We demonstrate that metalloproteases in breast tumor microenvironments can inactivate reovirus. Our findings expose that tumor microenvironment proteases could have a negative impact on proteinaceous cancer therapies, such as reovirus, and that modification of such therapies to circumvent inactivation by tumor metalloproteases merits consideration.
KEYWORDS: mammalian orthoreovirus, metalloproteins, proteases, reovirus, σ1, tumor microenvironment, virus attachment, virus structure
ABSTRACT
Reovirus is undergoing clinical testing as an oncolytic therapy for breast cancer. Given that reovirus naturally evolved to thrive in enteric environments, we sought to better understand how breast tumor microenvironments impinge on reovirus infection. Reovirus was treated with extracellular extracts generated from polyomavirus middle T-antigen-derived mouse breast tumors. Unexpectedly, these breast tumor extracellular extracts inactivated reovirus, reducing infectivity of reovirus particles by 100-fold. Mechanistically, inactivation was attributed to proteolytic cleavage of the viral cell attachment protein σ1, which diminished virus binding to sialic acid (SA)-low tumor cells. Among various specific protease class inhibitors and metal ions, EDTA and ZnCl2 effectively modulated σ1 cleavage, indicating that breast tumor-associated zinc-dependent metalloproteases are responsible for reovirus inactivation. Moreover, media from MCF7, MB468, MD-MB-231, and HS578T breast cancer cell lines recapitulated σ1 cleavage and reovirus inactivation, suggesting that inactivation of reovirus is shared among mouse and human breast cancers and that breast cancer cells by themselves can be a source of reovirus-inactivating proteases. Binding assays and quantification of SA levels on a panel of cancer cells showed that truncated σ1 reduced virus binding to cells with low surface SA. To overcome this restriction, we generated a reovirus mutant with a mutation (T249I) in σ1 that prevents σ1 cleavage and inactivation by breast tumor-associated proteases. The mutant reovirus showed similar replication kinetics in tumorigenic cells, toxicity equivalent to that of wild-type reovirus in a severely compromised mouse model, and increased tumor titers. Overall, the data show that tumor microenvironments have the potential to reduce infectivity of reovirus.
IMPORTANCE We demonstrate that metalloproteases in breast tumor microenvironments can inactivate reovirus. Our findings expose that tumor microenvironment proteases could have a negative impact on proteinaceous cancer therapies, such as reovirus, and that modification of such therapies to circumvent inactivation by tumor metalloproteases merits consideration.
INTRODUCTION
Mammalian orthoreovirus serotype 3 (reovirus T3D, for type 3 Dearing) has been known for some time to possess oncolytic activity and is being investigated as a treatment against numerous cancers (1–3). In immunocompetent hosts, reovirus is asymptomatic and therefore a safe candidate for oncolytic therapy, showing minimal side effects in numerous human clinical trials (4–8). Reovirus oncotherapy shows promise in, for example, improving overall survival from 10.4 months to 17.4 months when combined with paclitaxel in an ongoing breast cancer trial (9). Nevertheless, reovirus therapy fails to cure the majority of immunocompetent tumor-bearing animals and human patients (10–13), indicating that we must better understand the restrictions to reovirus treatment and ways to improve the potency of this therapeutic agent.
We posited whether the heterogeneity of tumor environments could contribute to differential activity of reovirus among tumors. The natural route of infection for reovirus is predominantly enteric, and consequently reovirus has evolved to be exquisitely well adapted to the human gastrointestinal tract. For example, reovirus evolved a very stable double-layered proteinaceous capsid that withstands harsh conditions of the environment and enteric tract. The outer capsid is composed of structural proteins μ1 and σ3 and a third protein, σ1, that protrudes from viral vertices and facilitates cell attachment. The inner capsid core is composed of additional structural proteins, transcription factors, and the double-stranded RNA genome (14–16). Importantly, to mediate efficient disassembly of the outer capsid, which is required for reovirus to establish infection, reovirus exploits the gut proteases chymotrypsin and trypsin. Specifically, these proteases generate infectious subvirion particles (ISVPs) by degrading the outermost capsid protein σ3 and cleaving the underlying μ1 protein into multiple fragments that promote membrane penetration and entry of reovirus particles into the cell (17–20). Membrane penetration results in disassociation of the remaining μ1 outer capsid fragments and σ1 (21), delivering transcriptionally competent core particles. In the absence of trypsin or chymotrypsin, reovirus conversion to ISVPs can still occur within endolysosomes, mediated by lysosomal cathepsins B and L (22). The dependency of reovirus on gut and lysosomal proteases provoked important questions for reovirus infectivity in tumors: what sort of proteases are in tumor environments, and do they support reovirus uncoating?
The tumor microenvironment presents a distinct set of proteases relative to the natural enteric route of reovirus infection. Although chymotrypsin and trypsin are restricted to the gut, tumors are rich in other proteases, such as various cathepsins and metalloproteases, which have been shown to play important roles in cancer progression (23–25). For example, tumor invasiveness and metastasis require degradation of the extracellular matrix (ECM), which is facilitated by proteolytic pathways involving multiple families of proteases (26, 27). Cathepsin B is classically considered a lysosomal protease; however, extracellular and pericellular translocation of the enzyme was observed in cancers such as breast and brain cancers and can correlate with poor prognosis (23, 28–31). Cathepsin B is implicated in the activation of other ECM degradation-associated proteases, such as urokinase plasminogen activator, which, in turn, proteolytically activates numerous metalloprotease downstream targets that contribute to ECM degradation (32). Whether tumor proteases can target reovirus proteins, and whether they are in sufficient concentration to modulate reovirus infectivity, is unknown.
The current study aimed to determine if breast tumor extracellular proteases have any effect on reovirus, and if so, whether the effect is positive, negative, or neutral toward reovirus infectivity of tumor cells. We prepared tumor extracellular extracts (T.E.E.) from murine breast tumors to mimic the tumor microenvironment and then treated reovirus with T.E.E. under various conditions. The three key discoveries of our study are (i) T.E.E. contains zinc-dependent metalloprotease activity that cleaves the cell attachment protein σ1 and (sometimes) σ3 yet does not produce the beneficial cleavage of μ1C to δ, leading to a 100-fold reduction of reovirus infectivity in many cell types; (ii) T.E.E proteases reduce binding of reovirus to cancer cells that have low sialic acid (SA) levels, because cleavage of σ1 removes the JAM-A binding capacity and forces a dependence on sialic acids for attachment; and (iii) inactivation of reovirus by breast tumor proteases can be circumvented by mutation of the σ1 protein.
(This article was submitted to a preprint archive [33].)
RESULTS
Breast tumor extracellular protease(s) cleave reovirus σ1 protein but do not produce ISVPs.
The fate of reovirus when exposed to intestinal digestive enzymes is well characterized. Degradation of outer capsid protein σ3 and cleavage of μ1C to δ generates infectious subviral particles (ISVPs) with the ability to directly penetrate membranes, thereby enhancing infectivity (19, 34, 35). For some strains of reovirus, the cell attachment protein σ1 is also cleaved in half, maintaining the sialic acid (SA)-binding σ1-tail fragment (σ1N), which permits attachment to the sialic acid-rich luminal face of intestinal cells. However, with reovirus currently in phase III clinical trials as an oncolytic therapy, it is important to know whether tumors also release proteases that digest reovirus proteins and, if so, whether tumor-associated proteases have positive, negative, or neutral effects on reovirus infection of tumor cells.
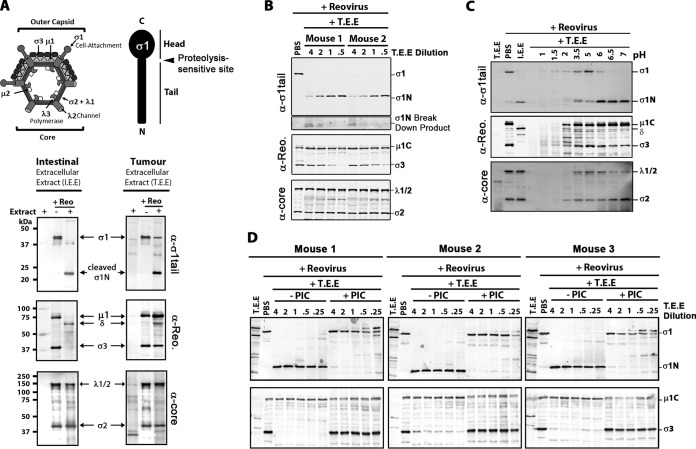
To determine the effects of tumor extracellular proteases on reovirus, we generated protease-enriched tumor extracellular extract (T.E.E.) from polyomavirus middle T-antigen-derived mouse breast tumors by diffusion of extracellular content into phosphate-buffered saline (PBS). We then exposed purified reovirus to T.E.E. and monitored reovirus protein processing by Western blotting. As a positive control for protease-mediated reovirus uncoating, we also exposed purified reovirus to intestinal extracellular extract (I.E.E.) prepared by flushing murine intestines with PBS. As anticipated, I.E.E. treatment resulted in hallmarks of ISVP formation, including degradation of outer capsid protein σ3 and cleavage of μ1C to δ (Fig. 1A). I.E.E. treatment also led to cleavage of σ1 to the σ1N-tail fragment. Despite processing of the outer capsid, the inner core proteins λ1/2 and σ2 remained intact; this was anticipated, since the reovirus core becomes transcription competent and does not further uncoat but rather synthesizes and expels positive-sense RNA into the cytoplasm during virus replication (36). Importantly, following treatment with T.E.E., σ1 was cleaved to the 22-kDa σ1N fragment. Cleavage of outer capsid μ1, which is a hallmark of ISVP generation, was, however, limited. Core viral proteins were unaffected by T.E.E. treatment. The effects of T.E.E. were therefore distinct from those of I.E.E., with T.E.E. causing cleavage of σ1 but failing to generate ISVPs.
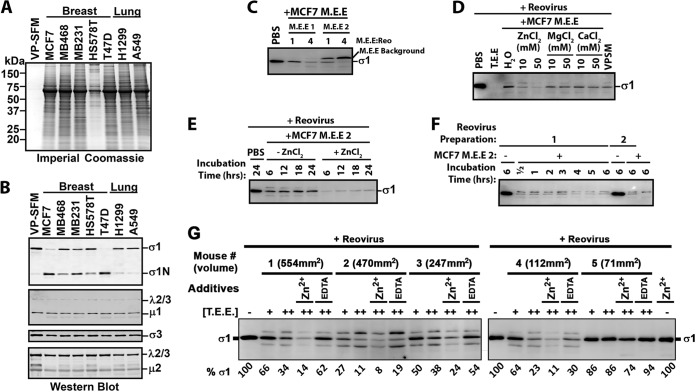
FIG 1.
Breast tumor extracellular protease activity cleaves reovirus σ1 protein but does not produce ISVPs. Extracellular contents of murine breast tumors (tumor extracellular extract, or T.E.E.) were used to mimic the tumor microenvironment. To mimic the intestinal environment, the protease-rich luminal contents were obtained from murine intestines (intestinal extracellular extract, or I.E.E.). (A, top) Diagrammatical depiction of reovirus outer capsid and core proteins, along with a diagram of σ1 tail and head domains. (Bottom) Reovirus (Reo) was treated with T.E.E. or I.E.E. at 37°C for 24 h, and viral proteins were subjected to Western blot analysis using antibodies against σ1-tail, outer capsid proteins (polyclonal antireovirus serum), or viral core proteins (polyclonal antireovirus core serum). (B) As described for panel A, but the activity of T.E.E. from two additional and independent murine breast tumors was assessed. (C) Reovirus was treated with T.E.E. under different pH conditions, ranging from pH 1 to pH 7. (D) Reovirus was treated with T.E.E. from 3 murine breast tumors in the absence or presence of protease inhibitor cocktail (PIC).
