Abstract
Background
Limb-girdle muscular dystrophies (LGMDs) are large group of heterogeneous genetic diseases, having a hallmark feature of muscle weakness. Pathogenic mutations in the gene encoding the giant skeletal muscle protein titin (TTN) are associated with several muscle disorders, including cardiomyopathy, recessive congenital myopathies and limb-girdle muscular dystrophy (LGMD) type10. The phenotypic spectrum of titinopathies is expanding, as next generation sequencing (NGS) technology makes screening of this large gene possible.
Aim
This study aimed to identify the pathogenic variant in a consanguineous Pakistani family with autosomal recessive LGMD type 10.
Methods
DNA from peripheral blood samples were obtained, whole exome sequencing (WES) was performed and several molecular and bioinformatics analysis were conducted to identify the pathogenic variant. TTN coding and near coding regions were further amplified using PCR and sequenced via Sanger sequencing.
Results
Whole exome sequencing analysis revealed a novel homozygous missense variant (c.98807G > A; p.Arg32936His) in the TTN gene in the index patients. No heterozygous individuals in the family presented LGMD features. The variant p.Arg32936His leads to a substitution of the arginine amino acid at position 32,936 into histidine possibly causing LGMD type 10.
Conclusion
We identified a homozygous missense variant in TTN, which likely explains LGMD type 10 in this family in line with similar previously reported data. Our study concludes that WES is a successful molecular diagnostic tool to identify pathogenic variants in large genes such as TTN in highly inbred population.
Keywords: LGMD, Consanguineous family, TTN, Whole exome sequencing
Background
Limb-girdle muscular dystrophies (LGMDs) are clinically and genetically heterogeneous muscle disorders inherited as an autosomal recessive or dominant pattern. Clinically, patients are characterized by symmetrical weakness of pelvic, scapular and trunk muscles [1, 2]. LGMDs also show clinical overlapping with other muscle disorders like Emery-Dreifuss Muscular Dystrophy (EDMD; MIM: 310300), recessive congenital myopathy [MIM: 612540], myofibrillar myopathy (MFM; MIM: 601419) and late onset dominant distal myopathy [MIM: 604454] [3, 4]. More than 30 recessively and dominantly inherited forms have been identified for LGMDs [3]. The prevalence of LGMDs is about 4–7/100,000 and may have childhood, teenage or adulthood onset [3, 4]. The prevalence of autosomal recessive muscle disorders like LGMD and congenital muscular dystrophies are rare in Pakistani populations. LGMD shows severe clinical manifestations such as proximal muscle weakness, loss of ambulation between third and sixth decade, severe disability within 20 years of onset, and muscle biopsy might reveal dystrophic changes [3, 4]. Patient with LGMD had a similar disease course as Duchene muscular dystrophy (DMD), had calf hypertrophy and were non-ambulatory after age 15. Pathogenic mutations in TTN has also been associated with other severe disorders such as cardiomyopathy, dilated, 1G (MIM:604145), cardiomyopathy, familial hypertrophic 9 (MIM:613765), muscular dystrophy, limb-girdle, autosomal recessive 10 (MIM:608807), myopathy, proximal, with early respiratory muscle involvement (MIM:603689), salih myopathy (MIM:611705), tibial muscular dystrophy, tardive (MIM:600334) [5–10] .
In this study, we documented a clinical and molecular investigation of a consanguineous Pakistani family segregating LGMD in an autosomal recessive form and identified a novel homozygous missense mutation in the TTN gene located on chromosome 2q31.2. To the best of our knowledge the molecular studies on mutation in the TTN gene is reported for the first time from Pakistan.
