Abstract
Recent interest in enlarged perivascular spaces (ePVS) in the brain, which can be visualized on MRI and appear isointense to cerebrospinal fluid on all sequence weightings, has resulted in the necessity of reliable algorithms for automated segmentation to allow for whole brain assessment of ePVS burden. However, several publicly available datasets do not contain sequences required for recently published algorithms. This prospective study presents a method for identification of enlarged perivascular spaces (ePVS) in white matter using 3T T1 and FLAIR MR imaging (MAPS-T1), making the algorithm accessible to groups with valuable sets of limited data. The approach was applied identically to two datasets: 1) a repeated measurement in a dementia-free aged human population (N=14), and 2) an aged sample of multisite ADNI datasets (N=30). ePVS segmentation was accomplished by a stepwise local homogeneity search of white matter-masked T1-weighted data, constrained by FLAIR hyperintensity, and further constrained by width, volume, and linearity measurements. Pearson’s r was employed for statistical testing between visual (gold standard) assessment and repeated measures in cohort one. Visual ePVS counts were significantly correlated with MAPS-T1 (r = .72, P < .0001). Correlations between repeated measurements in cohort one were significant for both visual and automated methods in the single visually-rated slice (MAPS-T1: r = .87, P < .0001, visual: (r = .86, P < .0001) and for whole brain assessment (MAPS-T1: r = .77, P = .001,). Results from each cohort were manually inspected and found to have positive predictive values of 77.5% and 87.5%, respectively. The approach described in this report is an important tool for detailed assessment of ePVS burden in white matter on routinely acquired MRI sequences.
Keywords: enlarged perivascular space, MRI, segmentation, Alzheimer’s, vascular disease, Virchow-Robin
1. Introduction
Enlarged perivascular spaces (ePVS), commonly termed Virchow-Robin spaces, are visible on standard magnetic resonance imaging protocols at clinical field strength and have been grossly associated with several neurological disease processes (Achiron and Faibel, 2002; Banerjee et al., 2017; Gutierrez et al., 2017; Kuribara et al., 2017). The detection of ePVS in MR imaging has classically included the necessity of visually inspecting both T1 and T2-weighted sequences to ensure that the space in question is isointense to ventricular cerebrospinal fluid (Wardlaw et al., 2013). Manual ePVS burden assessment can be onerous and time-consuming in clinical and in research settings, which may preclude whole brain assessment, and is typically done on axial images for which burden can vary based on the position of the slice and the orientation of vascular structures at that position. A recently published automated method utilizes spatially coregistered T1, T2, fluid attenuated inversion recovery (FLAIR) and proton density (PD) for whole brain burden assessment (Boespflug et al., 2018). Acquisition time of all four sequences may be prohibitive in certain clinical or research settings and the requirement of four separate sequences may preclude the use of the multimodal algorithm in some legacy longitudinal datasets, which are of particular interest in the context of neurodegenerative disease and normal aging (Gao et al., 2011; van den Heuvel et al., 2006). Finally, movements between sequences that are more than one millimeter and/or imperfect post hoc coregistrations may limit the sensitivity of the multimodal algorithm. ePVS detection is particularly vulnerable to small errors in coregistration due to the linear shape and small size of ePVS. An algorithm requiring fewer sequences increases sensitivity by requiring less coregistration and interpolation while shorter acquisition time minimizes opportunities for subject motion to compromise data.
These limitations drive the necessity of an algorithm that uses fewer volumes or weightings as a basis set. The most commonly acquired weightings performed at clinical MRI field strengths are T1 and T2. This report describes a segmentation algorithm that utilizes T1 and T2-weighted FLAIR volumes (MAPS-T1) and compares results in a repeated measures cohort to expert visual ratings, as well as a separate multisite validation of MAPS-T1 in the publicly available Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. We present a method for identification of enlarged perivascular spaces using only commonly clinically acquired T1 and FLAIR MR imaging, making the algorithm accessible to groups with valuable sets of limited data.
