Macrophages are professional immune cells that ingest and kill microbes. In this study, we show that different pathogenic bacteria induce the expression of cystathionine γ-lyase (CTH) in macrophages. This enzyme is involved in a metabolic pathway called the reverse transsulfuration pathway, which leads to the production of numerous metabolites, including cystathionine. Phagocytized bacteria use cystathionine to better survive in macrophages. In addition, the induction of CTH results in dysregulation of the metabolism of polyamines, which in turn dampens the proinflammatory response of macrophages. In conclusion, pathogenic bacteria can evade the host immune response by inducing CTH in macrophages.
KEYWORDS: Helicobacter pylori, immune evasion, immunometabolism, innate immunity, macrophages, pathogenic bacteria, polyamines
ABSTRACT
The reverse transsulfuration pathway is the major route for the metabolism of sulfur-containing amino acids. The role of this metabolic pathway in macrophage response and function is unknown. We show that the enzyme cystathionine γ-lyase (CTH) is induced in macrophages infected with pathogenic bacteria through signaling involving phosphatidylinositol 3-kinase (PI3K)/MTOR and the transcription factor SP1. This results in the synthesis of cystathionine, which facilitates the survival of pathogens within myeloid cells. Our data demonstrate that the expression of CTH leads to defective macrophage activation by (i) dysregulation of polyamine metabolism by depletion of S-adenosylmethionine, resulting in immunosuppressive putrescine accumulation and inhibition of spermidine and spermine synthesis, and (ii) increased histone H3K9, H3K27, and H3K36 di/trimethylation, which is associated with gene expression silencing. Thus, CTH is a pivotal enzyme of the innate immune response that disrupts host defense. The induction of the reverse transsulfuration pathway by bacterial pathogens can be considered an unrecognized mechanism for immune escape.
INTRODUCTION
Gastric cancer is the third leading cause of cancer deaths worldwide (http://globocan.iarc.fr/old/FactSheets/cancers/stomach-new.asp). The main etiologic factor is the bacterium Helicobacter pylori, the most common infectious agent associated with cancer. H. pylori infection exclusively develops in the human stomach, and it is estimated that it currently infects 4.4 billion persons worldwide (1). The pathogen incites chronic active gastric inflammation, and disease progresses along a histological cascade from gastritis to atrophic gastritis, intestinal metaplasia, and gastric adenocarcinoma (2). The underlying factor driving these events is the survival of the pathogen due to dysregulated innate and adaptive immune responses.
During infection with H. pylori, gastric epithelial cells (GECs) and myeloid cells develop an innate immune/inflammatory response characterized by the expression of inducible genes encoding chemokines and cytokines, which recruit and activate other myeloid cells as well as lymphocytes to the site of infection, with the latter leading to an adaptive immune response (3, 4). These events support the chronicity of inflammation and, consequently, the risk for neoplastic transformation. GECs and myeloid cells express enzymes producing important effector molecules that can limit the success of the bacterium in its gastric niche but also enhance the immunopathogenesis of the infection, since the immune response generally fails to eradicate the pathogen (3, 4). We have demonstrated that two gasotransmitters are produced and have major effects on the pathophysiology of H. pylori infection: (i) inducible nitric oxide synthase (NOS2)-derived nitric oxide (NO), which is mainly produced by macrophages and affects H. pylori growth (5–7) but has also been associated with gastric carcinogenesis (8), and (ii) carbon monoxide (CO), synthesized by heme-oxygenase 1, which impairs the immune response of macrophages by dampening the M1 phenotype (9) but also dampens signaling by SRC tyrosine kinase, thus inhibiting the phosphorylation of the H. pylori oncoprotein CagA in gastric epithelial cells (10). Epithelial and immune cells of the gastrointestinal tract can produce hydrogen sulfide (H2S), the third gaseous signaling molecule associated with physiological properties (11), including vasorelaxation (12), neuroprotection (13), oxidant regulation (14), and anti-inflammatory action (15). In addition, H2S has been shown to signal in pathogenic bacteria (16). H2S is synthesized in biological milieus by three enzymes, namely, cystathionine β-synthase (CBS), cystathionine γ-lyase (CTH), and mercaptopyruvate sulfurtransferase (MPST). CBS and CTH are involved in the mammalian reverse transsulfuration pathway (RTP); these enzymes catalyze the formation of H2S by cleavage of the sulfur-gamma carbon bond of either l-cysteine, homocysteine, or cystathionine (see Fig. S1 in the supplemental material) (17–19). MPST converts 3-mercaptopyruvate, which is synthesized from cysteine by cysteine aminotransferase, into pyruvate and H2S (Fig. S1) (20).
The reverse transsulfuration and polyamine pathways. AHCY, S-adenosylhomocysteine hydrolase; AMD1, adenosylmethionine decarboxylase 1; BHMT, betaine homocysteine methyltransferase; CBS, cystathionine β-synthase; CTH, cystathionine γ-lyase; dcSAM, S-adenosylmethioninamine (decarboxylated S-adenosylmethionine); H3, histone 3; MAT, methionine adenosyltransferase; MTR, methionine synthase; MTs, methyltransferases; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; SMS, spermine synthase; SRM, spermidine synthase. Download FIG S1, PDF file, 0.05 MB (52.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
CBS and CTH use homocysteine as a first substrate in the transsulfuration metabolic pathway. Homocysteine is generated in mammalian cells from methionine (Fig. S1): first, methionine is converted by methionine adenosyltransferase (MAT1A and MAT2A) into S-adenosylmethionine (SAM); second, SAM is metabolized by histone-lysine N-methyltransferases (MTs) into S-adenosylhomocysteine (SAH) plus dimethylated histones; and finally, the S-adenosylhomocysteine hydrolase (AHCY) converts SAH to homocysteine. Of importance, SAM is also a precursor for the synthesis of the polyamines spermidine and spermine (Fig. S1): SAM is first converted by the enzyme adenosylmethionine decarboxylase 1 (AMD1) into S-adenosylmethioninamine (also known as decarboxylated S-adenosylmethionine [dcSAM]), and this product is required for the sequential conversion of putrescine to spermidine by spermidine synthase (SRM) and of spermidine to spermine by spermine synthase (SMS) (21). Thus, SAM is a common substrate for the transsulfuration and the polyamine metabolic pathways, but the interconnection between these pathways is unknown. This can be critical because putrescine impairs the M1 macrophage response during bacterial infections (22), and spermine modulates the translation of proinflammatory mediators in H. pylori-infected macrophages (23).
The induction and role of the RTP during bacterial infection remain unknown. In the present report, we found that CTH, but not CBS, is induced by H. pylori and other pathogens in macrophages and supports bacterial growth by the generation of cystathionine. In addition, CTH affects the synthesis of spermidine and spermine by depletion of dcSAM, which results in an increase of the putrescine concentration in macrophages. Finally, the induction of the RTP increases histone modifications, thus inhibiting gene transcription and macrophage activation. Together, these data emphasize a novel strategy used by bacterial pathogens to dysregulate the innate immune response to favor their survival in the infected host.
RESULTS
H. pylori induces CTH in murine macrophages by a PI3K/MTOR/SP1 pathway.
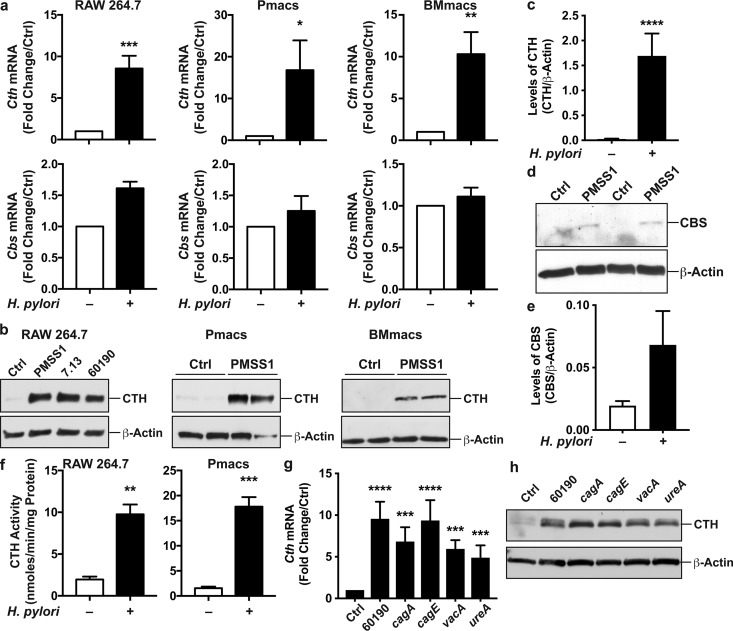
In response to infection with H. pylori PMSS1, mRNA levels of Cth were increased in RAW 264.7 cells as well as in peritoneal macrophages (Pmacs) and bone marrow-derived macrophages (BMmacs) from C57BL/6 mice compared to uninfected cells (Fig. 1a). In contrast, the expression of the other gene of the RTP, Cbs, was not induced by H. pylori in these cells (Fig. 1a). Moreover, the expression of Mpst, which encodes the third H2S-producing enzyme MPST, was not enhanced by H. pylori infection; in fact, the expression levels in H. pylori-infected RAW 264.7 cells and BMmacs were 0.52- ± 0.12- and 0.08- ± 0.05-fold lower than those in uninfected cells, respectively. By Western blotting, CTH protein levels were induced by three different strains of H. pylori in RAW 264.7 cells (Fig. 1b), shown to exhibit a 66-fold increase by densitometry (Fig. 1c), and also upregulated in infected primary macrophages (Fig. 1b). By a proteomic approach using isobaric tag for relative and absolute quantification (iTRAQ) technology, we also found that the CTH level was increased by 1.968-fold in infected macrophages compared to that in control cells (P = 2.8 × 10−5). The CBS level was not significantly increased by Western blotting in infected macrophages (Fig. 1d and e) and also was not detected in the iTRAQ analysis. The increased CTH expression in H. pylori-infected macrophages was associated with an enhancement of endogenous CTH activity (Fig. 1f). However, Cth mRNA expression was not induced by H. pylori in the murine gastric epithelial cell line ImSt (mRNA expression in H. pylori-infected cells, 1.01- ± 0.48-fold versus uninfected cells).
FIG 1.
CTH and CBS expression in macrophages. (a to f) RAW 264.7 cells, Pmacs, and BMmacs were infected with H. pylori PMSS1, 7.13, or 60190. (a) After 6 h, Cth and Cbs mRNA expression levels were analyzed by RT–real-time PCR. (b) CTH protein levels were assessed by Western blotting after 24 h. Shown are representative data from 4 experiments for RAW 264.7 cells and from 4 mice for Pmacs and BMmacs. (c) Densitometric analysis of CTH in RAW 264.7 cells. (d and e) The level of CBS protein was determined by Western blotting (d), and densitometric analysis was performed in 4 independent experiments (e). (f) CTH activity was analyzed in RAW 264.7 cells and in Pmacs after a 24-h infection. Shown are averages from 3 duplicate experiments. (g and h) RAW 264.7 cells were infected with H. pylori 60190 or with the cagA, cagE, vacA, and ureA isogenic mutants. After 6 and 24 h, Cth mRNA expression (g) and CTH protein (h) levels were analyzed by RT–real-time PCR and Western blotting (representative blot from 2 different experiments), respectively. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (versus uninfected cells) (n = 3 to 8 for RAW 264.7 cells and n = 4 mice for Pmacs and BMmacs).
To address the role of H. pylori virulence factors, RAW 264.7 macrophages were infected with mutant strains. Notably, the same level of CTH induction was observed in macrophages infected with H. pylori 60190 or with the isogenic mutants lacking the main virulence factors cagA, cagE, vacA, and ureA (Fig. 1f and g).
To determine whether Cth expression is specific for H. pylori infection, we also stimulated macrophages with the Gram-negative pathogenic bacteria Campylobacter jejuni, Salmonella enterica serovar Typhimurium, and Shigella flexneri or with the Gram-positive bacterium Staphylococcus aureus; we also used two commensal bacteria isolated from human gastric tissues, Streptococcus salivarius and Staphylococcus epidermidis (24). As expected for professional phagocytes, Cth was induced by all bacteria (see Fig. S2 in the supplemental material).
