Abstract
Background
Klebsiella pneumoniae-induced pyogenic liver abscess (KP-PLA) has emerged as a life-threatening disease worldwide. However, to date, a limited number of scholars have attempted to systematically elucidate the characteristics of KP-PLA. The aim of the present study was to analyze clinical, microbiological, and molecular epidemiological characteristics of KP-PLA patients in Southeastern China.
Methods
The KP-PLA cases from a tertiary teaching hospital in China from January 2016 to December 2017 were systemically studied and elucidated comprehensively. The virulence factors, resistant spectrum, and clones of K. pneumoniae isolates were identified with string test, polymerase chain reaction (PCR), antimicrobial susceptibility test, and multilocus sequence typing. Moreover, the characteristics in KP-PLA patients with and without other hepatobiliary diseases (OHD) were also been compared.
Results
A total of 163 KP-PLA cases were enrolled, in which the majority of those cases were senior males, and often associated with multiple underlying diseases, including diabetes (49.7%). The remaining cases belonged to healthy individuals (50.3%). The clinical symptoms were common but nonspecific, characterized by increased inflammatory parameters and abnormal liver function parameters. The abscess was often right-sided solitary presentation (58.3%). Cephalosporin or carbapenem plus metronidazole combined with percutaneous puncture or catheter drainage were favorable therapeutics. Although low resistance rates of commonly used antimicrobial drugs (< 10%) were observed, twelve strains were identified as multidrug resistant (MDR) strains, and were mainly isolated from the OHD patients. The hypermucoviscosity, as well as K1 and K2 serotypes accounted for 30.7, 40.5, and 19.0%, respectively. Except for iroN (24.5%) and magA (45.4%), the high prevalence of virulence genes (e.g. aerobactin, rmpA, mrkD, fimH, uge, ureA, entB, ybtA, kfuBC, and wcaG) was identified (68.7–100.0%). Additionally, ST23 was found as a predominant sequence type (ST; 38.7%), and three novel STs (ST3507, ST3508 and ST3509) were noted as well.
Conclusions
The present study reported the abundant hvKp strains in KP-PLA, as well as convergence of hypervirulent and MDR K. pneumoniae isolates from the KP-PLA patients, particularly those cases with OHD. Given the various clinical manifestations and destructive pathogenicity, determination of the comprehensive characteristics of such isolates is highly essential to effectively carry out for optimal management and treatment of KP-PLA.
Keywords: Pyogenic liver abscess, Clinical characteristics, Hypervirulent K. pneumoniae, Virulence factors, Sequence type, Multidrug resistant
Background
Pyogenic liver abscess (PLA) is a potentially life-threatening suppurating infection of hepatic parenchyma disease, which was frequently observed worldwide [1–3]. The morbidity of PLA has remarkably increased in Asian regions, e.g. Taiwan, where the annual incidence has gradually increased from 10.83 to 15.45 cases per 100,000 population since 2000 to 2011, and the mortality is up to 2–19% [4, 5]. PLA is often concomitant with numerous underlying hepatobiliary diseases that may lead to a higher overall rate of bacterial colonization [6]. Presenting symptoms of PLA are multiple and low specific, including fever, right upper abdominal pain, vomiting, nausea, and asthenia, thereby indicating a major medical challenge in diagnosis and therapeutic management of PLA [4, 6].
In terms of causative pathogens, Klebsiella pneumoniae has found as a predominant pathogen, accounting for 50–88% of PLA patients who lived Asia during the past two decades [2, 7]. Notably, K. pneumoniae isolating from PLA mainly associates with hypervirulence, with distinct clinical manifestations, as well as phenotypic and genotypic characteristics [1]. In a recently conducted study, Ye et al. reported that 90.9% of the pathogens causing PLA were hypervirulent K. pneumoniae (hvKp) strains, and there were correlations between the incidences of PLA and high prevalence of hvKp strains [8, 9]. In contrast to classic K. pneumoniae (cKp), the emerging variant, which was first reported in Taiwan in 1986, exhibited hypermucoviscosity, unique capsular serotype, virulence gene, sequence type (ST) and resistant spectrum [3]. In addition, hvKp-induced PLA may occur in young and healthy individuals, and then, migrate to distant sites, thereby leading to extrahepatic complications, such as endophthalmitis, meningitis, and necrotizing fasciitis [1, 3]. To date, K. pneumoniae-induced pyogenic liver abscess (KP-PLA) has become a global destructive disease. Therefore, systemic investigations on the clinical and microbiological characteristics of recently emerged KP-PLA population are highly essential, for making a comparison with the previously reported ones.
The hypermucoviscosity confirmed by a positive string test is one of the major virulence factors of K. pneumoniae in PLA, and the isolates are mainly taken as hvKp strains into consideration. Moreover, K. pneumoniae strains isolated from PLA are often K1 or K2 capsular serotype [8, 9]. Capsule, consisting of polysaccharide termed K antigen, is associated with virulence. K1 and K2 serotype isolates are especially hypervirulent due to their capability to confer resistance to phagocytosis and intracellular killing by neutrophils in serum. In addition to these two serotypes, K5, K20, K54, and K57 capsular serotype isolates are also hypervirulent as well [10, 11]. Furthermore, multiple virulence genes regarded as severe virulence determinants of K. pneumoniae are highly prevalent in KP-PLA. In previous studies, rmpA was shown to act on synthesis of capsular polysaccharide in order to regulate mucoid phenotype [1, 12]. This gene was reported present in 87.5% of K. pneumoniae strains in the PLA [12]. Additionally, aerobactin, a dominant iron siderophore, was taken as a critical virulence factor into account in K. pneumoniae, as well as being further prevalent in KP-PLA [8, 13]. Moreover, it was found in 93–100% of hvKp strains, and could enhance the virulence by 100-fold according to the mouse lethality test. As a result, it was even regarded as a marker gene of hvKp [14]. Apart from these two genes, other virulence genes of K. pneumoniae isolates include iroN, kfuBC, ybtA, entB (encoding siderophores), wcaG, magA, uge (capsule-associated genes), fimH (encoding type 1 fimbriae), mrkD (encoding type 3 fimbriae), and ureA (encoding nitrogen) [15–18]. However, the prevalence of these multitudinous virulence factors in KP-PLA has not been systematically evaluated. Moreover, multilocus sequence typing (MLST) is advantageous for categorizing strains in molecular epidemiological studies, and ST23 has been found as the most common sequence type in KP-PLA [3, 16]. In addition, K. pneumoniae strains in PLA are sensitive to the majority of antimicrobial agents [19]. However, antibiotics-resistant hvKp strains have been reported by numerous studies [6, 19]. The convergence of virulence and resistance in isolates from KP-PLA is still little-known, which might potentially lead to a poor prognosis and even serious clinical crises.
Despite the growth of prevalence and strong pathogenicity of KP-PLA, a limited number of cases have been reported in China, and the existing ones only described a certain aspect of characteristics [16, 20]. In the present study, a retrospective study was carried out, which systemically investigated and comprehensively assessed the clinical, microbiological, and molecular epidemiological characteristics of 163 KP-PLA cases from a tertiary teaching hospital in Southeastern China from January 2016 to December 2017. Furthermore, we compared the characteristics between KP-PLA patients with and without other hepatobiliary diseases (OHD). The present study may contribute to improve our understanding on KP-PLA, and also provide significant insights for the development of further effective therapeutic strategies for clinical trials.
