Abstract
Pancreatic cancer is an aggressive malignancy that often goes undiagnosed in the early stages. Non-invasive, early, and accurate diagnosis is therefore undoubtedly the “holy grail” of pancreatic cancer research. However, despite extensive research efforts, there is no definitive biomarker for this cancer. Previously, we identified alkaline phosphatase placental-like 2 (ALPPL2) as a diagnostic biomarker for pancreatic ductal adenocarcinoma and developed a 2′-fluoro modified RNA aptamer toward it. In this study, we show that ALPPL2 is present in pancreatic cancer extracellular vesicles (EVs) and therefore has potential application in liquid biopsy-based diagnostic strategies. We also developed ALPPL2 direct and sandwich aptamer-linked immobilized sorbent assay (ALISA) for EVs, which could sensitively and specifically detect the protein. We believe that our ALISA format may have a potential diagnostic utility in screening pancreatic-cancer-derived EVs.
Keywords: aptamer, pancreatic cancer, ALPPL2, extracellular vesicles, exosomes, ALISA
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is a very aggressive and lethal cancer. The high mortality rate associated with this cancer is mostly a consequence of the asymptomatic nature of the disease, resulting in diagnosis often at a later stage when the tumor can no longer be surgically removed.1, 2 In addition, despite extensive research efforts and many potential candidates, no definitive biomarkers have emerged that can demonstrate real clinical utility in the timely diagnosis of PDAC. Given the situation, currently, accurate and early pancreatic cancer diagnosis mostly relies on invasive imaging methods like endoscopic retrograde cholangiopancreatography or ultrasound-guided fine-needle aspiration.2, 3 However, being too invasive and costly, they lack patients’ compliance and hence cannot be used for widespread screening of pancreatic cancer.4 Therefore, it is imperative to develop new and improved non-invasive strategies to detect the early stages of pancreatic cancer with higher diagnostic sensitivity.5
Recent studies have shown that extracellular vesicles (EVs) derived from body fluids may serve as a promising alternative for disease diagnosis.6 EVs basically represent membrane-encapsulated cell-derived cargos ranging from 50 to 5,000 nm in diameter.7 Of these, the small-size EVs comprise mostly of exosomes and microvesicles (MVs), the former originating from the multi-vesicular endosome and the latter from the plasma membrane. They are finally released from the cells as circulating vesicles. Their distinctive biogenesis makes it possible for EVs to include the cell-type-specific biomarkers such as proteins, mRNA, microRNA, or non-coding RNAs, serving as a “fingerprint” of the original cells.8 Studies have shown that EVs are secreted from cancer cells at a higher rate than from healthy cells and are important in facilitating cancer progression and metastasis.9 In addition, their stability and abundance in a wide range of biological fluids6 makes EVs a viable substitute to non-invasive biopsy of the tumor mass. Recent studies on exosome-based diagnosis of pancreatic cancer clearly suggest that exosome transcriptomic and proteomic biomarkers can tremendously increase the likelihood of early and sensitive detections of cancer.10, 11 In particular, glypican 1 (GPC)12, 13 and macrophage migration inhibitory factor (MIF)14 were identified as potential exosome-associated biomarkers that could detect pancreatic cancer long before the early lesions could be seen. In addition, pancreatic cancer stem cell (CSC)-associated proteins have been shown to be present in exosomes15, 16 and can be used for detecting pancreatic-cancer-initiating cells.
We previously reported that alkaline phosphatase placental-like 2 (ALPPL2), a glycosyl phosphatidyl inositol (GPI)-anchored membrane protein,17 could be a potential biomarker for early diagnosis of PDAC.18 We also developed 2′-fluoro RNA aptamer (SQ2) by Cell-SELEX,18 which binds to the membrane-bound and secreted forms of ALPPL2. As GPI-anchored proteins are highly enriched in glycolipid domains of the cell membrane and form a part of exosomal proteins, we investigated the presence of ALPPL2 in EVs. Herein, we show that ALPPL2 protein is present in EVs derived from pancreatic cancer cells and truly represent the quantitative level of cellular ALPPL2 protein. We also developed an SQ2 aptamer-based ELISA (ALISA) for quantitative detection of PDAC-derived EVs. SQ2 aptamer-based direct or sandwich ALISA could detect ALPPL2-positive EVs with high specificity and sensitivity. Using exosome-specific capture antibody, the sandwich ALISA could also detect ALPPL2 from human serum spiked with the cancer exosomes. We believe that our ALPPL2 ALISA for exosome-based diagnosis may be a suitable option to sensitively detect pancreatic cancer in earlier stages.
