Abstract
Background
Bladder cancer (BC) is a common urothelial malignancy, characterized by a high recurrence rate. The biology of bladder cancer is complex and needs to be deciphered. The latest evidence reveals the critical role of the non-coding RNAs, particularly microRNAs (miRNAs), as vital regulatory elements in cancer.
Method
We performed a miRNAs microarray using paired tissues (tumor and adjacent normal bladder tissue), followed by the validation with qRT-PCR of five selected transcripts. Additional next-generation sequencing investigation established the interconnection among the altered miRNAs and mutated genes. Based on the overlapping between TCGA data and data obtained in the study, we focused on the systematic identification of altered miRNAs and genes mutated involved in bladder cancer tumorigenesis and progression.
Results
By overlapping the miRNAs expression data, the two patient cohorts, we identified 18 miRNAs downregulated and, 187 miRNAs upregulated. qRT-PCR validation was completed using a selected panel of two downregulated (miR-139-5p and miR-143-5p) and three up-regulated miRNAs (miR-141b, miR-200 s or miR-205). Altered miRNAs patterns are interrelated to bladder tumorigenesis, allowing them to be used for the development of novel diagnostic and prognostic biomarkers. Three EMT-related upregulated miRNAs have an essential role in the molecular mechanisms, specifically key processes underlying tumorigenesis, invasion and metastasis. Using the Ampliseq Cancer Panel kit and Ion Torrent PGM Next-Generation Sequencing an increased mutation rate for TP53, FGFR3, KDR, PIK3CA and ATM were observed, but the mutational status for only TP53 was correlated to the survival rate. The miRNAs pattern, along with the gene mutation pattern attained, can assist for better patient diagnosis.
Conclusion
This study thereby incorporates miRNAs as critical players in bladder cancer prognosis, where their altered gene expression profiles have a critical biological function in relationship with tumor molecular phenotype. The miRNA-mRNA regulatory networks identified in BC are ripe for exploitation as biomarkers or targeted therapeutic strategies.
Keywords: Bladder cancer, miRNA, Mutation, miRNA-mRNA network
Background
Bladder cancer (BC) represents a global problem for the urinary tract, statistically situated in fifth place in terms of mortality and morbidity [1]. BC incidence is higher in aged populations and is correlated with toxic environmental agents, in particular with smoking [2]. The prognosis of BC is unfavorable, with a high percentage of disease recurrence regardless of treatment; these treatments include surgery, chemotherapy or, a combination of these two [1]. About 70–75% of BC cases are defined as non-muscle invasive tumors, subclassified into low or high-grade tumors [3–5]. The identification of high-risk tumors is imperative and requires intravesical therapy as a standard practice [6].
BC is a multifactorial disease where both exogenous and endogenous factors are eessential in early carcinogenesis, disease progression but also responsible for the high recurrence rate, correlating to a specific mutation pattern [7]. Therefore, new biomarkers for early diagnosis of bladder cancer, prognostic markers for its recurrence and predictive markers for response/overall survival are needed with great urgency [8]. Molecular markers can provide vital information to refine the optimal treatment, which in turn permits a good patient prognosis [5].
MicroRNAs (miRNAs) are a novel class of short non-coding RNA sequences that have around 19–25 nucleotide length [8, 9]. These short non-coding structures can regulate gene expression and interfere with vital cellular pathways without being translated into proteins, including in cancer biology. A miRNA can target multiple genes, several miRNAs can target a gene and this gives rise to complex interaction networks; presently, the exact interactions have yet to be determined especially in relationship to the mutational status [8, 10]. Accordingly, miRNAs are essential candidates for both diagnosis and prognosis due to their oncogenic or tumor suppressor functions. Hence, miRNAs profiling studies from different tissues represent an excellent alternative application for these short sequences as biomarkers with clinical significance [11]. The main advantage is the high stability of these transcripts, remaining unaltered during the surgical resection procedure (TURB), which is often associated with a high degradation rate [12, 13]. Another advantage is the full range of tools and methods for miRNAs profiling (microarray, next-generation sequencing or Nanostring) or validation (qRT-PCR or in situ hybridization) [8].
Previous studies have examined miRNAs in BC; however, very few cases examine the global miRNA expression patterns by microarray in paired samples with subsequent overlapping using TCGA miRNA data. We utilized this combined data set to identify specific pathways associated with BC. Following the profiling of bladder cancer samples, we used Ion Torrent Next Generation Sequencing Cancer panel to determine the most relevant mutations in our patient cohort and TCGA dataset. The correlation of this data permits a better understanding of the interconnected regulatory networks that appear to have significant biological meaning in terms of tumor molecular phenotype and gene expression profiles; all this to be exploited as candidates for future therapies or as prognostic/diagnostic biomarkers.
Material and methods
Sample collection
Between 2014 and 2016, we collected the tumoral BC tissue along with the adjacent healthy tissue from patients, only after obtaining informed consent from each patient and approval by the institutional ethics committee of the Iuliu Hatieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania (UMPh), with the authorization no. 673A/20.11.2012. We stored transurethral resections of bladder tumor (TURBT) tissues in liquid nitrogen until sample processing and RNA extraction. When surgical and pathological procedures permitted, surgeons collected the healthy tissue, adjacent from the tumor, from each patient. We initially evaluated the expression of Her2 and TP53 using the standard immunohistochemistry staining protocol. The paired (healthy and tumor) tissue samples later used for the microarray and next-generation sequencing analyses are referred to as UMPh patient cohort. The second group of samples collected was for the qRT-PCR validation and named the validation set; we collected this additional patient cohort.
Sample processing and microarray evaluation
The total RNA extraction and isolation from 23 paired samples (normal and tumoral bladder tissue) is done using the TriReagent (Sigma-Aldrich) protocol. NanoDrop-1000 spectrophotometer was used to measure the concentration of RNA. The microarray probes were synthesized from equal quantities of 100 ng of total RNA, by using miRNA microarray protocol based on version 3.1 of September 2015 (Agilent Technologies) which included complete labeling and hybridization kit (cat no. 5190–0456 Agilent) and a purification step with Micro Bio-Spin P-6 Gel Column (Biorad). The SureScan Microarray Scanner (Agilent Technologies) scanned the microarray slides and Feature Extraction 12.0 software performed data extraction. The last step in the microarray evaluation was to identify the primary altered miRNAs. Gene Spring GX v.13.0, applying a fold change (FC) threshold of 2 moderated t-test and False Discovery Rate correction (p-valued ≤0.05), analyzed the microarray data and generated the comparisons of low-grade versus high-grade tumor tissues; low-grade tumor versus healthy tissues; high-grade tumors versus healthy tissues (data available on Arrayexpress, ID: E-MTAB-8356).
Bladder cancer TCGA data analysis
We performed a supplementary analysis at the third level of miRNAs-sequencing from 409 bladder tumors and 19 healthy tissues, adjacent to the tumors, obtained from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). The data were analyzed in GeneSpring GX v.13.0, applying the previous defined cut-off value.
miRNA qRT-PCR evaluation on tissue samples
We selected to validate three upregulated transcripts (miR-23a, miR-141-3p and miR-205-5p) and two downregulated transcripts (miR-139-5p, and miR-143-5p) from the paired tissue samples. RNA was extracted using TriReagent based method for qRT-PCR validation was performed on 18 healthy bladder tissues and 18 bladder tumor tissues. We performed the cDNA synthesis using a 7.5 μl of reverse transcription mixture containing 0.72 μl of RT primer, 50 ng of total RNA and 0.5 μl of MultiScribe Reverse Transcriptase, 0.75 μl Reverse Transcription Buffer (10×), 0.075 μl dNTPs (100 mM), 0.1 μl of RNase Inhibitor according to Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems) protocol. The cDNA mixture is incubated in PCR tubes for 30 min at 16 °C, 30 min at 42 °C and 5 min at 85 °C. qRT-PCR was performed using the in ViiA7 (Applied Biosystems) PCR machine with a total volume of reaction mix of 12.5 μl; this reaction mix consists of 6.25 μl of cDNA (diluted 1:6 with nuclease-free water), 5.63 μl of SSoAdvanced Universal Probe Supermix (Bio-Rad) and 0.73 μl primers for each miRNA. The reactions were set up as follows: initial denaturation step at 95 °C for 180 s, followed by 39 cycles of 95 °C for 5 s and, lastly, 60 °C for the 30s. The expression level of each miRNA is calculated by the threshold cycle (CT). The relative expression level was calculated using –ΔΔCT method and U6 for normalization.
TP53 evaluation by qRT-PCR on bladder tissue samples
Lab technicians synthesized the cDNA using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). The reaction preparation of qRT-qPCR used the SYBR Select Master Mix (Life Technologies) and executed using ViiA7. The following conditions were used: 95 °C for 2 min, 40 cycles of 95 °C for 10 s and 60 °C for 1 min. The FC of gene expression was calculated with the ΔΔCT method, using B2M as the housekeeping gene.
Next-generation sequencing of bladder cancer samples
A number of 22 bladder cancer samples, the same ones analyzed by microarray (except one samples from microarray patien cohort with low DNA concentration), were sequenced using Ion Ampliseq Cancer Panel and Ion Torrent PGM Next Generation Sequencing (Thermo Fischer Scientific); this panel contains the most relevant hot spot mutation. The amplicon libraries were prepared with 20 ng of DNA and the Ion Ampliseq™ Library Kit 2.0 (Life Technologies) and this was followed by a purification step using AMpure XP Beads (Beckman Coulter). Lastly, Qubit 2.0 was used for the quantification using Qubit HS DNA kit. For sequencing, four bar-coded 100pM-diluted libraries were used for each Ion 316 Chip (Thermo Fischer Scientific). Ion Torrent PGM Machine (Thermo Fischer Scientific) performed the sequencing, using the Ion PGM HI-Q Sequencing 200 kit. The software Torrent Suit 5.6 and Ion Reporter 5.6 executed the bioinformatics analysis, specifically for data trimming alignment and variant calling.