Methods
Family recruitment and DNA isolation
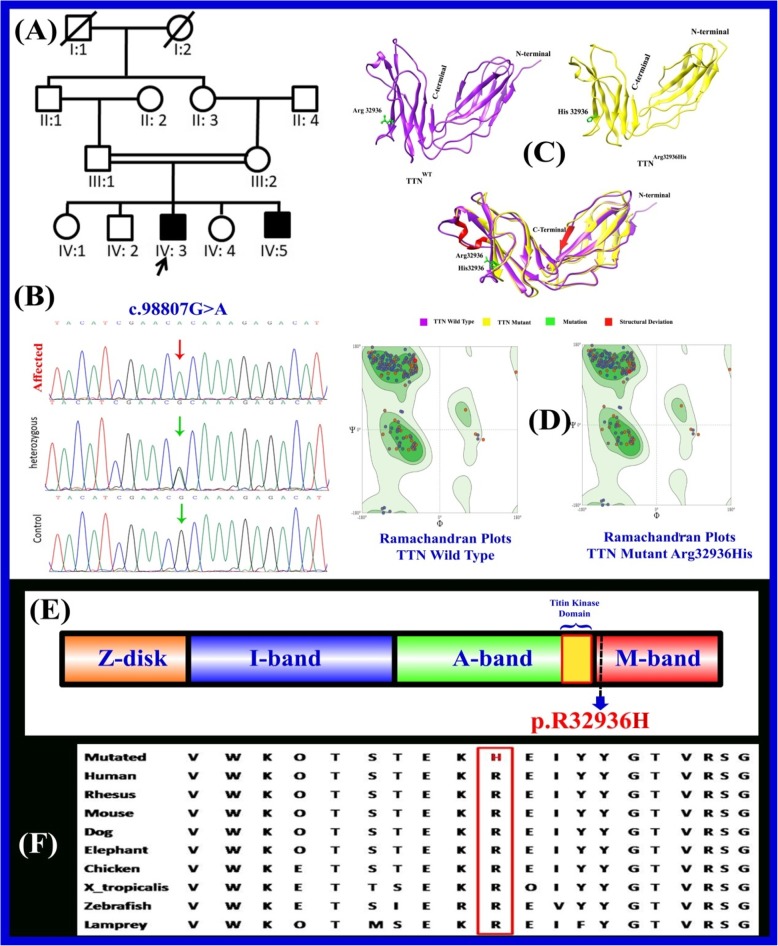
The present family has two affected individuals lives in the Bannu district of Khyber Pakhtunkhwa province, Pakistan. Pedigree was drawn (Fig. 1a) and the affected individuals were thoroughly examined by a local medical doctor. Clinical information including age, gender, family history and consanguinity was recorded. Blood samples were drawn from the two affected (IV: 3, IV: 5) and normal individuals of the family. Genomic DNA was extracted using the QIAquick DNA extraction kit (QIAamp, Qiagen, Valencia, CA, USA) and quantified using Nanodrop-2000 spectrophotometer (Thermo Scientific, Schaumburg, IL, USA).
Fig. 1.
a A consanguineous pedigree showing two affected members (IV:3 and IV:5) in the fourth generation having limb girdle muscular dystrophy. Affected individuals in the pedigree are shown with shaded symbols and unaffected with open symbols. Double lines indicate consanguineous union. b Sequence chromatogram of the TTN gene is showing segregation of c.98807G > A; p. Arg32936His in all family members c Ribbon representation of three-dimensional structure of human titin with close-up view of mutant (right) and wild type (left) at position 32,936 showing the local conformation induced by the substitution of arginine by histidine. d Ramachandran plots of wild and mutant types. e Schematic view of the functional domain of the TTN gene and localization of known mutation (Arg32936His). The novel missense variant p. Arg32936His reported here is indicated in red localized in the M domain. f The panel also shows the evolutionary conservation of Arg32936 across different species
Library preparation and whole-exome sequencing
A 100 ng of genomic DNA were needed to amplify the targeted amplicon. Exome libraries of the DNA product were created using the Ion AmpliSeq™ Exome Panel [11–13]. Emulsion polymerase chain reaction (emPCR) was performed using a OneTouch 2 instrument with an Ion PI Template OT2 200 Kit V3.The Ion OneTouch ES enrichment system (Life Technologies, Carlsbad, USA) was used for ISP enrichment step. The manufacturer’s instructions of Life Technologies company were followed to prepare and load the Ion Proton I chip [11–13].
Data processing
Sequencing data were aligned to the Homo sapiens hg 19 (GRCh37/hg19). Torrent Variant Caller software (version 4.4.3) was used to analyze the genotyping data and call the multi-allelic variations and indels. Post detection of variant was performed using wANNOVAR (http://wannovar.usc.edu/). An Integrative Genome Viewer (IGV, http://www.broadinstitute.org/igv/) was used to visualize sequencing data. Variant frequencies were obtained from various databases such as the 1000 Genomes Project, dbSNP142, Exome Aggregation Consortium (ExAC) and gnomAD (Additional file 1: Table S1).