2. Materials and Methods
2.1. Participants
2.1.1. Cohort one - repeated measures
Neuroimaging data were acquired as part of an ongoing prospective research study. Study participants signed informed consent and HIPAA authorization, approved by the Institutional Review Board, prior to participation in the study. Neuroimaging sessions were performed during two distinct sessions on 14 participants (Table 1, characteristics of cohort one are at visit one). The mean inter-scan interval was 366 days (range: 280-441 days, st. dev.=49). Participants were older dementia-free adults living independently. Visual ratings were made by three independent raters on each dataset using an established visual rating scale (Wardlaw et al., 2013). One rater is a board certified neuroradiologist (JMP) with 10.5 years of experience and the other two raters each have at least 10 years of MRI experience (ELB and DLL). Spatially co-registered T1-weighted and FLATR images were made available and each rater chose an axial slice superior to the lateral ventricles that subjectively represented the slice with the highest number of ePVS. Each rater identified the location of each distinct ePVS on the entire slice by drawing a single pixel region of interest. Segmented ePVS were overlaid on T1-weighted and FLAIR volumes for each subject and an author (DLS) screened the results for false alarms.
Table 1.
Dataset Characteristics
| Cohort 1 (N=14) | Cohort 2 (N=30) | |
|---|---|---|
| Age, years (range) | 85.3 (70-101) | 74.3 (58-92) |
| Gender (% female) | 57.8 | 40.0 |
| MMSE (range) | 28.36 (22-30) | n/a |
| CDR %0 (range) | 83 (0.0-0.5) | 16.7 (0.0-2.0) |
| ICV (cm3) | 1871.6 ± 202.8 | 1855.4 ± 219.1 |
| WMH (cm3) | 12.3 ± 10.2 | 10.5 ± 10.1 |
| Hachinski %>0 (range) | 64 (0-8) | 90 (1-3) |
| History of stroke (%) | 29 | 7 |
2.1.2. Cohort two – ADNI
Thirty datasets that was acquired as part of the multisite ADNT2 imaging protocol were downloaded as a part of a larger cohort from the ADNT website subject to the following constraints: 1) near isotropic T1-weighted and FLATR MRI, 2) Hachinski Ischemia Score (HTS) >0 and clinically reported presence of hypertension, 3) age was normally distributed in the cohort (range: 58-92 years, Table 1). Seventeen sites were represented in the final dataset. Segmented ePVS were overlaid on T1-weighted and FLATR volumes for each subject and two authors (ELB and DLS) screened the results for false alarms.
2.2. Image acquisition and preprocessing
Cohort one MRI data were obtained using a 3.0T Siemens Trio MRI (TIM Trio System, Siemens Healthineers, Erlangen, Germany). Two sequences were acquired: 3D T1-weighted magnetization prepared rapid gradient echo ([MPRAGE/T1]: TR/TE/TI/FA=2300ms/3.41ms/1200ms/12°, 128 sagittal 1mm slices with no gap, FOV=256x192mm, imaging matrix=256x192); 2D fluid attenuated inversion recovery ([FLAIR]: TR/TE /TI/FA=9000 ms/87ms/2500ms/100°, 95 axial 2mm slices with no gap, FOV=228x248 mm, imaging matrix=236x256, 2 averages). Cohort 2 MRI data were acquired as described at http://adni.loni.usc.edu/methods/mri-tool/mri-analysis/.
DICOM files were converted to NIFTI using mcverter (MRIConvert, http://lcni.uoregon.edu/downloads/mriconvert/mriconvert-and-mcverter). T1-weighted images were segmented into tissue types using Freesurfer v5.1, which yielded masks of white matter (WM), cortical gray matter (GM), subcortical gray matter (BG), and ventricular CSF. WM masks were corrected for tissue misclassification due to white matter hyperintensities (WMH), meaning that WMH were included in the WM mask, by the method described in (Promjunyakul et al., 2015). Briefly, clusters of contiguous voxels of intensity 35% higher than the peak of the distribution of intensities in FLAIR WM were used as seed clusters for a custom cluster growing algorithm. The mean intensity of each cluster was calculated and then all nearest neighbor voxels of intensity exceeding 95% of the mean cluster intensity were iteratively added to the cluster until no additional voxels met threshold. White matter masks were eroded by a single voxel to avoid the potential of partial volume effects. Separately, FLAIR and T1 volumes were bias field corrected using slicer3 (https://www.slicer.org/) in preparation for ePVS segmentation. The MAPS-T1 algorithm was accomplished in two phases, intensity-based and morphology-based, as follows. Note that the cohort one data used in this report is confidential and cannot be shared publically; however, the code that executes the segmentation of ePVS can be made available upon direct request to the corresponding author (DLS).
