Abstract
Background
Cryptosporidium viatorum is a minor Cryptosporidium pathogen in humans. Currently, there is limited information regarding the prevalence and genotypes of C. viatorum in animals in China.
Methods
In this study, 228 faecal samples were collected from two wild rat species (Leopoldamys edwardsi and Berylmys bowersi) in Chongqing Municipality and Guangdong Province, China. These specimens were analyzed for C. viatorum and then subtyped it using PCR and sequence analysis of the small subunit ribosomal RNA (SSU rRNA) and 60-kilodalton glycoprotein (gp60) genes, respectively.
Results
A total of 25 (11.0%) faecal samples were tested positive for C. viatorum by SSU rRNA assay. Of these samples, 4 (3.6%) came from L. edwardsi and 21 (18.0%) from B. bowersi. Of the 25 C. viatorum-positive samples, 17 were successfully amplified at the gp60 gene locus, which represented four subtypes belonging to two subtype families, including XVa (XVaA6, XVaA3g, XVaA3h) and XVc (XVcA2G1). Phylogenetic analysis based on the gp60 amino acid sequences indicated that all of the C. viatorum isolates grouped together, supporting the conclusion that C. viatorum from the wild rats represent two subtype families.
Conclusions
These results indicate an occurrence of C. viatorum XVa subtype family from rats which is genetically identical to those found in humans. Our findings suggest that wild rats may be a potential source of human cryptosporidiosis.
Keywords: Cryptosporidium viatorum, Cryptosporidiosis, Zoonosis, Wild rats, China
Background
Cryptosporidiosis is caused by Cryptosporidium spp. and is a serious enteric disease of humans and animals worldwide [1, 2]. Humans can become infected by ingesting food and water contaminated with Cryptosporidium oocysts [3]. To date, more than 78 species/genotypes of Cryptosporidium are known to infect humans and animals [4]. Cryptosporidium viatorum is a novel parasite that has emerged in the last decade, and is a minor Cryptosporidium species to infect humans [5].
Cryptosporidium viatorum was first found in the faecal samples of travellers returning to Britain from the Indian subcontinent in 2012 [6]. Later, C. viatorum was found in two Swedish patients in 2013 [7]. Since then, C. viatorum infection has been recorded in patients with underdeveloped or impaired immunity (including children and AIDS patients) living in developing countries such as Nigeria, Ethiopia, Colombia and India [8–13]. Recently, there was also one case of C. viatorum found in western Australian human populations (2015–2018) [14]. The clinical symptoms of C. viatorum infection in humans include diarrhoea, fever, headache, abdominal pain, nausea, vomiting and marked weight loss [1].
There have been reports of C. viatorum infection (XVa subtype family) in humans, but very limited information is available regarding C. viatorum infection in animal hosts [15, 16]. Recently, Koehler et al. [16] reported C. viatorum infection (XVb subtype family) in Australian swamp rats, suggesting that it is endemic to native rats in Australia. In addition, the identification of C. viatorum in urban wastewater and combined sewer overflows indicated that this human pathogen may be common in China as well [17]. Very recently, Zhao et al. [15] found that C. viatorum (XVc and XVd subtype families) was prevalent (7.3%) in wild rats in Hainan Province of China. However, to date, it is unknown whether C. viatorum XVa subtype family which is in humans is present in animal hosts, including in wild rats. Wild rodents are one of the largest of the mammalian groups [18]. They have a wide distribution and can transmit numerous pathogens to other animals and to humans [19].
In China, Leopoldamys edwardsi and Berylmys bowersi are two common rat species with a wide distribution. To determine whether C. viatorum XVa subtype family which is in humans is present in L. edwardsi and B. bowersi, we performed an investigation of the prevalence and the subtypes of C. viatorum in these two wild rat species in China.
