis (ICA) (Richiardi et al., 2011; Shirer et al., 2012). However, time windows with a properly defined width are required in order to reliably estimate the functional connectivity patterns. Deep belief neural network (DBN) has been adopted to learn a low-dimension representation of 3D fMRI volume for the brain decoding (Jang et al., 2017), where 3D images are flatten into 1D vectors as features for learning the DBN, losing spatial information of the 3D images. More recently, 3D convolutional neural networks (CNNs) are adopted to learn a latent representation for decoding functional brain task states (Wang et al., 2018). Although the CNNs could learn discriminative representations effectively, it is nontrivial to interpret biological meanings of the learned features.
Brain decoding models are typically built on functional signatures computed at individual time points or temporal windows with a fixed length using conventional classification techniques, such as support vector machine (SVM) (Shen et al., 2014) and logistic regression (Huth et al., 2016; Loula et al., 2017). These classifiers do not take into consideration the temporal dependency, which is inherently present in sequential fMRI data and may boost the brain decoding performance if properly explored. Though functional signatures extracted from time windows may help capture the temporal dependency implicitly (Loula et al., 2017; Shirer et al., 2012; Wang et al., 2018), time windows with a fixed width are not necessarily optimal over different brain states since they may change at unpredictable intervals. Meanwhile, recurrent neural networks (RNNs) with long short-term memory (LSTM) (Hochreiter and Schmidhuber, 1997) have achieved remarkable advances in sequence modeling (Lipton et al., 2015), and these techniques might be powerful alternatives for the brain decoding tasks. Recent studies have applied RNNs to intra-subject EEG and ECoG based brain decoding focusing on single region of interest and obtained better brain decoding performance than conventional brain decoding models (Glaser et al., 2017; Schwemmer et al., 2018). RNNs has also been adopted for classification and modeling functional dynamics based on fMRI data (Dvornek et al., 2017; Güçlü and van Gerven, 2017; Hjelm et al., 2018; Li and Fan, 2018b). However, RNNs based techniques have been rarely applied to fMRI based brain decoding tasks across subjects from a large cohort.
In this study, we develop a deep learning based framework for decoding the brain states from task fMRI data, building upon our recent study (Li and Fan, 2018a). Particularly, we learn mappings between functional signatures and brain states by adopting LSTM RNNs which could capture the temporal dependency adaptively by learning from data. Instead of selecting functional signatures using feature selection techniques or a priori knowledge of problems under study, we compute functional profiles of task functional imaging data based on subject-specific intrinsic functional networks (FNs) (Li et al., 2016; Li et al., 2017) and the functional profiles are used as features to build brain decoding models using LSTM RNNs. Our method has been evaluated for predicting brain states based on working memory (WM) task fMRI data obtained from the human connectome project (HCP) (Glasser et al., 2013), and experimental results have demonstrated that the proposed method could obtain better brain decoding performance than the conventional methods, such as Random Forest (RF) (Breiman, 2001). Informative FNs of the brain decoding models were largely overlapped with the WM evoked brain activations, indicating that the LSTM RNNs model captured the functional dynamics of the WM related brain states for the decoding task. Our experiments have also demonstrated that the LST RNNs also obtained promising decoding results on motor and social cognition tasks.
Methods
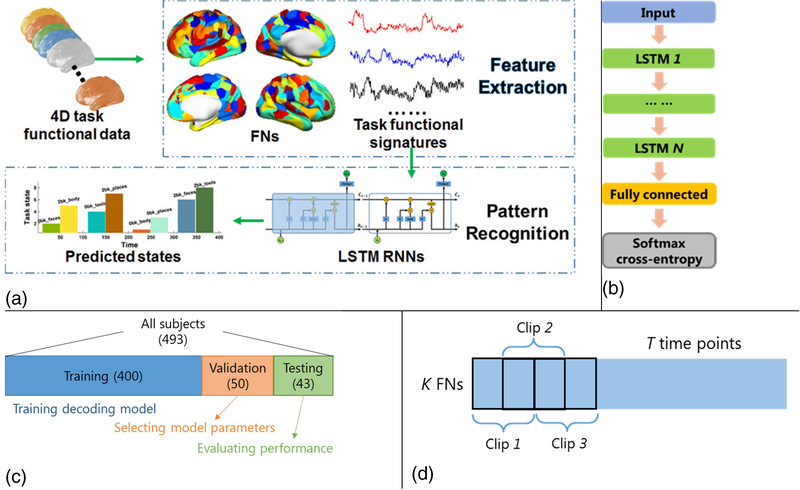
To decode the brain state from task fMRI data, a prediction model of LSTM RNNs (Hochreiter and Schmidhuber, 1997) is trained based on functional signatures extracted using a functional brain decomposition technique (Li et al., 2016; Li et al., 2017). The overall framework is illustrated in Fig. 1(a), consisting of feature extraction and pattern recognition.
Fig. 1.
Schematic illustration of the proposed brain decoding framework. (a) The overall architecture of the proposed model; (b) LSTM RNNs with two LSTM layers used in this study; (c) Data split used for model training, parameter selection, and testing; (d) Functional profile clipping for data augmentation and model training.
Imaging data
Task and resting-state fMRI data of 493 subjects from the HCP (Glasser et al., 2013) were included in this study. Particularly, task fMRI data were used to build and evaluate the proposed decoding framework, and the corresponding resting-state fMRI were used to obtain subject-specific intrinsic functional networks (FNs) for task functional signature extraction. We focused on the working memory, motor, and social cognition tasks in this study. The fMRI data acquisition and task paradigm were detailed in (Glasser et al., 2013), and brief data characteristics are summarized in table 1. In this study, the brain states refer to the task events in the task fMRI experimental paradigm, if not further specified.
Table 1.
fMRI data characteristics included in this study.
| fMRI data information | |||||
|---|---|---|---|---|---|
| # of time points | TR(s) | # of events | Duration of event block(s) | Task events | |
| Working memory | 405 | 0.72 | 8 | 27.5 | 2-back and 0-back task blocks of tools, places, faces, and body |
| Motor | 284 | 0.72 | 5 | 12 | Left foot, left hand, right foot, right hand, tongue |
| Social cognition | 274 | 0.72 | 2 | 16.6 | Mental, random |
| Resting-state | 1200 | 0.72 | N.A. | N.A. | N.A. |
Feature extraction: functional signature based on intrinsic functional networks
With good correspondence to the task activations(Smith et al., 2009), FNs provided an intuitive and generally applicable means to extract functional signatures for the brain state decoding. Using the FNs, 3D fMRI data could be represented by a low-dimension feature vector, which could alleviate the curse of dimensionality, be general to different brain decoding tasks, and provide better interpretability. Instead of identifying ROIs at a group level (Shirer et al., 2012), we applied a collaborative sparse brain decomposition model (Li et al., 2016; Li et al., 2017) to the resting-state fMRI data of all the subjects used for the brain decoding to identify subject-specific FNs.