2.3.1. Local voxel intensity inhomogeneity
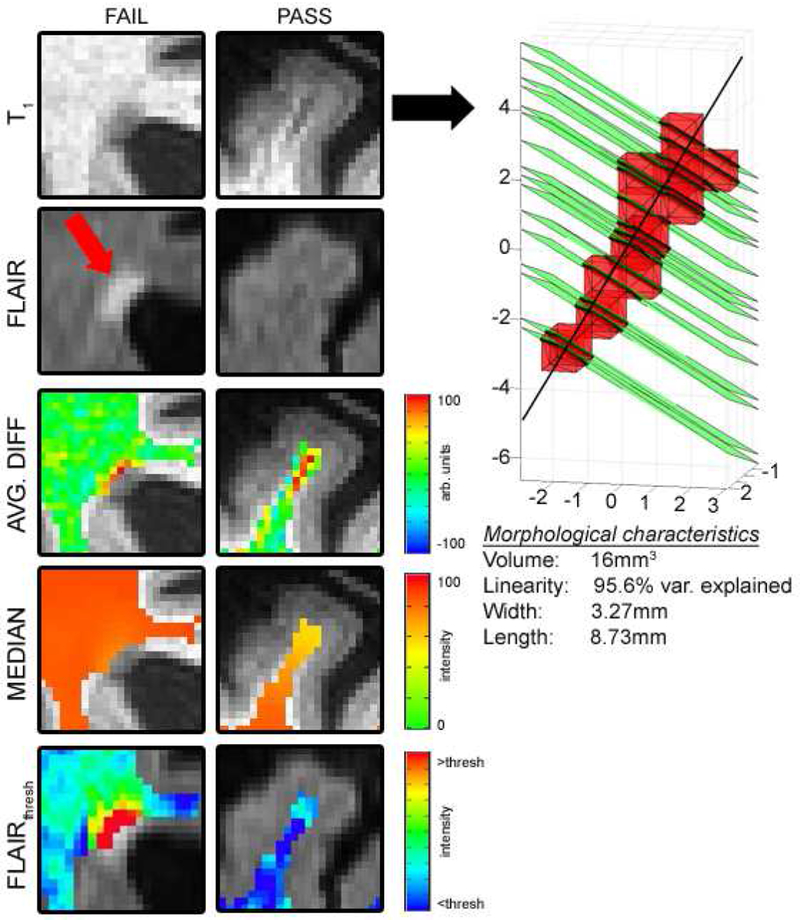
Analysis of the intensity-based phase was accomplished in AFNI (Cox, 1996) using the following methods. For voxels within the white matter (WM) mask, the local homogeneity of each voxel is assessed to identify clusters of interest. Specifically, in the T1-weighted images, a search field was defined as all voxels within 3.5 mm from the ith voxel and within the WM mask. The median intensity value of voxels in the search field neighborhood is computed and recorded as the ith voxel’s median score. Separately, the mean difference between the ith voxel and its neighbors in a field of radius 5.5 mm was calculated (its “difference score”). In order for a voxel to meet criteria for a likely ePVS on isotropic T1-weighted images, it must a) have an intensity that is less than 90% of the median score, b) be in the WM mask, and c) have a difference score that is larger than 5% of its intensity value. Each of these thresholds are generally governed by the SNR of the data and CNR of ePVS relative to normal appearing white matter (NAWM), respectively, and may be changed if data are noisy or if the weighting is tuned such that ePVS are less visible in contrast to NAWM. Separately, voxels that met criteria for being non-white matter hyperintensity on FLAIR were those for which the FLAIR intensity was lower than the sum of the mean and standard deviation of all voxels in the WM mask. The voxels that met both the FLAIR and the T1 criteria were constructed into voxel clusters larger than 5 (corners touching) and those clusters were submitted to the morphological constraint. See Figure 1 for a brief schematic of the algorithm.
Figure 1.
A periventricular FLAIR-screened object that resembles an ePVS but is a WMH edge (left, “FAIL”) and successful segmentation of an ePVS in the superior frontal gyrus (right, “PASS”). The isolated cluster is submitted to the morphological constraint (far right).
2.3.2. Cluster morphology
The morphological constraint was performed in MATLAB (Natick, MA). The minimum cluster size was set at 5 voxels with corners touching in order to ameliorate the likelihood of a configuration of voxels being collinear with respect to a best fit line through a longitudinal axis. Linearity was defined as the percent explained variance of the fit longitudinal vector; the inclusion constraint was set at >80%. The width was defined as the maximal distance between vertices that describe the intersection of a plane that is normal to the fit longitudinal vector (used in the linearity calculation) for each cluster and upon which the center of a voxel lies. This plane was constructed for each voxel in each cluster (green, Figure 1), and maximal vertex distances calculated, the largest of which defined the width of the cluster; clusters with a larger width than 15 voxels (16.41 mm, corner to corner) were excluded. The length of the cluster was calculated as the furthest distance between intersected vertices on the fit longitudinal vector; there were no inclusion or exclusion criteria for length.