Methods
Specimen collection and host identification
From November 2017 to January 2018, 111 Edwardʼs long-tailed rats (L. edwardsi) were collected from Chongqing Municipality, China (28°10′–32°13′N, 105°11′–110°11′E) and 117 Bower’s white-toothed rat (B. bowersi) were collected from Guangdong Province, China (20°13′–25°31′N, 109°39′–117°19′E). A total of 228 faecal samples from these rats were separately collected and stored in 2.5% potassium dichromate solution at − 20 °C for future analysis.
Specific identification of the two wild rat species was determined by PCR-based sequencing of the mitochondrial cox1 gene based on a previous method [20]. The obtained sequences were compared with sequences in GenBank using the BLASTn algorithm. The cox1 sequences of the two wild rat species were found to have a sequence identity of more than 99% to previously published sequences for L. edwardsi and B. bowersi from China and Vietnam (GenBank: KM434322 and JN105105, respectively).
DNA extraction and genotyping of C. viatorum
Fresh faecal specimens were subjected to direct DNA extraction using the E.Z.N.A.® Stool DNA Kit (Omega Biotek Inc., Norcross, GA, USA) according to the manufacturer’s protocols. The extracted DNA was stored at − 20 °C for subsequent molecular analysis. Nested PCR was used to amplify and sequence regions of the small subunit ribosomal RNA (SSU rRNA) gene from Cryptosporidium spp. [21]. The 60-kilodalton glycoprotein (gp60) gene was used to classify the subtype levels of C. viatorum [22]. Positive and negative controls were included in each amplification. Amplification products were examined using 1.5% agarose gel containing GoldViewTM (Solarbio, China) and observed under UV light.
Sequence and phylogenetic analysis
Positive PCR products were sent to Sangon Biotech (Shanghai, China) for sequencing in both directions. The sequences were aligned with known reference sequences available on GenBank using the Basic Local Alignment Search Tool (BLAST). The genotypes of C. viatorum were identified using Clustal X 1.83 [23]. Available gp60 amino acid sequences of C. viatorum were aligned using MAFFT 7.122 [24], and ambiguous sites and regions were excluded using Gblocks 0.91b with default parameters [25]. Phylogenetic analysis was performed using MEGA 5.0 [26]. The p-distance model was selected as the most suitable one. Neighbor-joining (NJ) tree was calculated based on 1000 bootstrap replicates.
Statistical analysis
Statistical analysis was performed using SPSS V20.0 (IBM, Chicago, IL, USA). The following variables were analysed using the Chi-square test: variation in the prevalence of C. viatorum (y), wild rat species (x1), gender (x2) and different geographic region (x3). Each variable was included in a Binary Logit Model and as an independent variable in a multivariate regression analysis. The best model was judged by Fisher’s scoring algorithm. All tests were two-sided. P < 0.05 was considered statistically significant. Odds ratios (ORs) and 95% confidence intervals (95% CIs) were estimated to explore the strength of the association between C. viatorum-positivity and the test conditions.
Results
Of the 228 wild rats collected, 25 (11.0%) were found to be C. viatorum-positive based on PCR amplification of the partial, small subunit (SSU) rRNA gene (Table 1). The SSU rDNA sequences from the two wild rat species exhibited more than 99.4% identity to previously published sequence from a human in Kenya (GenBank: JX978271). The overall prevalence of C. viatorum in L. edwardsi was 3.6% (4/111) and in B. bowersi it was 18.0% (21/117). The prevalence of C. viatorum in female rats (8.9%) was somewhat lower than in male rats (13.3%), although this difference was not statistically significant (P > 0.05).
Table 1.