Proteolytic processing of reovirus outer capsid proteins can depend on the concentration of proteases; therefore, we assessed the fate of reovirus proteins by Western blotting after exposing purified reovirus particles to increasing amounts of T.E.E. (Fig. 1B). Furthermore, to determine if reovirus processing was consistent among distinct T.E.E. samples, we obtained T.E.E. from two independent mouse breast tumor explants. Cleavage of σ1 to σ1N was observed for T.E.E. obtained from both mice in a concentration-dependent manner. Specifically, while all T.E.E. concentrations effectively cleaved σ1 to σ1N, high T.E.E.-to-reovirus ratios caused further degradation of the σ1N 22-kDa fragment. While core proteins were unaffected by T.E.E., outer capsid σ3 was degraded under low ratios of T.E.E. to reovirus. This could be a consequence of protease inactivation at high concentrations due to self-cleavage or cleavage by other proteases. Notably, however, under no condition was μ1C processed to δ, suggesting that T.E.E. factors from multiple mouse tumors did not yield ISVPs. This is the first time, to our knowledge, that processing of reovirus by T.E.E. has been characterized.
Since breast tumors have slightly acidic environments (37, 38), we hypothesized that incomplete processing of reovirus σ3 or μ1C by T.E.E. could be a result of poorly optimized pH conditions. Therefore, reovirus was treated with T.E.E. at a range of pH conditions (Fig. 1C). At highly acidic pH (≤2), reovirus particles were degraded completely, as seen by the absence of all viral proteins, including core proteins λ1 and σ2. Cleavage of σ1 was most efficient above pH 6, with complete conversion of σ1 to σ1N. Importantly, under no conditions that preserved reovirus core proteins (pH ≥ 2) was μ1C processed to δ. Overall, it seems that the optimal pH for reovirus processing was 6 to 7, consistent with the environmental pH of most tumors (39, 40).
To confirm that processing of reovirus proteins by T.E.E. was attributed to a protease, reovirus was treated with T.E.E. from three independent mouse tumors but in the presence or absence of a protease inhibitor cocktail (PIC) (Fig. 1D). Processing of σ1 and σ3 was strongly impaired in the presence of the PIC at all T.E.E./reovirus ratios tested, indicating that a secreted protease in mouse breast tumors is involved in cleavage of σ1 and occasional degradation of σ3.
Breast tumor extracellular protease(s) reduces reovirus attachment and decreases infectivity by 100-fold.
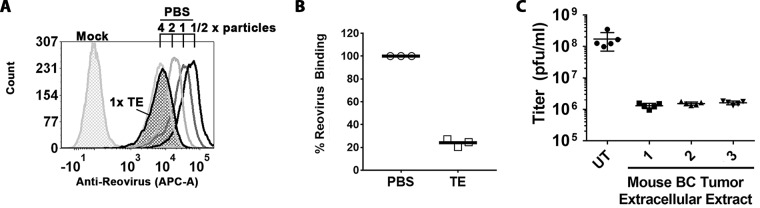
Having discovered that T.E.E. cleaves σ1 but does not produce ISVPs, it became important to determine whether this partial processing of reovirus by T.E.E. affects infectivity. Specifically, the cleavage of σ1 could be an important impediment to virus-cell attachment. Normally reovirus σ1 mediates attachment to cells through two domains: the head domain of σ1 mediates high-affinity interaction with junction adhesion molecules (JAM-A), while the tail domain of σ1 supports association with α2,3-, α2,6-, or α2,8-linked sialylated glycans (41). In the intestine, the luminal surface of intestinal cells displays SA but not JAM-A. Therefore, cleavage of σ1 and removal of the JAM-binding head of reovirus by gut proteases trypsin and chymotrypsin is unlikely to impede virus binding to the SA-rich and JAM-A-deficient luminal face of intestinal epithelial cells, but JAM-A binding may instead be necessary to mediate attachment at the JAM-rich basolateral side (42–44) of the intestinal epithelium, where absence of proteases would maintain full-length σ1. Distinct from gut epithelial cells, the levels of SA and JAM-A vary among tumor cells (45–49). Accordingly, we queried whether cleavage of σ1 to σ1N and consequential loss of JAM binding affected attachment of reovirus to nonepithelial cells. Equivalent particles of PBS- or T.E.E-treated reovirus were exposed to tumorigenic L929 mouse fibroblast cells at 4°C to permit attachment without entry. Cell-associated particles were then enumerated by flow cytometric analysis using antireovirus antibodies. A serial dilution of PBS-treated particles was used for quantitative purposes and demonstrated a linear relationship between particle number and cell attachment (Fig. 2A). T.E.E. treatment reduced binding of reovirus to L929 cells by ∼80% (Fig. 2B).
FIG 2.
Breast tumor extracellular protease activity reduces reovirus infectivity of L929 cells by 100-fold. (A) Attachment to L929 cells assessed by flow cytometry. Cells were exposed to PBS (mock) or reovirus treated with PBS or Tris-EDTA (TE) at equivalent doses (1× particles) for 1 h at 4°C, washed, and stained with antireovirus and fluorescence-conjugated anti-rabbit antibodies. One-half serial dilutions of PBS-treated reovirus show a linear relationship between mean fluorescence intensity and virus amount and subsaturation of conditions. (B) As described for panel A but repeated in 3 independent experiments. To determine relative binding, the MFI of the peak from TE-treated T3D at 1× dilution was extrapolated based on the standard curve generated for serial dilutions of PBS-treated T3D. P < 0.0001 by Student t test. BC, breast cancer. (C) Reovirus was treated with PBS or T.E.E. as described for Fig. 1 and subjected to plaque titration on L929 cells. The titer (PFU/ml) is presented for three independent mouse tumors, each treated with PBS or T.E.E. five independent times. P < 0.0001 between untreated (UT) and T.E.E. samples, determined by analysis of variance (ANOVA) statistical analysis and Dunnett’s multiple-comparison test.
To determine the impact of T.E.E. on infectivity, we then treated reovirus with T.E.E. or PBS and compared infectious titers (PFU) on L929 cells commonly used to propagate reovirus. T.E.E. from three independent mouse tumors reduced reovirus titers by ∼100-fold relative to the mock (PBS) treatment (Fig. 2C). This finding was striking, as it suggests that tumor proteases pose a barrier to reovirus oncolytic potency by reducing virus infectivity.
Mechanistic explanation for loss of infectivity: truncation of σ1 reduces binding by 100-fold in sialic acid-low cells.
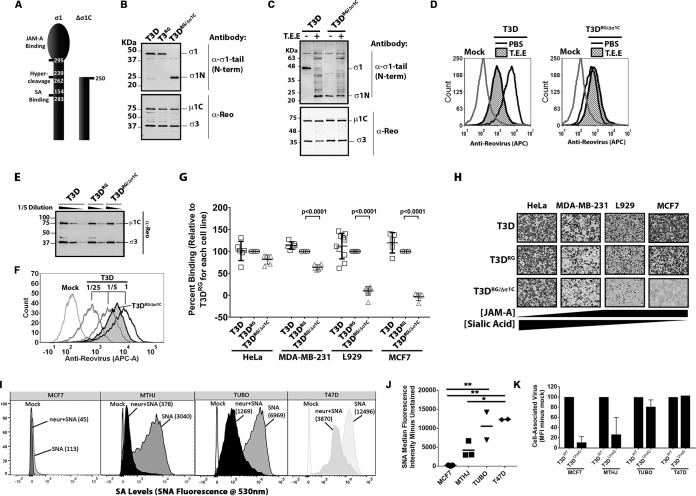
Since T.E.E. treatment not only caused cleavage of σ1 but also sometimes caused degradation of σ3 and perhaps other effects on protein conformations, it was important to directly determine if σ1 cleavage was necessary and sufficient for decreased reovirus binding. We therefore used reverse genetics to generate reovirus particles with truncated σ1N fibers (Fig. 3A, T3DRG/Δσ1C) that mimic σ1-cleavage by T.E.E. treatment without effects on σ3. We also generated wild-type reovirus (T3DRG) by reverse genetics to control for potential secondary effects of the reverse genetics system. In all experiments, T3D and T3DRG behaved the same, demonstrating that reverse genetics-generated virions faithfully recapitulate the parental phenotype. Western blot analysis of CsCl-purified virus showed that T3DRG/Δσ1C had truncated σ1N while T3D and T3DRG had full-length σ1 (Fig. 3B). All three viruses had equivalent relative levels of μ1C and σ3; therefore, any differences between these viruses can be attributed to σ1 status.
FIG 3.
Truncation of σ1 reduces binding of reovirus to sialic acid-low cells by 100-fold. (A) Depiction of σ1 truncation in T3DRG/Δσ1C. (B) Western blot analysis of σ1 and outer capsid proteins of wild-type viruses T3D and T3DRG versus mutant reovirus T3DRG/Δσ1C, showing that T3DRG/Δσ1C contains truncated σ1N. (C) T3D and T3DRG/Δσ1C were treated with PBS or T.E.E., similar to procedures described for Fig. 1, and subjected to Western blot analysis for cleaved or truncated σ1 versus other capsid proteins. (D) Attachment of T3D and T3DRG/Δσ1C treated with PBS or T.E.E. to L929 cells assessed by flow cytometry, as described for Fig. 2A. The image is representative of 3 independent experiments. (E) Western blot analysis was used to confirm that at the same dilution, there were equivalent levels of virus proteins and therefore equivalent numbers of T3D-, T3RG-, and T3DRG/Δσ1C-purified viruses used for binding assessment. (F) Example of flow cytometry to quantify attachment of T3D and T3DRG/Δσ1C to MDA-MB-231 cells. To determine relative binding of T3DRG/Δσ1C to T3D, the MFI of the peak from T3DRG/Δσ1C at 1× dilution produced was extrapolated based on the standard curve generated for T3D. (G) Binding assay with T3DPL and T3RG versus T3DRG/Δσ1C reovirus on HeLa, MDA-MB-231, L929, and MCF7 cells. MFI was set to 100% for T3DRG on a given cell line, and remaining virus MFIs are presented as a percentage of binding relative to that of T3DRG. Results depict five independent experiments, with 2-way ANOVA statistical analysis and Dunnett’s multiple-comparison test. (H) Infectivity of T3D, T3RG, and T3DRG/Δσ1C in cancer cell lines with different expression levels of JAM and SA, as indicated. Reovirus infection was detected by immunocytochemistry using a polyclonal antireovirus serum. Panels shown are representative of two independent experiments. (I) Sialic acid surface expression detected with fluorophore-labeled SNA for MCF7, MTHJ, TUBO, and T47D breast cancer cells. Neuraminidase (neur) treatment versus untreated cells was used to confirm specificity of SNA staining for sialic acids. (J) Mean fluorescence intensity for SNA staining relative to that of unstained controls (SNA minus mock), similar to that shown in panel I, for 2 to 3 independent experiments. *, P < 0.05; **, P < 0.001; both determined by ANOVA with Tukey’s multiple-comparison test. (K) Mean fluorescence intensity measures the relative particle attachment for T3D versus T3DRG/Δσ1C on MCF7, MTHJ, TUBO, and T47D breast cancer cells. The calculations, similar to those shown in panel G, are from 2 independent experiments.
If cleavage of σ1 by T.E.E. alone was sufficient for reduced binding and infectivity observed in Fig. 2, then we expected that T3DRG/Δσ1C should show (i) reduced binding to L929 cells and (ii) should not be affected by T.E.E. treatment. Equivalent particles of T3DRG/Δσ1C versus T3D were therefore assessed for cell attachment in the presence or absence of T.E.E treatment using flow cytometric analysis (Fig. 3C and D). First, compared to that of T3D, T3DRG/Δσ1C exhibited reduced attachment that mimicked levels observed with T3D treated with T.E.E. Second, while T3D binding reduced following T.E.E. treatment, attachment of T3DRG/Δσ1C to L929 cells was unaffected. The data implicate cleavage of σ1 as the dominant contributing mechanism for reduced binding of reovirus following T.E.E. treatment.
