Abstract
Background
The consequence of simultaneous and sequential inoculation of T. asperellum and B. amyloliquefaciens cultures with respect to growth rate, differential expression of vital genes and metabolites were examined.
Results
The competition was observed between T. asperellum and B. amyloliquefaciens under co-cultivation. The proliferation of Trichoderma was reduced in the simultaneous inoculation (TB1) method, possibly due to the fastest growth of Bacillus. Both T. asperellum and B. amyloliquefaciens were proliferated in sequential inoculation method (TB2). The sequential inoculation method (TB2) upregulated the expression of metabolites and vital genes (sporulation, secondary metabolites, mycoparasitism enzymes and antioxidants) in Trichoderma and downregulated in Bacillus and vice versa in co-inoculation method (TB1). The metabolic changes in the co-culture promoted the maize plant growth and defense potential under normal and biotic stress conditions.
Conclusion
The metabolites produced by the co-culture of T. asperellum and B. amyloliquefaciens improved the maize plant growth and defense potential under normal and biotic stress conditions.
Keywords: Simultaneous inoculation, Sequential inoculation, Co-cultivation, T. asperellum, B. amyloliquefaciens, Metabolomics, Plant growth, Biocontrol
Background
Plant growth-promoting and biocontrol microorganisms survive together in the soil and use various strategies to enhance the plant growth and defense potential [1]. Considering the microbial living together strategy, blending of different microbial communities were developed to fulfill the agricultural needs [2]. Application of microbial consortium on plants were considered as an advanced method to control the infections. Further, microbial consortium increased the efficiency and stability of the microbes in different soil and ecological conditions [2]. The microbes such as Trichoderma, Bacillus, Pseudomonas, Rhizobium, Glomus, etc., were utilized to develop the microbial consortia [3]. Jain et al. [3] designed a viable microbial consortia to upregulate the plant defense genes. While, co-inoculation of Azospirillum and Pseudomonas fluorescens on cotton increased the yield [4].
The advancement of microbial fertilizer involved the development of microbial consortia by mixing two independent microbial culture [2, 5]. In this way, we aimed to find an optimal condition for co-culturing the two distinctive microbes and to identify their efficacy on plant growth and bio-control activity. In addition, Co-cultivation has turned into a specific inducer of activating the silent genes through the intercommunication and competition between each microbe [6].
The genus Trichoderma is a filamentous soil and plant root associated fungi. It is one of the important biocontrol species, demonstrating over 60% of all the listed biocontrol agents (BCA) used to reduce the plant infectious diseases [7–9]. The mechanism behind the plant growth and disease control by Trichoderma involves (1) the secretion of cell wall hydrolytic enzymes; (2) conversion of large substrates into the smaller and available forms to be utilized by plants; (3) holding an extraordinary resistance against the chemical fungicides; (4) reduce the amount of pathogens surrounding the plant roots by making the competition for nutrients and space; (5) inhibition of pathogen growth by mycoparasitism and secondary metabolite production; (6) increase the antioxidant and systemic resistance of plants; (7) induce the plant growth by the secretion of plant growth promoting molecules [10–12].
Previous research has been reported that B. amyloliquefaciens is an effective BCA to fight against the different range of plant pathogens [13, 14]. The mechanism likely involves the production of various antimicrobial compounds, competition for nutrients and space, and increase the plant resistance [15, 16]. In addition, disease reduction has been attributed by promoting the growth of plant beneficial micro-organisms in the rhizosphere, thereby it inhibit the growth and survival of pathogens and increase the soil enzyme activity [14, 17–19].
The application of multiple microbes is a simple way to improve the biocontrol effects [5]. However, in most cases, the simple mixing of Trichoderma and Bacillus at a certain ratio were used as the biocontrol agents. While, the co-cultivation of two distinctive microbes for agricultural applications were less studied. Previously, the co-culture of Trichodema and Bacillus were studied to understand the changes in the metabolism and gene expression [20, 21]. Metabolomics study confirmed the stimulation of different metabolites in co-inoculation based co-culture of Trichodema and Bacillus [20]. Karuppiah et al. [21] reported that sequential inoculation based co-culture had induced the differential gene expression of vital genes and secondary metabolites to enhance the antagonistic and bio-control activity. These studies did not explore the impact of inoculation time of Trichoderma and Bacillus on the differential expression of metabolites and genes under the co-culture conditions. Since the inoculation sequence is essential for growth of both microbes and production of agriculturally important metabolites, the impact of simultaneous and sequential inoculation of Bacillus amyloliquefaciens 1841 into the Trichoderma asperellum GDFS1009 were explored to understand the interaction, differential expression of vital genes and metabolites on the plant growth and biocontrol potential.
Results
Co-cultivation of T. asperellum GDFS1009 and B. amyloliquefaciens 1841
In the present investigation, the growth of B. amyloliquefaciens 1841 and T. asperellum GDFS1009 was evaluated using co-inoculation and sequential inoculation based techniques. After fermentation, the axenic culture of B. amyloliquefaciens in YMC medium was found to be creamy yellow with 3.7 × 1011 CFU/ml. The color of the YMC medium inoculated with the axenic culture of T. asperellum GDFS1009 (8 × 108) was changed into tremendously lighter and beige with an even dissemination of mycelia with low viscosity. The co-inoculation of B. amyloliquefaciens ACCC11060 and T. asperellum GDFS1009 changed the medium into creamy yellow with high viscosity. The co-inoculation was dominated with Bacillus (5 × 1011) and limited T. asperellum GDFS1009 (8 × 103) mycelia. In sequential inoculation method, the T. asperellum (4 × 108) was enriched along with the B. amyloliquefaciens (3 × 1010). The fermentation medium of TB2 became creamy yellow after inoculation of Bacillus.
Effect of sequential and co-inoculation on the Differential Gene Expression of T. asperellum and B. amyloliquefaciens
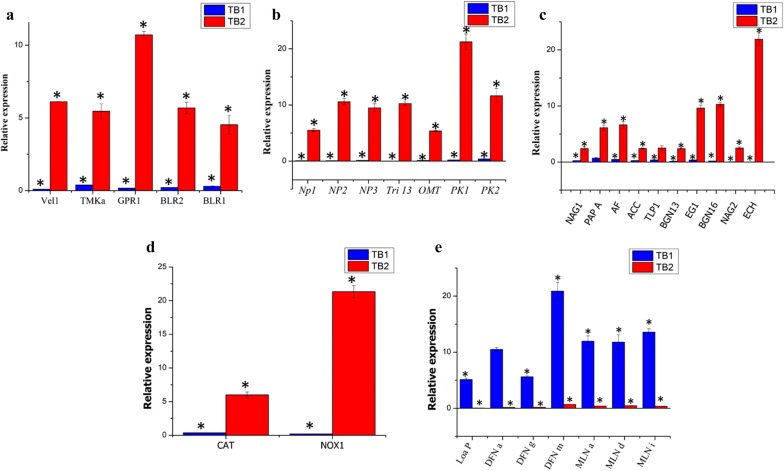
Distinct molecular expressions were observed between sequential and co-inoculation based co-cultivation of T. asperellum and B. amyloliquefaciens. The differences in the fold of gene expression under co-cultivations were evaluated using qRT-PCR with reference to their axenic controls. The cQ value of each gene category were normalized with 18S rRNA and 16S rRNA genes for T. asperellum and B. amyloliquefaciens, respectively. The analysis of the transcriptional response of T. asperellum under co-cultivation revealed that sequential inoculation upregulated the expression of genes related to the sporulation, secondary metabolism, antagonism and plant growth promoting related enzymes and antioxidants, while these genes were downregulated in co-inoculation based co-culture. The genes such as velvet (Vel 1), mitogen-activated protein kinase (TMKa), G protein receptor 1 (GPR1), blue-light-regulated genes (BLR1 and BLR2) involved in the Trichoderma sporulation were induced in the sequential inoculation based co-cultivation. Similarly, non-ribosomal peptide synthetase (NP1 and NP2), Putative ferrichrome synthetase (NP3), Cytochrome P450 (Tri 13) 1,O-methyl transferase (OMT) and Polyketide synthetase (PK1 and PK2) were induced in the sequential inoculation based co-culture.
The expression of genes involved in the mycoparasitism were estimated in both axenic (T and B) and co-culture (TB1 and TB2). The expression of chitinase (Ech) was upregulated 21.9 fold in sequential inoculation based co-cultivation, whereas it was downregulated (0.038578 fold) in co-inoculation technique (Fig. 1). Similarly, the expression of β-1,3-glucanase (BGN13), β-1,6-glucanase (BGN16), β-1,4-glucanase (Egl), N-acetyl-glucosaminidases (NAG1 and NAG2), aspartyl protease (PAP A), trypsin-like protease (TLP 1) and 1-aminocyclopropane-1-carboxylate deaminase (ACC) in sequential inoculation method was increased 0.15, 0.2, 0.3, 0.05, 0.2, 0.7, 0.35 and 0.26 folds, respectively, as compared to the axenic culture. The genes involved in the antioxidant such as NADPH oxidases (NOX) and catalase (CAT) were up-regulated 21.3, and 5.9 folds in the TB2. As shown in Fig. 1, 2−ΔΔCT of the genes involved in growth, secondary metabolites, mycoparasitism and plant growth promoting related enzymes and antioxidants in TB1 were lesser than 1, it indicates that expression of some Trichoderma genes were down-regulated in TB1.
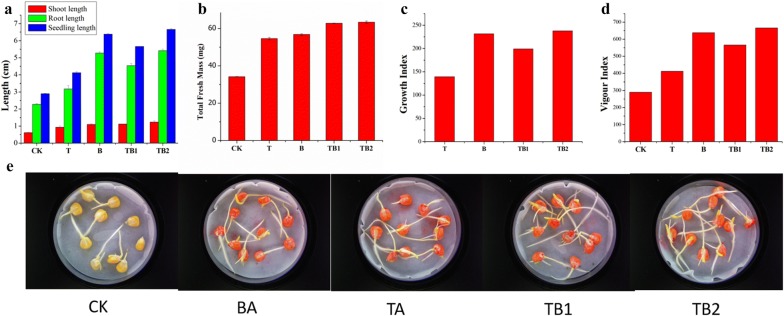
Fig. 1.