Materials and methods
Study subjects
This retrospective study was conducted on KP-PLA patients at the First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China) from June 1, 2016 to December 31, 2017. This hospital is a 4100-bed major tertiary teaching hospital located in Southeast China (Wenzhou, China) with an annual admission of more than 160,000 inpatients. The diagnosis of KP-PLA was conducted based on the following criteria: (i) clinical features; (ii) imaging evidences; (iii) presence of K. pneumoniae in blood or pus culture; (iv) evidence of percutaneous puncture or surgical treatment, and (v) exclusion of amoebic, tuberculous liver abscess [21]. Then, the enrolled cases were divided into two groups: OHD and non-OHD patients. OHD included any hepatobiliary diseases except for PLA.
Study data
The medical records were reviewed to integrally collect the data of KP-PLA patients during the study period. The study data included the following variables: demographic characteristics (age and gender), underlying or concomitant conditions (diabetes mellitus, biliary disease, hepatitis and cirrhosis, fatty liver, history of intra-abdominal trauma or surgery, malignancy, and hypertension), clinical symptoms, length of stay in hospital, admission temperature, admission to intensive care unit (ICU), imaging findings, laboratory values (C-reactive protein [CRP], white blood cell [WBC] count, red blood cell [RBC] count, alanine aminotransferase [ALT], aspartate aminotransferase [AST], albumin [ALB]), invasive procedure, details of therapeutic approach, drainage ways, abscess prognosis, and outcomes at discharge. The abscess prognosis was determined according to the criteria based on clinical symptoms and abscess changes, which were stipulated by the Chinese Academy of Medical Sciences.
Clinical K. pneumoniae isolates
Initial strains were isolated from sterile fluid including pus, blood, and drainage fluid of KP-PLA patients and identified as K. pneumoniae by a matrix-assisted laser desorption/ionization time-of-flight mass spectrometry system. Antimicrobial susceptibility test for K. pneumoniae isolates was conducted by bioMerieux VITEK-2 (BioMérieux, Marcy-l’Étoile, France). The tested antibiotics included ampicillin, ampicillin/sulbactam, cefazolin, cefotetan, aztreonam, ceftriaxone, ceftazidime, cefepime, cefoperazone/sulbactam, ertapenem, imipenem, ciprofloxacin, levofloxacin, gentamicin, tobramycin, amikacin, sulfamethoxazole/trimethoprim, and nitrofurantoin. The results were interpreted by the latest guidelines published by the Clinical and Laboratory Standards Institute (CLSI; Pittsburgh, PA, USA). Multidrug resistant (MDR) strains were defined as non-susceptible to three or more different antimicrobial categories. Then, these strains were stored at − 80 °C for further research.
String test
The bacterial colony of K. pneumoniae strains on the agar plate was stretched by an inoculation loop. The string test would be considered as positive, representing hypermucoviscosity when a strain generated a viscous string with a length of > 5 mm [22].
Polymerase chain reaction (PCR) for capsular serotypes and virulence genes
Crude genomic DNA was extracted from K. pneumoniae strains. Subsequently, capsular serotype-specific genes (for serotypes of K1, K2, K5, K20, K54, and K57) and virulence genes (e.g. aerobactin, rmpA, iroN, kfuBC, ybtA, entB, wcaG, magA, uge, fimH, mrkD, ureA) were amplified by PCR using specific primes as previously described [15–18, 23, 24]. In addition, strains with these genes determined by PCR and DNA sequencing were selected as positive control for the subsequent PCR experiments.
MLST
In the present study, seven housekeeping genes of K. pneumoniae (gapA, mdh, phoE, tonB, infB, pgi, and rpoB) were amplified and sequenced to characterize the genotypes of all isolates according to the provided protocols (http://bigsdb.pasteur.fr/klebsiella/klebsiella.html/). The alleles and STs were assigned according to the online database of the Pasteur Institute MLST for K. pneumoniae. In accordance with the genetic similarity diagram using the eBURSTv3 program, the clonal complexes (CCs) were analyzed to identify the molecular epidemiological relationships.
Statistical analysis
All statistical analyses were performed using SPSS 22.0 software (IBM, Armonk, NY, USA). The categorical variables were listed as percentages and evaluated using the Chi-square test or Fisher’s exact test. The continuous data were expressed as mean ± standard deviation (mean ± SD) or median (25th - 75th percentile) appropriately and analyzed using the Student’s t test or Mann-Whitney U test. P-value < 0.05 was considered statistically significant. All tests were two-tailed.
Results
Clinical characteristics
The results showed that the annual morbidity of PLA was 17.68 to 20.62 cases per 10,000 inpatients since 2007 to 2018. From January 2016 to December 2017, a total of 163 KP-PLA cases that met the inclusion criteria were investigated in the current study. The clinical characteristics are summarized in Table 1. These cases were male-dominated (61.3%, 100/163) and had a median age of 63.0 (52.3–70.0) years old. The median length of stay in hospital was 17 (12–26) days. There were 82 cases (50.3%) in healthy individuals, while 81 cases (49.7%) had underlying or concomitant diseases, including 81 cases (49.7%) with diabetes mellitus, 46 cases (28.2%) with intra-abdominal trauma or surgery history, and 29 cases (17.8%) with OHD (i.e. hepatitis, cirrhosis, gallstones, choledocholithiasis, chronic cholecystitis or postcholecystectomy). The most common clinical symptoms were fever (84.0%, 137/163) and chill (81.0%, 132/163), with a median admission temperature of 38.9 (37.8–39.5) °C, followed by abdominal pain (39.9%, 65/163), frail (15.3%, 25/163), nausea or vomit (14.7%, 24/163), and abdominal distension (3.1%, 5/163). Additionally, four cases were found with metastatic infections, including endophthalmitis, cephalomeningitis, spontaneous bacterial peritonitis, and pneumonia, respectively.
Table 1.
