In this article, we consider a role for bacteria in shaping coronavirus infection. Taking cues from studies of enteric viruses, we initially investigated how bacterial surface components might improve CoV infection. Instead, we found that peptidoglycan-associated surfactin is a potent viricidal compound that disrupts virion integrity with broad activity against enveloped viruses. Our results indicate that interactions with commensal bacterial may improve or disrupt viral infections, highlighting the importance of understanding these microbial interactions and their implications for viral pathogenesis and treatment.
KEYWORDS: MERS-CoV, SARS-CoV, coronavirus, cyclic lipopeptide, microbiome, surfactin
ABSTRACT
Enteric viruses exploit bacterial components, including lipopolysaccharides (LPS) and peptidoglycan (PG), to facilitate infection in humans. Because of their origin in the bat enteric system, we wondered if severe acute respiratory syndrome coronavirus (SARS-CoV) or Middle East respiratory syndrome CoV (MERS-CoV) also use bacterial components to modulate infectivity. To test this question, we incubated CoVs with LPS and PG and evaluated infectivity, finding no change following LPS treatment. However, PG from Bacillus subtilis reduced infection >10,000-fold, while PG from other bacterial species failed to recapitulate this. Treatment with an alcohol solvent transferred inhibitory activity to the wash, and mass spectrometry revealed surfactin, a cyclic lipopeptide antibiotic, as the inhibitory compound. This antibiotic had robust dose- and temperature-dependent inhibition of CoV infectivity. Mechanistic studies indicated that surfactin disrupts CoV virion integrity, and surfactin treatment of the virus inoculum ablated infection in vivo. Finally, similar cyclic lipopeptides had no effect on CoV infectivity, and the inhibitory effect of surfactin extended broadly to enveloped viruses, including influenza, Ebola, Zika, Nipah, chikungunya, Una, Mayaro, Dugbe, and Crimean-Congo hemorrhagic fever viruses. Overall, our results indicate that peptidoglycan-associated surfactin has broad viricidal activity and suggest that bacteria by-products may negatively modulate virus infection.
IMPORTANCE In this article, we consider a role for bacteria in shaping coronavirus infection. Taking cues from studies of enteric viruses, we initially investigated how bacterial surface components might improve CoV infection. Instead, we found that peptidoglycan-associated surfactin is a potent viricidal compound that disrupts virion integrity with broad activity against enveloped viruses. Our results indicate that interactions with commensal bacterial may improve or disrupt viral infections, highlighting the importance of understanding these microbial interactions and their implications for viral pathogenesis and treatment.
INTRODUCTION
Commensal bacteria inhabit nearly every surface of the human body, influencing numerous host processes (1, 2). While these bacteria are considered to serve a protective role, recent studies that indicate enteric viruses exploit bacterial envelope components to facilitate infection (3). Poliovirus was found to bind both lipopolysaccharides (LPS) and peptidoglycan (PG) to enhance its thermostability and receptor affinity, facilitating in vivo infection (3). Antibiotic depletion of commensal bacteria inhibited oral poliovirus infection, but was rescued by recolonization, pretreatment of virus with LPS, or bypassing the enteric system through intraperitoneal injection (3). Other viruses, including reovirus, mouse mammary tumor virus, and murine norovirus, have been shown to use similar mechanisms to facilitate infection (3, 4). Together, these results indicate a key role for commensal bacteria in improving infectivity and pathogenesis of enteric viruses.
Like the enteric system, the respiratory tract harbors high levels of commensal bacteria, particularly in the upper respiratory tract, including the nasal cavity, nasopharynx, and oropharynx (1). While poorly understood, the respiratory microbiome is complex, with differentiated bacterial communities inhabiting each niche (1). Like the enteric version, the respiratory microbiome plays a protective role in immunity (1). Nevertheless, a recent study demonstrated that influenza can interact with several pathogenic bacterial infections, increasing their adherence to respiratory cells and increasing bacterial colonization and disease in vivo (5). These results provide evidence that viral pathogens can interact with bacteria in the respiratory tract as well as in the gut.
Although they are human pathogens, severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) have their evolutionary origins in the bat enteric system (6) and may have, like human enteric viruses, exploited commensal bacteria. Given the high levels of commensals in the respiratory tract (1), it is possible that such interactions may have been maintained during emergence of CoV strains. Thus, we wondered if CoVs utilized bacterial components to facilitate infection. Previous work had identified a key role for the toll-like receptor (TLR) pathways in immunity to SARS-CoV, with the absence of LPS binding TLR4 or its downstream adaptors resulting in augmented disease (7–9). Given the interactions observed between enteric viruses and bacterial components, CoVs may also use similar microbial components to improve infectivity and subsequently stimulate the TLR4 response.
In this study, we explored the relationship between bacterial surface components and CoV infection. Surprisingly, we found that PG from Bacillus subtilis reduced CoV infectivity. Using mass spectrometry, we identified a cyclic lipopeptide (CLP), surfactin, as the molecule responsible for CoV inhibition. The inhibitory effect of surfactin was dose and temperature dependent, with treatment disrupting the integrity of the CoV particle. Notably, surfactin treatment of the inoculum ablated CoV infection in vivo, but prophylactic treatment had no effect. Other similar CLPs had no effect on CoV infectivity, suggesting that the viricidal properties of surfactin were unique. Importantly, surfactin treatment reduced the infectivity of several other enveloped viruses, including that of influenza A, Zika, Dugbe, Nipah, Crimean-Congo hemorrhagic fever, chikungunya, Mayaro, Una, and Ebola viruses. Together, these results demonstrate the efficacy of surfactin as a viricidal compound and highlight the potential for microbial environment to modulate virus infection.
