Abstract
Clathrin mediated endocytosis is one pathway for internalization of extracellular nano materials into cells [1, 2]. In this pathway, proteins attached to receptors and the internalized materials are encapsulated in clathrin coated membrane vesicles that subsequently fuse with or transform into intracellular compartments (early and late endosomes) as their contents are being directed to the lysosomes for degradation. The following proteins are commonly used to mark the pathway at various stages: Rab5 (early endosome), Rab7 (late endosome), and LAMP-1 (lysosome). In this work, we studied the distribution and co-localization of these marker proteins in two cell lines (C2C12 and A549) to determine whether these markers are unique for specific endosome types or whether they can co-exist with other markers. We estimate the densities and sizes of the endosomes containing the three markers, as well as the number of marker antibodies attached to each endosome. We determine that the markers are not unique to one endosome type but that the extent of co-localization is different for the two cell types. In fact, we find endosomes that contain all three markers simultaneously. Our results suggest that the use of these proteins as specific markers for specific endosome types should be reevaluated. This was the first successful use of triple image cross correlation spectroscopy to qualitatively and quantitatively study the extent of interaction among three different species in cells and also the first experimental study of three-way interactions of clathrin mediated endocytic markers.
Keywords: Analytical chemistry, Physical chemistry, Biophysics, Cell biology, Cell imaging, Image correlation spectroscopy, Endocytosis, Fluorescence microscopy
1. Introduction
Clathrin mediated endocytosis is one process known to take up extracellular, nanosized, materials through membrane invaginations that rely on the recruitment of clathrin from the cytoplasm to the membrane [1, 2, 3, 4, 5, 6]. This endocytic pathway then involves the formation of intracellular vesicles that transform into various endocytic compartments [7]. There are a series of maturation and fusion steps that are hypothesized to occur as the endosomes transform from early to late endosomes and then to lysosomes [8]. Two models have been proposed; the vesicle shuttle model and the maturation model [9, 10]. The vesicle shuttle model proposes that the endosomes are of fixed size, location, and morphology and transport of cargo occurs by smaller vesicles transporting the cargo between the endosomes. In this model specific endosomal markers could be uniquely associated with a particular endosome compartment. The maturation model proposes that the endosomes and their contents remain together as the endosomes undergo maturation and subsequent fusion with the lysosome. In this model, intracellular protein markers for the various endosomes could co-exist during the maturation and fusion process. The latter model appears to be the most commonly supported by evidence to date [11, 12, 13, 14, 15], but more specific, direct evidence for the simultaneous presence of two or more markers on the same endosomes is missing.
The Rab-GTPase (Rab) family of proteins are associated with intracellular membrane trafficking and Rab proteins have been identified to localize to specific domains on endocytic compartments [16, 17]. These specific Rab proteins are therefore referred to as markers for early and late endosomes. In particular, Rab5 is used as a marker for early endosomes [18, 19, 20, 21, 22, 23] and Rab7 is used as a marker for late endosomes [24, 25, 26, 27, 28]. No Rab proteins have been identified to localize specifically to the lysosome, however the lysosomal associated membrane protein (LAMP-1) is known as a marker for the lysosomes [29, 30, 31, 32, 33]. In this work, we used antibodies against Rab5, Rab7, and LAMP-1 to study their distribution and possible co-localization within the cell.
Fluorescence microscopy has been used to visualize the co-localization of cargo with intracellular proteins and membranes [14, 34, 35]. In addition, it has also been used to determine the distribution of various endocytic organelles based on association of Rab proteins with intracellular membranes [36], the dynamics of clathrin recruitment to the cell membrane [37, 38], the dynamics of adapter protein recruitment and removal [39], and the dynamics of dynamin assisted pinching of the invaginated pit [40, 41]. However, no quantitative studies have been performed to determine the extent of co-localization and clustering of Rab5, Rab7, and LAMP-1 markers in cells.
The purpose of this work was to determine as quantitatively as possible, the density of markers on the endosomes, the size of the endosomes, whether markers for the early endosome (Rab5), the late endosome (Rab7) and the lysosome (LAMP-1) can coexist on the endosomes and whether these observations depend on the type of cell being studied.
2. Methods
Cell culture and sample labelling experiments were done at the National Institute for Nanotechnology (NINT) in Edmonton, Alberta. Fluorescent imaging was done at the Cell Imaging Facility at the Cross Cancer Institute in Edmonton, Alberta.
2.1. Cell culture
Mouse muscle myoblastoma (C2C12) cells were cultured in Dulbecco's modified eagle medium (DMEM), supplemented with 10 % fetal bovine serum (FBS). Human alveolar adenocarcinoma (A549) cells were cultured in Ham's F-12 K medium (F-12 K) supplemented with 10 % FBS. The cells were grown in an incubator chamber maintained at 37 °C and with 5 % Carbon Dioxide (CO2) atmosphere and were passaged every five days using 0.25 % Trypsin-EDTA. At 80% cell surface confluence, the cells were passaged 1:5 onto 35 mm (mm) glass bottom dishes and maintained in a 37 °C, 5 % CO2 incubator for about two days when approximately 60 % cell confluency was reached for experimentation. At this time the media was replaced with 1 mL of fresh media and treated as indicated below.
2.2. Immunofluorescent labelling
Labeling of the various endocytic compartments was accomplished with antibodies specific for Rab5, Rab7, and LAMP-1 protein endocytic markers. The primary antibodies used were monoclonal and originated from three different species (mouse, rabbit, and rat) to avoid potential crosslinking in experiments with multiple labels. Polyclonal secondary antibodies labeled with different fluorescent probes were added to bind with the primary antibodies. The fluorescent probes of the polyclonal antibodies were labelled and identified by the manufacturer as Cy3, Alexa Fluor 488 nm, Alexa Fluor 555 nm, and Alexa Fluor 647 nm.
2.2.1. Labeling one endocytic marker
At 60% cell surface confluency, the cells were exposed to 1 mL of 4 % paraformaldehyde (PFA) at room temperature for 15 min for cell fixation. They were then washed with 1X phosphate buffered saline (PBS) and permeabilized with 1 mL of a 0.3% solution of Triton X-100 detergent in 1X PBS for 15 min at 37 °C. The cells were washed with 1X PBS, and exposed to 100 L of a 3 % solution of Bovine Serum Albumin (BSA) in 1X PBS to minimize non-specific intracellular binding of the antibodies.