Clinical characteristics of K. pneumoniae-induced pyogenic liver abscess
| Clinical characteristic | Value (n = 163) |
|---|---|
| Age, years | 63.0 (52.3–70.0) |
| Gender | |
| Male | 100 (61.3) |
| Female | 63 (38.7) |
| Underlying or concomitant conditions | |
| Diabetes mellitus | 81 (49.7) |
| History of intra-abdominal trauma or surgery | 46 (28.2) |
| Hypertension | 41 (25.2) |
| Malignancy | 7 (4.3) |
| Biliary disease | 23 (14.1) |
| Hepatitis and cirrhosis | 8 (4.9) |
| Fatty liver | 14 (8.6) |
| No underlying diseases | 82 (50.3) |
| Admission temperature (°C) | 38.9 (37.8–39.5) |
| Clinical Symptoms | |
| Fever (> 37.5 °C) | 137 (84.0) |
| Chill | 132 (81.0) |
| Abdominal pain | 65 (39.9) |
| Vomit | 24 (14.7) |
| Frail | 25 (15.3) |
| Abdominal distension | 5 (3.1) |
| Invasive procedure | 141 (86.5) |
| Use of hormones and/or immunosuppressants | 52 (31.9) |
| Multiplicity of the abscess | |
| Single | 117 (71.8) |
| Multiple (≥2 abscesses) | 46 (28.2) |
| Site of Single abscess | |
| Right hepatic lobe | 95 (58.3) |
| Left hepatic lobe | 22 (13.5) |
| Laboratory examination | |
| WBC count, × 109/L | 10.8 (7.9–13.4) |
| RBC count, × 1012/L | 3.9 (3.5–4.2) |
| ALT(U/L) | 39.0 (24.0–78.0) |
| AST(U/L) | 37.0 (24.0–68.5) |
| Albumin, g/L | 29.4 ± 5.1 |
| Method of abscess treatment | |
| Simple antibacterial | 30 (18.4) |
| Abscess drainage | 129 (79.1) |
| Surgical removal | 4 (2.5) |
| Prognosis of abscess | |
| Effective | 116 (71.2) |
| Ineffective | 47 (28.8) |
| Metastatic infections | |
| Endophthalmitis | 1 (0.6) |
| Cephalomeningitis | 1 (0.6) |
| Bacterial peritonitis | 1 (0.6) |
| Pneumonia | 1 (0.6) |
| Clinical outcomes | |
| Length of stay in hospital, days | 17.0 (12.0–26.0) |
| Admission to ICU | 16 (9.8) |
| Septic shock | 15 (9.2) |
| Septicemia | 20 (12.3) |
| In-hospital deaths | 1 (0.6) |
Values are presented as median (25th - 75th percentile), mean ± SD or No. (%) of patients
WBC White blood cell, RBC Red blood cell, ALT Alanine aminotransferase, AST Aspartate aminotransferase, ICU Intensive care unit
Medical imaging analysis revealed that solitary abscess accounted for 71.8% (117/163), of which 81.2% (95/117) were located in the right hepatic lobe, while multiple abscess only accounted for 28.2% (46/163). Significant laboratory abnormalities of inflammatory biomarkers and liver function indexes were observed, including increased CRP (100%, 163/163), WBC count (69.9%, 114/163), ALT (49.1%, 80/163), and AST (52.1%, 85/163). Moreover, serum hypoalbuminemia occurred in 96.9% (158/163) of cases, of whom, 43.0% (68/158) underwent severe serum hypoalbuminemia (Alb< 28 g/L).
In terms of clinical treatment, antibiotic therapy was applied to all the patients, 90.8% of cases (148/163) received a combination antibiotic therapy. The most common antimicrobial regimen was a third- or fourth-generation cephalosporin or carbapenem plus metronidazole. Drainage was implemented as the common treatment of abscess, in which 79.1% of cases (129/163) received percutaneous puncture or catheter drainage combined with antibiotic treatment, while 18.4% (30/163) received antibiotics-only treatment. The remaining four patients were surgically treated. However, there were 28.8% of patients (47/163) with ineffective prognosis of abscess. It is noteworthy that the majority of the patients with effective prognosis of abscess underwent drainage (P = 0.027), while the patients with ineffective prognosis of abscess mainly received antibiotics-only treatment (P = 0.005), as well as had the higher incidence of admission to ICU, septicemia, and septic shock. Overall, 9.2% of the patients (15/163) had septic shock, 12.3% of the patients (20/163) had septicemia, and 9.8% of the patients (16/163) were transferred to the ICU due to worsening condition, in which one case died during hospitalization.
Microbiological and molecular epidemiological characteristics
Herein, K. pneumoniae strains were isolated from various clinical specimens, including pus (53.4%, 87/163), drainage fluid (24.5%, 40/163), blood (18.4%, 30/163), hydrothorax (1.2%, 2/163), and other sterile fluid (2.5%, 4/163). The microbiological characteristics of the 163 isolates are listed in Table 2. The majority of isolates were rarely resistant to commonly used antimicrobial drugs with low rates of resistance (< 10%), except for nitrofurantoin (20.9%), ampicillin/sulbactam (10.4%), and ampicillin (100%; intrinsic resistance). However, twelve isolates were still identified as MDR strains. In addition, two CRKP strains belonged to ST11 and were resistant to almost all antibiotics tested. Regrettably, the outcomes of patients isolated the CRKP strains were poor.
Table 2.
Microbiological and molecular-epidemiological characteristics of K. pneumoniae strains isolated from KP-PLA patients
| Characteristic | Value (n = 163) |
|---|---|
| Hypermucoviscosity | 50 (30.7) |
| Capsular serotypes | |
| K1 | 66 (40.5) |
| K2 | 31 (19.0) |
| K5 | 8 (4.9) |
| K20 | 3 (1.8) |
| non-type | 54 (33.1) |
| Virulence genes | |
| aerobactin | 139 (85.3) |
| rmpA | 155 (95.1) |
| iroN | 40 (24.5) |
| kfuBC | 116 (71.2) |
| wcaG | 112 (68.7) |
| ybtA | 120 (73.6) |
| magA | 74 (45.4) |
| fimH | 156 (95.7) |
| mrkD | 163 (100.0) |
| uge | 147 (90.2) |
| entB | 144 (88.3) |
| ureA | 145 (89.0) |
| Antimicrobial resistance | |
| Ampicillin | 163 (100.0) |
| Ampicillin/sulbactam | 17 (10.4) |
| Piperacillin/tazobactam | 2 (1.2) |
| Cefazolin | 10 (6.1) |
| Cefotetan | 2 (1.2) |
| Aztreonam | 5 (3.1) |
| Ceftriaxone | 6 (3.7) |
| Ceftazidime | 3 (1.8) |
| Cefepime | 4 (2.5) |
| Cefoperazone/sulbactam | 0 (0.0) |
| Ertapenem | 3 (1.8) |
| Imipenem | 5 (3.1) |
| Ciprofloxacin | 5 (3.1) |
| Levofloxacin | 4 (2.5) |
| Gentamicin | 2 (1.2) |
| Tobramycin | 2 (1.2) |
| Amikacin | 1 (0.6) |
| SMZ-TMP | 6 (3.7) |
| Nitrofurantoin | 34 (20.9) |
| MDR strains | 12 (7.4) |
| CRKP strains | 6 (3.7) |
| Multilocus sequence typing | |
| ST23 | 63 (38.7) |
| ST29 | 10 (6.1) |
| ST65 | 14 (8.6) |
| ST86 | 10 (6.1) |
Values are presented as No. (%) of isolates
MDR Multi-drug resistant, CRKP Carbapenems-resistant K. pneumoniae, SMZ-TMP Sulfamethoxazole and trimethoprim
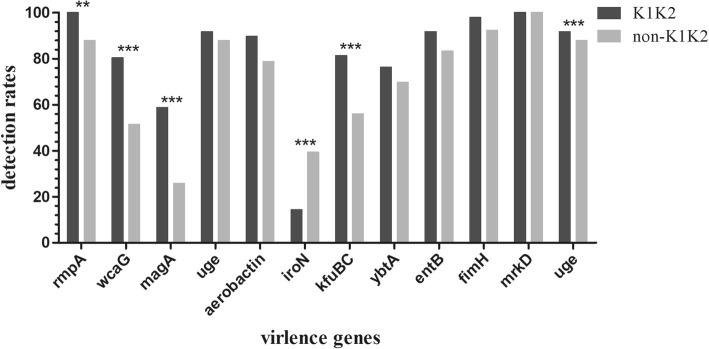
The string test disclosed that 30.7% (50/163) of strains exhibited hypermucoviscosity. As shown in Table 2, the dominant capsular serotype was serotype K1 (40.5%, 66/163), followed by serotype K2 (19.0%, 31/163), K5 (4.9%, 8/163), K20 (1.8%, 3/163), and non-type (33.1%, 54/163). Except for iroN and magA, all remaining virulence genes were presented in more than half of 163 strains. The prevalence of rmpA and aerobactin was up to 95.1 and 85.3%, respectively. Moreover, 88.0% (44/50) of hypermucoviscous K. pneumoniae isolates harbored rmpA and aerobactin, and 89.7% (87/97) of serotype K1K2 isolates possessed these two genes. Additionally, the prevalence of numerous virulence genes in the serotype K1K2 strains was notably higher than that in the non-K1K2 strains (Fig. 1).
