Abstract
Glioma is the most common brain tumor with a dismal prognosis. While temozolomide (TMZ) based chemotherapy significantly improves survival in glioma patients, resistance against this compound commonly leads to glioma treatment failure. Overexpression of long-noncoding RNA (LncRNA) FoxD2 adjacent opposite strand RNA 1 (FoxD2-AS1) was identified to promote glioma development, but the role in TMZ resistance remains unclear. In this paper, we found that FoxD2-AS1 was overexpressed in recurrent glioma, high FoxD2-AS1 expression was significantly correlated with poor patient outcome. Methylation of O6-methylguanine-DNA methyltransferase (MGMT) is significantly less frequent in high FoxD2-AS1 expression patients. Knockdown of FoxD2-AS1 decreased the proliferation, metastatic ability of glioma cells and promote the sensitivity to TMZ in glioma cells. Furthermore, knockdown of FoxD2-AS1 induced hypermethylation of the promoter region of MGMT. Our data suggested that FoxD2-AS1 is a clinical relevance LncRNA and mediates TMZ resistance by regulating the methylation status of the MGMT promoter region.
Keywords: Drug Resistance, Glioma, Long-noncoding RNA, Methylation, Temozolomide
INTRODUCTION
Glioma is the most common form of brain malignancy that produces severe damage to the brain leading to a very poor survival prognosis [1,2]. Currently, temozolomide (TMZ)-based chemotherapy significantly improves prognosis, and is recommended as a standard care for glioma patients [3]. TMZ is an alkylating agent which cause lethal DNA damage (O6-methylation-mediated DNA base mismatching) and subsequent induce cell death and apoptosis [4]. However, a majority of patients with glioma gradually develop resistance to TMZ during treatment. The most popular mechanism of TMZ resistance is the expression of O6-methylguanine-DNA methyltransferase (MGMT) which removes the cytotoxic O6-methylguanine-DNA adducts [5,6]. The expression of MGMT is regulated by MGMT promoter methylation. Normally, high promoter methylation status which means low MGMT activity predicts good response to TMZ chemotherapy and results in a longer survival period in glioma patients [7,8]. This observation led to the hypothesis that MGMT inhibition may be a plausible strategy for sensitizing TMZ therapy.
In recent years, emerging evidence has regarded the functional role of dysregulated long-noncoding RNAs (LncRNAs) in the cancer formation and progression, as well as the resistance to chemotherapy [9,10]. The LncRNA AC003092.1 promotes TMZ chemosensitivity through miR-195/TFPI-2 signaling pathway in glioblastoma [11]. LncRNA XIST can amplify the chemoresistance of glioma cell lines to TMZ through directly targeting miR-29c via SP1 and MGMT [12]. Additionally, LncRNA H9 and RP11-838N2.4 have been reported to enhance cytotoxic effects of TMZ in glioma cell lines [13,14]. Thus, genomic characterization of LncRNA alterations may provide potential therapeutic strategy for TMZ resistant in glioma.
LncRNA FoxD2 adjacent opposite strand RNA 1 (FoxD2-AS1) is a newly identified LncRNA which locate on chromosome 1p33 with a transcript 2527 nucleotides in length. FoxD2-AS1 was identified to be overexpressed in esophageal squamous cell carcinoma, nasopharyngeal carcinoma, bladder cancer, colorectal cancer and glioma [15,16,17,18,19]. FoxD2-AS1 can regulate cancer progression and recurrence, but the functional role of FoxD2-AS1 in chemoresistance remains unclear. In the present study, we analyzed the expression of FoxD2-AS1 in glioma tissues obtained from glioma gene expression datasets, and found that FoxD2-AS1 was a clinically relevant LncRNA, as high expression was associated with poor patient outcome. Moreover, we demonstrated that methylation of MGMT is significantly less frequent in high FoxD2-AS1 expression patients and downregulation of FoxD2-AS1 decreased TMZ resistance in glioma cells through regulating the methylation status of the MGMT promoter region. Our findings revealed that the dysregulation of FoxD2-AS1 is a potential component of glioma pathogenesis and TMZ resistance, which might become a new therapeutic target for patients with glioma.
METHODS
Cell culture
The human glioma cell lines U251 and A172 were obtained from Cell Resource Center of Shanghai and cultured in Dulbecco's Modified Eagle Medium (DMSO; Gibco, Grand Island, NY, USA) supplemented with 10% fetal calf serum (FBS; Gibco), 100 U/ml penicillin and 100 mg/ml streptomycin (Life Technologies, Grand Island, NY, USA) at 37℃ in 5% CO2.
Bioinformatics analysis
Glioma gene expression arrays with survival data were obtained from the National Cancer Institute Repository for Molecular Brain Neoplasia Data (NCI REMBRANDT), the Cancer Genome Atlas (TCGA), and gene expression omnibus datasets. TCGA data were extracted directly from the web site (https://portal.gdc.cancer.gov/). For assessing the overall survival (OS) of glioma patients included in REMBRANDT, we used project Betastasis (http://www.betastasis.com). For assessing the OS of glioma patients included in GSE16011, we used R2: Genomics Analysis and Visualization Platform (http://r2.amc.nl). OS was defined as the period from the date of the pathological diagnosis to death.