(This article was submitted to an online preprint archive [10].)
RESULTS
Peptidoglycan derived from B. subtilis reduces with coronavirus infectivity.
Given their origins in bat enteric systems, we wondered if CoVs might be stabilized by bacterial components (6). To test this possibility, human CoV-229E, a common cold-associated CoV, and MERS-CoV were treated with control (phosphate-buffered saline [PBS]), LPS (Escherichia coli), or PG (Bacillus subtilis), and viral infectivity was determined (Fig. 1A). In contrast to its effect on enteric viruses, LPS had no effect on CoV infectivity; however, the presence of PG from B. subtilis dramatically reduced the infectivity of both HCoV-229E and MERS-CoV (Fig. 1B). The structure of PG varies considerably between bacterial species (11), suggesting that PG from different bacteria may have distinct effects on CoV infectivity. To explore this, we tested a diverse set of bacterially derived PGs for the ability to modulate CoV infection (Fig. 1C). Notably, only PG derived from B. subtilis reduced HCoV-229E and MERS-CoV infection, suggesting that interference with CoV infectivity is not shared by PG from all bacterial species.
FIG 1.
Peptidoglycan from Bacillus subtilis reduces coronavirus infectivity. (A) Bacterial envelope components such as LPS are bound to CoVs, increasing their thermostability (right) relative to that of untreated samples (left). (B) Relative infectivity of HCoV-229E (n = 4) and MERS-CoV (n = 5) after treatment with PBS alone (black), 1 mg/ml LPS from E. coli (gray), or 1 mg/ml PG from B. subtilis (green) following 2 h of incubation at 37°C. (C) HCoV-299E (circles) and MERS-CoV (triangles) infectivity after treatment for 2 h at 37°C with peptidoglycan from the indicated bacterial species (n = 3). (D) HCoV-229E and (E) MERS-CoV after treatment with 1 mg/ml PG from B. subtilis at room temperature (RT), 32°C, and 37°C (n = 3). For all dot plots, the centered bar represents the group mean, while the error bars represent standard deviation (SD). P values are based on the two-tailed Student’s t test, indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Next, we wondered if incubation temperature also played a role in B. subtilis PG reduction of CoV infectivity. To investigate, HCoV-229E and MERS-CoV stocks were treated with B. subtilis PG at room temperature (RT), 32°C, or 37°C (Fig. 1D and E). Interestingly, PG disruption of viral infectivity was reduced at lower temperatures. For HCoV-229E, infectivity had a stepwise reduction with increasing temperature (Fig. 1D). In contrast, PG reduction of MERS-CoV infectivity was ablated at lower temperatures, with no significant loss of viral infectivity at either RT or 32°C (Fig. 1E). Together, these data indicate that the inhibitory effect of B. subtilis PG is influenced by incubation temperature.
Infectivity inhibition can be disassociated from PG.
Two possible scenarios explain why only B. subtilis PG reduces CoV infectivity: (i) B. subtilis PG reduces infectivity directly, using unique structural features absent in PG from other bacteria, or (ii) the PG preparation contains another compound that mediates inhibition. To differentiate these possibilities, we exploited the poor solubility of PG, washing it in a variety of solvents to separate its inhibitory effect (Fig. 2A). After three washes in PBS, PG maintained its reduction of HCoV-229E infectivity (Fig. 2A). In contrast, PG washed with either 100% ethanol or dimethyl sulfoxide (DMSO) lost the ability to inhibit HCoV-229E infectivity (Fig. 2A). These results suggest that the washes either modified the inhibitory capacity of PG or removed a soluble compound responsible for reducing CoV infectivity. To explore this, the supernatants from clarified PG samples were incubated with HCoV-229E (Fig. 2B). While the PBS, PBS control, and ethanol control had no inhibitory effect, the ethanol supernatant from PG potently reduced viral infectivity of HCoV-229E (Fig. 2B). Together, these data indicate that a soluble compound distinct from, but present in, the PG sample is responsible for reducing CoV infectivity.
FIG 2.
Identification of surfactin from B. subtilis peptidoglycan. (A and B) PG from B. subtilis in a PBS solution was clarified, washed with the indicated solvents, and clarified again. Supernatants were decanted and retained, while the insoluble fractions were resuspended in PBS. The (A) insoluble fraction and (B) supernatants were then used to treat HCoV-229E, and relative infectivity was determined (n = 3). (C, D, and E) Mass spectrometry was performed on PG (C) and ethanol wash (D). The peak corresponding to the molecular mass of 1,058 kDa in the ethanol wash was then further fragmented (E) to determine the identity of the molecule. Representative spectra are shown (n = 3). For all dot plots, the centered bar represents the group mean and error bars the SD.
Mass spectrometry identifies the inhibitor as surfactin.
Having isolated the inhibitory molecule, we utilized mass spectrometry to determine its identity. Unwashed B. subtilis PG and ethanol supernatants were analyzed using matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF/TOF) mass spectrometry (MS) (Fig. 2C). In the PG samples, three prominent peaks were observed with respective molecular masses of 1,010.5, 1,058.7, and 1,238.6 kDa (Fig. 2C). While all of these peaks were present in the ethanol supernatant, the compound with a mass of 1,058.7 was enriched nearly 10-fold (Fig. 2D). Further analysis of this peak by fragmentation produced a spectrum matching that of the cyclic lipopeptide surfactin (12), a potent biosurfactant produced naturally by B. subtilis and shown previously to have antimicrobial and antiviral properties (13, 14) (Fig. 2E; for structure, see Fig. 6A). Given its abundance and enrichment in the ethanol wash, as well as its described antiviral properties, we concluded that surfactin likely conferred the B. subtilis PG with the ability to interfere with CoV infection.
