Due to the promiscuous exchange of genetic material and asexual reproduction, delineating microbial species (and, by extension, populations) remains challenging. Because of this, the vast majority of microbial studies assessing population structure often compare divergent strains from disparate environments under varied selective pressures. Here, we investigated the population structure within a single bacterial ecotype, a unit equivalent to a eukaryotic species, defined as highly clustered genotypic and phenotypic strains with the same ecological niche. Using a combination of genomic and computational analyses, we assessed the phylogenetic structure, extent of recombination, and flexible gene content of this genomic diversity to infer patterns of gene flow. To our knowledge, this study is the first to do so for a dominant soil bacterium. Our results indicate that bacterial soil populations, similarly to those in other environments, are structured by gene flow discontinuities and exhibit distributional patterns consistent with both isolation by distance and isolation by environment. Thus, both dispersal limitation and local environments contribute to the divergence among closely related soil bacteria as observed in macroorganisms.
KEYWORDS: Curtobacterium, population structure, gene flow, microbial ecology, ecotype
ABSTRACT
For free-living bacteria and archaea, the equivalent of the biological species concept does not exist, creating several obstacles to the study of the processes contributing to microbial diversification. These obstacles are particularly high in soil, where high bacterial diversity inhibits the study of closely related genotypes and therefore the factors structuring microbial populations. Here, we isolated strains within a single Curtobacterium ecotype from surface soil (leaf litter) across a regional climate gradient and investigated the phylogenetic structure, recombination, and flexible gene content of this genomic diversity to infer patterns of gene flow. Our results indicate that microbial populations are delineated by gene flow discontinuities, with distinct populations cooccurring at multiple sites. Bacterial population structure was further delineated by genomic features allowing for the identification of candidate genes possibly contributing to local adaptation. These results suggest that the genetic structure within this bacterium is maintained both by ecological specialization in localized microenvironments (isolation by environment) and by dispersal limitation between geographic locations (isolation by distance).
INTRODUCTION
In eukaryotes, populations are typically defined as groups of interbreeding individuals within a species residing in the same geographic area (1). Geographically distinct (i.e., allopatric) populations are also often genetically distinct because of reduced gene flow, or the exchange of genetic variation, between populations of the same species. However, for microorganisms, the equivalent of the biological species concept does not exist, creating several obstacles to the study of the fine-scale genetic structure of microbial populations and, thus, the processes contributing to microbial diversification (2–4).
The first of these obstacles is that the degree of genetic relatedness delineating a microbial population is unclear. In eukaryotes, populations are, by definition, genetic units belonging to the same species, but defining a prokaryotic species remains challenging (5). Nonetheless, there is evidence for geographically distinct, genetically diverged groups of bacteria and archaea. Several studies have shown that the genetic similarity of closely related microbial individuals is negatively correlated with geographic distance across continental and global scales (6–9). This pattern is consistent with isolation by distance, whereby dispersal limitation contributes to reproductive isolation over geographic distances (10). Further, in some cases, these geographically localized genetic clades appear to be adapted to local environmental conditions, as individuals within these clades can differ in their temperature (11), nutrient (12), or habitat (13) preferences. However, the degree of divergence between genetic clades in such studies is usually quite high (<95% genome-wide average nucleotide identity [ANI]), indicating that they may not represent intraspecies relationships (14). These genetic units seem to be much broader than populations, or groups of individuals with the potential for contemporary interactions and exchange of genetic material (15). Therefore, a focus on much more closely related microorganisms is needed to investigate the processes responsible for initial diversification.
A second, related obstacle is recovering genetically similar individuals of the same species, however defined. Population genetic studies of eukaryotes typically characterize the genetic diversity among many individuals from a variety of geographic locations. For microbes, this sampling design requires reliable isolation of closely related strains (but see reference 16), which can be difficult in highly diverse microbial communities, such as soil. Finally, even if a sample of closely related individuals can be collected, a third obstacle is quantifying the exchange of genetic variation (i.e., gene flow) between individuals. For prokaryotes, the horizontal exchange of genetic material is mediated through genetic recombination, whether by homologous replacement of short gene segments or by transfer of entirely new genes. However, the asexual nature of prokaryotes makes it a challenge to quantify this process, particularly among closely related individuals. The more closely related that two genomes are, the more difficult it is to distinguish between differences caused by vertical inheritance and recombination (17).
For aquatic (18) and host-associated (19) systems, many of these obstacles have been addressed. In these environments, geographic proximity does not appear to be the most important factor in structuring microbial populations as typically observed in plants and animals. Indeed, an increasing number of studies find several, distinct genetic clades cooccurring in the same geographic location (20–23). For instance, the thermophilic archaeon Sulfolobus exhibited strong barriers to recombination between sympatric clades within a hot spring (24). Such evidence suggests that the genetic structure of microbial populations is influenced less by divergence among geographically distinct (allopatric) groups and more by ecological differentiation (isolation by environment [25]) among cooccurring (sympatric) groups (26). Thus, we might need to abandon the idea of defining microbial populations a priori based on geography (as done for larger organisms) and, instead, focus first on the emerging genetic structure among closely related individuals (27).
Soils are highly heterogeneous systems where differences in microhabitats can contribute to environmental variation over many spatial scales (28, 29). For this reason, one might expect that allopatric differentiation is more evident in soil bacteria than in microorganisms in aquatic or host-associated environments. Indeed, some soil fungi exhibit strong population structure at regional spatial scales (30, 31). Therefore, we asked whether population structure in a free-living soil bacterium was consistent with spatial patterns of allopatric or sympatric distributions. To do so, we investigated the abundant leaf litter taxon Curtobacterium (32), which is relatively easy to culture from the leaf litter layer of soil. Previously, we demonstrated that Curtobacterium encompasses multiple ecotypes, or fine-scale genetic clades that correspond to ecologically relevant phenotypes (33). Here, we concentrated on the genetic diversity within a single ecotype, Curtobacterium subclade IB/C, a unit that might be considered equivalent to a species designation (33). Specifically, we examined 26 strains (with identical full-length 16S rRNA regions and >97% mean genome-wide average amino acid identity) from a regional climate gradient, along with two closely related strains isolated across continental distances. We hypothesized that soil bacteria exhibit a pattern intermediate to those of aquatic free-living bacteria, archaea, and soil fungi. In particular, we expect that sympatric populations of soil bacteria exist within a particular geographic location, while also exhibiting a pattern consistent with allopatric differentiation among locations. Such a pattern would indicate that the genetic structure within this bacterium is maintained both by specialization in localized microenvironments (isolation by environment) and by dispersal limitation between geographic locations (isolation by distance).