Cell transfection
Small interfering RNA (siRNA) targeting FoxD2-AS1 and negative control siRNA were synthesized by Biomics Biotechnologies Co., Ltd. (Nantong, China). Sense siFoxD2-AS1: 5′-GCGAAGAGUACGUUGCUAUTT-3′, antisense siFoxD2-AS1 : 5′-AUAGCAACGUACUCUCGCTT-3′; Sense siNC: 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense siNC: 5′-ACGUGACACGUUCGGAGAATT-3′. Cells were grown on six-well plates to confluency and transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Briefly, each well was supplemented with 5 µl Lipofectamine 2000 and 100 pmol siRNA. About 48 h later, cells were acquired for the following experiments.
Western blot analysis
siFoxD2-AS1 or siNC transfected cells were treated with TMZ for 48 h, and then were washed once with phosphate buffer saline (PBS) and dissolved in Protein Extraction Reagent (Boster Bioengineering, Wuhan, China) containing 1 mM phenylmethanesulfonyl fluoride (PMSF; Roche Molecular Biochemicals, Indianapolis, IN, USA). Protein samples were separated by 10%–12% SDS-PAGE, transferred onto the surface of polyvinylidene fluoride membrane and immunoblotted with the indicated primary antibodies. Using the enhanced chemiluminescence reagent (Amersham Biosciences, Piscataway, NJ, USA), visualization of the protein bands was conducted in the Omega Lum G system (Aplegen, Pleasanton, CA, USA). The primary antibodiy to β-actin was purchased from Hua Bio (Hangzhou HuaAn Biotechnology Co., Ltd., Hangzhou, China), and the antibodys to Caspase-3, Bax, MGMT were purchased from (Proteintech Group, Chicago, IL, USA).
Isolation of DNA and methylation-specific PCR
The DNA was extracted using Takara MiniBEST Universal Genomic DNA Kit (Takara, Shiga, Japan) and then 1 µg of extracted DNA underwent bisulfite modification using a DNA Methylation Kit (CoWin Biosciences, Beijing, China) according to the manufacturer's instructions. The MS-PCR was performed under the following conditions: 95℃ for 10 min; 35 cycles of 95℃ for 30 sec, the annealing temperature 60℃ for 30 sec, and 72℃ for 30 sec; and a final extension of 10 min at 72℃. Primers for the methylated MGMT were 5′-TTTCGACGTTCGTAGGTTTTCGC-3′ (forward) and 5′-GCACTCTTCCGAAAACGAAACG-3′ (reverse); for the un-methylated MGMT were 5′-TTTGTGTTTTGATGTTTGTAGGTTTTT GT-3′ (forward) and 5′-AACTCCACACTCTTCCAAAAACAAAACA-3′ (reverse). The PCR product was directly loaded onto 2.5% agarose gels, stained with ethidium bromide, and visualized with the aid of ultraviolet light. The density of each band was quantified using imaging analysis and the relative band density values were calculated as the ratio of methylated MGMT to that of methylated plus un-methylated MGMT.
MTT assay
Cell viability was evaluated by using MTT assay. Glioma cells were seeded into 96-well plates at the concentration of 2 × 103 cells/well. Cells were treated with different concentrations of TMZ (MedChem Express, Monmouth Junction, NJ, USA) for 48 h. Then, 10 µl MTT (5 mg/ml) was added to each well and incubated in the dark at 37℃ for another 4 h. Absorbance was determined at a wavelength of 570 nm using a SpectraMax M3 microplatereader (Molecular Devices, Sunnyvale, CA, USA).
Colony formation assay
Cells were seeded (3–5 × 105 cells/well) and then transfected with siFoxD2-AS1 or siNC alone. After 24 h post-transfection, cells were treated with TMZ for another 24 h, washed, serially diluted, plated in triplicate into 6 well plates, allowed 6–8 days of cell growth for colony formation, stained with 5% crystal violet. Finally, colonies were photographed, and the number of colonies was counted manually.
Migration and invasion assays
In the migration assay, cells were suspended in DMEM and the density was adjusted to approximately 1 × 105 cells/ml. Cell suspension was then seed to the upper chamber (Corning, Corning, NY, USA) of the inserts (0.2 ml/well), and DMEM containing negative control siRNA were synthesized by Biomics Biotechnologies Co., Ltd. (Nantong, China). Sense siFoxD2-AS1: 5′-GCGAAGAGUACGUUGCUAUTT-3′, antisense siFoxD2-AS1 : 5′-AUAGCAACGUACUCUCGCTT-3′; Sense siNC: 5′-UUCUCCGAACGUGUCACGUTT-3′, antisense siNC: 5′-ACGUGACACGUUCGGAGAATT-3′. Cells were grown on six-well plates to confluency and transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Briefly, each well was supplemented with 5 µl Lipofectamine 2000 and 100 pmol siRNA. About 48 h later, cells were acquired for the following experiments.