2.4. Statistical Analysis
We evaluated the performance of this segmentation algorithm relative to visual counts in each dataset by three raters in cohort one. Reported correlations of manual counts are the average counts over three raters; in the case of the automated algorithm, average counts were assessed in the slices which were visually rated. In an effort to report the unedited result of the automated algorithm, these visual validation metrics were calculated on the output of MAPS-T1 before manual removal of false alarms. Within-subject false alarm rate was calculated by dividing the number of false alarms in a subject’s dataset by the total number of objects segmented by the algorithm. Signal-to-noise ratio (SNR) and ePVS contrast-to-noise (CNR) for both T1-weighted and FLAIR volumes were also calculated.
Statistical analysis of cohort two results included the calculation of within-subject false alarm rates, SNR and ePVS CNR for both T1-weighted and FLAIR volumes, and count and volume summary variables for each subject. ePVS SNR and CNR were calculated on true positives only.
Positive predictive value was calculated for both cohorts over all segmented ePVS. SNR for T1 and FLAIR was calculated as the mean intensity of identified ePVS or NAWM divided by the standard deviation of NAWM. CNR measurements were calculated as the absolute value of the difference between the mean of identified ePVS and NAWM (i.e. a volume from which segmented ePVS and hyperintense FLAIR voxels were subtracted) and divided by the standard deviation of NAWM.
3. Results
3.1. Cohort one
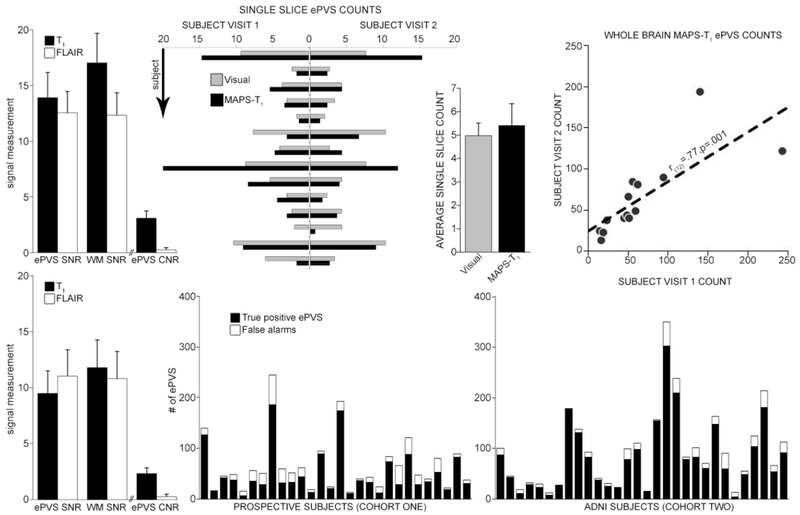
Visual ePVS counts were significantly correlated with MAPS-T1 (r(26)=.72, p<0.0001). Correlations between repeated measurements were significant for both methods in the single visually-rated slice (MAPS-T1: r(12)=.87, p<.0001, visual: r(12)=.86, p<.0001) and for whole brain assessment (MAPS-T1: r(12)=.77, p=.001). T1 ePVS and NAWM SNR mean (st. dev. over datasets) were 13.9 (2.2) and 17.0 (2.7), respectively. FLAIR ePVS and NAWM SNR mean (st. dev. over datasets) were 12.5 (1.9) and 12.3 (2.0), respectively. T1 and FLAIR ePVS CNR mean (st. dev.) were 3.1 (0.6) and 0.3 (0.2), respectively. The mean subject-wise false alarm frequency (st. dev., [range]) was 25% (16%, [6-56%]), which corresponded to an overall positive predictive value in cohort one of 77.5%. An example segmentation, with highlighted correct rejections and an example of a “miss”, can be found in Figure 3.
Figure 3.
A representative segmentation. Top row: T1-weighted image; bottom row: results of segmentation in red. Green circle = a deep WMH that has been avoided by the algorithm; green arrow = motion artifact avoided by the algorithm; blue arrow = a “missed” ePVS; yellow arrow = an example of the advantage of a three dimensional algorithm (3 ePVS oriented AP are identified proximal to an ePVS oriented obliquely in the coronal plane).