Fig. 1.
Comparing the detection rates of virulence genes between K1K2 strains and non-K1K2 strains. *, P < 0.05; **, P < 0.01; ***, P < 0.001
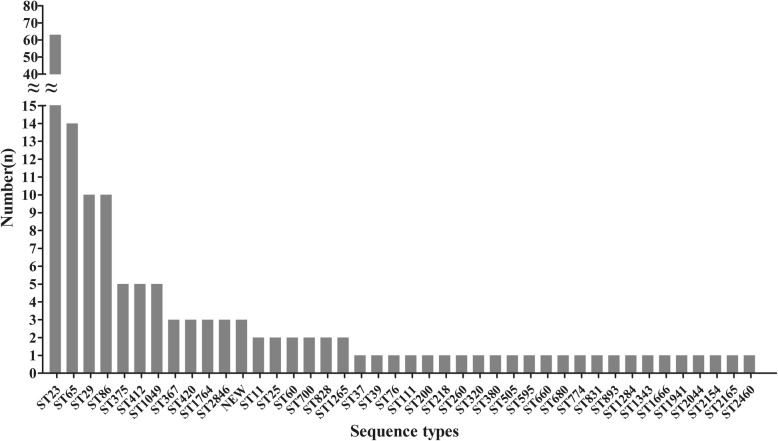
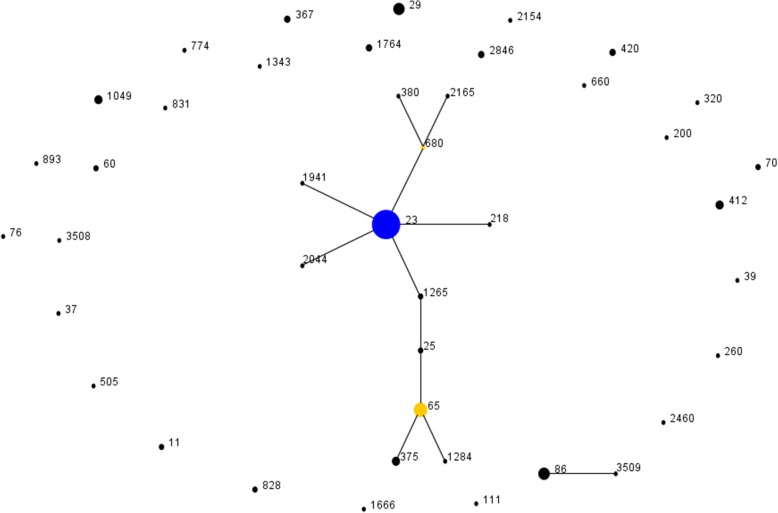
Furthermore, MLST revealed 44 sequence types among the 163 K. pneumoniae strains, involving three newly identified STs (ST3507, ST3508, and ST3509). The predominant type was ST23 (38.7%, 63/163), followed by ST65 (8.6%, 14/163), ST29 (6.1%, 10/163), ST86 (6.1%, 10/163), and other STs (Fig. 2). Besides, eBURST analysis showed that those 44 STs were grouped into clonal complex (CC23, n = 1), doublet (D, n = 1) and singleton (S, n = 28) (Fig. 3). The CC23 included 57% (93/163) isolates covering 12 STs, and ST23 was defined as founder of the homologous complex. The D included ST86 and ST3509, in which those were only inconsonant with pgi allele. In addition, 69.8% (44/63) of ST23 strains belonged to K1 isolates, while 77.4% (24/31) of K2 isolates belonged to ST65- and ST86-like isolates.
Fig. 2.
Specific types and quantities of all sequence types among the 163 K. pneumoniae strains
Fig. 3.
Performing eBURST analysis on molecular epidemiological characteristics of 163 K. pneumoniae isolates. The population snapshot indicates the clonal assignment of the STs presented in this study. Each black dot represents one ST, and blue dots indicate individual founders, while yellow spots denote sub-founders
Comparing characteristics between KP-PLA patients with or without OHD
A total of 29 OHD patients were included in this study, while the remaining 134 cases were non-OHD patients. The differences in characteristics between these two groups are summarized in Tables 3 and 4. The median age was 67.0 (62.0–73.0) years old in the OHD group, which was older than that in the non-OHD group (P = 0.053). Furthermore, the OHD patients had a lower prevalence of diabetes (24.1% vs. 55.2%, P = 0.002) and more likely had the history of intra-abdominal trauma or surgery (89.7% vs. 14.9%, P = 0.000). Moreover, the length of stay in hospital was significantly longer in the OHD group than that in the non-OHD group (23 vs. 16 days, P = 0.045), and the clinical symptoms were inconspicuous. Notably, the prevalence of ST23 and rmpA in the OHD group was lower than that in the non-OHD group (P = 0.028, 0.049, respectively). On the contrary, the incidence of MDR strains in the OHD group was markedly higher than that in the non-OHD group (24.1% vs. 3.7%, P = 0.001). Therefore, the present study further analyzed the differences in resistant spectrum between these two groups. Surprisingly, the resistance rates of the majority of tested antimicrobials in the OHD group were remarkably higher than those in the non-OHD group, and the differences were statistically significant (Table 4).
Table 3.