Given a group of n subjects, each having a resting-state fMRI scan Di ∈ RT×S, i = 1,2, …, n, consisting of S voxels and T time points, we first obtain K FNs and its corresponding functional time courses Ui∈ RT×K for each subject using the collaborative sparse brain decomposition model (Li et al., 2016; Li et al., 2017), which could identify subject-specific functional networks with inter-subject correspondence and better characterize the intrinsic functional representation at an individual subject level. Based on the subject-specific FNs, the functional signatures Fi∈ RT×K used for the brain decoding are defined as weighted mean time courses of the task fMRI data within individual FNs, and are calculated by
| (1) |
where is the full-length task fMRI data of subject i for the brain decoding, is the row-wise normalized Vi with row-wise sum equal to one. Example FNs used in this study are illustrated in Fig. 2.
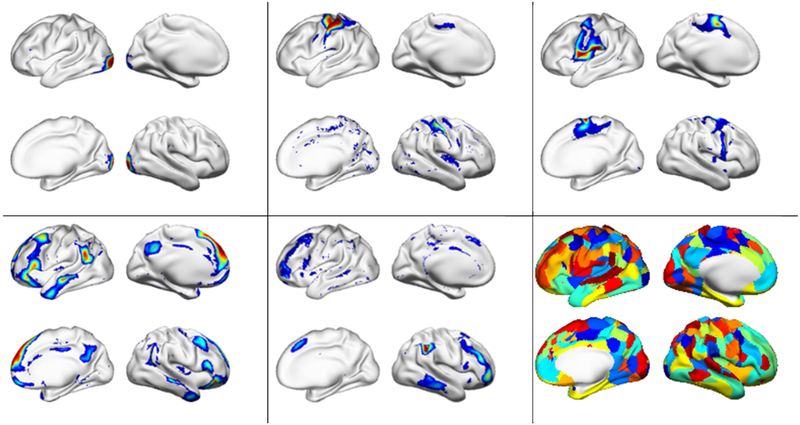
Fig. 2.
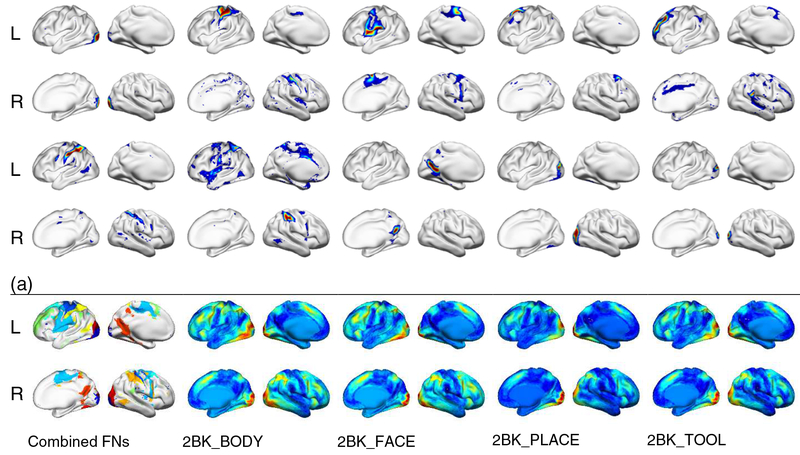
Five example functional networks used to extract task functional signatures for the brain decoding and all functional networks encoded in different colors (bottom right).
Pattern recognition: decoding brain states using LSTM RNNs
Given the functional signatures Fi of a group of subjects, i = 1,2, …, n, a LSTM RNNs (Hochreiter and Schmidhuber, 1997) model is built to predict the brain state of each time point based on its functional profile and temporal dependency on its preceding time points. The architecture of the LSTM RNNs used in this study is illustrated in Fig. 1(b), including two hidden LSTM layers and one fully connected layer. Multiple hidden LSTM layers could be used to encode the functional information with temporal dependency for each time point, and the fully connected layer is used to learn a mapping between the learned feature representation and the brain states. The functional representation encoded in each LSTM layer is calculated as
| (2) |
where , , , , and denote the output of forget gate, input gate, cell state, hidden state, and the input feature vector of the l-th LSTM layer (l = 1, 2) at the t-th time point respectively, and σ denotes the sigmoid function. The input features to the first LSTM layer are the functional signatures derived from FNs, and the input to the second LSTM layer is a hidden state vector obtained by the first LSTM layer. A fully connected layer with output nodes is adopted for predicting the brain state as
| (3) |
where S is the number of brain states to be decoded, and is the hidden state output of the second LSTM layer which encodes the input functional signature at the t-th time point and the temporal dependency information encoded in the cell state from its preceding time points. Softmax cross-entropy between real and predicted brain states is used as the objective function to optimize the LSTM RNNs model.
Validation and comparisons
We applied the collaborative sparse brain decomposition model (Li et al., 2016; Li et al., 2017) to the resting-state fMRI data of 493 subjects for identifying 90 subject-specific FNs. The number of FNs was determined automatically based on resting-state fMRI data using MELODIC’s LAP criteria (Jenkinson et al., 2012). The decomposition was performed using cortical surface data, which did not include whiter matter, ventricle, etc. Therefore, we did not exclude any components in current study. It is worth noting that the subject-specific FNs were identified based on resting state fMRI data. The subject-specific FNs were then used to extract functional signatures of task fMRI data for each subject, which were matrices of 405 by 90, 284 by 90, and 274 by 90 for characterizing working memory, motor, and social cognition tasks, respectively. The magnitude of functional signatures was normalized using z-score and then used as the input to the LSTM RNNs decoding model to predict their corresponding brain states for each task separately. Particularly, we split the whole cohort into training, validation, and testing cohorts as shown in Fig. 1(c). The training cohort included data of 400 subjects for training the LSTM RNNs model, the validation cohort included data of 50 subjects for determining the optimal hyper-parameters involved in the model and early-stop of the training procedure, and data of the remaining 43 subjects were used as an external testing cohort. The same split was used for all the tasks. To further investigate the robustness of the decoding performance, a 5-fold cross-validation was also carried out for all the tasks.
Due to the delay of blood oxygen level dependent (BOLD) response observed in fMRI data, the occurrence of brain response is typically not synchronized with the presentation of stimuli, so the brain state for each time point was adjusted according to the task paradigm and the delay of BOLD signal before training the brain decoding models. Based on an estimated BOLD response delay of 6s (Liao et al., 2002), we shifted the task paradigms forward by 8 time points and used them to update the ground truth brain states for training and evaluating the proposed brain state decoding model.