Attachment efficiency then was tested on a panel of cancer cell lines that differentially express surface JAM and SA; for example, human cervical carcinoma (HeLa) cells and human breast cancer (MDA-MB-231) cells were previously shown to have low JAM but high SA surface expression, while L929 cells and the human breast cancer cell line MCF-7 have low SA expression (49, 50). Figure 3E shows a representative standardization of virus particles, and Fig. 3F is a representative analysis of T3D versus T3DRG/Δσ1C binding to MDA-MB-231. The binding assay was then repeated in five independent experiments and mean fluorescent intensity (MFI) for a given cell line normalized to that of T3DRG (Fig. 3G). T3DRG/Δσ1C exhibited a statistically significant reduction (∼100-fold) in binding to SA-low L929 and MCF7 cells relative to that of control viruses, suggesting that loss of the σ1 head domain is sufficient to make reovirus dependent on SA for binding. Moreover, eliminating the JAM-binding σ1 domain in T3DRG/Δσ1C caused reduced infectivity (i.e., frequency of cells stained positive for reovirus antigen) toward SA-low L929 and MCF-7 cells but not toward SA-high HeLa cells (Fig. 3H).
The relationship between SA levels and infectivity of reovirus with truncated σ1 was important to validate, as it suggests that not only the protease status of tumors but also the SA status of the tumor cells can impact reovirus infectivity. We therefore tested two mouse (MTHJ and TUBO) and two human (MCF7 and T47D) breast cancer cell lines for levels of SA versus reovirus attachment. Levels of SA were measured by flow cytometry using fluorescence-labeled Sambucus nigra lectin (SNA). Specificity of fluorescence for SA was confirmed by pretreatment of cells with neuraminidase (Fig. 3I). Note that in all cases, neuraminidase strongly reduced SNA labeling but did not abolish signal completely, which was expected because neuraminidase activity is rarely complete. The four breast cancer cell lines varied dramatically in SA levels, with MCF7 representing minimal SA and T47D maximal SA levels relative to the rest (Fig. 3J). Importantly, T3DRG/Δσ1C exhibited reduced binding relative to that of T3D only when SA levels were low (Fig. 3K, MCF7 and MTHJ cells). Together, the findings strongly support that truncation of σ1 reduces attachment of reovirus toward SA-low cells.
Reovirus-inactivating breast cancer proteases are metalloproteases.
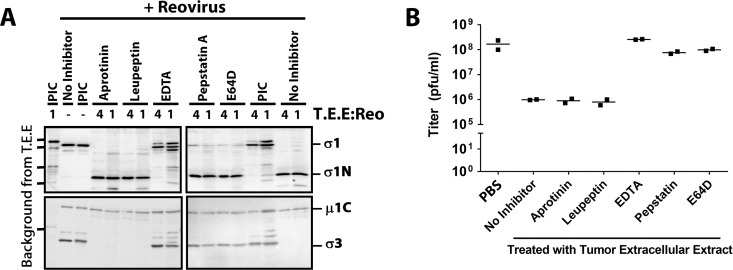
Considerable research has demonstrated that tumor environments are rich in proteases of all classes and that proteases can impact the fate of tumor growth and metastasis (25, 51). To elucidate the class of protease(s) present in T.E.E. that acts on σ1 and σ3, reovirus was treated with T.E.E. in the presence of protease inhibitors that target specific classes of proteases. Aprotinin, leupeptin, pepstatin A, and E64D were specifically used to inhibit serine, cysteine/serine/threonine, aspartyl, and cysteine proteases, respectively (Fig. 4A). Since metalloprotease activity depends on metals as cofactors (52, 53), EDTA was used to chelate metal ions and therefore inhibit metalloproteases. The PIC was able to impair cleavage of σ1 and degradation of σ3, as demonstrated in Fig. 1D. Neither aprotinin nor leupeptin was capable of impairing σ1 cleavage and σ3 degradation. Interestingly, pepstatin A and E64 very minimally impaired σ1 cleavage but strongly impaired degradation of σ3. EDTA most drastically impaired proteolysis of both σ1 and σ3, suggesting that metal ions are involved, and the dominant protease is a metalloprotease.
FIG 4.
Breast cancer proteases that inactivate reovirus are metalloproteases. (A and B) Reovirus was treated with 1× T.E.E. (+) in the presence of various protease inhibitors (as indicated) and analyzed by Western blotting as described for Fig. 1 (representative of two independent experiments) (A) or by plaque titration on L929 cells as described for Fig. 2 (two independent experiments) (B).
We next examined which protease inhibitors could reverse the loss of reovirus infectivity caused by T.E.E. treatment (Fig. 4B). Plaque titration was conducted on L929 cells similar to that shown in Fig. 2. As seen previously, exposure of reovirus to T.E.E. caused a 100-fold decrease in infectious titers. Neither aprotinin nor leupeptin was capable of rescuing infectivity, as expected given their inability to prevent σ1 cleavage. Pepstatin A and E64 treatments partially rescued infectivity, which correlates with their ability to partially prevent cleavage of σ1. We and others previously demonstrated that although reovirus particles can hold up to 12 σ1 fibers, 3 or more fibers are necessary and sufficient to mediate virus attachment (54, 55). Accordingly, the maintenance of some full-length σ1 during pepstatin A and E64 treatment (Fig. 4A) explains why infectious titers remain high (Fig. 4B). EDTA was able to fully rescue infectivity, reinforcing that metal ions play a key role in T.E.E.-mediated proteolysis of reovirus σ1 and subsequent loss of infectivity toward SA-low cells.
Together, the data suggest that pepstatin A- and E64-sensitive aspartyl and cysteine proteases (respectively) partially contribute to σ1 cleavage, but that metalloproteinase(s) are the main culprits in cleaving σ1 and reducing reovirus infectivity toward SA-low cells.
Breast cancer cells secrete proteases capable of cleaving σ1.
Proteases in the tumor microenvironment can come from multiple sources, such as from tumor cells, supporting fibroblasts, or innate and adaptive immune cells (25). To determine if tumor cells directly release a protease that cleaves reovirus σ1, we collected the conditioned media from a panel of cancer cell lines, filtered and concentrated the medium 10× using 10-kDa-cutoff Centricon columns, and exposed reovirus to these medium extracellular extracts (M.E.E.). The rationale for concentrating the medium was that cell culturing artificially dilutes extracellular factors by at least 100-fold and therefore can strongly underestimate factor activity. For example, we cultured 106 cells in 1 ml of medium, while T.E.E. and I.E.E. were collected in 1 ml for more than 108 cells in tumors (56), which by itself is a dilution of factors that would naturally be more concentrated. Therefore, we initially concentrated M.E.E. by 10-fold, and since we noted activity, we did not pursue further concentration. Also key to the success of this assay was the use of virus production serum-free medium (VP-SFM) to overcome the quenching effects of fetal bovine serum (FBS) proteins on protease activity. Also, the medium was collected after 3 days of culture, when cell death (i.e., floating cells) was minimal. The VP-SFM was largely clear of proteins, as visualized by SDS-PAGE electrophoresis and Coomassie staining (Fig. 5A). Conversely, M.E.E. from 5 breast (MCF-7, MDA-MB-468, MDA-MB-231, Hs578T, and T-47D) and 2 lung (H1299 and A549) cancer cell lines contained a spectrum of secreted proteins (Fig. 5A).
FIG 5.
Breast cancer cells secrete proteases capable of cleaving σ1. (A) Medium extracellular extract (M.E.E.) collected from breast and lung cancer cell lines (as indicated) was subjected to SDS-PAGE gel electrophoresis and Coomassie staining to visualize total secreted proteins. (B) Reovirus was treated with M.E.E. from the indicated cell lines for 24 h at 37°C and subjected to Western blot analysis as described for Fig. 1 to monitor the fates of reovirus proteins. Results are representative of 7 independent experiments and 2 to 4 independent M.E.E. extractions from each cell line. (C) Focusing on MCF-7 cells, the reproducibility of M.E.E. activity on reovirus σ1 was evaluated by treatment of reovirus with two additional and independently obtained MCF7 M.E.E. solutions (M.E.E. 1 and M.E.E. 2) at two different ratios of M.E.E to Reo, and preservation of full-length σ1 was assessed by Western blotting. (D) To establish which ions contributed to proteolysis of σ1 by MCF-7 M.E.E., reovirus was treated with MCF-7 M.E.E. in the absence or presence of ZnCl2, MgCl2, or CaCl2 at the indicated concentrations, and preservation of full-length σ1 was assessed by Western blotting. (E) To determine how rapidly reovirus σ1 is digested by MCF-7 M.E.E., reovirus was treated with MCF-7 M.E.E. in the absence or presence of ZnCl2 for 6, 12, 18, and 24 h, and preservation of full-length σ1 was assessed by Western blotting. (F) To ensure that susceptibility to MCF-7-mediated digestion of σ1 was independent of the reovirus preparation, two independently purified reovirus stocks were treated with MCF-7 M.E.E. or PBS for 0.5 to 6 h (as indicated), and preservation of full-length σ1 was assessed by Western blotting. (G) MCF7 tumors implanted into NSG mice were excised after 45 days of in vivo tumor growth, and T.E.E. was prepared. Reovirus was treated with 1× and 2× T.E.E. or 2× T.E.E in the presence of EDTA or ZnCl2 and subjected to Western blot analysis. Densitometric analysis of full-length σ1 is provided below the blot relative to that of PBS-treated reovirus.
When treated with M.E.E. from all 5 breast cancer cell lines, reovirus σ1 underwent cleavage to σ1N (Fig. 5B), but limited change was observed for μ1C. MCF7 M.E.E. collections from two independent cultures showed cleavage of σ1 to different extents, suggesting that the concentration of proteases varies between cultures (Fig. 5C). Overall, breast cancer cell M.E.E. recapitulated the effects of T.E.E., indicating that tumor cells alone can secrete proteases that target reovirus σ1. Our findings of course do not eliminate the possibility that supporting fibroblasts or innate and adaptive immune cells in tumor microenvironments also contribute additional reovirus-inactivating proteases. It was interesting that neither lung cancer cell M.E.E. cleaved reovirus σ1. Although it is tempting to speculate that tumor cell type (e.g., breast versus lung) influences differential ability to inactivate reovirus through secreted proteinases, a larger panel of lung cancer cells would be essential to make such an interpretation.
Reovirus was also treated with MCF-7 M.E.E. in the absence or presence of ZnCl2, MgCl2, or CaCl2 to determine which metal ion promoted σ1 cleavage. With this stock of MCF-7 M.E.E., σ1 was completely degraded rather than producing a stable σ1N fragment, as was described with high T.E.E. concentrations (Fig. 1B); therefore, we focused on the loss of full-length σ1 as our readout. Importantly, degradation of σ1 was reproducibly increased in the presence of Zn2+, implicating a Zn-dependent metalloprotease (MP) in reovirus processing (Fig. 5D). A time course analysis showed that σ1 proteolysis occurred rapidly by M.E.E. (Fig. 5E), reaching maximum loss of full-length σ1 by 6 h of treatment, with Zn2+ again promoting σ1 processing. A more detailed time course analysis shows maximal proteolysis achieved already by 30 min of exposure of two independent preparations of reovirus to MCF-7 M.E.E. (Fig. 5F). Together, the data suggest that breast cancer cells can themselves secrete Zn-dependent metalloproteases that cleave σ1 with rapid kinetics.