Functional analysis and target genes identification
The IPA analysis determined the miRNAs with altered expression levels to identify the most relevant networks, altered pathways and their respective biological significance. For the identification of the target genes most relevant to our miRNAs, the following database and webservers were used: miRTarBase (https://bio.tools/mirtarbase); miRNet (https://www.mirnet.ca/); and miRtargetLink (https://ccb-web.cs.uni-saarland.de/mirtargetlink/).
Results
Bladder cancer patients’ statistics
We determined the initial miRNA profiling using the WHO-2016 classification and the most frequent histological type, urothelial carcinoma. More specifically for the microarray study, the patients’ samples were graded as low or high and, additionally, through immunohistochemistry (IHC), the expression level of Her2 and TP53 was determined. Figure 1 exemplifies a low grade negative stained case for TP53 and Her2 compared to a high grade positive stained case for TP53 and Her2.
Fig. 1.
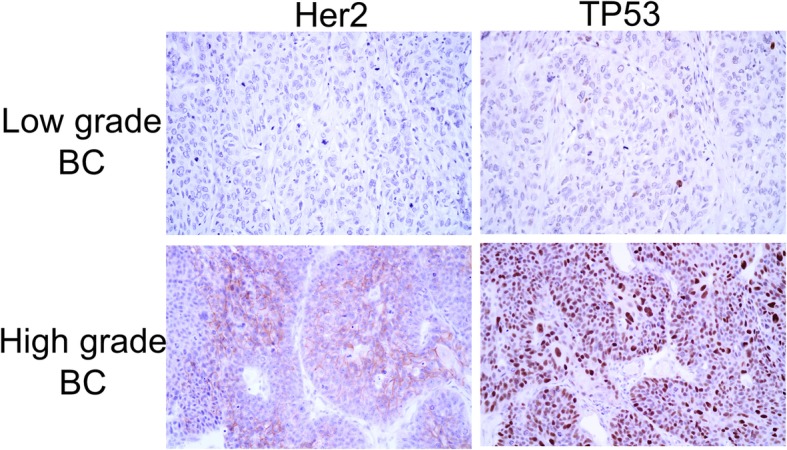
Expression of Her2 and TP53 in low grade and high bladder tumor samples using the standard immunohistochemistry staining protocol (400x magnification)
Table 1 presents the demographics and patient characteristic selected for the microarray. Of the 23 patients included in the microarray cohort, the average age is 64.04 ± 11.72, consisting of 5 females with an age average of 56.80 ± 6.30 and 18 males with an age average of 66.05 ± 12.20. The average age for the qRT-PCR cohort of 18 patients (5 females and 13 males), is respectively 66 ± 9,59 (57,2 ± 7,98 for females, respectively 69,38 ± 8.03 for males), which can be seen in Table 2. We classified the patient cohort selected for the profiling and qRT-PCR validations into two main subtypes: low-grade non-invasive tumors and high grade advanced tumors.
Table 1.
Demographic and histopathological characteristics of the 23 participants used for miRNA microarray evaluation
| No. | Sex | Age | Histopathological diagnosis | Histopatological stage |
|---|---|---|---|---|
| 1 | M | 81 | high grade urothelial carcinoma | pT2 |
| 2 | M | 71 | low grade urothelial carcinoma | pTa |
| 3 | M | 61 | high grade urothelial carcinoma | pT1 |
| 4 | F | 53 | low grade urothelial carcinoma | PTa |
| 5 | F | 50 | low grade urothelial carcinoma | pTa |
| 6 | M | 78 | low grade urothelial carcinoma | pTa |
| 7 | F | 54 | high grade urothelial carcinoma | pT2 |
| 8 | M | 45 | high grade urothelial carcinoma | pT4 |
| 9 | M | 67 | low grade urothelial carcinoma | pT1 |
| 10 | F | 63 | high grade urothelial carcinoma | pT2 |
| 11 | M | 43 | low grade urothelial carcinoma | pTa |
| 12 | M | 60 | low grade urothelial carcinoma | pTa |
| 13 | M | 75 | low grade urothelial carcinoma | pTa |
| 14 | M | 75 | low grade urothelial carcinoma | PTis |
| 15 | M | 61 | high grade urothelial carcinoma | pT4 |
| 16 | M | 93 | high grade urothelial carcinoma | pT3a |
| 17 | M | 65 | high grade urothelial carcinoma | pT2 |
| 18 | M | 58 | high grade urothelial carcinoma | pT2 |
| 19 | M | 57 | high grade urothelial carcinoma | pTa |
| 20 | F | 64 | low grade urothelial carcinoma | pTa |
| 21 | M | 61 | low grade urothelial carcinoma | pT1 |
| 22 | M | 70 | high grade urothelial carcinoma | pT2 |
| 23 | M | 68 | high grade urothelial carcinoma | pT2 |
Table 2.
Demographic and histopathological characteristics of the eighteen participants used for qRT-PCR data validation
| Patient code | Sex | Age | Histopathological diagnosis | Histopatological stage |
|---|---|---|---|---|
| 1 | M | 82 | high grade urothelial carcinoma | pT2 |
| 2 | F | 68 | high grade urothelial carcinoma | pT1 |
| 3 | M | 59 | low grade urothelial carcinoma | pT1 |
| 4 | M | 77 | high grade urothelial carcinoma | pT1 |
| 5 | M | 65 | high grade urothelial carcinoma | pTa |
| 6 | F | 54 | high grade urothelial carcinoma | pT4a |
| 7 | M | 58 | high grade urothelial carcinoma | pT2 |
| 8 | M | 75 | high grade urothelial carcinoma | pT2 |
| 9 | M | 67 | low grade urothelial carcinoma | pTa |
| 10 | F | 52 | high grade urothelial carcinoma | pTa |
| 11 | M | 73 | high grade urothelial carcinoma | pT1 |
| 12 | F | 63 | high grade urothelial carcinoma | pT2 |
| 13 | M | 67 | high grade urothelial carcinoma | pT2 |
| 14 | M | 58 | high grade urothelial carcinoma | pTa |
| 15 | M | 68 | high grade urothelial carcinoma | pT1a |
| 16 | M | 74 | high grade urothelial carcinoma | pT1 |
| 17 | M | 79 | high grade urothelial carcinoma | pT3b |
| 18 | F | 49 | high grade urothelial carcinoma | pTa |
Evaluation of tissue miRNA patterns in bladder cancer patients
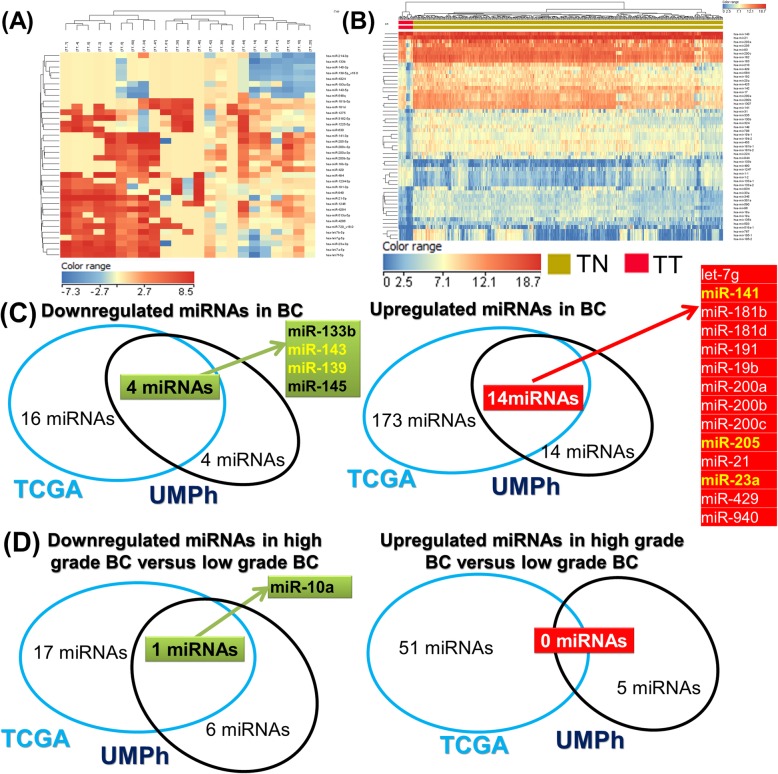
The present profiling investigation uses Agilent 60-mer SurePrint microarray technology that contains capture probes to characterize the expression of 2549 mature human miRNAs, obtained from the annotated miRbase 16. This profiling investigation leads to the identification of the most relevant altered miRNAs based on an analysis of paired samples: bladder cancer tumor tissues (TT) versus adjacent healthy tissues (TN). It identified 8 down-regulated and 28 up-regulated miRNAs in the UMPh patient cohort (Additional file 1: Table S1). The Heatmap analysis presenting the altered miRNAs in tumor tissue for UMPh patient cohort is showed in Fig. 2a; the blue color denoting downregulation while the red color signifies overexpression.
Fig. 2.
The tissue-specific signatures of miRNAs in bladder cancer. a miRNA Heatmap emphasizing the altered miRNA signatures in 409 tumor tissue versus 19 healthy tissue taken from the TCGA bladder cancer patient cohort. b miRNA Heatmap highlighting the modified miRNA pattern when comparing tumor versus healthy tissues, representing the 23 paired bladder tissues from the UMPh patients’ cohort. c Venn diagram of the statistically determined (FC ± 2 and p-value ≤0.05) upregulated and downregulated miRNA expressions by overlapping the UMPh and TCGA patient cohorts; d Venn diagram of the UMPh and TCGA patient cohorts from the statistically determined (FC ± 2 and p-value≤0.05) upregulated and downregulated miRNA expressions exhibited in high grade tumor versus healthy tissues
To strengthen and improve the accuracy of the miRNAs profiling investigation, we performed an additional TCGA analysis of 409 tumors and 19 tumor-adjacent healthy tissues (Additional file 2: Table S2). The cut-off value was FC ± 2 and p-value≤0,05 for this analysis, from which 18 downregulated and 187 upregulated miRNAs were identified (Additional file 3: Table S3). The Heatmap for this data is shown in Fig. 2b. We overlapped the miRNAs profiling data obtained from the two datasets mentioned above. The Venn diagram represented in Fig. 2c shows 14 upregulated and 4 downregulated miRNAs. For the analysis of the high versus low-grade bladder cancers (UMPh: Additional file 4: Table S4; TCGA: Additional file 5: Table S5), only one commonality, the downregulation of miR-10a, was identified (Fig. 2d).