Cell apoptosis analysis
An apoptosis analysis kit (NanJing KeyGen Biotech Co., Ltd., Nanjing, China) was utilized to detect the cell apoptotic rates. Briefly, the Annexin V and PI were firstly added into the binding buffer, and the treated cells were resuspended in the binding buffer. After incubating in the dark for 15 min, the cells were washed and analyzed using a Guava easyCyte 5 Flow Cytometer (EMD Millipore, Bellerica, MA, USA).
Statistical analysis
All statistical data were analyzed using the SPSS 21.0 software (IBM Co., Armonk, NY, USA) and GraphPad Prism 5 (GraphPad Software, San Diego, CA, USA). p-values < 0.05 were considered statistically significant. Data are expressed as mean ± standard deviation. Statistical significance was determined using Student's t-test or one-way analysis of variance. The Kaplan–Meier method with the log-rank test was applied for OS.
RESULTS
FoxD2-AS1 is associated with clinical outcome of patients with glioma
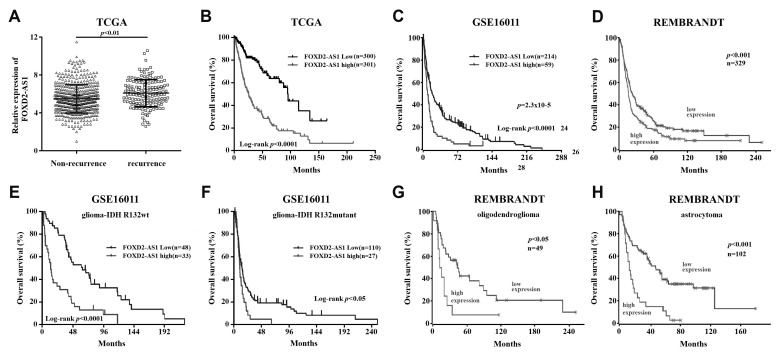
Three independent glioma gene expression datasets (TCGA, GSE16011, and REMBRANDT) were employed to examine the association between FoxD2-AS1 expression levels and clinical outcome of patients with glioma. First, we compared the FoxD2-AS1 expression in recurrent glioma tissues with non-recurrent tissues obtained from TCGA and found that FoxD2-AS1 expression is significantly higher in recurrent than in non-recurrent tissues (p < 0.01, Fig. 1A). Further Kaplan–Meier and log-rank test analysis showed that FoxD2-AS1 was associated with OS of glioma patients included in TCGA, GSE16011, and REMBRANDT, patients with FoxD2-AS1 higher expression had a significant shorter overall survival time than those with FoxD2-AS1 lower expression (Fig. 1B–D). Moreover, high expression of FoxD2-AS1 is significantly correlated with poor patient outcome in all tested cases, namely, IDH wildtype and mutant tumors, astrocytoma, and oligodendroglioma (Fig. 1E–H). Collectively, these data suggest that high FoxD2-AS1 expression is associated with poor outcome in patients with glioma and potentially contributing to the aggressive tumor biology of glioma.
Fig. 1. FoxD2-AS1 is associated with clinical outcome of patients with glioma.
(A) FoxD2-AS1 expression in recurrent glioma tissues and non-recurrent tissues obtained from the Cancer Genome Atlas (TCGA). (B–D) Kaplan–Meier plots of patient outcome based on FoxD2-AS1 expression. (E–H) Kaplan–Meier plots of patient outcome based on FoxD2-AS1 expression in glioma, stratified according to the annotated IDH mutation status and glioma subtype. Data, statistical evaluation, and visualization were obtained using the R2 website “R2: Genomics Analysis and Visualization Platform” (http://r2.amc.nl) and http://www.betastasis.com/. REMBRANDT, Repository for Molecular Brain Neoplasia Data.
FoxD2-AS1 is contributed to TMZ response of patients with glioma
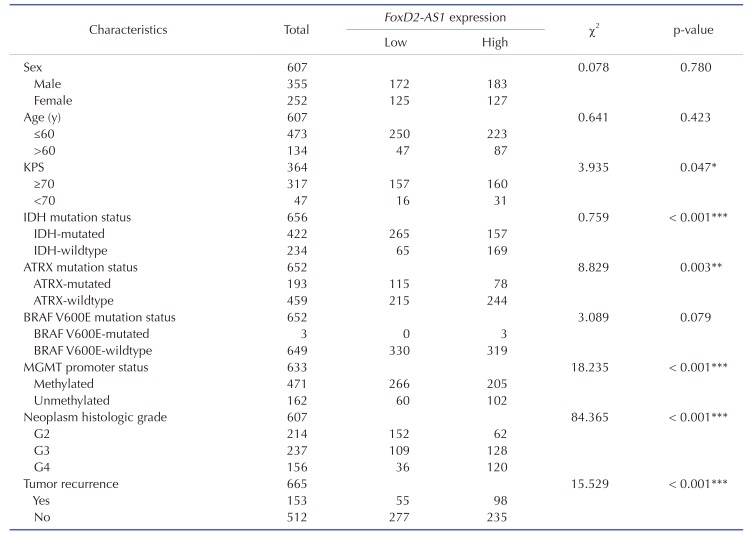
Evidence of a correlation between FoxD2-AS1 expression and the clinicopathological status of patients with glioma was searched in TCGA. The results of a chi-square test indicated that methylation of MGMT is significantly less frequent in high FoxD2-AS1 expression patients. As shown in Table 1, methylation of MGMT was found in 81.6% (266/326) of patients with low FoxD2-AS1 expression, but in 66.7% (205/307) of patients with high FoxD2-AS1 expression (p < 0.001). Significant correlations were also detected between FoxD2-AS1 expression and certain clinicopathological features, including IDH mutation status and neoplasm histologic grade (Table 1).