FIG 6.
Surfactin, but not other cyclic lipopeptides, broadly reduces the infectivity of enveloped viruses. (A) Biochemical models of each of the seven cyclic lipopeptides tested. The number of amino acids present in the cyclic ring is shown in parentheses. (B) HCoV-229E (blue) and MERS-CoV (gray) were treated with PBS or 100 μg/ml of the indicated cyclic lipopeptides in PBS and incubated for 2 h at 37°C. Viral infectivity was then determined (n = 3). (C) The indicated viruses were diluted in PBS (black) or 100 μg/ml of surfactin (red), incubated for 2 h at 37°C, and viral infectivity was determined (n = 3). Viruses are as follows: coxsackievirus (CVB3), Dugbe (DUGV), Crimean-Congo hemorrhagic fever virus (CCHFV), Zika virus (ZIKV), Nipah virus (NiV), chikungunya virus (CHIKV), Una virus, Mayaro virus, influenza A strains H1N1 and H3N2, and Ebola virus (EBOV). Bar graph bars represent the group means (n = 3). Error bars represent SD. ND, no titers were detected. The Student’s t test was used to calculate P values, with *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
Reduction of CoV infectivity by surfactin is temperature- and dose-dependent.
To confirm its inhibitory effect, we characterized the ability of purified surfactin to reduce CoV infectivity. HCoV-229E, MERS-CoV, or SARS-CoV were treated with PBS or surfactin at either RT, 32°C, or 37°C. For all three CoVs, surfactin reduced infectivity after treatment at 37°C, with a nearly complete loss of infectious virus (Fig. 3A to C). Similarly to B. subtilis PG (Fig. 1D and E), the degree of reduction varied based on incubation temperature and varied between the CoVs (Fig. 3A to C). To further characterize the kinetics of inhibition, HCoV-229E and MERS-CoV were treated with surfactin at 4°C, RT, 32°C, or 37°C, and sampled to measure time-dependent neutralization (Fig. 3D and E). At both 32°C and 37°C, surfactin rapidly reduced HCoV-229E and MERS-CoV, with a nearly complete loss of infectivity after 2 h of treatment (Fig. 3D and E). In contrast, RT incubation reduced CoV infectivity more slowly, and the effects of surfactin were ablated at 4°C. We also observed dose-dependent changes in surfactin activity against HCoV-229E, MERS-CoV, and SARS-CoV (Fig. 3F). Interestingly, higher concentrations of surfactin were required for inhibition of HCoV-229E compared to that for inhibition of either SARS-CoV or MERS-CoV, whose inhibition curves were nearly identical. Together, these data indicate that both temperature and dose impact the inhibitory effects of surfactin.
FIG 3.
Characterization of CoV inhibition by surfactin. (A) HCoV-229E, (B) MERS-CoV, and (C) SARS-CoV (MA15) were treated with PBS alone (black) or 100 μg/ml surfactin (red) and at room temperature (RT), 32°C, and 37°C, and infectivity was determined (n = 3). HCoV-229E (D) and MERS-CoV (E) were treated for the indicated time at 4°C (blue), RT (red), 32°C (green), or 37°C (purple), and infectivity was determined (n = 3). (F) HCoV-229E (blue), MERS-CoV (orange), and SARS-CoV MA15 (green) were diluted in concentrations ranging from 0.5 to 200 μg/ml, and viral infectivity was determined (n = 3). For dot plots, each point represents the titer from an independent experiment, while the group mean is indicated by a line. Each point on the line graph represents the group mean. All error bars represent SD. The two-tailed Student’s t test was used to determine P values, indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Surfactin reduces CoV infectivity by disrupting the structural integrity of viral particles.
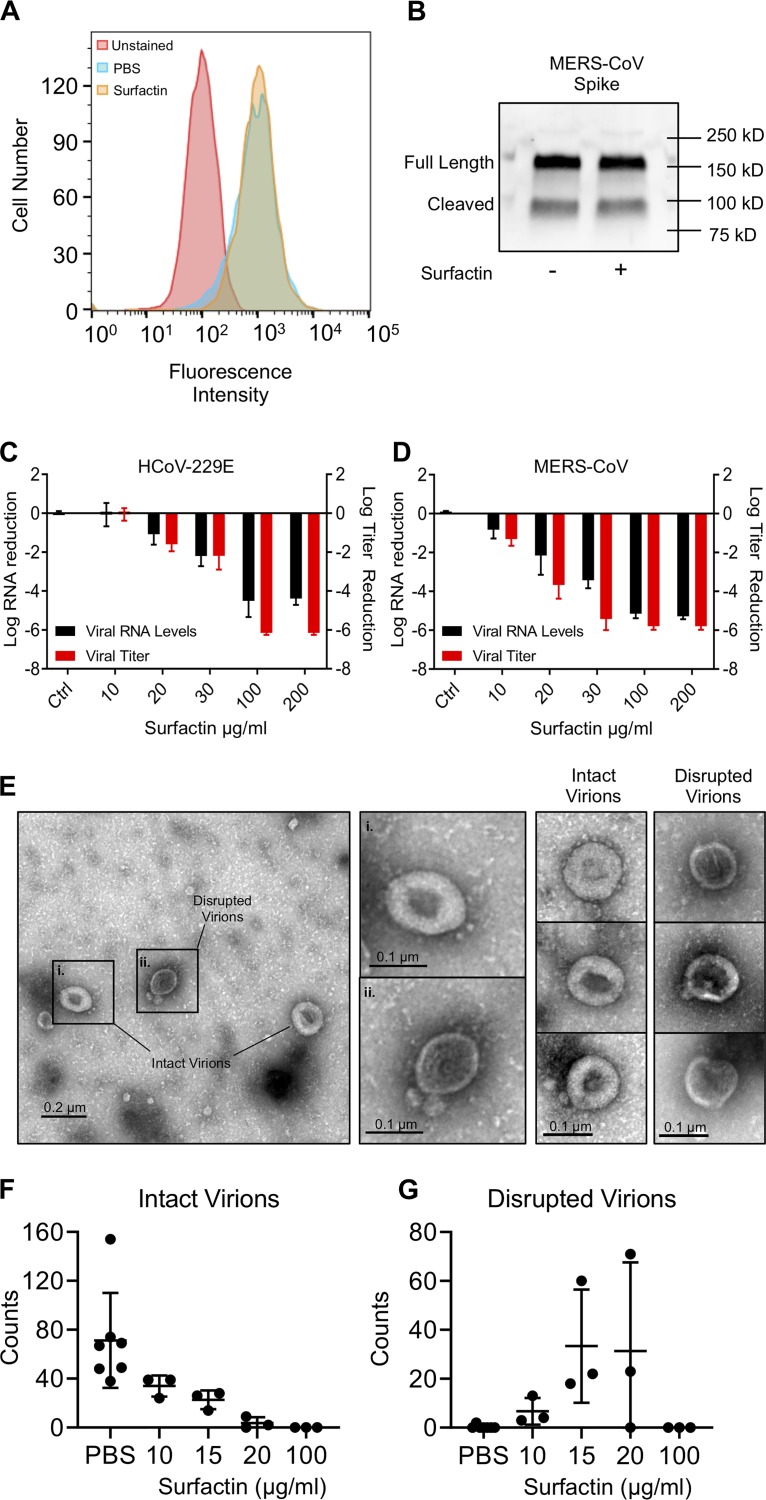