Results and Discussion
Characterization of EV Purity, Size, and Number
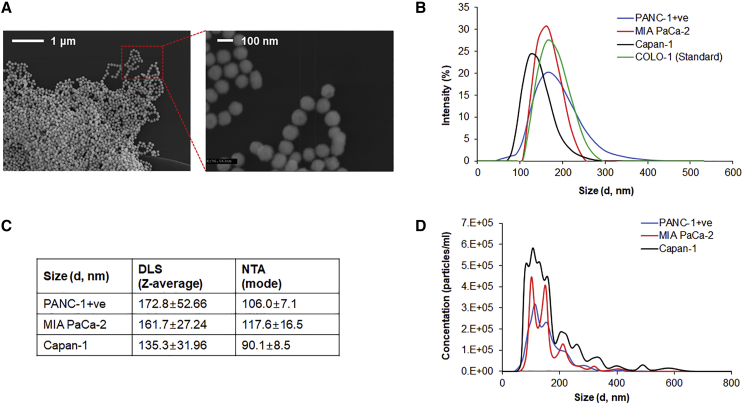
EVs were isolated from ALPPL2-positive PANC-1 (PANC-1+), MIA PaCa-2, and Capan-1 cell lines by ultracentrifugation. Scanning electron microscopy analysis of EVs showed a very homogeneous EV preparation with particle size around 100 nm in diameter (Figure 1A). Sizes were further analyzed by dynamic light scattering (DLS) and nanoparticle tracking analysis (NTA). DLS showed the particle size range of 50 to 300 nm for all the three cell lines under study, including the commercial COLO-1 exosomes (Figure 1B) with Z-average values ranging from 135 to 172 nm (Figure 1C). However, NTA revealed EVs size ranging from 90 to 106 nm (Figure 1D), which are smaller than those observed by DLS and was in agreement with the microscopic analysis. Although many studies have employed DLS for exosome size measurement, several drawbacks have been highlighted when analyzing polydispersed mixtures.19 This is mainly because larger particles scatter more light making the smaller particles relatively undetectable, skewing the distribution of data toward the size of larger particles.20 This is also evident from our DLS and NTA data comparison for the three cell types (Figure 1C). Particle size ≤ 100 nm indicates that most prepared EVs are composed of exosomes. The number of particles was calculated by NTA, and 6.37 × 107, 3.34 × 107, and 7.26 × 107 EVs/μg of protein was found to be present in PANC-1+, MIA PaCa-2, and Capan-1 secretions, respectively. Immunoblot analysis of protein markers for endoplasmic reticulum, mitochondria, and Golgi complex confirmed that the isolated EVs were devoid of any contamination from cellular material (Figure S1).
Figure 1.
Characterization of PDAC Cell-Derived Extracellular Vesicles
(A) SEM micrograph of PANC-1+-derived EVs acquired at 20k (left) and 100k (right) magnifications. (B) Size distribution of EVs by DLS assay. Commercial EV preparations from COLO-1 cells were used as a control. (C) NanoSight analysis (NTA) of EVs showing size distribution and (D) particle concentrations of cell culture supernatant (1 μg/mL) without ultracentrifugation. Data is represented as mean ± SD of three technical replicates.
ALPPL2 Is Present in Exosomes Secreted by Pancreatic Cancer Cells
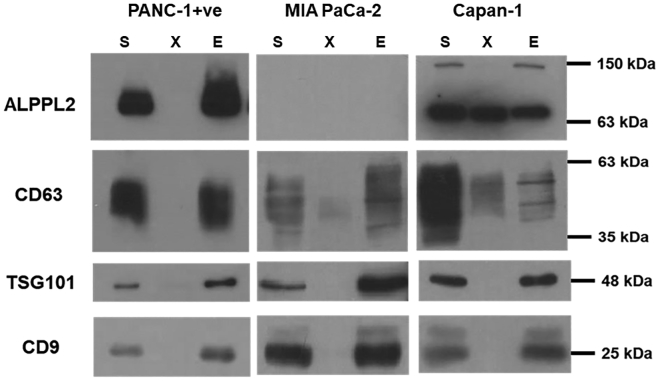
In our earlier studies, we showed the presence of ALPPL2 on cell membrane as well as in cell secretions of pancreatic cancers.18 This germ cell-associated alkaline phosphatase family of proteins are known to be present on cell membrane via GPI anchor21 and are often seen in the serum of patients suffering from germ cell cancers and pregnant females.17, 22 Until now, it was perceived that like most of the GPI-APs, the GPI anchor of these phosphatases is cleaved by enzyme phospholipase C, resulting in their presence in body fluids as cell-free proteins.23 However, recent studies have indicated that a number of GPI-anchored proteins are released from the cells in membrane-bound forms as a part of exosomes and MVs.24 These not only include prion proteins and major histocompatibility complex (MHC) molecules, but also proteins that are differentially expressed under disease conditions and are potential disease biomarkers.25 Herein, we wanted to see if the ALPPL2 protein is present in pancreatic cancer cell-derived EVs. Immunoblot analysis with EVs isolated from PANC-1+ and Capan-1 cell secretions clearly showed presence of a 63-kDa ALPPL2 protein band (Figure 2). CD9, CD63, and TSG101 as exosomal markers confirmed the presence of EVs in the sample. Complete secretome and EV-depleted secretome samples from the same lot of preparation were also analyzed simultaneously. The absence of ALPPL2 in EV-depleted secretome of PANC-1+ cells clearly showed that at least in the case of PANC-1+, the secreted form of ALPPL2 is exclusively present in EVs. On the other hand, the same was not the case in Capan-1 cells, where both free and EV-associated ALPPL2 seems to be present in equal amounts. Interestingly, the 135-kDa dimeric form of ALPPL2 was exclusively present in the Capan-1 EVs. As expected from our earlier findings on ALPPL2 expression profiles of these cell lines18 MIA PaCa-2 cell secretions did not show any detectable ALPPL2 protein levels (Figure 2). All together, these results indicate that the ALPPL2 protein profile of cell-secreted EVs resembles the cellular protein expression; hence, pancreatic-cancer-derived EVs are a reliable source for the estimation of ALPPL2 biomarker.