Table 1. Correlation between FoxD2-AS1 expression and glioma clinic-pathological features in TCGA.
Values are presented as number. TCGA, the Cancer Genome Atlas; KPS, Karnofsky performance score; MGMT, O6-methylguanine-DNA methyltransferase. *p < 0.05, **p < 0.01, ***p < 0.001.
FoxD2-AS1 is contributed to TMZ resistance of glioma cells
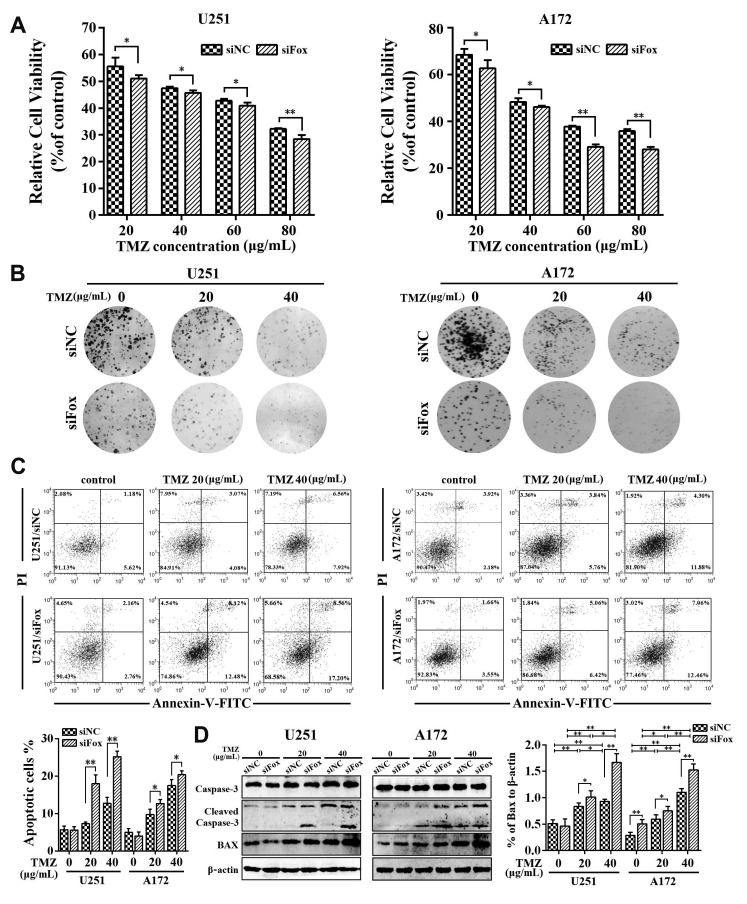
To asked whether FoxD2-AS1 could modulate the sensitivity of glioma cells to TMZ, the siNC or siFoxD2-AS1 transduced A172 and U251 cells were treated with different doses of TMZ, respectively. The results of the MTT assay shown that down-regulation of FoxD2-AS1 reduced the viability of glioma cells after treatment with TMZ (Fig. 2A). Moreover, we found that the IC50 values for TMZ in siFoxD2-AS1 transduced glioma cells were much lower than that in their control cells (56.66 µg/ml in A172 cells, 52.27 µg/ml in A172/siFox cells, 49.15 µg/ml in U251 cells, and 45.31 µg/ml in U251/siFox cells). Next, the colony formation assay, flow cytometry, and western blot analysis were performed to further determine the effect of FoxD2-AS1 on TMZ resistance of glioma cells. As shown in Fig. 2B, the colony formation for siFoxD2-AS1 transduced cells was lower than that of the siNC group by treatment with 20 and 40 µg/ml TMZ. The results of flow cytometry analysis revealed that FoxD2-AS1 knockdown significantly increased glioma cell apoptosis caused by TMZ treatment (Fig. 2C). Furthermore, Western blot analysis showed that TMZ-induced cellular apoptosis was greatly enhanced by FoxD2-AS1 knockdown compared to NC (Fig. 2D). Above all, it was implied that down-regulation of FoxD2-AS1 increased the sensitivity of glioma cells to TMZ.