Next, the cells were washed with 1X PBS and labelled with the primary antibody of interest using 100 L of the diluted (1:100) primary antibody solution overnight at 4 degrees Celsius. Finally, cells were washed with 1X PBS and labelled with the diluted (1:400 for Alexa Fluor 647 nm, 1:500 for Cy3, Alexa Fluor 488 nm, and Alexa Fluor 555 nm) secondary antibody for 90 min at room temperature and then washed with 1X PBS for the last time before imaging.
2.2.2. Labeling two endocytic markers
To study the extent of colocalization of two markers, the following three combinations were studied: Rab5 and Rab7 markers; Rab5 and LAMP-1 markers; and Rab7 and LAMP-1 markers.
Pairs of primary and secondary antibodies were carefully selected to avoid potential for cross-reactions. Specifically, we used the following combinations in this sequence; 1) Mouse-Anti Rab5 and Goat Anti-Mouse Cy3 with Rabbit Anti-Rab7 and Goat Anti-Rabbit Alexa Fluor 647, 2) Mouse-Anti Rab5 and Goat Anti-Mouse Cy3 with Rat Anti-LAMP-1 and Goat Anti-Rat Alexa Fluor 647, 3) Rabbit Anti-Rab7 and Goat Anti-Rabbit Alexa Fluor 647 with Rat Anti-LAMP-1 and Goat Anti-Rat Alexa Fluor 555.
Once samples were labelled with one primary and secondary antibody pair as described for one endocytic marker, the cells were washed with 1X PBS at room temperature, and then exposed to the second primary and secondary antibody pair of interest using the same procedure.
2.2.3. Labeling three endocytic markers
To determine the extent of co-localization of all three markers, the cells were labeled with primary and secondary antibody pairs for Rab5, then Rab7, and finally Lamp-1 using Mouse-Anti Rab5 and Goat Anti-Mouse Cy3; Rabbit Anti-Rab7 and Goat Anti-Rabbit Alexa Fluor 647; and Rat Anti-LAMP-1 and Goat Anti-Rat Alexa Fluor 488.
2.3. Image acquisition
Images of the cells in a sample were obtained with a Carl Zeiss 710 Laser Scanning Confocal Microscope with a 63 × 1.4 Numerical Aperture Oil DIC Plan-Apochromatic lens. ZEN 2011 was the software used to adjust and select parameters for image acquisition.
Lasers used for image acquisition were Argon Ion for Fluorescein Isothiocyanite (FITC) configuration (488 nm excitation of the Alexa Fluor 488 nm probe), Solid State for Cyanine-3 (Cy3) Configuration (561 nm excitation of the Cy3 and Alexa Fluor 555 nm probes), and HeNe for Cyanine-5 (Cy5) Configuration (633 nm excitation of the Alexa Fluor 647 nm probe). Complementary DIC was used for focusing.
The detectors for each configuration were adjusted in such a way that the emission from each channel was unique to a specific wavelength range. In addition, as a control measure, samples labelled with one fluorescent probe, such as Cy3, were illuminated with the FITC configuration, and then with the Cy5 configuration separately, to establish that potential cross talk was minimized (see supplemental). In addition, controls showed that there was little non-specific binding of secondary antibodies (see supplemental).
The pixel dwell time was set to 3.15 μs (μs) with an average of 2 line scans, and the pinhole was set to 1 airy-unit (AU). A Zoom factor of 14 was used to achieve a pixel size of approximately 20 nm. Each image comprised a square of 512 × 512 pixels, resulting in a 10 × 10 micron image for image correlation spectroscopy analysis. For each experiment, 25–40 images were obtained; each image recorded from a different cell in the sample.
2.4. Image J & ICS software for ICS family analysis
In this study, the image correlation toolkit in ImageJ software was utilized and the following pieces of information from images were obtained: the normalized auto and cross correlation function amplitudes, the average image intensity, and the laser beam radius in microns. Regions of interest were selected to 256 × 256 of the 512 × 512 image to avoid dark regions of the image where there is no cell. Cross correlation of images from two channels from two different cells to determine the cross-correlation amplitude that can arise from uncorrelated fluctuations. This represents a lower limit for the significance of cross correlation amplitude arising from two channels from the same cell.
For TRICCS measurements, images were analyzed using image correlation spectroscopy software written by Dr. Max Anikovsky (manuscript in preparation). Parameters for the range of data points, triple cross correlation amplitude, laser beam radius, and correlation value at infinite limits were estimated to obtain the auto, cross, and triple correlation functions amplitudes, and intensities for single, double, and triple color images, in various combinations.
3. Materials
C2C12 and A549 cells were obtained from American Type Culture Collection™. Dulbecco's modified eagle medium (DMEM), fetal bovine serum (FBS), and 0.25% Trypsin-EDTA were obtained from Invitrogen Life Technologies™.
PFA was obtained from EMD®.
Primary antibodies specific for Rab5 and LAMP-1 were obtained from Abcam®. Primary antibodies specific for Rab7 were obtained from Cell Signaling Technology®. All primary antibodies used were monoclonal. Secondary antibodies for Rab7, and LAMP-1 were obtained from Cell Signaling Technology®. Secondary antibody specific for Rab5 was obtained from Abcam®.
4. Theory
Image correlation spectroscopy (ICS) is a quantitative method used to analyze the fluorescent intensity fluctuations in an image obtained from a laser scanning confocal microscope. This analysis allows one to retrieve: the average intensity of markers, the number of marked endosomes, the number of marked endosomes per square micron, the relative degree of aggregation of markers per endosome, and the extent of co-localization between two or three markers of interest on an endosome.
4.1. Image correlation spectroscopy (ICS) theory
Image Correlation Spectroscopy (ICS) is an intensity fluctuation analysis of images obtained using a laser scanning confocal microscope in which relative fluctuations in intensity, , are used to calculate a normalized auto correlation function, where and are lag distances in the images [42].