Finally, we determined if xenograft tumors of MCF7 cells in severely immunocompromised NOD scid gamma mice (lacking mature T cells, B cells, and natural killer cells) produced a tumor microenvironment capable of cleaving reovirus σ1 (Fig. 5G). At 45 days postimplantation, excised tumors were rinsed twice in PBS, cut into 4 pieces, and incubated at 4°C in 1 ml (total) PBS for 2 h to diffuse extracellular content. These tumor extracellular extracts (T.E.E.) were clarified by centrifugation and 0.45-μm filtration. Reovirus was then exposed to the T.E.E.s and loss of full-length σ1 was monitored by Western blotting (Fig. 5G). Degradation of σ1 was T.E.E. dose dependent and varied from ∼10 to 90% depending on the tumor. Four of the five T.E.E.s showed >60% cleavage. Moreover, the smallest tumor exhibited limited cleavage activity, although more samples would be required to determine if tumor size correlates with cleavage potential. Since cleavage was increased in the presence of Zn2+ and decreased in the presence of EDTA, a Zn-dependent metalloprotease is likely functioning in these tumors. However, since EDTA treatment did not completely prevent cleavage, it is also possible that additional ion-independent σ1-degrading proteases were active in MCF7 tumors. These experiments confirm the presence of σ1-cleaving metalloproteases in tumors in vivo and in the absence of T, B, or natural killer (NK) cell sources.
Mutation of the σ1 neck domain overcomes reovirus cleavage by breast cancer metalloprotease.
Given that proteolysis of reovirus σ1 by Zn-dependent metalloproteases secreted by human breast cancer cells and present in mouse breast cancer tumors diminished reovirus binding and infection toward SA-low tumor cells, we next sought to modify reovirus σ1 to overcome cleavage. Proteolysis of reovirus by intestinal proteases chymotrypsin and trypsin was previously shown to occur in the flexible protease-hypersensitive neck region (residues 219 to 264) between the tail and the head domains of σ1 (57, 58) (Fig. 6A). We reasoned that if tumor-associated metalloproteases also cleaved in the σ1 neck domain, then the σ1N fragments generated by T.E.E., I.E.E., trypsin, and chymotrypsin should share a similar molecular weight. Indeed, the tail (σ1N) and head (σ1C) fragments detected by Western blotting with σ1N- and σ1C-specific antibodies (respectively) were similar when reovirus was treated with T.E.E., I.E.E., trypsin, or chymotrypsin (Fig. 6B). The shorter fragments of σ1 under some conditions of I.E.E. and chymotrypsin treatment likely reflect further cleavage at the extreme N terminus as described previously (58, 59). Importantly, since chymotrypsin and trypsin cleavage sites are six amino acids apart but the size of their σ1 cleavage fragments are not resolved by our assay, we can infer that the tumor-associated MP cleaves in the same general vicinity as the gut proteases.
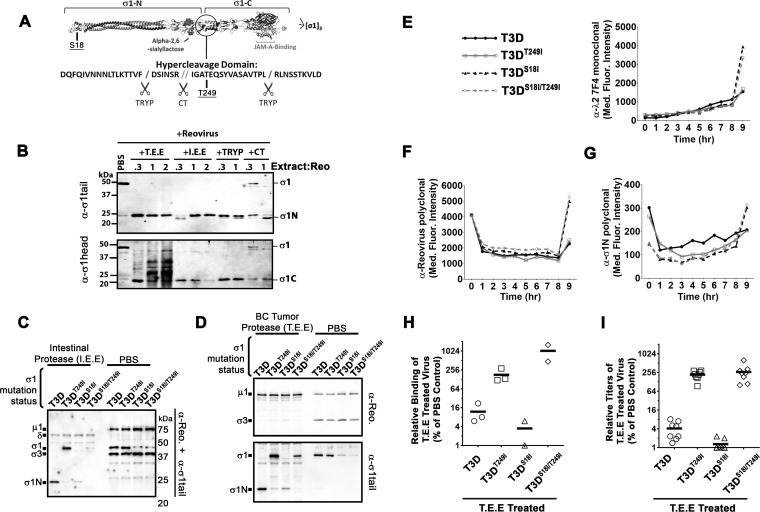
FIG 6.
Mutation in the σ1 neck domain can overcome σ1 proteolysis by breast cancer metalloprotease. (A) Diagrammatic depiction of σ1 with the protease-hypersensitive neck domain. (B) Reovirus was treated with either T.E.E., I.E.E., chymotrypsin (CT), or trypsin (TRYP) for 24 h at 37°C and subjected to Western blot analysis with tail (top)- or head (bottom)-specific antibodies. (C and D) CsCl-purified T3D, T3DT249I, T3DS18I, or T3DS18I/T249I was treated with PBS or with I.E.E. (C) or T.E.E. (D) for 24 h at 37°C. Western blot analysis with both polyclonal antireovirus antibodies and σ1N-specific antibodies demonstrates the levels of full-length σ1 and σ1N. (E to G) Reovirus infection dynamics of T3D, T3DT249I, T3DS18I, and T3DS18I/T249I viruses. Flow cytometry was used to evaluate expression of reovirus proteins λ2 (E), σ3 (F), and σ1N (G), from 0 to 8 h postinfection. (H) Binding assay as described for Fig. 3 with reovirus mutant T3D, T3DT249I, T3DS18I, or T3DS18I/T249I treated with T.E.E. on L929 cells. (I) Plaque titration of T.E.E.-treated reovirus mutants (T3D, T3DT249I, T3DS18I, or T3DS18I/T249I) as described for Fig. 2.
It was previously observed that a change from threonine to isoleucine at position 249 of σ1 can prevent cleavage by both chymotrypsin and trypsin despite their different cleavage locations in the neck domain (57). These findings suggested that the T249I modification eliminated cleavage susceptibility by altering the secondary structure of the neck domain, thereby altering the exposure of the hypercleavage domain. Accordingly, we tested if mutation of T249 to isoleucine could also prevent cleavage by tumor-associated metalloproteases. Using reverse genetics, we introduced the T249I mutation into T3D and assessed the fate of σ1T249I after treatment with I.E.E. (Fig. 6C) or T.E.E. (Fig. 6D). The T249I mutation successfully impeded cleavage of σ1 by both I.E.E. and T.E.E.
Our previous studies showed that a mutation in the domain that anchors σ1 in virions, σ1-S18I, reduces the number of σ1 fibers per reovirus particle to ∼4 (instead of 12 on wild-type T3D). We and others further showed that 3 σ1 trimers were sufficient to allow maximal binding to L929 and other tumorigenic cells. Moreover, having 4 to 7 (but fewer than 12) σ1 trimers promotes uncoating of σ1 during virus entry into tumor cells and thereby increases reovirus oncolysis in vitro and in vivo (13, 54). Having now learned that σ1 undergoes cleavage by breast tumor-associated metalloproteases, we reasoned that having fewer σ1 fibers would make T3DS18I hypersensitive to tumor-associated protease inactivation; in other words, that maintaining full-length σ1 would become less likely if there were fewer σ1 fibers to begin with. We therefore also incorporated the T249I mutation into T3DS18I to generate a double mutant, T3DS18I/T249I. As expected, both T3DS18I and T3DS18I/T249I showed lower σ1 levels than T3D or T3DT249I (Fig. 6C and D). Importantly, however, while σ1 of T3DS18I was cleaved by I.E.E. and T.E.E., the σ1 T3DS18I/T249I was refractory to cleavage by either extracellular extract. In summary, localizing the σ1 cleavage site to the neck domain, and subsequently introducing a T249I mutation, allowed us to successfully generate T3D and T3DS18I variants that withstand proteolysis by breast tumor-associated metalloproteases.
Chappell et al. previously found that clinical reovirus strains exhibit either a threonine or isoleucine at position 249 of σ1 (57), but it was unknown whether a T249I mutation would have undesired detrimental effects on virus entry, endocytosis, or uncoating. To test this, L929 cells were exposed to equivalent particle doses of T3D, T3DT249I, T3DS18I, or T3DS18I/T249I at 4°C, washed extensively, and then incubated at 37°C for 0 to 9 h. At every hour, cells were fixed, stained for specific reovirus proteins, and then analyzed by flow cytometry to monitor the fate of input reovirus particles versus de novo reovirus protein expression. First, flow cytometry with λ2-specific antibodies, which cannot detect input virions (i.e., λ2 epitopes are hidden in the virion) but can detect de novo λ2 protein expression, demonstrated new virus protein expression at 8 h postinfection (hpi). Importantly, T3D and T3DT249I demonstrated similar de novo protein synthesis levels, suggesting similar kinetics of infection (Fig. 6E). T3DS18I was similar to T3DS18I/T249I with respect to de novo λ2 expression. As expected from previous studies showing that S18I increases reovirus infectivity (13, 54), both T3DS18I and T3DS18I/T249I exhibited ∼3-fold more de novo λ2 expression than T3D and T3DT249I.
Polyclonal antireovirus antibodies and monoclonal antibodies toward σ3 that detect both input virions and de novo virus protein synthesis confirmed equivalent input levels for all four viruses yet increased infectivity (or rate of infectivity) of variants containing the previously characterized S18I mutation (Fig. 6F). Again, it is important to note that the T249I mutation did not impact the efficiency of establishing infection. Finally, antibodies directed to the tail domain of σ1 confirmed that input virions containing the S18I mutation contained ∼3-fold less σ1 but produced more de novo proteins (Fig. 6G). Altogether, these results indicate that the T249I mutation does not negatively affect T3D reovirus infection, whether in the context of wild-type T3D or the more oncolytic T3DS18I variant.
Since cleavage of σ1 by T.E.E. reduced attachment to SA-low cells and inhibited infectivity (Fig. 2 and 3), we evaluated if the T249I mutation that prevents σ1 cleavage can facilitate reovirus infectivity in the presence of MPs. Accordingly, L929 cells were exposed to equivalent doses of T3D, T3DT249I, T3DS18I, or T3DS18I/T249I that had been pretreated with T.E.E., and then binding was evaluated by flow cytometry (Fig. 6H) and infectivity was evaluated by plaque assays (Fig. 6I). T3DT249I bound to L929 cells 16× more than T3D (Fig. 6H), indicating that it was resistant to protease cleavage. This increase in binding correlated with 16× higher virus production than T3D (Fig. 6I). T3DS18I showed lower binding and virus production than T3D, probably because this mutant has reduced σ1 levels and is therefore hypersensitive to σ1 cleavage. Importantly, the dysfunction in binding and virus production was overcome by combining with the T249I mutation in T3DS18I/T249I. These results suggest that incorporating the T249I mutation into T3D generates a virus capable of resisting T.E.E.
T3DT249I and T3DS18I/T249I do not have added toxicity relative to that of T3D in vivo.
Clinical trials using T3D as a monotherapy in several cancers have shown that T3D is a safe therapy but would benefit from enhanced efficacy (60). Little is known about the effects of the tumor environment on reovirus oncolytic performance. Having found that T.E.E. cleaves σ1 and reduces infectivity toward tumor cells with low sialic acid levels (Fig. 3) and having developed σ1-uncleavable variants of T3D (T3DT249I and T3DS18I/T249I) (Fig. 6) suggest that uncleavable reovirus warrant consideration for testing as oncolytic agents. The first priority, however, was to determine if these mutants were safe. Previous studies found that JAM-A on endothelial cells contributed to bloodstream dissemination of serotype 1 reovirus (61), so it seemed essential to test if T3DT249I and T3DS18I/T249I, by virtue of retaining JAM-A binding, pose any toxicity/safety concerns relative to those of T3D.
While reovirus is restricted to tumors and safety has been demonstrated in immunocompetent mice and in humans in clinical trials, reovirus shows visible signs of toxicity in severely immunocompromised NOD scid gamma (NSG) mice. NGS mice lack mature T cells, B cells, and NK cells. In the absence of immune restrictions, T3D reovirus can disseminate from tumors to heart and circulatory systems, impair circulation, and cause blackfoot syndrome (62). We therefore tested the toxicity of T3D strains when injected into MCF7 tumor xenografts in the background of NSG mice. Specifically, MCF7 cells were implanted into the mammary fat pads of NSG mice. When tumors became palpable, five mice were injected intratumorally with either PBS or 106 PFU of T3D, T3DT249I, T3DS18I, or T3DS18I/T249I. The dose was chosen based on our preliminary dose escalation experiments with T3D, which defined 106 as an optimal dose to see toxicity within guidelines of strict animal care protocols (data not shown). Reovirus injections were repeated a total of three times over a 1-week period. Mice were monitored and euthanized based on humane endpoints (first sign of blacktail/blackfoot, or over 15% weight loss). The onset of symptoms of toxicity for the different treatment groups is shown in Fig. 7A. Although time to symptoms varied between 25 and 32 days after the first virus inoculation, there were no significant differences among the virus treatments with respect to the appearance of symptoms. PBS-treated animals did not exhibit symptoms of toxicity and were euthanized at 45 days. Reovirus titers in the hearts were not significantly different between groups (Fig. 7B). Therefore, in the NSG model of reovirus toxicity in the absence of sufficient immune restriction, the σ1-uncleavable T3D variants did not pose additional toxicity or safety concerns relative to those of wild-type T3D.