Figure 2.
ALPPL2 Is Present in PDAC Cell-Derived EVs
Immunoblot analysis of EVs extracted from the serum-free conditioned media of PANC-1+ and Capan-1 showed significant ALPPL2 expression, whereas MIA PaCa-2 showed no expression of the protein. 15 μg of secretome (S), 15 μg of EV-depleted secretome (X), and 2.5 μg EVs (E) were used for immunoblotting. EV markers CD9, CD63, and TSG101 confirm the presence of EVs in samples S and E.
ALPPL2 Aptamer-Based Direct and Sandwich-Type Diagnostic ELISA
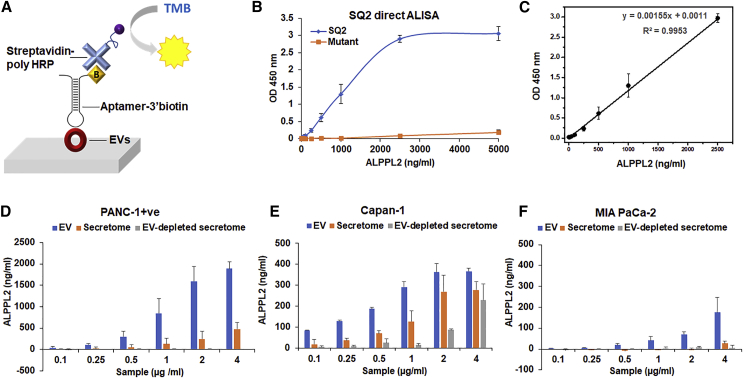
We next sought to develop an aptamer-based ELISA (ALISA) for the quantitative analysis of ALPPL2 in pancreatic cancer cell-derived EVs, using the truncated version of aptamer SQ2, earlier selected to bind to PANC-1+, Capan-1 cells, and their cell secretome. We set up a colorimetric direct ELISA where the samples were immobilized on the ELISA well plate, and the presence of ALPPL2 was detected using the biotinylated aptamer and streptavidin-poly horseradish peroxidase (HRP) (Figure 3A). SQ2 mutant was used as a control for non-specific oligonucleotide interactions. Linearity, sensitivity, and specificity of the assay were also determined using the full-length ALPPL2 recombinant protein, expressed in E. coli and HEK293T cells. Protein was tested at various concentrations ranging from 1 ng to 2.5 μg. No signal was seen with the protein expressed in E. coli (data not shown), while the protein expressed in HEK293T cells showed signals with linearity from 10 to 2,500 ng/mL of protein (Figures 3B and 3C). This clearly suggests that post-translational modifications of ALPPL2 are important for its recognition by the aptamer. No signals were seen with BSA and lysozyme protein controls (data not shown). The limit of detection (LOD) for ALPPL2 in the direct ALISA assay was 10 ng/mL.
Figure 3.
Aptamer SQ2-Based Direct ALISA for Quantitative Analysis of PDAC-Derived EVs
(A) Schematic illustration of SQ2 aptamer-based direct ALISA for EV detection. (B) SQ2-based ALISA can detect recombinant ALPPL2 protein with a sensitivity of 1 ng (10 ng/mL). (C) Standard curve showing linearity in the broad range of 10 to 2,500 ng/mL of protein. (D) ALPPL2 estimation in the secretomes and EVs of (D) PANC-1+, (E) Capan-1, and (F) MIA PaCa-2 cells using SQ2-based ALISA. ALISA could detect ALPPL2 in EVs with much higher sensitivity than in the secretome. Results are mean ± SD of more than three independent experiments.