Fig. 2. FoxD2-AS1 knockdown sensitizes glioma cells to temozolomide (TMZ) treatment.
(A) Cell viabilities of siFoxD2-AS1 or siNC transducted glioma cells under treatment of indicated doses of TMZ. (B) Colony formation of siFoxD2-AS1 or siNC transducted glioma cells after TMZ treatment. (C) Flow cytometry analysis shown the cell apoptosis of siFoxD2-AS1 or siNC transducted glioma cells after TMZ treatment. (D) Western blot analysis of cleaved caspase-3 and Bax in glioma cells transfected with siFoxD2-AS1 or siNC after TMZ treatment. siFox, siFoxD2-AS1. *p < 0.05, **p < 0.01.
FoxD2-AS1 is contributed to migration and invasion ability of glioma cells
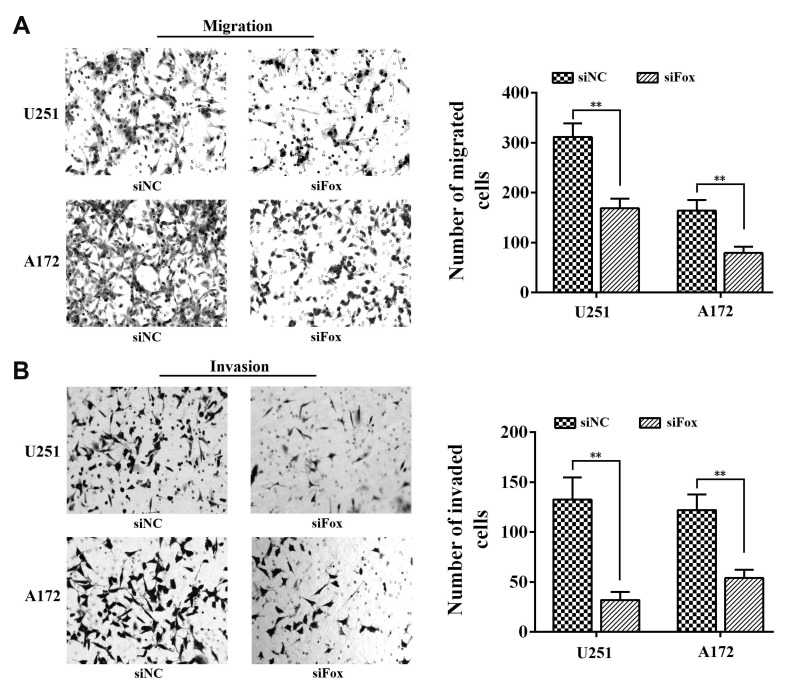
Previous studies reported that the arisen of chemoresistance may associated with the high metastatic ability of cancer cells [20], so we used transwell system to investigate the role of FoxD2-AS1 on the migration and invasion ability of glioma cells. As shown in Fig. 3, the cell counts of U251 and A172 were significantly decreased in both migration and invasion systems after transfection with siFoxD2-AS1, which suggested that FoxD2-AS1 knockdown could inhibit the metastatic ability of glioma cells.
Fig. 3. FoxD2-AS1 knockdown inhibits the metastatic ability of glioma cells.
(A) Effect of FoxD2-AS1 knockdown on the migration of glioma cells. (B) Effect of FoxD2-AS1 knockdown on the invasion of glioma cells. Cells were counted after staining with crystal violet (magnification, ×200). Data represent means ± standard deviation. of at least three independent experiments. siFox, siFoxD2-AS1. **p < 0.01.
FoxD2-AS1 mediated TMZ resistance is associated with methylation status of the MGMT promoter region in glioma cells
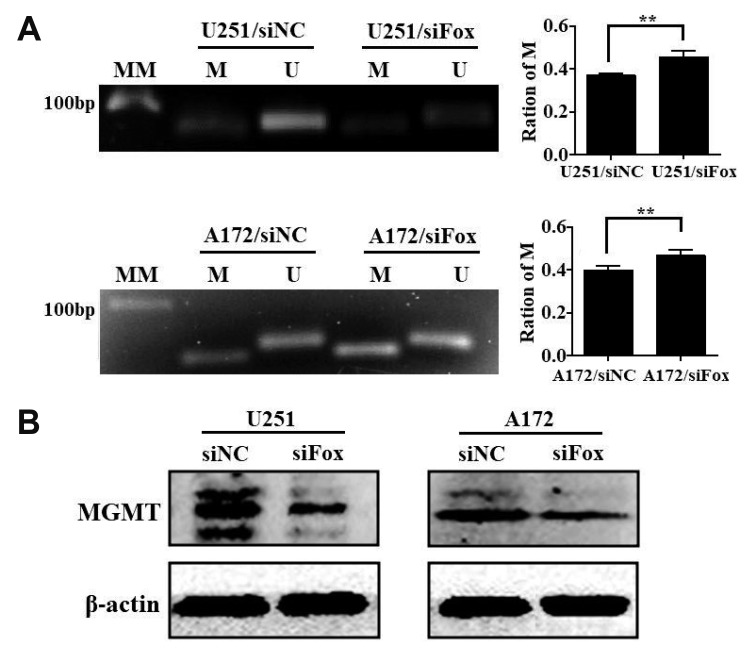
MGMT promoter methylation is the key mechanism of MGMT gene silencing and predicts a favorable TMZ chemotherapy outcome in glioblastoma (GBM) patients. To further clarify the relationship between FoxD2-AS1 expression and methylation status of the MGMT promoter in glioma cells, DNA was isolated from siNC or siFoxD2-AS1 transduced A172 and U251 cells and MGMT methylation was determined by MS-PCR. As shown in Fig. 4A, the ratio of methylated promoter DNA was much higher in siFoxD2-AS1 transduced than siNC transduced glioma cells. We further detected the MGMT protein expression in siNC or siFoxD2-AS1 transduced glioma cells by western blot, the results shown that the MGMT protein level was much lower in siFoxD2-AS1 transduced cells than that in siNC group (Fig. 4B). These results demonstrated that FoxD2-AS1 mediated TMZ resistance in glioma cells by regulating the methylation status of the MGMT promoter region.