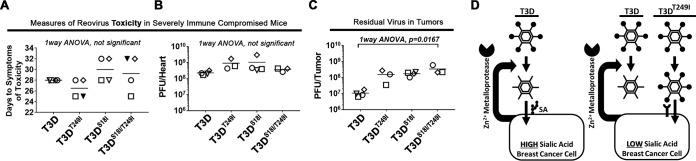
FIG 7.
Uncleavable reovirus does not increase toxicity in NSG mice when injected intratumorally. (A) Toxicity of T3D, T3DT249I, T3DS18I, or T3DS18I/T249I was assessed in severely immune-compromised (NSG) mice. When tumors became palpable, five mice were injected intratumorally with 106 plaque-forming units (PFU) of virus. Mice were euthanized when reovirus toxicity was observed (days to symptoms of toxicity), specifically when they showed first signs of black-foot (and/or tail or ears), indicating circulation deficiencies, or lost more than 15% of body weight. We were unable to obtain tissues from the 2 mice indicated by solid black triangles. In panels B to D, individual mice within each group have a unique symbol. PBS control-injected mice were symptom free at day 45, when the experiment was terminated. (B) Whole hearts were homogenized and subjected to plaque titration. Titers of reovirus in the heart provide a secondary measure of reovirus-induced toxicity. (C) Reovirus titers in whole tumors as described for panel B. (D) Final model. Some breast cancer cells secrete zinc-dependent metalloproteases that cleave σ1, removing the JAM-A-binding head domain and thereby making reovirus dependent on the presence of sialic acids for cell attachment. In breast cancer cells with low levels of sialic acid, the cleavage of σ1 strongly suppresses virus attachment and infection. Introduction of a T249I mutation into σ1 overcame the inactivation of reovirus by tumor-associated proteases, permitting efficient binding and entry.
The onset of black-foot syndrome necessitates euthanasia, which prevented assessment of virus-mediated increase in long-term survival of MCF7 tumor-bearing mice. However, since tumors were available to us upon euthanasia, we did assess the relative infectious virus titers of σ1-uncleavable T3D variants versus wild-type T3D in tumors. The mean reovirus titers in homogenized tumors were 1.0 × 107, 1.6 × 108, 1.8 × 108, and 3.5 × 108 PFU for T3D, T3DT249I, T3DS18I, and T3DS18I/T249I, respectively (Fig. 7C). While the trend suggested increasing titers for progressive addition of T249I and S18I mutations, statistical significance was only achieved for T3DS18I/T249I versus T3D, which showed a 30-fold difference (P = 0.0167). As explored further in Discussion, these studies were designed to monitor safety, and therefore distinct in vivo experimental designs beyond the scope of this study would be necessary in the future to compare oncolytic properties of these T3D variants. The current animal experiment supports that T3DS18I and T3DS18I/T249I do not exhibit gain of function with respect to virulence when introduced intratumorally.
DISCUSSION
While reovirus undergoes clinical testing as a cancer therapy, it is important to gain a fundamental understanding of factors in the virus and tumor environments that could alter reovirus behavior in tumors. Such factors might contribute to heterogenous response between patients and/or could help guide strategies to improve the potency of reovirus and other virus cancer therapeutics. Being a proteinaceous entity, we wondered if reovirus was processed by proteases that exist in breast tumor microenvironments. Moreover, since reovirus naturally exploits the gut proteases chymotrypsin and trypsin, or lysosomal cathepsins, to partially uncoat into membrane-penetrating ISVPs, we wondered what effect tumor proteases would have on reovirus infectivity toward tumor cells. Our studies show, for the first time, that proteases in breast tumor microenvironments have the potential to restrict reovirus infectivity (Fig. 7D). Specifically, proteases from in vivo breast tumors (T.E.E.) and from breast cancer cell lines (M.E.E.) fail to generate ISVPs. Instead, the T.E.E.- and M.E.E.-associated proteases cleave the reovirus cell attachment protein σ1 into two portions, removing the C-terminal domain that can bind to JAM-A and other host proteins while generating reovirus particles that retain only the sialic acid (SA)-binding tail domain. In other words, the tumor proteases make reovirus exclusively dependent on sialic acids for binding. Accordingly, T.E.E.- and M.E.E.-treated reovirus particles have ∼100-fold less attachment and infectivity toward tumor cells with reduced sialic acid levels. Our analysis further demonstrated that reovirus-inactivating proteases are commonly secreted by breast cancer cell lines and that the proteases are of the zinc-dependent metalloprotease (MP) family. Moreover, the inactivation of reovirus by breast cancer-associated MPs could be fully overcome by introducing a mutation (T249I) into σ1. Reovirus with the σ1T249I mutation did not exhibit added toxicity in immunocompromised NSG mice and therefore can proceed to testing for oncolytic activity in immunocompetent breast cancer models. Conceptually, our findings extend beyond reovirus, suggesting that tumor-associated proteases warrant consideration when using proteinaceous therapies in general.
Proteases are prevalent components of the tumor microenvironment, participating in extracellular matrix degradation and tumor cell invasiveness and altering activities of important signaling molecules (25). Many cells contribute to the pool of proteases in tumors, including the tumor cells themselves, mesenchymal stem cells, tumor-supporting fibroblasts and endothelial cells, and both innate and adaptive immune cells. In our studies, we discovered that breast cancer cells can secrete reovirus-inactivating proteases into the medium, but our findings do not preclude the possibility that other cells contribute additional proteases that act on reovirus in vivo. It would be interesting to expose reovirus to medium from tumor-supporting cells and immune cells to determine whether the composition of a specific tumor could predict reovirus processing/infectivity at the tumor site. Nevertheless, the extracellular extracts from two distinct in vivo breast tumor models (i.e., MCF7-derived tumors and polyomavirus middle T-antigen-derived tumors) produced the same outcome as media derived from breast cancer cells: an absence of the beneficial processing of reovirus to ISVPs but rather inactivation of reovirus infectivity toward sialic acid-low tumor cells by cleavage of σ1. Our preliminary analysis showed that unlike breast cancer cells, two lung cancer cell lines failed to secrete proteases that process reovirus. An analysis of a larger panel of lung and other cancer cell lines would be necessary, however, to determine if the inactivation of reovirus is breast cancer specific or can extend to other cancers.
Human proteases are categorized into aspartic, cysteine, metallo, serine, or threonine protease classes on the basis of the catalytic amino acid (or ion) in the active site. To distinguish which class(es) of proteases cleave σ1, reovirus processing by breast cancer proteases was tested in the presence of pharmacological inhibitors or divalent ions. The experiments indicate that the primary protease activity that cleaves σ1 is a zinc-dependent metalloprotease. There are hundreds of possible candidates and specific pharmacological inhibitors are only available for a select few, and this requires a proteomics approach in the future to identify the precise protease that cleaves σ1. The identity of the protease cannot be determined from the sequence of σ1 due to lack of defined linear consensus sequences for most candidates and unclear preferences for specific substrate secondary and tertiary structures. Moreover, it is important to note that our experiments also implicated aspartyl and cysteine proteases as having partial effects on σ1 cleavage and reovirus infectivity (Fig. 4). A remarkable feature of the tumor proteolytic network is the large extent to which proteases cleave other proteases and thereby affect their activities. For example, many metalloproteases are themselves substrates of aspartic, cysteine, and serine proteases (25). It is therefore possible that aspartyl and cysteine proteases cleave σ1 directly, or that they contribute to activation of the key metalloprotease. If the precise protease(s) that cleaves σ1 is identified, it would be interesting to test if it as a predictive marker for the extent of reovirus amplification in breast tumors. Given the complexity of the proteolytic network, we instead focused our attention on characterizing the effects of the protease on reovirus and modifying reovirus to withstand the protease(s) irrespective of identity.
Another interesting detail from our studies is that breast cancer MPs make reovirus dependent on SA and therefore reduce reovirus binding and infection, specifically in SA-low breast cancer cells such as MCF-7. SAs are frequently modified in breast cancer (63), for example, with overexpressed complex β-1,6-branched glycans (64), incomplete glycan structures (65), or nonmammalian xenoglycans (66). Hypersialylation is frequently seen in highly invasive breast cancers, which increases shedding of SAs that could occlude SA binding sites on the virion. To fully appreciate the effects of σ1 cleavage on reovirus oncolysis in breast cancer, it seems necessary to better understand which sialic acid modifications impact reovirus infection and whether shed SAs compete for reovirus attachment to cells. The nature of SAs in specific cancers then could serve as predictive markers for reovirus amplification. In any case, by overcoming σ1 cleavage with a T249I mutation, the retention of JAM-A binding capacity may overcome barriers posed by SA insufficiency or soluble SA competition.
As for application of knowledge toward improving reovirus oncolysis, our findings suggest a compendium of possibilities to be tested in the future. First, the T3DT249I and T3DS18I/T249I variants need to be thoroughly compared to T3D parental virus in various in vivo animal models to appreciate the extent to which cleavage of σ1 by tumor proteases affects oncolysis. We next propose that modification of T3D to transition to ISVPs in the context of well-characterized tumor proteases could be dually beneficial. Others have previously proposed using ISVPs (rather than whole reovirus particles) for cancer therapy (67), but such a procedure would only benefit infection for the incoming inoculum. Having reovirus consistently transition to ISVPs in tumor environments should produce a benefit at every round of virus amplification. Another idea is to generate reovirus particles that encode natural metalloprotease inhibitors, such as the small tissue inhibitors of metalloproteinase (TIMPs). Metalloproteases have served as possible cancer therapeutic targets but unfortunately they also play important roles in healthy tissues, and therefore global inhibition can be toxic. Perhaps a virus that directly delivers TIMPs to tumor microenvironments could serve two functions, inhibit inactivation of the virus and prevent protumor activities of MPs. Finally, our findings could extend beyond reovirus, as there are many oncolytic viruses and other proteinaceous treatments, such as antibodies undergoing clinical testing and use for cancer therapy. An expanded understanding of the effect(s) of tumor proteases on proteinaceous treatments could help develop second-generation therapies with augmented potencies.
Finally, beyond implications in reovirus oncolysis, our finding that proteases in tumor versus intestinal microenvironments exhibit differently in reovirus demonstrates that virus-host interactions are not just limited to the target infected cells but to the entire environment of a niche. In other words, if we better understand extracellular host factors that impact viruses, we may better appreciate why viruses thrive in specific host niches. In addition to proteases, extracellular factors such as the microbiota and immunological host factors can impact viruses. It will be interesting to know whether additional extracellular factors, such as the extracellular matrix, secreted lipids, carbohydrates, enzymes, or other soluble factors, impact the dynamics of viruses and therefore their oncolytic or pathogenic potential.
MATERIALS AND METHODS
Reovirus production.
Mammalian orthoreovirus serotype 3 Dearing (T3D) was propagated in L929 spinner cultures, extracted, and CsCl purified as previously described (68). Virus titers were determined by plaque assay on L929 cells as previously described (69). The titer of the stock virus used in most experiments was 1.01 × 1010 PFU/ml.
Cell culture.