Comparing clinical characteristics between OHD patients and non-OHD patients from KP-PLA
| Characteristic | OHD patients (n = 29) | non-OHD patients (n = 134) | P Value |
|---|---|---|---|
| Age, years | 67.0 (62.0-73.0) | 61.0 (51.0-70.0) | 0.053 |
| Gender | 0.451 | ||
| Male | 16 (55.2) | 84 (62.7) | |
| Female | 13 (44.8) | 50 (37.3) | |
| Underlying or concomitant conditions | |||
| Diabetes mellitus | 7 (24.1) | 74 (55.2) | 0.002* |
| History of intra-abdominal trauma or surgery | 26 (89.7) | 20 (14.9) | 0.000* |
| Hypertension | 5 (17.2) | 45 (33.6) | 0.084 |
| Malignancy | 2 (6.9) | 5 (3.7) | 0.797 |
| Admission temperature (°C) | 38.0 (37.3-39.0) | 39.0 (38.0-39.5) | 0.019* |
| Clinical Symptoms | |||
| Fever (>37.5°C) | 20 (69.0) | 117 (87.3) | 0.030* |
| Chill | 23 (79.3) | 109 (81.3) | 0.800 |
| Abdominal pain | 14 (48.3) | 51 (38.1) | 0.308 |
| Vomit | 4 (13.8) | 20 (14.9) | 1.000 |
| Frail | 3 (10.3) | 22 (16.4) | 0.590 |
| Abdominal distension | 2 (6.9) | 3 (2.2) | 0.216 |
| Invasive procedure | 25 (86.2) | 116 (86.6) | 1.000 |
| Use of hormones and/or immunosuppressants | 11 (37.9) | 41 (30.6) | 0.442 |
| Multiplicity of the abscess | 0.409 | ||
| Single | 19 (65.5) | 98 (73.1) | |
| Multiple (≥2 abscesses) | 10 (34.5) | 36 (26.9) | |
| Laboratory examination | |||
| WBC count, ×109/L | 8.9 (5.9-11.6) | 11.1 (8.2-14.0) | 0.014* |
| RBC count, ×1012/L | 3.9 (3.5-4.1) | 3.9 (3.6-4.2) | 0.667 |
| ALT(U/L) | 33.0 (14.0-49.0) | 43.0 (27.0-82.8) | 0.033* |
| AST(U/L) | 35.0 (26.0-50.0) | 37.0 (24.0-70.8) | 0.621 |
| Albumin, g/L | 29.8±5.0 | 29.3±5.1 | 0.609 |
| Method of abscess treatment | |||
| Simple antibacterial | 6 (20.7) | 24 (17.9) | 0.726 |
| Abscess drainage | 20 (69.0) | 109 (81.3) | 0.137 |
| Surgical removal | 3 (10.3) | 1 (0.7) | 0.021* |
| Prognosis of abscess | 0.233 | ||
| Effective | 18 (62.1) | 98 (73.1) | |
| Ineffective | 11 (37.9) | 36 (26.9) | |
| Metastatic infections | 1.000 | ||
| Endophthalmitis | 0 (0.0) | 1 (0.7) | |
| Cephalomeningitis | 0 (0.0) | 1 (0.7) | |
| Bacterial peritonitis | 0 (0.0) | 1 (0.7) | |
| Pneumonia | 0 (0.0) | 1 (0.7) | |
| Clinical outcomes | |||
| Length of stay in hospital, days | 23.0 (15.0-28.0) | 16.0 (12.0-25.0) | 0.045* |
| Admission to ICU | 1 (3.4) | 15 (11.2) | 0.354 |
| Septic shock | 3 (10.3) | 12 (9.0) | 1.000 |
| Septicemia | 5 (17.2) | 15 (11.2) | 0.557 |
| In-hospital deaths | 0 (0.0) | 1 (0.7) | 1.000 |
Values are presented as median (25th - 75th percentile), mean ± SD or No. (%) of patients
WBC white blood cell, RBC red blood cell, ALT alanine aminotransferase, AST aspartate aminotransferase, ICU intensive care unit *P < 0.05
Table 4.
Comparing microbiological and molecular-epidemiological characteristics of K. pneumoniae strains between OHD patients and non-OHD patients
| Characteristic | Kp from OHD patients (n = 29) | Kp from non-OHD patients (n = 134) | P Value |
|---|---|---|---|
| Hypermucoviscosity | 8 (27.6) | 42 (31.3) | 0.691 |
| Capsular serotypes | |||
| K1 | 11 (37.9) | 55 (41.0) | 0.757 |
| K2 | 5 (17.2) | 26 (19.4) | 0.788 |
| K5 | 1 (3.4) | 7 (5.2) | 1.000 |
| K20 | 1 (3.4) | 2 (1.5) | 0.447 |
| non-type | 11 (37.9) | 43 (32.1) | 0.545 |
| Virulence genes | |||
| aerobactin | 22 (75.9) | 117 (87.3) | 0.197 |
| rmpA | 25 (86.2) | 130 (97.0) | 0.049* |
| iroN | 4 (13.8) | 36 (26.9) | 0.138 |
| kfuBC | 17 (58.6) | 99 (73.9) | 0.100 |
| wcaG | 18 (62.1) | 94 (70.1) | 0.395 |
| ybtA | 23 (79.3) | 97 (72.4) | 0.443 |
| magA | 9 (31.0) | 65 (48.5) | 0.087 |
| fimH | 29 (100.0) | 127 (94.8) | 0.451 |
| mrkD | 29 (100.0) | 134 (100.0) | NA |
| uge | 26 (89.7) | 121 (90.3) | 1.000 |
| entB | 24 (82.8) | 120 (89.6) | 0.475 |
| ureA | 27 (93.1) | 118 (88.1) | 0.646 |
| Antimicrobial resistance | |||
| Ampicillin | 29 (100.0) | 134 (100.0) | NA |
| Ampicillin/sulbactam | 7 (24.1) | 10 (7.5) | 0.020* |
| Piperacillin/tazobactam | 2 (6.9) | 0 (0.0) | 0.031* |
| Cefazolin | 6 (20.7) | 4 (3.0) | 0.001* |
| Cefotetan | 2 (6.9) | 0 (0.0) | 0.031* |
| Aztreonam | 5 (17.2) | 0 (0.0) | 0.000* |
| Ceftriaxone | 5 (17.2) | 1 (0.7) | 0.000* |
| Ceftazidime | 3 (10.3) | 0 (0.0) | 0.005* |
| Cefepime | 4 (13.8) | 0 (0.0) | 0.001* |
| Cefoperazone/sulbactam | 0 (0.0) | 0 (0.0) | NA |
| Ertapenem | 2 (6.9) | 1 (0.7) | 0.082 |
| Imipenem | 3 (10.3) | 2 (1.5) | 0.040* |
| Ciprofloxacin | 5 (17.2) | 0 (0.0) | 0.000* |
| Levofloxacin | 4 (13.8) | 0 (0.0) | 0.001* |
| Gentamicin | 1 (3.4) | 1 (0.7) | 0.325 |
| Tobramycin | 1 (3.4) | 1 (0.7) | 0.325 |
| Amikacin | 0 (0.0) | 1 (0.7) | 1.000 |
| SMZ-TMP | 4 (13.8) | 2 (1.5) | 0.008* |
| Nitrofurantoin | 13 (44.8) | 21 (15.7) | 0.000* |
| MDR strains | 7 (24.1) | 5 (3.7) | 0.001* |
| CRKP strains | 3 (10.3) | 3 (2.2) | 0.119 |
| Multilocus sequence typing | |||
| ST23 | 6 (20.7) | 57 (42.5) | 0.028* |
| ST29 | 3 (10.3) | 7 (5.2) | 0.538 |
| ST65 | 2 (6.9) | 12 (9.0) | 1.000 |
| ST86 | 2 (6.9) | 8 (6.0) | 1.000 |
Values are presented as No. (%) of isolates
MDR Multi-drug resistant, CRKP Carbapenems-resistant K. pneumoniae, SMZ-TMP sulfamethoxazole and trimethoprim, NA not applicable, *P < 0.05
Discussion
The incidence of PLA has notably elevated in recent years and accordingly, K. pneumoniae emerged as a life-threatening bacterial pathogen across Asian and European countries, as well as the United States [3, 25]. To our knowledge, KP-PLA has still remained as a mortality-associated infectious disease worldwide [3, 25]. To date, a limited number of studies have concentrated on characterization of the clinical and pathogenic features from one KP-PLA population simultaneously. Hence, in the present retrospective study, we systematically analyzed the 163 KP-PLA cases in a tertiary teaching hospital in Southeast China, which made the parallel comparability of clinical and microbiological data, and further supplemented the current data of KP-PLA worldwide. Moreover, we compared the characteristics between KP-PLA patients with or without OHD. To the best of our knowledge, this study was the first systematic analysis on these two groups of KP-PLA.