Fig. 4. FoxD2-AS1 is associated with the methylation status of the O6-methylguanine-DNA methyltransferase (MGMT) promoter in glioma cells.
(A) MS-PCR analysis depicts the methylation status of the MGMT promoter in siFoxD2-AS1 or siNC transducted glioma cells. The density of each band was quantified using imaging analysis and the relative band density values were calculated as the ratio of methylated MGMT to that of methylated plus un-methylated MGMT. MGMT unmethylated: 92 bp; MGMT methylated: 8l bp. (B) Western blot analysis of MGMT expression in siFoxD2-AS1 or siNC transducted glioma cells. U, unmethylated; M, methylated; MM, molecular marker; siFox, siFoxD2-AS1. **p < 0.01.
DISCUSSION
Although previous studies have demonstrated the overexpression of FoxD2-AS1 in glioma tissues and cells, the biological role and the clinical relevance of FoxD2-AS1 in glioma are only emerging. Therefore, we interrogated three glioma gene expression datasets to clarify the clinical relevance of FoxD2-AS1 in patients with glioma and found that high FoxD2-AS1 expression was significantly correlated with poor patient outcome. Furthermore, we pointed out the higher expression of FoxD2-AS1 in recurrent glioma compared with primary glioma. Both Shen and Ni groups [19,21] reported that the expression of FoxD2-AS1 was upregulated as the grade of glioma increased. Taken together, these results suggest that FoxD2-AS1 are overexpressed in glioma with more aggressive phenotypes and could function as a prognostic marker for glioma patients.
De novo and acquired resistance to TMZ in glioma cells have emerged as a challenging problem in clinical practice [22]. To date, the bulk of evidence suggests that epigenetic silencing of the MGMT gene through hypermethylation of the cytidine phosphate guanosinedinucleotides in the promoter region is associated with greater response to the TMZ treatment of glioma patients. So we detected the relationship of FoxD2-AS1 expression and glioma patient's MGMT promoter status. Our results shown that methylation of MGMT is significantly less frequent in high FoxD2-AS1 expression patients, suggesting the promoting function of FoxD2-AS1 overexpression on TMZ resistance.
To further determine whether FoxD2-AS1 could modulate the sensitivity of glioma cells to TMZ, we knocked down the FoxD2-AS1 expression level in two glioma cell lines using siRNA. Our results shown that downregulation of FoxD2-AS1 could decrease the cell proliferation and promote the sensitivity to TMZ in a dose dependent manner both in A172 and U251 cells. Further western blot and flow cytometric analysis found that downregulation of FoxD2-AS1 could promote TMZ induced activation of apoptosis. The metastasis and chemoresistance have been reported to be closely related, the arisen of chemoresistance could promote metastasis of cancer cells and the metastasis contributed to chemoresistanc [23,24,25]. After transfection of siFoxD2-AS1, we found that the migration and invasion ability were significantly decreased in both glioma cell lines. Finally we evaluated whether FoxD2-AS1 mediated TMZ resistance is associated with methylation status of the MGMT promoter region in glioma cells, and we found that downregulation of FoxD2-AS1 could induce hypermethylation of the promoter region of MGMT.
We also investigated the methylation status of MGMT promoter and the expression of FoxD2-AS1 in glioma cells after treated with TMZ. The results shown that neither the methylation status of MGMT promoter nor the level of FoxD2-AS1 in U251 and A172 cells changed significantly after treatment with 20 and 40 µg/ml TMZ for 48 h. The methylation status of MGMT promoter and the expression of FoxD2-AS1 were associated with TMZ resistance in glioma cells. A short time (48 h) treatment with TMZ can't induce the chemoresistance of glioma cells; therefore, it is reasonable that 48 h TMZ treatment can't control MGMT methylation or FoxD2-AS1 expression. In the future, we will develop TMZ-resistant glioma cell lines to further explore the relationship between the expression of FoxD2-AS1 and the chemoresistance of glioma cell to TMZ.