Influence of simultaneous (TB1) and sequential (TB2) inoculation based co-cultivation on the gene expression of (a) the T. asperellum morphology related genes [Velvet (Vel1), mitogen-activated protein Kinase (TMKa), G protein receptor 1 (GPR1), blue-light-regulated genes (BLR1 and BLR2) and ENVOY (ENV1)], (b) secondary metabolism related genes [non-ribosomal peptide synthetase (NP1 and NP2), Putative ferrichrome synthetase (NP3), Cytochrome P450 (Tri 13) 1,O-methyl transferase (OMT) and Polyketide synthetase (PK1 and PK2)]; (c) mycoparasitism-related genes [chitinase (ECH), β-1,3-glucanase (BGN13), β-1,6-glucanase (BGN16), β-1,4-glucanase (Egl), N-acetyl-glucosaminidases (NAG1 and NAG2), aspartyl protease (PAP A), trypsin-like protease (TLP 1) and α-l-arabinofuranosidases (AF)]; and plant growth promoting enzyme [1-Aminocyclopropane-1-carboxylate deaminase (ACC)]. d Anti-oxidant genes [NADPH oxidase (NOX), catalase (CAT)] and (e) genes encoding B. amyloliquefaciens macrolactin and difficidin in the co-culture [intrinsic terminators located within the polyketide synthase (PKS) gene cluster encoding for the antibiotic difficidin (DFN) (Loa P), beginning, middle and end of the difficidin operon (DFN A, DFN G, and DFN M); beginning, middle and end of the macrolactin operon (MLN a, MLN d, and MLN i)]. Bars represent the standard error of the mean values of three biological replicates. ∗ represent significant differences between the axenic and co-culture (P < 0.05)
The production of bioactive substances by B. amyloliquefaciens under co-cultivation was analyzed by the expression of macrolactin (MLN a, MLN d and MLN i) and difficidin genes (DFN a, DFN g and DFN m). Figure 1 shows that co-inoculation based co-cultivation (TB1) significantly increased the expression of macrolactin and difficidin genes, while sequential inoculation (TB2) technique significantly reduced the expression of macrolactin and difficidin genes.
Enzyme assay
In order to examine differences in the mycoparasitism related enzyme activity in the co-culture, the enzymes such as β-1,3 glucanase, chitinase, cellulase and protease were accessed in the axenic (T and B) and co-culture (TB1 and TB2) (Fig. 2). The activities of β-1,3 glucanase, chitinase, cellulase and protease in the TB2 were elevated to 17.9, 24.1, 15, and 18.7 IU/mL, respectively. The increment of enzyme activity in TB2 was concurred with the upregulation of mycoparasitism related gene expression. Protease activity of TB1 was increased to 16.1 IU/mL, while the other enzyme activity such as β-1,3 glucanase, chitinase, and cellulase were not detected in the TB1 and B. The enzyme activities detected in the TB1 and TB2 were higher than the axenic cultures (Fig. 2).
Fig. 2.
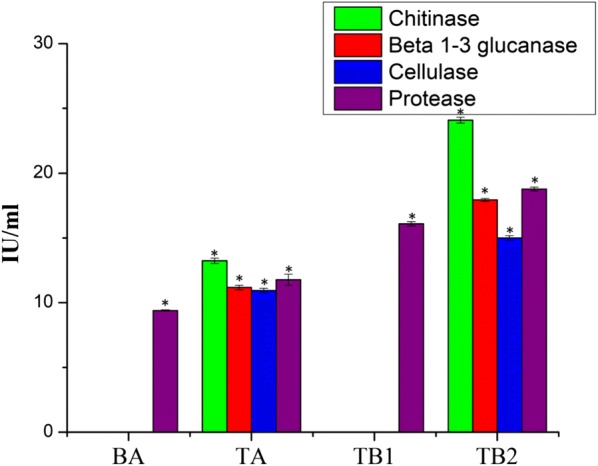
Enzyme activities associated with culture filtrates from the axenic (T and B) and co-culture (TB1 and TB2) of T. asperellum and B. amyloliquefaciens grown on YMC medium. (BA) axenic culture of B. amyloliquefaciens (TA) axenic culture of T. asperellum (TB1) simultaneous inoculation based co-culture (TB2) sequential inoculation based co-culture. Bars represent the standard error of the mean values of five biological replicates. ∗ represent significant differences between the axenic and co-culture (P < 0.05)
Metabolic profiles of axenic and co-culture
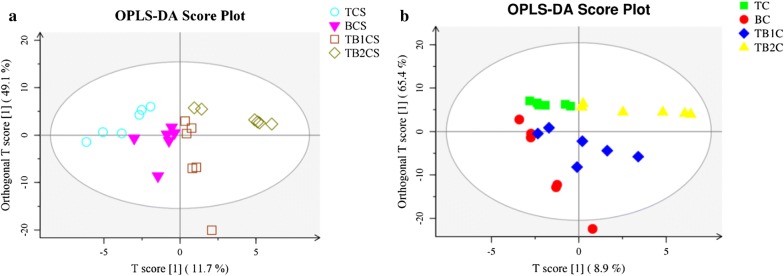
The modifications in the metabolic patterns of the co-culture and axenic culture were studied using the LC–MS technique. The OPLS-DA investigation of metabolite contents in both extracellular and intracellular showed the significant differences between the metabolites of axenic (T and B) and co-culture (TB1 and TB2) (Fig. 3a, b). This results was reliable with principal component analysis (Additional file 1: Fig. S1).
Fig. 3.
OPLS-DA analysis of the metabolic differences between the axenic (T and B) and co-culture (TB1 and TB2) of and T. asperellum and B. amyloliquefaciens based on LC–MS. a Extracellular based metabolic differences. b Intracellular based metabolic differences. (TCS) Culture supernatant of T. asperellum axenic culture (BCS) Culture supernatant of B. amyloliquefaciens axenic culture (TB1CS) Culture supernatant of simultaneous inoculation based co-culture (TB2CS) Culture supernatant of sequential inoculation based co-culture (TC) Culture pellet of T. asperellum axenic culture (BC) Culture pellet of B. amyloliquefaciens axenic culture (TB1C) Culture pellet of simultaneous inoculation based co-culture (TB2C) Culture pellet of sequential inoculation based co-culture
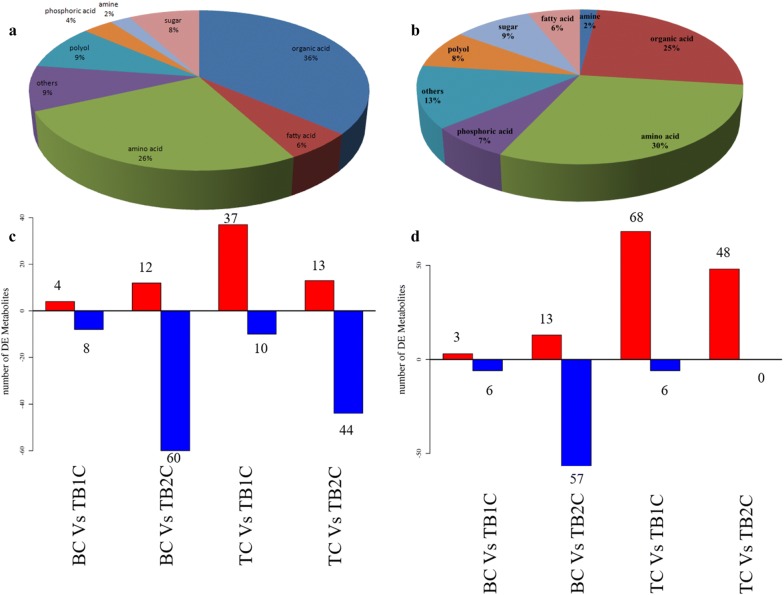
Hierarchical cluster analysis was carried out to assess the level of metabolites changes between the axenic (T and B) and co-culture (TB1 and TB2). This method explained variations between metabolic processes of each group (Additional file 1: Fig. S2). Totally, 101 and 89 metabolites were detected in the intracellular and extracellular level, respectively. These metabolites include amino acids, organic acids, carbohydrates, fatty acids, polyol, phosphoric acid, amines and others (Fig. 4). In extracellular, 37 and 4 metabolites from TB1 were upregulated compared to the T and B, respectively. While, 12 and 13 metabolites of TB2 were up regulated compared to the T and B, respectively. In contrast, 68 and 48 intracellular metabolites of TB1 and TB2 were up regulated compared to the T. 3 and 13 metabolites of TB1 and TB2 were upregulated compared to the B. In TB1, 6 metabolites were down regulated when compared to the axenic culture. None of the metabolites of TB2 were downregulated when compared to the T.
Fig. 4.