Prevalence (in %) and factors risk of Cryptosporidium viatorum infection in wild rats in China
| Factor | Category | No. tested | No. positive | Prevalence (95% CI) | OR (95% CI) | P-value |
|---|---|---|---|---|---|---|
| Region | Chongqing | 111 | 4 | 3.6 (0.14–7.1) | Reference | |
| Guangdong | 117 | 21 | 18.0 (11.0–24.9) | 5.9 (1.9–17.7) | 0.001 | |
| Gender | Female | 113 | 10 | 8.9 (3.6–14.1) | Reference | 0.40 |
| Male | 115 | 15 | 13.3 (6.9–19.2) | 1.6 (0.7–3.6) | ||
| Species | Leopoldamys edwardsi | 111 | 4 | 3.6 (0.1–7.1) | Reference | |
| Berylmys bowersi | 117 | 21 | 18.0 (11.0–24.9) | 5.9 (1.9–17.7) | 0.001 | |
| Total | 228 | 25 | 11.0 (6.9–15.0) |
Cryptosporidium viatorum was subtyped by gp60 gene sequence analysis and 17 of the 25 specimens were successfully amplified. Further analysis of these sequences suggests that four subtypes belonged to two subtype families, including XVa (XVaA6, n = 2; XVaA3g, n = 7; and XVaA3h, n = 7) and XVc (XVcA2G1, n = 1). The XVaA6 (two L. edwardsi) had 100% homology to that of the XVaA6 subtype from wastewater in China (GenBank: KX190061) [17]. Other three subtypes (XVaA3g, XVaA3h and XVcA2G1) were identified in the 15 B. bowersi. The XVaA3g had 100% homology to that of the XVaA3g subtype from human in Western Ausralia (GenBank: MK165991) [14]. Single nucleotide changes were observed between XVaA3g and XVaA3h. The XVcA2G1 had 99.2% identity with the XVcA2G1a (GenBank: MK433562) subtype identified from China.
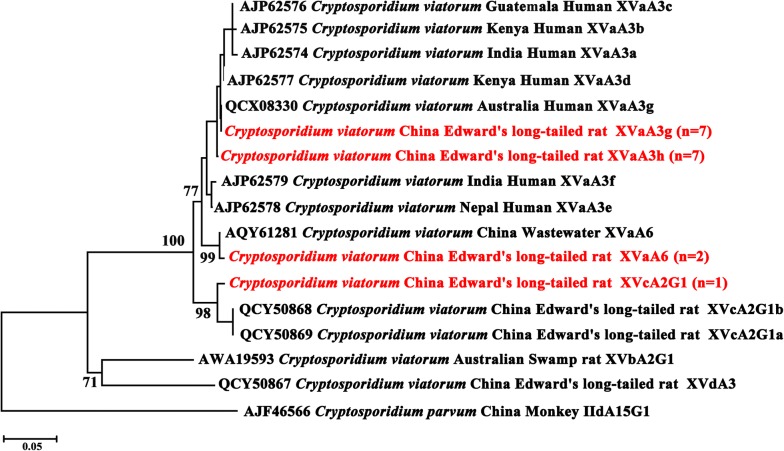
Phylogenetic analysis based on SSU rDNA sequences demonstrated that all of the C. viatorum isolates (including human isolates) grouped together with high statistical support (bootstrapping frequencies = 100) (not shown), indicating that all of the isolates from the present study represent C. viatorum. In addition, the gp60 gene provided clear evidence that the present four subtypes (XVaA6, XVaA3g, XVaA3h and XVcA2G1) of C. viatorum from the wild rats represented two subtype families (XVa and XVc) with strong bootstrap support (Fig. 1).
Fig. 1.
Phylogenetic analyses of gp60 amino acid data of C. viatorum and selected Cryptosporidium taxa using neighbor-joining (NJ) method
Discussion
Cryptosporidium viatorum has been documented in many countries [6–13], and infection can cause persistent gastrointestinal diseases in humans. The prevalence of C. viatorum in patients may be caused by their lack of natural or acquired resistance, as well as by behavioural and habitual activities related to environmental and socio-economic determinants [10]. Recently, C. viatorum was found in rats in Australia [16] and Hainan Province of China [15], indicating that these rats can act as a natural host. In the present study, C. viatorum was found in wild rats in China, which warrants further consideration into whether wild rats will become a potentially important avenue for C. viatorum transmission to humans and other animals.