The data of annual morbidity in the current investigation confirmed high incidence of PLA in the last decade. Male gender, patients with diabetes, OHD and intra-abdominal trauma or surgery were more susceptible to have KP-PLA, which was comparable with the latest epidemiological trends [5–7]. Diabetes is known as an important risk factor for KP-PLA. A poor glycemic control could impair the neutrophil phagocytosis and promote growth of pathogen in tissues, while metabolic disorders could negatively influence the liver [21, 26]. Therefore, the blood sugar level should be strictly monitored and controlled for KP-PLA patients. Additionally, the KP-PLA patients with OHD or intra-abdominal trauma or surgery were not uncommon in the present study. A possible explanation was that K. pneumoniae easily entered into the liver via direct spreading, portal circulation or cross the intestinal barrier to cause KP-PLA and severe K. pneumoniae infection in those patients [27–29]. It indicated that such patients should be strictly followed-up and reviewed regularly in order to detect any possible signs and treat KP-PLA in time. In addition, about half of the cases occurred in healthy individuals, demonstrating a strong possibility of having an invasive fatal PLA caused by hvKp [1, 8]. The clinical manifestations and imaging findings of 163 KP-PLA cases were consistent with previous studies [8, 9, 20, 21]. Furthermore, the laboratory outcomes revealed that the majority of patients underwent inflammation and impaired liver function. However, the definite diagnosis of KP-PLA is often delayed due to lack of typical symptoms, and further progressed to fearful septicemia or septic shock [29]. It indicated that physicians are encouraged to make a high clinical suspicion on KP-PLA during presentation of the above-mentioned signs.
For patients who diagnosed with KP-PLA, percutaneous drainage under ultrasonography or computed tomography guidance combined with a proper antimicrobial therapy has been taken as a standard treatment into consideration [30]. The data of drainage rate and abscess prognosis existing in the current study re-verified the necessity of drainage for KP-PLA patients. Drainage may contribute to a better control of infection source, accurate identification of pathogens, and rational application of antibiotics. Although fatal metastatic infections were seldom observed in the present investigation, those extrahepatic invasive complications, especially in eye, lung or central nervous system, were required to be observed prudently for site-directed treatment in-time. The in-hospital mortality rate (0.6%) in the present investigation was lower than a previous study (5%) [1], which may be due to accurate and timely implementation of interventions for KP-PLA. In addition, a fraction of patients with serious conditions gave up treatment, it may lead to underestimate the mortality rate. Taken together, it is imperative for KP-PLA patients to perform a reasonable management in advance to attenuate morbidity, complications, and mortality.
In terms of microbiological characteristics of isolates from KP-PLA, virulence and antibiotic resistance were considered to play a substantial role in bacterial pathogenesis. The hypermucoviscosity was considered as a surrogate marker for hvKp and a significant contributor to the virulence for invasive KP-PLA infections [1, 3]. However, only a third of isolates possessed hypermucoviscosity in the present research. The rate of hypermucoviscosity was compatible with another recent study conducted in East China [20], while that was remarkably lower than that reported in other Asian countries (more than 70%) [3, 16, 19]. The majority of recently conducted studies suggested that the correspondence between hypermucoviscosity and hypervirulence was variable, and therefore, the virulence of isolates from KP-PLA should be assessed by a combination of genotypic and clinical features [22]. A polysaccharide capsule acts as a major virulence factor for hvKp by protecting K. pneumoniae from phagocytosis of immune cells and bactericidal action of complement or antimicrobial peptides [10]. K. pneumoniae strains are presented in at least 78 capsular serotypes, in which K1 and K2 are related to hvKp, as well as being pathogenic to humans strongly [11]. In the present research, the prevalence of K1 and K2 was 40.5 and 19.0%, respectively, suggesting that substantial percentages of isolates from KP-PLA were hypervirulent. Compared to previous reports (K1 ranged from 46.6 to 63.4%, and K2 ranged from 14.2 to 20.5%) [16, 31], it needs to be characterized whether geographical differences accounted for this distinction. Meanwhile, the non-K1K2 isolates also played a pivotal role in KP-PLA and should not be overlooked because these isolates may be hypervirulent as well.
To rule out one-sidedness in assessment of hypervirulence based on hypermucoviscosity or K1K2 serotypes [3, 19], the virulence genes of all the 163 strains were tested. Thereinto, rmpA and aerobactin are the most important genes for hypervirulence. rmpA regulates the synthesis of extracellular polysaccharide capsule and is responsible for hypermucoviscosity, the ablation of rmpA may lead to the loss or thinning of capsule, thereby weakening the ability to evade immune responses, in turn causing virulence of K. pneumoniae to be markedly reduced [12, 14]. The high prevalence of rmpA (95.1%) was observed in the current research. However, a remarkable number of rmpA-positive strains did not show hypermucoviscosity, reflecting that there may be other regulatory mechanisms for expression of hypermucoviscosity. For instance, wcaG, magA, and uge genes associate with biosynthesis of the capsule. These genes were also found to be prevalent in KP-PLA isolates. Another important virulence gene is aerobactin, which is essential for the growth and virulence of K pneumoniae in the host via regulation of iron supply. The aerobactin-positive strains were up to 85.3% in the current study. In addition, aerobactin and rmpA are often concomitant with hypermucoviscosity and have been extensively used to define hvKp [13, 32, 33]. In the study, 84.0% of isolates simultaneously carried aerobactin and rmpA, which were more likely to be hvKp strains, while did not always belong to K1K2 strains or possessed hypermucoviscosity. These observations were consistent with findings of previous studies [8, 32]. Other siderophores genes (iroN, kfuBC, ybtA and entB), mediating uptake of ferric iron were reported as virulence genes in KP-PLA [13, 15, 18]. In addition to iroN, the other three siderophores genes were possessed by the majority of KP-PLA isolates. Furthermore, fimH and mrkD mediate adhesion through encoding K. pneumoniae type 1 and type 3 fimbriae, and mrkD may promote development of biofilms. Besides, ureA, an α-subunit of the urease, is associated with invasion [18, 34]. In the present study, fimH and mrkD were observed in almost all isolates except for seven fimH-negative strains, and 89.0% of isolates possessed ureA. Therefore, clinicians are advised to consider phenotypes and genotypes of pathogen, and also carefully select an appropriate catheter and more frequent catheter irrigation or replacement to reduce bacterial adhesion and colonization. Altogether, all the 163 isolates harbored various virulence genes, reflecting that the majority of K. pneumoniae isolates from PLA exhibited hypervirulence and strong pathogenicity. Additionally, the majority of virulence genes, as well as the hypermucoviscosity, were notably widespread in K1K2 strains, suggesting that K1K2 isolates from KP-PLA typically carried various hypervirulent factors.
Although virulence factors did not always singularly determine hvKp, certain intrinsic correlations were noted, which required further investigation and characterization. In particular, virulence factors acted as a warning of hvKp-PLA, which recommended prolonged treatment properly and long-term follow-up to maximize the possibility of a successful treatment and minimize relapse rate [19, 22]. Regrettably, the current study could not present accurate numbers of hvKp in the 163 KP-PLA cases due to the fact that a reference standard for hvKp has not been defined yet [19, 22]. Nevertheless, from the microbiological perspective, clinicians are recommended to do not ignore the detection value of these virulence factors in KP-PLA isolates, especially after combining with antibiotic resistance. Meanwhile, the exploration of virulence factors may provide novel therapeutic targets and develop novel vaccines for hvKp infections, which are not limited to only the management of KP-PLA [35].