With clear inhibition of coronavirus infectivity by surfactin treatment, we next explored mechanism of action. We initially considered the possibility that surfactin disrupted spike/receptor binding activity, thus disrupting infection. We explored whether levels of the MERS-CoV glycoprotein spike or its receptor DDP4 were affected by surfactin treatment. DPP4 levels were assayed by flow cytometry in HUH7 cells, a line known to be permissive to MERS-CoV infection. Briefly, HUH7 cells were treated with surfactin. Cells were then incubated with a recombinant MERS-S1-Fc, stained with an anti-Fc-fluorescein isothiocyanate (FITC) secondary antibody, and total FITC staining was measured by flow cytometry (Fig. 4A). Surfactin- and PBS-treated cells had nearly identical fluorescent intensities, suggesting that surfactin treatment has no effect on DPP4 levels. Similarly, Western blot analysis of treated stock virus demonstrated that surfactin treatment had no effect on MERS-CoV spike glycoprotein protein concentrations (Fig. 4B). Together, these data suggested that both the host receptor and the viral spike glycoprotein were not disturbed by surfactin treatment.
FIG 4.
Surfactin disrupts CoV structural integrity. (A) HUH7 cells were first treated with either PBS or 100 μg/ml surfactin solution. Cells were then incubated with recombinant MERS-CoV S1-Fc protein and then stained with an anti-human IgG-FITC secondary in order to examine DPP4 levels. Unstained (red), PBS treated (blue), and surfactin treated (gold). Shown is a representative figure from 3 independent experiments. (B) MERS-CoV were treated with 100 μg/ml surfactin or PBS, as indicated, and then immunoblotted for MERS-CoV spike. Shown is a representative figure from 3 independent experiments. (C) HCoV-229E and (D) MERS-CoV were treated with 0 (ctrl) 10, 20, 30, 100, and 200 μg/ml of surfactin, as indicated. Viral infectivity was then determined (red) or samples were then treated with RNase I, RNA was extracted, and viral genome copy number determined by RT-qPCR (black). Bar graph bars represent the group mean for each experiment, and error bars represent SD (n = 3). (E to G) PBS- or 100 μg/ml surfactin-treated HCoV-229E samples were negatively stained and examined by TEM, and intact and disrupted virions were counted. A representative micrograph shown in panel E, with intact (i) and disrupted (ii) virions (left). Additional examples of intact and disrupted virions are also shown (right). Total counts of intact (F) and disrupted (G) virions are shown at various concentrations, with no less than 3 independent experiments having been performed for each concentration of surfactin. Horizontal lines represent group mean, while error bars represent SD. A two-tailed Student’s t test determined significance. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Using prior studies, we identified two other mechanisms for viricidal activity, namely, disruption of the viral membrane or inhibition of host-virus membrane fusion (13–15). Therefore, we examined if virion integrity was maintained by performing RNase I protection assays. Following surfactin treatment, particles were exposed to RNase I to digest exposed viral RNA; samples were subsequently extracted for RNA, and relative viral RNA was determined by quantitative reverse transcription real-time PCR (RT-qPCR). Increasing surfactin concentrations correlated with a decrease in viral RNA and viral titer for both HCoV-229E (Fig. 4C) and MERS-CoV (Fig. 4D), despite having no effect on MERS-CoV spike protein levels (Fig. 4B). These results indicate that disruption of virion integrity is the primary mechanism by which surfactin inhibits CoV infection. To confirm these results, we performed transmission electron microscopy (TEM) on HCoV-229E treated with 10, 15, 20, and 100 μg/ml surfactin or PBS. In control-treated samples, numerous intact HCoV-229E particles could be visualized (Fig. 4E to G). As surfactin concentration increased, fewer virions were observed by TEM, with few intact virions being observed in 20 μg/ml and none in 100 μg/ml surfactin-treated samples (Fig. 4F). Notably, numerous round, deflated structures of similar diameter as the HCoV-229E virions could be found in the 15- and 20-μg/ml samples, which may represent disrupted virions (Fig. 4G). Taken together, these results demonstrate that surfactin inhibits CoV infection primarily through the disruption of viral particles.