To train a LSTM RNNs model, we have generated training samples by splitting the functional signatures of each training subject into clip matrices of 40 by 90 with overlap of 20 time points between temporally consecutive training clips, as illustrated in Fig. 1(d). We adopted the cropped dataset for training our model for following reasons. Firstly, the task paradigms of most subjects from the HCP dataset shared almost the identical temporal patterns. In other words, the ground truth brain states of most subjects were the same, which may mislead the model training to generate the same output regardless of the functional signatures fed into the LSTM RNNs model if we used their full-length data for training the brain decoding model. In our study, the length of data clips was set to 40 so that each clip contained 2 or 3 different brain states and such randomness could eliminate the aforementioned bias. Secondly, the data clips with temporal overlap also served as data augmentation of the training samples for improving the model training. When evaluating our LSTM RNNs model, we applied the trained model to the full-length functional signatures of the testing subjects to predict brain states of their entire task fMRI scans. We implemented the proposed method using Tensorflow (Abadi et al.). Particularly, we adopted the ADAM optimizer with a learning rate of 0.001, which was updated every 50,000 training steps with a decay rate of 0.1, and the total number of training steps was set to 200,000. Batch size was set to 32 during the training procedure. Parameters including number of hidden layers ({1, 2, 3) and number of nodes in hidden layer ({32, 64, 128, 256, 512, 1024}), were selected based on their decoding performance on the validation dataset. The parameter selection was performed on working memory task fMRI data only, and the selected parameters were used for all other experiments without further optimization.
We compared the proposed model with a brain decoding model built using RF (Breiman, 2001), which used the functional signatures at individual time point or within a fixed time-window as features. The random forests classifier was adopted due to its inherent feature selection mechanism and its capability of handling multi-class classification problems. For the random forests based brain decoding model, the number of decision trees, the minimum leaf size of the tree, and the length of time-window were selected from a set of parameters ({100, 200, 500, 1000} for the number of trees, {3, 5, 10} for the minimum leaf size, and {5, 10, …, 35, 40, with an increment of 5} for the length of time-window) to optimize its brain decoding performance on the validation dataset. The maximum of window length was set to a similar value to the length of training data clips used by LSTM RNNs, so that the temporal dependency that could be encoded by different decoding models stay at the same level.
To investigate the capability of LSTM based model for learning informative representations for decoding, we measure the association between the hidden cell states (from the 2nd LSTM layer) of the trained LSTM model and different brain states on the WM dataset. As the states of the hidden LSTM cells contribute to the decoding jointly, we embed the output of the hidden cells from the testing subjects into a 2D plane using t-SNE (Maaten and Hinton, 2008), and compare the embedding result with embeddings obtained based on functional profiles of single time points and time-windows.
To investigate association between the hidden LSTM cells and brain states of the different tasks, we visualize activities of the hidden cells of the second LSTM layer using two complementary methods. First, activities of all the 128 hidden cells of the second LSTM layer are shown as a data matrix with each row containing values of one hidden cell for all time points, and the cells are sorted according to absolute values of their corresponding weights in the trained prediction model with a descending order for each task event separately. Second, we identify the hidden LSTM cell whose values are maximally correlated with the onset of each task event.
Performance analysis of the decoding model
To further evaluate the importance of temporal dependency of functional profiles for brain decoding, we have evaluated one brain decoding model obtained using functional signatures without temporal dependency. Particularly, the temporal dependency of task functional signatures was manually removed by randomly permuting the data along the temporal dimension on the training dataset before generating training samples, from which the LSTM based brain decoding model were trained. As the decoding model could not learn any temporal dependency information from the training samples, its prediction performance on the testing dataset is expected to decrease if the temporal dependency does facilitate the brain decoding.
In addition, we have performed more analysis regarding the following two aspects. First, we looked into the prediction performance at each time-point across subjects, in order to unveil how the decoding model works within the task event blocks and transition zones between task blocks. Second, the subjects’ in-scanner performance and level of involvement could also impact the prediction performance, due to that the brain states were labeled based on task paradigms other than the BOLD signal. Therefore, we further investigated the association between the decoding performance and subject’s in-scanner performance. The mean decoding accuracy and in-scanner accuracy measures of two task runs (LR and RL) of each subject were adopted in this study. We also investigated the association between the brain decoding performance and individual subject’s task involvement based on the testing data. Particularly, each individual test subject’s task involvement was measured as the similarity of task-evoked brain activation patterns identified by GLM between the testing and training subjects. For the training subjects, a mean activation pattern across all the training subjects was computed for each task event. The spatial correlation between each testing subject’s brain activation pattern and the mean activation pattern of all the training subjects was used to measure their similarity, and a mean value of all similarity measures across all the task events was used to measure the overall task-evoked activation similarity between the individual testing subject and all the training subjects. The task-evoked activation patterns of all the subjects used in this study were obtained from the HCP dataset.
Moreover, we have also investigated the impact of different task fMRI acquisitions to the brain decoding model. Particularly, two runs of each task are available in the HCP dataset, with phase encodings of LR and RL respectively. Three decoding models were built upon phase encoding LR, RL, and mixed LR & RL training functional signatures respectively, and their prediction performance were evaluated on the corresponding testing datasets. It is worth noting that the experimental results demonstrated was based on phase encoding LR dataset if not specified.
Sensitivity analysis of the decoding model
To understand the LSTM RNNs based decoding model, we have carried out a sensitivity analysis to determine how changes in the functional signatures affect the decoding model based on the 43 testing subjects using a principal component analysis (PCA) based sensitivity analysis method (Koyamada et al., 2015). Particularly, with the trained LSTM RNNs model fixed, functional signatures of 90 FNs were excluded (i.e., their values were set to zero) one by one from the input and changes in the decoding accuracy were recorded. Once all the changes in the brain decoding accuracy with respect to all FNs were obtained for all testing subjects, we obtained a change matrix of 90 × 43, encapsulating changes of the brain decoding. We then applied PCA to the change matrix to identify principle components (PCs) that encoded main directions of the prediction changes with respect to changes in the functional signatures of FNs. The sensitive analysis revealed FNs whose functional signatures were more sensitive than others to the brain decoding. The sensitivity analysis was carried out on the working memory and motor task fMRI data separately.
Results
Training brain decoding models based on training and validation data
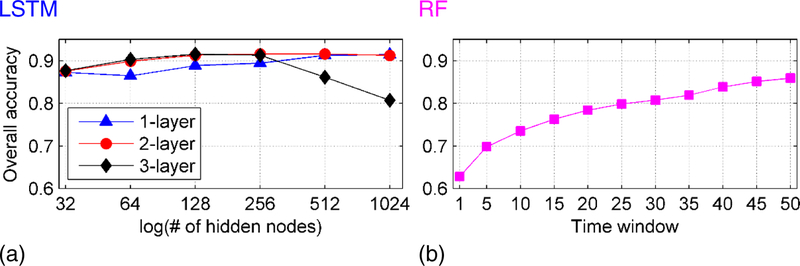
Brain decoding models were built and optimized based on fMRI data of 400 training subjects and 50 validation subjects, obtained from the Human Connectome Project (HCP). Particularly, optimal parameters of the brain decoding models were determined to optimize their overall prediction accuracy on the validation dataset. The overall accuracy was calculated as the mean prediction accuracy across all WM states, including 2-back and 0-back tasks of tools, places, faces and body, as well as cue and fixation. As illustrated in Fig. 3 (a), for the LSTM RNNs based model, the brain decoding accuracy improved with the increase of number of hidden nodes for the brain decoding models with 1 layer and 2 layers of LSTM RNNs before reaching a plateau, while the brain decoding performance degraded when the number of hidden nodes exceeded 256 for the brain decoding model with 3 layers of LSTM RNNs, indicating that this model overfitted the training data when the model became more complicated. We adopted the 2-layer brain decoding model with 128 hidden nodes in each LSTM layer for all the LSTM RNNs based model in all the remaining experiments, due to its relatively high accuracy and small number of trainable parameters, compared with 1-layer model with 512 or 1024 hidden nodes. As illustrated in Fig. 3 (b), the RF based brain decoding model’s performance improved with the increase of the length of the time-window, indicating that longer time-window might contain more temporal information for the brain decoding. However, only marginal improvement could be obtained when time windows larger than 40 time points were used. It is noteworthy that adopting time windows larger than 40 time points will exclude the first task event entirely (the event Left_foot in Motor task for example) and decrease the computational efficiency for training RF based decoding models. We adopted the time-window of 40 time points for all RF based models in the remaining experiments.