3.2. Cohort two
T1 ePVS and NAWM SNR mean (st. dev. over datasets) were 9.5 (2.2) and 11.8 (2.5), respectively. FLAIR ePVS and NAWM SNR mean (st. dev. over datasets) were 11.0 (2.3) and 10.8 (2.4), respectively. T1 and FLAIR ePVS CNR mean (st. dev.) were 2.3 (0.5) and 0.2 (0.2), respectively. The mean subject-wise false alarm frequency (st. dev., [range]) was 15% (13%, [0-61%]; note that the cohort two subject with the highest error rate had a relatively low overall burden, the stacked bar sixth from the right in the bottom right graph in Figure 2), which corresponded to an overall positive predictive value in cohort two of 87.5%.
Figure 2.
Results (Top row: cohort one; bottom row: cohort two). Top left: SNR and CNR measurements for T1 and FLAIR. Top middle left: Average of single slice counts by visual raters (gray) and single slice MAPS-T1 counts for each of the 14 subjects. Top middle right: Single slice averages over subjects (bars are st. dev.). Top right: Whole brain within subject correlation for MAPS-T1. Bottom left: SNR and CNR measurements. Bottom middle and right: True positive and false alarm rates for MAPS-T1 in cohort one and in cohort two.
4. Discussion
This report introduces a method of segmenting ePVS based on the relative intensity of a voxel’s neighbors and the morphological properties of isolated clusters on T1-weighted imaging using hyperintensity on FLAIR as a rule-out. Comparison of the burden metric (ePVS counts) in a single slice to gold standard visual assessment yields significant and reliable estimates over repeated acquisitions <2 years apart. The choice of the modifiable thresholds for search fields, voxel median score (in these data, 10% smaller than the median) and mean difference score (5%) are driven by NAWM homogeneity, SNR, and ePVS CNR, respectively, and different thresholds may easily be chosen and implemented on a dataset-wide basis, though the settings used in this analysis appeared to be applicable to subjects with a wide range of WM pathology and spatially variable SNR and CNR, as well as in a separately acquired and analyzed multisite cohort from ADNI. These thresholds were chosen after qualitatively acceptable segmentations on the “SUBJECT VISIT 1” subset (Figure 2, top middle left graph, left side). The maximum volume of the search field is informed by NAWM homogeneity in a dataset; if tissue homogeneity is very low (as in data for which B1 inhomogeneity is high), the search field may be reduced, though minimum search field volumes are constrained by the physiological size of ePVS. The FLAIR is used for the screening of WMH that are falsely identified on T1 as putative ePVS due to their similar hypointensity relative to NAWM (Wardlaw et al., 2013); manually edited WMH masks can be applied in place of the voxelwise FLAIR constraint, though these masks may remove ePVS that are proximal to or contained by a WMH.
4.1. Related work
There have been several recently published articles describing various methods for automated or semi-automated ePVS segmentation on MRI. Reliable segmentation of these structures with clinically relevant sensitivity and specificity can be difficult owing to their small size relative to standard clinical imaging voxel sizes, low CNR compared to surrounding NAWM, morphological discontinuity due to both physiological variability and to partial volume effects, and shared intensity profiles over some or all commonly acquired MRI weightings with other MRI-visible pathology such as lacunar infarcts and WMH. Some methods have overcome many of these obstacles by utilizing high field (7 Tesla) acquisitions (Park et al., 2016; Zhang et al., 2017), machine learning (Dubost et al., 2018; Zhang et al., 2017), binary classifiers (Gonzalez-Castro et al., 2017), Frangi (Ballerini et al., 2018) or object-based filtering (Descombes et al., 2004), or software designed for lesion exploration that has been modified to detect ePVS (Ramirez et al., 2015). All of these methods appear to have some measure of success when comparing them to categorical visual ratings. The method described herein is relatively accessible compared to approaches which may be less relevant in the clinical context (e.g. high field acquisition or acquisition with specialized hardware a la the Human Connectome Project), due to other methods being computationally expensive, requiring a considerable amount of human interaction, being study-specific (i.e. classifiers and optimization schemes in machine learning paradigms are often driven by within-study data), or perhaps most importantly requiring multiple acquisitions (i.e. T2-weighted spin echo and/or proton density) that may not be acquired as a matter of course. Notably, this method has not yet been applied to ePVS that are located in the basal ganglia or in the midbrain, two regions which can exhibit high burden in aged brains (Saeki et al., 2005); the method specifically relies on local intensity contrast which is different in subcortical gray matter and in the brainstem, and can vary widely across subjects and acquisition parameters due to heavy metal concentration in the basal ganglia and substantia nigra, especially in the aging population (Brar et al., 2009; Zhu et al., 2009).