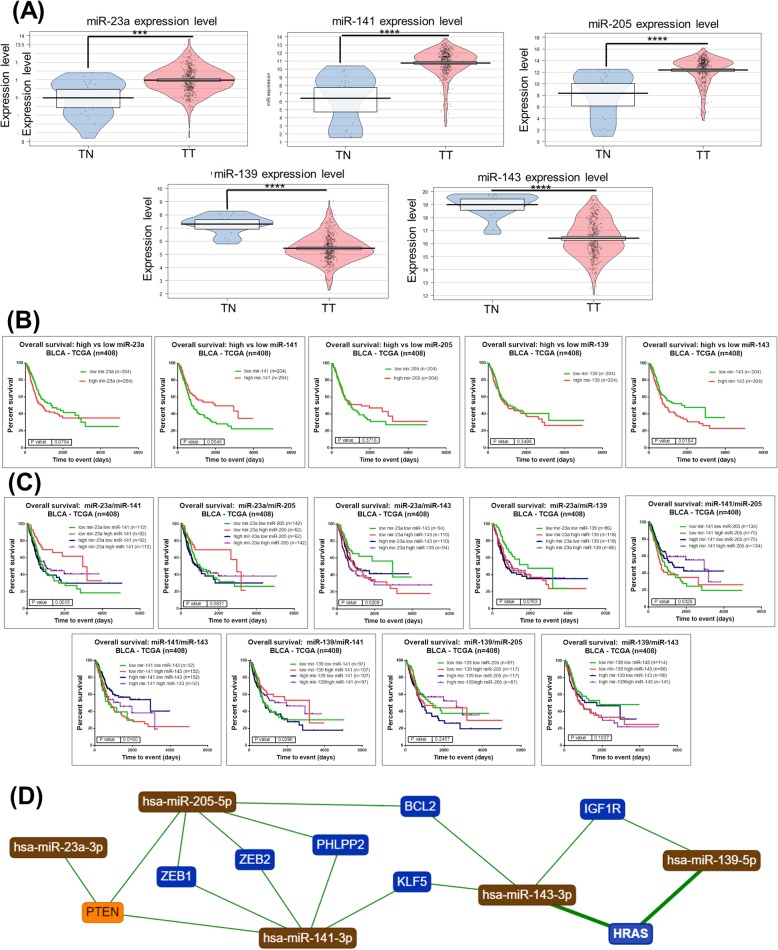
Given the context from the information displayed in Fig. 3, five miRNAs were selected for further validation based on the statistically significant FC obtained from our paired bladder tissues samples and the following criteria: the two most downregulated miRNAs (miR-143 and miR-139); the highly upregulated miRNAs related to EMT that have been less studied (miR-141 and miR-205); and one intermediary upregulated miRNA (miR-23a).
Fig. 3.
The respective miRNA expression level determined from the TCGA data on bladder cancer. a Expression levels presented as pirate plots based on TCGA data for five selected altered miRNA transcripts. b Kaplan–Meier survival curve for each selected miRNA transcript altered in bladder cancer based on TCGA data; c Kaplan–Meier survival curve for combinations of two miRNA transcripts from the five selected (*p ≤ 0.05, **p ≤ 0.01, ***P ≤ 0.001, Ns: not statistically significant, p > 0.05). d The network interconnection among the selected miRNA transcripts and genes generated using miRtargetLink (https://ccb-web.cs.uni-saarland.de/mirtargetlink/)
Figure 3a shows the Pirate plots that were generated using the TCGA data for the five selected miRNAs (overexpressed transcripts: miR-23a, miR-141 and miR-205; downregulated transcripts: miR-139 and miR-143). The related survival curves for each miRNA transcript can be seen Fig. 3b while Fig. 3c displays the survival curves for combinations of two miRNAs. We observed an overall superior survival rate for the following three cases: high expression of miR-141 (p = 0.0045); low expression of miR-143 (p = 0.0184); and low expression of miR-143 combined with high expression of miR-141 (p = 0.0163). These altered miRNAs are directly interconectd via key genes regulated apoptosis or invasion related genes as can be emphases by network generated using miRtargetlink (Fig. 3d).
qRT-PCR data validation
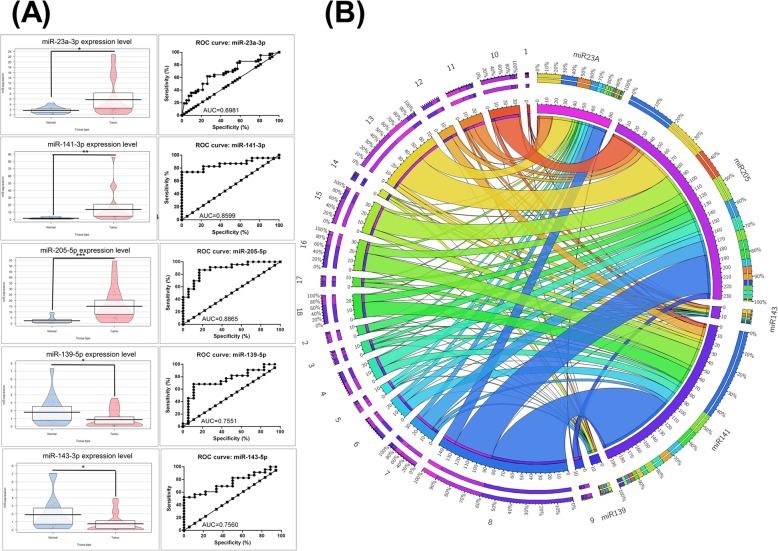
qRT-PCR is one of the most relevant and standardized approaches to validate microarray data. The new patient cohort used for the qRT-PCR consisted of 18 healthy tissues and 18 tumor tissues. From the common list of miRNAs between UMPh patients’ cohort and TCGA data, we selected two downregulated (miR-139-5p and miR-143-5p) and three up-regulated (miR-23a-3p, miR-141-3p, and miR-205-5p) to validate as independent prognostic markers. For miRNAs normalization, the ΔΔCt method with U6 as reference/normalizer; qRT-PCR validation data of the new additional patient cohort is presented in Fig. 4a. A ROC curve was constructed to estimate the sensitivity and specificity of these transcripts as biomarkers for bladder cancer, the highest AUC value being for miR-141 (0.8599) and miR-205 (0.8865). Circos diagram can be seen in Fig. 4b and the following observations were made: miR-205-5p and miR-141-3p are very heterogeneous providing expression level value, that should be considered as possible biomarker. miR-143-5p and miR-139-5p are very homogenous offering a potential advantage as biomarkers; and as expected with our relatively intermediary-expressed upregulated microRNA, miR-23a lies between the previously mentioned miRNAs.
Fig. 4.
qRT-PCR data validation. a Left panel of Pirate Plots presents the expression level for miR-23a-3p, miR-139-5p, miR-141-3p, miR-143-5p and miR-205-5p in the new validation set comprised of healthy (n = 18) and tumor tissues (n = 18). The data was normalized using U6 based on ΔΔCt method (*p < 0.05, **p < 0.01, ***p < 0.001). Right panel of graphs presents ROC curves for each selected miRNA’s specificity and sensitivity (ROC: receiver-operating characteristic, AUC: area under ROC curve). b Circos diagram representing the expression level for the validated miRNA transcripts in tumoral tissues of the new patient cohort
Mutation signatures are characteristic of bladder cancer tissues
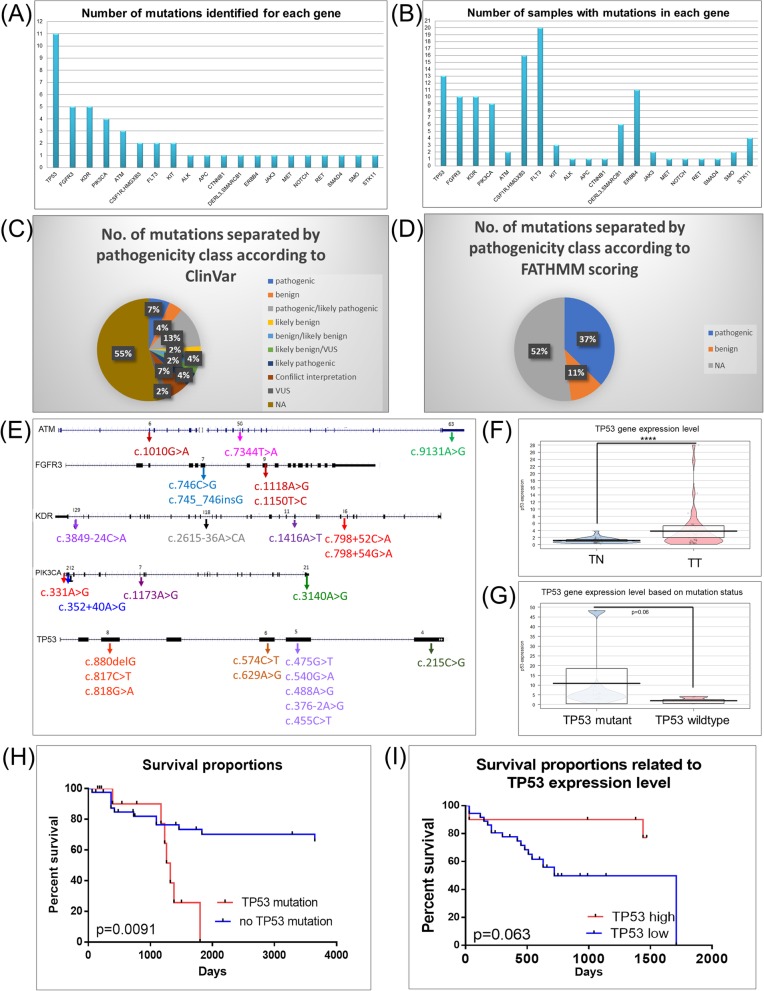
We analyzed the mutation patterns for tumor tissues from the UMPh cohort of 22 patients diagnosed with bladder cancer (Additional file 9: Table S6). An increased mutation rate is shown for TP53, FGFR3, KDR, PIK3CA and ATM. Figure 5a presents the number of mutations identified in each gene; meanwhile, Fig. 5b shows the mutations frequency for each gene in the analyzed samples. Using NCBI ClinVar or FATHMM, the mutations exhibited were classified based on pathogenicity, meaning as pathogenic, benign/non-pathogenic or not applicable/unknown (NA) (Fig. 5c). A significant percentage of the mutations were classified with unknown (NA) role (55%) and 7% are pathogenic based on the ClinVar classification. In comparison, using the FATHMM scoring 52% were classified as unknown (NA) while 37% of the mutations were classified as pathogenic (Fig. 5d). The mutated genes in bladder cancer have both intronic and exonic localizations. The mutation localizations for the most relevant genes (ATM, FGFR3, KDR, TP53 and PIK3CA) are presented in Fig. 5e. For the case of TP53, an increased survival rate for patients expressing TP53 wild-type versus those expressing mutant was observed; for the rest of the frequently mutated genes there was no statistically significant data. The frequency of mutations in our pattern exhibited is consistent with the frequent mutations in 200 bladder cancer cases from the TCGA; OncoGrid summarizes the 200 most mutated bladder cancer cases and top 50 mutated genes. From the TGCA data and the frequent mutated genes obtained, the overall survival rate for the case of patients expressing mutated and wild-type genes can be seen Additional file 6: Figure S1. No statistically significant correlation between the survival rate and mutational status was observed for AKT, ATM, CTNNB1, FGFR3, TP53, KIT, MET, PIK3CA and SMO.