Secretome, EVs, and EV-depleted secretome isolated from PANC-1+ cells were analyzed for ALPPL2 using this direct ELISA format. Although ALISA could detect ALPPL2 from both secretome and EVs, the complete absence of signal in EV-depleted secretome indicates that ALPPL2 in PANC-1+ is exclusively present in EVs (Figure 3D). While the same was not the case with Capan-1, as even the EV-depleted secretome showed a considerable signal, indicating the presence of free ALPPL2 protein in Capan-1 secretions (Figure 3E). Based on our earlier studies on Mia PaCa-2 ALPPL2 expression and the immunoblot analysis (Figure 2), no ALPPL2 was expected in MIA PaCa-2 cells. At high exosome concentration, however, a very low but concentration-dependent signal was seen in ALISA, suggesting that these cells might not be completely devoid of ALPPL2 expression (Figure 3F). This also indicates that SQ2-ALISA is sensitive enough to detect low copy number protein as well. Altogether, SQ2 ALISA not only was in complete agreement with the immunoblot analysis, but also accurately reflected the overall ALPPL2 expression levels in the cells and cell-derived secretions. Across all three cells, EV-based ALPPL2 detection was more sensitive and specific than the secretome. This clearly indicates that, for ALPPL2, quantitative ALISA EVs could be a more reliable diagnostic sample than serum or plasma.
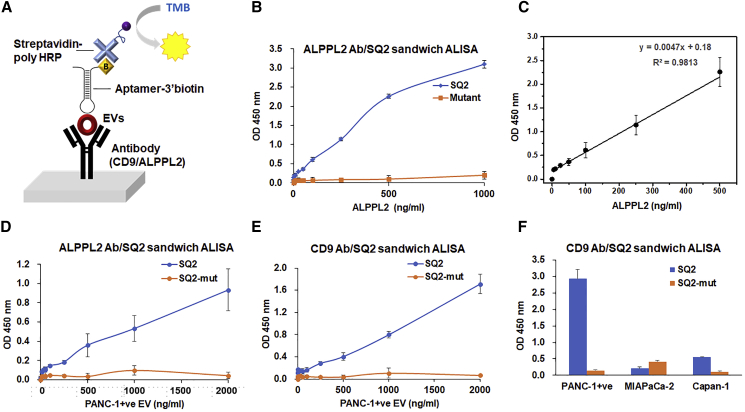
To further improve the sensitivity of ALISA and its applicability to complex samples such as serum, plasma, and exosomes isolated from other body fluids, we set up a sandwich ALISA, using commercial ELISA wells coated with an ALPPL2-capturing antibody (Figure 4A). This ALPPL2 antibody/SQ2 aptamer sandwich ALISA could detect ALPPL2 protein as low as 125 pg/mL (Figure 4B), which is comparable to the commercial ALPPL2 antibody-based sandwich ELISA kit (120 pg/mL). However, the assay showed linearity only in the range of 25 to 500 ng/mL (Figure 4C). Nevertheless, the ALPPL2 antibody/SQ2 assay did not work with the same efficiency in the EVs. As shown in Figure 4D, ALISA signals were low, with maximum optical density around 1, with even 2 μg/mL of PANC-1+ EVs. This clearly suggested that ALPPL2 antibody binding to EVs is not optimal. Also, the LOD for PANC-1+ EVs was 35 ng/mL, which is higher than even the direct ALISA. A similar problem was encountered in the commercial ALPPL2 sandwich ELISA, which showed efficient binding to the ALPPL2 proteins or cell secretome, however, showed no binding to the EVs (Figure S2). Therefore, to sensitively detect the EVs secreted from pancreatic cell secretions, we developed a CD9 antibody/SQ2 aptamer sandwich ALISA. CD9 tetraspanin is a canonical marker for exosome and is commonly used for exosome purification from biologically complex samples.26, 27 This sandwich ALISA assay could detect as low as 100 pg/mL of PANC-1+ EVs with high specificity (Figure 4E). In this platform, both MIA PaCa-2 and Capan-1-derived EVs showed similar results to those measured by the direct SQ2-ALISA (Figure 4F).
Figure 4.
Quantitative Detection in ALPPL2 or CD9 Antibody/SQ2 Aptamer Sandwich ALISA
(A) Scheme of sandwich ALISA. (B) ALPPL2 antibody/SQ2 aptamer-based sandwich ALISA for detecting recombinant ALPPL2 proteins with sensitivity of 3.5 ng (= 35 ng/mL) of ALPPL2. (C) The standard curve showed linearity in the range from 5 to 500 ng/mL. (D) ALPPL2 antibody/SQ2-based detection of ALPPL2 in PANC-1+ EVs (E) CD9 antibody/SQ2 aptamer-based sandwich ALISA for detecting PANC-1+ EVs. Sensitivity of detection for PANC-1+ EVs was as low as 0.1 ng (= 1 ng/mL). (F) CD9 antibody/SQ2 sandwich ALISA for detecting ALPPL2 in three PDAC-derived EVs (5 μg/mL) showed signals comparable to direct ALISA.