In vivo characterization of surfactin on CoV infection.
With no approved therapeutics (16), emerging, zoonotic CoVs pose a significant threat to public health (17, 18). Numerous in vivo studies have demonstrated that surfactin is tolerated at higher concentrations than in vitro cytotoxicity experiments with erythrocytes would suggest (19, 20). Additionally, our own in vitro cytotoxicity experiments have demonstrated that other cell lines tolerate surfactin at higher levels than those tolerated by erythrocytes. Together, this suggests that surfactin may have therapeutic potential. Therefore, we wanted to examine the potential of surfactin to treat infections in vivo. We reasoned that virions must interact with the respiratory microbiome upon release and transmission. To model this interaction, we tested whether direct treatment of the inoculum reduced in vivo infection and disease. SARS-CoV (104 PFU) was treated with PBS or surfactin and used to infect BALB/c mice intranasally (i.n.). Mice were monitored over 4 days for weight loss and lethality, with lung titers determined at 2 and 4 days postinfection. As expected, animals infected with PBS-treated virus experienced rapid weight loss and exhibited high lung titers at both 2 and 4 days postinfection (Fig. 5A). In contrast, mice infected with surfactin-treated SARS-CoV lost no weight, and no infectious virus was detected in their lungs (Fig. 5B). Additionally, mock-infected mice receiving surfactin alone demonstrated no signs of disease or weight loss, suggesting that surfactin treatment alone does not have any pathological effects (Fig. 5A).
FIG 5.
In vivo characterization of surfactin treatment on SARS-CoV infection. (A and B) BALB/c mice were then intranasally infected with 104 PFU of PBS- (black) or 100 μg/ml surfactin (red)-treated SARS-CoV MA15 and (A) monitored for weight loss over 4 days. Dotted lines and triangles represent mock-infected animals receiving PBS alone (black) or surfactin alone (red). (B) Lung tissue was harvested and viral titer determined at days 2 and 4. n = 4 for all infected groups, n = 2 for mock groups. (C and D) BALB/c mice were pretreated intranasally with 50 μl of either PBS (black) or surfactin in PBS (red). Eighteen h later, BALB/c mice were infected with 104 PFU of SARS-CoV (MA15) and (C) monitored for weight loss over 4 days. (D) Lung titer determined 2 (n = 5) and 4 days postinfection (n = 10). Dots on line graphs and bars on bar graphs represent the group mean. ND, no titers were detected. All error bars represent SD. P values were calculated using the two-tailed Student’s t test, with *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
To examine therapeutic potential, we next evaluated if pretreatment with surfactin could reduce respiratory CoV disease. BALB/c mice were treated i.n. with 50 μl of either PBS control or surfactin daily, starting 18 h prior to infection and continuing over the first 2 days of infection. Mice were subsequently infected with 104 PFU of SARS-CoV (MA15) and monitored for weight loss and lethality, with lung titer determined at 2 and 4 days postinfection. In contrast to the surfactin-treated inoculum, prophylactic surfactin treatment had no effect on weight loss (Fig. 5C) or viral titer in the lung (Fig. 5D). These results indicate that prophylactic surfactin treatment by this route does not reduce SARS-CoV disease in this mouse model.
Effects of other cyclic lipopeptides on CoV infectivity.
Surfactin belongs to a family of 80 natural antibiotic compounds referred to as cyclic lipopeptides (CLPs) (13). While structurally diverse, all CLPs share two key features, a nonpolar hydrocarbon tail and a non-ribosomally produced peptide ring (13, 14). While many CLPs have been found to be antifungal and antibacterial, antiviral properties have not been described except for surfactin, (13, 14). Therefore, we tested six additional CLPs for the ability to reduce CoV infectivity (Fig. 6A). Despite similar biochemical structures, none of the CLPs tested had a significant effect on HCoV-229E or MERS-CoV infection (Fig. 6B). These results suggest that unique features allow surfactin to reduce CoV infectivity.
Surfactin broadly reduces viral infectivity.
With its potent antiviral properties against CoVs, we next tested the effect of surfactin against other highly pathogenic viruses. Given its ability to disrupt virion integrity, we focused on enveloped viruses from diverse families, including two influenza A strains (H1N1 and H3N2), Zika virus (ZIKV), Dugbe virus (DUGV), Nipah virus (NiV), Crimean-Congo hemorrhagic fever virus (CCHFV), chikungunya virus (CHIKV), Mayaro virus, Una virus, and Ebola virus (EBOV). As a negative control, we tested the nonenveloped coxsackievirus B3 (CVB3). Each virus was treated with either PBS or surfactin, and viral infectivity was determined. As expected, surfactin had no effect on the nonenveloped CVB3 (Fig. 6C). In contrast, surfactin significantly reduced infectivity in each of the enveloped viruses (Fig. 6C), but the magnitude of effect was not uniform. Most enveloped viruses were reduced either to below their limit of detection or more than 100,000-fold. In contrast, Mayaro virus, both influenza strains, and EBOV exhibited some resistance, having their infectious titer reduced by only 2.6, 2.7, 2.4, and 1.6 logs, respectively. These data suggest that while surfactin treatment broadly reduced the infectivity of enveloped viruses, factors beyond the mere presence or absence of an envelope may govern overall sensitivity.