RESULTS
Evolutionary history within a Curtobacterium ecotype.
We identified 26 strains from a Curtobacterium ecotype, subclade IB/C, that share ecologically relevant genotypic and phenotypic characteristics. These traits include the ability to degrade polymeric carbohydrates (i.e., cellulose and xylan), the degree of biofilm formation, and temperature preference for both growth and carbon degradation (33). These strains were previously isolated from leaf litter, the top layer of soil, at four geographic locations from a regional climate gradient in southern California (see Table S1 in the supplemental material). The isolation sites span an elevation gradient covarying in mean annual precipitation (ranging from 84.4 to 402 mm) and temperature (ranging from 10.3 to 24.6°C) (Fig. S1) (33). All analyzed strains have identical full-length 16S rRNA regions and share high sequence identity across their genomes, with >95% average nucleotide identity (ANI) and >97% average amino acid identity (AAI), congruent with previous observations for defining discrete sequence clusters within natural microbial communities (34). We also included two additional strains from subclade IB/C that were isolated from grassland leaf litter in Boston, MA (strains MCBA), to provide various geographic scales (ANImean similarity = 95.1%).
Detailed abiotic characteristics of five of the sites along the climate gradient. (A) Mean leaf litter chemistry of non-ash (dry weight); (B) mean surface soil temperature and precipitation. For more information, see the work of Chase et al. (33). Download FIG S1, TIF file, 1.1 MB (1.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genomic and geographic characteristics of strains. Download Table S1, DOCX file, 0.01 MB (14.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
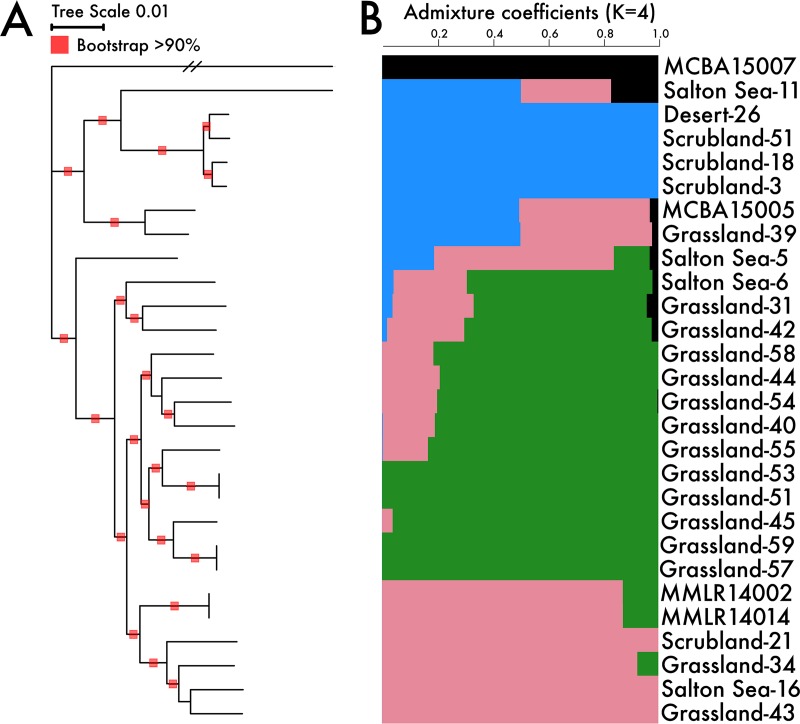
To examine whether genetically similar strains within the IB/C subclade clustered by geographic location, we reconstructed the phylogenetic relationship among the strains using the core genome (Fig. 1A). The core genome phylogeny revealed highly structured genetic lineages; however, clusters contained strains isolated from a variety of geographic locations. While one strain from Boston, MA, formed the outgroup, the other Boston strain was highly similar to a grassland strain from Loma Ridge, CA. At the regional scale within the climate gradient, most of the grassland strains clustered together, while strains from the scrubland and Salton Sea leaf litter communities were dispersed throughout the tree. Phylogenetic distance was negatively correlated with geographic distance (adjusted R2 = 0.26, P value < 0.001) suggesting that isolation by distance is a contributing factor in the spatial distribution among closely related strains (Fig. S2A).
FIG 1.
(A) Phylogeny of the Curtobacterium ecotype, subclade IB/C, from a core genome alignment; (B) ancestral population structure estimated from admixture analysis. Bar plots reflect the proportion of an individual genome that originates from estimated ancestral gene pools (K = 4). Genome names designate the site of isolation along the climate gradient except for MCBA (Boston, MA) and MMLR (grassland isolate from 2010).
Geographic distance (in kilometers) in relation to phylogenetic distance (A) and flexible gene content dissimilarity (B). Each point represents a pairwise genome comparison and is colored by population assignment. The total regression line is black. Download FIG S2, PDF file, 0.7 MB (758.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Phylogenetic analyses alone cannot delineate population structure, as it is necessary to account for both vertical descent and contributions from shared ancestral gene pools. Therefore, we supplemented the phylogenetic analysis by computing ancestry coefficients for each strain across the core genome using a structure-like (35) analysis (Fig. 1B). The most probable number of ancestral gene pools (K) contributing to the proportion of an individual genome (see Materials and Methods) was 4, demonstrating high congruence between the admixture coefficients and phylogenetic analysis. For example, an outgroup strain originating from Boston, MA, exhibited little evidence for mixing with most of the climate gradient strains in California across continental scales (Fig. 1B). Within the regional climate gradient, we detected three ancestral gene pools that may represent finer population structure across ecologically similar strains in ecotype IB/C.
Gene flow delineates bacterial populations.
Although structure-like analyses can provide insights into the ancestral population structure among divergent lineages, contemporary bacterial population boundaries (defined as groups with the potential to exchange genetic material) must be resolved by examining patterns of gene flow (i.e., recombination). However, in asexual organisms, measurements of homologous recombination can be overestimated when individuals are closely related, as distinguishing between recombination and point mutations is difficult (17). Further, other forms of horizontal gene transfer can be ecologically relevant as well (36). To address these limitations, we employed a novel method, PopCOGenT, that attempts to detect all recent recombination events between pairs of strains (27).