Induction of Cth in macrophages in response to different bacteria. RAW 264.7 cells were infected with H. pylori PMSS1 (Hp), C. jejuni (Cj), S. enterica serovar Typhimurium (ST), S. flexneri (Sf), S. aureus (Sa), S. salivarius (Ss), or S. epidermidis (Se). After 6 h, Cth mRNA expression was analyzed by RT–real-time PCR. ***, P < 0.001; ****, P < 0.0001 (versus uninfected cells) (n = 3). Download FIG S2, PDF file, 0.4 MB (431.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
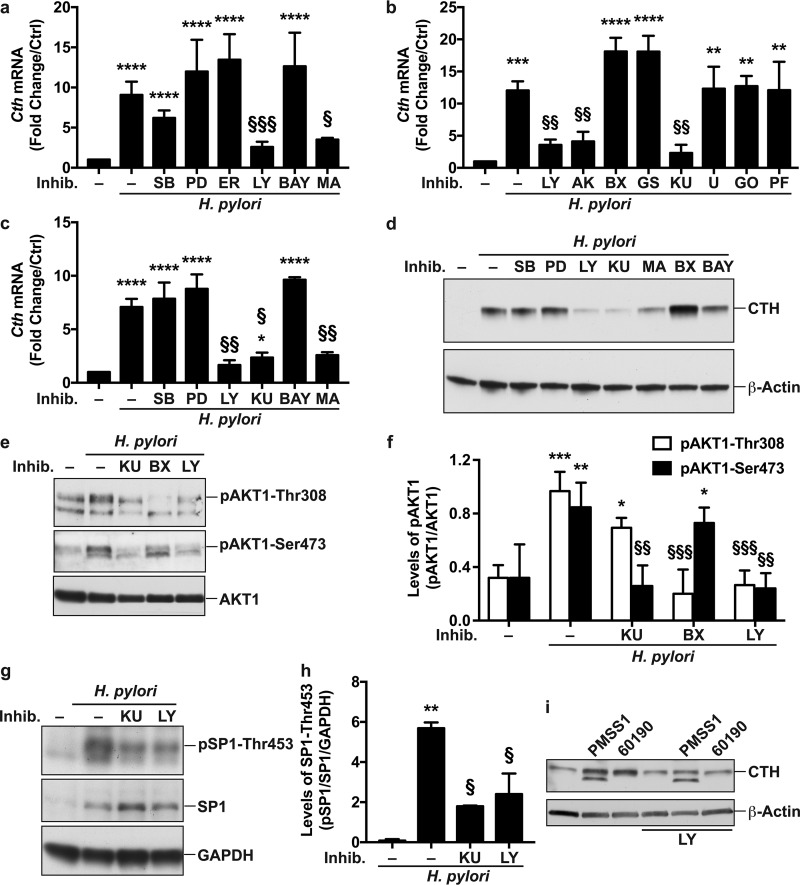
We then analyzed the molecular mechanism by which H. pylori stimulates Cth gene expression. A pharmacological approach, aiming at inhibiting the main signaling events in macrophages (Fig. S3), evidenced that blocking phosphatidylinositol 3-kinase (PI3K) with LY294002 or SP1 with mithramycin A led to a significant reduction in Cth mRNA levels in infected macrophages (Fig. 2a). However, inhibitors of mitogen-activated protein kinase 14 (MAPK14) (also known as P38 MAPK), MAP2K1 (MEK1), MAPK1/3 (extracellular signal-regulated kinase 1/2 [ERK1/2]), or NF-κB had no effect on H. pylori-induced Cth transcription. We verified the specificity of these results by analyzing Nos2 mRNA expression, which was induced by H. pylori and inhibited by the NF-κB inhibitor but not by the other inhibitors (Fig. S4). To gain further insight into the molecular signaling initiated by PI3K and leading to Cth expression, we used other pharmacological inhibitors related to the PI3K pathway (Fig. S3) and found that blocking AKT1 or mechanistic target of rapamycin kinase (MTOR) reduced H. pylori-stimulated Cth expression similarly to the PI3K inhibitor, whereas inhibition of pyruvate dehydrogenase kinase 1 (PDK1), serum/glucocorticoid-regulated kinase 1 (SGK1), phospholipase C beta 2 (PLCB2), protein kinase C (PKC), or ribosomal protein S6 kinase B1 (RPS6KB1) did not affect Cth induction (Fig. 2b). The essential role of PI3K/MTOR/SP1 signaling in H. pylori-mediated Cth gene expression was then recapitulated in BMmacs; there was decreased expression of Cth in infected cells treated with the PI3K, MTOR, or SP1 inhibitors but not in those treated with MAPK14, MAP2K1, or NF-κB inhibitors (Fig. 2c). Accordingly, inhibition of PI3K, MTOR, or SP1 led to a reduction of CTH protein expression induced by H. pylori, whereas blockade of MAPK14, MAP2K1, or PDK1 signaling did not affect the level of CTH in infected macrophages (Fig. 2d).
FIG 2.
Signal transduction involved in CTH induction. (a to c) RAW 264.7 cells (a and b) or BMmacs (c) were pretreated or not for 30 min with SB203580 (SB), PD98059 (PD), an ERK inhibitor (ER), LY294002 (LY), Bay 11-7082 (BAY), mithramycin A (MA), an AKT inhibitor (AK), BX795 (BX), GSK650394 (GS), KU0063794 (KU), U-73122 (U), GO6983 (GO), or PF-4708671 (PF) for 30 min prior to infection with H. pylori PMSS1. Cth mRNA expression was analyzed after 6 h. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 (versus uninfected cells); §, P < 0.05; §§, P < 0.01; §§§, P < 0.001 (versus H. pylori-infected cells) (n = 3 to 6 for panels a and b, and n = 4 mice for panel c). (d) CTH and β-actin levels were assessed by Western blotting from macrophages pretreated with the pharmacological inhibitors and infected with H. pylori for 24 h. Shown are representative data from 3 independent experiments. (e) Representative Western blot (n = 3) of pAKT1-Thr308, pAKT1-Ser473, and AKT1 in macrophages 1 h after infection with H. pylori PMSS1. (f) Densitometric analysis of pAKT1-Thr308, pAKT1-Ser473, and AKT1 levels. (g) Representative Western blot (n = 3) of pSP1-Thr453, SP1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in macrophages after 3 h of infection with H. pylori PMSS1. (h) Densitometric analysis of pSP1-Thr453. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (versus uninfected cells); §, P < 0.05; §§, P < 0.01; §§§, P < 0.001 (versus H. pylori-infected cells) (for panels f [n = 3] and h [n = 3]). (i) CTH and β-actin levels in PMA-differentiated THP-1 cells, pretreated or not for 30 min with LY294002 and then infected with H. pylori PMSS1 or 60190 for 24 h. Shown is a representative blot from 2 independent experiments.
Signaling pathways and inhibitors. The inhibitors of each of the signaling molecules are indicated in red. Download FIG S3, PDF file, 0.3 MB (324.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Expression of Nos2 in response to H. pylori and to different inhibitors. RAW 264.7 cells were pretreated with SB203580 (SB), PD98059 (PD), an ERK inhibitor (ER), LY294002 (LY), or Bay 11-7082 (BAY) for 30 min prior to infection with H. pylori PMSS1; Nos2 mRNA expression was analyzed after 6 h. **, P < 0.01; ****, P < 0.0001 (versus uninfected cells); §§, P < 0.01 (versus H. pylori-infected cells) (n = 3 to 6). Download FIG S4, PDF file, 0.10 MB (102.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
AKT1 is a signaling protein downstream of PI3K; AKT1 is phosphorylated by PDK1 on Thr308 (25) and on Ser473 by MTORC2 (26) (Fig. S3). Thus, to further demonstrate the activation of the PI3K pathway in macrophages in response to H. pylori, we analyzed AKT1 activation. We found that AKT1 was phosphorylated on Thr308 and Ser473 in response to H. pylori infection and that this was inhibited by the PI3K inhibitor LY294002 (Fig. 2e and f). Moreover, blocking MTOR by KU0063794 resulted in a significant reduction of AKT1 phosphorylation on Ser473 (pAKT1-Ser473; Fig. 2e and f); the PDK1 inhibitor BX795 had no effect on pAKT1-Ser473 levels (Fig. 2e and f). Inversely, AKT1 phosphorylation on Thr308 was suppressed by the PDK1 inhibitor but not by the MTOR inhibitor (Fig. 2e and f). Moreover, we observed that the level of pSP1-Thr453 was increased with H. pylori infection (Fig. 2g and h). Surprisingly, the total SP1 level was also increased with infection (Fig. 2g), suggesting that H. pylori induces SP1 expression. H. pylori-stimulated SP1 activation was inhibited by 68.4% ± 2.2% and by 58.5% ± 15.9% in the presence of MTOR and PI3K inhibitors, respectively (Fig. 2g and h).
Finally, we also found that CTH was induced by 4.14- and 5.49-fold, by densitometry analysis of Western blots, in the human macrophage cell line THP-1 differentiated with phorbol 12-myristate 13-acetate (PMA) and infected by H. pylori PMSS1 and 60190, respectively (Fig. 2i). These inductions were repressed by ∼30% and ∼69%, respectively, using the AKT1 inhibitor (Fig. 2i).
These results demonstrate that H. pylori induces Cth expression through a PI3K/MTOR/SP1 signaling pathway. This was present in both murine and human macrophages.
Infection with H. pylori results in enhanced CTH levels in gastric macrophages.
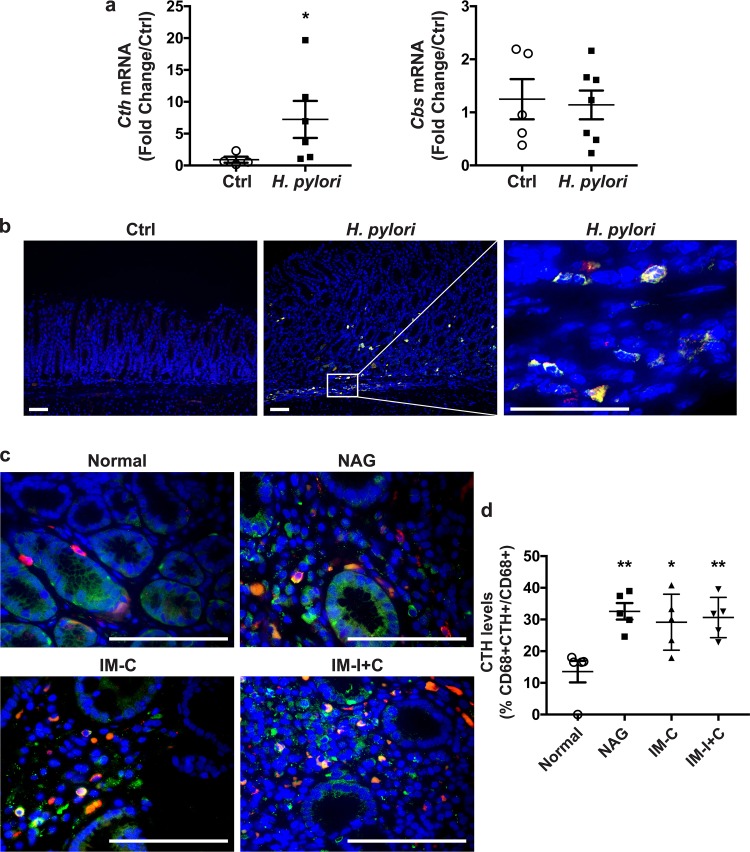
To demonstrate the in vivo relevance of our findings, we analyzed the expression of CTH in gastric tissues of C57BL/6 mice infected or not with H. pylori PMSS1. Levels of Cth mRNA in infected mice were significantly increased compared with those in uninfected animals (Fig. 3a). However, the expression of the gene Cbs was not modulated by H. pylori infection. In addition, we observed that CTH levels were increased in C57BL/6 mice infected with H. pylori compared with those in uninfected mice (Fig. 3b) and that CTH staining colocalized to cells that were positive for the macrophage marker CD68 (Fig. 3b).
FIG 3.