Classification and regulation of the metabolites in both extracellular and intracellular level of axenic and co-culture of and T. asperellum and B. amyloliquefaciens based on LC–MS. a Extracellular metabolic classification, b Intracellular metabolic classification, c extracellular metabolic regulation between the axenic and co-culture (d) intracellular metabolic regulation between the axenic and co-culture. (TCS) Culture supernatant of T. asperellum axenic culture (BCS) Culture supernatant of B. amyloliquefaciens axenic culture (TB1CS) Culture supernatant of simultaneous inoculation based co-culture (TB2CS) Culture supernatant of sequential inoculation based co-culture (TC) Culture pellet of T. asperellum axenic culture (BC) Culture pellet of B. amyloliquefaciens axenic culture (TB1C) Culture pellet of simultaneous inoculation based co-culture (TB2C) Culture pellet of sequential inoculation based co-culture
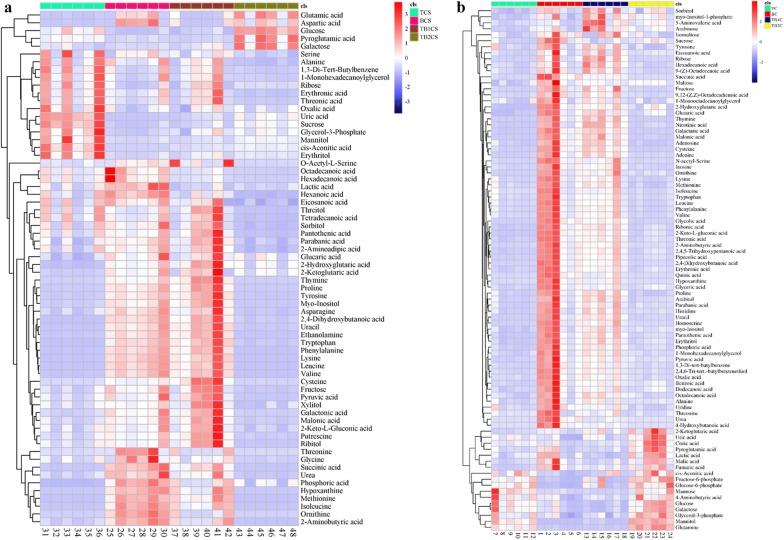
Heat maps showed the differential expression of metabolites between the axenic and co-culture of both intra and extra cellular level (Fig. 5). It was generated using the normalized data of the axenic and co-culture. The results showed that the metabolites of B. amyloliquefaciens and T. asperellum were induced in the co-culture of TB1 and TB2, respectively. In the supernatant of TB1, cis-Aconitic acid, uric acid, cysteine and citric acid were increased compared to the B. In intracellular, mannitol, 5-aminovaleric acid, and arabinose were increased compared to the B. Glutamic acid, aspartic acid, phosphoric acid, 2,4-dihydroxybutanoic acid, 2-hydroxyglutaric acid, 2-ketoglutaric acid, galactose, tryptophan, pyroglutamic acid, maltose, uracil, glucose and glucaric acid were increased in TB2 compared to the T, whereas 48 different metabolites were upregulated in the TB2 compared to T at intracellular level.
Fig. 5.
Metabolic differences between the axenic (T and B) and co-culture (TB1 and TB2) of and T. asperellum and B. amyloliquefaciens based on LC–MS. a Heat map of extracellular metabolites. b Heat map of intracellular metabolites. (TCS) Culture supernatant of T. asperellum axenic culture (BCS) Culture supernatant of B. amyloliquefaciens axenic culture (TB1CS) Culture supernatant of simultaneous inoculation based co-culture (TB2CS) Culture supernatant of sequential inoculation based co-culture (TC) Culture pellet of T. asperellum axenic culture (BC) Culture pellet of B. amyloliquefaciens axenic culture (TB1C) Culture pellet of simultaneous inoculation based co-culture (TB2C) Culture pellet of sequential inoculation based co-culture
Investigation on KEGG was carried out to compare changes between the metabolic pathway of axenic (T and B) and co-culture (TB1 and TB2) in both extracellular and intracellular level (Additional file 1: Fig. S3). 54 metabolic pathways showed major variations between TB2 and T. Among them, 12 were associated with amino acid metabolism, and others such as 4 carbohydrate metabolism, 2 nucleic acid metabolism, tropane, piperidine and pyridine alkaloid biosynthesis, C5-Branched dibasic acid metabolism, 2-Oxocarboxylic acid metabolism, carbon metabolism, pantothenate and CoA biosynthesis, inositol phosphate metabolism, streptomycin biosynthesis, taurine and hypotaurine metabolism, glycerophospholipid metabolism, sulfur metabolism, methane metabolism, biosynthesis of antibiotics, butanoate metabolism, carbon fixation pathways in prokaryotes, microbial metabolism in diverse environments, xylene degradation, atrazine degradation, phenylpropanoid biosynthesis, biosynthesis of secondary metabolites, glyoxylate and dicarboxylate metabolism, benzoate degradation, aminoacyl-tRNA biosynthesis, nicotinate and nicotinamide metabolism and galactose metabolism. Similarly, In TB1 55 metabolic pathway including 7 carbohydrate metabolism, 16 amino acid metabolism, tropane, piperidine and pyridine alkaloid biosynthesis, C5-Branched dibasic acid metabolism, pyrimidine metabolism, inositol phosphate metabolism, streptomycin biosynthesis, carbon metabolism, butanoate metabolism, glycerophospholipid metabolism, pantothenate and CoA biosynthesis, microbial metabolism in diverse environments, sulfur metabolism, xylene degradation, atrazine degradation, biosynthesis of antibiotics, carbon fixation pathways in prokaryotes, 2-oxocarboxylic acid metabolism, glyoxylate and dicarboxylate metabolism, biosynthesis of secondary metabolites, novobiocin biosynthesis, benzoate degradation, methane metabolism, purine metabolism, carbon fixation in photosynthetic organisms, taurine and hypotaurine metabolism, ascorbate and aldarate metabolism, monobactam biosynthesis, isoquinoline alkaloid biosynthesis, glycerolipid metabolism, biosynthesis of vancomycin group antibiotics, degradation of aromatic compounds, propanoate metabolism, aminoacyl-tRNA biosynthesis and nicotinate and nicotinamide metabolism were upregulated compared to the Trichoderma metabolic pathway. 21 and 64 pathways of TB1 and TB2 were upregulated when compared to the Bacillus axenic culture.
Inhibition of F. graminearum growth by the axenic and co-culture
The effect of axenic and co-culture fermentation liquor on the growth inhibition of F. graminearum was defined by the growth inhibition assay. The growth of F. graminearum in the PDA medium supplemented with TB2 liquor was decreased to 80.1%, followed by TB1 (71.5%), T (48.6%) and B (45.3%) (Additional file 1: Fig. S4).
Maize seed germination assay
Our results highlight the significant role of co-culture on maize seed germination. The positive effects of the axenic and co-culture of T. asperellum and B. amyloliquefaciens on seed germination are shown in Fig. 6. The parameters such as root length, shoot length, total seedling length, total seedling fresh mass, growth index and vigor index were improved significantly compared to the control. The seedling length of the seeds treated with TB2 and TB1 were higher than the other treatments, whose root length, shoot length and total seedling length values ranged from 4.54 to 5.42 cm, 1.12 to 1.24 cm and 5.66 to 6.66 cm, respectively. The fresh mass of the seeds treated with TB1 and TB2 were superior then the axenic culture, presenting 62.8 and 63.4 mg, respectively (Fig. 6). The control showed the lowest weight. The vigor index of the seeds treated with TB1 (566) and TB2 (666) were higher than other treatments. In this sequence, the axenic cultures of T and B were superior to the negative control (290) with vigor index ranging from 412 to 638 (Fig. 6).
Fig. 6.
Effect of axenic (T and B) and co-culture (TB1 and TB2) of T. asperellum and B. amyloliquefaciens on maize seed germination. a Shoot, root and total seedling length of germinated seeds. b Total fresh weight of the germinated seeds. c Growth index of the germinated seeds. d Vigour index of the germinated seeds. e Maize seed germination differences between each treatment. (CK) control; (BA) axenic culture of B. amyloliquefaciens (TA) axenic culture of T. asperellum (TB1) simultaneous inoculation based co-culture (TB2) sequential inoculation based co-culture. Results are means of 10 replicates for each treatment; the value is the standard error of the mean. Different letters above the bars are significantly different (P < 0.05) based on the ANOVA
Enhancement of biocontrol activity and Maize Plant growth by the co-culture of T. asperellum and B. amyloliquefaciens
Growth-promoting and bio-control effects of the axenic and co-culture on maize plants were observed based on the root length, shoot length, fresh and dry weight of root and shoot and disease reduction. As shown in Table 1, the maize plants treated with the axenic (T and B) and co-culture of T. asperellum and B. amyloliquefaciens (TB1 and TB2) either infested or un-infested with F. graminearum significantly improved the plant growth compared to control. Besides, co-culture (TB1 and TB2) significantly triggered the maize growth and development compared to treatment of axenic culture (T and B). The plants treated with both axenic and co-culture improved the height of maize plant under infected and un-infected soil. The co-culture increased the shoot and root length compared to axenic culture. Though, the sequential inoculation based co-culture showed the maximum shoot length either in infected (117.6 cm) or in un-infected soil (119.8 cm). The co-inoculation based co-culture showed the maximum root length in both infected (59.1 cm) and un-infected soil (57.8 cm). As shown in Table 1, the shoot and root biomass of the plants treated with axenic and co-culture increased significantly compared to the control. Likewise, co-culture (TB1 and TB2) significantly increased wet and dry weight of plant shoot and root compare to the axenic culture. However, the TB2 provided the highest wet and dry weight of the shoot either in infected (77.08 g and 6.02 g, respectively) or in un-infected soil (75.89 g and 6.95 g, respectively), while the treatment of TB1 provided the superior wet and dry weight of the root either in infected (25.60 g and 2.54 g, respectively) or in un-infected soil (24.04 g and 2.74 g, respectively). The treatment of maize infected with F. graminearum by the co-culture significantly increased the disease reduction compared to the axenic culture and infected control (FG). Consequently, T and B significantly reduced the infection of F. graminearum in maize and improved the plant growth.
Table 1.