Fig. 5.
Mutation patterns in bladder cancer patients evaluated using Ion Ampliseq Cancer panel kit. a Bar Graph of the number of mutations identified for each gene in the Ion Ampliseq Cancer kit; b Bar Graph of the mutation frequency of each gene in the Ion Torrent Cancer panel from the analyzed UMPh patient samples; c Pie graph of the identified mutations separated by pathogenicity according to ClinVar classification; d Pie graph of the identified mutations separated by pathogenicity according to FATHMM scoring and classification; e Localization of mutations in each of the gene sequences for the most frequent mutated genes in bladder cancer patients; f qRT-PCR evaluation of TP53 gene expression level in all healthy tissues (n = 41) and bladder tumor tissues (n = 41); this cohort means the 23 samples from the UMPh patient cohort and 18 samples from the validation set. g Pirate Plot of the TP53 expression level in wild-type and TP53-mutated samples; h Kaplan-Meier curve of overall survival rate based on mutation status for TP53 mutated gene from the UMPh patient cohort; i Kaplan-Meier curve of overall survival rate based on TP53 expression levels from the UMPh patient cohort
Evaluation of TP53 expression using qRT-PCR
The calculation of gene expression FC used the ΔΔCT method and B2M as the housekeeping gene (Fig. 5f). Figure 5f presents relative expression level of TP53 when comparing bladder tumor and normal tissues. There was no correlation among the expression level of TP53and their status (Fig. 5g). An additional graph of TP53 expression level using a median cut-off (50% above or low) to separate the expression levels into high or low and TP53 mutational status can be seen in Fig. 5e or based on the expression level (Fig. 5i). We observed an increase in survival rate for the case of TP53 wild type patients versus mutant TP53 (p = 0.0091).
Identification of miRNAs functional pathways related to bladder cancer tumorigenesis
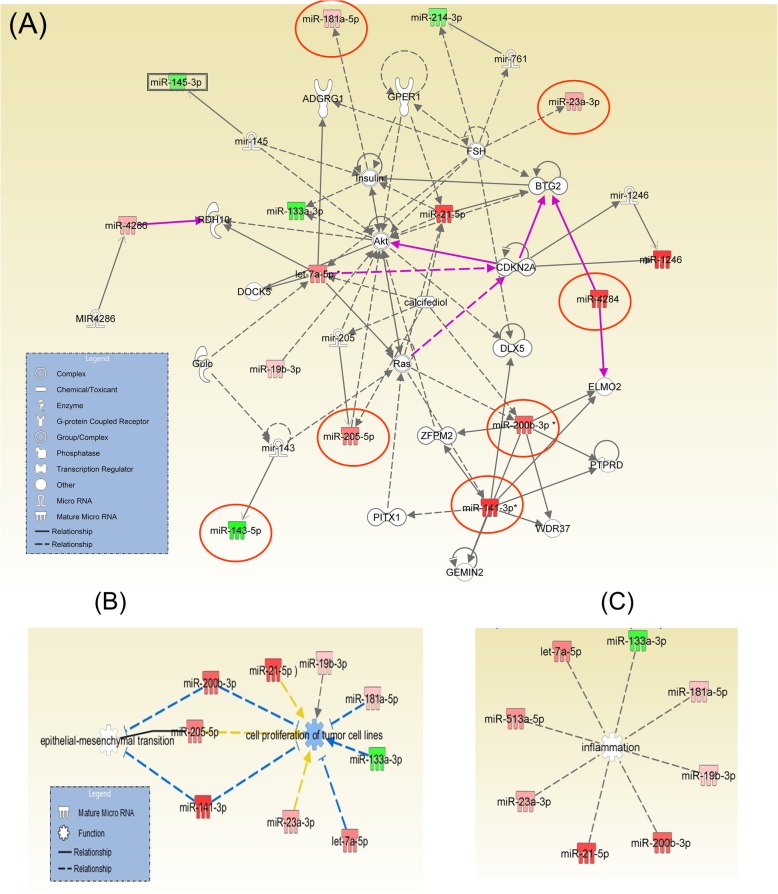
The miRNAs with an altered expression level were input into the specialized software IPA (Ingenuity Pathways Analysis). This analysis highlights the main altered networks indicating disease or biological function based on the miRNA signatures from the bladder cancer patients and the interconnection with target genes. Therefore, 9 relevant molecular networks were identified for bladder cancer as can be seen in Table 3. Furthermore, Table 4 shows the statistically significant biological, cellular and molecular functions as well as diseases that these altered miRNAs are associated to. This statistical correlation is based on the number of molecules altered within a given network. The data from Table 4 was used to generate graphical representation of molecular networks using IPA, which displays the nodal genes for the altered miRNAs in bladder cancer. This improves identification of miRNA-gene interactions and determines their nature as direct or indirect. Figure 6a presents an overlap of the most important networks in terms of the connection between the altered miRNAs and target genes. Altered miRNAs target genes critical to regulating tumour progression and cell fate are particularly important for patient prognosis. Once again, this highlights the connection of the uncovered genes TP53, AKT, RAS, CDKN2A or CDK2 to cell fate, as important regulators. These genes are interconnected with the validated miRNAs which are highlighted with red circles. Another important mechanism in carcinogenesis is the transition from epithelial to mesenchymal (EMT) phenotype (Fig. 6b). It was determined that AKT is directly related to ZEB2 that interacts with miR-205-5p, miR-141-3p and miR-200b-3p. These miRNAs have highest expression levels, extracted from the top altered transcripts of both patient cohorts (found in the TCGA as well as our experimental data). Thus, genes were identified that are involved in carcinogenesis or responsible to apoptotic resistance. These genes are recognized to have prognostic role in colorectal cancer (RAS) or related to the progression of bladder cancer (AKT). RAS and AKT have been shown to be related to miR-21. Figure 6c highlights the relevant transcripts related to inflammation, based on the literature information including the IPA.
Table 3.
The altered networks based on the miRNA signature of the bladder cancer participants, where the relevance score was generated using IPA. Each network integrates miRNAs along with the most relevant target genes, indicating disease or biological function, based on which the score is calculated
| ID | Top Diseases and Functions | Score | Focus Molecules | Molecules in Network |
|---|---|---|---|---|
| 1 | Organismal Injury and Abnormalities, Reproductive System Disease, Cancer | 29 | 12 | ↑let-7a-5p*, mir-143, mir-145, mir-205, mir-761, ↓miR-133a-3p, ↑miR-141-3p*, ↓miR-143-5p, ↓miR-145-3p, ↑miR-181a-5p, ↑miR-19b-3p, ↑miR-200b-3p*,↑ miR-205-5p, ↑miR-21-5p, ↓miR-214-3p, ↑miR-23a-3p, PITX1, PTPRD, Ras, RDH10, resolvin D1, WDR37, ZFPM2 |
| 2 | Cancer, Organismal Injury Abnormalities, Cell Cycle | 6 | 3 | BRF1, CDK2, Ck2, MIR3162, mir-25, mir-193, mir-506, ↓ miR-193a-5p, ↑miR-3162-5p, ↑miR-513a-5p, RNA polymerase iii, SSB, TP53 |
| 3 | Organismal Injury and Abnormalities, Reproductive System Disease, Developmental Disorder | 3 | 1 | mir-548, ↓miR-548q |
| 4 | Organismal Injury and Abnormalities, Reproductive System Disease, Cancer | 3 | 1 | mir-191, ↑miR-191-3p |
| 5 | Developmental Disorder, Hereditary Disorder, Organismal Injury, Abnormalities | 3 | 1 | miR-1225, ↑mir-1225-5p |
| 6 | Cancer, Cell Cycle, Cell Death and Survival | 3 | 1 | CDKN2A, mir-1246, ↑miR-1246 |
| 7 | Cancer, Gastrointestinal Disease, Organismal Injury and Abnormalities | 3 | 1 | MIR1234, MIR7170, ↑miR-1234-3P |
| 8 | Cancer, Gastrointestinal Disease, Organismal Injury and Abnormalities | 3 | 1 | mir-630, ↑miR-630, TMEM8B |
| 9 | Cancer, Connective Tissue Disorders, Hematological Disease | 2 | 1 | miR-4665, miR-6941, miR-7023, miR-7024, miR-7119, miR-1275, ↑miR-1275, YBX1 |
Table 4.