Expression of CTH in gastric tissue during H. pylori infection. (a) Cth and Cbs mRNA expression in the gastric tissue of C57BL/6 mice infected or not (control [Ctrl]) with H. pylori PMSS1 for 2 months. *, P < 0.05 (versus uninfected cells). (b) The macrophage marker CD68 (red), CTH (green), and nuclei (blue) were detected by immunofluorescence. Merged images are shown, with cells double positive for CD68 and CTH depicted in yellow. Representative images for 5 mice in each group are shown. (c) Expression of CTH and CD68 in patients infected or not (normal) with H. pylori, as described above for panel b. (d) Quantification of CTH staining in macrophages. NAG, nonatrophic gastritis; IM-C, complete intestinal metaplasia; IM-I+C, incomplete and complete intestinal metaplasia. *, P < 0.05; **, P < 0.01 (versus normal patients).
Similarly, increased double staining for CTH and CD68 was observed in gastric tissues from patients infected with H. pylori compared to that in tissues from individuals without infection (Fig. 3c and d). CTH staining was similar in patients with nonatrophic gastritis, complete intestinal metaplasia, and complete plus incomplete intestinal metaplasia (Fig. 3c and d), demonstrating that infection is associated with CTH induction in vivo and persists during progression along the histological cascade toward carcinogenesis.
CTH supports H. pylori growth by generating cystathionine.
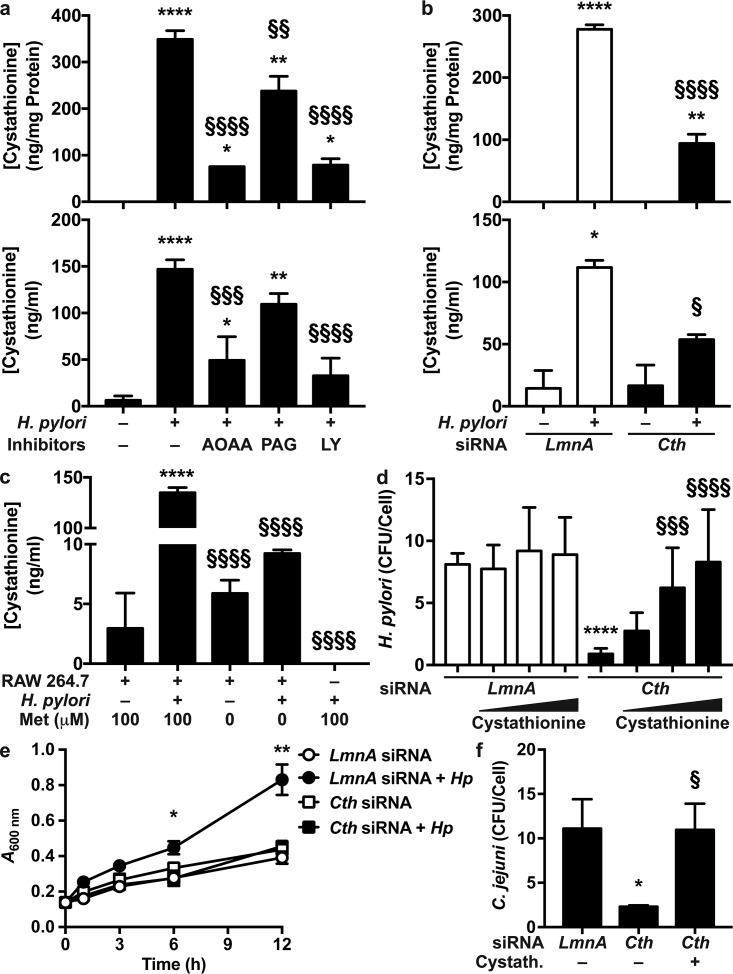
Because cystathionine has been shown to regulate bacterial growth, we reasoned that CTH induction may affect H. pylori survival in macrophages (27). We thus first analyzed cystathionine synthesis by macrophages. The intracellular and extracellular concentrations of cystathionine were dramatically increased in H. pylori-infected macrophages (Fig. 4a and b). In addition, cystathionine generation was significantly reduced by a general inhibitor of pyridoxal phosphate (PLP)-dependent enzymes, aminooxyacetic acid (AOAA), which thus also inhibits CTH and CBS activity (28), and by the more specific CTH inhibitor propargylglycine (PAG) (Fig. 4a). Note that the production of cystathionine was more inhibited by AOAA, which is consistent with the greater potency of AOAA in inhibiting CTH than of PAG (28). Of importance, the PI3K inhibitor LY294002, which suppressed CTH induction (Fig. 2a), also blocked cystathionine production by H. pylori-infected macrophages (Fig. 4a). These data were then confirmed by a molecular approach: the intracellular content of cystathionine and its concentration in the extracellular milieu were reduced when macrophages were transfected with Cth small interfering RNA (siRNA) before infection with H. pylori (Fig. 4b). Furthermore, we also evidenced in another set of experiments that Cth siRNA, but not Cbs siRNA, reduced H. pylori-induced synthesis of cystathionine and lanthionine (Fig. S5a), demonstrating that H. pylori enhances RTP activity by stimulating CTH specifically. The efficacy of the knockdown of CTH expression by Cth siRNA, but not by Cbs siRNA, is depicted in Fig. S5b.
FIG 4.
Effect of CTH on H. pylori survival. (a) Macrophages were treated with LY294002 (LY) for 30 min and infected or not with H. pylori for 24 h. Cells were washed, and fresh medium containing antibiotics and AOAA or PAG was added. After 24 h, the cystathionine concentration was determined in cells (top) and supernatants (bottom). *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (versus uninfected cells); §§, P < 0.01; §§§, P < 0.001; §§§§, P < 0.0001 (versus infected cells without an inhibitor) (n = 5 to 7). (b) Macrophages were transfected with LmnA or Cth siRNA and infected or not with H. pylori for 24 h. Cells were washed, and fresh medium containing antibiotics was added. The cystathionine concentration was measured in cells (top) and supernatants (bottom) after 24 h. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (versus uninfected cells); §, P < 0.05; §§§§, P < 0.0001 (versus cells transfected with LmnA siRNA and infected with H. pylori) (n = 3). (c) Macrophages were infected or not with H. pylori PMSS1, in the presence or absence of 100 μM methionine (Met). Bacteria were also cultured in the same medium without cells. After 24 h, the cystathionine concentration was determined in macrophages or bacteria. ****, P < 0.0001 (versus uninfected cells); §§§§, P < 0.0001 (versus infected cells) (n = 3). (d) Macrophages were transfected with LmnA or Cth siRNA and then infected with H. pylori for 24 h, with or without cystathionine (50 to 1,000 ng/ml). The number of CFU in macrophages was determined by a gentamicin assay. ****, P < 0.0001 (versus cells transfected with LmnA siRNA); §§§, P < 0.001; §§§§, P < 0.0001 (versus cells transfected with Cth siRNA without cystathionine treatment) (n = 3). (e) Macrophages were transfected with LmnA or Cth siRNA and then infected or not with H. pylori (Hp) for 24 h. Cells were washed, and fresh medium was added. After 24 h, supernatants were collected, filtered, and used to grow H. pylori. *, P < 0.05; **, P < 0.01 (versus all other conditions) (n = 5). (f) Macrophages were transfected with LmnA or Cth siRNA and then infected with C. jejuni with or without 100 ng/ml cystathionine for 24 h. The number of CFU in macrophages was determined by a gentamicin assay. *, P < 0.05 (versus cells transfected with LmnA siRNA); §, P < 0.05 (versus cells transfected with Cth siRNA without cystathionine) (data are from 4 experiments).
Effect of Cth and Cbs silencing on metabolites of the RTP. (a) RAW 264.7 macrophages transfected with LmnA, Cth, or Cbs siRNA were infected or not with H. pylori for 24 h. Cells were washed, and fresh medium containing antibiotics was added. The cystathionine concentration was measured in cells by LC-MS after 24 h. *, P < 0.05; **, P < 0.01 (versus uninfected cells); §, P < 0.05 (compared to cells transfected with LmnA siRNA and infected with H. pylori) (n = 4). (b) The CTH protein level was assessed by Western blotting after 24 h. Shown are representative data from 2 experiments. Download FIG S5, PDF file, 0.5 MB (486.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
We ensured that cystathionine was neosynthesized by macrophages and not by H. pylori itself by showing that the bacteria did not generate detectable levels of cystathionine when grown without eukaryotic cells (Fig. 4c). We also observed that cystathionine was not released by infected cells in a medium devoid of methionine (Fig. 4c), evidencing the important role of the methionine cycle in CTH-dependent cystathionine synthesis.
To determine the role of CTH induction in H. pylori survival, we first used AOAA, the best inhibitor of cystathionine production in our model, in macrophage-bacterium cocultures. However, we observed that H. pylori growth was directly inhibited by AOAA in a concentration-dependent manner (Fig. S6a). Next, we thus assessed the effect of Cth silencing on H. pylori survival. Blocking Cth expression using siRNA resulted in a 9.1-fold reduction in the number of live H. pylori bacteria per macrophage (Fig. 4d). This effect was reversed by the addition of increasing concentrations of cystathionine in the coculture (Fig. 4d). Note that the addition of cystathionine to macrophages transfected with LmnA siRNA as a negative control did not result in further survival of intracellular H. pylori bacteria (Fig. 4d). To confirm this result and to determine the effect of extracellular cystathionine on H. pylori growth, we generated conditioned medium from macrophages transfected with LmnA or Cth siRNA and infected or not with H. pylori. These conditioned media were filtered and used to culture H. pylori. H. pylori grew more efficiently in the conditioned medium obtained from RAW 264.7 cells transfected with LmnA siRNA and infected with H. pylori than in (i) the medium of uninfected macrophages transfected with LmnA siRNA and (ii) the medium of cells transfected with Cth siRNA, infected or not with H. pylori (Fig. 4e). Together, these data suggest that induction of CTH expression/activity by H. pylori enhances its intra- or extracellular viability. We also found that the H2S donor sodium hydrosulfide (NaHS) does not affect H. pylori growth (Fig. S6b), suggesting that H2S does not exhibit a major effect on H. pylori survival. Thus, CTH-mediated H. pylori survival is due to cystathionine.
Effect of AOAA and NaHS on H. pylori growth. H. pylori PMSS1 was grown in Bertani broth with 10% FBS with various concentrations of AOAA (a) or 500 μM NaHS (b). The optical density at 600 nm was monitored at the indicated times (n = 3 independent experiments). Download FIG S6, PDF file, 0.2 MB (169.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Finally, the effect of CTH/cystathionine on the survival of C. jejuni was assessed. Blocking of Cth expression by siRNA resulted in a significant decrease in the number of live C. jejuni bacteria in macrophages compared to cells with control knockdown (Fig. 4f). This effect was reversed when exogenous cystathionine was added to the coculture medium (Fig. 4f), evidencing that CTH also supports the growth of C. jejuni.