4.2. Error assessment and methods comparison
The present method overcomes potential errors resulting from requisite voxelwise assessment in >2 separately acquired MRI contrasts. Coregistration between sequences that have been acquired with different weightings can be inaccurate; the width of ePVS can be as small as 1mm, and slight errors in coregistration can result in reduced sensitivity when implementing algorithms using this approach. A more logistical concern is that multiple sequences may not be available in legacy data, and may be precluded by scan time constraints in prospective experiments. High resolution (1-2mm isotropic voxels) T1 and FLAIR weighted data can each be acquired in less than 5 minutes, reducing the effects of motion, which can be substantial in aged subjects in whom this algorithm is often applied. Using two commonly acquired sequences alleviates some concerns related to registration and the necessity of multiple weightings in a single subject, making this segmentation algorithm more widely applicable.
Any algorithm used to assess WM ePVS will suffer from incomplete or imperfect white matter segmentations. Many intensity-based tissue-type segmentation algorithms, including Freesurfer, have difficulty classifying non-NAWM (Caligiuri et al., 2015), and older subjects who may be more likely to have a larger number of ePVS (Zhu et al., 2010) are more likely to have a higher burden of non-NAWM than younger subjects (Meyer et al., 1992). We use in house software (Promjunyakul et al., 2015) to correct defects due to white matter hyperintensities (on FLAIR or T2, and hypointense on T1) in white matter segmentations. However, partial volume effects inherent in MRI can smooth white matter hyperintensity/NAWM and NAWM/gray matter interfaces, especially in cases of marked neocortical atrophy which can make the cortical ribbon difficult to delineate from NAWM. Often, false positive errors are made at these interfaces; thin clusters that track the interface are identified as possible ePVS and will pass the morphological constraint. A specific false positive has been observed by our group in datasets for which the posterior horn of the lateral ventricle is not contiguous with the rest of the lateral ventricle; in the absence of partial volume effects, ePVS are by definition isointense to cerebrospinal fluid and these objects are shaped as an elongated teardrop. Often this structure is incorrectly classified as white matter by automated tissue segmentation algorithms and so is identified as a relatively large ePVS.
ePVS can occur in clusters following the course of penetrating vessels and appear as such in elderly without other signs of disease (Zhu et al., 2011). As with other ePVS segmentation algorithms, if ePVS are closely grouped in such a way that partial volume effects cause them to appear as a large cluster of heterogeneous intensity, the object will be removed as insufficiently linear. However, if there is sufficient resolution, contrast-to-noise, and NAWM between individual ePVS, they may be properly segmented with this algorithm even though they may be quite close to one another (<2mm, see Figure 3, yellow arrow).
Finally, lacunes are difficult to differentiate from ePVS on T1, and though they may often have a hyperintense rim on FLAIR, some lacunes do not. The morphological component of this automated approach serves to reduce the frequency of these types of false positives. Figure 4 illustrates an example of the difficulty of parsing lacunes from ePVS in a single subject imaged twice; at the first scan (top), the identified object met the linearity criteria (r>.8), while in the second scan it did not (r<.8). Although Wardlaw et al. (Wardlaw et al., 2013) identify lacunes by their “round or ovoid” morphology, a recent report that specifically addresses the morphology of lacunes finds that lacunes can be relatively linear and that they follow the geometry of perforating arteries (Gesierich et al., 2016). The Wardlaw report also seeks to differentiate ePVS from lacunes on the basis of their width, with PVS defined as having a diameter generally smaller than 3mm, and lacunes having a diameter from 3mm to 15mm. This criterion applied in this example would have screened out the lacune (widths = 7.1 and 6.6mm on the two measurements, Figure 4). Though a relatively small share of overall identified ePVS in this population had a width >3mm, ePVS that have a width larger than 3 mm have an outsized relative contribution to overall burden as measured by volume, and so a width threshold of <3mm for ePVS may lead to consequential misses. Conversely, the omission of a conservative width threshold may lead to the inclusion of outsized objects such as lacunes, as in the top row of Figure 4. As with any automated segmentation algorithm, visual inspection of the results of this algorithm is necessary to ensure accurate segmentation of true ePVS, especially in the case of lesions that can mimic ePVS intensity and morphology. While manual inspection and removal of false alarm clusters can be time-consuming, we found that high positive predictive values for the algorithm (77.5% in cohort one and 87.5% in cohort two) made this process manageable, as the low incidence of false alarms decreased the amount of time necessary to perform quality control.