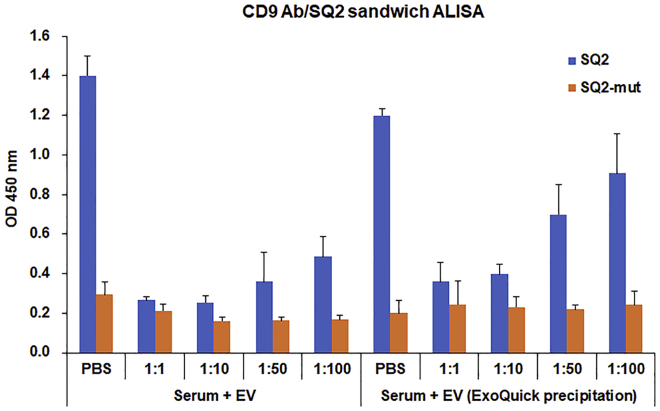
Both the direct and CD9 antibody/SQ2 sandwich ALISA worked well with the cell-derived EVs. However, studies have shown that EVs derived from clinical liquid biopsies such as serum or plasma are often contaminated with serum proteins that interfere with the ELISA.28 In particular, albumin and immunoglobulins, which make up almost half of the human serum protein composition, are the main causes of non-specific binding to capturing antibodies in ELISA, blocking the positive interactions between capture antibodies and target antigens.29 Therefore, to determine the clinical applications of this CD9 antibody/SQ2 aptamer sandwich ALISA to complex samples such as serum and serum-derived EVs, we conducted a proof-of-principle liquid biopsy test where human serum negative for ALPPL2 was diluted in PBS and spiked with ALPPL2-positive EVs. First, ALISA for PANC-1+ EV-spiked serum was conducted without dilution but gave no signals. As the serum dilution increased, detectable differences between SQ2 and SQ2 mutant were observed, suggesting that the other protein components in the serum blocked the positive binding (Figure 5). Next, we tried to isolate EVs from these spiked samples using ExoQuick exosome precipitation reagent. As can be seen in Figure 5, signals improved with EVs isolated from complex mixtures. However, nearly 100-fold serum dilution was required for optimal binding. Alternative precipitation reagents were also tested, but none gave an EV preparation devoid of serum proteins (data not shown). Altogether, this suggests that the EV isolation method is critical to the successful application of the CD9 antibody-SQ2 ALISA to clinical samples of pancreatic cancer, and the absence of suitable isolation methods currently may pose a practical limitation in its further studies with clinical samples.
Figure 5.
Proof-of-Principle Liquid Biopsy Test Using ALPPL2 Negative Human Serum Spiked with PANC-1+ EVs in CD9 Antibody/SQ2 Aptamer-Based Sandwich ALISA
Serum was diluted in a ratio of 1:1 to 1:100 in PBS and then spiked with 0.5 μg of EVs. Samples were directly tested or EVs were isolated by Exoquick-based precipitation method and then tested with this sandwich ALISA.
In conclusion, we found that ALPPL2, a potential biomarker of pancreatic cancer, is present in pancreatic cancer cell-secreted EVs. Using pancreatic cancer cells with variable expression of ALPPL2, we showed that the amount of ALPPL2 in the EVs secreted by these cells correlates with their overall cellular expression of the protein. This clearly shows the implications of ALPPL2 in EV-based diagnosis of pancreatic cancer. Toward this direction, we used the ALPPL2 binding aptamer, earlier developed by us, to set up a diagnostic quantitative ALISA for liquid biopsy. Direct and ALPPL2 or CD9 antibody-based sandwich ALISA were established, which could detect both free and EV-bound forms of ALPPL2 with high specificity and sensitivity. However, our preliminary data on EVs isolated from spiked serum showed that the presence of serum proteins negatively affects the assay, interfering with sensitive detection of ALPP2. Hence, the purity of the EVs is a critical factor in the optimal working of ALISA. Nonetheless, this study clearly shows the diagnostic utility of the aptamer in ALPPL2-based non-invasive detection of pancreatic cancer. Alternative diagnostic strategies not limited to ALISA, like microfluidic-based on-chip devices30 and plasmon sensors chips,31 can be used, where the aptamer-mediated ALPPL2-positive EVs can be detected with minimal interference of serum proteins.