Effect of axenic (T and B) and co-culture (TB1 and TB2) of T. asperellum and B. amyloliquefaciens on the plant growth and biological control of F. graminearum under green-house conditions
| Treatments | Shoot length (cm) | Root length (cm) | Wet weight (shoot) (g) | Wet weight (root) (g) | Dry weight (shoot) (g) | Dry weight (root) (g) | Disease reduction (%) |
|---|---|---|---|---|---|---|---|
| T1 | 93.43 ± 0.14f | 52.1 ± 0.39c | 59.77 ± 0.34d | 22.272 ± 0.37cd | 5.586 ± 0.24bcd | 3.73 ± 0.36a | NA |
| T2 | 92.22 ± 0.17 g | 54.76 ± 0.44b | 53.692 ± 0.34e | 22.64 ± 0.26cd | 5.28 ± 0.24cde | 2.81 ± 0.18a | NA |
| T3 | 110.68 ± 0.17c | 57.88 ± 0.35a | 66.894 ± 0.38c | 26.334 ± 0.30a | 5.768 ± 0.23bcd | 2.886 ± 0.21a | NA |
| T4 | 119.82 ± 0.38a | 56.2 ± 0.32b | 75.898 ± 0.44a | 24.048 ± 0.38abc | 6.956 ± 0.37a | 2.74 ± 0.27a | NA |
| T5 | 98.3 ± 0.23e | 48.42 ± 0.32d | 56.438 ± 0.47e | 23.664 ± 0.40bcd | 6.26 ± 0.14bcd | 2.364 ± 0.14a | 70b |
| T6 | 89.54 ± 0.24h | 55.72 ± 0.52b | 52.12 ± 0.39f | 21.602 ± 0.22d | 4.89 ± 0.15d | 2.386 ± 0.22a | 70b |
| T7 | 107.86 ± 0.3d | 59.12 ± 0.48a | 59.646 ± 0.27d | 25.602 ± 0.48ab | 6.334 ± 0.41abc | 2.544 ± 0.30a | 90a |
| T8 | 117.66 ± 0.27b | 55.48 ± 0.32b | 72.084 ± 0.33b | 26.318 ± 0.54a | 6F.026 ± 0.35ab | 3.48 ± 0.24a | 90a |
| T9 | 43.48 ± 0.17i | 36.7 ± 0.34f | 12.462 ± 0.30 h | 4.724 ± 0.44f | 1.354 ± 0.22f | 0.898 ± 0.43c | 40c |
| T10 | 76.22 ± 0.24j | 41.18 ± 0.57e | 47.348 ± 0.42g | 13.268 ± 0.4e | 3.8 ± 0.38e | 1.692 ± 0.19b | NA |
Results are means of 5 replicates for each treatment; the value is the standard error of the mean
NA represents not applicable
Different superscripts in the same column are significantly different (P < 0.05) based on the ANOVA. (T1)—T (Trichoderma asperellum GDFS1009); (T2)—B (Bacillus amyloliquefaciens 1841); (T3)—TB1 (co-inoculated T. asperellum GDFS1009 + B. amyloliquefaciens 1841); (T4)—TB2 (Sequential inoculated T. asperellum GDFS1009 + B. amyloliquefaciens 1841); (T5)—T + FG (Challenged with Fusarium graminearum); (T6)—B + FG; (T7)—TB1 + FG; (T8)—TB2 + FG; (T9)—FG; (T10)—Control
Consistently, a large number of Trichoderma and Bacillus were developed under the root surface. The highest number of Bacillus spores was re-isolated from the rhizosphere soil of plants treated with B in un-infected soils. Followed by, TB1 and TB2 treatment on both infected and un-infected soil showed the presence of Bacillus in the rhizosphere. Similarly, the Trichoderma was found highest in the plants treated with T in un-infected soil (TA), followed by TB1, TB2, TB1 + FG and TB2 + FG. Whereas, the highest number of Fusarium colonization was observed in the control of infected soil (FG). The lowest number of Fusarium colonization was observed in TB1 + FG and TB2 + FG (Table 2). Whole together, the in vivo experiments revealed the root colonization of Trichoderma and Bacillus could induce the maize systemic disease resistance against the F. graminearum. The proportion of bio-control agent relative to pathogen in the rhizosphere of the plants treated with axenic and co-cultures under biotic stress was calculated. The results showed that the proportion of BC: P was higher in the plants treated with the TB2. While, TB1 was lesser than the B (Table 2).
Table 2.
Re-isolation of Trichoderma, Bacillus and Fusarium from the soils treated with the axenic (T and B) and co-culture (TB1 and TB2) of T. asperellum and B. amyloliquefaciens under normal and biotic stress conditions
| T (CFU) | B (CFU) | TB1 (CFU) | TB2 (CFU) | T + FG (CFU) | B + FG (CFU) | TB1 + FG (CFU) | TB2 + FG (CFU) | FG (CFU) | Control (CFU) | |
|---|---|---|---|---|---|---|---|---|---|---|
| Bacillus count | ND | 13 × 108 ± 0.31a | 11 × 107 ± 0.31b | 6.6 × 106 ± 0.48c | ND | 2 × 1080.31d | 8 × 106 ± 0.31c | 3.3 × 106 ± 0.31d | ND | ND |
| Other bacterial count | 4.5 × 107 ± 0.15 cd | ND | ND | 24.3 × 106 ± 0.48a | 13 × 106 ± 0.31b | ND | ND | 5.4 × 106 ± 0.49c | 8 × 108 ± 0.31d | 3 × 108 ± 0.31d |
| Trichoderma count | 4.1 × 105 ± 0.3b | ND | 1.63 × 103 ± 0.09c | 3 × 105 ± 0.31b | 6 × 106 ± 0.31a | ND | 3.4 × 104 ± 0.23b | 3.2 × 105 ± 0.22b | ND | ND |
| Other fungal count | 1.3 × 102 ± 0.18e | 3.1 × 102 ± 0.23d | 8.6 × 103 ± 0.23a | 8.6 × 102 ± 0.18a | 5.9 × 102 ± 0.11b | 3.2 × 102 ± 0.14d | 5.0 × 102 ± 0.09c | 6.2 × 102 ± 0.04b | 8.2 × 105 ± 0.1c | 4.2 × 104 ± 0.09c |
| Fusarium count | ND | ND | ND | ND | 4 × 102 ± 0.31a | 2.9 × 102 ± 0.22b | 0.3 × 101 ± 0.031c | 0.15 × 101 ± 0.02bc | 8.6 × 106 ± 0.48d | ND |
| BC: P proportion | NA | NA | NA | NA | 15,000 ± 0.45d | 689,655.1724 ± 0.37b | 267,800 ± 0.24c | 2,413,333.333 ± 0.11a | NA | NA |
Results are means of 3 replicates for each treatment; the value is the standard error of the mean. Values below the detectable levels are indicated in the graphs as ND (not detected); NA (not applicable)
ND not detectable, NA not applicable
Different superscripts in the same column are significantly different (P < 0.05) based on the ANOVA
The expression of maize defense genes on the roots were investigated to know the effect of axenic and co-culture on regulation of maize disease resistance against pathogen (Fig. 7). The expression of allene oxide synthase (AOS) gene belongs to the jasmonic acid pathway was significantly changed among the different treatments. Expression of AOS gene was higher in the plants infested with F. graminearum (T7) and it was steadily decreased in the infested plants treated with co-culture (TB2 and TB1) and axenic culture (T and B). The Cq for the AOS gene was not detected in the non-infested plants. The expression of genes such as AOC and ACS involved in the ethylene pathway was studied. The AOC and ACS genes were up-regulated in TB1 + FG and TB2 + FG, followed by TB1 and TB2. Similarly, the axenic cultures (T and B) also induced the expression of AOC and ACS genes in both infested and un-infested plants, while the expression of these genes were downregulated in the FG. The expression of PR1 and PR10 involved in the systemic acquired resistance pathway (SAR) was studied. Among the different treatments, PR1 and PR10 were up-regulated in the T8 (FG + TB2) T7 (FG + TB1), T3 (TB1) and T4 (TB2) followed by T5 (FG + T), T6 (FG + B), T1 (T) and T2 (B). Similarly, the PAL and PAL1 genes involved in salicylic acid pathway was also induced in the T8 (FG + TB2) T7 (FG + TB1), T3 (TB1) and T4 (TB2), followed by T5 (FG + T), T6 (FG + B), T1 (T) and T2 (B). The gene expressions of HPL, lectin, lipase, MFS, Cyst2, PX5, Cyst and thiolase were also induced in T8 and T7 (Fig. 7), while these genes were downregulated in T9.
Fig. 7.
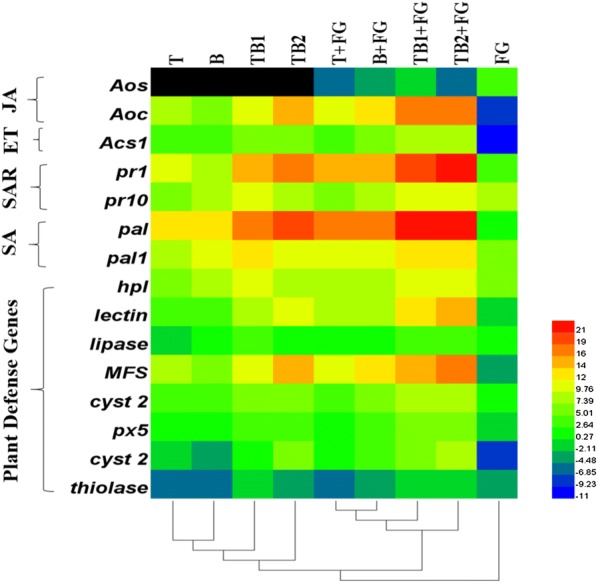
Differential expression profiles of defense-related genes in maize root tissues treated with axenic (T and B) and co-culture (TB1 and TB2) of T. asperellum and B. amyloliquefaciens under normal and biotic stress (F. graminearum infected soil (FG)) conditions. Relative transcript abundance was determined using RT-qPCR. Data are expressed as mean ± standard error of three replicates. JA jasmonic acid related response; ET ethylene related response; SAR systematic acquired resistance related response; SA salicylic acid related response
Discussion
Trichoderma and Bacillus have been proved as an effective candidate for controlling the plant diseases and growth promotion. Several research has defined the application of Bacillus either independently or in blend with additional microbes like Trichoderma to control the plant pathogens [22–24]. Trichoderma and Bacillus could propagate in a broad range of pH and soil. They produce hydrolytic enzymes, secondary metabolites and plant growth promoting components to enhance the plant growth and defense potential. Microbial genome mining revealed that secondary metabolite biosynthetic gene cluster remains silent in most of the microorganisms. This statement stimulates the idea to activate these silent gene using different methods such as modification of culturing technology, ribosome engineering, exchange and regulation of promoters, and synthetic biology-based pathway alteration [25, 26]. Modification in the culture conditions has been considered as the modest approach [25]. Hence, In the present investigation the co-cultivation of Trichoderma and Bacillus were carried out using sequential and co-inoculation based method to induce the production of secondary metabolites and plant growth promoting compounds.