List of diseases and functions at the biological, molecular or cellular level that are the most representative of the altered molecules exhibited in bladder cancer. The selected IPA score based on p-value and number of molecules altered in bladder cancer; more specifically, p-values is calculated based on the number of molecules altered in a given pathway divided by the total number of molecules from a specific pathway or biological processes
| Function | Name | p-value | #Molecules |
|---|---|---|---|
| Top Diseases and Bio Functions | Organismal Injury and Abnormalities | 4.85E-02 – 5.80E-14 | 16 |
| Reproductive System Disease | 3.30E-02 – 5.80E-14 | 12 | |
| Cancer | 4.62E-02 – 1.75E-13 | 14 | |
| Inflammatory Disease | 1.49E-02 – 2.42E-08 | 9 | |
| Inflammatory Response | 4.64E-03 – 2.42E-08 | 8 | |
| Molecular and Cellular Functions | Cellular Development | 4.99E-02 – 1.60E-05 | 9 |
| Cellular Growth and Proliferation | 4.99E-02 – 1.60E-05 | 9 | |
| Cellular Movement | 2.15E-02 – 4.20E-05 | 5 | |
| Cell Death and Survival | 4.99E-02 – 1.55E-03 | 7 | |
| Cell Morphology | 2.15E-02 – 1.55E-03 | 3 |
Fig. 6.
The bladder cancer specific miRNA-gene regulatory network of the genes found by NGS and their miRNA target. a Overlapping of most relevant miRNA molecular networks involved in bladder cancer. The overexpressed miRNAs are highlighted in red, the downregulated miRNAs are marked in green, and the most relevant target genes determined from the literature are in grey. A solid connecting line represents a direct action while a dotted line signifies an indirect action. b IPA analysis for the identification of miRNAs related to epithelial to mesenchymal transition (EMT) (c) IPA analysis for identification of miRNAs related to inflammation
miRNA target genes involved in bladder cancer
In order to identify the biological significance of the altered miRNA expression for bladder cancer, miRTarBase was used to identify validated target genes. The target genes relating to the most upregulated miRNAs were grouped into specific families. These miRNA families have an important role in the regulation of translational processes and, through their identification, relevant hallmarks of cancer can be correlated. Using the miRnet database, we determined the initial miRNA-target interactions and represented the network in Additional file 7: Figure S2. Following this initial inquiry, we looked at every individual interaction between miRNA-target and these can be seen in Additional file 7: Figure S2.
In Additional file 8: Figure S3A, the target genes for miR-23a-3p involved in PI3K-AKT signaling pathway and tight junctions are presented, and respectively emphasized in red and blue. Additional file 7: Figure S2B shows the target genes for miR-139-5p involved in Chemokine signaling pathway colored as blue, while the targeted genes involved focal adhesion colored as red. Additional file 8: Figure S3C presents the target genes for miR-141-3p that regulate the cell cycle in blue and the most frequently mutated miRNA-regulated genes in cancer in red. Additional file 8: Figure S3D presents the target genes for miR-143-5p involved in focal adhesion in blue and the most frequently mutated miRNA-regulated genes in cancer in red. Additional file 8: Figure S3E presents the target genes for miR-205-5p involved in adherent junctions in yellow, focal adhesion in green, the most frequently modified cancer pathways in blue and the most frequently mutated miRNA-regulated genes in cancer in red. In its entirety, Additional file 8: Figure S3 emphasizes the important relationship between EMT and miRNAs in terms of the frequently mutated genes or dysregulated pathways; with the chosen miRNAs, governing some aspect of cell adhesion. Multiple core regulatory processes or signaling pathways have demonstrated to be of functionally relevance to bladder cancer tumor progression, particularly in terms of invasiveness and reoccurrence. This has essential impact for bladder cancer in terms of diagnosis and/or prognosis.
Network analysis of altered miRNA and frequent mutated genes
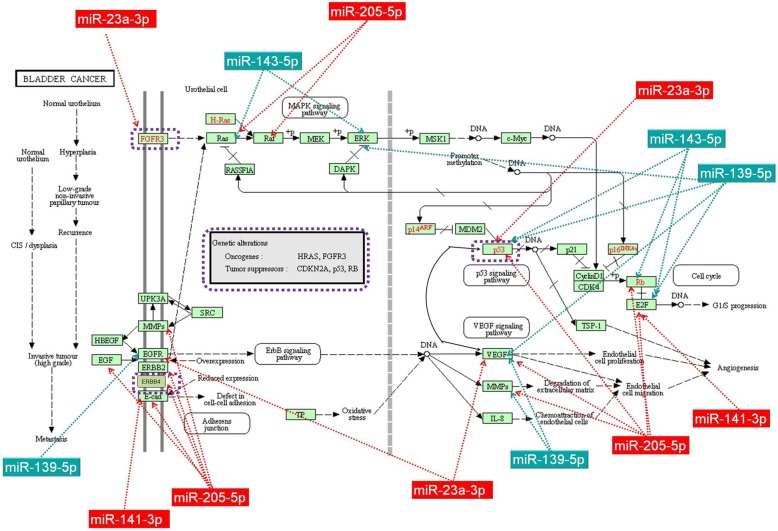
The interconnection between mutated genes and altered miRNAs evaluated can been seen in Fig. 7. As was expected, miRNAs are not isolated transcripts. This is exemplified by the large number of miRNAs, with altered expression levels in bladder cancer, connected to the most frequently mutated genes (TP53, FGFR3, KDR, PIK3CA and ATM). Figure 7 also emphasizes the direct or indirect mode of affecting the transcriptomic pattern, which includes other miRNAs. Alterations in some genes, including TP53, FGFR3, ATM, KDR and PIK3CA, have been shown to trigger carcinogenesis and become intertwined with a high number of altered miRNA transcripts in bladder cancer (Additional file 8: Figure S3).
Fig. 7.
MicroRNAs downregulated (miR-139-5p and miR-143-5p) and overexpressed (miR-23a-3p, miR-141-3p, and miR-205-5p) in bladder cancer target multiple genes involved in the bladder cancer carcinogenesis network as displayed by KEEG. The red rectangles represent the upregulated miRNAs while the dark green rectangles represent the downregulated miRNAs. This Fig. was developed based on the output of KEEG database
Discussions
Bladder cancer is a very heterogeneous disease, which makes the matched/paired samples advantageous in analyzing their molecular profile. This allows a miRNAs profiling evaluation from a limited number of patients; however, it is the overlapping with TCGA data that specifically permits the identification of the most representative altered transcripts. The importance to understanding any disease is not in the specific quantifications of certain transcriptomic information (like miRNA). Rather in how this transcriptomic information relates to specific genetic alterations and their intrinsic cellular mechanisms, which affect the equilibrium therein facilitating the progression of or toward the disease.
It was identified a specific miRNA pattern in bladder cancer, from which a selected subset of individual miRNAs and their combinations were associated with overall survival rate. This study represents the basis for developing miRNA expression signatures as diagnostic tools for BC and also encourages our comprehension of the miRNA function in the initiation or progression of disease. The integrated miRNA profiling of bladder carcinomas reported here extends our knowledge based on building a comprehensive characteristic miRNAs signature with clinical importance for prognosis or diagnosis.
Our data demonstrates consistent dysregulation of miRNAs in bladder cancer. Some of the altered miRNAs are exhibited in all the cancer types like the cases of let-7 family members [14–18]. Also, miR-21 is a commonly up-regulated miRNA in human cancers, which also appears to be consistent in our case. In bladder cancer in vitro models, miR-21 is able to regulate cell proliferation and migration via its cross talk with PTEN and TP53 [19], this is one of the most important pathways related to bladder carcinogenesis [20]. A recent study confirmed the altered expression level for miR-21, miR-205 and miR-200c in bladder cancer when analyzing data generated by pairing the tissue to plasma samples [21]. As the previous authors concluded, we did find miR-205 to be specific to bladder tumor tissue and, in addition, this miRNA was consistently upregulated.
In addition, we were able to identify specific miRNAs signatures for epithelial transition to mesenchymal state (EMT). EMT is a well-known process related to tumor invasion, drug resistance and potential to be stem cell-like [22]. These EMT related transcripts particularly regulate loss of adhesion, leading to increasing cell motility which facilitates metastasis. It was demonstrated that genes BCL2, CDH11, ZEB1/ZEB2 and TIMP2 are targeted respectively by miR-200b/c, miR-200c and miR-200c causing this regulatory effect on adhesion and indirect regulation of EMT [23–26], also emphasis in Fig. 3d.
There appears to be an inverse relationship between the expression of the miR-200 family, particularly miR-200c and miR-141, and the expression of E-cadherin and concurrently exhibiting significant downregulation of ZEB1 expression [27]. This provides an important therapeutic target worth investigating further explicitly for bladder cancer to re-enforce the adhesion between cells and hinder EMT. Detailed evaluation of the expression level of the miR-200 family (miR-141/200c and miR-200a/200b/429) has shown frequent upregulation in BC, but are frequently downregulated in other cancer types, like renal cancer [28]. MiR-200c expression level is associated with early stage T1 bladder tumor progression [29]. More specifically, miR-200c downregulation was connected with progression to muscle invasive bladder cancer and unfavorable prognosis [29]. Other miRNAs with an altered expression level in bladder cancer involved in EMT and invasion is miR-141. In multiple other pathologies [30], not only bladder cancer [31], miR-200c and miR-141 were considered prognostic markers [32]. It should be noted that for the range of bladder cancer samples studied, the expression levels of miR-205 and miR-200c are highly disputed [22, 33, 34], often reported as both downregulated [35] and overexpressed [17].