The gp60 subtyping tool has been used to divide Cryptosporidium into 15 subtype families (numbered I-XV) according to their gp60 sequence [21, 27]. Stensvold et al. in 2015 [22] has classified a number of C. viatorum sequences isolated from humans into six subtypes (XVaA3a-XVaA3f). Recently, a new subtype (XVaA3g) of C. viatorum has been found in an Australian patient [14]. To our knowledge, to date, only the XVa subtype family has found in C. viatorum-positive patients [22] and wastewater in China [17]. Interestingly, the results of the present study provide clear evidence that the three subtypes (XVaA6, XVaA3g and XVaA3h) from the wild rats belong to the XVa subtype family which is genetically identical to those found in humans, suggesting that wild rats may have a potential for zoonotic transmission, and must be considered as a potential threat to human health. Our results indicate an occurrence of C. viatorum XVa subtype family in wild rats, indicating that C. viatorum XVa subtype family may be an extensive host range. In fact, in previous studies, XVa subtype family of C. viatorum were not found in rats (Edwardʼs long-tailed rats and Australian swamp rats) [15, 16]. However, we should be cautious whether the finding of XVa subtype family in rats represents a natural infection needs to be further confirmed with additional datasets. Unfortunately, we are unable to determine the source of infection and transmission dynamics of C. viatorum XVa subtype family in investigated rats due to the lack of C. viatorum data from humans in the investigated areas. In addition, three novel subtypes (XVcA2G1a, XVcA2G1b and XVdA3) of C. viatorum were identified in Edwardʼs long-tailed rats in Hainan Province, China. However, these subtypes of C. viatorum were not found in the present investigation. The difference may be related to the geographical origin [22], the health status of the animals at the time of sampling, and the overall sample sizes.
A previous study [15] and the present study provide strong evidence that C. viatorum infection is prevalent in wild rats in China, suggesting that wild rats infected with C. viatorum may pose a threat to human health. Importantly, the C. viatorum XVa subtype family identified in the present study has significant public health implications, suggesting that these animals represent a potential zoonotic risk for the transfer of the pathogen in China. More in-depth studies are needed to understand the transmission of this C. viatorum XVa subtype family in rats. In addition, we believe that an increase in the number of wild rats from broader geographical locations and diversity of rat species surveyed for C. viatorum may help gain a much-improved understanding of the role of wild rats in the zoonotic transmission of C. viatorum.
Conclusions
The present results indicate an occurrence of C. viatorum XVa subtype family in rats which are genetically identical to those found in humans. Our findings suggest that L. edwardsi and B. bowersi may play a potential role in the transmission of C. viatorum to other animals and humans. Future studies investigating the molecular epidemiology of C. viatorum in wild rats are needed to better clarify the risks and modes of transmission.
Acknowledgements
Not applicable.
Abbreviations
- AIDS
Acquired Immune Deficiency Syndrome
- BS
bootstrap support
- BLAST
Basic Local Alignment Search Tool
- CI
confidence interval
- cox1
cytochrome c oxidase subunit 1
- gp60
60-kilodalton glycoproteingene
- HIV
Human Immunodeficiency Virus
- NJ
neighbor-joining
- OR
odds ratio
- PCR
polymerase chain reaction
- SSU rRNA
small subunit ribosomal RNA
Authors’ contributions
G-HL and QZ conceived and designed the study, and critically revised the manuscript. Y-WC, W-BZ, and B-ZG performed the experiments. G-HL and Y-WC analyzed the data. Y-WC and G-HL drafted the manuscript. N-ZZ, Q-YL, and J-QY helped in study design, study implementation, and manuscript preparation. All authors read and approved the final manuscript.
Funding
This study was supported by the Planned Programme of Hunan Province Science and Technology Innovation (Grant no. 2018RS3085) and the Training Programme for Excellent Young Innovators of Changsha (Grant No. KQ1802035).