In the limit as α and β approach zero, the amplitude of the normalized auto correlation function, denoted by , becomes the variance of the intensity fluctuations [43];
| (1) |
Whenever the intensity is proportional to the concentration of fluorescent probes (here fluorescent markers), the variance can be shown to be the inverse of the average number of fluorescent markers or clusters of fluorescent markers – here the marked endosomes, - in the observation region;
| (2) |
The cross-sectional area of the observation volume is given by , where ω is the beam radius. Thus, we define the average number of marked endosomes per square micron, the Endosome Density (ED), as:
| (3) |
The average intensity is proportional to the number of fluorescent markers, , where is a proportionality constant reflecting spectroscopic and optical parameters such as emission collection efficiency, quantum yields, and molar adsorption coefficients [42]. We define the degree of aggregation, as:
| (4) |
This is an estimate of the average number of fluorescent markers per endosome.
4.1.1. Image cross correlation spectroscopy (ICCS) theory
Image cross correlation spectroscopy (ICCS) is an extension of ICS that enables the analysis of images collected from two markers labelled with two different colors, say green (g) and red (r) [44].
Using ICCS we calculate the auto correlation amplitudes, and, and the cross-correlation amplitude. The average number of endosomes that contain both green and red markers, can be estimated from the cross correlation and individual auto correlation amplitudes:
| (5) |
The extent of co-localization can be represented as fractions of endosomes with one marker that also contain the other:
| (6) |
| (7) |
Eq. (6) defines the fraction of endosomes containing green markers that also have red markers. Correspondingly, Eq. (7) defines the fraction of endosomes containing red markers that also have green markers.
4.1.2. Triple image cross correlation spectroscopy (TRICCS)
Triple Image Cross Correlation Spectroscopy (TRICCS) is a further extension of ICCS that enables the analysis of images of three colored markers to estimate the extent of co-localization of all three species, green (g), red (r), and blue (b).
The average number of endosomes that contain green, red, and blue markers, can be estimated from the triple cross correlation amplitude and the individual auto correlation amplitudes [36]:
| (8) |
The fraction of endosomes containing one marker that contain both of the other markers are:
| (9) |
| (10) |
| (11) |
As an example, Eq. (9) defines the fraction of endosomes that contain green markers that also contain both red and blue markers.
5. Results
5.1. Distribution of markers and endosome size is cell type specific
We examined endosome markers on singly labeled cells to understand their distribution using imaging and ICS analysis. We observed the average intensity and the amplitude of the correlation functions were both orders of magnitude greater than for the control samples for which either the primary or the secondary antibody was omitted in the labelling steps. Thus, the measurement of the intensity of markers in the observation area is specific and well above background intensity.
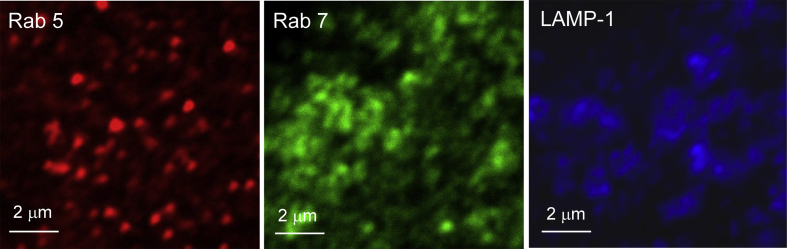
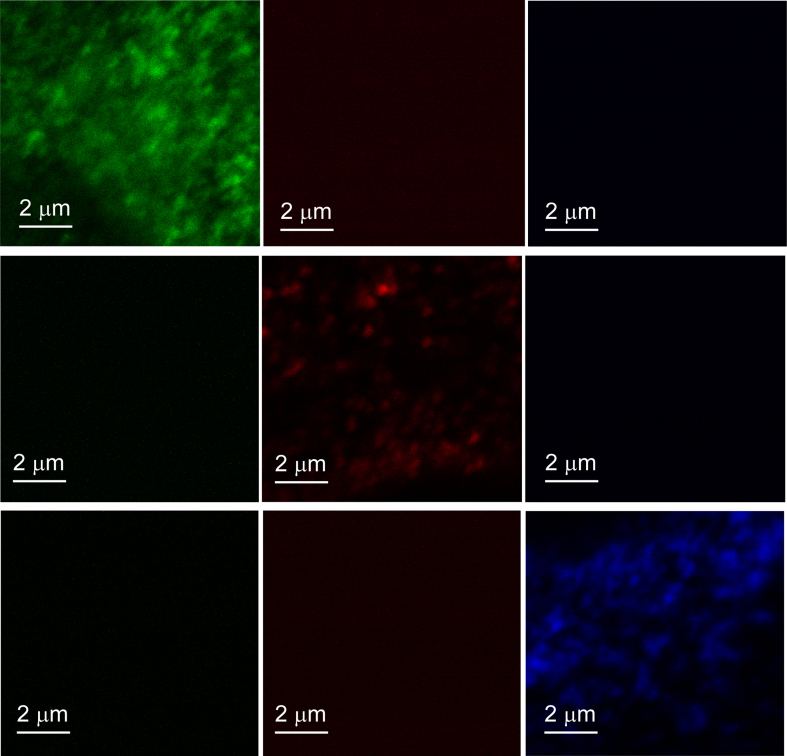
Fig. 1 shows contrast enhanced fluorescence microscopy images of three different C2C12 cells. Each cell was labelled individually with antibodies for Rab5, Rab7, or LAMP-1. Approximately 40 images (from 40 different cells) were obtained for each of the three single label experiments.
Fig. 1.
Rab5, Rab7, and LAMP-1 Marked Endosomes in C2C12 cells.
The images Fig. 1 show that the bright regions of Rab7 and LAMP-1 appear larger than those of Rab5. It is important to note that the quality of the images is determined by the resolution of the microscope and by the contrast of the objects. These objects are small and do not appear to have sharp edges. Therefore, they may appear blurry but that is the nature of the objects. In addition, these images are obtained at a high resolution so most of the objects are on the micron scale. Finally, the images are oversampled so that they can be used for image correlation analysis which makes them appear blurry.