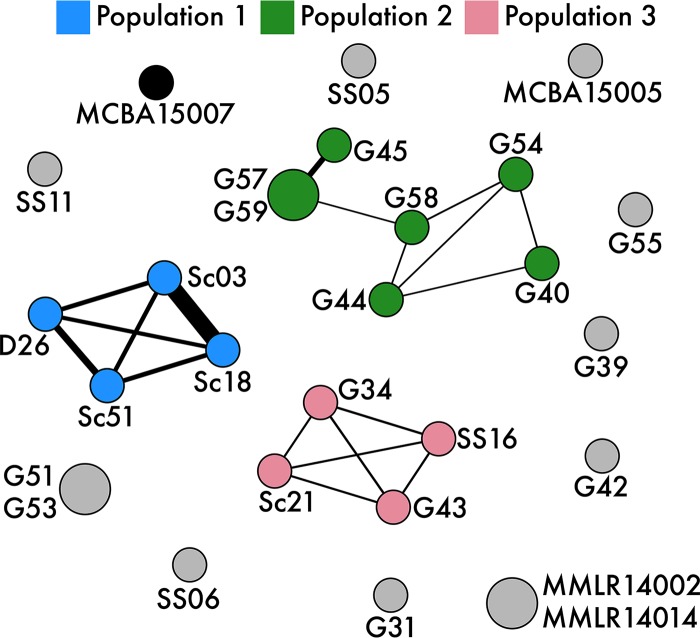
To distinguish between vertical descent and homologous recombination in structuring populations, we used PopCOGenT to estimate the degree of recombination among the genomes. This analysis revealed three recombining populations that are evident as isolated clusters in the network (Fig. 2). One of the populations (population 2) was restricted to a single location (in the grassland site). The other two populations included strains from multiple sites along the climate gradient; for example, population 3 contained strains isolated from the grassland, scrubland, and Salton Sea leaf litter communities, which are geographically separated by 177 km.
FIG 2.
Recombination network across all pairwise strains. Thicker edges represent increased recombination between strains. Nodes are colored by population designation, and node size indicates the number of clonal clusters (strains too closely related to differentiate recombination). D, desert; Sc, scrubland; G/MMLR, grassland; SS, Salton Sea; MCBA, Boston, MA.
This approach enabled the identification of recombining populations that would otherwise be masked with traditional phylogenetic analyses. For example, two strains (MMLR14002 and MMLR14014) isolated from a grassland site 5 years prior share no recent recombination events (Fig. 2), despite sharing a high degree of phylogenetic relatedness and a common ancestral gene pool with strains within population 3 (Fig. 1). Additionally, the analysis revealed that the highly similar strains isolated across the continent from one another (from Boston, MA, and a California grassland) (Fig. 1) were not connected by recent recombination events. Indeed, this conservative approach to estimate recombination events reduced most strains within the IB/C subclade to singleton nodes, suggesting that no recent recombination events connect these individuals to the three identified populations (Fig. 2) and that these strains are probably representatives of other, unsampled populations.
To confirm the effect of homologous recombination on the genetic diversity within subclade IB/C, we employed ClonalFrameML (37). Specifically, we concentrated on the ratio at which nucleotides are replaced by either recombination or point mutations (r/m). Throughout the evolution of the IB/C subclade, recombination rates were generally low (r/m = 0.94), indicating barriers to gene flow and the occurrence of mutation accumulation within the subclade. However, when we assessed the rates of recombination within each population assignment, we found homologous recombination rates to be high in populations 1 and 3 (r/m = 3.34 and 2.75, respectively), while population 2 (r/m = 1.62) had an intermediate recombination rate (Table S2). The observed r/m values are especially notable, as terrestrial free-living bacteria have previously been shown to have low r/m values (r/m < 1) (38), although others have noted high recombination metrics in Streptomyces species (39).
Ratios of nucleotide substitutions from recombination to point mutations (r/m). Download Table S2, DOCX file, 0.01 MB (12.8KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Population differentiation of the flexible genome.
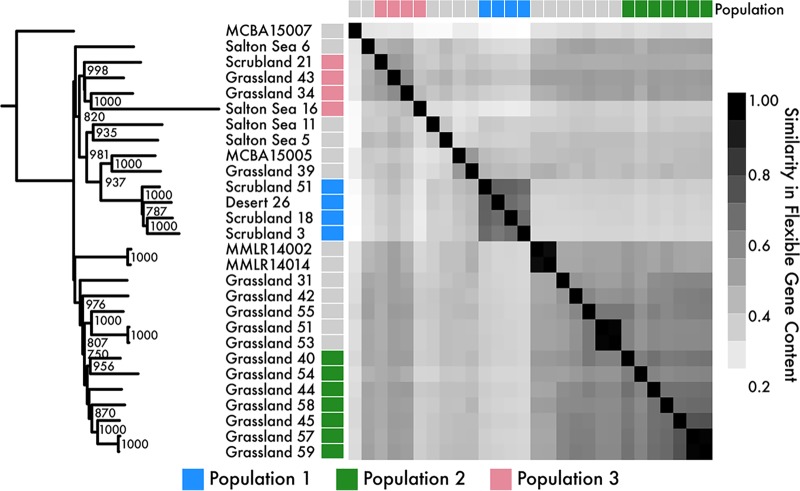
Based on the recombination networks, we expected individuals within the same population to also share more flexible genes (genes not present in all strains) than individuals between different populations. The similarity between flexible gene contents among strains was highly congruent with the population assignments (Fig. 3); strains within a population (analysis of similarity [ANOSIM], R = 0.88, P = 0.001) shared more flexible genes than expected by chance. We also observed that flexible gene content differed significantly by site (ANOSIM, R = 0.81, P < 0.01), suggesting that processes within and across locations structure the differences in the flexible genome within subclade IB/C. Indeed, flexible gene content similarity was negatively correlated with geographic distance (adjusted R2 = 0.14, P value < 0.001), providing additional evidence for isolation by distance (Fig. S2B).
FIG 3.
Flexible gene content similarity between strains. The tree is derived from a consensus neighbor-joining analysis showing only nodes with ≥750 support. Strains are colored by population assignments identified from the recombination network (Fig. 2).