DISCUSSION
In this study, we explored the relationship between bacterial components and CoV infection. While initially predicting enhanced infection, treatment with B. subtilis PG reduced CoV infectivity, while envelope components from other bacteria had no effect. Separating the inhibitory effect using solvent washes, we used mass spectrometry to identify that the CLP surfactin was responsible for reduced CoV infectivity and disruption of virion integrity. Unfortunately, despite efficacy against the inoculum, prophylactic surfactin treatment prior to infection had no effect on CoV-related disease in vivo. Notably, other CLPs had no effect on CoV infectivity despite having similar biochemical structures. Finally, we found that surfactin treatment was efficacious against many enveloped viruses in vitro, including influenza A virus (IAV) strains H1N1 and H3N2, ZIKV, DUGV, NiV, CCHFV, CHIKV, Una virus, Mayaro virus, and EBOV. Together, these data demonstrate that surfactin is a potent virucide and highlight that interactions with bacterial derived compounds can also negatively modulate virus infection.
Over the last two decades, surfactin has been shown to be antibacterial, antifungal, and antiviral (13, 14, 19, 21, 22). Mechanistically, the broad antimicrobial efficacy of surfactin has been linked to disruption of lipid membranes (15). However, more recently, researchers describing the efficacy of surfactin against the animal CoV porcine epidemic diarrhea virus (PEDV) suggested that surfactin inhibited viral-host membrane fusion (19). Concentration of surfactin may explain the distinct mechanisms. In our studies, a higher surfactin dose (100 μg/ml) corresponded to virion destruction and a >10,000-fold reduction in CoV infectivity (Fig. 4C and D). In contrast, the PEDV study used a lower surfactin concentration (20 μg/ml), which corresponded to fusion inhibition based on intact particles and membrane integration assays (19). However, they reported only limited changes in viral load based on percent reduction in PFU. Our examination at the lower surfactin dose (20 μg/ml) found intermediate results with less reduction in CoV infectivity (10-fold reduction) but also a corresponding loss of viral RNA following RNase I treatment (Fig. 4C and D). TEM at this dose indicated the presence of both intact and disrupted virions. Together, the results suggest that virion structure may be maintained at this lower concentration but that virion integrity has been compromised in a subset of particles. Notably, the 50% inhibitory concentration (IC50) for surfactin varies within the human respiratory CoVs, with HCoV-229E (32.5 μg/ml) requiring a higher dose than SARS-CoV and MERS-CoV (18.9 and 16.8 μg/ml, respectively). This fact suggests that PEDV, an enteric CoV, may have increased resistance to surfactin-like molecules due to posttranslational modifications, including lipid composition or glycosylation state, possibly contributing to differences in mechanistic findings.
Similarly to the question of mechanism, in vivo efficacy of surfactin also varied between PEDV and human CoVs. While surfactin ablates SARS-CoV disease when treating the inoculum, prophylactic treatment was not protective. The failure of surfactin to protect mice against SARS-CoV is puzzling, given the efficacy of prophylactic oral surfactin treatment against PEDV disease (19). One explanation is that intranasal (i.n.) surfactin administration is not efficient in delivery to the lower respiratory tract. Branching of the airway may result in uneven distribution of surfactin, and prior studies have shown that intranasal instillation achieves only 50% efficiency at delivery to the lower respiratory tract (23, 24). In contrast, oral administration efficiently delivers compounds to the gastrointestinal system and is especially effective for lipopeptides, which persist within the intestines (25, 26). Dosing may also play an important role in distinguishing the differences in disease outcome, with oral treatment of the gut permitting a much higher dose (20 mg/kg every 3 hours) than intranasal delivery (0.5 mg/kg 2 times a day) (19). To overcome these problems, futures studies should use higher doses or more effective delivery methods, such as intubation or the inhalation of an aerosolized surfactin. Alternatively, several surfactin derivatives exist that enhance its viricidal activity and decrease its hemolytic activity (13, 14, 27). Using a derivative with higher activity and specificity may compensate for inefficient delivery or dose restriction. It is also possible that tissue-specific differences between respiratory tract and gut may limit access of surfactin to free virions. However, without further manipulation and understanding, surfactin is not yet a tangible target for therapeutic treatments of respiratory CoV infection.
In addition to CoVs, we examined the viricidal efficacy of surfactin against other enveloped viruses, discovering broad efficacy but wide variation. While all tested enveloped viruses were sensitive to surfactin treatment, IAV strains H1N1 and H3N2, Mayaro virus, and EBOV demonstrated a degree of resistance. These data suggest that factors beyond the mere presence of a viral envelope regulate surfactin efficacy. One possible factor is the lipid content of the viral envelope. Previous studies have shown that membranes enriched in cholesterol and phosphatidylethanolamines (PE) are resistant to surfactin permeabilization, while membranes containing phosphatidylcholines (PC) are more sensitive (28). The envelope of influenza A viruses have been reported to be enriched for both cholesterol and PE (29), providing support for this hypothesis. Unfortunately, the lipid content of the other viruses tested have not been determined, preventing direct comparison. Nevertheless, some broad observations are worth mentioning. CoVs, ZIKV, and bunyaviruses (CCHFV and DUGV) derive their envelopes from either the Golgi apparatus or endoplasmic reticulum, organelles enriched in surfactin-sensitive PC (30). NiV (31), IAV (32), and EBOV (33) are thought to derive their envelopes from lipid rafts of the plasma membrane, which could specify their lipid content and thus surfactin sensitivity. Alphaviruses such as CHIKV, Mayaro, and Una also bud from the plasma membrane, although neither the lipid content nor the involvement of lipid rafts has been explored (34). Together, these observations suggest that the lipid content of enveloped viruses may explain their differential sensitivity to surfactin.