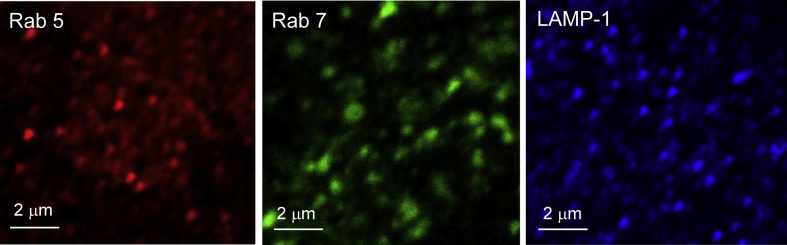
Fig. 2 shows contrast enhanced fluorescence microscopy images of three different A549 cells. Each cell was labelled individually with antibodies for Rab5, Rab7, and LAMP-1. Approximately 30 images were obtained for each of the three single label experiments.
Fig. 2.
Rab5, Rab7, and LAMP-1 Marked Endosomes in A549 cells.
The images shown in Fig. 2 show that each marked endosome appears to be similarly distributed over the entire image.
Table 1 shows the averages of the fitted laser beam width, ω ( and average number of clusters (endosomes), per unit area for C2C12 and A549 cells. These parameters were obtained by ICS analysis and averaged over the number of images indicated for each.
Table 1.
Summary of averaged ICS parameters for cells labelled for marked endosomes in C2C12 and A549 cells.
| Marker | C2C12 |
A549 |
||||
|---|---|---|---|---|---|---|
| Rab-5 | 38 | 0.32 ± 0.011 | 5.7 ± 0.4 | 31 | 0.31 ± 0.01 | 4.8 ± 0.6 |
| Rab-7 | 37 | 0.48 ± 0.06 | 4.2 ± 0.4 | 26 | 0.43 ± 0.06 | 1.9 ± 0.2 |
| LAMP-1-488 | 35 | 0.47 ± 0.01 | 2.2 ± 0.1 | 30 | 0.28 ± 0.01 | 4.9 ± 0.4 |
| LAMP-1-561 | 41 | 0.50 ± 0.01 | 2.9 ± 0.2 | 32 | 0.36 ± 0.01 | 2.2 ± 0.2 |
| LAMP-1-633 | 41 | 0.51 ± 0.01 | 2.6 ± 0.1 | 26 | 0.33 ± 0.01 | 11 ± 1 |
Standard error.
The average number of endosomes per unit area ranges from 2-6, consistent with a highly clustered pattern of fluorescence. There does not appear to be a consistent pattern of densities between the different markers or the two cell types.
The average width of the correlation function is slightly larger for the Rab7 and LAMP-1 in C2C12 cells but is smaller and more variable in A549 cells. Theoretically, we expect the laser beam width to range from 0.17 to 0.22 m for the 488, 561, and 633 nm lasers with the optics used in these experiments. In all cases, the beam widths presented in Table 1, which refer to the widths of the correlation functions, are larger than the expected beam widths. This suggests that there may be a convolution effect between finite sized endosomes and the laser beam. We can model this convolution (see supplemental) and use it to estimate the actual endosome sizes as shown in Table 2.
Table 2.
Measured beam width and object radius in C2C12 and A549 Cells.
| Marker | C2C12 |
A549 |
||
|---|---|---|---|---|
| Rab-5 | 0.32 | 0.34 | 0.31 | 0.31 |
| Rab-7 | 0.48 | 0.62 | 0.43 | 0.52 |
| LAMP-1 | 0.49 | 0.64 | 0.33 | 0.42 |
These values compare well with other work, in which the endocytic vesicles have been determined to range in size from 0.2-0.6 m using confocal and electron microscopy [45, 46].
Because the intensity of fluorescence emitted from a sample labelled with only secondary antibodies should correspond to non-specific binding of individual secondary antibodies, we measured the number of individual antibodies per cluster (see supplemental) using the intensities and amplitudes of the autocorrelation functions corresponding to samples labeled with only secondary antibodies [47, 48]. We estimated that there are approximately 1000 Rab5 and Rab7 antibodies per endosome in C2C12 cells and approximately 500 Rab5 and Rab7 antibodies per endosome in A549 cells. We did not get reliable estimates for the number of LAMP-1 antibodies because the non-specific binding of that secondary antibody was too low.
5.2. Endosomal markers are not unique to one compartment
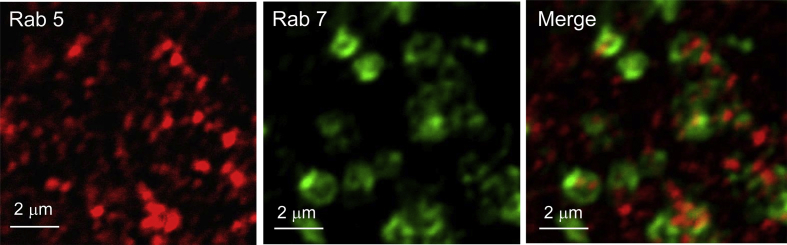
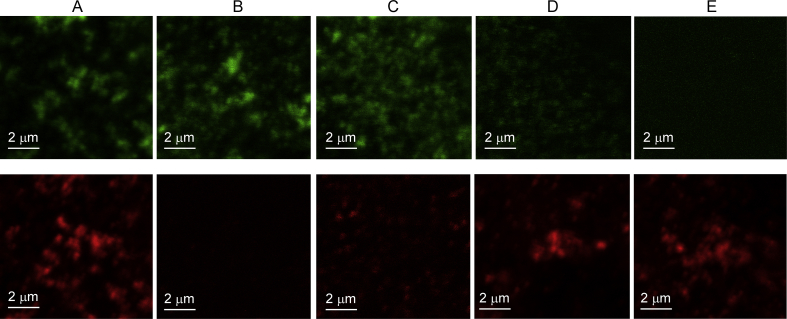
Fig. 3 shows contrast enhanced images of antibodies to Rab5 (Rab5-561, in red) and antibodies to Rab7 (Rab7-633, in green) in the same cell. The Merge image is an overlay of the Rab5-561 and Rab7-633 contrast enhanced images.
Fig. 3.
Rab5-561 & Rab7-633 marked endosomes in C2C12 cells.