It is worth to note growth rate of T. asperellum and B. amyloliquefaciens in co-inoculation and sequential inoculation treatments. The results showed a negative effect on the growth of T. asperellum in the co-inoculation. Faster growth of Bacillus over Trichoderma resulted in declined growth rate of Trichoderma in the co-inoculation method. The inoculation of Bacillus in the 48 h grown Trichoderma pre-culture promoted the growth of both microbes.
Interactions of Trichoderma with other microorganisms could alter the pattern of gene expression. Karuppiah et al. [21] reported that several genes have been upregulated during co-cultivation. Hence in the present investigation, the gene expression of Trichoderma spp. under the different co-cultivation conditions have been studied. The genes such as BLR1 and BLR2, ENV1, Vel1, TMKa and GPR1 involved in the sporulation’s were upregulated in the sequential based co-culture, while these genes were downregulated in the co-inoculation based co-culture. These results are in agreement with the spore count of axenic, sequential and co-inoculation based co-culture. The regulation between the expression of blue-light-regulated genes (BLR1 and BLR2), ENVOY (ENV1), velvet (Vel1) mitogen-activated protein kinase (TMKa) and G protein receptor 1 (GPR1) are in agreement with the Karuppiah et al. [21] and Carreras-Villasenor et al. [27].
Secondary metabolites produced by microbes could be affected under the laboratory culture condition [28]. Depending on the communication between the microbes in co-culture, several genes could activate the production secondary metabolites. In the present investigation, non-ribosomal peptide synthetase (NP1 and NP2), Putative ferrichrome synthetase (NP3), Cytochrome P450 (Tri 13) 1, OMT and Polyketide synthetase (PK1 and PK2) genes of Trichoderma involved in the secondary metabolite synthesis were up-regulated in sequential based co-culture, while these genes were downregulated in the co-inoculation based co-culture. B. amyloliquefaciens synthesize the polyketide compounds and involved in biocontrol activity [29]. In the present investigation Loa P, DFN a, DFN g, DFN m (difficidin operon) MLN a, MLN d, MLN i (macrolactin operon) were induced in the co-inoculation based co-culture. This results revealed the co-cultivation of T. asperellum and B. amyloliquefaciens was an appropriate technique to induce the secondary metabolites.
In-addition, the secretion of antioxidnats by Trichoderma in response to the Bacillus have been accessed using the quantification of catalase and NADPH oxidase gene. It has been established that NOX proteins are an important enzyme for sporulations [30]. The ROS scavenging systems such as ascorbate peroxidases, glutathione, superoxide dismutases, and catalases of Trichoderma are associated with NADPH oxidases to preserve the ROS homeostasis [30]. In the present study sequential inoculation based co-culture induced the expression of catalase and NADPH oxidase gene compared to the axenic culture. Further, the NOX also involved in the activation of proteases, together with aspartic, subtilisin serine, and trypsin-like proteases in T. harzianum T34 [30]. Similarly, we observed the induction of genes encoding aspartyl protease, trypsin-like protease, chitinase, β-1, three-glucanase, β-1,6-glucanase, β-1,4-glucanase and N-acetyl-glucosaminidases in the sequential based co-culture.
Co-cultivation influences the metabolic profile of the another microbe. The present investigation demonstrated that co-culture not only generate new metabolites, it also generates the metabolites of both microbes with different quantity. In TB1, the Bacillus reduced the quantity of some Trichoderma metabolites. Similarly, in TB2, Trichoderma downregulated the production of some Bacillus metabolites. Pyroglutamic acid, a non-protein amino-acid was induced in TB2. The induced production of glutamic acid and glutamine in the TB2 might be responsible for the synthesis of pyroglutamic acid [31].
Amino acids have several uses in pharmaceutical and agriculture application [32–34]. Interestingly, in the present investigation, amino acids covered up to 26–39% of the total metabolites. 5 amino acids including aspartic acid, homoserine, alanine, isoleucine and methionine were significantly increased in B. Similarly, asparagine and serine were significantly increased in T. The amino acids such as histidine, lysine, phenyl alanine, proline, tryptophan and tyrosine were significantly increased in TB1. Glutamine and aspartic acid was found to be significantly high in TB2. The amino-acids produced by Bacillus were detected in TB2, but the level of production was significantly reduced compared to the TB1 and B.
Atilio and Causin et al. [35] reviewed that amino acid is an essential component for the plant growth and metabolism. They also help the plants to grow under the biotic and abiotic stresses [36–38]. Under stress conditions, amino acids were catabolized through tricarboxylic acid (TCA) cycle and generate energy requisite for the growth and development [39]. In TB2, the metabolites of TCA cycle, glycolysis and sugars such as glucose and galactose were increased be due to the catabolism of amino acids generated by the Bacillus in co-culture (TB2) and this might be the reason for the lowest concentration of amino acids in TB2. Further, the maize growth under greenhouse conditions showed that these amino acids have been utilized by the plants to grow higher than that of axenic culture under normal and biotic stress conditions.
Some bacteria synthesize vitamin B to stimulate the plant growth [40–42]. In the present study, TB1 produced higher amount of pantothenic acid, and thiamine compared to the B, while in TB2 it was reduced. Thiamine is an important cofactor for the biosynthesis of indole acetic acid in plants and also required for several essential metabolisms [43, 44]. Foliar application of thiamine increased the vegetative growth and chemical constituents of plants [45]. The nicotinic acid improved the root length of the plant [46]. Similarly, in the present investigation the root length of the maize plants treated with TB1 was increased under both normal and biotic stress conditions.
Pipecolic acid detected in TB1 was abundant compared to B. It was found to be a key element to establish the systemic acquired resistance pathway in plants [47]. The 4-aminobutyric acid was induced in the both TB1 and TB2. 4-aminobutyric acid is a non-protein amino acid, which was regularly accumulated in plants during ecological stress and there by induce ethylene production [48, 49]. It also helps the plant to grow under environmental stresses [49, 50]. It also reduced the growth rate of larvae and stimulated the plant growth [51]. Similarly, the trans ferulic acid was found to be induced in TB1 compared to other groups. The trans ferulic acid had a potential to induce plant resistance [52]. Moussa et al. [53] observed that the plants treated with ethanolamine under salt stress were highly adapted through the highest ROS scavenging photosynthetic activity. In present study the ethanolamine was detected in both TB1 and TB2. The presence of these metabolites are correlated well with the induction of maize plant defense genes related to SAR and salicylic acid pathway. The organic acids such as lactic acid, malic acid and citric acid were found to be highest in TB2. These organic acids have the antimicrobial activity against Listeria monocytogenes, Escherichia coli, and Salmonella gaminara [54]. In addition, these organic acids solubilize the soil inorganic phosphates to induce the plant growth [55, 56]. These organic acids present in the co-cultures might be involved in the reduction of F. graminearum in the soils treated with co-cultures (Table 2). Further, the highest production of organic acids in TB2 increased the proportion of BC: P of plants treated with TB2 under biotic stress conditions. The results revealed that the TB2 was more active against the soil pathogens compared to the TB1.
Conclusions
The inoculation sequence proved the competitive interaction between T. asperellum and B. amyloliquefaciens, which also influenced growth rate, differential expression of vital genes and metabolites during the fermentation. The competitive interaction between the T. asperellum and B. amyloliquefaciens in co-inoculation method downregulated the expression of Tricoderma vital genes and metabolites, while in sequential inoculation the didicidin and macrolactin gene expression and metabolites of Bacillus were downregulated. On the other hand, it has been predicted that aminoacids produced by Bacillus might be utilized by Trichoderma for its energy through TCA cycle in TB2. From this study it has been observed that metabolic and genetic expression of T. asperellum and B. amyloliquefaciens could be induced by the simultaneous and sequential inoculation based co-cultivation method, respectively. Even though the competitive interaction occurred in the simultaneous and sequential inoculation, it favored the maize growth and bio-control activity in different ways. Taken together, the results suggested that the metabolites generated by the co-culture helps the maize plant to grow under normal and biotic stress conditions by (1) providing the nutrients such as carbon and nitrogen for plant growth, (2) improve the plant resistance by the activation of the defense related genes and (3) inhibit the growth of soil pathogens through the secretion of antimicrobial metabolites. Thus in future, the co-cultivation based microbial consortia could be used as a new technology to increase the plant growth and development.
Methods
Microbes and culture conditions
The T. asperellum GDFS1009 and B. amyloliquefaciens 1841 were procured from China General Microbiological Culture Collection Center and Laboratory of Microbial Fermentation, Sichuan University, China, respectively. The fungus was recovered and grown on Potato Dextrose (PD) medium at 28 °C. The B. amyloliquefaciens 1841 was recovered and grown on nutrient medium at 37 °C. The T. asperellum GDFS1009 and B. amyloliquefaciens 1841 grown in PD and nutrient medium were used as inoculum for all investigations. Fusarium graminearum (maize stacks and root rot pathogen) was used as the test pathogen for biocontrol activity.
Co-cultivation of T. asperellum (GDFS1009) and B. amyloliquefaciens 1841
Sequential inoculation: 1% of T. asperellum GDFS1009 was pre-cultured at 28 °C for 48 h in 100 mL of YMC broth [21]. Consequently, 0.1% of the B. amyloliquefaciens 1841 (1.0 OD at 600 nm) was inoculated into the pre-culture medium and continued the co-cultivation for 2 days at 28 °C. Co-inoculations: 1% of T. asperellum GDFS1009 and 0.1% of B. amyloliquefaciens 1841 were co-inoculated into 100 mL YMC broth and incubated in shaker for 4 days at 28 °C. After incubation, the broth cultures were serially diluted and plated in the PDA containing streptomycin and chloramphenicol to estimate the growth of fungus. The bacterial growth was estimated using the nutrient agar containing nystatin and cycloheximide [21].