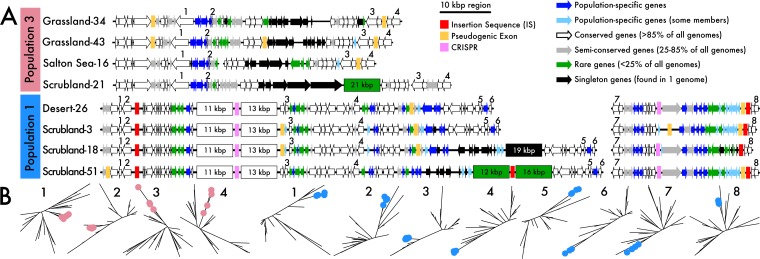
The flexible genome also provides insights into the traits that distinguish populations. For example, flexible genes present only in all individuals within a particular population may have swept through the population by positive selection (26). We searched for population-specific genes shared among all members and discovered that many were highly localized to a limited number of genomic regions. Specifically, 16 of 48 population-specific genes in population 1 were highly localized in the genome, while 4 of 6 population-specific genes in population 3 were localized (Fig. 4A). Additionally, these population-specific genes had reduced nucleotide diversity compared to that of whole-genome measurements (Fig. SS3), which can be indicative of relatively recent selective sweeps. These putative sweep regions may have arrived prior to population diversification and subsequently codiversified but, nonetheless, represent genomic regions harboring population-specific flexible genes. We did not detect any localization of population-specific genes in population 2, perhaps due to its lower rate of homologous recombination (Table S2).
FIG 4.
Highly structured genomic backbones across strains. (A) Population-specific genomic backbones within all individuals in populations 1 and 3. Population-specific genes (colored in blue) are consistently flanked by highly conserved regions (in white). Putative mobile elements are also designated in boxes along the chromosome. (B) Phylogenies of a subset of conserved genes (white arrows in panel A) flanking the population-specific regions colored by the strains in each respective population.
The flanking genomic regions surrounding the population-specific genes exhibited high levels of synteny across all members in the population as well, suggesting that these genomic regions may be hot spots for genetic exchange within the populations (Fig. 4A). While we did not detect phage or integrative and conjugative elements (ICEs), we did identify other mobile genetic elements, such as insertion sequences and clustered regularly interspaced short palindromic repeats (CRISPRs). Further, the regions were littered with pseudogenic exons, indicating the interruption of functional proteins due to recombining genomic segments (Table S3). The genomic regions also contained rare (<25% of all members within subclade IB/C) or strain-specific genes. In contrast to these variable regions, the flanking genes were highly conserved (shared by >85% of all members within subclade IB/C) in nearly identical genetic architectures (Fig. S4). Many of the conserved flanking core genes supported a strict monophyletic division of the population (Fig. 4B), suggesting that integration of population-specific genes is mediated by homologous recombination of the conserved flanking homologous gene regions (4).
Boxplots show nucleotide diversity (π) across all population-specific genes (present in all members within the population). Each point is a population-specific gene and is colored if the gene is localized in the genomic region displayed in Fig. 4. The dashed line shows the genome-wide average (πmean) of each strain in the population across all core genes within subclade IB/C. Download FIG S3, TIF file, 0.4 MB (416.9KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Mauve alignments showing population-specific genomic backbones depicted in Fig. 4. Colored blocks represent shared locally collinear blocks (LCBs) with similarity profiles of the genome sequences. Gapped regions with low similarity profiles in LCBs correspond to rare or singleton genes in Fig. 4. Black boxes show open reading frames, and bold red lines designate contig edges. Download FIG S4, PDF file, 1.3 MB (1.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Averages number of mobile elements (i.e., insertion sequences and clustered regularly interspaced short palindromic repeats [CRISPRs]), pseudogenic exons, and rare and unique genes (genes present in <25% of all genomes in subclade IB/C) for each population-specific genomic backbone. Download Table S3, DOCX file, 0.01 MB (12.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Most of the population-specific genes within the variable regions annotate as hypothetical proteins with some transcriptional regulators; however, other genes may be involved in differential use of environmental resources. For example, the regions contained a high number of metal uptake and transport proteins, along with glycoside hydrolase (GH) enzymes and glycosyltransferases, which contribute to the breakdown of carbohydrates commonly found in leaf litter. To that end, we also observed a difference in the full genomic potential to degrade various carbohydrates in leaf litter between populations (Fig. S5C) (analysis of variance [ANOVA], P < 0.01). However, other predicted genomic traits (i.e., minimum generation time and optimal growth temperature) were indistinguishable between populations, most likely due to the calculation incorporating full genome-wide codon usage biases (Fig. S5).
Distributions of predicted genomic traits in strains belonging to populations. Traits include minimum generation time (in hours) (A), optimal growth temperature (in degrees Celsius) (B), and total abundance of glycoside hydrolase (GH) and carbohydrate binding module (CBM) proteins (C). P < 0.05. Download FIG S5, TIF file, 1.5 MB (1.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
Our study demonstrates that the population structure of a free-living terrestrial bacterium can encompass related strains across geographic locations (although analyses of recombination suggest that gene flow occurs at only short scales), while less related strains of different populations can coexist within the same site. This genetic resolution was possible by isolating a variety of Curtobacterium strains from the same habitat (leaf litter) across geographic locations (33). Within the most abundant ecotype, subclade IB/C, we quantified gene flow among closely related, cooccurring lineages to identify distinct genetic populations of Curtobacterium across geographic distances. An analysis of the flexible genome confirmed that these populations are structured by gene flow discontinuities and provided additional evidence for population-specific genes that might be involved in local adaptation. Finally, the distributional patterns of the populations suggest that both isolation by distance and isolation by environment contribute to Curtobacterium population structure. Thus, both dispersal limitation and local environments contribute to differentiation among closely related soil bacteria, as observed in macroorganisms (40).
Previously, studies of two soil bacteria, Streptomyces and Bradyrhizobium, found continent-scale patterns consistent with allopatric diversification over distantly related strains (<95% ANI) (11, 13). Further, clonal sympatric strains of the social bacterium Myxococcus were found to have barriers to recombination over centimeter distances in soil (41). By isolating strains within a single Curtobacterium ecological cluster at various geographic scales, we could characterize the processes driving recent population divergence both between cooccurring strains and across regional spatial scales. As a comparison, we included two strains within this ecotype that were isolated from Boston, MA, and found no recent recombination events connecting strains across continental scales (Fig. 2). Notably, along the regional climate gradient, we found that closely related strains isolated from similar leaf litter communities were constrained in their geographic extent (mean geographic range of populations, 20.6 ± 49.7 km), suggesting that observed gene flow patterns are consistent with allopatric differentiation (see Fig. S2 in the supplemental material). However, we also observed multiple, genetically distinct populations overlapping at three of the sites. Two of these populations were comprised of individuals from spatially distinct sites that remained connected by gene flow, suggesting that isolation by distance is reduced at regional spatial scales. In contrast, other studies conducted at similar spatial scales found that genomic differences among fungal populations strongly reflected local site adaptations, a pattern consistent with strictly allopatric differentiation (30, 31).