Materials and Methods
Cell Culture
PANC-1 (ATCC CRL1469), MIAPaCa-2 (ATCC CRL-1420), and Capan-1 (ATCC HTB-79) were obtained from the American Tissue Culture Collection and cultured at 37°C and 5% CO2. PANC-1 and MIA PaCa-2 cell lines were grown in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen), and Capan-1 was cultured in Iscove Modified Dulbecco Media (Invitrogen) supplemented with 20% FBS. PANC-1+ cells were enriched as described earlier.18
Preparation of Secretome and Isolation of Extracellular Vesicles
EVs were isolated by differential centrifugation of conditioned media obtained from cells following the standard protocols.32 Cells were grown up to nearly 80% confluency in their respective culture media. For conditioned media preparation, cells were first washed three times with Dulbecco’s PBS supplemented with MgCl2 and CaCl2 (Invitrogen), followed by incubation in FBS-free culture media with 100 U/mL penicillin and 100 mg/mL streptomycin at 37°C and 5% CO2 for 36 h. The conditioned media was collected and clarified by centrifugation at 2,000 × g for 10 min to remove dead cells and cell debris. This secretome was then centrifuged at 10,000 × g for 30 min to remove large vesicles and apoptotic bodies. The supernatant was collected and processed in an ultracentrifuge (Beckman Coulter, Optima MAX-XP, MLA-50 rotor, USA) at 100,000 × g for 70 min at 4°C. The supernatant was collected, and the pellet containing EVs were washed once with PBS and centrifuged again at 100,000 × g for 70 min at 4°C. The final EV pellets were resuspended in PBS. Both the initial secretome and EV-depleted secretome collected above were concentrated by ultrafiltration using 10 kDa centrifugal filters (Amicon ultra-15 centrifugal filter, Millipore) and dialyzed overnight at 4°C against Dulbecco’s PBS. The EVs, concentrated cell secretome, and EV-depleted secretome were stored at −80°C. To isolate EVs in serum, human serum sample of a healthy control was obtained from Discovery Life Sciences (CA, USA). Serum was diluted in the ratio of 1:1 to 1:100 in PBS followed by addition of 0.5 μg of PANC-1+ EVs. EVs from the spiked serum were isolated using ExoQuick exosome precipitation solution (System Biosciences, EXOQ20A-1) in accordance with the manufacturer’s protocol.
DLS
Size distributions of the isolated EVs were measured with DLS using Zetasizer Nano ZS90 (Malvern Instruments, Germany) equipped with a 532-nm laser wavelength. Freshly prepared EVs and commercial exosome from COLO-1 cells (#U0341, Abnova, Taiwan) were diluted 1,000-fold in PBS. 3 × 10 measurement runs were performed with standard settings (dispersant refractive index: 1.331; viscosity (cP), 0.89; temperature, 25°C). The EVs sizes (Z average) refer to the scattering distribution by % intensity.
Nanoparticle Tracking Analysis (NTA)
Size distributions and particle concentrations were measured using NanoSight NTA (NS300, Malvern Instruments). Conditioned media obtained after 24 h incubation was serially diluted in PBS to obtain the recommended measurement range of concentration (2 × 108 to 10 × 108 particles/mL). The samples were analyzed at 22°C under standard settings. Each measurement was carried out in triplicate. The particle concentrations of the final concentrated samples were normalized by protein amounts of the same media.
Scanning Electron Microscope
EVs resuspended in PBS were fixed with a 2% paraformaldehyde solution for 2 h at 25°C, followed by immobilization on the clean silicon chip and allowed to air dry in a clean bench. The silicon chips were washed gently in PBS and dried. To make the surface conductive, Au-Pd alloy was applied by sputtering before imaging. Scanning electron microscopy (JSM-7100F) was performed under 5.0 kV of beam energy. Images were acquired at 20,000 or higher magnifications.
Immunoblotting
Samples were lysed in RIPA (radioimmunoprecipitation assay) buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl [pH 8.0]) for 15 min at 4°C. Total protein amounts of EVs and other secretome samples were estimated using a micro BCA protein assay reagent kit (Thermo Fisher Scientific) in accordance with the manufacturer’s specifications. Proteins were separated on SDS-PAGE, transferred to polyvinylidene fluoride (PVDF) membrane, and probed with respective antibodies: CD9 (Santa Cruz, sc-13118), TSG101 (Santa Cruz, sc-6037), CD63 (Santa Cruz, sc-15363), ALPPL2 (Abcam, ab54780), calnexin (Santa Cruz, sc-23954), GM-130 (Santa Cruz, sc-55591), cytochrome c (Santa Cruz, sc-13156), nucleoporin p62 (Santa Cruz, sc-48393), and secondary anti-mouse immunoglobulin HRP (Santa Cruz, sc-516102) according to standard protocols. Protein bands were detected on an X-ray film using clarity western ECL substrate (Bio-Rad).
Aptamer-Based Direct and Sandwich ELISA
SQ2 aptamer-based direct ELISA was performed with pancreatic cancer cell-derived EVs, concentrated secretome, or human recombinant ALPPL2 protein expressed in HEK293 (OriGene, TP304330) or E. coli (OriGene, SC123000). Samples were diluted in PBS and coated on Nunc-Immuno 96 MicroWell plates (Thermo Scientific) overnight at 4°C. The plate was washed once with wash buffer (PBS with 0.05% Tween-20) and then blocked with 2% BSA in PBS for 2 h at room temperature (RT). After blocking, 20 pmol of SQ2 and SQ2 mutant aptamers33 (St Pharma, South Korea) in binding buffer (PBS with 5 mM MgCl2, 0.5% BSA, 0.1 mg/mL yeast tRNA, 0.05% Tween-20) were added onto each well and kept for 1 h at RT. Wells were washed four times with washing buffer, and 2,000-fold diluted streptavidin-poly-HRP conjugate (Pierce) in 0.5% BSA-PBS was added and incubated for 30 min at RT. After four washes in wash buffer, ultra TMB-ELISA reagent (Thermo Scientific) was added and incubated for 5–15 min to allow blue color expression. The reaction was stopped with 1 M sulfuric acid, and absorbance was measured at 450 nm using Multiskan microplate photometer (Thermo Scientific).