Figure 4.
An example of an unsuccessful elimination of a lacune by the morphology step (top, timepoint 1) and a successful elimination (bottom, timepoint 2) in the same subject in cohort one. Metrics shown are [volume, linearity, maximum width x length]. “pre” and “post” signify pre- and post- morphology constraint.
4.3. Clinical potential of MAPS-T1
In addition to linearity measurements, the morphological step of this algorithm provides information regarding the length and width of these structures in three dimensions at the resolution of acquisition. These particular metrics may provide additional insight into the clinical relevance of ePVS, and this tool makes these measurements readily available for large scale clinical studies that represent the power to overcome small effect sizes. For example, though this study was not powered or designed for such a comparison, it is perhaps notable that cohort two, a subject pool that was younger, more male, and had higher clinical dementia ratings than cohort one, as well as lower overall SNR, had 64% more ePVS/subject than in cohort one (50.6 vs. 82.8 ePVS/subject, bottom of Figure 2) after manual omission of false alarms. The accurate characterization of previously considered subpathological enlarged Virchow-Robin spaces has the potential to introduce a research path into a multitude of disease processes, and this method will make these investigations immediately available to prospective investigations that have limited imaging time available and to retrospective and/or longitudinal studies with limited datasets.
Acknowledgments
5. Funding sources
This project was supported by the NIH/NIA (K01AG059842, U01AG016976, P30AG008017, and R01AG036772).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- Achiron A, Faibel M, 2002. Sandlike appearance of Virchow-Robin spaces in early multiple sclerosis: a novel neuroradiologic marker. AJNR Am J Neuroradiol 23, 376–380. [PMC free article] [PubMed] [Google Scholar]
- Ballerini L, Lovreglio R, Valdes Hernandez MDC, Ramirez J, MacIntosh BJ, Black SE, Wardlaw JM, 2018. Perivascular Spaces Segmentation in Brain MRI Using Optimal 3D Filtering. Sci Rep 8, 2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee G, Kim HJ, Fox Z, Jager HR, Wilson D, Charidimou A, Na HK, Na DL, Seo SW, Werring DJ, 2017. MRI-visible perivascular space location is associated with Alzheimer’s disease independently of amyloid burden. Brain 140, 1107–1116. [DOI] [PubMed] [Google Scholar]
- Boespflug EL, Schwartz DL, Lahna D, Pollock J, Iliff JJ, Kaye JA, Rooney W, Silbert LC, 2018. MR Imaging-based Multimodal Autoidentification of Perivascular Spaces (mMAPS): Automated Morphologic Segmentation of Enlarged Perivascular Spaces at Clinical Field Strength. Radiology 286, 632–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar S, Henderson D, Schenck J, Zimmerman EA, 2009. Iron accumulation in the substantia nigra of patients with Alzheimer disease and parkinsonism. Arch Neurol 66, 371–374. [DOI] [PubMed] [Google Scholar]
- Caligiuri ME, Perrotta P, Augimeri A, Rocca F, Quattrone A, Cherubini A, 2015. Automatic Detection of White Matter Hyperintensities in Healthy Aging and Pathology Using Magnetic Resonance Imaging: A Review. Neuroinformatics 13, 261–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RW, 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res 29, 162–173. [DOI] [PubMed] [Google Scholar]
- Descombes X, Kruggel F, Wollny G, Gertz HJ, 2004. An object-based approach for detecting small brain lesions: application to Virchow-Robin spaces. IEEE Trans Med Imaging 23, 246–255. [DOI] [PubMed] [Google Scholar]
- Dubost F, Yilmaz P, Adams H, Bortsova G, Ikram MA, Niessen W, Vernooij M, de Bruijne M, 2018. Enlarged perivascular spaces in brain MRI: Automated quantification in four regions. Neuroimage. [DOI] [PubMed] [Google Scholar]
- Gao FQ, Swartz RH, Scheltens P, Leibovitch FS, Kiss A, Honjo K, Black SE, 2011. Complexity of MRI white matter hyperintensity assessments in relation to cognition in aging and dementia from the Sunnybrook Dementia Study. J Alzheimers Dis 26 Suppl 3, 379–388. [DOI] [PubMed] [Google Scholar]