H. pylori-induced CTH regulates polyamine synthesis.
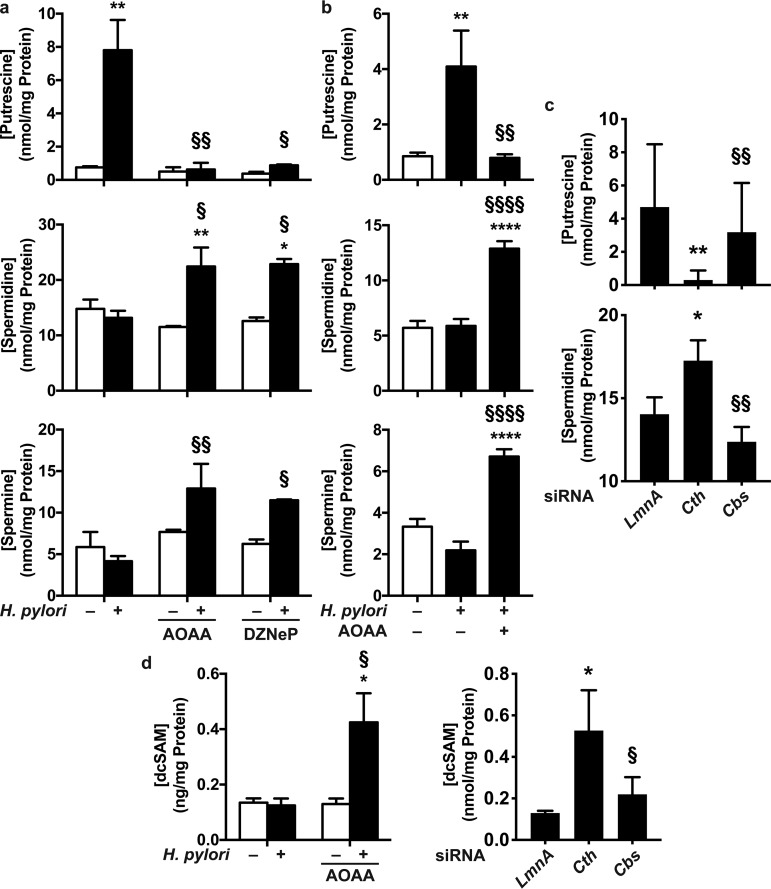
Because SAM is a common substrate for MTs and AMD1 (Fig. S1), we hypothesized that increased expression of CTH in H. pylori-infected macrophages may favor the use of SAM toward homocysteine synthesis, resulting in a diminution of dcSAM synthesis and, thus, a decrease in spermine and spermidine production. To test this postulate, RAW 264.7 macrophages were first infected or not with H. pylori for 24 h to induce CTH expression in infected cells; cells were then washed; fresh medium with or without AOAA or an inhibitor of MTs, 3-deazaneplanocin (DZNeP), was added to the cells for 24 h; and polyamine concentrations were analyzed in these macrophages. We observed that the putrescine level was increased in H. pylori-infected macrophages, whereas spermine and spermidine levels were not significantly modified by infection (Fig. 5a), as we previously reported (22). In the presence of AOAA or DZNeP, the putrescine concentration was markedly reduced in infected macrophages (Fig. 5a); this was concomitantly associated with a significant increase in spermidine and spermine concentrations in cells infected with H. pylori (Fig. 5a). The concentrations of all three polyamines were not affected by AOAA or DZNeP in uninfected macrophages (Fig. 5a). Similarly, the concentration of putrescine, but not spermidine or spermine, was increased in BMmacs infected with H. pylori compared to that in uninfected cells (Fig. 5b); again, in the presence of AOAA, the putrescine concentration was significantly reduced, whereas the generation of spermidine and spermine was restored (Fig. 5b). Moreover, silencing of Cth in infected RAW 264.7 cells led to a significant reduction in the putrescine concentration and an increase of the spermidine level (Fig. 5c); these changes were not observed with Cbs siRNA (Fig. 5c). Importantly, both AOAA and Cth siRNAs, but not Cbs siRNA, significantly enhanced the intracellular concentration of dcSAM in H. pylori-infected macrophages (Fig. 5d), demonstrating that blocking of CTH activity reestablishes the metabolism of SAM through the spermidine/spermine pathway.
FIG 5.
Role of CTH in polyamine production. (a and b) RAW 264.7 macrophages (a) or BMmacs (b) were infected or not with H. pylori for 24 h. Cells were washed, and fresh medium containing antibiotics with or without AOAA or DZNeP was added. Polyamine concentrations were measured in macrophages after 24 h. *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (compared to uninfected cells); §, P < 0.05; §§, P < 0.01; §§§§, P < 0.0001 (versus H. pylori-infected macrophages) (n = 3). (c) RAW 264.7 macrophages were transfected with LmnA, Cth, or Cbs siRNA and then infected with H. pylori for 24 h. Cells were washed, and fresh medium containing antibiotics was added. Polyamine concentrations were measured in macrophage lysates after 24 h. *, P < 0.05; **, P < 0.01 (versus infected cells with LmnA siRNA); §§, P < 0.01 (compared to cells transfected with Cth siRNA) (n = 4). (d) Intracellular concentrations of dcSAM were determined in RAW 264.7 macrophages stimulated as described above for panels a and c.
The effect of the CTH inhibitor on polyamine synthesis in macrophages was then confirmed by metabolomics. The 7 databases that were interrogated evidenced that polyamine metabolism is significantly modulated by H. pylori and by CTH inhibition (Table S1); furthermore, metabolism of polyamines was the pathway that was the most significantly regulated according to the Edinburgh Human Metabolic Network (EHMN), the Integrating Network Objects with Hierarchies (INOH) database, the Kyoto Encyclopedia of Genes and Genomes (KEGG), the Reactome database, Wikipathways, and the Small Molecule Pathway Database (SMPDB) (Table S1). As a proof of principle that the metabolism of CTH was also affected by infection and by AOAA, the metabolism of methionine and cysteine was evidenced by 6 of the 7 databases (Table S1).
The metabolomics pathways significantly regulated by H. pylori infection and AOAA. Macrophages were infected or not with H. pylori with or without AOAA for 24 h. The metabolome of each group was subsequently assessed, and the data were analyzed using IMPaLA. For each database, P denotes the statistical confidence that the metabolic pathway is altered. Only pathways that were determined to be significant are included. The rows underlined in blue represent the pathways related to methionine/cysteine metabolism, the rows in red depict the polyamine pathways, and the rows in green represent the arginine/ornithine metabolism pathway, which is related to the polyamine pathway (see http://www.genome.jp/kegg-bin/show_pathway?map00330). Note that the KEGG glutathione pathway (row in yellow) includes cysteine, methionine, and polyamine pathways (see http://www.genome.jp/kegg/pathway/map/map00480.html) (n = 2 independent experiments assessed in triplicate). Download Table S1, PDF file, 0.2 MB (165KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Macrophage activation is regulated by CTH.
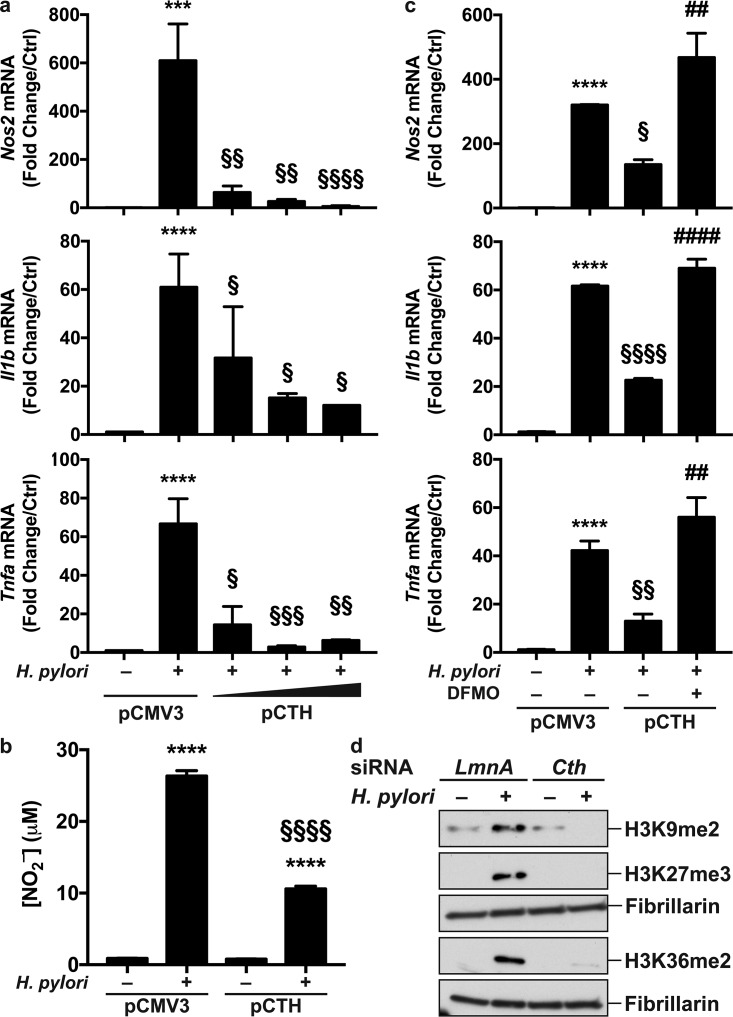
We recently reported that putrescine generated by increased ornithine decarboxylase (ODC) expression dampens macrophage activation through histone modifications and altered euchromatin formation (22). Because we observed that H. pylori-mediated CTH induction increases putrescine concentrations, we hypothesized that CTH is also involved in macrophage activation. Since (i) transfection of cells with Cth siRNA diminishes the number of live H. pylori bacteria in infected cells (Fig. 4d), which would limit macrophage activation, (ii) AOAA directly kills H. pylori, and (iii) H. pylori-induced M1 genes are expressed before the increase of CTH activity, we therefore transfected macrophages with a pCMV3 plasmid harboring the human CTH gene (pCTH) before infection. Cells transfected with pCTH had increased CTH mRNA expression compared to cells transfected with the empty vector (Fig. S7). In accordance with our hypothesis, H. pylori-induced Nos2, Il1b, and Tnfa mRNA expressions were markedly inhibited in macrophages expressing the human CTH gene compared to pCMV3-tranfected cells infected with H. pylori (Fig. 6a). Likewise, the production of NO2– by H. pylori-infected cells was reduced by pCTH transfection (Fig. 6b). The decrease of gene expression in cells transfected with pCTH was reversed by the ODC inhibitor difluoromethylornithine (DFMO) (Fig. 6c). Note that the levels of putrescine, the product of ODC, were as follows: 0.71 nmol/mg protein in uninfected cells, 3.79 nmol/mg protein in H. pylori-infected cells, and 0 nmol/mg protein in H. pylori-infected cells plus DFMO (n = 2 independent experiments). This demonstrates that CTH dampens macrophage activation by increasing putrescine accumulation in macrophages.
FIG 6.
CTH regulates macrophage activation. (a and b) RAW 264.7 cells were transfected with pCMV3 (0.5 μg/ml) or pCTH (0.05, 0.1, and 0.5 μg/ml) and then infected or not with H. pylori for 6 h. (a) The levels of Nos2, Il1b, and Tnfa mRNAs were analyzed by RT–real-time PCR. (b) The NO2– concentration was assessed after 24 h. ***, P < 0.01; ****, P < 0.0001 (versus uninfected cells); §, P < 0.05; §§, P < 0.01; §§§, P < 0.001; §§§§, P < 0.0001 (versus cells transfected with pCMV3 and infected with H. pylori) (n = 5). (c) Macrophages expressing pCMV3 (0.5 μg/ml) or pCTH (0.5 μg/ml) were treated for 24 h with DFMO and then infected with H. pylori for 6 h. Gene expression was assessed by RT–real-time PCR. ****, P < 0.0001 (versus uninfected cells); §, P < 0.05; §§, P < 0.01; §§§§, P < 0.0001 (versus infected cells transfected with pCMV3); ##, P < 0.01; ####, P < 0.0001 (versus infected pCTH-expressing macrophages) (n = 3). (d) Macrophages transfected with LmnA or Cth siRNA were then infected or not with H. pylori for 24 h. Nuclear proteins were extracted, and H3 methylation was assessed by Western blotting; shown are representative data from 3 independent experiments.
Expression of CTH mRNA. RAW 264.7 cells were transfected with pCMV3 (0.5 μg/ml) or pCTH (0.05, 0.1, and 0.5 μg/ml). After 24 h, the level of CTH mRNA was analyzed by RT–real-time PCR (n = 5). NA, not amplified. Download FIG S7, PDF file, 0.1 MB (53.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Finally, since (i) increased CTH activity in H. pylori-infected macrophages favors the metabolism of SAM toward the RTP (Fig. 5c) and (ii) SAM is first converted into SAH by MTs, thus supporting the di-/trimethylation of histones, we reasoned that CTH activity might alter immune gene expression by regulating histone methylation. Using an enzyme-linked immunosorbent assay (ELISA)-like assay, we found that the levels of H3K9me2, H3K27me3, H3K36me2, and H3K79me3 were increased in cells transfected with pCTH compared to those in cells transfected with the empty vector and that this increase was completely reversed by AOAA (Fig. S8). Furthermore, we confirmed by Western blotting that H3K9, H3K27, and H3K36 were di-/trimethylated in macrophages infected with H. pylori (Fig. 6d), and this effect was eliminated in cells transfected with Cth siRNA (Fig. 6d).