EVs were added to CD9 antibody-coated plate (AVIVA, OKCD00751) or human ALPPL2 antibody-coated plate (Abbexa, abx150631) in 1 mM HEPES buffer at 37°C for 2 h. Unbound ones were removed, and the plates were further probed using the aptamer and detection reagent. The rest of the protocol was same as aptamer-direct ELISA mentioned above. Commercial ALPPL2 sandwich ELISA kit (Dl Develop, DL-ALPPL2-Hu) was also used for comparisons. Recombinant ALPPL2 protein expressed in HEK293T cells was used to determine the specificity and linearity in the assay. BSA and lysozyme were used as protein controls. The sensitivity of the assay was calculated as the lowest concentration, which showed values of OD450 nm (SQ2)/OD450 nm (SQ2 mutant) more than 3.
Author Contributions
Conception and design: Pooja Dua Dong ki Lee; data acquisition: Pooja Dua, Hye-su Shin, Sang Baek Jung, Sungho Park; data analysis and interpretation: Hye-su Shin, Pooja Dua, Dong-ki Lee; writing and review of manuscript: Pooja Dua, Hye-su Shin, Dong ki Lee; technical and material support: Dong ki Lee; study spervision: Pooja Dua Dong ki Lee.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the Basic Science Research Programs through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF 2013 R1A1A2062908) and a Global Research Laboratory grant from the Ministry of Education, Science, and Technology of Korea (no. 2008-00582). We also thank Dr. Changjin Lee (Rosetta Exosome, Korea), who provided insight and expertise in exosome isolation that greatly assisted this study.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.08.016.
Contributor Information
Pooja Dua, Email: pooja.dua@unigoa.ac.in.
Dong ki Lee, Email: dklee@skku.edu.
Supplemental Information
References
- 1.Ilic M., Ilic I. Epidemiology of pancreatic cancer. World J. Gastroenterol. 2016;22:9694–9705. doi: 10.3748/wjg.v22.i44.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L., Sanagapalli S., Stoita A. Challenges in diagnosis of pancreatic cancer. World J. Gastroenterol. 2018;24:2047–2060. doi: 10.3748/wjg.v24.i19.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tummala P., Junaidi O., Agarwal B. Imaging of pancreatic cancer: An overview. J. Gastrointest. Oncol. 2011;2:168–174. doi: 10.3978/j.issn.2078-6891.2011.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herreros-Villanueva M., Bujanda L. Non-invasive biomarkers in pancreatic cancer diagnosis: what we need versus what we have. Ann. Transl. Med. 2016;4:134. doi: 10.21037/atm.2016.03.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X., Shi S., Zhang B., Ni Q., Yu X., Xu J. Circulating biomarkers for early diagnosis of pancreatic cancer: facts and hopes. Am. J. Cancer Res. 2018;8:332–353. [PMC free article] [PubMed] [Google Scholar]
- 6.Boukouris S., Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics Clin. Appl. 2015;9:358–367. doi: 10.1002/prca.201400114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szatanek R., Baj-Krzyworzeka M., Zimoch J., Lekka M., Siedlar M., Baran J. The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int. J. Mol. Sci. 2017;18:1153. doi: 10.3390/ijms18061153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raposo G., Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tickner J.A., Urquhart A.J., Stephenson S.A., Richard D.J., O’Byrne K.J. Functions and therapeutic roles of exosomes in cancer. Front. Oncol. 2014;4:127. doi: 10.3389/fonc.2014.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moutinho-Ribeiro P., Macedo G., Melo S.A. Pancreatic Cancer Diagnosis and Management: Has the Time Come to Prick the Bubble? Front. Endocrinol. (Lausanne) 2019;9:779. doi: 10.3389/fendo.2018.00779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan Y., Fu G., Ming L. Role of exosomes in pancreatic cancer. Oncol. Lett. 2018;15:7479–7488. doi: 10.3892/ol.2018.8348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herreros-Villanueva M., Bujanda L. Glypican-1 in exosomes as biomarker for early detection of pancreatic cancer. Ann. Transl. Med. 2016;4:64. doi: 10.3978/j.issn.2305-5839.2015.10.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diamandis E.P., Plebani M. Glypican-1 as a highly sensitive and specific pancreatic cancer biomarker. Clin. Chem. Lab. Med. 2016;54:e1–e2. doi: 10.1515/cclm-2015-0773. [DOI] [PubMed] [Google Scholar]