The presence of sympatric Curtobacterium populations can indicate the presence of an isolating mechanism to maintain the cohesiveness of cooccurring genetic lineages (1). Alternatively, spatial barriers between the populations may have existed previously but have since been removed without sufficient time for genetic homogenization. The flexible genome of Curtobacterium provides two lines of evidence for the former and, specifically, that the identified populations have remained genetically isolated due to ecological differentiation, as others have observed in bacterial populations (42). First, Curtobacterium populations shared more flexible genes within populations than between populations, suggesting that the populations represent cohesive, ecologically differentiated clusters (Fig. 3). Flexible genes are thought to contribute to differences in niche exploitation (43) and can contribute to small fitness differences among microhabitats (15). For example, in the marine bacterium Vibrio, sympatric populations encode habitat-specific genes (44) between free-living and particle-associated populations (45). At a similar microscale, Curtobacterium populations may differentiate between leaf litter microhabitats caused by variability in resources, such as metal and carbohydrate availability. Accordingly, we observed differences in carbohydrate degradation potential (Fig. S5C) and observed population-specific genomic islands bearing genes related to physiological features.
The second line of evidence that sympatric populations are maintained by ecological differences is that all individuals within populations shared highly conserved genomic backbones containing population-specific genes (Fig. 4). The population-specific genomic backbones consisted of both core genes exhibiting a strict monophyletic division and population-specific flexible genes indicating recent selective sweeps within a population. These patterns have been previously identified in marine bacterial populations of Vibrio (44) and Prochlorococcus (16) and in the archaeon Sulfolobus (24), in which population-specific genomic regions were linked to small fitness differences and niche exploitation, contributing to the coexistence of sympatric populations. Similarly, increased homologous recombination among strains of Curtobacterium populations may enable the rapid exchange of niche-adaptive genes for differential microhabitat specialization on leaf litter. This observation is consistent with isolation by environment when gene exchange rates among similar environments is higher than within geographic locations (25). Thus, the populations along the regional climate gradient seem to represent genetically isolated lineages that have ecologically diverged because of their partitioning microhabitats (within a location).
Conclusions.
A major gap in our understanding of microbial diversity is the mechanisms contributing to the origin and maintenance of microbial diversification. Collectively, our results suggest a model for the recent microevolution of a soil bacterium. As with macroorganisms, free-living soil bacterial populations are geographically restricted. At the same time, distinct Curtobacterium populations may also have diverged to specialize on different leaf litter microhabitats, causing a reduction in gene flow between populations. Thus, overlapping populations are maintained within the same location, while also being connected via dispersal to individuals in other locations.
MATERIALS AND METHODS
Field sites and Curtobacterium strains.
We downloaded 28 Curtobacterium genomes (see Table S1 in the supplemental material) from the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/) database that were previously isolated from leaf litter (32), including a robust genomic data set consisting of 26 strains from a climate gradient in southern California (33). We included two additional strains within the same ecotype from outside Boston, MA, to provide various spatial scales. Protein-coding regions and gene annotations were derived from the NCBI prokaryotic genome annotation pipeline (46). Genomes were further screened for the presence of mobile genetic elements by identifying integrating and conjugative elements (ICEs) with the ICEberg database (47), prophage sequences using PhiSpy (48), insertion sequences (IS) with ISfinder (49), and CRISPRs with CRISPRCasFinder (50). Whole-genome pairwise average nucleotide identity was performed with FastANI (https://github.com/ParBLiSS/FastANI).
Evolutionary history of the core genome.
We aligned all genomes using progressiveMauve (51) to identify locally colinear blocks (LCBs) of genomic data. We identified 49,610 LCBs of >1,500 bp found across all 28 genomes that represented 1.28 Mbp of the core genome. This core genome alignment was used to perform a maximum likelihood bootstrap analysis using RAxML v8.2.10 (52) under the general time reversal model with a gamma distribution for 100 replicates.
Using the core genome, we performed an initial analysis to infer the relative effects of recombination and mutation rates using ClonalFrameML v1.11 (37). Specifically, we attempted to reconstruct phylogenetic relationships by detecting regions of recombination across the phylogeny to provide an initial estimate for clonal genealogy. Due to the weak clonal structure among strains, we sought to infer population structure from multilocus genotype data. First, we converted the core genome sequence data to a genotype matrix reflecting the distance between polymorphic sites of all individuals (https://github.com/xavierdidelot). We then used this genotype matrix to compute ancestry coefficients to delineate genetic clusters. Specifically, we employed sparse nonnegative matrix factorization algorithms to estimate the cross-entropy parameter (53). Based on the cross-entropy criterion which best fit the statistical model, we designated the number of ancestral populations (K) as equal to 4 to estimate individual admixture coefficients using the LEA package (35) in the R software environment (54).
Gene flow and recombination networks.
To differentiate between vertical transmission and recent recombination, we identified recent transfer events across all pairs of genomes using PopCOGenT (https://github.com/philarevalo/PopCOGenT) (27). Briefly, we used a null model of sequence divergence to calculate the expected length distribution of identical genomic regions between strain pairs. Recently exchanged genes enrich this distribution by introducing identical genomic regions that are longer and more frequent than expected. The extent of this enrichment is our measurement of recent transfer. Strains that were too closely related (<0.035% ANI divergence) to accurately assess recombination transfers were collapsed into clonal complexes. Finally, strains that were connected to any other strain in the recombination network were considered to be a part of the same recombining population. To confirm the importance of recombination events in structuring populations, we inferred the relative effects of recombination and mutations rates of the core genome (see above) within each population using ClonalFrameML.
Population genetic analyses.