In Fig. 3, several yellow and orange regions are observed in the Merge image, implying that there is some degree of co-localization of Rab5-561 and Rab7-633 markers.
Table 3 shows the average number of clusters containing Rab5, Rab7, and both, as well as the fractions as defined by Eqs. (6) and (7) obtained by ICCS analysis from measurements on 30–40 cells.
Table 3.
Average number of endosomes containing markers and fractions of Co-Localization of Rab5 & Rab7 markers in C2C12 cells.
| Marker | |||
|---|---|---|---|
| Rab5-561 | 5.4 ± 0.51 | ||
| Rab7-633 | 5.5 ± 0.9 | ||
| Rab5-561 & Rab7-633 | 2.2 ± 0.3 | 0.46 ± 0.05 | 0.53 ± 0.06 |
Standard error.
The average number of endosomes containing Rab5 or Rab7 are observed to be just over twice the number of endosomes that contain both Rab5 and Rab7 markers, suggesting that there are a significant number of endosomes that contain both markers. The average fractions of co-localization represented in Table 3 are calculated as an average of 30–40 individual fractions (as compared to a fraction of the averages) and reflect that on average approximately 46% of the endosomes that contain Rab5-561 markers also contain Rab7-633 markers and approximately 53% of the endosomes containing Rab7-633 markers also contain Rab5-561 markers. The observation that about half of the endosomes contain both markers show that these markers are not unique to a specific endosome type since they can coexist on the same endosomes. While this was suggested by the images, the more quantitative ICCS approaches gives better insight into the extent of co-existence.
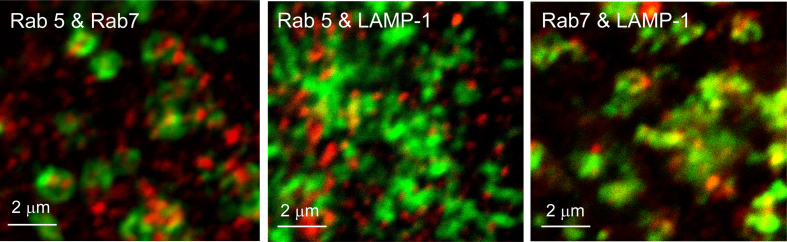
Fig. 4 shows examples of three merged images of C2C12 cells doubly labelled for Rab5-561 and Rab7-633; Rab5-561 and LAMP-1-633; and Rab7-633 and LAMP-1-561 markers.
Fig. 4.
Images of pairwise interaction of endocytic markers in C2C12 cells.
The images in Fig. 4 show various degrees of yellow and orange indicating some degree of co-localization of all pairs of markers. For example, there are a significant number of yellow regions in the image corresponding to Rab7 and LAMP-1.
Table 4 summarizes the calculated average values of fractions of co-localization for each pairwise interaction studied in C2C12 cells (including the data from Table 3).
Table 4.
Summary of fractions of co-localization in C2C12 cells.
| Rab5 | Rab7 | LAMP-1 | |
|---|---|---|---|
| Rab5 | – | 0.46 ± 0.051 | 0.32 ± 0.05 |
| Rab7 | 0.53 ± 0.06 | – | 0.74 ± 0.04 |
| LAMP-1 | 0.31 ± 0.04 | 0.86 ± 0.03 | – |
Standard error.
The results presented in Table 4 show that there is some co-localization of all the endosome and lysosome markers, but to different extents. Of those endosomes containing Rab5, 46% also contain Rab7, and 32% also contain LAMP-1. Of those endosomes containing Rab7, 53% also contain Rab5 and 74% also contain LAMP-1. Of those lysosomes that contain LAMP-1, 31% also contain Rab5 and 86% also contain Rab7.
The observation that Rab5 and Rab7 markers co-localize is consistent with the early endosomes maturing into the late endosomes; it may take some time before the Rab5 is cleared from the endosomes. Those endosomes that contain Rab5 but not Rab7 may then be early endosomes prior to maturation while those that contain Rab7 but not Rab5 may be even later stage endosomes from which the Rab5 has been cleared.
The observation that Rab7 and LAMP-1 markers co-localize to between 74% and 86% is consistent with the process of fusion of the late endosome into a lysosome and that the process to clear all the Rab7 from the lysosome is very slow.
The observation that Rab5 and LAMP-1 markers co-localize to a significant extent is surprising and suggests that the Rab5 is not cleared completely from the late endosome before it fuses with the lysosome. This observation coupled with the nearly complete co-localization of Rab7 and LAMP-1, further suggests that there must be endosomes which contain all three markers simultaneously. To test this, we performed triple labeling experiments and analyzed these using triple cross-correlation spectroscopy (vide infra).
Table 5 summarizes the calculated averaged fractions of co-localization for each pairwise interaction in A549 cells.
Table 5.
Summary of fractions of co-localization in A549 cells.
| Rab5 | Rab7 | LAMP-1 | |
|---|---|---|---|
| Rab5 | – | 0.50 ± 0.031 | 0.82 ± 0.04 |
| Rab7 | 0.39 ± 0.03 | – | 0.26 ± 0.06 |
| LAMP-1 | 0.80 ± 0.05 | 0.37 ± 0.06 | – |
Standard error.
Table 5 shows once again that there is a significant co-localization of all the endosome and lysosome markers, but the extent of co-localization is different in the A549 cells when compared to C2C12 cells. In the A549 cells, the co-localization between Rab5 and LAMP-1 is much higher (about 80%) and the co-localization between Rab7 and LAMP-1 is much smaller (about 25–35%). This suggests that in these cells some of the early endosomes that contain only Rab5 may fuse directly with the lysosomes prior to the maturation process that form the late endosomes. This is different from the observations for C2C12 cells which may reflect the difference in the way these cell types process internalized materials. We note that the A549 cell is derived from pulmonary type II cells which have the function of recycling lipids from the pulmonary air-water interface and forming lamellar bodies that are exported [49, 50]. Therefore, there is an important recycling function in addition to a degradation function in these cells. This could mean that more of the late endosomes are converted to recycling endosomes rather than fusing with lysosomes in the A549 cells.
Fig. 5 shows contrast enhanced images of C2C12 and A549 cells each labelled with antibodies against Rab5-561 (red), Rab7-633 (blue), and LAMP-1-488 (green).