Bioinformatics analysis
Different prediction programs including Polyphen-2 (http://genetics.bwh.harvard.edu/pph2/), SIFT (http://sift.jcvi.org/), PROVEAN (http://provean.jcvi.org) and Mutation Taster (http://www.mutationtaster.org/) predicted this mutations to be probably damaging. Finally, for the interpretation of variants, American College of Medical Genetics and Genomics (ACMG) 2015 guidelines were used [14].
Mutation confirmation
To validate the detected variant, specific fragments were PCR-amplified using site-specific primers using primer3 software (http://primer3.ut.ee) and analyzed by Sanger sequencing (Fig. 1b). The identified variant was analyzed in 200 ethnically matched control individuals (Fig. 1).
Protein modeling
The primary sequence of TTN was retrieved in FASTA format through UniProtKB/SwissProt database (https://www.uniprot.org/uniprot/O95672). Retrieved sequence was used to predict the three-dimensional (3D) protein structure using I-TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). The three-dimensional model of mutated TTN protein (p.Arg32936His) was generated by MODELLER 9.17 (https://salilab.org/modeller/9.17/release.html). The recognition of errors in experimental and theoretical models of protein structures is a major problem in structural bioinformatics. Different evaluation tools were used for the assessment of protein structure. The model was further processed by RAMPAGE (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php) ERRAT (https://servicesn.mbi.ucla.edu/ERRAT/) and Protein Structure Analysis (ProSA; https://prosa.services.came.sbg.ac.at/prosa.php). RAMPAGE generates Ramachandran plot for the assessment of models along with distribution of residues in favoured, allowed and outlier regions. ERRAT generated a plot indicating the confidence and overall quality of model. ProSA calculated an overall quality score of the predicted structure (Fig. 1c & d).
Results
Clinical description of patients
Clinical examination was performed for both affected individuals (IV: 3; IV: 5). They were born to first-cousin parents with a normal pregnancy and delivery. They were 20–25 years old, and had severe LGMD. Notable clinical findings include difficulty in rising from the floor, delay in motor milestones, and muscle weakness. They had mild microcephaly, intellectual disability (ID), generalized muscle hypertrophy and developmental delay (Table 1). Follow up clinical examination of the patients revealed cardiomyopathy, proximal and distal weakness, inability to stand, loss of ambulation, and both were confined to a wheelchair. They also had a triangular face, low set of ears, and clinodactyly in lower limb digits. In addition, both were suffering pelvic and shoulder girdle muscular dystrophy, muscular pain and also facial muscles weakness when doing a usual muscle exercise. Features such as skin, teeth, nails, eyes, reproductive and cardiac deformities were not observed in both of them. Their parents showed no abnormalities and were healthy.
Table 1.
Clinical features of the affected individuals
| Variable | Subject (IV:3) | Subject (IV:5) |
|---|---|---|
| Sex | Male | Male |
| Age | 25 | 20 |
| Microcephaly | + | + |
| Wheelchair | + | + |
| Scoliosis | + | + |
| Synophrys | + | + |
| Hearing impairment | – | – |
| Intellectual disability | + | + |
| Pelvic girdle weakness | + | + |
| Skeletal abnormalities | ++ | ++ |
| Difficulty in rising from the floor | ++ | ++ |
| Syncope attack | + | + |
| Scapular and trunk muscles weakness | + | + |
| Cardiac impairment | + | + |
| Muscle pain and stiffness | – | – |
| Seizures | – | – |
| Cancer | – | – |
| Narrow shoulder | + | + |
| Skin | Normal | Normal |
| Eye sight | Normal | Normal |
| Behavior | Nervous/forgetful | Nervous/forgetful |
| Pregnancy event | Normal | Normal |
+, present; ++, severe phenotype; −, absent
Whole exome sequencing
In the present study, clinical diagnosis was confirmed by genetic analysis. Of these, both patients (IV: 3; IV: 5) and their parents (III-1;III-2) were subjected to whole exome sequencing (WES) as described earlier using Ion Torrent platform [11–13]. WES results indicated a novel homozygous missense variant (c.98807G > A; p.Arg32936His) in TTN (MIM: 188840) gene responsible for LGMD phenotype (Table 2). Sanger sequencing perfectly confirmed segregation of the disease phenotype. The variation G-to-A transversion results into the substitution of arginine (R) to histadine residues (Arg32936His). This mutation is conserved across different species and can affect greatly the amino acid (aa) sequence located in the M domain of TTN gene that might change the protein features and also affect the splice site (Fig. 1 e& f). Different online bioinformatics tools were used to analyze the pathogenicity of the variant (Table 2).