To perform within-population genetic analyses, we identified all orthologous protein-coding genes (orthologs) shared across all strains. Orthologs were initially predicted using ROARY (55), with a minimum sequence identity of 90% to ensure that all possible orthologs were included across populations (Fig. S6A). The resulting 2,193 orthologs shared across all strains were individually aligned with Clustal O v1.2.3 (56) and used to create a 2.14-Mbp concatenated nucleotide alignment. Note, the size of this alignment differs from that of the core genome alignment since genes do not necessarily need to be located on LCBs. To verify the effects of using a gene-by-gene approach on the core genome, we reconstructed the phylogenetic relationship of the concatenated alignment of all orthologous protein-coding genes, using RAxML v8.2.10 (52) under the general time reversal model with a gamma distribution for 100 replicates, and compared the phylogenies derived from the Mauve core genome alignment (Fig. S6B). Next, all individual ortholog alignments were screened for complete codon reading frames (i.e., multiple of 3 bp), and the resulting 2,137 genes were individually used to calculate nucleotide diversity within populations using the PopGenome package (57) in R, as previously outlined (58).
Breakdown of orthologous protein groups derived from all strains. (A) Numbers of identified orthologous protein groups in both the core and flexible genome based on initial clustering of proteins; (B) cladogram comparison of core genes (2,137 orthologous proteins) and core genome alignment (defined as locally collinear blocks). Terminal branches are colored by geographic location, with lines connecting identical strains in each respective cladogram. Download FIG S6, TIF file, 2.4 MB (2.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Predicted orthologs that were not shared across all strains represent the flexible genome (Fig. S6A). Using all identified orthologs, we computed a Jaccard distance between pairs of strains to estimate shared gene content. The distance matrix was used to generate a neighbor-joining tree based on 1,000 resamplings and to create a heatmap showing gene content similarity across strains. We tested the significance of gene content using an analysis of similarities (ANOSIM) for populations and the site of isolation for 9,999 permutations. In addition, we looked for orthologs that were unique to our populations. Specifically, we identified orthologs that were carried by every member within a population and were not found in any member outside the population. To reduce this list even further, we identified population-specific orthologs that were localized in genomic regions (<10-kbp separation).
Analysis of genomic traits.
We analyzed all genomic sequences for specific ecological traits that may contribute to population divergence. We concentrated on genomic traits related to growth strategies and substrate (i.e., carbohydrate) utilization that may be advantageous on leaf litter.
To infer growth strategies, we estimated minimum generation times (MGTs) and optimal growth temperature (OGT). We predicted MGTs by comparing codon usage biases between highly expressed ribosomal proteins and all other carried genes by following a linear regression model (59) (equation 1).
| (1) |
where ENC is the effective number of codons given the percent GC (60).
We analyzed each strain for the genomic potential to degrade various carbohydrates by searching the predicted coding regions against the Pfam-A v30.0 database (61) using HMMER (62). Identified protein families were reduced to only known protein families that encode glycoside hydrolase (GH) and carbohydrate-binding module (CBM) proteins as described in reference 32.
Availability of data.
Relevant data and code used can be found at https://github.com/alex-b-chase/curto-popgen. Biosample identification numbers are available in Table S1. Additional data sets used and/or analyzed during the current study are available from the corresponding author upon request.
ACKNOWLEDGMENTS
We thank Claudia Weihe, Chamee Moua, and Michaeline Albright for their assistance in strain isolation. We also thank Brandon Gaut for invaluable insight into data analysis and interpretation, Sarai Finks, Kendra Walters, Cynthia Rodriguez, and the rest of the Martiny lab for helpful comments, and Xavier Didelot and Kevin Bonham for software assistance.
We declare that we have no competing interests.
This work was supported by a U.S. Department of Education Graduate Assistance in Areas of National Need (GAANN) fellowship (A.B.C.), a U.S. Department of Energy, Office of Science Graduate Student Research (SCGSR) fellowship (A.B.C.), and grant DE-SC0016410 from the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (J.B.H.M.).
A.B.C., E.L.B., U.K., and J.B.H.M. designed the research project; A.B.C., P.A., and U.K. analyzed data; and A.B.C., M.F.P., and J.B.H.M. wrote the manuscript, with input from all authors.
Footnotes
Citation Chase AB, Arevalo P, Brodie EL, Polz MF, Karaoz U, Martiny JBH. 2019. Maintenance of sympatric and allopatric populations in free-living terrestrial bacteria. mBio 10:e02361-19. https://doi.org/10.1128/mBio.02361-19.
REFERENCES
- 1.Mayr E. 2001. What evolution is. Science Masters Series. Basic Books, New York, NY. [Google Scholar]
- 2.Chase AB, Martiny J. 2018. The importance of resolving biogeographic patterns of microbial microdiversity. Microbiol Aust 39:5–8. [Google Scholar]
- 3.Shapiro BJ, Leducq J-B, Mallet J. 2016. What is speciation? PLoS Genet 12:e1005860. doi: 10.1371/journal.pgen.1005860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rocha E. 2018. Neutral theory, microbial practice: challenges in bacterial population genetics. Mol Biol Evol 35:1338–1347. doi: 10.1093/molbev/msy078. [DOI] [PubMed] [Google Scholar]
- 5.Ward DM, Cohan FM, Bhaya D, Heidelberg JF, Kühl M, Grossman A. 2008. Genomics, environmental genomics and the issue of microbial species. Heredity (Edinb) 100:207–219. doi: 10.1038/sj.hdy.6801011. [DOI] [PubMed] [Google Scholar]
- 6.Andam CP, Doroghazi JR, Campbell AN, Kelly PJ, Choudoir MJ, Buckley DH. 2016. A latitudinal diversity gradient in terrestrial bacteria of the genus Streptomyces. mBio 7:e02200-15. doi: 10.1128/mBio.02200-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choudoir MJ, Doroghazi JR, Buckley DH. 2016. Latitude delineates patterns of biogeography in terrestrial Streptomyces. Environ Microbiol 18:4931–4945. doi: 10.1111/1462-2920.13420. [DOI] [PubMed] [Google Scholar]
- 8.Whitaker RJ, Grogan DW, Taylor JW. 2003. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science 301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 9.Zwirglmaier K, Jardillier L, Ostrowski M, Mazard S, Garczarek L, Vaulot D, Not F, Massana R, Ulloa O, Scanlan DJ. 2008. Global phylogeography of marine Synechococcus and Prochlorococcus reveals a distinct partitioning of lineages among oceanic biomes. Environ Microbiol 10:147–161. doi: 10.1111/j.1462-2920.2007.01440.x. [DOI] [PubMed] [Google Scholar]