Fig. 5.
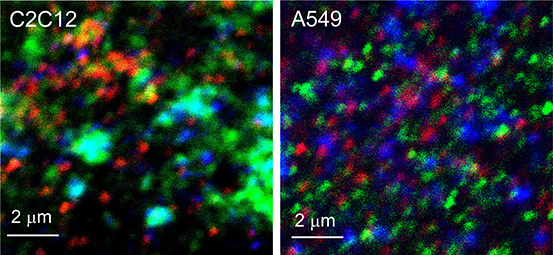
Rab5, Rab7, and LAMP-1 marked endosomes in C2C12 & A549 cells.
It is evident from the appearance of yellow-orange regions and turquoise regions that there is some co-localization of these markers. Perfect co-localization of all three markers should lead to white regions, which are hard to observe in either image.
Table 6 summarizes the fractions of three-way co-localization as calculated from Eqs. (9), (10), and (11) using TRICCS.
Table 6.
Extent of association of one marker with two markers of endosomes in C2C12 and A549 cells.
| C2C12 | A549 | |
|---|---|---|
| 0.56 0.061 | 0.16 0.05 | |
| 0.42 0.06 | 0.11 0.03 | |
| 0.28 0.05 | 0.08 0.03 |
Standard error.
These data show that the extent of three-way co-localization is much greater in C2C12 cells than in A549 cells. Among those endosomes containing Rab5, about 56% also contain both Rab7 and LAMP-1 in C2C12 cells compared to about 16% in A549 cells. Likewise, among those endosomes containing Rab7, about 42% also contain both Rab5 and LAMP-1 compared to about 11% in A549 cells. Finally, among those lysosomes that contain LAMP-1, about 18% also contain both Rab5 and Rab7 compared to about 8% in A549 cells. It should be noted that the extent of three-way co-localization observed in the A549 cells is close to the limit of reliable detection and there may not be much, if any, co-localization when the number is less than about 10%. Nevertheless, these experiments confirm that in C2C12 cells there is a significant number of endosomes or lysosomes that contain all three markers, while in A549 cells there are only a few that may contain all three markers. This is consistent with the notion that in A549 cells many of the late endosomes are diverted to recycling endosomes rather than lysosomes and that the early lysosomes can fuse directly with the lysosomes.
6. Discussion
The quantitative analysis of the distribution and extent of clustering of the antibodies against the intracellular proteins Rab5, Rab7, and LAMP-1 reveal that they are highly clustered (on the order of 500–1000 antibodies per cluster), that these clusters are large (0.2–0.6m), and that they are distributed evenly intracellularly. These observations are consistent with each of these proteins being associated with organelles such as the endosomes and the lysosomes [51, 52, 53].
It is clear from the data presented here that the markers purported to represent early endosomes (Rab5), late endosomes (Rab7) and lysosomes (LAMP-1) are not as specific as previously reported with at least 30% and up to 80% co-localization of some of these markers on the same endosomes or lysosomes. This is not surprising if the maturation model is correct, since it is reasonable to expect that during a maturation process where Rab5 proteins are being cleared and Rab7 proteins are emerging, there will be a period when both proteins can co-exist in the same endosomes [54]. Likewise, if there is fusion of late endosomes with lysosomes, there may well be a period before the Rab7 proteins are fully cleared from the lysosomes after the fusion event. Out findings are therefore consistent with a maturation-fusion process.
With live cell imaging, Rink et al and Poteryaev et al. both successfully showed that Rab5 is replaced with Rab7 during endocytosis [26, 27]. In both works, images of cells were co-expressed with Rab5 and Rab7 fluorescent proteins and were imaged over time. In both studies, an increase of Rab7 fluorescent signal with a simultaneous decrease of Rab5 fluorescent signal was observed. Thus, these findings suggest a substitution of Rab5 by Rab7, but also show evidence for a point in time in which there is some degree of co-localization of the two markers on some endosomes. The work presented in this manuscript supports the notion that Rab5 is replaced with Rab7, and has quantified the extent of the coexistence of the two proteins. Our findings suggest that these markers are not as unique to a single compartment as commonly thought.
The observation that a significant amount of Rab5 is co-localized with LAMP-1 suggests that the maturation and fusion processes are less linear than the model might imply. In other words, it is possible that fusion of endosomes with lysosomes can occur well before the maturation process from early to late endosomes is complete. This is even more pronounced in the A549 cells, where there appears to be more lysosomes that contain Rab5 than contain Rab7, suggesting that the fusion process can precede the maturation process in these cells and supporting the notion that the late endosomes are targeted more to become recycling endosomes.
That the complete maturation from early to late endosomes is not a prerequisite for fusion is further supported by the evidence that all three proteins can co-exist to a significant extent, at least in the C2C12 cells. The smaller extent of three-way co-localization in A459 cells coupled with the higher degree of co-localization of Rab5 and LAMP-1 than Rab7 and LAMP-1 may indicate that there is a higher probability for early endosomes to fuse with lysosomes in these cells. Whether this is related to their function as cells that recycle liposome materials in the lung remains to be investigated [55, 56].
The application of triple cross correlation spectroscopy allowed us to study the extent of Rab5, Rab7, and LAMP-1 in three-way fashion, which to the best of our knowledge has not previously been attempted. As a result, we learned Rab5, Rab7, and LAMP-1 can co-exist in the same intracellular organelles and that the extent to which they associate can vary significantly between different cell types.
Since the three proteins (Rab5, Rab7, and LAMP-1) can co-exist in the same intracellular organelles, it may be prudent to reevaluate their use as specific markers for specific endosomal or lysosomal entities. Moreover, if completion of the maturation process from early to late endosomes is not required for their fusion with the lysosomes, the linearity of the maturation-fusion model is in question.
We note that in this study we are not directly studying the dynamics of the endocytosis process but rather focus on the steady state distribution of the classical markers for components of the endocytosis pathway. Our conclusions are therefore limited to an understanding of their distribution in normally growing cells that have been fixed at an arbitrary point of time in the cell cycle.