Table 2.
Homozygous variant on chromosome 2 from exome data of TTN family
| Family | Individuals (IV:3 and IV:5) |
|---|---|
| Chr. Position (hg19) | chr2:179403855 |
| Reference allele | G |
| Alternate allele | A |
| Gene | TTN |
| MIM | 188,840 |
| Gene Bank | NM_001267550.2 |
| ExonicFunc.refgene | nonsynonymous SNV |
| cDNA Change | c.98807G > A |
| Amino Acid change | p.Arg32936His |
| 1000G_ALL | 0.00 |
| ExAC_Freq | 0.0001019 |
| dbSNP | rs774296358 |
| ClinVar_Status | – |
| SIFT Score &prediction | 0.044/D |
| Polyphen2 score & prediction | 0.99/PD |
| Mutation taster score &predict | 0.99/D |
| FATHMM_score & prediction | 0.7881/D |
| CADD score | 24.3/D |
| ACMG Classification | PM2 |
| Variant Status | Novel |
| Other Information’s | Homozygous |
*SNV Single Nucleotide Variant, D Damaging, PD Probably Damaging,PM2 Pathogenic Moderate 2
Using homology modelling, 3D models of wild type and mutated TTN protein (p.Arg32936His) were predicted and evaluated by online structure analysis tools as described above. Ramachandran plot generated by RAMPAGE indicated that approximately 93% of residues in the model lie in allowed regions of torsion angles. ERRAT and ProSA showing overall quality of model and quality score of the predicted structure (Fig. 1 c & d).
Discussion
LGMD is an inherited genetic disorder characterized by limb and girdle weakness and transmitted in either an autosomal recessive or an autosomal dominant pattern [1, 2]. Several genes are associated with the LGMD phenotype and the next generation sequencing (NGS) technology can be the best choice for definitive diagnosis of LGMD [15, 16]. The affected individuals reported here, exhibit several phenotypes such as difficulty in rising from the floor, delay in motor milestones, and muscle weakness, mild microcephaly, intellectual disability, generalized muscle hypertrophy and developmental delay (Table 1). Such features were also reported previously [15, 16]. Cardiomyopathy also was observed in our patients [17]. Recently, Younus et al (2019) reported a nonsense mutation in the SGCD gene among Pakistani population having LGMD features that shows variability with features in comparison the cases reported here [18]. Through WES, we detected a homozygous missense mutation (c.98807G > A; p.Arg32936His) in the exon 353 of the TTN gene known to be associated with LGMD phenotypes.
The titin protein is organized into four structurally and functionally distinct regions that correlate with the muscle sarcomere [19–21]. These regions, located at the amino terminus to the carboxy terminus of the protein, include the Z-disk, I-band, A-band, and the M-line [21–23] (Fig. 1). Carriage of the mutation c.98807G > A which is very close to the M domain of the TTN gene, results in amino acid change of the Arg32936 residue into the His32936 and alter the secondary structure of the TTN protein causing protein instability. Using homology modelling; three-dimensional models of wild-type and the mutated TTN protein (p.Arg32936His) revealed a Z scores between 0.5–1.0, indicating no significant deviation from the scores determined for proteins of similar size. The entire TTN gene consists of 364 exons, located on chromosome 2q31, and transcribes an mRNA over 100 kb long that could hypothetically produce around 38,138 residues and 4200 kDa proteins [24].
TTN has multiple key roles in all striated muscle cells, well suited for its role as an architectural protein and provide specific attachment to a plethora of essential proteins [23]. A total of 346 TTN disease-causing mutations (259 missense/nonsense, 23 splicing, 13 small insertions, 47 small deletions, 1 small indels and 2 gross deletions) have been reported in the human gene mutation database (HGMD) with at least 10 different conditions, including isolated cardiomyopathies, purely skeletal muscle phenotypes and infantile diseases affecting both types of striated muscles (Table 3) [17, 18]. A majority of patients with TTN mutations have normal intelligence quotient (IQ), but our patients showed poor language development, mild microcephaly and developmental delay (Table 1).
Table 3.