Availability of data and materials
The small subunit ribosomal RNA gene and gp60 gene sequences of Cryptosporidium viatorum from the wild rats have been deposited in the GenBank database under the accession numbers MK522269–MK522270 and MK796003–MK796005.
Ethics approval and consent to participate
All procedures involving animals in the present study were approved and this study was approved by the Animal Ethics Committee of Hunan Agricultural University (No. 43321503).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yi-Wei Chen and Wen-Bin Zheng contributed equally to this work
Contributor Information
Yi-Wei Chen, Email: 375798729@qq.com.
Wen-Bin Zheng, Email: wenbinzheng1@126.com.
Nian-Zhang Zhang, Email: zhangnianzhang@caas.cn.
Bin-Ze Gui, Email: 571644024@qq.com.
Qiu-Yan Lv, Email: 1328372661@qq.com.
Jia-Qi Yan, Email: 137629383@qq.com.
Quan Zhao, Email: zhaoquan0825@163.com.
Guo-Hua Liu, Email: liuguohua5202008@163.com.
References
- 1.Caccio SM, Pozio E. Advances in the epidemiology, diagnosis and treatment of cryptosporidiosis. Expert Rev Anti Infect Ther. 2006;4:429–443. doi: 10.1586/14787210.4.3.429. [DOI] [PubMed] [Google Scholar]
- 2.Caccio SM. Molecular epidemiology of human cryptosporidiosis. Parassitologia. 2005;47:185–192. [PubMed] [Google Scholar]
- 3.Xiao L, Fayer R, Ryan U, Upton SJ. Cryptosporidium taxonomy: recent advances and implications for public health. Clin Microbiol Rev. 2004;17:72–97. doi: 10.1128/CMR.17.1.72-97.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng Y, Ryan UM, Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;34:997–1011. doi: 10.1016/j.pt.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Z, Wang R, Zhao W, Qi M, Zhao J, Zhang L, et al. Genotyping and subtyping of Giardia and Cryptosporidium isolates from commensal rodents in China. Parasitology. 2015;142:800–806. doi: 10.1017/S0031182014001929. [DOI] [PubMed] [Google Scholar]
- 6.Elwin K, Hadfield SJ, Robinson G, Crouch ND, Chalmers RM. Cryptosporidium viatorum n. sp. (Apicomplexa: Cryptosporidiidae) among travellers returning to Great Britain from the Indian subcontinent, 2007–2011. Int J Parasitol. 2012;42:675–682. doi: 10.1016/j.ijpara.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 7.Lebbad M, Beser J, Insulander M, Karlsson L, Mattsson JG, Svenungsson B, et al. Unusual cryptosporidiosis cases in Swedish patients: extended molecular characterization of Cryptosporidium viatorum and Cryptosporidium chipmunk genotype I. Parasitology. 2013;140:1735–1740. doi: 10.1017/S003118201300084X. [DOI] [PubMed] [Google Scholar]
- 8.Adamu H, Petros B, Zhang G, Kassa H, Amer S, Ye J, et al. Distribution and clinical manifestations of Cryptosporidium species and subtypes in HIV/AIDS patients in Ethiopia. PLoS Negl Trop Dis. 2014;8:e2831. doi: 10.1371/journal.pntd.0002831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Lucio A, Amor-Aramendia A, Bailo B, Saugar JM, Anegagrie M, Arroyo A, et al. Prevalence and genetic diversity of Giardia duodenalis and Cryptosporidium spp. among school children in a rural area of the Amhara region, North-West Ethiopia. PLoS ONE. 2016;11:e0159992. doi: 10.1371/journal.pone.0159992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez A, Munoz M, Gomez N, Tabares J, Segura L, Salazar A, et al. Molecular epidemiology of Giardia, Blastocystis and Cryptosporidium among indigenous children from the Colombian Amazon Basin. Front Microbiol. 2017;8:248. doi: 10.3389/fmicb.2017.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khalil S, Mirdha BR, Panda A, Singh Y, Makharia G, Paul J. Cryptosporidium species subtypes and associated clinical manifestations in Indian patients. Gastroenterol Hepatol Bed Bench. 2017;10:311–318. [PMC free article] [PubMed] [Google Scholar]