Fig. 3.
Parameter selection for LSTM RNNs and random forests (RF) based brain decoding models on the validation dataset. (a) Prediction performance of the LSTM RNNs based decoding model with different numbers of layers and hidden nodes; (b) Prediction performance of the RF based decoding model with time-windows of different lengths.
Decoding accuracy and efficiency on working memory fMRI
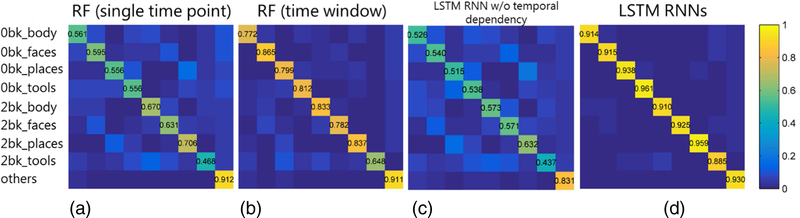
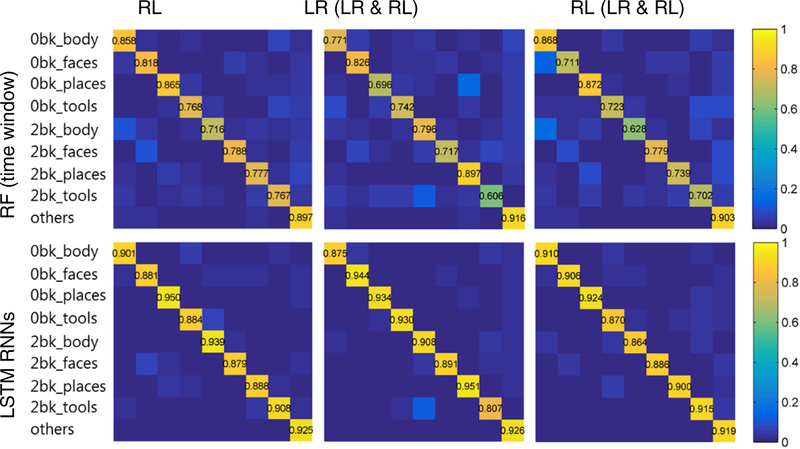
Mean normalized confusion matrices of brain decoding accuracy of the working memory tasks on 43 testing subjects obtained by the brain decoding models built using RF and LSTM RNNs are shown in Fig. 4. The LSTM RNNs based model outperformed the RF based model for all the task events. The overall accuracy obtained by the LSTM RNNs model was 0.926 ± 0.024, while the random forests models built upon functional signatures of time windows and individual time points obtained overall accuracy of 0.807 ± 0.073 and 0.628 ± 0.128 respectively, demonstrating that the LSTM RNNs based model performed significantly better than the RF based models (Wilcoxon signed rank test, p < 1 × 10−10). The decoding performance obtained under a 5-fold cross validation setting also demonstrated that the LSTM RNNs based decoding model was more accurate than the RF based models, as shown in Fig. S5 (a). Brain decoding models built on functional profiles without temporal dependency using LSTM RNNs obtained much worse decoding performance, as shown in Fig. 4 (c), further demonstrating the importance of temporal dependency for the brain decoding. An off-diagonal pattern can be observed in Fig. 4, especially in Fig. 4 (a) and (c), that the events of 0-back category were mainly misclassified into their counterparts of 2-back category. Moreover, a certain portion of wrongly classified time points of each task event were misclassified into the class ‘other’, mainly due to that they located within the transition zones between task events. Computational time for the prediction of brain states of an individual time point using the LSTM RNNs based brain decoding model was less than 0.5 ms on average on a TITAN Xp GPU.
Fig. 4.
Performance of random forest (RF) and LSTM RNNs based brain decoding models on the testing dataset of working memory task fMRI. (a) The RF based model was built on functional signatures of individual time points; (b) The RF based model was built on functional signatures within a window of time points; (c) The LSTM RNNs based brain decoding model was built on training data with temporal dependency removed; (d) The LSTM RNNs model was built on the original training data. The colorbar indicates mean decoding accuracy on the 43 testing subjects. The working memory task events included 2-back and 0-back tasks of tools, places, faces and body, as well as cue and fixation (others).
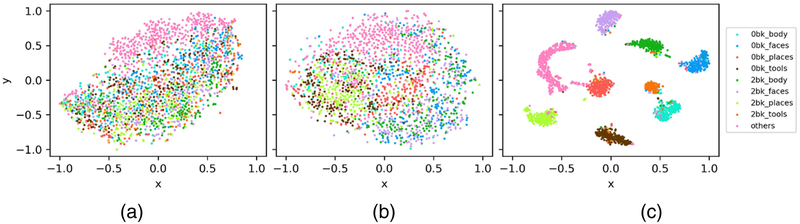
Fig. 5 shows 2D embeddings of individual time points of different WM task events of testing subjects, obtained using t-SNE based on original fMRI profiles at a single time point, within a time window of 40 time points, and latent representations learned by the LSTM RNNs. These results have demonstrated that the LSTM based representation was more effective to distinguish different task events.
Fig. 5.
2D embeddings of individual time points of different WM task events of testing subjects, obtained using t-SNE based on original fMRI profiles at a single time point (a), within a time window of 40 time points (b), and latent representations learned by the LSTM RNNs (c). Time points of different task events are shown in different colors.