Declarations
Author contribution statement
Lindsay Shearer: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Nils Petersen: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was supported by a discovery grant from the Natural Sciences and Engineering Research Council of Canada to NOP.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We would like to thank Dr. Xuejun Sun and Geraldine Baron from the Department of Experimental Oncology in the Cross Cancer Institute for assistance with image correlation spectroscopy. We would like to thank Angela Brigley and Cathy DeGuzman from the National Institute for Nanotechnology for providing use of facilities for immunofluorescence. For analysis with this image correlation spectroscopy software, images were converted with the help from Dr. Nick Smisdom, from.lsm to.bmp.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Figure S3.
Figure S4.
References
- 1.Kaksonen M., Roux A. Mechanisms of clathrin-mediated endocytosis. Nat. Rev. Mol. Cell Biol. 2018;19:313–326. doi: 10.1038/nrm.2017.132. [DOI] [PubMed] [Google Scholar]
- 2.Ng C.T. Clathrin-mediated endocytosis of gold nanoparticles in vitro. Anat. Rec. 2015;298:418–427. doi: 10.1002/ar.23051. [DOI] [PubMed] [Google Scholar]
- 3.Lee H.-J. Assembly-dependent endocytosis and clearance of extracellular alpha-synuclein. Int. J. Biochem. Cell Biol. 2008;40:1835–1849. doi: 10.1016/j.biocel.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 4.Chithrani B.D., Ghazani A.A., Chan W.C.W. Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 5.Hillaireau H., Couvreur P. Nanocarriers’ entry into the cell: relevance to drug delivery. Cell. Mol. Life Sci. 2009;66:2873–2896. doi: 10.1007/s00018-009-0053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mettlen M., Chen P.-H., Srinivasan S., Danuser G., Schmid S.L. Regulation of clathrin-mediated endocytosis. Annu. Rev. Biochem. 2018;87:871–896. doi: 10.1146/annurev-biochem-062917-012644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutta D., Donaldson J.G. Rab and Arf G proteins in endosomal trafficking. Methods Cell Biol. 2015;130:127–138. doi: 10.1016/bs.mcb.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gautreau A., Oguievetskaia K., Ungermann C. Function and regulation of the endosomal fusion and fission machineries. Cold Spring Harb. Perspect. Biol. 2014;6 doi: 10.1101/cshperspect.a016832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landes Bioscience/Eurekah.Com. Springer Science+Business Media; 2006. Endosomes. [Google Scholar]
- 10.Helenius A., Mellman I., Wall D., Hubbard A. Endosomes. Trends Biochem. Sci. 1983;8:245–250. [Google Scholar]
- 11.Huotari J., Helenius A. Endosome maturation. EMBO J. 2011;30:3481–3500. doi: 10.1038/emboj.2011.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pryor P.R., Luzio J.P. Delivery of endocytosed membrane proteins to the lysosome. Biochim. Biophys. Acta Mol. Cell Res. 2009;1793:615–624. doi: 10.1016/j.bbamcr.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 13.Le Roy C., Wrana J.L. Clathrin- and non-clathrin-mediated endocytic regulation of cell signalling. Nat. Rev. Mol. Cell Biol. 2005;6:112–126. doi: 10.1038/nrm1571. [DOI] [PubMed] [Google Scholar]
- 14.Liewen H. Characterization of the human GARP (Golgi associated retrograde protein) complex. Exp. Cell Res. 2005;306:24–34. doi: 10.1016/j.yexcr.2005.01.022. [DOI] [PubMed] [Google Scholar]
- 15.Podinovskaia M., Spang A. The endosomal network: mediators and regulators of endosome maturation. In: Lamaze C., Prior I., editors. Endocytosis and Signaling. Springer International Publishing; 2018. pp. 1–38. [DOI] [PubMed] [Google Scholar]
- 16.Pfeffer S.R. Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 2013;25:414–419. doi: 10.1016/j.ceb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenmark H., Olkkonen V.M. The Rab GTPase family. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-5-reviews3007. reviews3007.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gorvel J.P., Chavrier P., Zerial M., Gruenberg J. rab5 controls early endosome fusion in vitro. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 19.Hutagalung A.H., Novick P.J. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bucci C. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 21.Barbieri M.A. Evidence for a symmetrical requirement for rab5-GTP in in vitro endosome-endosome fusion. J. Biol. Chem. 1998;273:25850–25855. doi: 10.1074/jbc.273.40.25850. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen E., Severin F., Backer J.M., Hyman A.A., Zerial M. Rab5 regulates motility of early endosomes on microtubules. Nat. Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- 23.Goto-Silva L. Retrograde transport of Akt by a neuronal Rab5-APPL1 endosome. Sci. Rep. 2019;9:1–14. doi: 10.1038/s41598-019-38637-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epp N., Rethmeier R., Krämer L., Ungermann C. Membrane dynamics and fusion at late endosomes and vacuoles--Rab regulation, multisubunit tethering complexes and SNAREs. Eur. J. Cell Biol. 2011;90:779–785. doi: 10.1016/j.ejcb.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 25.Wang T., Ming Z., Xiaochun W., Hong W. Rab7: role of its protein interaction cascades in endo-lysosomal traffic. Cell. Signal. 2011;23:516–521. doi: 10.1016/j.cellsig.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Rink J., Ghigo E., Kalaidzidis Y., Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 27.Poteryaev D., Datta S., Ackema K., Zerial M., Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141:497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Cioni J.-M. Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell. 2019;176:56–72. doi: 10.1016/j.cell.2018.11.030. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bucci C., Thomsen P., Nicoziani P., McCarthy J., van Deurs B. Rab7: a key to lysosome biogenesis. Mol. Biol. Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jäger S. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 31.Eskelinen E.-L. Roles of LAMP-1 and LAMP-2 in lysosome biogenesis and autophagy. Mol. Asp. Med. 2006;27:495–502. doi: 10.1016/j.mam.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Kornfeld S., Mellman I. The biogenesis of lysosomes. Annu. Rev. Cell Biol. 1989;5:483–525. doi: 10.1146/annurev.cb.05.110189.002411. [DOI] [PubMed] [Google Scholar]