HGMD reported mutations in TTN gene associated with LGMD disorders
| Gene Name | DNA Variation |
Protein Variation |
Mutation type | Reported phenotype |
|---|---|---|---|---|
| TTN | c.187G > A | p.A63T | Missense | Muscular dystrophy, limb-girdle |
| TTN | c.3100G > A | p.V1034 M | Missense | Muscular dystrophy |
| TTN | c.7961G > A | p.R2654K | Missense | Muscular dystrophy |
| TTN | c.22771A > T | p.K7591* | Nonsense | Muscular dystrophy |
| TTN | c.28730C > T | p.P9577L | Missense | Muscular dystrophy |
| TTN | c.46363C > T | p.R15455* | Nonsense | Muscle weakness |
| TTN | c.49243G > A | p.A16415T | Missense | Muscular dystrophy |
| TTN | c.63658G > A | p.A21220T | Missense | Muscle weakness |
| TTN | c.76850G > A | p.R25617Q | Missense | Muscle weakness |
| TTN | c.78320C > T | p.P26107L | Missense | Muscle weakness |
| TTN | c.87483G > C | p.W29161C | Missense | Muscle weakness |
| TTN | c.97332C > A | p.Y32444* | Nonsense | Muscular dystrophy |
| TTN | c.98456C > G | p.S32819* | Nonsense | Muscle weakness |
| TTN | c.99274C > T | p.Q33092* | Nonsense | Muscular dystrophy |
| TTN | c.100133A > C | p.H33378P | Missense | Tibial muscular dystrophy |
| TTN | c.100136 T > A | p.I33379N | Missense | Tibial muscular dystrophy |
| TTN | c.100163 T > C | p.L33388P | Missense | Tibial muscular dystrophy |
| TTN | c.100186C > T | p.Q33396* | Nonsense | Tibial muscular dystrophy |
| TTN | c.57871 + 2 T > G | – | Splice site | Muscular dystrophy |
| TTN | c.99673 + 1G > C | – | Splice site | Muscle weakness |
| TTN | c.6379_6380delTA | p.(Tyr2127Leufs*8) | Deletion | Muscular dystrophy |
| TTN | c.43733-4_43740del12 | – | Deletion | Muscular dystrophy |
| TTN | c.59385delT | p.(Lys19796Argfs*24) | Deletion | Tibial muscular dystrophy |
| TTN | c.90401delC | p.(Pro30134Leufs*15) | Deletion | Tibial muscular dystrophy |
| TTN | c.93409delT | p.(Ser31137Leufs*4) | Deletion | Muscular dystrophy, limb girdle 2 J |
| TTN | c.98807G > A | p.Arg32936His | Missense | Muscular dystrophy, limb-girdle |
| TTN | c.99943delT | p.(Ser33315Glnfs*10) | Deletion | Tibial muscular dystrophy |
| TTN | c.100185delA | p.(Lys33395Asnfs*9) | Deletion | Tibial muscular dystrophy |
| TTN | c.32190dupT | – | Duplication | Muscular dystrophy, limb girdle 2 J |
| TTN | c.92854_92857dupACTG | – | Duplication | Tibial muscular dystrophy |
| TTN | c.100076_100086delins11 | – | Indels | Tibial muscular dystrophy |
| TTN | c.1662 + 15_3101–3 | – | Gross deletion | Muscular dystrophy |
| TTN | ex. 34–41 | – | Gross deletion | Muscle weakness |
Homozygous knockdown mice (ttn−/−) had a degeneration of both distal and proximal skeletal muscles by 2–3 weeks of age [25]. ttn−/−mice developed a rigid gait, kyphosis due to axial skeletal muscle association and normally does not survive long. Histological studies indicated that degeneration was specific to skeletal muscles with no other symptoms such as cardiomyopathy or impairment of the central or peripheral nervous system [25]. Both fore and hind limbs skeletal muscles had severe and progressive dystrophic phenotypes indicating that TTN plays a pivotal role in skeletal system development [25].
Conclusion
This is the first report of TTN pathogenesis causing LGMD type 10 from Pakistani population. Failure for detection of c.98807G > A (p. Arg32936His) in 200 ethnically matched control individuals chromosomes outside of the family or in the public databases, designate that this homozygous missense mutation (is probably pathogenic and deleterious. However, further studies regarding LGMDs among large number of Pakistani population might lead to a deeper understanding, genetic mechanisms and future therapeutic interventions.
Supplementary information
Additional file 1: Table S1. Filtering steps followed to search for the candidate variant.
Acknowledgments
We thank all individuals for their participation in this study. We wish to thank Dr. Peter Gergics, the University of Michigan for the helpful comments on the manuscript.