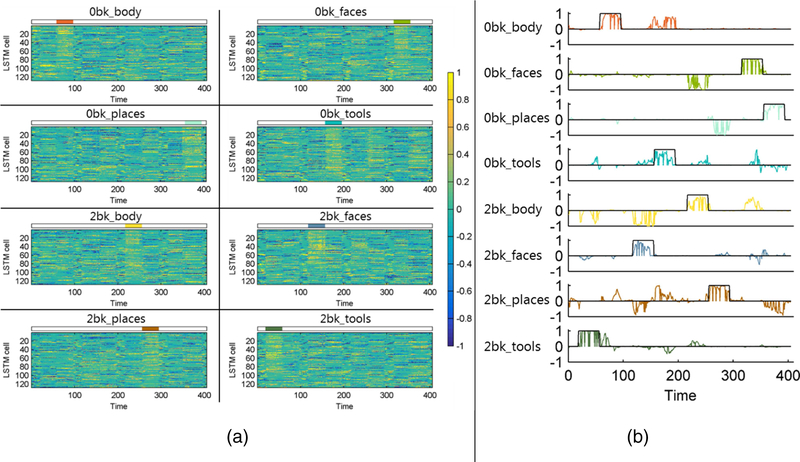
Fig. 6 shows visualization results of association between the hidden LSTM cells and brain states of the different tasks. As illustrated by Fig. 6(a), different cells contributed collaboratively to each of the decoding tasks. Particularly, the activities of each cell varied with the task onset, and cells with larger predictive weights (absolute values) in the prediction model had relatively stronger activities (larger values) on each onset task, demonstrating that the hidden cells reacted to changes of task states and were capable of distinguishing different task states. Fig. 6(b) illustrates that certain hidden cells reacted positively to different task states. These complementary visualization results have demonstrated that RNNs were capable of capturing the temporal dynamics of fMRI data associated with different tasks.
Fig. 6.
Activities of hidden cells in the second LSTM layer of one randomly selected testing subject on the working memory task fMRI dataset. (a) Activities of all 128 hidden cells, the order of cells in each subplot is sorted according to absolute values of their prediction weights (with a descending order) in the prediction model for each task event, the colored block above each subplot indicates the onset interval of each task event, and the colorbar indicates activities (values) of each cell on different time points; (b) Hidden cells with activities (values) maximally correlated with onset of each task event, the black line indicates the onset of each task event, and the colored curve is the activity of the LSTM hidden cell. The activity (value) of the hidden cell is scaled using its maximal absolute value for the visualization.
Performance analysis of the decoding model on working memory fMRI
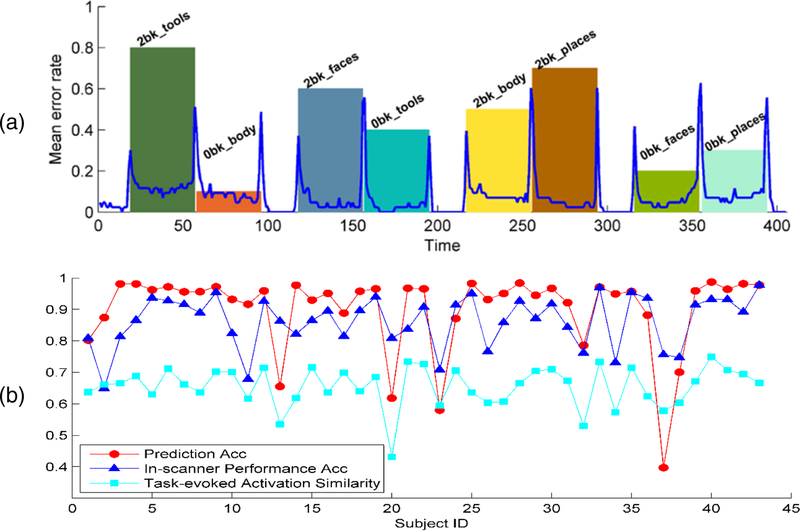
Beyond overall accuracy, the decoding performance at each individual time point across subjects obtained by the LSTM RNNs model is illustrated in Fig. 7(a). The mean brain decoding error rates on time points around task event block onsets and offsets were much larger than on the other time points. Fig. 7(b) shows association of the overall brain decoding accuracy with the in-scanner performance and similarity of task-evoked brain activation patterns between training data and testing data across all the testing subjects. Particularly, the overall decoding accuracy was significantly correlated with the in-scanner performance (r=0.626, p=7.13 × 10−6, Spearman’s rank correlation) and the similarity of task-evoked brain activation patterns between training data and testing data (r=0.521, p=3.39 × 10−4, Spearman’s rank correlation).
Fig. 7.
Performance analysis of the LSTM RNNs based brain decoding model on working memory task fMRI data. (a) Most of the misclassified time points located at inter-state regions; (b) The prediction accuracy is significantly correlated with the subjects’ in-scanner performance accuracy (r=0.626, p=7.13 × 10−6) and task-evoked activation similarity between training data and testing data (r=0.521, p=3.39 × 10−4). The bars with different colors and heights in the background correspond to the different task events in (a).
The brain decoding accuracy measures of WM tasks obtained based on fMRI data acquired using different phase encoding directions are shown in Fig. 8. The overall decoding accuracy obtained by the LSTM RNNs based model was 0.906 ± 0.027, 0.907 ± 0.045, and 0.899 ± 0.022 when the model was trained on RL encoding data (tested on RL encoding data), trained on combined LR & RL encoding data and tested on LR encoding data and RL encoding data respectively, while that obtained by random forests model (using time-window based functional signatures) was 0.806 ± 0.058, 0.774 ± 0.098, and 0.769 ± 0.093 respectively.
Fig. 8.
Brain decoding performance of the random forests (RF, top row) and LSTM RNNs (bottom row) models on the testing dataset of working memory task fMRI with different phase encoding acquisitions. (left) Decoding model trained and tested on phase encoding RL data, (middle) decoding models trained on combined phase encoding LR and RL data and tested on LR data, (right) decoding models trained on combined phase encoding LR and RL data and tested on RL data. The colorbar indicates mean decoding accuracy on the 43 testing subjects. The working memory task events included 2-back and 0-back tasks of tools, places, faces and body, as well as cue and fixation (others).
Sensitivity analysis of the decoding model on working memory fMRI
Sensitivity analysis was adopted to understand how the LSTM RNNs model worked. The sensitive analysis revealed that FNs whose functional signatures were more sensitive than others to the brain decoding on the WM task fMRI data. As shown in Fig. 9 (a), top 10 ranked sensitive FNs, including dorsolateral and anterior prefrontal, inferior frontal, precentral gyrus, anterior cingulate, dorsal parietal, and visual cortex, largely overlapped with the working memory evoked activation patterns identified using GLM (Barch et al., 2013) as demonstrated in Fig. 9 (b). The associations between the top ranked FNs and task evoked activation have also been investigated quantitatively. As the FNs contributed to the decoding jointly, we built one linear regression model to explore the relationship between the 10 selected FNs and the evoked activation maps for each task condition of the testing subjects identified using GLM, and R2 was adopted to measure the variance explained by the FNs. The mean and standard deviation of R2 across subjects for each condition are listed in table 2. F-tests on the regression models confirmed that the model fitting was statistically significant (p<0.05). Moreover, we also built one linear regression model to explore the relationship between the functional profiles of the 10 selected FNs and task event labels for the WM task fMRI data of each testing subjects. The mean R2 was 0.419 (standard deviation 0.077). F-tests on the regression models demonstrated that the model fitting was statistically significant (p<0.05). All these results indicated that the LSTM RNNs model captured the functional activation patterns of the WM related brain states.
Fig. 9.