Enzyme activity
The axenic (T and B), sequential (TB2) and co-inoculation (TB1) of co-cultures were centrifuged and the supernatants were used as the crude protein extract. The enzyme activity such as chitinase, neutral protease, β-1,3-glucanase and cellulase were estimated using Enzyme Activity Determination Kits (Shanghai Cablebridge Biotechnology Co., Ltd.), according to the manufacturer’s protocol [21].
Gene expression
The expression of genes associated with sporulation (VEL 1, TMK, GPR 1, BLR 1, BLR 2, and ENV1), mycoparasitism related enzymes (NAG 1, NAG 2, PAP A, PAP B, ECH, AF, ACC, BGN 13, BGN 16, and EG1) secondary metabolism (genes encoding three NRPSs, two PKSs, O-methyltransferase B, and cytochrome P450) and antioxidants (NOX and CAT) of Trichoderma [21], macrolactin and difficidin genes [29] of B. amyloliquefaciens were studied in the axenic and co-culture of the T. asperellum and B. amyloliquefaciens grown in the YMC medium at 4th day.
RNA was extracted as defined previously [21]. For expression analysis, RNA was extracted using TRIeasy total RNA extraction reagent (YEASEN). cDNA was generated according to the procedure of Prime Script RT reagent kit with DNA eraser (Takara). The real time PCR and relative quantification was performed as described by Karuppiah et al. [21]. The primers used for gene quantification was given in Additional file 1: Table S1.
Preparation of samples for LC–MS/MS
50 mL of each culture supernatant BCS, TCS, TB1CS and TB2CS, and its pellet containing microbes BC, TC, TB1C and TB2C alongside 50 mL of YMC medium as controls, were snap-frozen with fluid nitrogen and transported on dry ice to the Suzhou BioNovoGene Metabolomics Platform. All of them were defrosted at 4 °C and blended consistently. 200 µL of each sample was taken in a 1.5 mL microcentrifuge tube, to which 800 µL of methanol was added and vortexed for 60 s and centrifuged at 12,000 RPM at 4 °C for 10 min. The supernatants were exchanged to another 1.5 mL micro-centrifuge tube, dried and mixed in 300 µL of 80% methanol. Next, the supernatants were separated through a 0.22 µm film to acquire the examples for analysis. The samples were analyzed through Waters ACQUITY UPLC with ACQUITY UPLC® BEH C18 1.7 µm (2.1 × 100 mm) chromatography column and mass spectrometry using Thermo LTQ Orbitrap XL instrument with electrospray ionization (ESI) and cation–anion ionization mode as described by Wu et al. [20].
Analysis of metabolome data
The information on the data preprocessing were followed as described by Wu et al. [20], The raw data acquired was changed into the mzXML format by utilizing a Proteowizard program (v3.0.8789). The XCMS tool of R (v3.3.2) was used for peak detection, filtration and alignment. The significant parameters included bw = 5, ppm = 15, peak width = c (10, 120), mzwid = 0.015, mzdiff = 0.01 and strategy = ”centWave”. The data matrix containing the data of mass to charge proportion (m/z), retention time and peak area were obtained. The positive and negative particle models were assembled for 3680 and 3566 precursor molecules separately. The information was gathered and the accompanying investigations were carried out. The batch normalization of peak area was directed. Few multivariate statistical investigations such as principal component analysis (PCA) and Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) were directed to uncover the distinctions in the metabolic creations between various groups. The metabolites with precise changes were analyzed in KEGG. The correlation between the metabolites were dissected by figuring the Pearson or Spearman rank correlation coefficient between any two metabolites. At the point when the straight connection between two metabolites expanded, the relationship coefficient drifted toward 1 or − 1 i.e., 1 represents a positive correlation and − 1 represents negative correlation. The statistical check was analyzed using cor.test in R package. Also, the false positive p-value was analyzed. FDR p-value ≤ 0.05 was utilized as the significant correlation [20].
Determination of pathogen inhibition
The effect of axenic and co-culture on the inhibition of pathogen growth was determined according to the method of karuppiah et al. [21]. The PDA agar plates were prepared by mixing the filtered fermented broth of T, B, TB1 and TB2 in the ratio of 1:5. The Fusarium graminearum was loaded into the center of plates to access its growth inhibition by the axenic and co-cultures.
Seed germination
Maize seeds were disinfected with 70% ethanol and 0.2% sodium hypochlorite for 2 min, followed by it was washed with sterile distilled water and air dried. 50 grams of maize seeds were treated with 2 mL of axenic and co-culture suspension (5 × 108/mL) complemented with 0.15% of seed coating agent (Jilin Bada Pesticide Co. Ltd.). After the treatment, the seeds were distributed on the petri dish containing Whatman filter paper and germinated in the dark at 25 °C. The treatment with sterile distilled water was used as the control. The germination parameters such as root length, shoot length, total seedling length, total seedling fresh mass and vigor index were documented after 7 days.
Plant growth and defense assay
Maize Seeds were planted into 30 cm diameter pots containing horticulture soil with 5 seeds per pot in 5 replications. Seedlings were thinned to two plants per pot after 1 week. The greenhouse experiment was carried out at day, night conditions; 28 °C for 12 h under lights, and at 24 °C in the dark for 12 h. The experiment was organized in a randomized complete blocks plan. After 2 weeks the plants were treated as follows: (T1)—T (Trichoderma asperellum GDFS1009); (T2)—B (Bacillus amyloliquefaciens 1841); (T3)—TB1 (co-inoculated T. asperellum GDFS1009 + B. amyloliquefaciens 1841); (T4)—TB2 (Sequential inoculated T. asperellum GDFS1009 + B. amyloliquefaciens 1841); (T5)—T + FG (Challenged with Fusarium graminearum); (T6)—B + FG; (T7)—TB1 + FG; (T8)—TB2 + FG; (T9)—FG; (T10)—Control. The T. asperellum and F. graminearum inoculum were set at 1 × 106 spores/mL while B. amyloliquefaciens inoculum was set at 1 × 108 spores/mL. The axenic, sequential and co-inoculation of co-cultures were inoculated 5 days before and after inoculation with F. graminearum. The soil was moistened regularly. The plants were carefully uprooted and length of the root and shoot, wet and dry weight of the shoot and root were estimated.
The evaluation of disease was based on a scale of leaf spot disease from grade zero to grade five. Grade zero: no disease spot; Grade 1: no more than 10%; Grade 2: 11–30%; Grade 3: 31–50%; Grade 4: 51–70%; and Grade 5: more than 70% and the disease index was calculated according to the formula: Disease index = Σ (number of plants in each disease stage × levels value)/(total number of plants × the highest levels × 100). The disease reduction was computed with the following formula: Disease reduction = disease index of CK − disease index after treatment [57].
Total RNA was extracted from the plant root using the TRIeasy total RNA extraction reagent (YEASEN) and cDNA was generated according to the procedure of Prime Script RT reagent kit with DNA eraser (Takara). The expression of genes related to the jasmonic acid, ethylene (ET), systemic acquired resistance (SAR), salicylic acid (SA), plant innate immunity (PII) and other defense related genes were analyzed according to the method of Saravanakumar et al. [58]. The primers used for gene quantification is given in Additional file 1: Table S1. Relative gene expression was estimated by 2ΔΔCT method using actin as a reference gene.
The inoculated Bacillus, Trichoderma and Fusarium were re-isolated from the rhizosphere soil of the treated plants. The rhizospheric soil of all treated plants were collected carefully after uprooting the plants. The rhizospheric soil was serially diluted and plated in the potato dextrose agar and nutrient agar to enumerate the fungi and bacteria, respectively. The percentage proportion of biocontrol agents (Bacillus and Trichoderma) relative to a pathogen (Fusarium) was calculated as follows:
Proportion of biocontrol agents: pathogen (BC: P) = Total CFU of bio-control agents/CFU of pathogen.
Statistical analysis
All experiments were studied based on different replication and were repeated at least three times, with reproducible results. The graphs were constructed by means of Microsoft office excel and origin 6.0 with standard error bars. Results shown were average of replicates along with the standard error of mean values. For multiple comparisons, two-way ANOVA with post hoc LSD and Duncan were carried out using the SPSS 2.0. Student’s t-test was conducted to examine the differences between the gene expression using the SPSS 2.0. P < 0.05 was considered as significant. The heat map of defense gene expressions was made by means of HemI: A Toolkit for Illustrating Heat maps [59].
Supplementary information
Additional file 1. Additional figures and table.