- 14.Costa-Silva B., Aiello N.M., Ocean A.J., Singh S., Zhang H., Thakur B.K., Becker A., Hoshino A., Mark M.T., Molina H. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015;17:816–826. doi: 10.1038/ncb3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z., Sun H., Provaznik J., Hackert T., Zöller M. Pancreatic cancer-initiating cell exosome message transfer into noncancer-initiating cells: the importance of CD44v6 in reprogramming. J. Exp. Clin. Cancer Res. 2019;38:132. doi: 10.1186/s13046-019-1129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z., von Au A., Schnölzer M., Hackert T., Zöller M. CD44v6-competent tumor exosomes promote motility, invasion and cancer-initiating cell marker expression in pancreatic and colorectal cancer cells. Oncotarget. 2016;7:55409–55436. doi: 10.18632/oncotarget.10580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millán J.L., Fishman W.H. Biology of human alkaline phosphatases with special reference to cancer. Crit. Rev. Clin. Lab. Sci. 1995;32:1–39. doi: 10.3109/10408369509084680. [DOI] [PubMed] [Google Scholar]
- 18.Dua P., Kang H.S., Hong S.M., Tsao M.S., Kim S., Lee D.K. Alkaline phosphatase ALPPL-2 is a novel pancreatic carcinoma-associated protein. Cancer Res. 2013;73:1934–1945. doi: 10.1158/0008-5472.CAN-12-3682. [DOI] [PubMed] [Google Scholar]
- 19.Filipe V., Hawe A., Jiskoot W. Critical evaluation of Nanoparticle Tracking Analysis (NTA) by NanoSight for the measurement of nanoparticles and protein aggregates. Pharm. Res. 2010;27:796–810. doi: 10.1007/s11095-010-0073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stetefeld J., McKenna S.A., Patel T.R. Dynamic light scattering: a practical guide and applications in biomedical sciences. Biophys. Rev. 2016;8:409–427. doi: 10.1007/s12551-016-0218-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger J., Howard A.D., Gerber L., Cullen B.R., Udenfriend S. Expression of active, membrane-bound human placental alkaline phosphatase by transfected simian cells. Proc. Natl. Acad. Sci. USA. 1987;84:4885–4889. doi: 10.1073/pnas.84.14.4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iles R.K., Ind T.E., Chard T. Production of placental alkaline phosphatase (PLAP) and PLAP-like material by epithelial germ cell and non-germ cell tumours in vitro. Br. J. Cancer. 1994;69:274–278. doi: 10.1038/bjc.1994.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura T., Nakamura K., Stinson R.A. Release of alkaline phosphatase from human osteosarcoma cells by phosphatidylinositol phospholipase C: effect of tunicamycin. Arch. Biochem. Biophys. 1988;265:190–196. doi: 10.1016/0003-9861(88)90384-0. [DOI] [PubMed] [Google Scholar]
- 24.López-Cobo S., Campos-Silva C., Valés-Gómez M. Glycosyl-Phosphatidyl-Inositol (GPI)-Anchors and Metalloproteases: Their Roles in the Regulation of Exosome Composition and NKG2D-Mediated Immune Recognition. Front. Cell Dev. Biol. 2016;4:97. doi: 10.3389/fcell.2016.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Gassart A., Geminard C., Fevrier B., Raposo G., Vidal M. Lipid raft-associated protein sorting in exosomes. Blood. 2003;102:4336–4344. doi: 10.1182/blood-2003-03-0871. [DOI] [PubMed] [Google Scholar]
- 26.Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreu Z., Yáñez-Mó M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014;5:442. doi: 10.3389/fimmu.2014.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Webber J., Clayton A. How pure are your vesicles? J. Extracell. Vesicles. 2013;2:3402. doi: 10.3402/jev.v2i0.19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selby C. Interference in immunoassay. Ann. Clin. Biochem. 1999;36:704–721. doi: 10.1177/000456329903600603. [DOI] [PubMed] [Google Scholar]
- 30.Mazaafrianto D.N., Maeki M., Ishida A., Tani H., Tokeshi M. Recent Microdevice-Based Aptamer Sensors. Micromachines (Basel) 2018;9:E202. doi: 10.3390/mi9050202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vance S.A., Sandros M.G. Zeptomole detection of C-reactive protein in serum by a nanoparticle amplified surface plasmon resonance imaging aptasensor. Sci. Rep. 2014;4:5129. doi: 10.1038/srep05129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thery C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;30:3.22.1–3.22.29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 33.Dua P., S S., Kim S., Lee D.K. ALPPL2 Aptamer-Mediated Targeted Delivery of 5-Fluoro-2′-Deoxyuridine to Pancreatic Cancer. Nucleic Acid Ther. 2015;25:180–187. doi: 10.1089/nat.2014.0516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.