H3 methylation. Macrophages were transfected with pCMV3 (0.5 μg/ml) or pCTH (0.5 μg/ml). After 24 h, medium was removed, and fresh medium containing or not AOAA was added. The levels of H3 mono-, di-, or trimethylation were evaluated by an ELISA. Shown are representative data from two experiments. Download FIG S8, PDF file, 0.2 MB (204KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
The emerging picture of our investigation is that we have evidenced a novel strategy by which bacterial pathogens escape innate immunity. CTH expression through a PI3K/MTOR/SP1 signaling pathway enhances the survival of bacteria in macrophages by generating cystathionine and also dampens the inflammatory and antimicrobial response by modulating polyamine metabolism and by supporting histone H3 di-/trimethylation. CBS is not significantly induced in macrophages infected with H. pylori. It should be noted that because CBS has similar metabolic functions as CTH, a potential increase of CBS under other conditions would be expected to yield the same overall biological effects.
In gastric epithelial cells, the PI3K/AKT1 pathway can be induced by CagA-dependent induction of the tyrosine-protein kinase MET (29, 30), through a direct interaction of native CagA with PI3K (31), and/or by epidermal growth factor receptor (EGFR) signaling (32). Moreover, activation of the MTOR pathway has also been shown to occur in epithelial cells but independently of CagA signaling (32). The activation of the classical PI3K/PDK1 pathway in H. pylori-induced macrophages has also been described (33), and AKT1 phosphorylation on Thr308 was shown to be involved in the phagocytosis of H. pylori and in NF-κB activation (33, 34). In the present report, we demonstrate that H. pylori signals in macrophages to activate the PI3K and MTOR pathways, leading to the phosphorylation of AKT1 on Ser473. Interestingly, blocking of PI3K, MTOR, or AKT1 results in decreased phosphorylation/activation of the transcription factor SP1 and inhibition of Cth mRNA expression; however, the inhibition of PDK1 did not result in a decrease of Cth mRNA expression, suggesting that only the phosphorylation of Ser473 is required for the AKT1-dependent induction of Cth gene transcription. Finally, pharmacological inhibition of SP1 also reduces H. pylori-mediated Cth induction. We therefore propose that the PI3K/MTOR/AKT1/SP1 signal is the main transduction pathway stimulated by H. pylori involved in the regulation of the innate function of macrophages through CTH induction. Supporting this concept, the activation of SP1 by PI3K has been described in murine macrophages stimulated with IFN-γ (35) and in the human macrophage cell line THP-1 activated by heat shock protein 27 (36). In addition, SP1 can be also activated by MTOR (37). Finally, the SP1 binding site (GC box) exists in the promoter region of the murine Cth gene (position −120), and it has been reported that Cth expression in tumor necrosis factor alpha (TNF-α)-stimulated macrophages depends on SP1 activation (38). Conversely, it has been described that lipopolysaccharide (LPS) induces CTH expression in macrophages through a MAPK1/3- and NF-κB-dependent pathway (39). Since H. pylori does not signal through Toll-like receptor 4 (TLR4) in macrophages, this can explain the discrepancy between our results and those of that study. Moreover, the signaling molecule MTOR is associated with different partners to form the MTORC1 and MTORC2 complexes. MTORC1 is activated by AKT1 (40), whereas the PI3K-dependent stimulation of MTORC2 facilitates the phosphorylation of AKT1 on Ser473 (41). We therefore propose that MTORC1 and MTORC2 are involved in the transcription of Cth by an SP1-dependent pathway.
Most microbes use only the forward transsulfuration pathway that converts cysteine to cystathionine and to homocysteine using sequentially cystathionine γ-synthase and cystathionine β-lyase (27, 42). Comparably to other pathogens from the genera Listeria, Mycobacterium, and Enterococcus (43), H. pylori encodes both CBS-like (CysKHp or MccA) and CTH-like (MetBHp or MccB) enzymes that catalyze the RTP as in mammals (27). It also harbors a luxSHp gene encoding an enzyme that converts S-ribosylhomocysteine into homocysteine, which is then metabolized into cystathionine by MccA (27). It is essential to note that strains of H. pylori that lack the mccA or luxSHp gene cannot synthesize cystathionine and fail to grow without cysteine (27); however, the growth of the mutants can be restored by providing exogenous cystathionine (27), demonstrating the essential role of the RTP for H. pylori. In this context, the CTH-dependent generation of cystathionine by macrophages increases homocysteine and cysteine syntheses in H. pylori, which are essential for the resistance of H. pylori to oxidative and nitrosative stresses (44, 45). These data support our results showing that blocking of CTH induction by Cth siRNA inhibits the ability of H. pylori to survive in macrophages and that this was reversed by the addition of cystathionine to the coculture supernatant. We thus propose that H. pylori favors its own intracellular survival by forcing macrophages to produce cystathionine via CTH to fight the detrimental milieu of the phagosome. We propose to define by “nutritional escape” the mechanism by which a pathogen induces the production by the host of a metabolite required for its survival and its evasion of the innate immune response.
Although we found that H2S does not affect H. pylori growth, it has been described that H2S can signal in bacteria such as Staphylococcus aureus through S-sulfhydration (16). This mechanism, which can be potentiated by the gasotransmitter NO, results in the regulation of protein activities, an increase of the antioxidant capacity, and virulence gene expression (16, 46). Consequently, the effect of macrophage CTH-derived H2S on H. pylori gene expression deserves further investigation.
In the present study, and in our previous report (22), we observed that the level of putrescine, but not spermidine and spermine, was increased in H. pylori-infected macrophages. Because the RTP and the polyamine anabolic pathway share SAM as a common precursor, we reasoned that H. pylori-induced CTH in macrophages may increase the use of SAM toward the RTP, thus limiting the synthesis of dcSAM and, consequently, spermidine and spermine. Supporting this idea, we found that spermidine and spermine concentrations were increased when CTH/CBS or AHCY inhibitors were used. Concomitantly, the intracellular pool of dcSAM was enhanced with CTH inhibition, further evidencing that CTH activity regulates SAM availability for spermidine and spermine synthesis. Because spermidine was not synthesized in CTH-expressing cells, due to a lack of dcSAM, we also observed that putrescine accumulated in H. pylori-infected macrophages as a result of ODC activity (22, 47). Hence, putrescine was completely depleted from infected cells in the presence of AOAA or DZNeP, further demonstrating that induction of the RTP not only reduces spermidine/spermine synthesis but also leads to increased putrescine levels. We recently demonstrated that putrescine supports heterochromatin formation and repression of M1 gene expression (22). Similarly, when CTH was overexpressed in infected cells, we found an attenuation of macrophage activation, which was abrogated by the use of the ODC inhibitor DFMO. In this context, we now propose that the CTH-dependent increase in the putrescine concentration is also involved in an attenuation of the innate immune response of myeloid cells. While we propose in this study that the induction of the RTP during infection regulates the macrophage response through a polyamine-dependent pathway, it is also probable that other metabolites of the RTP, such as lanthionine, α-ketobutyrate, or H2S, may affect the physiology and activation of myeloid and other somatic cells.
The conversion of SAM into SAH is performed by MTs. These enzymes include DNA MTs, arginine MTs, and histone MTs (48). The level of histone methylation is tightly associated with the level of transcription (49, 50). Because CTH metabolizes homocysteine, which is generated from SAH by AHCY, we reasoned that H. pylori-induced CTH expression can increase MT activity and promote histone methylation. Using a screening approach and confirmation by Western blotting, we determined that H3K9, H3K27, and H3K36 are di- or trimethylated in response to H. pylori and that these epigenetic modifications are suppressed with Cth knockdown. Of note, methylation of the lysine residues K9 (51), K27 (52, 53), and K36 (54) has been associated with reduced transcription activity in macrophages. Accordingly, we found that the increased methylation of these residues by a CTH-mediated pathway led to a striking reduction of the expression of the genes encoding the proinflammatory markers NOS2, interleukin-1β (IL-1β), and TNF-α in H. pylori-infected macrophages. Supporting our findings, Liu et al. showed that CTH expression in LPS-treated macrophages and mice is associated with enhanced trimethylation of H3K27 levels and a concomitant decrease of NOS2, COX2, IL-1β, and IL-6 induction (55).
In conclusion, CTH expression in macrophages (i) supports the growth of pathogenic bacteria by increasing cystathionine synthesis; (ii) leads to an increased level of intracellular putrescine, which has been shown to dampen innate immune function during bacterial infection (22), by depletion of SAM/dcSAM; and (iii) enhances histone H3 methylation, which in turn inhibits the transcription of proinflammatory genes. In this context, induction of CTH may ultimately lead to decreased inflammation and to increased bacterial survival. Thus, we speculate that CTH induction represents an unrecognized strategy for pathogenic bacteria to escape the host response. More specifically, we suggest that CTH is a critical mediator of the mucosal immune response that helps H. pylori persist in the stomach. Investigation of the effect of CTH on H. pylori-mediated gastric inflammation and carcinogenesis is now warranted by using Cth-deficient mice and is under way in our laboratory. The use of AOAA may have important therapeutic applications, by not only inhibiting macrophage CTH but also directly affecting H. pylori growth, as demonstrated in the present study.
MATERIALS AND METHODS
Reagents.
The H2S donor NaHS was purchased from Cayman. l-Cystathionine was purchased from Sigma and resuspended in 0.5 M HCl. The pharmacological inhibitors are described in Table S2 in the supplemental material.
Pharmacological inhibitors, primers, and antibodies. Download Table S2, PDF file, 0.1 MB (81.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Human tissues.
Biopsy specimens from gastric tissues were obtained from human subjects in Colombia as described previously (56), under protocols approved by the ethics committees of the local hospitals and of the Universidad del Valle in Cali, Colombia, as well as the Institutional Review Board at Vanderbilt University. Histology was determined by a gastrointestinal pathologist (M. B. Piazuelo).
Bacteria.
The cagA+ H. pylori strains PMSS1 (57), 7.13 (58), and 60190 (59) as well as the cagA, cagE, vacA, and ureA isogenic mutants constructed in strain 60190 (9); C. jejuni; S. aureus strain LAC USA300 (60) (obtained from James E. Cassat, Vanderbilt University Medical Center, Nashville, TN); S. salivarius MIT14-1770-C1; and S. epidermidis MIT14-1777-C6 (both obtained from James G. Fox, Massachusetts Institute of Technology, Cambridge, MA) (24) were grown on blood agar plates. S. Typhimurium and S. flexneri were maintained on Luria-Bertani agar plates. Bacteria were harvested from plates and grown in Bertani broth containing 10% fetal bovine serum (FBS) (for H. pylori, C. jejuni, S. salivarius, and S. epidermidis) or in LB liquid medium (for S. Typhimurium, S. flexneri, and S. aureus) overnight. Bacteria were then diluted to an A600 of 0.1 in defined Dulbecco’s modified Eagle’s medium (DMEM) (100 μM methionine and 50 μM cysteine) containing 5% FBS and harvested at the exponential phase to infect the cells. To infect mice, a culture of H. pylori grown overnight was diluted to an A600 of 0.1 in Bertani broth containing 10% FBS and harvested at the exponential phase.
Infection of mice.
Animals were used under protocol M/05/176 approved by the Institutional Animal Care and Use Committee at Vanderbilt University. C57BL/6 male mice (6 to 8 weeks old), bred in our facility, were randomly distributed into two groups: mice infected intragastrically three times, every 2 days, with 109 CFU of H. pylori PMSS1 and control mice treated with Bertani broth containing 10% FBS. Animals were sacrificed after 2 months. Sample sizes were based on previous studies (21, 22). Animals were randomly selected for infection or the broth control.
Immunostaining.
Immunofluorescence staining for CTH and the macrophage marker CD68 was performed on murine and human gastric tissues as described previously (22, 61), using the antibodies (Abs) listed in Table S2. Slides were reviewed and scored by a gastrointestinal pathologist (M. B. Piazuelo). The percentage of macrophages staining positively for CTH was determined for each patient by counting cells with moderate- or high-intensity staining on antral biopsy specimens.
Cells, transfections, and infections.