L929 cells were cultured in minimal essential medium (MEM) (Sigma) supplemented with 10% FBS (Sigma), 1× sodium pyruvate (Sigma), nonessential amino acids (NEAA) (Sigma), and 1× antibiotic-antimycotic (catalog number 15240062; Gibco). HeLa cells were cultured in Dulbecco’s minimal essential medium (DMEM) and 1× antibiotic-antimycotic. MCF-7, T47D, MB-468, MB-231, H1299, and A549 cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (supplemented as described for MEM). L929, HeLa, and A549 cells were laboratory stocks obtained from Patrick Lee’s laboratory (Dalhousie University), while remaining cells were purchased from the American Type Culture Collection; all cells were confirmed mycoplasma free by Hoechst DNA staining. For experiments, cells were cultured in 1 ml per well of a 12-well plate, and medium was scaled according to the surface area (for example, in a well of a 6-well plate, we used 2 ml of medium).
Preparation of protease-rich extracts.
To mimic the tumor and intestinal environments reovirus would encounter, we obtained extracellular extracts from both environments. Tumor extracellular extract (T.E.E.) was generated by establishing breast tumors in mice from polyomavirus middle T-antigen-derived tumor cells (70). Tumors were explanted and incubated in 1 ml phosphate-buffered saline at 4°C for 2 h. The PBS solution was then centrifuged at 10,000 × g and filtered (0.45 μm) to reduce cellular debris. I.E.E. was made by flushing entire murine intestines with 1 ml PBS followed by filtering (0.45 μm) to remove debris. Polyoma virus middle T-antigen tumor sizes ranged from 700 to 2,000 mm2. The same method was used to extract T.E.E. from MCF7 tumors as that used for Fig. 5G, except tumors were smaller and therefore diffused into 0.5 ml of PBS (tumor sizes are indicated in the figure). To determine whether cancer cells alone are capable of secreting proteases that inactivate reovirus, protease-rich extracellular extract was obtained from a variety of breast and lung cancer cell lines (breast, MCF-7, T47D, MB-468, and MB-231; lung, H1299 and A549) for 24 h at 37°C (M.E.E., or medium extracellular extract) using the following protocol. First, cells were cultured to 80% confluence as described above. After an overnight incubation, the supplemented RPMI medium was replaced with protein-free VP-SFM (virus production serum-free medium) supplemented with l-glutamine, and the cells were then cultured for 3 days; health of cells with limited cell detachment was confirmed daily by microscopy. The medium then was collected and centrifuged at 1,000 × g for 7 min to remove any floating cells and debris. The supernatants were passed through a 0.45-μm low-protein-binding syringe filter. The filtrate then was centrifuged in a 5,000-NMWL (nominal molecular weight limit) centrifugal filter (UFV4BCC00; Millipore) at 3,000 rpm for 5 to 10 min until the volume reached 1/10 the initial volume. For use as a negative control, a plate containing supplemented RPMI medium without any cells was subjected to the same procedure. To avoid enzymatic inactivation as a result of freeze-thaw cycles, T.E.E., M.E.E., and I.E.E. were aliquoted before flash freezing in liquid N2 and stored at –80°C. A fresh aliquot was used for each experiment.
In vitro proteolysis assay with T.E.E., M.E.E., I.E.E., chymotrypsin, or trypsin.
Reovirus was incubated at 37°C for 24 h (unless indicated otherwise) with T.E.E., M.E.E., or I.E.E. in the presence or absence of different inhibitors. Volumes of these protease-rich extracts varied in each experiment, as indicated in figures, but ranged from 1 to 15 μl of extract at original concentration and 1.08 × 1010 to 5.40 × 1010 viral particles (1 to 5 μl of reovirus, at a concentration of 1.08 × 1013 particles/ml, as calculated based on the equivalency that a spectrophotometric optical density at 260 nm [OD260] of 5.42 = 1.13 × 1013 particles). The proteolysis assay was conducted in PCR tubes and incubated using a Bio-Rad T100 thermal cycler (with heated lid). For the time point digests, Bio-Rad S1000 and C1000 thermal cyclers were also used. Reaction mixtures were incubated at 37°C for 24 h or for the time lengths indicated during time course analyses. The inhibitors used were EDTA (ethylenediaminetetraacetic acid at 36.25 μM), EDTA-free protease inhibitor cocktail (PIC; 1× final concentration; number P8340; Sigma-Aldrich), and protease inhibitor cocktail with EDTA (CPIC; 1× final concentration; number 11-697-498-001; Roche). The following protease-class inhibitors were added: leupeptin (100 μM final concentration; Sigma), aprotinin (200 nM final concentration; Sigma), and pepstatin A (20 μM final concentration; Sigma). Chymotrypsin (Sigma) and trypsin (Sigma) were added at a 14-μg/ml final concentration. Following the proteolysis assay, samples were mixed with Laemmli sample buffer, heated in a thermal cycler to 95°C for 7 min, and subjected to SDS-PAGE for Western blotting.
Western blot analysis of in vitro proteolysis assays.
Following the 24-h incubation of reovirus and either T.E.E. or M.E.E. (as mentioned above) at 37°C, 1× protein sample buffer (PSB; Laemmli buffer) was added to each sample, which was then heated to 95°C for 7 to 10 min and run on a 12% SDS-PAGE gel. A Bio-Rad Trans-Blot Turbo system was used to transfer the SDS-PAGE gel to a nitrocellulose membrane (Amersham Hybond ECL; GE Healthcare Life Sciences). Following the transfer, the blots were then incubated overnight in 3% BSA–TBS-T (3% bovine serum albumin in Tris-buffered saline with 0.1% Tween 20) blocking buffer. This incubation was followed by a 1-h treatment with primary antibodies against reovirus capsid proteins (anti-reo; 1:1,000 dilution in 3% BSA–TBS-T), reovirus core proteins (anti-core; 1:200 dilution), or the reovirus cell attachment protein σ1 [anti-σ1N(tail), anti-σ1C(head); 1:500 dilution]. Primary antibodies (polyclonal mouse IgG) were kindly provided by Patrick Lee and Roy Duncan (Dalhousie University) as well as Terence Dermody (University of Pittsburgh). Goat anti-rabbit secondary antibodies conjugated with horseradish peroxidase (HRP; 1:10,000 dilution) or Alexa Fluor-647 (AF-647; 1:5,000 dilution) were used (Jackson ImmunoResearch). All antibodies were diluted in 3% BSA–TBS-T. Following application of each antibody, the blots were subjected to washing steps (totaling 15 min) prior to treatment with the next antibody or detection reagent. For all HRP antibodies, Pierce ECL (enhanced chemiluminescent) peroxidase substrate was used as a detection reagent. The blots were imaged and quantified using an ImageQuant LAS4000 biomolecular imager.
Binding assay.
HeLa, MDA-MB-231, L929, and MCF-7 cells were seeded in 100-mm plates. Cells were grown until confluent, washed with PBS, detached from the plates with cell stripper, and resuspended in PBS, and then 5 × 105 cells of each cell line were transferred to 1.5-ml Eppendorf tubes. All steps of the binding assay were carried out at 4°C to allow binding but prevent internalization. Cells were incubated for 30 min, followed by infection for 1 h on a rotator with the indicated reovirus variants. Cells were centrifuged at 550 × g for 5 min at 4°C, and pellets were washed twice with 500 μl 10% FBS in PBS solution. Final pellets were resuspended in 300 μl of antireovirus primary antibody (probes for μ1C, δ, and σ3, diluted 1:1,000 in 10% FBS in PBS solution), and rotated for 45 min. Cells were then centrifuged as described above, washed twice with 700 μl 10% FBS in PBS, resuspended with 300 μl of anti-rabbit AF-647 secondary antibody (diluted at 1:500 in 10% FBS in PBS; 1-495-144; Jackson Immunoresearch), and rotated for 45 min at 4°C. Cells were then washed as described above following primary antibody treatment. Final pellets were resuspended in 350 μl of 4% paraformaldehyde (Sigma) and incubated for 45 min. Cells then were centrifuged at 1,000 × g for 7 min, washed with cold 1× PBS, and resuspended in 300 μl PBS. Cells then were analyzed on the BD LSRFortessa and 10,000 cells gated using BD FACS Aria III for viable and singlet cells based on forward scatter and side scatter parameters. Mean fluorescence intensity was compared for samples exposed to virus relative to that of mock (PBS)-exposed cells and presented without further gating strategies. To compare relative binding efficiencies between samples, background MFI (mock) was always subtracted from remaining MFI values. A standard curve of particle dilution versus MFI then was generated for a positive control (wild-type T3D and corresponding cell line). MFI then was used to determine relative bound virus particles for remaining samples, keeping in mind that cells were exposed to equivalent virus particles of all treatments and variants in a given experiment. FCS Express 5 and FlowJo software programs were both used to present data. For binding assays with MCF7, MTHJ, TUBO, and T47D cells, a similar protocol was used with the following changes: 2 × 105 cells were used, washing was completed in 1 ml 2% FBS in PBS with 1 μM EDTA, and anti-σ3 primary antibody (4F2; Hybridoma Bank) and secondary goat anti-mouse Alexa 647 were used for detection of cell-bound virions.
Immunocytochemistry.
Cells were seeded in 24-well plates and infected with reovirus (MOI as indicated) at confluence, followed by incubation for 1 h at 37°C. Virus was removed by aspiration and replaced with fully supplemented MEM. Cells were incubated for 12 h at 37°C and then fixed with methanol for 5 min and washed with PBS. Cells were then blocked with 1 ml of blocking solution (PBS, 0.1% Triton X-100, 3% BSA). After 30 min of blocking, solution was aspirated and 400 μl of rabbit anti-reovirus primary antibody (diluted 1:10,000 in block solution) was added to each well and incubated for 1 h at room temperature. After removing the antibody, cells were washed 3× with wash solution and incubated with 400 μl of alkaline phosphatase-conjugated secondary antibody (111-055-144; Jackson Immunoresearch) (diluted 1:5,000 in blocking solution) for 1 h. After discarding the secondary antibody, cells were rinsed with alkaline phosphatase buffer (10 mM Tris, pH 9.5), and 400 μl BCIP–NBT (5-bromo-4-chloro-3′-indolyphosphate–nitroblue tetrazolium; B8503-500 [BCIP] and N6639-1G [NBT]; Sigma) was added to each well. The cells were incubated in the dark at room temperature for 10 to 30 min. The reaction was stopped with a stop solution (10 mM EDTA in PBS) and imaged on EVOS cell imaging systems (Invitrogen, Thermo Fisher Scientific).
SNA staining and neuraminidase treatments.
MCF7, MTHJ, TUBO, and T47D cells were grown until confluent, washed with PBS, detached with cell stripper, and resuspended in PBS, and then 6 × 105 cells of each cell line were added. Cells were centrifuged at 350 × g for 5 min, and pellets were washed twice with 1 ml flow buffer (0.1% BSA in 1× Hank’s balanced salt solution [HBSS] with magnesium chloride and calcium chloride), after which they were incubated with 120 mU neuraminidase (from Clostridium perfringens; Sigma) in PBS at 37°C for 30 min. Cells then were centrifuged twice with 1 ml flow buffer at 350 × g for 5 min and resuspended in 50 μl of fluorescein-labeled Sambucus nigra lectin (SNA diluted at 1:1,000 in washing solution; Vector Laboratories) in the dark for 20 min. All samples were fixed with 4% paraformaldehyde for 30 min, centrifuged at 700 × g for 3 min, and resuspended in flow buffer. Binding of SNA to sialic acid on cells was analyzed by flow cytometry.
In vivo tumor model.