Additionally, K. pneumoniae isolates from KP-PLA were highly susceptible to almost all kinds of antimicrobial agents, such as β-lactamase inhibitors, third generation cephalosporins, and carbapenems, which might be related to abundant hvKp in KP-PLA [1, 36]. Nevertheless, twelve MDR strains were detected in the present study, which rarely occurred in KP-PLA [8], and these MDR strains were more likely to occur in the OHD patients. Further analysis revealed that the resistance rates of almost all tested antibiotics in the OHD group were significantly higher than those in non-OHD group. These results indicated that the accelerated emergence of resistance and colonization could be induced by increasing hepatobiliary interventions with routine prophylactic antibiotics in such patients [6]. However, it also could not be neglected to prevent infections for those patients because of unimaginable outcomes once hvKp occurred with MDR. Moreover, two ST11 CRKP isolates from the current research simultaneously disclosed hypervirulence, extensively drug resistance, and terrible pathogenicity, highlighting the significant threat of such real superbugs. An emerged clinical challenge is the convergence of hypervirulence and extensively drug- and pan-drug-resistant in K. pneumoniae, which might lead to further emergence of a “post-antibiotic” scenario [37]. These findings demonstrated that clinicians should be highly prudent and cautious in prescribing antibiotics and dosage options, in which host, pathogen, and host-pathogen interactions need to be taken into account to prevent MDR-hvKp, especially in the OHD patients.
MLST analysis uncovered the molecular epidemiological characteristics of K. pneumoniae strains in the 163 KP-PLA cases. Additionally, ST23 was found as a predominant type of KP-PLA with a rate of 38.7%, which was similar to a previous report [16]. ST23 is one of the dominant clones of hvKp and defined as a founder of clonal lineage CC23, representing a specific genetic background that conferred hypervirulence and fitness [16, 19, 37]. Even though CC23 has a capacity to spread worldwide through multiple international transmission events rather than local expansions, CC23 isolates from KP-PLA were genotypically closely related [38]. It is noteworthy that ST23 was firmly linked to serotype K1, while ST65- and ST86-like isolates were associated with serotype K2 in KP-PLA as similar to other reports performed in Asia [20, 25], while the reason has still remained elusive. Those findings disclosed strict association of hypervirulent clones with capsular serotypes. Meanwhile, there still existed 43% of strains that belonged to clones other than CC23, including three novel sequence types. The novel ST3509 was a single locus variant of ST86, which expanded the cloned lineage of CC86. The existence of doublets and singletons demonstrated the genetic diversity in isolates from KP-PLA.
However, the current study contains a number of limitations. Firstly, it was designed as a retrospective study at a single center, that might increase selection and information biases. Secondly, for reasons already explained, no accurate data of hvKp were reported. Further in vitro and in vivo virulence-based experiments may be required for isolates from KP-PLA, such as galleria mellonella models, mice models, and neutrophil phagocytosis assay.
Conclusions
The present study may draw attention from clinicians to take into account KP-PLA in patients with non-specific symptoms, and indicate the necessity to fully combine the auxiliary examination for early diagnosis and appropriate treatment, in order to improve outcome and prevent severe metastatic complications. Management of the underlying diseases is decisive in KP-PLA patients, especially in OHD patients. There existed large quantities of hvKp in KP-PLA, and even MDR-hvKp. With the prospective convergence of virulence and resistance, it is essential to perform prompt identification of characteristics of K. pneumoniae, followed by effective therapeutic strategies. Hence, additional studies need to be conducted to elucidate the relationships between the phenotypes, genotypes, resistant spectrum and clones of K. pneumoniae isolates from KP-PLA, with specific focus on hypervirulence, MDR, and pathogenicity. Further researches can be helpful to raise the awareness of hvKp and provide effective treatments for KP-PLA patients.
Acknowledgements
We thank the team of the curators of the Institut Pasteur MLST system (Paris, France) for importing novel profiles and isolates at http://bigsdb.pasteur.fr/.
Abbreviations
- ALB
Albumin
- ALT
Alanine aminotransferase
- AST
Aspartate aminotransferase
- CCs
Clonal complexes
- cKp
classic K. pneumoniae
- CLSI
Clinical and Laboratory Standards Institute
- CRP
C-reactive protein
- hvKp
hypervirulent K. pneumoniae
- ICU
Intensive care unit
- KP-PLA
Klebsiella pneumoniae-induced pyogenic liver abscess
- MDR
Multidrug resistant
- MLST
Multilocus sequence typing
- OHD
Other hepatobiliary diseases
- PCR
Polymerase chain reaction
- PLA
Pyogenic liver abscess
- RBC
Red blood cell
- STs
Sequence types
- WBC
White blood cell
Authors’ contributions
ZSQ, ZXC, DGF, FRC and ZYZ contributed to the acquisition and analysis of the data. ZSQ wrote the initial draft of this paper. WQ and ZXK performed the statistical analyses. ZTL and CJM contributed to the concept of the study, the revision of this paper, and the final approval of the version to be published. All authors have read and approved the final manuscript.
Funding
This work was supported by the research grant from the National Natural Science Foundations of China (No.81741059 and 81971986) and the Health Department of Zhejiang Province of the People’s Republic of China (No. 2019KY098).
Availability of data and materials
All data generated or analysed during this study are included in this manuscript.
Ethics approval and consent to participate
This study was approved by the First Affiliated Hospital of Wenzhou Medical University Ethics Committee. Informed consent was not needed due to the retrospective nature of the study; additionally, the patient data accessed in this research was anonymous. Therefore, the First Affiliated Hospital of Wenzhou Medical University Ethics Committee waived the need for consent.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Siqin Zhang, Email: zhangsiqin347347@163.com.
Xiucai Zhang, Email: xczhang1994310@163.com.
Qing Wu, Email: wuqing830@163.com.
Xiangkuo Zheng, Email: kuo7436@163.com.
Guofeng Dong, Email: dongguofeng0112@163.com.
Renchi Fang, Email: fangrenchi123@163.com.
Yizhi Zhang, Email: zhangyizhi218@126.com.
Jianming Cao, Email: wzcjming@163.com.
Tieli Zhou, Email: wyztli@163.com.