Peritoneal cells from C57BL/6 mice were collected in phosphate-buffered saline (PBS) and layered onto 24-well plates, and macrophages were enriched by washing off nonadherent cells after a 1-h incubation. BMmacs were isolated and differentiated as described previously (61). We also used the murine macrophage cell line RAW 264.7 and the human macrophage cell line THP-1, maintained in DMEM and RPMI containing 10% FBS, 10 mM HEPES, and 1 mM sodium pyruvate, respectively; THP-1 cells were differentiated for 24 h with 10 ng/ml phorbol myristate acetate (PMA). The immortalized murine gastric epithelial cell line ImSt was cultured as described previously (62). Cells were stimulated with H. pylori at a multiplicity of infection (MOI) of 10 to 100 in defined DMEM (100 μM methionine and 50 μM cysteine) supplemented with 1% FBS, HEPES, and sodium pyruvate. All pharmacological inhibitors were added 30 min before infection. For transfections, RAW 264.7 cells (5 × 105) were maintained in Opti-MEM I reduced serum medium (Invitrogen) and transfected using Lipofectamine 2000 (Invitrogen) with (i) 100 nM On-Targetplus siRNAs (Dharmacon) directed against Cth or LmnA and (ii) 0.05 to 0.5 μg/ml of pCTH or the pCMV3 empty vector (Sino Biological). After 6 h (pCMV3 and pCTH) or 16 h (siRNA), cells were washed, maintained for 24 to 36 h in fresh medium devoid of antibiotics, and then infected.
To determine levels of phagocytosis, infected RAW 264.7 cells were washed thoroughly with PBS after infection, incubated for 1 h with 200 μg/ml gentamicin, and lysed in 0.1% saponin for 30 min at 37°C (9). The number of bacteria in each lysate was determined by counting the CFU after plating serial dilutions on agar plates.
All the cell lines tested negative for mycoplasma contamination using the Lookout mycoplasma detection kit (Sigma-Aldrich).
Determination of histone methylation.
Total core histone proteins were extracted from macrophages using the EpiQuik total histone extraction kit (Epigentek). Histone H3 methylation was analyzed using the EpiQuik histone H3 modification multiplex assay kit (Epigentek).
Analysis of mRNA levels.
Total RNA was isolated using TRIzol (Ambion). Reverse transcription (RT) was performed using Superscript II reverse transcriptase and oligo(dT) (Invitrogen). mRNAs were amplified by real-time PCR using the iQ SYBR green kit (Bio-Rad) (9) and the primers listed in Table S2.
Western blot analysis.
RAW 264.7 cells were lysed using radioimmunoprecipitation assay (RIPA) buffer or the NE-PER nuclear protein extraction kit (Pierce) containing a protease inhibitor cocktail (set III; Calbiochem) and a phosphatase inhibitor cocktail (set I; Calbiochem). Protein concentrations were determined using the bicinchoninic acid (BCA) protein assay (Pierce). Western blotting was performed using 10 μg of protein per lane. Primary and secondary Abs are listed in Table S2. Densitometric analysis of Western blots was performed with ImageJ 1.50i software.
CTH activity assay.
A method described previously by Manna et al. was used to determine CTH activity in macrophages (63). After infection, cells were lysed in 50 mM potassium phosphate buffer (pH 6.9) containing 1 mM EDTA and a protease inhibitor cocktail (set III; Calbiochem), and protein concentrations were determined. A 20-μl aliquot of the lysates, or murine CTH recombinant protein (MyBioSource), used as positive control, was added to 180 μl of a solution containing 100 mM potassium phosphate buffer (pH 7.4), 4 mM l-cystathionine, 0.125 mM pyridoxal-5′-phosphate (Sigma), 0.32 mM NADH (Roche Diagnostics), and 1.5 U lactate dehydrogenase (Roche Diagnostics). Reactions without l-cystathionine were performed as controls. The absorbance at 340 nm was monitored at 37°C in a Synergy 4 microplate reader (BioTek). Maximum velocities were calculated from the linear portion of the graphs, and the results are expressed as nanomoles per minute per milligram of protein.
Proteomics.
RAW 264.7 macrophages were infected with H. pylori PMSS1 for 24 h and lysed in a solution containing 50 mM Tris-HCl (pH 7.6), 150 mM NaCl, 1% NP-40, and 2 mM EDTA. After centrifugation at 16,000 × g for 15 min, the protein concentration in the supernatants was determined by a BCA assay. Protein samples were precipitated with ice-cold acetone overnight at −20°C, washed, and digested with sequencing-grade trypsin overnight. Next, proteomic analysis was performed using iTRAQ technology. Upon gradient elution, peptides were introduced via nanospray ionization into a Q Exactive HF mass spectrometer (Thermo Scientific). Peptide/protein identifications and quantitative analysis were performed using Spectrum Mill (Agilent). Tandem mass spectrometry (MS/MS) spectra were searched against a subset of the UniProt KB protein database containing Mus musculus protein sequences. Autovalidation procedures in Spectrum Mill were used to filter the data to <1% false discovery rates at the protein and peptide levels. The median log2 iTRAQ protein ratios were calculated over all peptides identified for each protein and fit using least-squares regression, and P values were corrected for multiple comparisons by the Benjamini-Hochberg method (64).
Metabolomics.
RAW 264.7 cells were infected with H. pylori PMSS1 for 24 h at an MOI of 100. Cells were then washed, and fresh DMEM containing antibiotics and 100 μM methionine was added in the presence and absence of AOAA (250 μM). Cells were harvested after 24 h. Macrophage lysis was performed as reported previously (65), with slight modifications. Snap-frozen cell pellets were reconstituted in 300 μl of a solution containing chloroform-methanol-water (3:4:3) with 1.5 μM carbamazepine as an internal standard. Samples were subsequently shaken at 4°C for 1 h, followed by centrifugation at 16,000 × g for 15 min. Samples (50 μl) were randomized and injected in triplicate onto a Vanquish ultrahigh-performance liquid chromatography (UHPLC) instrument (Thermo Fisher Scientific) interfaced with a Q Exactive HF high-resolution orbitrap mass spectrometer (Thermo Fisher Scientific). Chromatographic separation was performed with a reverse-phase Kinetex C18 column (1.7 μm, 100 by 2.1 mm; Phenomenex) at a flow rate of 300 μl/min. The spray voltage was set to 5 kV in positive mode with a capillary temperature of 320°C and an S-lens RF level of 80 V. Mass spectra were acquired over a scan range of m/z 70 to 1,050 at a resolution of 30,000, and top-two data-dependent MS/MS was conducted with an AGC target of 1 × e5, a maximum injection time of 50 ms, and a normalized collision energy of 30. Chromatographic alignment, peak picking, peak deconvolution, and statistical comparisons were performed using Progenesis QI (Nonlinear Dynamics). All features that were determined to have a P value of <0.05 and a fold change of >1.5 were then searched for matches in the Human Metabolome Database (HMDB) with a mass error cutoff of 5 ppm. All putative matches with a Progenesis score of >30 out of 99 (calculated by software using mass error, isotope abundance, and fragmentation) were tentatively accepted to be accurate. For increased confidence, reference standards for putrescine, spermidine, and spermine were used to confirm the retention time and fragmentation pattern. Metabolite matches were then filtered to exclude biologically irrelevant drug metabolites, and the finalized list of putative HMDB identifications was exported for further pathway overrepresentation analysis. integrated molecular pathway-level analysis (IMPaLA) was used to search for pathway hits in various databases, including EHMN, BioCyc, INOH, KEGG, Reactome, Wikipathways, and SMPDB. Pathway significance and false discovery rates were calculated by IMPaLA using Fisher’s method and the Benjamini-Hochberg procedure, respectively.
Metabolite analysis by LC-MS.
Amino acids and dcSAM were measured at the Vanderbilt Neurochemistry Core. Cell pellets were homogenized in 0.1 M trichloroacetic acid containing 10−2 M sodium acetate, 10−4 M EDTA, and 10.5% methanol (pH 3.8). After centrifugation at 10,000 × g for 20 min, the supernatants were used for protein assays using BCA and for liquid chromatography-mass spectrometry (LC-MS).
To prepare internal standards, 50-μl stock solutions of each amino acid (5 ng/μl) were diluted with 200 μl acetonitrile and 100 μl each of 500 mM NaCO3 (aq) and 2% isotopically labeled benzoyl chloride ([13C6]BZC) in acetonitrile. After 2 min, the reaction was stopped by the addition of 200 μl of 20% acetonitrile in water containing 3% sulfuric acid and 400 μl water. These solutions were diluted 100-fold with 20% acetonitrile in water containing 3% sulfuric acid to make the working internal standard solution used for sample analysis.
Cell extracts or cell supernatants (5 μl) were diluted in acetonitrile (20 μl), 500 mM NaCO3 (10 μl), and 2% benzoyl chloride in acetonitrile (10 μl). After 2 min, the reaction was stopped by the addition of 20 μl of the internal standard solution and 40 μl water.
LC was performed on a 2- by 50-mm, 1.7-μm-particle-size Acquity BEH C18 column (Waters Corporation) using a Waters Acquity ultraperformance liquid chromatography (UPLC) system. Mobile phase A was 15% aqueous formic acid, and mobile phase B was acetonitrile. Samples were separated by a gradient of 98 to 5% mobile phase A over 11 min at a flow rate of 600 μl/min prior to delivery to a Sciex 6500+ QTrap mass spectrometer. The peak height of the endogenous amino acids was compared to the peak height of the isotopically labeled internal standards for quantitation. All data were analyzed using MultiQuant software version 3.0 (Sciex).
Measurement of polyamines.
Concentrations of polyamines were measured in macrophages and in cell supernatants by LC-MS, as described previously (22).
Statistics.
All the data shown represent the means ± standard errors of the means (SEM). Data that were not normally distributed according to the D’Agostino-Pearson normality test were log or square-root transformed. Student’s t test or analysis of variance (ANOVA) with the Tukey test was used to determine significant differences between two groups or to analyze significant differences among multiple test groups, respectively. All statistical tests were two sided.
Data availability.
The mass spectrometry proteomics data that support the findings of this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with data set accession number PXD010082. Metabolomics data that support the findings of this study have been deposited in the EMBL-EBI MetaboLights database (66) with accession number MTBLS696 (https://www.ebi.ac.uk/metabolights/MTBLS696).
ACKNOWLEDGMENTS
This work was funded by NIH grants R01CA190612, P01CA116087, P01CA028842, and R21AI142042 (K.T.W.); Veterans Affairs Merit Review grant I01BX001453 (K.T.W.); the Thomas F. Frist, Sr., Endowment (K.T.W.); and the Vanderbilt Center for Mucosal Inflammation and Cancer (K.T.W.). This work was also supported by NIH grant R01AT006896 (C.S.). P.B.L. was supported by a postdoctoral fellowship award from the American Heart Association (16POST27250138). Mass spectrometry analyses were supported in part by core scholarships from the Vanderbilt University Medical Center Digestive Disease Research Center funded by NIH grant P30DK058404 and Vanderbilt Ingram Cancer Center support grant P30CA068485. A.P.G. was supported by the Philippe Foundation.
A.P.G., Y.L.L., M.A., T.G.V., J.L.F., D.P.B., P.B.L., C.S., G.L.M., E.S.R., K.L.-R., K.L.S., A.G.D., J.C.S., and M.B.P. performed the experiments; E.S.R., K.L.-R., K.L.S., and A.P.G. performed statistical analysis; A.P.G. and K.T.W. designed the study; and A.P.G. and K.T.W. wrote the paper.
We declare no competing interests.
Footnotes
Citation Gobert AP, Latour YL, Asim M, Finley JL, Verriere TG, Barry DP, Milne GL, Luis PB, Schneider C, Rivera ES, Lindsey-Rose K, Schey KL, Delgado AG, Sierra JC, Piazuelo MB, Wilson KT. 2019. Bacterial pathogens hijack the innate immune response by activation of the reverse transsulfuration pathway. mBio 10:e02174-19. https://doi.org/10.1128/mBio.02174-19.