Animal experiments were conducted with the approval of the University of Alberta Health Sciences Animal Care and Use Committee in accordance with guidelines from the Canadian Council for Animal Care. NOD-scid IL2Rgammanull (NSG) mice were bred in-house by Lynne Postovit. Mice were housed in a biosafety level 2 containment suite at the University of Alberta Health Sciences Laboratory Animal Services Facility. Six-week-old NSG mice were implanted with 17β-estradiol pellets (0.72 mg/pellet, 60-day release; no. SE-121; Innovative Research of America) at the back of the neck. One week later, MCF-7 cells (2 × 106 cells in 50 μl 50% Matrigel) were injected into the mammary fat pad, and when tumors were palpable, mice were sorted to give an even distribution of tumor sizes between groups. When tumors were palpable, mice were injected 3 times with PBS or 106 PFU of T3D, T3D(T249I), T3D(S18I), and T3D(S18I/T249I) viruses in 50 μl. After virus injections, mice were monitored twice daily and weighed every other day. Tumor growth was measured by caliper twice weekly. Mice were euthanized by CO2 inhalation when virus-treated animals presented initial symptoms of blackfoot/blacktail syndrome. Tumor and heart were excised and frozen at −80°C. To determine titers, samples were thawed, weighed, and resuspended in 2 ml of HBSS buffer (no. 14025092; ThermoFisher Scientific). Samples next were homogenized with a VDI 25 S41 homogenizer (no. 03231116; VWR), using 3 pulses of 10 s at the highest speed (24,000 rpm). Titers in L929 cells were determined by standard plaque assay (68).
Statistical analysis.
Statistical methods are described in the figure legends for respective experiments. Statistical analysis was performed using GraphPad Prism, version 7.05.
ACKNOWLEDGMENTS
We thank Matthew Macauley (University of Alberta) for providing expertise on sialic acids and the protocol for SNA staining. We thank Richard Schulz (University of Alberta) for providing expertise on metalloproteases.
This work was funded by a Cancer Research Society project grant to M.S., a grant to M.S. and M.H. from the Canadian Cancer Society, a project grant from the Li Ka Shing Institute of Virology to M.S. and M.H., a salary award to M.S. from the Canada Research Chairs (CRC) and infrastructure support from Canada Foundation for Innovation (CFI), along with salary support for H.E. from a Canadian Institutes of Health Research (CIHR) project grant to M.S. J.F. and S.H. received stipend funding from the University of Alberta Undergraduate Research Initiative (URI), and J.F. and I.B. also received stipend funding from Alberta Innovates Health Solutions (AIHS). F.C. received stipend funding from the University of Alberta Faculty of Medicine & Dentistry, the John & Rose McAllister Graduate Scholarship, the Faculty of Graduate Studies & Research at the University of Alberta, and the John Thibault Memorial Fund. Flow cytometry was performed at the University of Alberta, Faculty of Medicine and Dentistry Flow Cytometry Facility, which receives financial support from the Faculty of Medicine and Dentistry, and Canadian Foundation for Innovation (CFI) awards to contributing investigators.
Following discovery that mutation of reovirus σ1 overcomes inactivation by breast cancer metalloproteases, M.S. and M.H. filed a provisional patent for the modified reovirus as an improved oncolytic vector (serial no. 62/642,881). J.F., F.C., H.E., P.C., I.B., and S.H. have no potential conflict of interest to declare.
REFERENCES
- 1.Hashiro G, Loh PC, Yau JT. 1977. The preferential cytotoxicity of reovirus for certain transformed cell lines. Arch Virol 54:307–315. doi: 10.1007/bf01314776. [DOI] [PubMed] [Google Scholar]
- 2.Norman KL, Lee P. 2000. Reovirus as a novel oncolytic agent. J Clin Investig 105:1035–1038. doi: 10.1172/JCI9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clements D, Helson E, Gujar SA, Lee PW. 2014. Reovirus in cancer therapy: an evidence-based review. Oncolytic Virother 3:69–82. doi: 10.2147/OV.S51321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradbury PA, Morris DG, Nicholas G, Tu D, Tehfe M, Goffin JR, Shepherd FA, Gregg RW, Rothenstein J, Lee C, Kuruvilla S, Keith BD, Torri V, Blais N, Hao D, Korpanty GJ, Goss G, Melosky BL, Mates M, Leighl N, Ayoub JP, Sederias J, Feilotter H, Seymour L, Laurie SA. 2018. Canadian Cancer Trials Group (CCTG) IND211: a randomized trial of pelareorep (Reolysin) in patients with previously treated advanced or metastatic non-small cell lung cancer receiving standard salvage therapy. Lung Cancer 120:142–148. doi: 10.1016/j.lungcan.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 5.Jonker DJ, Tang PA, Kennecke H, Welch SA, Cripps MC, Asmis T, Chalchal H, Tomiak A, Lim H, Ko YJ, Chen EX, Alcindor T, Goffin JR, Korpanty GJ, Feilotter H, Tsao MS, Theis A, Tu D, Seymour L. 2018. A randomized phase II study of FOLFOX6/bevacizumab with or without pelareorep in patients with metastatic colorectal cancer: IND.210, a Canadian Cancer Trials group trial. Clin Colorectal Cancer 17:231–239. doi: 10.1016/j.clcc.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Mahalingam D, Fountzilas C, Moseley J, Noronha N, Tran H, Chakrabarty R, Selvaggi G, Coffey M, Thompson B, Sarantopoulos J. 2017. A phase II study of Reolysin (pelareorep) in combination with carboplatin and paclitaxel for patients with advanced malignant melanoma. Cancer Chemother Pharmacol 79:697–703. doi: 10.1007/s00280-017-3260-6. [DOI] [PubMed] [Google Scholar]
- 7.Morris DG, Feng X, DiFrancesco LM, Fonseca K, Forsyth PA, Paterson AH, Coffey MC, Thompson B. 2013. REO-001: a phase I trial of percutaneous intralesional administration of reovirus type 3 Dearing (Reolysin) in patients with advanced solid tumors. Investig New Drugs 31:696–706. doi: 10.1007/s10637-012-9865-z. [DOI] [PubMed] [Google Scholar]
- 8.Comins C, Spicer J, Protheroe A, Roulstone V, Twigger K, White CM, Vile R, Melcher A, Coffey MC, Mettinger KL, Nuovo G, Cohn DE, Phelps M, Harrington KJ, Pandha HS. 2010. REO-10: a phase I study of intravenous reovirus and docetaxel in patients with advanced cancer. Clin Cancer Res 16:5564–5572. doi: 10.1158/1078-0432.CCR-10-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein V, Ellard SL, Dent SF, Tu D, Mates M, Dhesy-Thind SK, Panasci L, Gelmon KA, Salim M, Song X, Clemons M, Ksienski D, Verma S, Simmons C, Lui H, Chi K, Feilotter H, Hagerman LJ, Seymour L. 2018. A randomized phase II study of weekly paclitaxel with or without pelareorep in patients with metastatic breast cancer: final analysis of Canadian Cancer Trials group IND.213. Breast Cancer Res Treat 167:485–493. doi: 10.1007/s10549-017-4538-4. [DOI] [PubMed] [Google Scholar]
- 10.Qiao J, Wang H, Kottke T, White C, Twigger K, Diaz RM, Thompson J, Selby P, de Bono J, Melcher A, Pandha H, Coffey M, Vile R, Harrington K. 2008. Cyclophosphamide facilitates antitumor efficacy against subcutaneous tumors following intravenous delivery of reovirus. Clin Cancer Res 14:259–269. doi: 10.1158/1078-0432.CCR-07-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandha HS, Heinemann L, Simpson GR, Melcher A, Prestwich R, Errington F, Coffey M, Harrington KJ, Morgan R. 2009. Synergistic effects of oncolytic reovirus and cisplatin chemotherapy in murine malignant melanoma. Clin Cancer Res 15:6158–6166. doi: 10.1158/1078-0432.CCR-09-0796. [DOI] [PubMed] [Google Scholar]
- 12.Yang WQ, Lun X, Palmer CA, Wilcox ME, Muzik H, Shi ZQ, Dyck R, Coffey M, Thompson B, Hamilton M, Nishikawa SG, Brasher PM, Fonseca K, George D, Rewcastle NB, Johnston RN, Stewart D, Lee PW, Senger DL, Forsyth PA. 2004. Efficacy and safety evaluation of human reovirus type 3 in immunocompetent animals: racine and nonhuman primates. Clin Cancer Res 10:8561–8576. doi: 10.1158/1078-0432.CCR-04-0940. [DOI] [PubMed] [Google Scholar]
- 13.Shmulevitz M, Gujar SA, Ahn DG, Mohamed A, Lee PW. 2012. Reovirus variants with mutations in S1 and L2 genome segments exhibit enhanced virion infectivity and superior oncolysis. J Virol 86:7403–7413. doi: 10.1128/JVI.00304-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borden EC. 2007. Augmentation of effects of interferon-stimulated genes by reversal of epigenetic silencing: potential application to melanoma. Cytokine Growth Factor Rev 18:491–501. doi: 10.1016/j.cytogfr.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Endo-Munoz L, Warby T, Harrich D, McMillan NA. 2005. Phosphorylation of HIV Tat by PKR increases interaction with TAR RNA and enhances transcription. Virol J 2:17. doi: 10.1186/1743-422X-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMillan NA, Chun RF, Siderovski DP, Galabru J, Toone WM, Samuel CE, Mak TW, Hovanessian AG, Jeang KT, Williams BR. 1995. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology 213:413–424. doi: 10.1006/viro.1995.0014. [DOI] [PubMed] [Google Scholar]
- 17.Joklik WK. 1972. Studies on the effect of chymotrypsin on reovirions. Virology 49:700–715. doi: 10.1016/0042-6822(72)90527-2. [DOI] [PubMed] [Google Scholar]
- 18.Bodkin DK, Nibert ML, Fields BN. 1989. Proteolytic digestion of reovirus in the intestinal lumens of neonatal mice. J Virol 63:4676–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nibert ML, Fields BN. 1992. A carboxy-terminal fragment of protein mu 1/mu 1C is present in infectious subvirion particles of mammalian reoviruses and is proposed to have a role in penetration. J Virol 66:6408–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odegard AL, Chandran K, Zhang X, Parker JSL, Baker TS, Nibert ML. 2004. Putative autocleavage of outer capsid protein 1, allowing release of myristoylated peptide 1N during particle uncoating, is critical for cell entry by reovirus. J Virol 78:8732–8745. doi: 10.1128/JVI.78.16.8732-8745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandran K, Parker JSL, Ehrlich M, Kirchhausen T, Nibert ML. 2003. The δ region of outer-capsid protein μ1 undergoes conformational change and release from reovirus particles during cell entry. J Virol 77:13361–13375. doi: 10.1128/JVI.77.24.13361-13375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebert DH, Deussing J, Peters C, Dermody TS. 2002. Cathepsin L and cathepsin B mediate reovirus disassembly in murine fibroblast cells. J Biol Chem 277:24609–24617. doi: 10.1074/jbc.M201107200. [DOI] [PubMed] [Google Scholar]
- 23.Jedeszko C, Sloane BF. 2004. Cysteine cathepsins in human cancer. Biol Chem 385:1017–1027. doi: 10.1515/BC.2004.132. [DOI] [PubMed] [Google Scholar]
- 24.Vasiljeva O, Turk B. 2008. Dual contrasting roles of cysteine cathepsins in cancer progression: apoptosis versus tumour invasion. Biochimie 90:380–386. doi: 10.1016/j.biochi.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 25.Mason SD, Joyce JA. 2011. Proteolytic networks in cancer. Trends Cell Biol 21:228–237. doi: 10.1016/j.tcb.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liotta LA. 2016. Adhere, degrade, and move: the three-step model of invasion. Cancer Res 76:3115–3117. doi: 10.1158/0008-5472.CAN-16-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liotta LA. 1986. Tumor invasion and metastases–role of the extracellular matrix: Rhoads Memorial Award lecture. Cancer Res 46:1–7. [PubMed] [Google Scholar]