- 10.Wright S. 1943. Isolation by distance. Genetics 28:114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choudoir MJ, Buckley DH. 2018. Phylogenetic conservatism of thermal traits explains dispersal limitation and genomic differentiation of Streptomyces sister-taxa. ISME J 12:2176–2186. doi: 10.1038/s41396-018-0180-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. 2006. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311:1737–1740. doi: 10.1126/science.1118052. [DOI] [PubMed] [Google Scholar]
- 13.VanInsberghe D, Maas KR, Cardenas E, Strachan CR, Hallam SJ, Mohn WW. 2015. Non-symbiotic Bradyrhizobium ecotypes dominate North American forest soils. ISME J 9:2435–2441. doi: 10.1038/ismej.2015.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 2017. High-throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. bioRxiv 225342 https://www.biorxiv.org/content/10.1101/225342v1.full. [DOI] [PMC free article] [PubMed]
- 15.Cordero OX, Polz MF. 2014. Explaining microbial genomic diversity in light of evolutionary ecology. Nat Rev Microbiol 12:263–273. doi: 10.1038/nrmicro3218. [DOI] [PubMed] [Google Scholar]
- 16.Kashtan N, Roggensack SE, Rodrigue S, Thompson JW, Biller SJ, Coe A, Ding H, Marttinen P, Malmstrom RR, Stocker R, Follows MJ, Stepanauskas R, Chisholm SW. 2014. Single-cell genomics reveals hundreds of coexisting subpopulations in wild Prochlorococcus. Science 344:416–420. doi: 10.1126/science.1248575. [DOI] [PubMed] [Google Scholar]
- 17.Ravenhall M, Škunca N, Lassalle F, Dessimoz C. 2015. Inferring horizontal gene transfer. PLoS Comput Biol 11:e1004095. doi: 10.1371/journal.pcbi.1004095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui Y, Yang X, Didelot X, Guo C, Li D, Yan Y, Zhang Y, Yuan Y, Yang H, Wang J, Wang J, Song Y, Zhou D, Falush D, Yang R. 2015. Epidemic clones, oceanic gene pools, and eco-LD in the free living marine pathogen Vibrio parahaemolyticus. Mol Biol Evol 32:1396–1410. doi: 10.1093/molbev/msv009. [DOI] [PubMed] [Google Scholar]
- 19.Sheppard SK, McCarthy ND, Falush D, Maiden M. 2008. Convergence of Campylobacter species: implications for bacterial evolution. Science 320:237–239. doi: 10.1126/science.1155532. [DOI] [PubMed] [Google Scholar]
- 20.Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- 21.Cohan FM. 2001. Bacterial species and speciation. Syst Biol 50:513–524. doi: 10.1080/10635150118398. [DOI] [PubMed] [Google Scholar]
- 22.Chase AB, Karaoz U, Brodie EL, Gomez-Lunar Z, Martiny AC, Martiny J. 2017. Microdiversity of an abundant terrestrial bacterium encompasses extensive variation in ecologically relevant traits. mBio 8:e01809-17. doi: 10.1128/mBio.01809-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitaker RJ, Grogan DW, Taylor JW. 2005. Recombination shapes the natural population structure of the hyperthermophilic archaeon Sulfolobus islandicus. Mol Biol Evol 22:2354–2361. doi: 10.1093/molbev/msi233. [DOI] [PubMed] [Google Scholar]
- 24.Cadillo-Quiroz H, Didelot X, Held NL, Herrera A, Darling A, Reno ML, Krause DJ, Whitaker RJ. 2012. Patterns of gene flow define species of thermophilic Archaea. PLoS Biol 10:e1001265. doi: 10.1371/journal.pbio.1001265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang IJ, Bradburd GS. 2014. Isolation by environment. Mol Ecol 23:5649–5662. doi: 10.1111/mec.12938. [DOI] [PubMed] [Google Scholar]
- 26.Polz MF, Alm EJ, Hanage WP. 2013. Horizontal gene transfer and the evolution of bacterial and archaeal population structure. Trends Genet 29:170–175. doi: 10.1016/j.tig.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arevalo P, VanInsberghe D, Elsherbini J, Gore J, Polz MF. 2019. A reverse ecology approach based on a biological definition of microbial populations. Cell 178:820–834. doi: 10.1016/j.cell.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 28.Ranjard L, Richaume A. 2001. Quantitative and qualitative microscale distribution of bacteria in soil. Res Microbiol 152:707–716. doi: 10.1016/s0923-2508(01)01251-7. [DOI] [PubMed] [Google Scholar]
- 29.Nannipieri P, Ascher J, Ceccherini M, Landi L, Pietramellara G, Renella G. 2003. Microbial diversity and soil functions. Eur J Soil Science 54:655–670. doi: 10.1046/j.1351-0754.2003.0556.x. [DOI] [Google Scholar]
- 30.Amend A, Garbelotto M, Fang Z, Keeley S. 2010. Isolation by landscape in populations of a prized edible mushroom Tricholoma matsutake. Conserv Genet 11:795–802. doi: 10.1007/s10592-009-9894-0. [DOI] [Google Scholar]
- 31.Branco S, Gladieux P, Ellison CE, Kuo A, LaButti K, Lipzen A, Grigoriev IV, Liao H-L, Vilgalys R, Peay KG, Taylor JW, Bruns TD. 2015. Genetic isolation between two recently diverged populations of a symbiotic fungus. Mol Ecol 24:2747–2758. doi: 10.1111/mec.13132. [DOI] [PubMed] [Google Scholar]
- 32.Chase AB, Arevalo P, Polz MF, Berlemont R, Martiny J. 2016. Evidence for ecological flexibility in the cosmopolitan genus Curtobacterium. Front Microbiol 7:1874 http://journal.frontiersin.org/article/10.3389/fmicb.2016.01874/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chase AB, Gomez-Lunar Z, Lopez AE, Li J, Allison SD, Martiny AC, Martiny JBH. 2018. Emergence of soil bacterial ecotypes along a climate gradient. Environ Microbiol 20:4112–4126. doi: 10.1111/1462-2920.14405. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-R LM, Konstantinidis KT. 2014. Bypassing cultivation to identify bacterial species. Microbe 9:111–118. doi: 10.1128/microbe.9.111.1. [DOI] [Google Scholar]
- 35.Frichot E, François O. 2015. LEA: an R package for landscape and ecological association studies. Methods Ecol Evol 6:925–929. doi: 10.1111/2041-210X.12382. [DOI] [Google Scholar]
- 36.van Elsas JD, Bailey MJ. 2002. The ecology of transfer of mobile genetic elements. FEMS Microbiol Ecol 42:187–197. doi: 10.1111/j.1574-6941.2002.tb01008.x. [DOI] [PubMed] [Google Scholar]
- 37.Didelot X, Wilson DJ. 2015. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol 11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vos M, Didelot X. 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J 3:199. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- 39.Doroghazi JR, Buckley DH. 2014. Intraspecies comparison of Streptomyces pratensis genomes reveals high levels of recombination and gene conservation between strains of disparate geographic origin. BMC Genomics 15:970. doi: 10.1186/1471-2164-15-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sexton JP, Hangartner SB, Hoffmann AA. 2014. Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution 68:1–15. doi: 10.1111/evo.12258. [DOI] [PubMed] [Google Scholar]