REFERENCES
- 1.Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, Malfertheiner P, Graham DY, Wong VWS, Wu JCY, Chan FKL, Sung JJY, Kaplan GG, Ng SC. 2017. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 153:420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Correa P. 1988. A human model of gastric carcinogenesis. Cancer Res 48:3554–3560. [PubMed] [Google Scholar]
- 3.Wilson KT, Crabtree JE. 2007. Immunology of Helicobacter pylori: insights into the failure of the immune response and perspectives on vaccine studies. Gastroenterology 133:288–308. doi: 10.1053/j.gastro.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Peek RM Jr, Fiske C, Wilson KT. 2010. Role of innate immunity in Helicobacter pylori-induced gastric malignancy. Physiol Rev 90:831–858. doi: 10.1152/physrev.00039.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson KT, Ramanujam KS, Mobley HL, Musselman RF, James SP, Meltzer SJ. 1996. Helicobacter pylori stimulates inducible nitric oxide synthase expression and activity in a murine macrophage cell line. Gastroenterology 111:1524–1533. doi: 10.1016/S0016-5085(96)70014-8. [DOI] [PubMed] [Google Scholar]
- 6.Fu S, Ramanujam KS, Wong A, Fantry GT, Drachenberg CB, James SP, Meltzer SJ, Wilson KT. 1999. Increased expression and cellular localization of inducible nitric oxide synthase and cyclooxygenase 2 in Helicobacter pylori gastritis. Gastroenterology 116:1319–1329. doi: 10.1016/s0016-5085(99)70496-8. [DOI] [PubMed] [Google Scholar]
- 7.Gobert AP, McGee DJ, Akhtar M, Mendz GL, Newton JC, Cheng Y, Mobley HL, Wilson KT. 2001. Helicobacter pylori arginase inhibits nitric oxide production by eukaryotic cells: a strategy for bacterial survival. Proc Natl Acad Sci U S A 98:13844–13849. doi: 10.1073/pnas.241443798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nam KT, Oh S-Y, Ahn B, Kim YB, Jang DD, Yang K-H, Hahm K-B, Kim D-Y. 2004. Decreased Helicobacter pylori associated gastric carcinogenesis in mice lacking inducible nitric oxide synthase. Gut 53:1250–1255. doi: 10.1136/gut.2003.030684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gobert AP, Verriere T, Asim M, Barry DP, Piazuelo MB, de Sablet T, Delgado AG, Bravo LE, Correa P, Peek RM, Chaturvedi R, Wilson KT. 2014. Heme oxygenase-1 dysregulates macrophage polarization and the immune response to Helicobacter pylori. J Immunol 193:3013–3022. doi: 10.4049/jimmunol.1401075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gobert AP, Verriere T, de Sablet T, Peek RM Jr, Chaturvedi R, Wilson KT. 2013. Heme oxygenase-1 inhibits phosphorylation of the Helicobacter pylori oncoprotein CagA in gastric epithelial cells. Cell Microbiol 15:145–156. doi: 10.1111/cmi.12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linden DR. 2014. Hydrogen sulfide signaling in the gastrointestinal tract. Antioxid Redox Signal 20:818–830. doi: 10.1089/ars.2013.5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang G, Wu L, Jiang B, Yang W, Qi J, Cao K, Meng Q, Mustafa AK, Mu W, Zhang S, Snyder SH, Wang R. 2008. H2S as a physiologic vasorelaxant: hypertension in mice with deletion of cystathionine gamma-lyase. Science 322:587–590. doi: 10.1126/science.1162667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paul BD, Sbodio JI, Xu R, Vandiver MS, Cha JY, Snowman AM, Snyder SH. 2014. Cystathionine gamma-lyase deficiency mediates neurodegeneration in Huntington’s disease. Nature 509:96–100. doi: 10.1038/nature13136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shirozu K, Tokuda K, Marutani E, Lefer D, Wang R, Ichinose F. 2014. Cystathionine gamma-lyase deficiency protects mice from galactosamine/lipopolysaccharide-induced acute liver failure. Antioxid Redox Signal 20:204–216. doi: 10.1089/ars.2013.5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferlito M, Wang Q, Fulton WB, Colombani PM, Marchionni L, Fox-Talbot K, Paolocci N, Steenbergen C. 2014. Hydrogen sulfide increases survival during sepsis: protective effect of CHOP inhibition. J Immunol 192:1806–1814. doi: 10.4049/jimmunol.1300835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng H, Zhang Y, Palmer LD, Kehl-Fie TE, Skaar EP, Trinidad JC, Giedroc DP. 2017. Hydrogen sulfide and reactive sulfur species impact proteome s-sulfhydration and global virulence regulation in Staphylococcus aureus. ACS Infect Dis 3:744–755. doi: 10.1021/acsinfecdis.7b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aitken SM, Kirsch JF. 2005. The enzymology of cystathionine biosynthesis: strategies for the control of substrate and reaction specificity. Arch Biochem Biophys 433:166–175. doi: 10.1016/j.abb.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 18.Singh S, Padovani D, Leslie RA, Chiku T, Banerjee R. 2009. Relative contributions of cystathionine beta-synthase and gamma-cystathionase to H2S biogenesis via alternative trans-sulfuration reactions. J Biol Chem 284:22457–22466. doi: 10.1074/jbc.M109.010868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiku T, Padovani D, Zhu W, Singh S, Vitvitsky V, Banerjee R. 2009. H2S biogenesis by human cystathionine gamma-lyase leads to the novel sulfur metabolites lanthionine and homolanthionine and is responsive to the grade of hyperhomocysteinemia. J Biol Chem 284:11601–11612. doi: 10.1074/jbc.M808026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura H. 2015. Physiological roles of hydrogen sulfide and polysulfides. Handb Exp Pharmacol 230:61–81. doi: 10.1007/978-3-319-18144-8_3. [DOI] [PubMed] [Google Scholar]
- 21.Pegg AE. 2016. Functions of polyamines in mammals. J Biol Chem 291:14904–14912. doi: 10.1074/jbc.R116.731661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hardbower DM, Asim M, Luis PB, Singh K, Barry DP, Yang C, Steeves MA, Cleveland JL, Schneider C, Piazuelo MB, Gobert AP, Wilson KT. 2017. Ornithine decarboxylase regulates M1 macrophage activation and mucosal inflammation via histone modifications. Proc Natl Acad Sci U S A 114:E751–E760. doi: 10.1073/pnas.1614958114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaturvedi R, Asim M, Hoge S, Lewis ND, Singh K, Barry DP, de Sablet T, Piazuelo MB, Sarvaria AR, Cheng Y, Closs EI, Casero RA, Gobert AP, Wilson KT. 2010. Polyamines impair immunity to Helicobacter pylori by inhibiting L-arginine uptake required for nitric oxide production. Gastroenterology 139:1686–1698. doi: 10.1053/j.gastro.2010.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang I, Woltemate S, Piazuelo MB, Bravo LE, Yepez MC, Romero-Gallo J, Delgado AG, Wilson KT, Peek RM, Correa P, Josenhans C, Fox JG, Suerbaum S. 2016. Different gastric microbiota compositions in two human populations with high and low gastric cancer risk in Colombia. Sci Rep 6:18594. doi: 10.1038/srep18594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. 1997. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol 7:261–269. doi: 10.1016/S0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 26.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. 2005. Phosphorylation and regulation of AKT/PKB by the Rictor-mTOR complex. Science 307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 27.Doherty NC, Shen F, Halliday NM, Barrett DA, Hardie KR, Winzer K, Atherton JC. 2010. In Helicobacter pylori, LuxS is a key enzyme in cysteine provision through a reverse transsulfuration pathway. J Bacteriol 192:1184–1192. doi: 10.1128/JB.01372-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asimakopoulou A, Panopoulos P, Chasapis CT, Coletta C, Zhou Z, Cirino G, Giannis A, Szabo C, Spyroulias GA, Papapetropoulos A. 2013. Selectivity of commonly used pharmacological inhibitors for cystathionine beta synthase (CBS) and cystathionine gamma lyase (CSE). Br J Pharmacol 169:922–932. doi: 10.1111/bph.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki M, Mimuro H, Kiga K, Fukumatsu M, Ishijima N, Morikawa H, Nagai S, Koyasu S, Gilman RH, Kersulyte D, Berg DE, Sasakawa C. 2009. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe 5:23–34. doi: 10.1016/j.chom.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 30.Li N, Tang B, Jia Y-P, Zhu P, Zhuang Y, Fang Y, Li Q, Wang K, Zhang W-J, Guo G, Wang T-J, Feng Y-J, Qiao B, Mao X-H, Zou Q-M. 2017. Helicobacter pylori CagA protein negatively regulates autophagy and promotes inflammatory response via c-Met-PI3K/AKT-mTOR signaling pathway. Front Cell Infect Microbiol 7:417. doi: 10.3389/fcimb.2017.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei J, Nagy TA, Vilgelm A, Zaika E, Ogden SR, Romero-Gallo J, Piazuelo MB, Correa P, Washington MK, El-Rifai W, Peek RM, Zaika A. 2010. Regulation of p53 tumor suppressor by Helicobacter pylori in gastric epithelial cells. Gastroenterology 139:1333–1343. doi: 10.1053/j.gastro.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabassam FH, Graham DY, Yamaoka Y. 2009. Helicobacter pylori activate epidermal growth factor receptor- and phosphatidylinositol 3-OH kinase-dependent AKT and glycogen synthase kinase 3 beta phosphorylation. Cell Microbiol 11:70–82. doi: 10.1111/j.1462-5822.2008.01237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen LA, Allgood JA, Han X, Wittine LM. 2005. Phosphoinositide 3-kinase regulates actin polymerization during delayed phagocytosis of Helicobacter pylori. J Leukoc Biol 78:220–230. doi: 10.1189/jlb.0205091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmed AU, Sarvestani ST, Gantier MP, Williams BR, Hannigan GE. 2014. Integrin-linked kinase modulates lipopolysaccharide- and Helicobacter pylori-induced nuclear factor kappa B-activated tumor necrosis factor-alpha production via regulation of p65 serine 536 phosphorylation. J Biol Chem 289:27776–27793. doi: 10.1074/jbc.M114.574541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris SM, Harvey EJ, Hughes TR, Ramji DP. 2008. The interferon-gamma-mediated inhibition of lipoprotein lipase gene transcription in macrophages involves casein kinase 2- and phosphoinositide-3-kinase-mediated regulation of transcription factors Sp1 and Sp3. Cell Signal 20:2296–2301. doi: 10.1016/j.cellsig.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuang H-J, Zhao G-J, Chen W-J, Zhang M, Zeng G-F, Zheng X-L, Tang C-K. 2017. Hsp27 promotes ABCA1 expression and cholesterol efflux through the PI3K/PKC zeta/Sp1 pathway in THP-1 macrophages. Eur J Pharmacol 810:57–62. doi: 10.1016/j.ejphar.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 37.Mireuta M, Darnel A, Pollak M. 2010. IGFBP-2 expression in MCF-7 cells is regulated by the PI3K/AKT/mTOR pathway through Sp1-induced increase in transcription. Growth Factors 28:243–255. doi: 10.3109/08977191003745472. [DOI] [PubMed] [Google Scholar]
- 38.Sen N, Paul BD, Gadalla MM, Mustafa AK, Sen T, Xu R, Kim S, Snyder SH. 2012. Hydrogen sulfide-linked sulfhydration of NF-kappaB mediates its antiapoptotic actions. Mol Cell 45:13–24. doi: 10.1016/j.molcel.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badiei A, Gieseg S, Davies S, Izani Othman M, Bhatia M. 2015. LPS up-regulates cystathionine gamma-lyase gene expression in primary human macrophages via NF-kappa B/ERK pathway. Inflamm Allergy Drug Targets 14:99–104. doi: 10.2174/1871528114666151201201719. [DOI] [PubMed] [Google Scholar]
- 40.Hay N. 2005. The AKT-mTOR tango and its relevance to cancer. Cancer Cell 8:179–183. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 41.Katholnig K, Linke M, Pham H, Hengstschlager M, Weichhart T. 2013. Immune responses of macrophages and dendritic cells regulated by mTOR signalling. Biochem Soc Trans 41:927–933. doi: 10.1042/BST20130032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong Y, Wu D, Bai H, Han C, Chen J, Chen L, Hu L, Jiang H, Shen X. 2008. Enzymatic characterization and inhibitor discovery of a new cystathionine gamma-synthase from Helicobacter pylori. J Biochem 143:59–68. doi: 10.1093/jb/mvm194. [DOI] [PubMed] [Google Scholar]