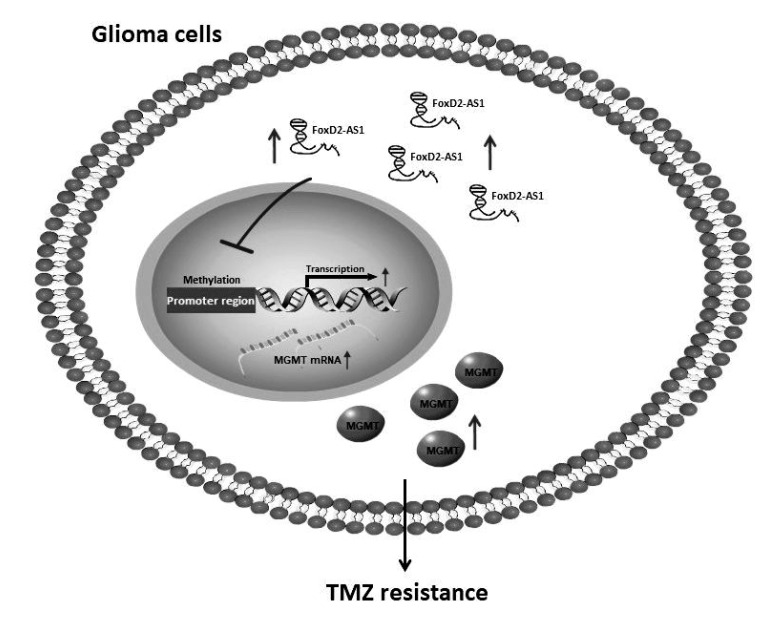
In summary, our results demonstrated that FoxD2-AS1 was a clinically relevant LncRNA conferring TMZ resistance in glioma. Downregulation of FoxD2-AS1 reduced the TMZ resistance of A172 and U251 cells by regulating the methylation status of the MGMT promoter region (Fig. 5). FoxD2-AS1 is a potential target for developing new therapeutic strategies against drug resistance in glioma.
Fig. 5. A diagram summarizing the main findings of this study.
FoxD2-AS1 contributes to temozolomide (TMZ) resistance of glioma cells by regulating the methylation status of the O6-methylguanine-DNA methyltransferase (MGMT) promoter region.
ACKNOWLEDGEMENTS
This study was supported in part by research fund for National Natural Science Foundation of China (81602624).
Footnotes
Author contributions: N.T. conceived and designed the experiments. W.S. performed the experiments. X.L. analyzed the data. N.T. wrote the paper.
CONFLICTS OF INTEREST: The authors declare no conflicts of interest.
References
- 1.Schijns VEJC, Pretto C, Strik AM, Gloudemans-Rijkers R, Deviller L, Pierre D, Chung J, Dandekar M, Carrillo JA, Kong XT, Fu BD, Hsu FPK, Hofman FM, Chen TC, Zidovetzki R, Bota DA, Stathopoulos A. Therapeutic immunization against glioblastoma. Int J Mol Sci. 2018;19:E2540. doi: 10.3390/ijms19092540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392:432–446. doi: 10.1016/S0140-6736(18)30990-5. [DOI] [PubMed] [Google Scholar]
- 3.Chua J, Nafziger E, Leung D. Evidence-based practice: temozolomide beyond glioblastoma. Curr Oncol Rep. 2019;21:30. doi: 10.1007/s11912-019-0783-5. [DOI] [PubMed] [Google Scholar]
- 4.Roos WP, Batista LF, Naumann SC, Wick W, Weller M, Menck CF, Kaina B. Apoptosis in malignant glioma cells triggered by the temozolomide-induced DNA lesion O6-methylguanine. Oncogene. 2007;26:186–197. doi: 10.1038/sj.onc.1209785. [DOI] [PubMed] [Google Scholar]
- 5.Sarkaria JN, Kitange GJ, James CD, Plummer R, Calvert H, Weller M, Wick W. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14:2900–2908. doi: 10.1158/1078-0432.CCR-07-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Storey K, Leder K, Hawkins-Daarud A, Swanson K, Ahmed AU, Rockne RC, Foo J. Glioblastoma recurrence and the role of O6-methylguanine-DNA methyltransferase promoter methylation. JCO Clin Cancer Inform. 2019;3:1–12. doi: 10.1200/CCI.18.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunn J, Baborie A, Alam F, Joyce K, Moxham M, Sibson R, Crooks D, Husband D, Shenoy A, Brodbelt A, Wong H, Liloglou T, Haylock B, Walker C. Extent of MGMT promoter methylation correlates with outcome in glioblastomas given temozolomide and radiotherapy. Br J Cancer. 2009;101:124–131. doi: 10.1038/sj.bjc.6605127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Nifterik KA, van den Berg J, van der Meide WF, Ameziane N, Wedekind LE, Steenbergen RD, Leenstra S, Lafleur MV, Slotman BJ, Stalpers LJ, Sminia P. Absence of the MGMT protein as well as methylation of the MGMT promoter predict the sensitivity for temozolomide. Br J Cancer. 2010;103:29–35. doi: 10.1038/sj.bjc.6605712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang FT, Chen WY, Gu ZQ, Zhuang YY, Li CQ, Wang LY, Peng JF, Zhu Z, Luo X, Li YH, Yao HR, Zhang SN. The novel long intergenic noncoding RNA UCC promotes colorectal cancer progression by sponging miR-143. Cell Death Dis. 2017;8:e2778. doi: 10.1038/cddis.2017.