Sensitivity analysis of the brain decoding model on the working memory task fMRI dataset. (a) The top 10 FNs with most sensitive functional signatures are illustrated; (b) The mean WM evoked activation patterns obtained using GLM for different task events. The top 10 FNs in the combined map are shown in different colors (b, upper left). L and R indicates left and right hemisphere.
Table 2.
Variance explained by the selected FNs with respect to the WM task evoked activation patterns.
| 2BK_BODY | 2BK_FACE | 2BK_PLACE | 2BK_TOOL | 0BK_BODY | 0BK_FACE | 0BK_PLACE | 0BK_TOOL | |
|---|---|---|---|---|---|---|---|---|
| R2 Mean±std | 0.186±0.048 | 0.170±0.053 | 0.208±0.063 | 0.186±0.049 | 0.191±0.063 | 0.188±0.068 | 0.256±0.078 | 0.240±0.069 |
More evaluation on motor and social cognition task fMRI
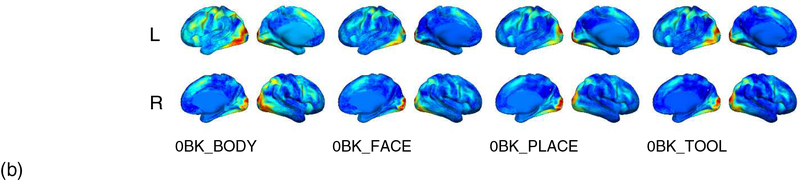
The LSTM RNNs decoding framework has also been evaluated on based motor and social cognition task fMRI data of the same training, validation, and testing subjects as used in the experiment of WM tasks. The mean normalized confusion matrices of the brain decoding accuracy of the 43 testing subjects on the motor task fMRI data obtained by the RF and the LSTM RNNs based models are shown in Fig. S1 (a, b, d). The overall accuracy obtained by the LSTM RNNs based model was 0.966 ± 0.026, while the overall accuracy obtained by the RF based model built on functional signatures of time windows and individual time points was 0.863 ± 0.066 and 0.742 ± 0.111, respectively, demonstrating the superiority of the LSTM RNNs based decoding model. As shown in Fig. S1 (c, d), the performance difference between decoding models built on functional profiles with and without temporal dependency using the LSTM RNNs further demonstrated the importance of modeling temporal dependency for brain decoding tasks. Sensitivity analysis results of the decoding model regarding motor task fMRI data as shown in Fig. S2 revealed that the LSTM RNNs based decoding model could effectively identify the task related brain regions, including regions involved in sensory motor network, visual network, and executive control network.
The mean normalized confusion matrices of the brain decoding accuracy of the testing subjects on the social cognition task fMRI data obtained by the RF and LSTM RNNs based models are shown in Fig. S3. The overall accuracy obtained by the LSTM RNNs based model was 0.929 ± 0.060, while the overall accuracy obtained by the RF based model built on functional signatures of time windows and individual time points was 0.920 ± 0.039 and 0.912 ± 0.65, respectively, demonstrating that the LSTM RNNs based decoding model was capable of capturing the functional dynamics underlying complex social cognitive processes. Sensitivity analysis results of the decoding model regarding social cognition task fMRI data, as shown in Fig. S4, revealed that the LSTM RNNs based decoding model could effectively identify the task related brain regions, including regions in visual cortex, auditory cortex, superior temporal sulcus, temporo-parietal junction, premotor cortex, and prefrontal cortex.
The decoding accuracy of the LSTM RNNs based model on both motor and social cognition task fMRI datasets also outperformed the RF based model under the 5-fold cross-validation setting, as demonstrated in Fig. S5 (b) and (c).
Discussion
In this study, we demonstrated that brain decoding models built on functional signatures of individualized FNs using LSTM RNNs better distinguished subtly distinct brain states based on task fMRI data than those built using RF. To build interpretable brain decoding models, we extracted functional signatures underlying different brain states from task fMRI data for individual subjects based on their individualized FNs, and built a LSTM RNNs based brain decoding model on these functional signatures to capture their temporal dynamics and learn the mapping between functional signatures and their corresponding brain states. The decoding performance on the working memory, motor, and social cognition task fMRI data from the HCP demonstrated that the proposed brain decoding framework was capable of decoding brain states across different categories of tasks with improved performance.
The performance of a decoding model is dependent on the functional profiles used to build the decoding model. Functional signals of brain regions identified based on prior knowledge or brain activation maps and whole-brain functional connectivity patterns modulated by task stimuli are widely adopted as functional profiles for the brain decoding in fMRI studies (Loula et al., 2017; Naselaris et al., 2011; Richiardi et al., 2011; Shirer et al., 2012). The brain activation maps and prior knowledge help identify task-specific brain regions, but may limit their general applications to decoding of brain states associated with other tasks. The computation of functional connectivity patterns requires to specify a time window with a certain length to reliably estimate the functional connectivity. However, it is nontrivial to determine a time window length that is optimal for different brain decoding tasks due to varying temporal durations and dependency among different cognitive processes. In this study, inspired by the good coincidence between the task evoked activation patterns and FNs (Li et al., 2016; Li et al., 2017), we utilized intrinsic FNs to extract task related functional signatures for brain decoding. The FNs based low-dimensional functional signatures could capture task-related functional dynamics with good fidelity, and also be able to generalize well across different cognitive tasks as intrinsic FNs are not task-specific. Experimental results have demonstrated that the FNs based functional signatures could achieve promising performance on working memory tasks, motor tasks, and social cognition tasks related brain decoding tasks, indicating that it could generalize well across distinct cognitive tasks, including those involving distributed brain functional networks such as working memory tasks as well as those involving only localized functional networks such as motor tasks. In this study, we used a computationally efficient method to compute fMRI signal of individual FNs. However, a least square optimization based projection could be adopted too. Moreover, we adopted an NMF based brain decomposition method that could identify functional networks without anti-correlated signals within each of them. Other brain decomposition methods, such as group information guided independent component analysis (Du and Fan, 2013), could also be adopted for the brain decoding. However, anti-correlated signals may present in the same component obtained by ICA methods, which could result in cancel-out effects in the computation of functional signals of FNs.
Temporal dependency is inherently present in sequential fMRI data and underlying brain activities and may boost the brain decoding performance if properly explored in brain decoding models. LSTM RNNs provide an ideal tool to model sequential data as it can learn to characterize the temporal dependency adaptively in a data-driven way. Recent deep learning studies have demonstrated that CNNs could achieve performance similar to or even better than RNNs in language understanding tasks such as language modeling, machine translation and question answering (Yin et al., 2017). However, the methods based on CNNs might be ineffective for real-time brain decoding applications with different task events due to their utilization of a fixed window size that is not necessarily optimal for different tasks. In contrast, RNNs could adaptively capture temporal dynamics. Improved brain decoding performance has been obtained by brain decoding models built on functional signals of single brain region of interest using LSTM RNNs for EEG/ECoG based intra-subject brain decoding tasks (Glaser et al., 2017; Schwemmer et al., 2018). RNNs have also been adopted to identify functional networks from fMRI data in a generative modeling setting to account for temporal dynamics and dependencies through recurrent parameters(Hjelm et al., 2018). Experiments with simulated and resting state fMRI data have demonstrated that this method could visualize the temporal dynamics of both first order (activity) and second order (directed connectivity) information in brain networks. This method is fundamentally different from ours in that we adopt RNNs in a supervised setting to model the functional dynamics of fMRI data associated with different task states. Interestingly, RNNs in both the generative and supervised settings have demonstrated encouraging results for capturing the temporal dynamics of fMRI data. These results support the feasibility and validity of RNNs to model the temporal dynamics and dependencies for brain decoding tasks.