References
- 1.Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12(11):881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 2.Tsai FC, Huang YT, Chang LY, Wang JT. Pyogenic liver abscess as endemic disease, Taiwan. Emerg Infect Dis. 2008;14(10):1592–1600. doi: 10.3201/eid1410.071254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerwenka H. Pyogenic liver abscess: differences in etiology and treatment in Southeast Asia and Central Europe. World J Gastroenterol. 2010;16(20):2458–2462. doi: 10.3748/wjg.v16.i20.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen YC, Lin CH, Chang SN, Shi ZY. Epidemiology and clinical outcome of pyogenic liver abscess: an analysis from the National Health Insurance Research Database of Taiwan, 2000-2011. J Microbiol Immunol Infect. 2016;49(5):646–653. doi: 10.1016/j.jmii.2014.08.028. [DOI] [PubMed] [Google Scholar]
- 6.Shi SH, Zhai ZL, Zheng SS. Pyogenic liver abscess of biliary origin: the existing problems and their strategies. Semin Liver Dis. 2018;38(3):270–283. doi: 10.1055/s-0038-1661363. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Wang JY, Jiang W. An increasing prominent disease of Klebsiella pneumoniae liver abscess: etiology, diagnosis, and treatment. Gastroenterol Res Pract. 2013;2013:258514. doi: 10.1155/2013/258514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jun JB. Klebsiella pneumoniae Liver Abscess. Infect Chemother. 2018;50(3):210–218. doi: 10.3947/ic.2018.50.3.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye M, Tu J, Jiang J, Bi Y, You W, Zhang Y, Ren J, Zhu T, Cao Z, Yu Z, et al. Clinical and genomic analysis of liver abscess-causing Klebsiella pneumoniae identifies new liver abscess-associated virulence genes. Front Cell Infect Microbiol. 2016;6:165. doi: 10.3389/fcimb.2016.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung CP, Chang FY, Lin JC, Ho DM, Chen CT, Chen JH, Yeh KM, Chen TL, Lin YT, Siu LK. Immune response and pathophysiological features of Klebsiella pneumoniae liver abscesses in an animal model. Lab Investig. 2011;91(7):1029–1039. doi: 10.1038/labinvest.2011.52. [DOI] [PubMed] [Google Scholar]
- 11.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603. doi: 10.1128/CMR.11.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu WL, Ko WC, Cheng KC, Lee HC, Ke DS, Lee CC, Fung CP, Chuang YC. Association between rmpA and magA genes and clinical syndromes caused by Klebsiella pneumoniae in Taiwan. Clin Infect Dis. 2006;42(10):1351–1358. doi: 10.1086/503420. [DOI] [PubMed] [Google Scholar]
- 13.Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun. 2015;83(8):3325–3333. doi: 10.1128/IAI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paczosa MK, Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol Mol Biol Rev. 2016;80(3):629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Candan ED, Aksoz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochim Pol. 2015;62(4):867–874. doi: 10.18388/abp.2015_1148. [DOI] [PubMed] [Google Scholar]
- 16.Luo Y, Wang Y, Ye L, Yang J. Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin Microbiol Infect. 2014;20(11):O818–O824. doi: 10.1111/1469-0691.12664. [DOI] [PubMed] [Google Scholar]
- 17.Wasfi R, Elkhatib WF, Ashour HM. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Sci Rep. 2016;6:38929. doi: 10.1038/srep38929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang S, Yang G, Ye Q, Wu Q, Zhang J, Huang Y. Phenotypic and genotypic characterization of Klebsiella pneumoniae isolated from retail foods in China. Front Microbiol. 2018;9:289. doi: 10.3389/fmicb.2018.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee CR, Lee JH, Park KS, Jeon JH, Kim YB, Cha CJ, Jeong BC, Lee SH. Antimicrobial resistance of Hypervirulent Klebsiella pneumoniae: epidemiology, Hypervirulence-associated determinants, and resistance mechanisms. Front Cell Infect Microbiol. 2017;7:483. doi: 10.3389/fcimb.2017.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu TT, Zhou JC, Jiang Y, Shi KR, Li B, Shen P, Wei ZQ, Yu YS. Clinical and microbiological characteristics of Klebsiella pneumoniae liver abscess in East China. BMC Infect Dis. 2015;15:161. doi: 10.1186/s12879-015-0899-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foo NP, Chen KT, Lin HJ, Guo HR. Characteristics of pyogenic liver abscess patients with and without diabetes mellitus. Am J Gastroenterol. 2010;105(2):328–335. doi: 10.1038/ajg.2009.586. [DOI] [PubMed] [Google Scholar]
- 22.Russo TA, Olson R, Fang CT, Stoesser N, Miller M, MacDonald U, Hutson A, Barker JH, La Hoz RM, Johnson JR. Identification of Biomarkers for Differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. pneumoniae. J Clin Microbiol. 2018;56(9):e00776–18. [DOI] [PMC free article] [PubMed]
- 23.Pan YJ, Lin TL, Chen YH, Hsu CR, Hsieh PF, Wu MC, Wang JT. Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. PLoS One. 2013;8(12):e80670. doi: 10.1371/journal.pone.0080670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45(3):284–293. doi: 10.1086/519262. [DOI] [PubMed] [Google Scholar]
- 25.Siu LK, Fung CP, Chang FY, Lee N, Yeh KM, Koh TH, Ip M. Molecular typing and virulence analysis of serotype K1 Klebsiella pneumoniae strains isolated from liver abscess patients and stool samples from noninfectious subjects in Hong Kong, Singapore, and Taiwan. J Clin Microbiol. 2011;49(11):3761–3765. doi: 10.1128/JCM.00977-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin JC, Siu LK, Fung CP, Tsou HH, Wang JJ, Chen CT, Wang SC, Chang FY. Impaired phagocytosis of capsular serotypes K1 or K2 Klebsiella pneumoniae in type 2 diabetes mellitus patients with poor glycemic control. J Clin Endocrinol Metab. 2006;91(8):3084–3087. doi: 10.1210/jc.2005-2749. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Du Z, Bi J, Wu Z, Lv Y, Zhang X, Wu R. The impact of previous abdominal surgery on clinical characteristics and prognosis of pyogenic liver abscess: a 10-year retrospective study of 392 patients. Medicine (Baltimore) 2018;97(39):e12290. doi: 10.1097/MD.0000000000012290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tu YC, Lu MC, Chiang MK, Huang SP, Peng HL, Chang HY, Jan MS, Lai YC. Genetic requirements for Klebsiella pneumoniae-induced liver abscess in an oral infection model. Infect Immun. 2009;77(7):2657–2671. doi: 10.1128/IAI.01523-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi S, Xia W, Guo H, Kong H, Zheng S. Unique characteristics of pyogenic liver abscesses of biliary origin. Surgery. 2016;159(5):1316–1324. doi: 10.1016/j.surg.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 30.Mezhir JJ, Fong Y, Jacks LM, Getrajdman GI, Brody LA, Covey AM, Thornton RH, Jarnagin WR, Solomon SB, Brown KT. Current management of pyogenic liver abscess: surgery is now second-line treatment. J Am Coll Surg. 2010;210(6):975–983. doi: 10.1016/j.jamcollsurg.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Fung CP, Chang FY, Lee SC, Hu BS, Kuo BI, Liu CY, Ho M, Siu LK. A global emerging disease of Klebsiella pneumoniae liver abscess: is serotype K1 an important factor for complicated endophthalmitis? Gut. 2002;50(3):420–424. doi: 10.1136/gut.50.3.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tan TY, Ong M, Cheng Y, Ng LSY. Hypermucoviscosity, rmpA, and aerobactin are associated with community-acquired Klebsiella pneumoniae bacteremic isolates causing liver abscess in Singapore. J Microbiol Immunol Infect. 2019;52(1):30–34. doi: 10.1016/j.jmii.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32(3):e00001–19. [DOI] [PMC free article] [PubMed]
- 34.Struve C, Bojer M, Krogfelt KA. Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect Immun. 2009;77(11):5016–5024. doi: 10.1128/IAI.00585-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russo TA, Gulick AM. Aerobactin synthesis proteins as Antivirulence targets in Hypervirulent Klebsiella pneumoniae. ACS Infect Dis. 2019;5(7):1052–1054. doi: 10.1021/acsinfecdis.9b00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore R, O'Shea D, Geoghegan T, Mallon PW, Sheehan G. Community-acquired Klebsiella pneumoniae liver abscess: an emerging infection in Ireland and Europe. Infection. 2013;41(3):681–686. doi: 10.1007/s15010-013-0408-0. [DOI] [PubMed] [Google Scholar]
- 37.Lam MMC, Wyres KL, Duchene S, Wick RR, Judd LM, Gan YH, Hoh CH, Archuleta S, Molton JS, Kalimuddin S, et al. Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat Commun. 2018;9(1):2703. doi: 10.1038/s41467-018-05114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, Engelthaler DM, Andersen PS, Driebe EM, Keim P, Krogfelt KA. Mapping the evolution of Hypervirulent Klebsiella pneumoniae. MBio. 2015;6(4):e00630. doi: 10.1128/mBio.00630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this manuscript.