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Y, Hu B, Wang Q, Ye M, Qiu Q, Zhou Y, Zeng F, Zhang X, Guo Y, Guo L. Long non-coding RNA HOTTIP promotes BCL-2 expression and induces chemoresistance in small cell lung cancer by sponging miR-216a. Cell Death Dis. 2018;9:85. doi: 10.1038/s41419-017-0113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu N, Liu B, Lian C, Doycheva DM, Fu Z, Liu Y, Zhou J, He Z, Yang Z, Huang Q, Zeng H, Guo H. Long noncoding RNA AC003092.1 promotes temozolomide chemosensitivity through miR-195/TFPI-2 signaling modulation in glioblastoma. Cell Death Dis. 2018;9:1139. doi: 10.1038/s41419-018-1183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Du P, Zhao H, Peng R, Liu Q, Yuan J, Peng G, Liao Y. LncRNA-XIST interacts with miR-29c to modulate the chemoresistance of glioma cell to TMZ through DNA mismatch repair pathway. Biosci Rep. 2017;37:BSR20170696. doi: 10.1042/BSR20170696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang P, Wang P, Sun X, Yuan Z, Zhan R, Ma X, Li W. Knockdown of long noncoding RNA H19 sensitizes human glioma cells to temozolomide therapy. Onco Targets Ther. 2016;9:3501–3509. doi: 10.2147/OTT.S96278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Xu N, Liu B, Huang Y, Zeng H, Yang Z, He Z, Guo H. Long noncoding RNA RP11-838N2.4 enhances the cytotoxic effects of temozolomide by inhibiting the functions of miR-10a in glioblastoma cell lines. Oncotarget. 2016;7:43835–43851. doi: 10.18632/oncotarget.9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An Q, Zhou L, Xu N. Long noncoding RNA FoxD2-AS1 accelerates the gemcitabine-resistance of bladder cancer by sponging miR-143. Biomed Pharmacother. 2018;103:415–420. doi: 10.1016/j.biopha.2018.03.138. [DOI] [PubMed] [Google Scholar]
- 16.Yang X, Duan B, Zhou X. Long non-coding RNA FoxD2-AS1 functions as a tumor promoter in colorectal cancer by regulating EMT and Notch signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21:3586–3591. [PubMed] [Google Scholar]
- 17.Bao J, Zhou C, Zhang J, Mo J, Ye Q, He J, Diao J. Upregulation of the long noncoding RNA FoxD2-AS1 predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Biomark. 2018;21:527–533. doi: 10.3233/CBM-170260. [DOI] [PubMed] [Google Scholar]
- 18.Chen G, Sun W, Hua X, Zeng W, Yang L. Long non-coding RNA FoxD2-AS1 aggravates nasopharyngeal carcinoma carcinogenesis by modulating miR-363-5p/S100A1 pathway. Gene. 2018;645:76–84. doi: 10.1016/j.gene.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 19.Shen F, Chang H, Gao G, Zhang B, Li X, Jin B. Long noncoding RNA FoxD2-AS1 promotes glioma malignancy and tumorigenesis via targeting miR-185-5p/CCND2 axis. J Cell Biochem. 2019;120:9324–9336. doi: 10.1002/jcb.28208. [DOI] [PubMed] [Google Scholar]
- 20.Villanueva MT. Cell signalling: stuck in the middle of chemoresistance and metastasis. Nat Rev Clin Oncol. 2012;9:490. doi: 10.1038/nrclinonc.2012.129. [DOI] [PubMed] [Google Scholar]
- 21.Ni W, Xia Y, Bi Y, Wen F, Hu D, Luo L. FoxD2-AS1 promotes glioma progression by regulating miR-185-5P/HMGA2 axis and PI3K/AKT signaling pathway. Aging (Albany NY) 2019;11:1427–1439. doi: 10.18632/aging.101843. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Li H, Chen L, Li JJ, Zhou Q, Huang A, Liu WW, Wang K, Gao L, Qi ST, Lu YT. miR-519a enhances chemosensitivity and promotes autophagy in glioblastoma by targeting STAT3/Bcl2 signaling pathway. J Hematol Oncol. 2018;11:70. doi: 10.1186/s13045-018-0618-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, Schwabe RF, Vahdat LT, Altorki NK, Mittal V, Gao D. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geldof AA, Rao BR. Doxorubicin treatment increases metastasis of prostate tumor (R3327-MatLyLu) Anticancer Res. 1988;8:1335–1339. [PubMed] [Google Scholar]