- 33.Wartosch L., Bright N.A., Luzio J.P. Lysosomes. Curr. Biol. 2015;25:R315–R316. doi: 10.1016/j.cub.2015.02.027. [DOI] [PubMed] [Google Scholar]
- 34.Ikari A. Clathrin-dependent endocytosis of claudin-2 by DFYSP peptide causes lysosomal damage in lung adenocarcinoma A549 cells. Biochim. Biophys. Acta. 2015;1848:2326–2336. doi: 10.1016/j.bbamem.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Studzian M., Bartosz G., Pulaski L. Endocytosis of ABCG2 drug transporter caused by binding of 5D3 antibody: trafficking mechanisms and intracellular fate. Biochim. Biophys. Acta. 2015;1853:1759–1771. doi: 10.1016/j.bbamcr.2015.04.011. [DOI] [PubMed] [Google Scholar]
- 36.Kasmapour B., Cai L., Gutierrez M.G. Spatial distribution of phagolysosomes is independent of the regulation of lysosome position by Rab34. Int. J. Biochem. Cell Biol. 2013;45:2057–2065. doi: 10.1016/j.biocel.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Rappoport J.Z., Simon S.M. Endocytic trafficking of activated EGFR is AP-2 dependent and occurs through preformed clathrin spots. J. Cell Sci. 2009;122:1301–1305. doi: 10.1242/jcs.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jouvenet N., Zhadina M., Bieniasz P.D., Simon S.M. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol. 2011;13:394–401. doi: 10.1038/ncb2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rappoport J.Z., Kemal S., Benmerah A., Simon S.M. Dynamics of clathrin and adaptor proteins during endocytosis. Am. J. Physiol. Cell Physiol. 2006;291:C1072–1081. doi: 10.1152/ajpcell.00160.2006. [DOI] [PubMed] [Google Scholar]
- 40.Rappoport J.Z., Heyman K.P., Kemal S., Simon S.M. Dynamics of dynamin during clathrin mediated endocytosis in PC12 cells. PLoS One. 2008;3:e2416. doi: 10.1371/journal.pone.0002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshida A. Morphological changes of plasma membrane and protein assembly during clathrin-mediated endocytosis. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2004786. e2004786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petersen N.O. Analysis of membrane protein cluster densities and sizes in situ by image correlation spectroscopy. Faraday Discuss. 1998:289–305. doi: 10.1039/a806677i. discussion 331-343. [DOI] [PubMed] [Google Scholar]
- 43.Kolin D.L., Wiseman P.W. Advances in image correlation spectroscopy: measuring number densities, aggregation states, and dynamics of fluorescently labeled macromolecules in cells. Cell Biochem. Biophys. 2007;49:141–164. doi: 10.1007/s12013-007-9000-5. [DOI] [PubMed] [Google Scholar]
- 44.Wang M., Petersen N.O. Lipid-coated gold nanoparticles promote lamellar body formation in A549 cells. Biochim. Biophys. Acta. 2013;1831:1089–1097. doi: 10.1016/j.bbalip.2013.01.018. [DOI] [PubMed] [Google Scholar]
- 45.Zhang W. Influence of cell physiological state on gene delivery to T lymphocytes by chimeric adenovirus Ad5F35. Sci. Rep. 2016;6:22688. doi: 10.1038/srep22688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vermeulen L.M.P. Endosomal size and membrane leakiness influence proton sponge-based rupture of endosomal vesicles. ACS Nano. 2018;12:2332–2345. doi: 10.1021/acsnano.7b07583. [DOI] [PubMed] [Google Scholar]
- 47.Abney J.R., Scalettar B.A., Hackenbrock C.R. On the measurement of particle number and mobility in nonideal solutions by fluorescence correlation spectroscopy. Biophys. J. 1990;58:261–265. doi: 10.1016/S0006-3495(90)82371-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.St-Pierre P.R., Petersen N.O. Average density and size of microclusters of epidermal growth factor receptors on A431 cells. Biochemistry. 1992;31:2459–2463. doi: 10.1021/bi00124a004. [DOI] [PubMed] [Google Scholar]
- 49.Foster K.A., Oster C.G., Mayer M.M., Avery M.L., Audus K.L. Characterization of the A549 cell line as a type II pulmonary epithelial cell model for drug metabolism. Exp. Cell Res. 1998;243:359–366. doi: 10.1006/excr.1998.4172. [DOI] [PubMed] [Google Scholar]
- 50.Yu S.-H., Possmayer F. Lipid compositional analysis of pulmonary surfactant monolayers and monolayer-associated reservoirs. J. Lipid Res. 2003;44:621–629. doi: 10.1194/jlr.M200380-JLR200. [DOI] [PubMed] [Google Scholar]
- 51.Cheng X.-T. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J. Cell Biol. 2018;217:3127–3139. doi: 10.1083/jcb.201711083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stroupe C. This is the end: regulation of Rab7 nucleotide binding in endolysosomal trafficking and autophagy. Front. Cell Dev. Biol. 2018;6 doi: 10.3389/fcell.2018.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimamura H., Nagano M., Nakajima K., Toshima J.Y., Toshima J. Rab5-independent activation and function of yeast Rab7-like protein, Ypt7p, in the AP-3 pathway. PLoS One. 2019;14 doi: 10.1371/journal.pone.0210223. e0210223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kiral F.R., Kohrs F.E., Jin E.J., Hiesinger P.R. Rab GTPases and membrane trafficking in neurodegeneration. Curr. Biol. 2018;28:R471–R486. doi: 10.1016/j.cub.2018.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Balis J.U., Bumgarner S.D., Paciga J.E., Paterson J.F., Shelley S.A. Synthesis of lung surfactant-associated glycoproteins by A549 cells: description of an in vitro model for human type II cell dysfunction. Exp. Lung Res. 1984;6:197–213. doi: 10.3109/01902148409109248. [DOI] [PubMed] [Google Scholar]
- 56.Shapiro D.L., Nardone L.L., Rooney S.A., Motoyama E.K., Munoz J.L. Phospholipid biosynthesis and secretion by a cell line (A549) which resembles type II alveolar epithelial cells. Biochim. Biophys. Acta Lipids Lipid Metab. 1978;530:197–207. doi: 10.1016/0005-2760(78)90005-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.