- 41.Wielgoss S, Didelot X, Chaudhuri RR, Liu X, Weedall GD, Velicer GJ, Vos M. 2016. A barrier to homologous recombination between sympatric strains of the cooperative soil bacterium Myxococcus xanthus. ISME J 10:2468–2477. doi: 10.1038/ismej.2016.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro BJ, Polz MF. 2014. Ordering microbial diversity into ecologically and genetically cohesive units. Trends Microbiol 22:235–247. doi: 10.1016/j.tim.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez-Valera F, Ussery DW. 2012. Is the pan-genome also a pan-selectome? F1000Research 1:16. doi: 10.12688/f1000research.1-16.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, Szabó G, Polz MF, Alm EJ. 2012. Population genomics of early events in the ecological differentiation of bacteria. Science 336:48–51. doi: 10.1126/science.1218198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yawata Y, Cordero OX, Menolascina F, Hehemann J-H, Polz MF, Stocker R. 2014. Competition—dispersal tradeoff ecologically differentiates recently speciated marine bacterioplankton populations. Proc Natl Acad Sci U S A 111:5622–5627. doi: 10.1073/pnas.1318943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bi D, Xu Z, Harrison EM, Tai C, Wei Y, He X, Jia S, Deng Z, Rajakumar K, Ou H-Y. 2012. ICEberg: a web-based resource for integrative and conjugative elements found in Bacteria. Nucleic Acids Res 40:D621–D626. doi: 10.1093/nar/gkr846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akhter S, Aziz RK, Edwards RA. 2012. PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity-and composition-based strategies. Nucleic Acids Res 40:e126. doi: 10.1093/nar/gks406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siguier P, Pérochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C. 2018. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res 46:W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frichot E, Mathieu F, Trouillon T, Bouchard G, François O. 2014. Fast and efficient estimation of individual ancestry coefficients. Genetics 196:973–983. doi: 10.1534/genetics.113.160572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pinheiro J, Bates D, DebRoy S, Sarkar D, R Development Core Team. 2010. nlme: linear and nonlinear mixed effects models. R package version 3.1-97 R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 55.Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pfeifer B, Wittelsbürger U, Ramos-Onsins SE, Lercher MJ. 2014. PopGenome: an efficient Swiss army knife for population genomic analyses in R. Mol Biol Evol 31:1929–1936. doi: 10.1093/molbev/msu136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lemieux JE, Tran AD, Freimark L, Schaffner SF, Goethert H, Andersen KG, Bazner S, Li A, McGrath G, Sloan L, Vannier E, Milner D, Pritt B, Rosenberg E, Telford S, Bailey JA, Sabeti PC. 2016. A global map of genetic diversity in Babesia microti reveals strong population structure and identifies variants associated with clinical relapse. Nat Microbiol 1:16079. doi: 10.1038/nmicrobiol.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vieira-Silva S, Rocha E. 2010. The systemic imprint of growth and its uses in ecological (meta) genomics. PLoS Genet 6:e1000808. doi: 10.1371/journal.pgen.1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Subramanian S. 2008. Nearly neutrality and the evolution of codon usage bias in eukaryotic genomes. Genetics 178:2429–2432. doi: 10.1534/genetics.107.086405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39(Suppl 2):29–37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Detailed abiotic characteristics of five of the sites along the climate gradient. (A) Mean leaf litter chemistry of non-ash (dry weight); (B) mean surface soil temperature and precipitation. For more information, see the work of Chase et al. (33). Download FIG S1, TIF file, 1.1 MB (1.1MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Genomic and geographic characteristics of strains. Download Table S1, DOCX file, 0.01 MB (14.4KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Geographic distance (in kilometers) in relation to phylogenetic distance (A) and flexible gene content dissimilarity (B). Each point represents a pairwise genome comparison and is colored by population assignment. The total regression line is black. Download FIG S2, PDF file, 0.7 MB (758.3KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Ratios of nucleotide substitutions from recombination to point mutations (r/m). Download Table S2, DOCX file, 0.01 MB (12.8KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Boxplots show nucleotide diversity (π) across all population-specific genes (present in all members within the population). Each point is a population-specific gene and is colored if the gene is localized in the genomic region displayed in Fig. 4. The dashed line shows the genome-wide average (πmean) of each strain in the population across all core genes within subclade IB/C. Download FIG S3, TIF file, 0.4 MB (416.9KB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Mauve alignments showing population-specific genomic backbones depicted in Fig. 4. Colored blocks represent shared locally collinear blocks (LCBs) with similarity profiles of the genome sequences. Gapped regions with low similarity profiles in LCBs correspond to rare or singleton genes in Fig. 4. Black boxes show open reading frames, and bold red lines designate contig edges. Download FIG S4, PDF file, 1.3 MB (1.3MB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Averages number of mobile elements (i.e., insertion sequences and clustered regularly interspaced short palindromic repeats [CRISPRs]), pseudogenic exons, and rare and unique genes (genes present in <25% of all genomes in subclade IB/C) for each population-specific genomic backbone. Download Table S3, DOCX file, 0.01 MB (12.7KB, docx) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Distributions of predicted genomic traits in strains belonging to populations. Traits include minimum generation time (in hours) (A), optimal growth temperature (in degrees Celsius) (B), and total abundance of glycoside hydrolase (GH) and carbohydrate binding module (CBM) proteins (C). P < 0.05. Download FIG S5, TIF file, 1.5 MB (1.6MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Breakdown of orthologous protein groups derived from all strains. (A) Numbers of identified orthologous protein groups in both the core and flexible genome based on initial clustering of proteins; (B) cladogram comparison of core genes (2,137 orthologous proteins) and core genome alignment (defined as locally collinear blocks). Terminal branches are colored by geographic location, with lines connecting identical strains in each respective cladogram. Download FIG S6, TIF file, 2.4 MB (2.5MB, tif) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.