WEB resources
1000 Genomes _ https://www.internationalgenome.org/
Exome Variant Server _ http://evs.gs.washington.edu/EVS/
ExAC_http://exac.broadinstitute.org/
dbSNP _ http://www.ncbi.nlm.nih.gov/SNP/
OMIM _ http://www.omim.org/
Abbreviations
- ACMG
American College of Medical Genetics and Genomics
- DMD
Duchene muscular dystrophy
- emPCR
Emulsion polymerase chain reaction
- ERB
Ethical Review Committee
- ExAC
Exome Aggregation Consortium
- HGMD
Human gene mutation database
- ID
Intellectual disability
- LGMDs
Limb-girdle muscular dystrophies
- NGS
Next generation sequencing
- TTN
Titin
- WES
Whole exome sequencing
Authors’ contributions
XZ, SA, and MIK were involved in the planning of the experiments. AK extracted DNA of the proband and her healthy parents’ samples and performed polymerase chain reaction. SH and MA1 performed the WES experiment. RW, AK, and MU carried out genetic experiments, analyzed obtained results, and performed bioinformatics analysis. MA2 and XZ supervised the findings of this work. AK wrote the manuscript with consultation and support from RW and SH. All authors read and approved the final manuscript.
Funding
This work was financially supported by the National Key Research and Development Program of China [grant number 2016YFC0905100], the CAMS Innovation Fund for Medical Sciences (CIFMS) [grant numbers2017-I2M-B&R- 05 and 2016-I2M-1-002], and the Central Research Institutes of Basic Research and Public Service Special Operations [grant number 2018PT32024]. The funding body had no influence in the in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its additional file.
Ethics approval and consent to participate
The study design and protocol was conducted in accordance with the guidelines of the Helsinki Declaration, and was approved by Ethical Review Committee (ERC) of Peking Union Medical College (Beijing, China), and China Medical University (Shenyang, China). Informed, written consent to participate was obtained from the parents or legal guardians of the participants under the age of 16 or the participants who are over the age of 16.
Consent for publication
Informed, written consent for publication of the participants (under the age of 18) clinical details and/or clinical images was obtained from their parents or legal guardians or the participants who are over the age of 18.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Amjad Khan and Rongrong Wang contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12881-019-0895-7.
References
- 1.Nigro V, Savarese M. Genetic basis of limb-girdle muscular dystrophies: the 2014 update. Acta Myol. 2014;33(1):1–12. [PMC free article] [PubMed] [Google Scholar]
- 2.Vissing J. Limb girdle muscular dystrophies: classification, clinical spectrum and emerging therapies. Curr Opin Neurol. 2016;29:635–641. doi: 10.1097/WCO.0000000000000375. [DOI] [PubMed] [Google Scholar]
- 3.Yiş U, Diniz G, Hazan F, et al. Childhood onset limb-girdle muscular dystrophies in the Aegean part of Turkey. Acta Myol. 2018;37(3):210–220. [PMC free article] [PubMed] [Google Scholar]
- 4.Bushby KM. The limb-girdle muscular dystrophies-multiple genes, multiple mechanisms. Hum Mol Genet. 1999;8(10):1875–1882. doi: 10.1093/hmg/8.10.1875. [DOI] [PubMed] [Google Scholar]
- 5.Evila A, Palmio J, Vihola A, Savarese M, Tasca G, Penttila S, et al. Targeted next-generation sequencing reveals novel TTN mutations causing recessive distal titinopathy. Mol Neurobiol. 2017;54(9):7212–7223. doi: 10.1007/s12035-016-0242-3. [DOI] [PubMed] [Google Scholar]
- 6.Pfeffer G, Barresi R, Wilson IJ, Hardy SA, Griffin H, Hudson J, et al. Titin founder mutation is a common cause of myofibrillar myopathy with early respiratory failure. J Neurol Neurosurg Psychiatry. 2014;85(3):331–338. doi: 10.1136/jnnp-2012-304728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evila A, Vihola A, Sarparanta J, Raheem O, Palmio J, Sandell S, et al. Atypical phenotypes in titinopathies explained by second titin mutations. Ann Neurol. 2014;75(2):230–240. doi: 10.1002/ana.24102. [DOI] [PubMed] [Google Scholar]