The failure of other CLPs to reduce CoV infectivity is also surprising, given the structural similarity to surfactin. In particular, iturin A is biochemically similar to surfactin and has also been reported to disrupt lipid membranes (Fig. 6A) (13). A possible explanation involves differences in their mechanisms of action. Surfactin penetrates lipid layers, alone solubilizing and permeabilizing them (15). In contrast, iturin A must interact with sterol components to cause membrane permeabilization, explaining its broad antifungal activity but only selective antibacterial activity (13). However, iturin A is also quite hemolytic (13, 14), making it unclear why the membranes of enveloped viruses grown in mammalian cells would not also be susceptible to this mechanism due to the presence of sterols. Compounding this mechanistic uncertainty, daptomycin’s permeabilization of membranes requires no such interaction, but CoVs are resistant to its effects as well (35) (Fig. 6B). The results argue that surfactin possesses unique properties that confer its viricidal activity.
The temperature sensitivity of the viricidal properties of surfactin is interesting, as it implies that temperature modulates the physical interaction between surfactin and the CoV particle. Molecular dynamics simulation suggest that temperature governs both the 3-dimensional “horse saddle” structure of individual surfactin molecules and shape of surfactin micelles (36). Thus, one explanation for the temperature-sensitive viricidal activity of surfactin is that the specific structural conformations necessary to disrupt virion integrity only occur at higher temperatures. Alternatively, temperature could modulate the specific structure of the CoV virus itself. Virions are dynamic structures that undergo “viral breathing,” whereby they undergo large structural fluctuations that are often required for infection (37). Importantly, viral breathing can be enhanced by incubation at higher temperatures (37). While viral breathing has not been examined in the context of CoVs, the receptor binding domain (RBD) of both the SARS-CoV and MERS-CoV S ectodomain trimer fluctuates between lying and standing positions, demonstrating the flexibility of CoV structural proteins (38). Thus, it is possible that, at lower temperatures, CoVs adopt a conformation that sterically hinders surfactin from accessing the viral membrane, preventing disruption. Future studies are necessary to differentiate between these possibilities in order to fully understand the implications of the temperature dependent effects of surfactin.
While the use of surfactin as a treatment proved ineffective for respiratory infection, surfactin may have potential utility as a disinfectant. Its rapid activity against an array of enveloped viruses offers a rapid and effective means to inactivate infectious samples. In contrast to other detergent- and heat-based approaches, inactivation with surfactin would be expected to only act on membranes, maintaining protein-protein interactions and allowing a range of further studies. Similarly, the relatively low cost and lack of corrosive effect could permit its use in a variety of locations, including health care, school, and home settings. While bacterial resistance might normally be a concern, the existence of several resistant species already limits its utility as an antibiotic for treatment (39). While dosing and efficacy against different virus families is required, these factors offer surfactin as a potent viricidal disinfectant moving forward.
Overall, while the microbiome has historically been thought to serve a protective role against pathogens (1, 2), recent studies with viruses complicate this view. Studies with poliovirus demonstrated that the presence of commensal bacteria is necessary for oral poliovirus infection in mice (40). Similar findings with other enteric viruses suggest that utilizing bacterial components is a common approach. In contrast, our results add further complexity, demonstrating that surfactin, a secondary metabolite of B. subtilis, can potently reduce CoV infectivity. Although B. subtilis is not generally part of the human microbiome (41), it is often used as an intestinal probiotic and has been found to transiently persist in the gut (42). Additionally, surfactin-like molecules are produced by a broad array of bacterial species (13, 43–45). For example, the novel surfactin-like CLP coryxin was recently found to be produced by Corynebacterium xerosis, a common member of the respiratory microbiome (45). These facts suggest that microbial components typically thought to work against bacterial competitors could also potentially disrupt viral infection. Thus, as the relationship between the microbiome and viral infections is further explored, the role that bacterial metabolites such as surfactin and other CLPs play in modulating infection must be considered in viral disease. Overall, these results highlight the dynamic microbial environment and its potential to impact viral pathogenesis, as well as for identifying novel inhibitory factors for therapeutic use.
MATERIALS AND METHODS
Viruses, cells, and in vitro infection.
HCoV-229E, provided by the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA), was propagated on HUH7 cells grown in Dulbecco’s minimal essential medium (DMEM; Gibco), 10% fetal bovine serum (HyClone), and 1% antibiotic-antimycotic (A/A) (Gibco). Titration was performed by 50% tissue culture infective dose (TCID50) in HUH7 cells and calculated by the Spearman-Karber method. MERS-CoV (EMC-2012 strain) (46) and recombinant SARS-CoV (MA15) (47) were titrated and propagated on VeroCCL81 and VeroE6 cells, respectively, which were grown in DMEM with 5% fetal bovine serum and 1% A/A. Standard plaque assays were used for SARS-CoV and MERS-CoV (48, 49). Coxsackievirus B3 (50) and chikungunya (51), Nipah (52), Dugbe (53), Zika (54), Crimean-Congo hemorrhagic fever (53), influenza A H1N1 (A/California/04/09) and H3N2 (A/Panama/2007/99) (55), and Ebola (56) viruses were propagated and quantitated via standard methods. All experiments involving infectious virus were conducted at the University of Texas Medical Branch (Galveston, TX) in approved biosafety level 2, 3, or 4 (BSL) laboratories and animal facilities, with routine medical monitoring of staff.
Treatment with bacterial surface components and cyclic lipopeptides.