In a recent review paper, miR-145 was presented as a frequently miRNA downregulated in bladder cancer [27, 36, 37], which was confirmed by our data. miR-143/145 is presented as clustered in the literature with tumor suppressor function in a wide range of cancers, including bladder cancer [28, 38]. We found that miR-143 had a downregulated expression in the bladder tumor tissue and that it interacts with RAS, ERK, P53, Rb or E2F (Fig. 8). Some authors argue that the downregulated miR-143 is regulated by the oncoprotein EZH2, frequently overexpressed in bladder cancer, representing an important therapeutic target not only a biomarker [39, 40].
Several studies present miR-133b to be downregulated in bladder cancer, but also in other cancers [36, 41]. It not only targets the mechanism of apoptosis and cell proliferation [42], but also migration and invasions via EGFR (epidermal growth factor receptor) and its downstream or upstream effectors [43, 44]. Another one of our downregulated transcripts is miR-139, whose expression level, interaction with MMPs (Fig. 8) and associated gene network of focal adhesion (Additional file 7: Figure S2B) are consistent with literature data [45, 46].
For bladder cancer, a number of the altered miRNAs were proven to regulate TP53 network at multiple levels [28, 50], emphasizing the importance of TP53 status. TP53 overexpression (by IHC) was proven to be associated with the presence of mutation in this gene that leads to its inactivation. In turn, inactivation of TP53 promotes tumorigenesis in cancer cells and, not coincidently, is correlated to poor survival in human tumors. A study done by Puzio-Kuter et al. (2009) demonstrated that the combined inactivation of TP53 and p10 in bladder epithelium lead to an invasive cancer characteristic in mouse models [51]. The authors claimed that the synergy of deleting both p53 and p10 is mediated by deregulation of mTOR signaling. Specifically, TP53 was observed to interact with miR-200a, miR-214, miR-513 and miR-1225. Additionally, miR-19 provides a possible link between the TP53, KDR, ERBB4 and PIK3CA. Low-grade carcinoma usually have PIK3CA mutation and it progresses into high grade tumor after in inactivation CDKN2A [52]. The presence of specific SNP on the level of PIK3CA was related with bladder cancer risk. Therefore, recognizing and targeting genetic variations of the PI3K/AKT/mTOR pathway has an important clinical implication for bladder cancer prognosis [53].
TP53 mutation alteration of the receptors tyrosine kinase pathways are observed in our investigation, which is in agreement with literature date. This pathway is activated in around 40% bladder cancer cases (FGFR3: > 10%, EGFR:> 10%, ERBB2:~ 10%, ERBB3:~ 10%, NRAS/HRAS/KRAS:~ 10%, PTEN: ~ 10%, AKT3:~ 10%) [54]. TCGA data revealed 58 significantly mutated genes and high incidence of several genetic pathways [52]. In our study, an important difference between the overall survival of TP53 mutant and wild-type was confirmed; a fact established by previous studies on bladder cancers [55–57]. Literature data reports frequent alterations on the expression levels of FGFRs [7, 58], which if further clarified and has significant role in the response to therapy [59, 58]. In our case, a single exonic mutation (exonic: NM_000142.4) in 13/24(54%) was identified and determined to be pathogenic using the FATHMM web server. FGFR3 and TP53 have proven to have a higher incidence of mutation in our analyzed samples. Using miRnet database, we demonstrated (see Additional file 8: Figure S3.) that these genes do interact with several miRNAs that were altered in our investigation, however, the specifics of the interaction has yet to be discovered. Based on our results and using the KEGG database, in Fig. 8 we proposed interactions between the genes associated to BC carcinogenesis and our altered miRNAs expressions (▼ miR-139-5p; ▼ miR-143-5p; ▲ miR-23a-3p; ▲ miR-141-3p; ▲ miR-205-5p). In the end, it is important to highlight the complexity of the transcriptome and its interactions with the genome, especially if mutational status is added as another level of analysis.
Lastly, ERBB4 (the epidermal growth factor receptor 4) belong to the ERB family of growth factor receptor tyrosine kinases (TKI) being involved in the regulation of cell proliferation, cell differentiation, migration and invasion related in mechanism by two major pathways Ras/Raf/MAPK and PI3K/Akt/mTOR signaling [60]. The presence of this type of mutation was identified in 11 of our 23 paired samples analyzed and has a clinically important application in predicting the response to TKI inhibitors. ERBB4 mutated gene has been shown to be interconnected with the aforementioned miR-145 and miR-193a, but we identified only an intronic mutation, with no pathogenic effect.
Conclusions
Alteration of the miRNAs expression level affects tumor molecular phenotype. These changes are detectable indirectly through the pathways with which they exhibit BC; for example, the presence of some specific mutations in key genes found in these pathways. Genes are acted upon by miRNAs where altering their expression acts on a variety of functions that impact cancer development and prognosis. More specific examples can be found with the EMT related miRNAs (miR-141b, miR-200 s or miR-205) that have an important role in the molecular mechanisms, underlying key processes related to tumorigenesis, invasion and metastasis.
To further extend the complexity of interactions, miRNAs can simultaneously target multiple components of the same signaling pathway or of multiple signaling pathways. By integrating of relevant mutations with the altered miRNA expressions, a better insight into this complex network is attained which contributes to an enhanced understanding of bladder cancer tumorigenesis, progression and recurrence. The combined understanding of molecular data facilitates the identification of important targets at cellular level that will lead to new clinical and biological strategies to manage a personalized treatment in BC [61]. Although the numbers of cases were limited in this NGS study, we were able to demonstrate the aberrations in genes disturbing different signaling pathways, particularly those relating to the regulation of EMT. Moreover, we were able to demonstrate the crosstalk among the frequently mutated genes TP53, FGFR3, PIK3CA or ATM from a representative list of the altered miRNA transcripts (presented in Fig. 6 and Figure S4), serving as therapeutic intervention points or as potential biomarkers.
Supplementary information
Additional file 1: Table S1. The main altered miRNAs in the case of 23 matched paired samples (fold change±2, p-valued ≤0.05).
Additional file 2: Table S2. Demographic and histopathological characteristics of TCGA bladder cancer patient cohort.
Additional file 3: Table S3. The main altered miRNAs 409 tumours and 19 tumour-adjacent normal tissues retrieved from TCGA (fold change±2, p-valued ≤0.05).
Additional file 4: Table S4. miRNA with and altered expression level in high grade versus low grade bladder cancer- UMPh patient cohort.
Additional file 5: Table S5. miRNA with and altered expression level in high grade versus low grade bladder cancer- TCGA patient cohort.
Additional file 6: Figure S1. Bladder cancer survival curves showing the overall survival rate for TCGA bladder cancer patients for the most frequent mutated genes retrieved in our study, bases on mutation status.
Additional file 7: Figure S2. Target gene network generated using String10.5 based on the interactions from the miRTarBase database for (A) miR-23a-3p (B) miR-139-5p, (C) miR-141-3p, (D) miR-143-5p and (F) miR-205-5p.
Additional file 8: Figure S3. A Network analysis of the mutated genes in relation to their targeted miRNAs in bladder cancer. Schematic representation was obtained from miRnet (https://www.mirnet.ca/).
Additional file 9: Table S9. NGS sequencing data for bladder cancer.
Acknowledgements
Not applicable.
Abbreviations
- BC
Bladder cancer
- EMT
Epithelial to mesenchymal transition
- ERBB4
Epidermal growth factor receptor 4
- miRNAs
MicroRNAs
- TCGA
The Cancer Genome Atlas
- TKI
Tyrosine kinases receptor
- TN
Normal adjacent tissues
- TT
Tumor tissues
- TURBT
The transurethral resection of bladder tumour
- UMPh
The Iuliu Hatieganu University of Medicine and Pharmacy, Cluj-Napoca, Romania
Authors’ contributions
CB and RB wrote the paper; CB performed the microarray evaluation; RL, LAP, SC, LB, AJ, CC and LM performed the experiments. CB, RP and MB analyzed the experimental data. FD and BP furnished the biological samples and clinical data based. AI, BP and IBN assisted in the preparation of the manuscript and editing. IBN coordinated all the research activities, from the experiments design to final correction of the manuscript. All authors have read and approved the final manuscript.
Funding
This study was financed by the PN-II-PT-PCCA-2011-3.1-1221: “Intelligent Systems for Reccurence and Progression Prediction in Superficial Bladder Cancer Based on Artificial Intelligence and Microarray Data: Tumor mRNA and Plasma microRNA-IntelUro.” and PNCDI III 2015–2020 “Increasing the performance of scientific research and technology transfer in translational medicine through the formation of a new generation of young researchers” – ECHITAS, no. 29PFE/ 18.10. 2018.
Availability of data and materials
The TCGA material is public available. In the case of microarray plasma data are available on arrayexpress database (ID: E-MTAB-8356), will be public released immediately after the publication.
Ethics approval and consent to participate
All the samples were collected according to the national and international legislation approved by ethical committee of The Oncology Institute “Prof. Dr. Ion Chiricuta”. All the patients and the healthy volunteers signed the informed consent form; the samples were stored and processed anonymously in according to the biobanks regulation.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Cornelia Braicu and Rares Buiga contributed equally to this work.
Contributor Information
Cornelia Braicu, Email: braicucornelia@yahoo.com, Email: cornelia.braicu@umfcluj.ro.
Rares Buiga, Email: rares.buiga@umfcluj.ro.
Roxana Cojocneanu, Email: roxana.cojocneanu@umfcluj.ro.
Mihail Buse, Email: mihail.buse@umfcluj.ro.
Lajos Raduly, Email: raduly.lajos@umfcluj.ro.
Laura Ancuta Pop, Email: laura.pop@umfcluj.ro.
Sergiu Chira, Email: chira.sergiu@umfcluj.ro.
Liviuta Budisan, Email: liviuta.budisan@umfcluj.ro.
Ancuta Jurj, Email: anca.jurj@umfcluj.ro.
Cristina Ciocan, Email: cristina.ciocan@umfcluj.ro.
Lorand Magdo, Email: lorand.magdo@gmail.com.
Alexandru Irimie, Email: airimie@umfcluj.ro.
Florentin Dobrota, Email: florentin.dobrota@umfcluj.ro.