The temporal dependency of functional signatures plays vital role for improving the decoding performance. The brain decoding models built using the LSTM RNNs outperformed those built on time-window based functional signatures using RF, which also outperformed those built on functional signatures without temporal dependency, as shown in Fig. 4 (a, b, d) and Fig. S1 (a, b, d), indicating that the temporal dependency of fMRI data could be effectively captured by the LSTM RNNs to provide robust and discriminative information for the brain decoding. The importance of temporal dependence of fMRI data for the brain decoding is also supported by the performance gap between brain decoding models built on data with and without temporal dependency, as shown in Fig. 4 (c, d) and S1 (c, d). The visualization results shown in Fig. 5 and Fig. 6 have demonstrated that the LSTM RNNs could learn discriminative patterns and effectively capture the temporal dynamics of fMRI data.
Not surprisingly, Fig. 7 (a) demonstrated that the brain decoding models had relatively low decoding accuracy on time points around task event block onsets and offsets, indicating that functional signals of time points around task event block onsets and offsets might not be reliable for the brain decoding. Fig. 7 (b) demonstrated that the inter-subject brain decoding models’ performance is dependent on individual subjects’ in-scanner task performance and similarity between brain activation patterns between the training and testing subjects. These results demonstrate that the LSTM RNNs brain decoding model indeed captured the task evoked brain functional activations to a certain extent, other than overfitted the fMRI data and mapped task irrelevant functional profiles (functional signatures of subjects who did not fully engage in the tasks) to brain states encoded by the task paradigms. In the current study, a common delay time of the BOLD response was adopted for all the subjects. It is worth noting that hemodynamic response (HRF) may vary across different subjects and across brain regions (Handwerker et al., 2004; Pedregosa et al., 2015). Such variations make it difficult to identify “ground-truth” brain states for fMRI data for training and evaluating the brain decoding models, especially for time points within the transition zone between task events. Such variations possibly contributed to the higher prediction error at the brain state transition zones as demonstrated in Fig. 6 (a). A data-driven method has been developed for joint estimation of brain activation and HRF in a general linear model (GLM) setting by forcing the estimated HRF to be equal across task events or experimental conditions, but permitting it to differ across voxels (Pedregosa et al., 2015). It merits further investigation to learn the BOLD response delay for better training and evaluating brain decoding models.
Sensitivity analysis of the brain decoding models of WM and motor tasks (Fig. 9 (a), Fig. S2, and Fig. S4) revealed that top ranked sensitive FNs were largely consistent with brain regions involved in WM and motor tasks reported in the literature, indicating the LSTM RNNs based brain decoding model could capture the brain state related functional dynamics.
The WM brain decoding models built on the fMRI data acquired using mixed, different phase encoding directions had similar performance as those build on the fMRI data acquired using the same phase encoding direction, indicating that the brain decoding models achieved relatively stable decoding performance, and the LSTM RNNs based models had better accuracy than the RF based models.
Although the proposed brain decoding method has achieved better performance than RF, further efforts are needed in following aspects. First, the brain decoding models were evaluated using block designed tasks, and it merits further evaluation based on event designed task fMRI to investigate how the temporal duration of brain states impact the decoding performance. Second, FNs at a single spatial scale were used to compute functional signatures for the brain decoding. Our recent study has demonstrated multiple scale, hierarchical FNs could better characterize functional network organization (Li et al., 2018), and therefore the multiple scale, hierarchical FNs could potentially improve the brain decoding performance too. Third, sensitive analysis was used to understand how different FNs contributed to the brain decoding models. Other techniques, such as attention model (Bahdanau et al., 2014) may help capture the co-activation among multiple FNs at the same time and provide complementary information to the interpretation of brain decoding models. Finally, other than LSTM RNNs adopted in the present study, gated recurrent neural networks could be adopted to build brain decoding models in the same framework (Chung et al., 2014).
In summary, we propose a deep learning based framework for decoding the brain states underlying different cognitive processes from task fMRI data. Subject-specific intrinsic functional networks are used to extract task related functional signatures, and the LSTM RNNs technique is adopted to adaptively capture the temporal dependency within the functional data as well as the relationship between the learned functional representations and the brain functional states. The experimental results on the brain decoding of working memory, motor, and social cognition tasks based on fMRI data have demonstrated that the proposed model could obtain improved brain decoding performance compared with those built without considering the temporal dependency explicitly.
Supplementary Material
Acknowledgements
This work was supported in part by the National Institutes of Health grants [EB022573, CA223358, and DK117297] and a NVIDIA Academic GPU grant.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, TensorFlow: A System for Large-Scale Machine Learning.