- Gesierich B, Duchesnay E, Jouvent E, Chabriat H, Schmidt R, Mangin JF, Duering M, Dichgans M, 2016. Features and Determinants of Lacune Shape: Relationship With Fiber Tracts and Perforating Arteries. Stroke 47, 1258–1264. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Castro V, Valdes Hernandez MDC, Chappell FM, Armitage PA, Makin S, Wardlaw JM, 2017. Reliability of an automatic classifier for brain enlarged perivascular spaces burden and comparison with human performance. Clin Sci (Lond) 131, 1465–1481. [DOI] [PubMed] [Google Scholar]
- Gutierrez J, Elkind MSV, Dong C, Di Tullio M, Rundek T, Sacco RL, Wright CB, 2017. Brain Perivascular Spaces as Biomarkers of Vascular Risk: Results from the Northern Manhattan Study. AJNR Am J Neuroradiol 38, 862–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuribara T, Mikami T, Komatsu K, Suzuki H, Ohnishi H, Houkin K, Mikuni N, 2017. Prevalence of and risk factors for enlarged perivascular spaces in adult patients with moyamoya disease. BMC Neurol 17, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JS, Kawamura J, Terayama Y, 1992. White matter lesions in the elderly. J Neurol Sci 110, 1–7. [DOI] [PubMed] [Google Scholar]
- Park SH, Zong X, Gao Y, Lin W, Shen D, 2016. Segmentation of perivascular spaces in 7T MR image using auto-context model with orientation-normalized features. Neuroimage 134, 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Promjunyakul N, Lahna D, Kaye JA, Dodge HH, Erten-Lyons D, Rooney WD, Silbert LC, 2015. Characterizing the white matter hyperintensity penumbra with cerebral blood flow measures. Neuroimage Clin 8, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J, Berezuk C, McNeely AA, Scott CJ, Gao F, Black SE, 2015. Visible Virchow-Robin spaces on magnetic resonance imaging of Alzheimer’s disease patients and normal elderly from the Sunnybrook Dementia Study. J Alzheimers Dis 43, 415–424. [DOI] [PubMed] [Google Scholar]
- Saeki N, Sato M, Kubota M, Uchino Y, Murai H, Nagai Y, Ishikura H, Nomura S, Matsuura I, Yamaura A, 2005. MR imaging of normal perivascular space expansion at midbrain. AJNR Am J Neuroradiol 26, 566–571. [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel DM, ten Dam VH, de Craen AJ, Admiraal-Behloul F, van Es AC, Palm WM, Spilt A, Bollen EL, Blauw GJ, Launer L, Westendorp RG, van Buchem MA, 2006. Measuring longitudinal white matter changes: comparison of a visual rating scale with a volumetric measurement. AJNR Am J Neuroradiol 27, 875–878. [PMC free article] [PubMed] [Google Scholar]
- Wardlaw JM, Smith EE, Biessels GJ, Cordonnier C, Fazekas F, Frayne R, Lindley RI, O’Brien JT, Barkhof F, Benavente OR, Black SE, Brayne C, Breteler M, Chabriat H, Decarli C, de Leeuw FE, Doubal F, Duering M, Fox NC, Greenberg S, Hachinski V, Kilimann I, Mok V, Oostenbrugge R, Pantoni L, Speck O, Stephan BC, Teipel S, Viswanathan A, Werring D, Chen C, Smith C, van Buchem M, Norrving B, Gorelick PB, Dichgans M, 2013. Neuroimaging standards for research into small vessel disease and its contribution to ageing and neurodegeneration. Lancet Neurol 12, 822–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Gao Y, Park SH, Zong X, Lin W, Shen D, 2017. Structured Learning for 3-D Perivascular Space Segmentation Using Vascular Features. IEEE Trans Biomed Eng 64, 2803–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu WZ, Zhong WD, Wang W, Zhan CJ, Wang CY, Qi JP, Wang JZ, Lei T, 2009. Quantitative MR phase-corrected imaging to investigate increased brain iron deposition of patients with Alzheimer disease. Radiology 253, 497–504. [DOI] [PubMed] [Google Scholar]
- Zhu YC, Dufouil C, Mazoyer B, Soumare A, Ricolfi F, Tzourio C, Chabriat H, 2011. Frequency and location of dilated Virchow-Robin spaces in elderly people: a population-based 3D MR imaging study. AJNR Am J Neuroradiol 32, 709–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YC, Tzourio C, Soumare A, Mazoyer B, Dufouil C, Chabriat H, 2010. Severity of dilated Virchow-Robin spaces is associated with age, blood pressure, and MRI markers of small vessel disease: a population-based study. Stroke 41, 2483–2490. [DOI] [PubMed] [Google Scholar]