- 43.Shatalin K, Shatalina E, Mironov A, Nudler E. 2011. H2S: a universal defense against antibiotics in bacteria. Science 334:986–990. doi: 10.1126/science.1209855. [DOI] [PubMed] [Google Scholar]
- 44.Bryk R, Griffin P, Nathan C. 2000. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407:211–215. doi: 10.1038/35025109. [DOI] [PubMed] [Google Scholar]
- 45.Lester J, Kichler S, Oickle B, Fairweather S, Oberc A, Chahal J, Ratnayake D, Creuzenet C. 2015. Characterization of Helicobacter pylori HP0231 (DsbK): role in disulfide bond formation, redox homeostasis and production of Helicobacter cystein-rich protein HcpE. Mol Microbiol 96:110–133. doi: 10.1111/mmi.12923. [DOI] [PubMed] [Google Scholar]
- 46.Fravega J, Álvarez R, Díaz F, Inostroza O, Tejías C, Rodas PI, Paredes-Sabja D, Fuentes JA, Calderón IL, Gil F. 2016. Salmonella Typhimurium exhibits fluoroquinolone resistance mediated by the accumulation of the antioxidant molecule H2S in a CysK-dependent manner. J Antimicrob Chemother 71:3409–3415. doi: 10.1093/jac/dkw311. [DOI] [PubMed] [Google Scholar]
- 47.Gobert AP, Cheng Y, Wang J-Y, Boucher J-L, Iyer RK, Cederbaum SD, Casero RA, Newton JC, Wilson KT. 2002. Helicobacter pylori induces macrophage apoptosis by activation of arginase II. J Immunol 168:4692–4700. doi: 10.4049/jimmunol.168.9.4692. [DOI] [PubMed] [Google Scholar]
- 48.Smith BC, Denu JM. 2009. Chemical mechanisms of histone lysine and arginine modifications. Biochim Biophys Acta 1789:45–57. doi: 10.1016/j.bbagrm.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 50.Alvarez-Errico D, Vento-Tormo R, Sieweke M, Ballestar E. 2015. Epigenetic control of myeloid cell differentiation, identity and function. Nat Rev Immunol 15:7–17. doi: 10.1038/nri3777. [DOI] [PubMed] [Google Scholar]
- 51.Hachiya R, Shiihashi T, Shirakawa I, Iwasaki Y, Matsumura Y, Oishi Y, Nakayama Y, Miyamoto Y, Manabe I, Ochi K, Tanaka M, Goda N, Sakai J, Suganami T, Ogawa Y. 2016. The H3K9 methyltransferase Setdb1 regulates TLR4-mediated inflammatory responses in macrophages. Sci Rep 6:28845. doi: 10.1038/srep28845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ishii M, Wen H, Corsa CAS, Liu T, Coelho AL, Allen RM, Carson WF, Cavassani KA, Li X, Lukacs NW, Hogaboam CM, Dou Y, Kunkel SL. 2009. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood 114:3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kruidenier L, Chung C, Cheng Z, Liddle J, Che K, Joberty G, Bantscheff M, Bountra C, Bridges A, Diallo H, Eberhard D, Hutchinson S, Jones E, Katso R, Leveridge M, Mander PK, Mosley J, Ramirez-Molina C, Rowland P, Schofield CJ, Sheppard RJ, Smith JE, Swales C, Tanner R, Thomas P, Tumber A, Drewes G, Oppermann U, Patel DJ, Lee K, Wilson DM. 2012. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature 488:404–408. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu G, Liu G, Xiong S, Liu H, Chen X, Zheng B. 2015. The histone methyltransferase Smyd2 is a negative regulator of macrophage activation by suppressing interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-alpha) production. J Biol Chem 290:5414–5423. doi: 10.1074/jbc.M114.610345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S, Wang X, Pan L, Wu W, Yang D, Qin M, Jia W, Xiao C, Long F, Ge J, Liu X, Zhu Y. 2018. Endogenous hydrogen sulfide regulates histone demethylase JMJD3-mediated inflammatory response in LPS-stimulated macrophages and in a mouse model of LPS-induced septic shock. Biochem Pharmacol 149:153–162. doi: 10.1016/j.bcp.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 56.Mera RM, Bravo LE, Camargo MC, Bravo JC, Delgado AG, Romero-Gallo J, Yepez MC, Realpe JL, Schneider BG, Morgan DR, Peek RM Jr, Correa P, Wilson KT, Piazuelo MB. 2018. Dynamics of Helicobacter pylori infection as a determinant of progression of gastric precancerous lesions: 16-year follow-up of an eradication trial. Gut 67:1239–1246. doi: 10.1136/gutjnl-2016-311685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arnold IC, Lee JY, Amieva MR, Roers A, Flavell RA, Sparwasser T, Müller A. 2011. Tolerance rather than immunity protects from Helicobacter pylori-induced gastric preneoplasia. Gastroenterology 140:199–209. doi: 10.1053/j.gastro.2010.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sierra JC, Asim M, Verriere TG, Piazuelo MB, Suarez G, Romero-Gallo J, Delgado AG, Wroblewski LE, Barry DP, Peek RM, Gobert AP, Wilson KT. 2018. Epidermal growth factor receptor inhibition downregulates Helicobacter pylori-induced epithelial inflammatory responses, DNA damage and gastric carcinogenesis. Gut 67:1247–1260. doi: 10.1136/gutjnl-2016-312888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peek RM, Blaser MJ, Mays DJ, Forsyth MH, Cover TL, Song SY, Krishna U, Pietenpol JA Jr.. 1999. Helicobacter pylori strain-specific genotypes and modulation of the gastric epithelial cell cycle. Cancer Res 59:6124–6131. [PubMed] [Google Scholar]
- 60.Boles BR, Thoendel M, Roth AJ, Horswill AR. 2010. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One 5:e10146. doi: 10.1371/journal.pone.0010146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hardbower DM, Singh K, Asim M, Verriere TG, Olivares-Villagómez D, Barry DP, Allaman MM, Washington MK, Peek RM, Piazuelo MB, Wilson KT. 2016. EGFR regulates macrophage activation and function in bacterial infection. J Clin Invest 126:3296–3312. doi: 10.1172/JCI83585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yan F, Cao H, Chaturvedi R, Krishna U, Hobbs SS, Dempsey PJ, Peek RM, Cover TL, Washington MK, Wilson KT, Polk DB. 2009. Epidermal growth factor receptor activation protects gastric epithelial cells from Helicobacter pylori-induced apoptosis. Gastroenterology 136:1297–1307.e1–e3. doi: 10.1053/j.gastro.2008.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manna P, Gungor N, McVie R, Jain SK. 2014. Decreased cystathionine-gamma-lyase (CSE) activity in livers of type 1 diabetic rats and peripheral blood mononuclear cells (PBMC) of type 1 diabetic patients. J Biol Chem 289:11767–11778. doi: 10.1074/jbc.M113.524645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 57:289–300. [Google Scholar]
- 65.Vincent IM, Ehmann DE, Mills SD, Perros M, Barrett MP. 2016. Untargeted metabolomics to ascertain antibiotic modes of action. Antimicrob Agents Chemother 60:2281–2291. doi: 10.1128/AAC.02109-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haug K, Salek RM, Conesa P, Hastings J, de Matos P, Rijnbeek M, Mahendraker T, Williams M, Neumann S, Rocca-Serra P, Maguire E, González-Beltrán A, Sansone SA, Griffin JL, Steinbeck C. 2013. MetaboLights—an open-access general-purpose repository for metabolomics studies and associated meta-data. Nucleic Acids Res 41:D781–D786. doi: 10.1093/nar/gks1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The reverse transsulfuration and polyamine pathways. AHCY, S-adenosylhomocysteine hydrolase; AMD1, adenosylmethionine decarboxylase 1; BHMT, betaine homocysteine methyltransferase; CBS, cystathionine β-synthase; CTH, cystathionine γ-lyase; dcSAM, S-adenosylmethioninamine (decarboxylated S-adenosylmethionine); H3, histone 3; MAT, methionine adenosyltransferase; MTR, methionine synthase; MTs, methyltransferases; SAH, S-adenosylhomocysteine; SAM, S-adenosylmethionine; SMS, spermine synthase; SRM, spermidine synthase. Download FIG S1, PDF file, 0.05 MB (52.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Induction of Cth in macrophages in response to different bacteria. RAW 264.7 cells were infected with H. pylori PMSS1 (Hp), C. jejuni (Cj), S. enterica serovar Typhimurium (ST), S. flexneri (Sf), S. aureus (Sa), S. salivarius (Ss), or S. epidermidis (Se). After 6 h, Cth mRNA expression was analyzed by RT–real-time PCR. ***, P < 0.001; ****, P < 0.0001 (versus uninfected cells) (n = 3). Download FIG S2, PDF file, 0.4 MB (431.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Signaling pathways and inhibitors. The inhibitors of each of the signaling molecules are indicated in red. Download FIG S3, PDF file, 0.3 MB (324.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Expression of Nos2 in response to H. pylori and to different inhibitors. RAW 264.7 cells were pretreated with SB203580 (SB), PD98059 (PD), an ERK inhibitor (ER), LY294002 (LY), or Bay 11-7082 (BAY) for 30 min prior to infection with H. pylori PMSS1; Nos2 mRNA expression was analyzed after 6 h. **, P < 0.01; ****, P < 0.0001 (versus uninfected cells); §§, P < 0.01 (versus H. pylori-infected cells) (n = 3 to 6). Download FIG S4, PDF file, 0.10 MB (102.1KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effect of Cth and Cbs silencing on metabolites of the RTP. (a) RAW 264.7 macrophages transfected with LmnA, Cth, or Cbs siRNA were infected or not with H. pylori for 24 h. Cells were washed, and fresh medium containing antibiotics was added. The cystathionine concentration was measured in cells by LC-MS after 24 h. *, P < 0.05; **, P < 0.01 (versus uninfected cells); §, P < 0.05 (compared to cells transfected with LmnA siRNA and infected with H. pylori) (n = 4). (b) The CTH protein level was assessed by Western blotting after 24 h. Shown are representative data from 2 experiments. Download FIG S5, PDF file, 0.5 MB (486.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effect of AOAA and NaHS on H. pylori growth. H. pylori PMSS1 was grown in Bertani broth with 10% FBS with various concentrations of AOAA (a) or 500 μM NaHS (b). The optical density at 600 nm was monitored at the indicated times (n = 3 independent experiments). Download FIG S6, PDF file, 0.2 MB (169.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The metabolomics pathways significantly regulated by H. pylori infection and AOAA. Macrophages were infected or not with H. pylori with or without AOAA for 24 h. The metabolome of each group was subsequently assessed, and the data were analyzed using IMPaLA. For each database, P denotes the statistical confidence that the metabolic pathway is altered. Only pathways that were determined to be significant are included. The rows underlined in blue represent the pathways related to methionine/cysteine metabolism, the rows in red depict the polyamine pathways, and the rows in green represent the arginine/ornithine metabolism pathway, which is related to the polyamine pathway (see http://www.genome.jp/kegg-bin/show_pathway?map00330). Note that the KEGG glutathione pathway (row in yellow) includes cysteine, methionine, and polyamine pathways (see http://www.genome.jp/kegg/pathway/map/map00480.html) (n = 2 independent experiments assessed in triplicate). Download Table S1, PDF file, 0.2 MB (165KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Expression of CTH mRNA. RAW 264.7 cells were transfected with pCMV3 (0.5 μg/ml) or pCTH (0.05, 0.1, and 0.5 μg/ml). After 24 h, the level of CTH mRNA was analyzed by RT–real-time PCR (n = 5). NA, not amplified. Download FIG S7, PDF file, 0.1 MB (53.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
H3 methylation. Macrophages were transfected with pCMV3 (0.5 μg/ml) or pCTH (0.5 μg/ml). After 24 h, medium was removed, and fresh medium containing or not AOAA was added. The levels of H3 mono-, di-, or trimethylation were evaluated by an ELISA. Shown are representative data from two experiments. Download FIG S8, PDF file, 0.2 MB (204KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Pharmacological inhibitors, primers, and antibodies. Download Table S2, PDF file, 0.1 MB (81.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The mass spectrometry proteomics data that support the findings of this study have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with data set accession number PXD010082. Metabolomics data that support the findings of this study have been deposited in the EMBL-EBI MetaboLights database (66) with accession number MTBLS696 (https://www.ebi.ac.uk/metabolights/MTBLS696).