- 8.Chauveau C, Bonnemann CG, Julien C, Kho AL, Marks H, Talim B, et al. Recessive TTN truncating mutations define novel forms of core myopathy with heart disease. Hum Mol Genet. 2014;23(4):980–991. doi: 10.1093/hmg/ddt494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ceyhan-Birsoy O, Agrawal PB, Hidalgo C, Schmitz-Abe K, DeChene ET, Swanson LC, et al. Recessive truncating titin gene, TTN, mutations presenting as centronuclear myopathy. Neurology. 2013;81(14):1205–14. doi: 10.1212/WNL.0b013e3182a6ca62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dabby R, Sadeh M, Hilton-Jones D, Plotz P, Hackman P, Vihola A, et al. Adult onset limb-girdle muscular dystrophy – a recessive titinopathy masquerading as myositis. J Neurol Sci. 2015;351(1–2):120–123. doi: 10.1016/j.jns.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Thermo Fisher Scientific . Ion AmpliSeqTM Library Preparation for Human Identification Applications, P/N MAN0010640, Rev. A.0. Massachusetts: Thermo Fisher Scientific; 2014. [Google Scholar]
- 12.Thermo Fisher Scientific . Ion PGMTM Template OT2 200 Kit for use with the Ion OneTouchTM 2 System, P/N MAN0007220, Rev. A.0. Massachusetts: Thermo Fisher Scientific; 2014. [Google Scholar]
- 13.Thermo Fisher Scientific . Ion PGMTM Sequencing 200 Kit v2, P/N MAN0007273, Rev. 3.0. Massachusetts: Thermo Fisher Scientific; 2013. [Google Scholar]
- 14.Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nigro V, Aurino S, Piluso G. Limb girdle muscular dystrophies: update on genetic diagnosis and therapeutic approaches. Curr Opin Neurol. 2011;24(5):429–436. doi: 10.1097/WCO.0b013e32834aa38d. [DOI] [PubMed] [Google Scholar]
- 16.Taghizadeh E, Abdolkarimi H, Boostani R, Sadrnabavi A. Limb-girdle muscular dystrophy with new mutation in Sarcoglycan Beta gene: a case report. Iran J Public Health. 2018;47(12):1953–1957. [PMC free article] [PubMed] [Google Scholar]
- 17.Neiva-Sousa M, Almeida-Coelho J, Falcão-Pires I, Leite-Moreira AF. Titin mutations: the fall of goliath. Heart Fail Rev. 2015;20(5):579–588. doi: 10.1007/s10741-015-9495-6. [DOI] [PubMed] [Google Scholar]
- 18.Younus M, Ahmad F, Malik E, et al. SGCD Homozygous Nonsense Mutation (p.Arg97∗) Causing Limb-Girdle Muscular Dystrophy Type 2F (LGMD2F) in a Consanguineous Family, a Case Report. Front Genet. 2019;9:727. doi: 10.3389/fgene.2018.00727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labeit S, Gautel M, Lakey A, Trinick J. Towards a molecular understanding of titin. EMBO J. 1992;11(5):1711–1716. doi: 10.1002/j.1460-2075.1992.tb05222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller G, Musa H, Gautel M. PeckhamM. A targeted deletion of the C-terminal end of titin, including the titin kinase domain, impairs myofibrillogenesis. J Cell Sci. 2003;116(Pt 23):4811–4819. doi: 10.1242/jcs.00768. [DOI] [PubMed] [Google Scholar]
- 21.Tskhovrebova L, Trinick J. Titin: properties and family relationships. Nat Rev Mol Cell Biol. 2003;4(9):679–689. doi: 10.1038/nrm1198. [DOI] [PubMed] [Google Scholar]
- 22.Meyer LC, Wright NT. Structure of giant muscle proteins. Front Physiol. 2013;4:368. doi: 10.3389/fphys.2013.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trinick J. Titin and nebulin: protein rulers inmuscle? Trends Biochem Sci. 1994;19(10):405–409. doi: 10.1016/0968-0004(94)90088-4. [DOI] [PubMed] [Google Scholar]
- 24.Hackman P, Vihola A, Haravuori H, Marchand S, Sarparanta J, De Seze J, et al. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am J Hum Genet. 2002;71(3):492–500. doi: 10.1086/342380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huebsch KA, Kudryashova E, Wooley CM, et al. Mdm muscular dystrophy: interactions with calpain 3 and a novel functional role for titin's N2A domain. Hum Mol Genet. 2005;14(19):2801–2811. doi: 10.1093/hmg/ddi313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Filtering steps followed to search for the candidate variant.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional file.