Bogdan Petrut, Email: bogdan.petrut@umfcluj.ro.
Ioana Berindan-Neagoe, Email: ioana.neagoe@umfcluj.ro.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13046-019-1406-6.
References
- 1.Enokida H, Yoshino H, Matsushita R, Nakagawa M. The role of microRNAs in bladder cancer. Investig Clin Urol. 2016;57:S60–S76. doi: 10.4111/icu.2016.57.S1.S60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Qie Y, Hu H, Tian D, Zhang Y, Xie L, Xu Y, Wu C. The value of extensive transurethral resection in the diagnosis and treatment of nonmuscle invasive bladder cancer with respect to recurrence at the first follow-up cystoscopy. Onco Targets Ther. 2016;9:2019–2025. doi: 10.2147/ott.s103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anastasiadis A, de Reijke TM. Best practice in the treatment of nonmuscle invasive bladder cancer. Ther Adv Urol. 2012;4:13–32. doi: 10.1177/1756287211431976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comperat EM, Burger M, Gontero P, Mostafid AH, Palou J, Roupret M, van Rhijn BWG, Shariat SF, Sylvester RJ, Zigeuner R, Babjuk M. Grading of urothelial carcinoma and the new “World Health Organisation classification of Tumours of the urinary system and male genital organs 2016”. Eur Urol Focus. 2018. 10.1016/j.euf.2018.01.003. [DOI] [PubMed]
- 6.Grignon DJ. The current classification of urothelial neoplasms. Mod Pathol. 2009;22:S60–SS9. doi: 10.1038/modpathol.2008.235. [DOI] [PubMed] [Google Scholar]
- 7.di Martino E, Tomlinson DC, Williams SV, Knowles MA. A place for precision medicine in bladder cancer: targeting the FGFRs. Future Oncol. 2016;12:2243–2263. doi: 10.2217/fon-2016-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braicu C, Cojocneanu-Petric R, Chira S, Truta A, Floares A, Petrut B, Achimas-Cadariu P, Berindan-Neagoe I. Clinical and pathological implications of miRNA in bladder cancer. Int J Nanomedicine. 2015;10:791–800. doi: 10.2147/ijn.s72904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braicu C, Calin GA, Berindan-Neagoe I. MicroRNAs and cancer therapy - from bystanders to major players. Curr Med Chem. 2013;20:3561–3573. doi: 10.2174/0929867311320290002. [DOI] [PubMed] [Google Scholar]
- 10.Braicu C, Catana C, Calin GA, Berindan-Neagoe I. NCRNA combined therapy as future treatment option for cancer. Curr Pharm Des. 2014;20:6565–6574. doi: 10.2174/1381612820666140826153529. [DOI] [PubMed] [Google Scholar]
- 11.Ling H, Pickard K, Ivan C, Isella C, Ikuo M, Mitter R, Spizzo R, Bullock MD, Braicu C, Pileczki V, Vincent K, Pichler M, Stiegelbauer V, et al. The clinical and biological significance of MIR-224 expression in colorectal cancer metastasis. Gut. 2016;65:977–989. doi: 10.1136/gutjnl-2015-309372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pop LA, Pileczki V, Cojocneanu-Petric RM, Petrut B, Braicu C, Jurj AM, Buiga R, Achimas-Cadariu P, Berindan-Neagoe I. Normalization of gene expression measurement of tissue samples obtained by transurethral resection of bladder tumors. Onco Targets Ther. 2016;9:3369–3380. doi: 10.2147/ott.s97519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kriebel S, Schmidt D, Holdenrieder S, Goltz D, Kristiansen G, Moritz R, Fisang C, Müller SC, Ellinger J. Analysis of tissue and serum MicroRNA expression in patients with upper urinary tract urothelial Cancer. PLoS One. 2015;10:e0117284. doi: 10.1371/journal.pone.0117284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Liu H, Lai C, Du X, Su Z, Gao S. The Lin28/let-7a/c-Myc pathway plays a role in non-muscle invasive bladder cancer. Cell Tissue Res. 2013;354:533–541. doi: 10.1007/s00441-013-1715-6. [DOI] [PubMed] [Google Scholar]
- 15.Kozinn SI, Harty NJ, Delong JM, Deliyiannis C, Logvinenko T, Summerhayes IC, Libertino JA, Holway AH, Rieger-Christ KM. MicroRNA Profile to Predict Gemcitabine Resistance in Bladder Carcinoma Cell Lines. Genes Cancer. 2013;4:61–69. doi: 10.1177/1947601913484495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boyerinas B, Park SM, Hau A, Murmann AE, Peter ME. The role of let-7 in cell differentiation and cancer. Endocr Relat Cancer. 2010;17:F19–F36. doi: 10.1677/erc-09-0184. [DOI] [PubMed] [Google Scholar]
- 17.Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, Gomella LG, Croce CM, Baffa R. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 18.Jérôme T, Laurie P, Louis B, Pierre C. Enjoy the Silence: The Story of let-7 MicroRNA and Cancer. Current Genomics. 2007;8:229–233. doi: 10.2174/138920207781386933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei M, Xie W, Sun E, Sun Y, Tian D, Liu C, Han R, Li N, Liu M, Han R, Liu L. microRNA-21 regulates cell proliferation and migration and cross talk with PTEN and p53 in bladder Cancer. DNA Cell Biol. 2015;34:626–632. doi: 10.1089/dna.2015.2868. [DOI] [PubMed] [Google Scholar]
- 20.Calderaro J, Rebouissou S, de Koning L, Masmoudi A, Herault A, Dubois T, Maille P, Soyeux P, Sibony M, de la Taille A, Vordos D, Lebret T, Radvanyi F, et al. PI3K/AKT pathway activation in bladder carcinogenesis. Int J Cancer. 2014;134:1776–1784. doi: 10.1002/ijc.28518. [DOI] [PubMed] [Google Scholar]
- 21.Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer. 2015;14:194. doi: 10.1186/s12943-015-0466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran MN, Choi W, Wszolek MF, Navai N, Lee IL, Nitti G, Wen S, Flores ER, Siefker-Radtke A, Czerniak B, Dinney C, Barton M, McConkey DJ. The p63 protein isoform DeltaNp63alpha inhibits epithelial-mesenchymal transition in human bladder cancer cells: role of MIR-205. J Biol Chem. 2013;288:3275–3288. doi: 10.1074/jbc.M112.408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J-Y, Yun SJ, Jeong P, Piao X-M, Kim Y-H, Kim J, Subramaniyam S, Byun YJ, Kang HW, Seo SP, Kim J, Kim JM, Yoo ES, et al. Identification of differentially expressed miRNAs and miRNA-targeted genes in bladder cancer. Oncotarget. 2018;9:27656–27666. doi: 10.18632/oncotarget.24441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinez-Fernandez M, Duenas M, Feber A, Segovia C, Garcia-Escudero R, Rubio C, Lopez-Calderon FF, Diaz-Garcia C, Villacampa F, Duarte J, Gomez-Rodriguez MJ, Castellano D, Rodriguez-Peralto JL, et al. A Polycomb-mir200 loop regulates clinical outcome in bladder cancer. Oncotarget. 2015;6:42258–42275. doi: 10.18632/oncotarget.5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, Bar-Eli M, Dinney C. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–5072. doi: 10.1158/1078-0432.ccr-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gulei D, Mehterov N, Ling H, Stanta G, Braicu C, Berindan-Neagoe I. The “good-cop bad-cop” TGF-beta role in breast cancer modulated by non-coding RNAs. Biochim Biophys Acta. 1861;2017:1661–1675. doi: 10.1016/j.bbagen.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Tamagawa S, Beder LB, Hotomi M, Gunduz M, Yata K, Grenman R, Yamanaka N. Role of miR-200c/miR-141 in the regulation of epithelial-mesenchymal transition and migration in head and neck squamous cell carcinoma. Int J Mol Med. 2014;33:879–886. doi: 10.3892/ijmm.2014.1625. [DOI] [PubMed] [Google Scholar]
- 28.Kurozumi A, Goto Y, Okato A, Ichikawa T, Seki N. Aberrantly expressed microRNAs in bladder cancer and renal cell carcinoma. J Hum Genet. 2016. 10.1038/jhg.2016.84. [DOI] [PubMed]
- 29.Wiklund ED, Bramsen JB, Hulf T, Dyrskjot L, Ramanathan R, Hansen TB, Villadsen SB, Gao S, Ostenfeld MS, Borre M, Peter ME, Orntoft TF, Kjems J, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011;128:1327–1334. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 30.Mahdavinezhad A, Mousavi-Bahar SH, Poorolajal J, Yadegarazari R, Jafari M, Shabab N, Saidijam M. Evaluation of miR-141, miR-200c, miR-30b Expression and Clinicopathological Features of Bladder Cancer. Int J Mol Cell Med. 2015;4:32–39. [PMC free article] [PubMed] [Google Scholar]
- 31.Liu G, Chen Z, Danilova IG, Bolkov MA, Tuzankina IA, Liu G. Identification of miR-200c and miR141-mediated lncRNA-mRNA Crosstalks in muscle-invasive bladder Cancer subtypes. Front Genet. 2018;9:422. doi: 10.3389/fgene.2018.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amuran GG, Eyuboglu IP, Tinay I, Akkiprik M. New Insights in Bladder Cancer Diagnosis: Urinary miRNAs and Proteins. Med Sci (Basel, Switzerland) 2018;6:113. doi: 10.3390/medsci6040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun X, Du P, Yuan W, Du Z, Yu M, Yu X, Hu T. Long non-coding RNA HOTAIR regulates cyclin J via inhibition of microRNA-205 expression in bladder cancer. Cell Death Dis. 2015;6:e1907. doi: 10.1038/cddis.2015.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahdavinezhad A, Mousavibahar SH, Poorolajal J, Yadegarazari R, Jafari M, Shabab N, Saidijam M. Association between tissue miR-141, miR-200c and miR-30b and bladder cancer: a matched case-control study. Urol J. 2015;12:2010–2013. [PubMed] [Google Scholar]
- 35.Catto JW, Alcaraz A, Bjartell AS, De Vere WR, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L, Schlomm T, Visakorpi T. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59:671–681. doi: 10.1016/j.eururo.2011.01.044. [DOI] [PubMed] [Google Scholar]