Abbreviations
- T
Trichoderma asperellum
- B
Bacillus amyloliquefaciens
- TB1
simultaneous inoculation based T. asperellum and B. amyloliquefaciens co-culture
- TB2
sequential inoculation based T. asperellum and B. amyloliquefaciens co-culture
- BCS
B. amyloliquefaciens culture supernatant
- TCS
T. asperellum culture supernatant
- TB1CS
TB1 culture supernatant
- TB2CS
TB2 culture supernatant
- BC
B. amyloliquefaciens culture pellet
- TC
T. asperellum culture pellet
- TB1C
TB1 culture pellet
- TB2C
TB2 culture pellet
- PR
pathogenesis related proteins
- BCA
biocontrol agents
- PD
potato dextrose medium
- YMC
yeast extract, molasses and corn gluten meal medium
- Vel 1
velvet
- TMKa
mitogen-activated protein kinase
- GPR1
G protein receptor 1
- BLR1
blue-light-regulated genes I
- BLR2
blue-light-regulated genes II
- NP1
non-ribosomal peptide synthetase I
- NP2
non-ribosomal peptide synthetase II
- NP3
putative ferrichrome synthetase
- Tri 13
cytochrome P450
- OMT
1,O-methyl transferase
- PK1
polyketide synthetase I
- PK2
polyketide synthetase II
- ENV1
ENVOY
- Ech
chitinase
- BGN13
β-1,3-glucanase
- BGN16
β-1,6-glucanase
- Egl
β-1,4-glucanase
- NAG1
N-acetyl-glucosaminidases 1
- NAG2
N-acetyl-glucosaminidases 2
- PAP A
aspartyl protease
- TLP 1
trypsin-like protease
- ACC
1-aminocyclopropane-1-carboxylate deaminase
- NOX
NADPH oxidases
- CAT
catalase
- Loa P
regulatory gene of difficidin and macrolactin operon
- DFN a
difficidin A
- DFN g
difficidin G
- DFN m
difficidin M
- MLN a
macrolactin A
- MLN d
macrolactin D
- MLN i
macrolactin i
- FG
Fusarium graminearum
- ET
ethylene
- SAR
systemic acquired resistance
- SA
salicylic acid
- PII
plant innate immunity
- P
pathogen
- AOS
allene oxide synthase
- AOC
allene oxide cyclase
- ACS
1-aminocyclopropane-1-carboxylic acid synthase
- PR1
pathogenesis-related protein 1
- PR10
pathogenesis-related protein 10
- PAL
phenylalanine ammonia-lyase
- PAL1
phenylalanine ammonia-lyase 1
- HPL
hydroperoxide lyase
- MFS
multiflux efflux synthase
- Cyst2
cystatin ii proteinase inhibitor
- PX5
peroxidase
- Cyst
cystatin proteinase inhibitor
Authors’ contributions
VK and JC conceived and designed the experiments. VK carried out the main work, analyzed the data and drafted the manuscript. MV and TL participated in the research. JC supervised the work and revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by National Key Research and Development Program of China (2017YFD0200403, 2017YFD0201108), National Natural Science Foundation of China (Nos. 31872015; 31672072); The Key Project of Science and Technology of Shanghai No. 18391902400); Earmarked Fund for China Agriculture Research System (CARS-02).
Availability of data and materials
All data generated during this study are included in this published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12934-019-1233-7.
References
- 1.Vacheron J, Desbrosses G, Bouffaud ML, Touraine B, Moenne-Loccoz Y, Muller D, Legendre L, Wisniewski-Dye F, Prigent-Combaret C. Plant growth-promoting rhizobacteria and root system functioning. Front Plant Sci. 2013;4:356. doi: 10.3389/fpls.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarma BK, Yadav SK, Singh S, Singh HB. Microbial consortium-mediated plant defense against phytopathogens: readdressing for enhancing efficacy. Soil Biol Biochem. 2015;87:25–33. doi: 10.1016/j.soilbio.2015.04.001. [DOI] [Google Scholar]
- 3.Jain A, Singh S, Kumar Sarma B, Bahadur Singh H. Microbial consortium-mediated reprogramming of defence network in pea to enhance tolerance against Sclerotinia sclerotiorum. J Appl Microbiol. 2012;112:537–550. doi: 10.1111/j.1365-2672.2011.05220.x. [DOI] [PubMed] [Google Scholar]
- 4.Marimuthu S, Ramamoorthy V, Samiyappan R, Subbian P. Intercropping system with combined application of Azospirillum and Pseudomonas fluorescens reduces root rot incidence caused by Rhizoctonia bataticola and increases seed cotton yield. J Phytopathol. 2013;161:405–411. doi: 10.1111/jph.12084. [DOI] [Google Scholar]
- 5.Ravnskov S, Jensen B, Knudsen IMB, Bødker L, Funck Jensen D, Karliński L, Larsen J. Soil inoculation with the biocontrol agent Clonostachys rosea and the mycorrhizal fungus Glomus intraradices results in mutual inhibition, plant growth promotion and alteration of soil microbial communities. Soil Biol Biochem. 2006;38:3453–3462. doi: 10.1016/j.soilbio.2006.06.003. [DOI] [Google Scholar]
- 6.Netzker T, Fischer J, Weber J, Mattern DJ, König CC, Valiante V, Schroeckh V, Brakhage AA. Microbial communication leading to the activation of silent fungal secondary metabolite gene clusters. Front Microbiol. 2015;6:299. doi: 10.3389/fmicb.2015.00299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species—opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2:43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- 8.Mukherjee PK, Horwitz BA, Herrera-Estrella A, Schmoll M, Kenerley CM. Trichoderma research in the genome era. Annu Rev Phytopathol. 2013;51:105–129. doi: 10.1146/annurev-phyto-082712-102353. [DOI] [PubMed] [Google Scholar]
- 9.Sharma V, Salwan R, Sharma PN, Gulati A. Integrated translatome and proteome: approach for accurate portraying of widespread multifunctional aspects of Trichoderma. Front Microbiol. 2017;8:1602. doi: 10.3389/fmicb.2017.01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerin D, Pollastro S, Raguseo C, De Miccolis Angelini RM, Faretra F. A ready-to-use single- and duplex-TaqMan-qPCR assay to detect and quantify the biocontrol agents Trichoderma asperellum and Trichoderma gamsii. Front Microbiol. 2018;9:2073. doi: 10.3389/fmicb.2018.02073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lorito M, Woo SL, Gary E, Harman GE, Monte E. Translational research on Trichoderma: from ‘Omics to the field. Annu Rev Phytopathol. 2010;48:395–417. doi: 10.1146/annurev-phyto-073009-114314. [DOI] [PubMed] [Google Scholar]
- 12.Singh A, Sarma BK, Singh HB, Upadhyay RS. Chapter 40—Trichoderma: a silent worker of plant rhizosphere. In: Gupta VK, Schmoll M, Herrera-Estrella A, Upadhyay RS, Druzhinina I, Tuohy MG, editors. Biotechnology and biology of Trichoderma. Amsterdam: Elsevier; 2014. pp. 533–542. [Google Scholar]
- 13.Wan T, Zhao H, Wang W. Effect of biocontrol agent Bacillus amyloliquefaciens SN16-1 and plant pathogen Fusarium oxysporum on tomato rhizosphere bacterial community composition. Biol Control. 2017;112:1–9. doi: 10.1016/j.biocontrol.2017.05.014. [DOI] [Google Scholar]
- 14.Han L, Wang Z, Li N, Wang Y, Feng J, Zhang X. Bacillus amyloliquefaciens B1408 suppresses Fusarium wilt in cucumber by regulating the rhizosphere microbial community. Appl Soil Ecol. 2019;136:55–66. doi: 10.1016/j.apsoil.2018.12.011. [DOI] [Google Scholar]
- 15.Martínez-Viveros O, Jorquera MA, Crowley DE, Gajardo G, Mora ML. Mechanisms and practical considerations involved in plant growth promotion by rhizobacteria. J Soil Sci Plant Nutr. 2010;10:293–319. doi: 10.4067/S0718-95162010000100006. [DOI] [Google Scholar]
- 16.Chen D, Liu X, Li C, Tian W, Shen Q, Shen B. Isolation of Bacillus amyloliquefaciens S20 and its application in control of eggplant bacterial wilt. J Environ Manag. 2014;137:120–127. doi: 10.1016/j.jenvman.2014.01.043. [DOI] [PubMed] [Google Scholar]
- 17.Han L, Pu T, Wang X, Liu B, Wang Y, Feng J, Zhang X. Optimization of a protective medium for enhancing the viability of freeze-dried Bacillus amyloliquefaciens B1408 based on response surface methodology. Cryobiology. 2018;81:101–106. doi: 10.1016/j.cryobiol.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Wu B, Wang X, Yang L, Yang H, Zeng H, Qiu Y, Wang C, Yu J, Li J, Xu D, et al. Effects of Bacillus amyloliquefaciens ZM9 on bacterial wilt and rhizosphere microbial communities of tobacco. Appl Soil Ecol. 2016;103:1–12. doi: 10.1016/j.apsoil.2016.03.002. [DOI] [Google Scholar]
- 19.Raza W, Yuan J, Ling N, Huang Q, Shen Q. Production of volatile organic compounds by an antagonistic strain Paenibacillus polymyxa WR-2 in the presence of root exudates and organic fertilizer and their antifungal activity against Fusarium oxysporum f. sp. niveum. Biol Control. 2015;80:89–95. doi: 10.1016/j.biocontrol.2014.09.004. [DOI] [Google Scholar]
- 20.Wu Q, Ni M, Dou K, Tang J, Ren J, Yu C, Chen J. Co-culture of Bacillus amyloliquefaciens ACCC11060 and Trichoderma asperellum GDFS1009 enhanced pathogen-inhibition and amino acid yield. Microb Cell Fact. 2018;17:155. doi: 10.1186/s12934-018-1004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karuppiah V, Sun J, Li T, Vallikkannu M, Chen J. Co-cultivation of Trichoderma asperellum GDFS1009 and Bacillus amyloliquefaciens 1841 causes differential gene expression and improvement in the wheat growth and biocontrol activity. Front Microbiol. 2019;10:1068. doi: 10.3389/fmicb.2019.01068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abdullah MT, Ali NY, Suleman P. Biological control of Sclerotinia sclerotiorum (Lib.) de Bary with Trichoderma harzianum and Bacillus amyloliquefaciens. Crop Protect. 2008;27:1354–1359. doi: 10.1016/j.cropro.2008.05.007. [DOI] [Google Scholar]
- 23.Chowdappa P, Mohan Kumar SP, Jyothi Lakshmi M, Upreti KK. Growth stimulation and induction of systemic resistance in tomato against early and late blight by Bacillus subtilis OTPB1 or Trichoderma harzianum OTPB3. Biol Control. 2013;65:109–117. doi: 10.1016/j.biocontrol.2012.11.009. [DOI] [Google Scholar]