- Bahdanau D, Cho K, Bengio Y, 2014. Neural machine translation by jointly learning to align and translate. arXiv preprint arXiv:1409.0473. [Google Scholar]
- Barch DM, Burgess GC, Harms MP, Petersen SE, Schlaggar BL, Corbetta M, Glasser MF, Curtiss S, Dixit S, Feldt C, Nolan D, Bryant E, Hartley T, Footer O, Bjork JM, Poldrack R, Smith S, Johansen-Berg H, Snyder AZ, Van Essen DC, Consortium WU-MH, 2013. Function in the human connectome: task-fMRI and individual differences in behavior. Neuroimage 80, 169–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L, 2001. Random forests. Machine learning 45, 5–32. [Google Scholar]
- Chung J, Gulcehre C, Cho K, Bengio Y, 2014. Empirical Evaluation of Gated Recurrent Neural Networks on Sequence Modeling. arXiv:1412.3555. [Google Scholar]
- Davatzikos C, Ruparel K, Fan Y, Shen DG, Acharyya M, Loughead JW, Gur RC, Langleben DD, 2005. Classifying spatial patterns of brain activity with machine learning methods: Application to lie detection. Neuroimage 28, 663–668. [DOI] [PubMed] [Google Scholar]
- Du Y, Fan Y, 2013. Group information guided ICA for fMRI data analysis. Neuroimage 69, 157–197. [DOI] [PubMed] [Google Scholar]
- Dvornek NC, Ventola P, Pelphrey KA, Duncan JS, 2017. Identifying autism from resting-state fMRI using long short-term memory networks International Workshop on Machine Learning in Medical Imaging. Springer, pp. 362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Shen D, Davatzikos C, 2006. Detecting Cognitive States from fMRI Images by Machine Learning and Multivariate Classification 2006 Conference on Computer Vision and Pattern Recognition Workshop (CVPRW’06), pp. 89–89. [Google Scholar]
- Glaser JI, Chowdhury RH, Perich MG, Miller LE, Kording KP, 2017. Machine learning for neural decoding. arXiv preprint arXiv:1708.00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, Xu J, Jbabdi S, Webster M, Polimeni JR, Van Essen DC, Jenkinson M, Consortium WU-MH, 2013. The minimal preprocessing pipelines for the Human Connectome Project. Neuroimage 80, 105–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Castillo J, Hoy CW, Handwerker DA, Robinson ME, Buchanan LC, Saad ZS, Bandettini PA, 2015. Tracking ongoing cognition in individuals using brief, whole-brain functional connectivity patterns. Proceedings of the National Academy of Sciences 112, 8762–8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güçlü U, van Gerven MA, 2017. Modeling the dynamics of human brain activity with recurrent neural networks. Frontiers in computational neuroscience 11, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerker DA, Ollinger JM, D’Esposito M, 2004. Variation of BOLD hemodynamic responses across subjects and brain regions and their effects on statistical analyses. Neuroimage 21, 1639–1651. [DOI] [PubMed] [Google Scholar]
- Haxby JV, Gobbini MI, Furey ML, Ishai A, Schouten JL, Pietrini P, 2001. Distributed and overlapping representations of faces and objects in ventral temporal cortex. Science 293, 2425–2430. [DOI] [PubMed] [Google Scholar]
- Hjelm RD, Damaraju E, Cho K, Laufs H, Plis SM, Calhoun VD, 2018. Spatio-Temporal Dynamics of Intrinsic Networks in Functional Magnetic Imaging Data Using Recurrent Neural Networks. Frontiers in neuroscience 12, 600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochreiter S, Schmidhuber J, 1997. Long short-term memory. Neural computation 9, 1735–1780. [DOI] [PubMed] [Google Scholar]
- Huth AG, Lee T, Nishimoto S, Bilenko NY, Vu AT, Gallant JL, 2016. Decoding the Semantic Content of Natural Movies from Human Brain Activity. Front Syst Neurosci 10, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H, Plis SM, Calhoun VD, Lee JH, 2017. Task-specific feature extraction and classification of fMRI volumes using a deep neural network initialized with a deep belief network: Evaluation using sensorimotor tasks. Neuroimage 145, 314–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM, 2012. Fsl. Neuroimage 62, 782–790. [DOI] [PubMed] [Google Scholar]
- Koyamada S, Koyama M, Nakae K, Ishii S, 2015. Principal Sensitivity Analysis. Pacific-Asia Conference on Knowledge Discovery and Data Mining. Springer, pp. 621–632. [Google Scholar]
- LaConte SM, 2011. Decoding fMRI brain states in real-time. Neuroimage 56, 440–454. [DOI] [PubMed] [Google Scholar]
- Li H, Fan Y, 2018a. Brain Decoding from Functional MRI Using Long Short-Term Memory Recurrent Neural Networks. Springer International Publishing, Cham, pp. 320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Fan Y, 2018b. Identification of Temporal Transition of Functional States Using Recurrent Neural Networks from Functional MRI. Springer International Publishing, Cham, pp. 232–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Satterthwaite T, Fan Y, 2016. Identification of subject-specific brain functional networks using a collaborative sparse nonnegative matrix decomposition method 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), pp. 984–987. [Google Scholar]
- Li H, Zhu X, Fan Y, 2018. Identification of Multi-scale Hierarchical Brain Functional Networks Using Deep Matrix Factorization. Springer International Publishing, Cham, pp. 223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Jing RX, Zhao RJ, Ding ZB, Shi L, Sun HQ, Lin X, Fan TT, Dong WT, Fan Y, Lu L, 2017. Electroconvulsive therapy-induced brain functional connectivity predicts therapeutic efficacy in patients with schizophrenia: a multivariate pattern recognition study. NPJ Schizophr 3, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao CH, Worsley KJ, Poline JB, Aston JA, Duncan GH, Evans AC, 2002. Estimating the delay of the fMRI response. Neuroimage 16, 593–606. [DOI] [PubMed] [Google Scholar]
- Lipton ZC, Berkowitz J, Elkan C, 2015. A critical review of recurrent neural networks for sequence learning. arXiv preprint arXiv:1506.00019. [Google Scholar]
- Loula J, Varoquaux G, Thirion B, 2017. Decoding fMRI activity in the time domain improves classification performance. Neuroimage. [DOI] [PubMed] [Google Scholar]
- Maaten L.v.d., Hinton G, 2008. Visualizing data using t-SNE. Journal of machine learning research 9, 2579–2605. [Google Scholar]
- Mumford JA, Turner BO, Ashby FG, Poldrack RA, 2012. Deconvolving BOLD activation in event-related designs for multivoxel pattern classification analyses. Neuroimage 59, 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naselaris T, Kay KN, Nishimoto S, Gallant JL, 2011. Encoding and decoding in fMRI. Neuroimage 56, 400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedregosa F, Eickenberg M, Ciuciu P, Thirion B, Gramfort A, 2015. Data-driven HRF estimation for encoding and decoding models. Neuroimage 104, 209–220. [DOI] [PubMed] [Google Scholar]
- Richiardi J, Eryilmaz H, Schwartz S, Vuilleumier P, Van De Ville D, 2011. Decoding brain states from fMRI connectivity graphs. Neuroimage 56, 616–626. [DOI] [PubMed] [Google Scholar]
- Rose NS, LaRocque JJ, Riggall AC, Gosseries O, Starrett MJ, Meyering EE, Postle BR, 2016. Reactivation of latent working memories with transcranial magnetic stimulation. Science 354, 1136–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemmer MA, Skomrock ND, Sederberg PB, Ting JE, Sharma G, Bockbrader MA, Friedenberg DA, 2018. Meeting brain–computer interface user performance expectations using a deep neural network decoding framework. Nature medicine, 1. [DOI] [PubMed] [Google Scholar]
- Shen G, Zhang J, Wang M, Lei D, Yang G, Zhang S, Du X, 2014. Decoding the individual finger movements from single-trial functional magnetic resonance imaging recordings of human brain activity. Eur J Neurosci 39, 2071–2082. [DOI] [PubMed] [Google Scholar]
- Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD, 2012. Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cereb Cortex 22, 158–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF, 2009. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci U S A 106, 13040–13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liang X, Zhou Y, Wang Y, Cui J, Wang H, Li Y, Nguchu BA, Qiu B, 2018. Task state decoding and mapping of individual four-dimensional fMRI time series using deep neural network. arXiv preprint arXiv:180109858. [Google Scholar]
- Watanabe T, Sasaki Y, Shibata K, Kawato M, 2017. Advances in fMRI Real-Time Neurofeedback. Trends Cogn Sci 21, 997–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W, Kann K, Yu M, Schütze H, 2017. Comparative study of cnn and rnn for natural language processing. arXiv preprint arXiv:1702.01923. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.