- 36.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, Kawahara K, Toki K, Kawakami K, Nishiyama K, Tsujimoto G, Nakagawa M, Seki N. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–352. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 37.Zaravinos A, Radojicic J, Lambrou GI, Volanis D, Delakas D, Stathopoulos EN, Spandidos DA. Expression of miRNAs involved in angiogenesis, tumor cell proliferation, tumor suppressor inhibition, epithelial-mesenchymal transition and activation of metastasis in bladder cancer. J Urol. 2012;188:615–623. doi: 10.1016/j.juro.2012.03.122. [DOI] [PubMed] [Google Scholar]
- 38.Wu J, Huang Q, Meng D, Huang M, Li C, Qin T. A functional rs353293 polymorphism in the promoter of miR-143/145 is associated with a reduced risk of bladder Cancer. PLoS One. 2016;11:e0159115. doi: 10.1371/journal.pone.0159115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Q, Zhao W, Ye C, Zhuang J, Chang C, Li Y, Huang X, Shen L, Li Y, Cui Y, Song J, Shen B, Eliaz I, et al. Honokiol inhibits bladder tumor growth by suppressing EZH2/miR-143 axis. Oncotarget. 2015;6:37335–37348. doi: 10.18632/oncotarget.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Motawi TK, Rizk SM, Ibrahim TM, Ibrahim IA. Circulating microRNAs, miR-92a, miR-100 and miR-143, as non-invasive biomarkers for bladder cancer diagnosis. Cell Biochem Funct. 2016;34:142–148. doi: 10.1002/cbf.3171. [DOI] [PubMed] [Google Scholar]
- 41.Chen X, Wu B, Xu Z, Li S, Tan S, Liu X, Wang K. Downregulation of miR-133b predict progression and poor prognosis in patients with urothelial carcinoma of bladder. Cancer Med. 2016;5:1856–1862. doi: 10.1002/cam4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen XN, Wang KF, Xu ZQ, Li SJ, Liu Q, Fu DH, Wang X, Wu B. MiR-133b regulates bladder cancer cell proliferation and apoptosis by targeting Bcl-w and Akt1. Cancer Cell Int. 2014;14:70. doi: 10.1186/s12935-014-0070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Wu D, Tao J, Qu P, Zhou Z, Hou J. MicroRNA-133 inhibits cell proliferation, migration and invasion by targeting epidermal growth factor receptor and its downstream effector proteins in bladder cancer. Scand J Urol. 2013;47:423–432. doi: 10.3109/00365599.2012.748821. [DOI] [PubMed] [Google Scholar]
- 44.Tao J, Wu D, Xu B, Qian W, Li P, Lu Q, Yin C, Zhang W. microRNA-133 inhibits cell proliferation, migration and invasion in prostate cancer cells by targeting the epidermal growth factor receptor. Oncol Rep. 2012;27:1967–1975. doi: 10.3892/or.2012.1711. [DOI] [PubMed] [Google Scholar]
- 45.Yonemori M, Seki N, Yoshino H, Matsushita R, Miyamoto K, Nakagawa M, Enokida H. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci. 2016;107:1233–1242. doi: 10.1111/cas.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jia AY, Castillo-Martin M, Domingo-Domenech J, Bonal DM, Sanchez-Carbayo M, Silva JM, Cordon-Cardo C. A common MicroRNA signature consisting of miR-133a, miR-139-3p, and miR-142-3p clusters bladder carcinoma in situ with normal umbrella cells. Am J Pathol. 2013;182:1171–1179. doi: 10.1016/j.ajpath.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ratert N, Meyer HA, Jung M, Mollenkopf HJ, Wagner I, Miller K, Kilic E, Erbersdobler A, Weikert S, Jung K. Reference miRNAs for miRNAome analysis of urothelial carcinomas. PLoS One. 2012;7:e39309. doi: 10.1371/journal.pone.0039309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding D, Zhang Y, Yang R, Wang X, Ji G, Huo L, Shao Z, Li X. miR-940 Suppresses Tumor Cell Invasion and Migration via Regulation of CXCR2 in Hepatocellular Carcinoma. Biomed Res Int. 2016;2016:7618342. doi: 10.1155/2016/7618342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou Q, Zheng X, Chen L, Xu B, Yang X, Jiang J, Wu C. Smad2/3/4 pathway contributes to TGF-beta-induced MiRNA-181b expression to promote gastric Cancer metastasis by targeting Timp3. Cell Physiol Biochem. 2016;39:453–466. doi: 10.1159/000445638. [DOI] [PubMed] [Google Scholar]
- 50.Liu J, Zhang C, Zhao Y, Feng Z. MicroRNA control of p53. J Cell Biochem. 2017;118:7–14. doi: 10.1002/jcb.25609. [DOI] [PubMed] [Google Scholar]
- 51.Puzio-Kuter AM, Castillo-Martin M, Kinkade CW, Wang X, Shen TH, Matos T, Shen MM, Cordon-Cardo C, Abate-Shen C. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes Dev. 2009;23:675–680. doi: 10.1101/gad.1772909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inamura K. Bladder Cancer: New Insights into Its Molecular Pathology. Cancers. 2018;10:100. doi: 10.3390/cancers10040100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bizhani F, Hashemi M, Danesh H, Nouralizadeh A, Narouie B, Bahari G, Ghavami S. Association between single nucleotide polymorphisms in the PI3K/AKT/mTOR pathway and bladder cancer risk in a sample of Iranian population. EXCLI J. 2018;17:3–13. doi: 10.17179/excli2017-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seok Ju Y. The mutational signatures and molecular alterations of bladder cancer. Translational Cancer Research. 2017;2017:S689–701.
- 55.Bodoor K, Al-Ghabkari A, Matalka I, Haddad Y, Alkhateeb A, Jaradat S, Jaradat ZW, Al-Ghazo M, Abu-sheikha A, Jalboush SA, Jarun Y. Assessment of p53 mutations, expression and prognosis in bladder cancer patients from Jordan: Identification of novel deletion mutations in the DNA-binding domain. Meta Gene. 2017;12:33–42. doi: 10.1016/j.mgene.2017.01.002. [DOI] [Google Scholar]
- 56.da Silva GN, Filoni LT, Salvadori MC, Salvadori DMF. Gemcitabine/cisplatin treatment induces concomitant SERTAD1, CDKN2B and GADD45A modulation and cellular changes in bladder Cancer cells regardless of the site of TP53 mutation. Pathol Oncol Res. 2018;24:407–417. doi: 10.1007/s12253-017-0255-x. [DOI] [PubMed] [Google Scholar]
- 57.Traczyk-Borszynska M, Borkowska E, Jablonowski Z, Jedrzejczyk A, Pietrusinski M, Kaluzewski B, Sosnowski M, Borowiec M. Genetic diversity of urinary bladder cancer and the risk of recurrence based on mutation analysis. Neoplasma. 2016;63:952–960. doi: 10.4149/neo_2016_614. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez-Vida A, Saggese M, Hughes S, Rudman S, Chowdhury S, Smith NR, Lawrence P, Rooney C, Dougherty B, Landers D, Kilgour E, Arkenau HT. Complexity of FGFR signalling in metastatic urothelial cancer. J Hematol Oncol. 2015;8:119. doi: 10.1186/s13045-015-0221-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kang HW, Kim YH, Jeong P, Park C, Kim WT, Ryu DH, Cha EJ, Ha YS, Kim TH, Kwon TG, Moon SK, Choi YH, Yun SJ, et al. Expression levels of FGFR3 as a prognostic marker for the progression of primary pT1 bladder cancer and its association with mutation status. Oncol Lett. 2017;14:3817–3824. doi: 10.3892/ol.2017.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Martino M, Zhuang D, Klatte T, Rieken M, Rouprêt M, Xylinas E, Clozel T, Krzywinski M, Elemento O, Shariat SF. Impact of ERBB2 mutations on in vitro sensitivity of bladder cancer to lapatinib. Cancer Biol Ther. 2014;15:1239–1247. doi: 10.4161/cbt.29687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopez-Beltran A, Santoni M, Massari F, Ciccarese C, Tortora G, Cheng L, Moch H, Scarpelli M, Reymundo C, Montironi R. Bladder cancer: molecular determinants of personalized therapy. Curr Drug Targets. 2015;16:115–124. doi: 10.2174/1389450116666150204115756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The main altered miRNAs in the case of 23 matched paired samples (fold change±2, p-valued ≤0.05).
Additional file 2: Table S2. Demographic and histopathological characteristics of TCGA bladder cancer patient cohort.
Additional file 3: Table S3. The main altered miRNAs 409 tumours and 19 tumour-adjacent normal tissues retrieved from TCGA (fold change±2, p-valued ≤0.05).
Additional file 4: Table S4. miRNA with and altered expression level in high grade versus low grade bladder cancer- UMPh patient cohort.
Additional file 5: Table S5. miRNA with and altered expression level in high grade versus low grade bladder cancer- TCGA patient cohort.
Additional file 6: Figure S1. Bladder cancer survival curves showing the overall survival rate for TCGA bladder cancer patients for the most frequent mutated genes retrieved in our study, bases on mutation status.
Additional file 7: Figure S2. Target gene network generated using String10.5 based on the interactions from the miRTarBase database for (A) miR-23a-3p (B) miR-139-5p, (C) miR-141-3p, (D) miR-143-5p and (F) miR-205-5p.
Additional file 8: Figure S3. A Network analysis of the mutated genes in relation to their targeted miRNAs in bladder cancer. Schematic representation was obtained from miRnet (https://www.mirnet.ca/).
Additional file 9: Table S9. NGS sequencing data for bladder cancer.
Data Availability Statement
The TCGA material is public available. In the case of microarray plasma data are available on arrayexpress database (ID: E-MTAB-8356), will be public released immediately after the publication.