- 28.Demchik LL, Sameni M, Nelson K, Mikkelsen T, Sloane BF. 1999. Cathepsin B and glioma invasion. Int J Dev Neurosci 17:483–494. doi: 10.1016/S0736-5748(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 29.Podgorski I, Linebaugh BE, Sameni M, Jedeszko C, Bhagat S, Cher ML, Sloane BF. 2005. Bone microenvironment modulates expression and activity of cathepsin B in prostate cancer. Neoplasia 7:207–223. doi: 10.1593/neo.04349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bervar A, Zajc I, Sever N, Katunuma N, Sloane BF, Lah TT. 2003. Invasiveness of transformed human breast epithelial cell lines is related to cathepsin B and inhibited by cysteine proteinase inhibitors. Biol Chem 384:447–455. doi: 10.1515/BC.2003.050. [DOI] [PubMed] [Google Scholar]
- 31.Rempel SA, Rosenblum ML, Mikkelsen T, Yan PS, Ellis KD, Golembieski WA, Sameni M, Rozhin J, Ziegler G, Sloane BF. 1994. Cathepsin B expression and localization in glioma progression and invasion. Cancer Res 54:6027–6031. [PubMed] [Google Scholar]
- 32.Kobayashi H, Moniwa N, Sugimura M, Shinohara H, Ohi H, Terao T. 1993. Effects of membrane-associated cathepsin B on the activation of receptor-bound prourokinase and subsequent invasion of reconstituted basement membranes. Biochim Biophys Acta 1178:55–62. doi: 10.1016/0167-4889(93)90109-3. [DOI] [PubMed] [Google Scholar]
- 33.Fernandes J, Cristi F, Eaton H, Chen P, Haeflinger S, Bernard I, Hitt M, Shmulevitz M. 2019. Breast tumor-associated metalloproteases restrict reovirus oncolysis by cleaving the σ1 cell-attachment protein, and can be overcome by mutation of σ1. bioRxiv 10.1101/742478. [DOI] [PMC free article] [PubMed]
- 34.Chandran K, Farsetta DL, Nibert ML. 2002. Strategy for nonenveloped virus entry: a hydrophobic conformer of the reovirus membrane penetration protein micro 1 mediates membrane disruption. J Virol 76:9920–9933. doi: 10.1128/jvi.76.19.9920-9933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dryden KA, Wang G, Yeager M, Nibert ML, Coombs KM, Furlong DB, Fields BN, Baker TS. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol 122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dermody T, Sherry B. 2013. Orthoreoviruses, p 1321–1322. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 37.Chen M, Chen C, Shen Z, Zhang X, Chen Y, Lin F, Ma X, Zhuang C, Mao Y, Gan H, Chen P, Zong X, Wu R. 2017. Extracellular pH is a biomarker enabling detection of breast cancer and liver cancer using CEST MRI. Oncotarget 8:45759–45767. doi: 10.18632/oncotarget.17404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gatenby RA, Gawlinski ET, Gmitro AF, Kaylor B, Gillies RJ. 2006. Acid-mediated tumor invasion: a multidisciplinary study. Cancer Res 66:5216–5223. doi: 10.1158/0008-5472.CAN-05-4193. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Lin Y, Gillies RJ. 2010. Tumor pH and its measurement. J Nucl Med 51:1167–1170. doi: 10.2967/jnumed.109.068981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rakashanda Rana SF, Rafiq S, Masood A, Amin S. 2012. Role of proteases in cancer: a review. Biotechnol Mol Biol Rev 7:90–101. doi: 10.5897/BMBR11.027. [DOI] [Google Scholar]
- 41.Reiter DM, Frierson JM, Halvorson EE, Kobayashi T, Dermody TS, Stehle T. 2011. Crystal structure of reovirus attachment protein sigma1 in complex with sialylated oligosaccharides. PLoS Pathog 7:e1002166. doi: 10.1371/journal.ppat.1002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barton ES, Forrest JC, Connolly JL, Chappell JD, Liu Y, Schnell FJ, Nusrat A, Parkos CA, Dermody TS. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441–451. doi: 10.1016/s0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- 43.Weiner DB, Girard K, Williams WV, McPhillips T, Rubin DH. 1988. Reovirus type 1 and type 3 differ in their binding to isolated intestinal epithelial cells. Microb Pathog 5:29–40. doi: 10.1016/0882-4010(88)90078-2. [DOI] [PubMed] [Google Scholar]
- 44.Rubin DH. 1987. Reovirus serotype 1 binds to the basolateral membrane of intestinal epithelial cells. Microb Pathog 3:215–219. doi: 10.1016/0882-4010(87)90098-2. [DOI] [PubMed] [Google Scholar]
- 45.Magara K, Takasawa A, Osanai M, Ota M, Tagami Y, Ono Y, Takasawa K, Murata M, Hirohashi Y, Miyajima M, Yamada G, Hasegawa T, Sawada N. 2017. Elevated expression of JAM-A promotes neoplastic properties of lung adenocarcinoma. Cancer Sci 108:2306–2314. doi: 10.1111/cas.13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao C, Lu F, Chen H, Zhao X, Sun J, Chen H. 2014. Dysregulation of JAM-A plays an important role in human tumor progression. Int J Clin Exp Pathol 7:7242–7248. [PMC free article] [PubMed] [Google Scholar]
- 47.McSherry EA, McGee SF, Jirstrom K, Doyle EM, Brennan DJ, Landberg G, Dervan PA, Hopkins AM, Gallagher WM. 2009. JAM-A expression positively correlates with poor prognosis in breast cancer patients. Int J Cancer 125:1343–1351. doi: 10.1002/ijc.24498. [DOI] [PubMed] [Google Scholar]
- 48.Vajaria BN, Patel KR, Begum R, Patel PS. 2016. Sialylation: an avenue to target cancer cells. Pathol Oncol Res 22:443–447. doi: 10.1007/s12253-015-0033-6. [DOI] [PubMed] [Google Scholar]
- 49.Cui H, Lin Y, Yue L, Zhao X, Liu J. 2011. Differential expression of the alpha 2,3-sialic acid residues in breast cancer is associated with metastatic potential. Oncol Rep 25:1365–1371. doi: 10.3892/or.2011.1192. [DOI] [PubMed] [Google Scholar]
- 50.Terasawa Y, Hotani T, Katayama Y, Tachibana M, Mizuguchi H, Sakurai F. 2015. Activity levels of cathepsins B and L in tumor cells are a biomarker for efficacy of reovirus-mediated tumor cell killing. Cancer Gene Ther 22:188. doi: 10.1038/cgt.2015.4. [DOI] [PubMed] [Google Scholar]
- 51.Voura EB, English JL, Yu HY, Ho AT, Subarsky P, Hill RP, Hojilla CV, Khokha R. 2013. Proteolysis during tumor cell extravasation in vitro: metalloproteinase involvement across tumor cell types. PLoS One 8:e78413. doi: 10.1371/journal.pone.0078413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagase H, Visse R, Murphy G. 2006. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc Res 69:562–573. doi: 10.1016/j.cardiores.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 53.Yang H, Makaroff K, Paz N, Aitha M, Crowder MW, Tierney DL. 2015. Metal ion dependence of the matrix metalloproteinase-1 mechanism. Biochemistry 54:3631–3639. doi: 10.1021/acs.biochem.5b00379. [DOI] [PubMed] [Google Scholar]
- 54.Mohamed A, Teicher C, Haefliger S, Shmulevitz M. 2015. Reduction of virion-associated σ1 fibers on oncolytic reovirus variants promotes adaptation toward tumorigenic cells. J Virol 89:4319–4334. doi: 10.1128/JVI.03651-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larson SM, Antczak JB, Joklik WK. 1994. Reovirus exists in the form of 13 particle species that differ in their content of protein sigma 1. Virology 201:303–311. doi: 10.1006/viro.1994.1295. [DOI] [PubMed] [Google Scholar]
- 56.Del Monte U. 2009. Does the cell number 10(9) still really fit one gram of tumor tissue? Cell Cycle 8:505–506. doi: 10.4161/cc.8.3.7608. [DOI] [PubMed] [Google Scholar]
- 57.Chappell JD, Barton ES, Smith TH, Baer GS, Duong DT, Nibert ML, Dermody TS. 1998. Cleavage susceptibility of reovirus attachment protein σ1 during proteolytic disassembly of virions is determined by a sequence polymorphism in the σ1 neck. J Virol 72:8205–8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Duncan R, Lee P. 1994. Localization of two protease-sensitive regions separating distinct domains in the reovirus cell-attachment protein σ1. Virology 203:149–152. doi: 10.1006/viro.1994.1465. [DOI] [PubMed] [Google Scholar]
- 59.Duncan R, Horne D, Strong JE, Leone G, Pon RT, Yeung MC, Lee PW. 1991. Conformational and functional analysis of the C-terminal globular head of the reovirus cell attachment protein. Virology 182:810–819. doi: 10.1016/0042-6822(91)90622-i. [DOI] [PubMed] [Google Scholar]
- 60.Phillips MB, Stuart JD, Rodriguez Stewart RM, Berry JT, Mainou BA, Boehme KW. 2018. Current understanding of reovirus oncolysis mechanisms. Oncolytic Virother 7:53–63. doi: 10.2147/OV.S143808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lai CM, Boehme KW, Pruijssers AJ, Parekh VV, Van Kaer L, Parkos CA, Dermody TS. 2015. Endothelial JAM-A promotes reovirus viremia and bloodstream dissemination. J Infect Dis 211:383–393. doi: 10.1093/infdis/jiu476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loken SD, Norman K, Hirasawa K, Nodwell M, Lester WM, Demetrick DJ. 2004. Morbidity in immunosuppressed (SCID/NOD) mice treated with reovirus (Dearing 3) as an anti-cancer biotherapeutic. Cancer Biol Ther 3:734–738. doi: 10.4161/cbt.3.8.963. [DOI] [PubMed] [Google Scholar]
- 63.Boligan KF, Mesa C, Fernandez LE, von Gunten S. 2015. Cancer intelligence acquired (CIA): tumor glycosylation and sialylation codes dismantling antitumor defense. Cell Mol Life Sci 72:1231–1248. doi: 10.1007/s00018-014-1799-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abbott KL, Aoki K, Lim JM, Porterfield M, Johnson R, O’Regan RM, Wells L, Tiemeyer M, Pierce M. 2008. Targeted glycoproteomic identification of biomarkers for human breast carcinoma. J Proteome Res 7:1470–1480. doi: 10.1021/pr700792g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Freire T, Bay S, von Mensdorff-Pouilly S, Osinaga E. 2005. Molecular basis of incomplete O-glycan synthesis in MCF-7 breast cancer cells: putative role of MUC6 in Tn antigen expression. Cancer Res 65:7880–7887. doi: 10.1158/0008-5472.CAN-04-3746. [DOI] [PubMed] [Google Scholar]
- 66.Marquina G, Waki H, Fernandez LE, Kon K, Carr A, Valiente O, Perez R, Ando S. 1996. Gangliosides expressed in human breast cancer. Cancer Res 56:5165–5171. [PubMed] [Google Scholar]
- 67.van Houdt WJ, Smakman N, van den Wollenberg DJ, Emmink BL, Veenendaal LM, van Diest PJ, Hoeben RC, Borel Rinkes I, Kranenburg O. 2008. Transient infection of freshly isolated human colorectal tumor cells by reovirus T3D intermediate subviral particles. Cancer Gene Ther 15:284–292. doi: 10.1038/cgt.2008.2. [DOI] [PubMed] [Google Scholar]
- 68.James KT, Cooney B, Agopsowicz K, Trevors MA, Mohamed A, Stoltz D, Hitt M, Shmulevitz M. 2016. Novel high-throughput approach for purification of infectious virions. Sci Rep 6:36826. doi: 10.1038/srep36826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shmulevitz M, Lee PW. 2012. Exploring host factors that impact reovirus replication, dissemination, and reovirus-induced cell death in cancer versus normal cells in culture. Methods Mol Biol 797:163–176. doi: 10.1007/978-1-61779-340-0_12. [DOI] [PubMed] [Google Scholar]
- 70.Desilva A, Wuest M, Wang M, Hummel J, Mossman K, Wuest F, Hitt MM. 2012. Comparative functional evaluation of immunocompetent mouse breast cancer models established from PyMT-tumors using small animal PET with [18F]FDG and [18F]FLT. Am J Nucl Med Mol Imaging 2:88–98. [PMC free article] [PubMed] [Google Scholar]