- 12.Ayinmode AB, Zhang H, Dada-Adegbola HO, Xiao L. Cryptosporidium hominis subtypes and Enterocytozoon bieneusi genotypes in HIV-infected persons in Ibadan, Nigeria. Zoonoses Public Health. 2014;61:297–303. doi: 10.1111/zph.12072. [DOI] [PubMed] [Google Scholar]
- 13.Ukwah BN, Ezeonu IM, Ezeonu CT, Roellig D, Xiao L. Cryptosporidium species and subtypes in diarrheal children and HIV-infected persons in Ebonyi and Nsukka, Nigeria. J Infect Dev Ctries. 2017;11:173–179. doi: 10.3855/jidc.8034. [DOI] [PubMed] [Google Scholar]
- 14.Braima K, Zahedi A, Oskam C, Reid S, Pingault N, Xiao L, et al. Retrospective analysis of Cryptosporidium species in Western Australian human populations (2015–2018), and emergence of the C. hominis IfA12G1R5 subtype. Infect Genet Evol. 2019;73:306–313. doi: 10.1016/j.meegid.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Zhao W, Zhou H, Huang Y, Xu L, Rao L, Wang S, et al. Cryptosporidium spp. in wild rats (Rattus spp.) from the Hainan Province, China: molecular detection, species/genotype identification and implications for public health. Int J Parasitol Parasites Wildl. 2019;9:317–321. doi: 10.1016/j.ijppaw.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koehler AV, Wang T, Haydon SR, Gasser RB. Cryptosporidium viatorum from the native Australian swamp rat Rattus lutreolus—an emerging zoonotic pathogen? Int J Parasitol Parasites Wildl. 2018;7:18–26. doi: 10.1016/j.ijppaw.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C, Hu Y, Wang L, Wang Y, Li N, Guo Y, et al. Environmental transport of emerging human-pathogenic Cryptosporidium species and subtypes through combined sewer overflow and wastewater. Appl Environ Microbiol. 2017;83:e00682. doi: 10.1128/AEM.00682-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandão J, Mayer J. Behavior of rodents with an emphasis on enrichment. J Exot Pet Med. 2011;20:256–269. doi: 10.1053/j.jepm.2011.07.001. [DOI] [Google Scholar]
- 19.Meerburg BG, Singleton GR, Kijlstra A. Rodent-borne diseases and their risks for public health. Crit Rev Microbiol. 2009;35:221–270. doi: 10.1080/10408410902989837. [DOI] [PubMed] [Google Scholar]
- 20.Robins JH, Hingston M, Matisoo-Smith E, Ross HA. Identifying Rattus species using mitochondrial DNA. Mol Ecol Notes. 2007;7:717–729. doi: 10.1111/j.1471-8286.2007.01752.x. [DOI] [Google Scholar]
- 21.Xiao L, Morgan UM, Limor J, Escalante A, Arrowood M, Shulaw W, et al. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stensvold CR, Elwin K, Winiecka-Krusnell J, Chalmers RM, Xiao L, Lebbad M. Development and application of a gp60-based typing assay for Cryptosporidium viatorum. J Clin Microbiol. 2015;53:1891–1897. doi: 10.1128/JCM.00313-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 26.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Y, Cebelinski E, Matusevich C, Alderisio KA, Lebbad M, McEvoy J, et al. Subtyping novel zoonotic pathogen Cryptosporidium chipmunk genotype I. J Clin Microbiol. 2015;53:1648–1654. doi: 10.1128/JCM.03436-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The small subunit ribosomal RNA gene and gp60 gene sequences of Cryptosporidium viatorum from the wild rats have been deposited in the GenBank database under the accession numbers MK522269–MK522270 and MK796003–MK796005.