- 24.Srinath J, Bagyaraj DJ, Satyanarayana BN. Enhanced growth and nutrition of micropropagated Ficus benjamina to Glomus mosseae co-inoculated with Trichoderma harzianum and Bacillus coagulans. World J Microbiol Biotechnol. 2003;19:69–72. doi: 10.1023/A:1022569702845. [DOI] [Google Scholar]
- 25.Romano S, Jackson SA, Patry S, Dobson ADW. Extending the “One Strain Many Compounds” (OSMAC) principle to marine microorganisms. Marine Drugs. 2018;16:244. doi: 10.3390/md16070244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reen FJ, Romano S, Dobson ADW, O’Gara F. The sound of silence: activating silent biosynthetic gene clusters in marine microorganisms. Marine Drugs. 2015;13:4754–4783. doi: 10.3390/md13084754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carreras-Villaseñor N, Sánchez-Arreguín JA, Herrera-Estrella AH. Trichoderma: sensing the environment for survival and dispersal. Microbiology. 2012;158:3–16. doi: 10.1099/mic.0.052688-0. [DOI] [PubMed] [Google Scholar]
- 28.De Roy K, Marzorati M, Van den Abbeele P, Van de Wiele T, Boon N. Synthetic microbial ecosystems: an exciting tool to understand and apply microbial communities. Environ Microbiol. 2014;16:1472–1481. doi: 10.1111/1462-2920.12343. [DOI] [PubMed] [Google Scholar]
- 29.Goodson JR, Klupt S, Zhang C, Straight P, Winkler WC. LoaP is a broadly conserved antiterminator protein that regulates antibiotic gene clusters in Bacillus amyloliquefaciens. Nat Microbiol. 2017;2:17003. doi: 10.1038/nmicrobiol.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montero-Barrientos M, Hermosa R, Cardoza RE, Gutiérrez S, Monte E. Functional analysis of the Trichoderma harzianum nox1 gene, encoding an NADPH oxidase, relates production of reactive oxygen species to specific biocontrol activity against Pythium ultimum. Appl Environ Microbiol. 2011;77:3009–3016. doi: 10.1128/AEM.02486-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiménez-Arias D, García-Machado FJ, Morales-Sierra S, Luis JC, Suarez E, Hernández M, Valdés F, Borges AA. Lettuce plants treated with L-pyroglutamic acid increase yield under water deficit stress. Environ Exp Bot. 2019;158:215–222. doi: 10.1016/j.envexpbot.2018.10.034. [DOI] [Google Scholar]
- 32.Singh R, Kumar M, Mittal A, Mehta PK. Microbial metabolites in nutrition, healthcare and agriculture. 3 Biotech. 2017;7:15. doi: 10.1007/s13205-016-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Souza PM, de Oliveira Magalhães P. Application of microbial α-amylase in industry—a review. Braz J Microbiol. 2010;41:850–861. doi: 10.1590/S1517-83822010000400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ivanov K, Stoimenova A, Obreshkova D, Saso L. Biotechnology in the production of pharmaceutical industry ingredients: amino acids. Biotechnol Biotechnol Equip. 2014;81:2050–2061. [Google Scholar]
- 35.Atilio JB, Causin HF. The central role of amino acids on nitrogen utilization and plant growth. J Plant Physiol. 1996;149:358–362. doi: 10.1016/S0176-1617(96)80134-9. [DOI] [Google Scholar]
- 36.Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 37.Van Oosten MJ, Pepe O, De Pascale S, Silletti S, Maggio A. The role of biostimulants and bioeffectors as alleviators of abiotic stress in crop plants. Chem Biol Technol Agric. 2017;4:5. doi: 10.1186/s40538-017-0089-5. [DOI] [Google Scholar]
- 38.Nardi S, Pizzeghello D, Schiavon M, Ertani A. Plant biostimulants: physiological responses induced by protein hydrolyzed-based products and humic substances in plant metabolism. Scientia Agricola. 2016;73:18–23. doi: 10.1590/0103-9016-2015-0006. [DOI] [Google Scholar]
- 39.Galili G, Avin-Wittenberg T, Angelovici R, Fernie AR. The role of photosynthesis and amino acid metabolism in the energy status during seed development. Front Plant Sci. 2014;5:447. doi: 10.3389/fpls.2014.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marek-Kozaczuk M, Skorupska A. Production of B-group vitamins by plant growth-promoting Pseudomonas fluorescens strain 267 and the importance of vitamins in the colonization and nodulation of red clover. Biol Fertil Soils. 2001;33:146–151. doi: 10.1007/s003740000304. [DOI] [Google Scholar]
- 41.Vogl C, Grill S, Schilling O, Stülke J, Mack M, Stolz J. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J Bacteriol. 2007;189:7367–7375. doi: 10.1128/JB.00590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.LeBlanc JG, Laiño JE, del Valle MJ, Vannini V, van Sinderen D, Taranto MP, de Valdez GF, de Giori GS, Sesma F. B-Group vitamin production by lactic acid bacteria—current knowledge and potential applications. J Appl Microbiol. 2011;111:1297–1309. doi: 10.1111/j.1365-2672.2011.05157.x. [DOI] [PubMed] [Google Scholar]
- 43.Palacios OA, Gomez-Anduro G, Bashan Y, de Bashan LE. Tryptophan, thiamine and indole-3-acetic acid exchange between Chlorella sorokiniana and the plant growth-promoting bacterium Azospirillum brasilense. FEMS Microbiol Ecol. 2016;92:flw077. doi: 10.1093/femsec/fiw077. [DOI] [PubMed] [Google Scholar]
- 44.Goyer A. Thiamine in plants: aspects of its metabolism and functions. Phytochemistry. 2010;71:1615–1624. doi: 10.1016/j.phytochem.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 45.Hossein Aminifard M, Jorkesh A, Fallahi H-R, Alipoor K. Foliar application of thiamin stimulates the growth, yield and biochemical compounds production of coriander and fenugreek. J Horticult Res. 2018;26:77–85. doi: 10.2478/johr-2018-0009. [DOI] [Google Scholar]
- 46.Satoh S, Nomura Y. Promotion of root elongation by pyridinecarboxylic acids known as novel cut flower care agents. Plant Root. 2017;11:40–47. doi: 10.3117/plantroot.11.40. [DOI] [Google Scholar]
- 47.Wang Y, Schuck S, Wu J, Yang P, Doring AC, Zeier J, Tsuda K. A MPK3/6-WRKY33-ALD1-pipecolic acid regulatory loop contributes to systemic acquired resistance. Plant Cell. 2018;30:2480–2494. doi: 10.1105/tpc.18.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Liu J, Ashraf U, Li G, Li Y, Lu W, Gao L, Han F, Hu J. Exogenous γ-aminobutyric acid (GABA) application improved early growth, net photosynthesis, and associated physio-biochemical events in maize. Front Plant Sci. 2016;7:919. doi: 10.3389/fpls.2016.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi S-Q, Shi Z, Jiang Z-P, Qi L-W, Sun X-M, Li C-X, Liu J-F, Xiao W-F, Zhang S-G. Effects of exogenous GABA on gene expression of Caragana intermedia roots under NaCl stress: regulatory roles for H2O2 and ethylene production. Plant Cell Environ. 2010;33:149–162. doi: 10.1111/j.1365-3040.2009.02065.x. [DOI] [PubMed] [Google Scholar]
- 50.Kinnersley AM, Turano FJ. Gamma aminobutyric acid (GABA) and plant responses to stress. Crit Rev Plant Sci. 2000;19:479–509. doi: 10.1080/07352680091139277. [DOI] [Google Scholar]
- 51.Ramputh AI, Bown AW. Rapid [gamma]-aminobutyric acid synthesis and the inhibition of the growth and development of oblique-banded leaf-roller larvae. Plant Physiol. 1996;111:1349–1352. doi: 10.1104/pp.111.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gan X, Hu D, Wang Y, Yu L, Song B. Novel trans-Ferulic Acid derivatives containing a chalcone moiety as potential activator for plant resistance induction. J Agric Food Chem. 2017;65:4367–4377. doi: 10.1021/acs.jafc.7b00958. [DOI] [PubMed] [Google Scholar]
- 53.Moussa HR, El-Sayed Mohamed Selem E, Ghramh HA. Ethanolamine affects physiological responses of salt-treated jute plants. Int J Veg Sci. 2019;25:1–9. doi: 10.1080/19315260.2019.1566187. [DOI] [Google Scholar]
- 54.Eswaranandam S, Hettiarachchy NS, Johnson MG. Antimicrobial Activity of citric, lactic, malic, or tartaric acids and nisin-incorporated soy protein film against Listeria monocytogenes, Escherichia coli O157:H7, and Salmonella gaminara. J Food Sci. 2004;69:FMS79–FMS84. [Google Scholar]
- 55.Vyas P, Gulati A. Organic acid production in vitro and plant growth promotion in maize under controlled environment by phosphate-solubilizing fluorescent Pseudomonas. BMC Microbiol. 2009;22:174. doi: 10.1186/1471-2180-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen W, Yang F, Zhang L, Wang J. Organic acid secretion and phosphate solubilizing efficiency of Pseudomonas sp psb12: effects of phosphorus forms and carbon sources. Geomicrobiol J. 2016;133:870–877. doi: 10.1080/01490451.2015.1123329. [DOI] [Google Scholar]
- 57.Li Y, Sun R, Yu J, Saravanakumar K, Chen J. Antagonistic and biocontrol potential of Trichoderma asperellum ZJSX5003 against the maize stalk rot pathogen Fusarium graminearum. Indian J Microbiol. 2016;56:318–327. doi: 10.1007/s12088-016-0581-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saravanakumar K, Dou K, Lu Z, Wang X, Li Y, Chen J. Enhanced biocontrol activity of cellulase from Trichoderma harzianum against Fusarium graminearum through activation of defense-related genes in maize. Physiol Mol Plant Pathol. 2018;103:130–136. doi: 10.1016/j.pmpp.2018.05.004. [DOI] [Google Scholar]
- 59.Deng W, Wang Y, Liu Z, Cheng H, Xue Y. HemI: a toolkit for illustrating heatmaps. PLoS ONE. 2014;9:e111988. doi: 10.1371/journal.pone.0111988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Additional figures and table.
Data Availability Statement
All data generated during this study are included in this published article.