CoVs were diluted 10% (vol/vol) in solutions with final concentrations of 1 mg/ml (PG and LPS) or 100 μg/μl (CLPs) unless otherwise specified in the text. For alcohol wash experiments, samples were instead diluted 5% (vol/vol). Treated samples were then incubated for 2 h at 37°C, after which they titer was determined. The following bacterial components were purchased from Sigma-Aldrich: lipopolysaccharides from Escherichia coli (catalog no. L4130), peptidoglycan Bacillus subtilis (catalog no. 69554), Staphylococcus aureus (catalog no. 77140), Streptomyces species (catalog no. 79682), and Micrococcus luteus (catalog no. 53243). Peptidoglycan from Escherichia coli (PGN-EB) was purchased from InvivoGen. For each surface component, stock solutions were created by suspending the component in PBS and then stored at –20°C. The cyclic lipopeptides surfactin (catalog no. S3523), iturin A (catalog no. I1774), fengycin (catalog no. SMB00292), polymyxin B (catalog no. P1004), colistin (catalog no. C4461), ramoplanin (catalog no. R1781), and daptomycin (catalog no. D2446) were also purchased from Sigma-Aldrich. PBS was selected as a vehicle based on solubility data from previous studies, which established that surfactin is soluble in PBS at levels well above the concentrations used here (57–59).
Mass spectrometry.
Stock peptidoglycan was centrifuged at 15,000 × g for 1 min in a tabletop centrifuge, and the insoluble PG fraction was then resuspended in 100% ethanol. Following a 5-min incubation at room temperature, samples were centrifuged, and the supernatant and insoluble fractions were used for treatment of viruses or delivered to the mass spectrometry core facility. Peptidoglycan (1 μl) was combined 1:1 with a 10 mg/ml α-cyano-4-hydroxycinnamic acid (60% acetonitrile) and spotted onto MALDI targets. All MALDI-MS experiments were performed using a 5800 MALDI-TOF/TOF system (Applied Biosystems). The MS data were acquired using the reflectron detector in positive mode (700 to 4,500 Da, 1,900 Da focus mass) using 300 laser shots (50 shots per subspectrum). Collision-induced dissociation tandem MS spectra were acquired using 1 kV of collision energy. Fragmentation data were analyzed manually to determine structural information.
Flow cytometry.
DPP4 levels were measured by first incubating HUH7 cells with 100 μg/ml of surfactin. Cells (2.5 × 105) were then incubated with 15 μg/ml of purified recombinant MERS-CoV S1 fused to the Fc region of human IgG, followed by incubation with anti-human IgG-FITC (catalog no. F9512; Sigma-Aldrich). Flow cytometry was performed on a Guava easyCyte 8HT system (Luminex), and data were analyzed using FlowJo 10.6.1.
Western blot analysis.
MERS-CoV was inactivated by diluting viral stock in AV lysis buffer and 10 mM EDTA-0.9% SDS in a 1:1:1 ratio before heating at 95°C for 15 min. Equal volumes of inactivated virus then underwent electrophoresis by SDS-PAGE and were transferred to polyvinylidene difluoride (PVDF) membranes. MERS-CoV spike levels were determined using a MERS-CoV (NCoV/novel coronavirus) spike antibody (catalog no. 40069-RP01; Sino Biologicals) and measured using horseradish peroxidase (HRP)-conjugated anti-rabbit antibody (catalog no. 7074S; Cell Signaling Technology). Blots were developed using enhanced chemiluminescence Clarity Max reagent (Bio-Rad) and imaged using the ChemiDoc MP imaging system (Bio-Rad). Image processing was done using ImageLab 6.0.1 (Bio-Rad).
Transmission electron microscopy.
HCoV-229E virions were visualized by transmission electron microscopy (TEM) through negative staining with 2% uranyl acetate (60). Briefly, 200-mesh Formvar carbon-coated copper grids (catalog no. FCF200-CU; Electron Microscopy Sciences) were treated for 20 min with HCoV-229E samples. Excess sample solution was then wicked off with filter paper, and each grid was then stained for 45 s with 2% uranyl acetate solution. Excess stain was again wicked off with filter paper. Grids were then dried and visualized on a Philips CM100 transmission electron microscope. Images were recorded with a Gatan Orius SC200 charge-coupled-device (CCD) camera. In order to ensure even counting, 10 pictures were taken on 3 different cells on each grid for each replicate. No more than 10 min was allotted for looking for virions in each cell.
RNase I protection assay.
Assays were performed in accordance with standard protocols described previously (61). Briefly, samples were treated with 250 U RNase I for 30 min. To halt RNA digestion and inactivate RNase I, a 2× volume of viral RNA buffer (catalog no. R1034-1-100; Zymo Research) with 2-mercaptoethanol was added. RNA was then extracted using the Quick-RNA viral kit (catalog no. R1035; Zymo Research). RNA was then converted into cDNA using the iScript cDNA synthesis kit (catalog no. 170-8891; Bio-Rad). Quantitative real-time PCR was performed using SsoAdvanced Universal SYBR Green Supermix (catalog no. 172-5271; Bio-Rad). HCoV-229E specific primer sequences were forward, 5-TGACATTCGCGACTACAAGC-3, and reverse, 5-TAACGGTGGTTTGGCTTTTC-3. MERS-CoV specific primer sequences were forward, 5-TCGCTTGGCAAATGAGTGTG-3, and reverse, 5-ACATTAGCAGTTGTCGCCTG-3.
Statistical analysis.
All statistical comparisons in this study involved the comparison between 2 groups, untreated control virus and peptidoglycan/surfactin-treated virus. Thus, significant differences in viral titer, RNA levels, and weight loss were determined by the unpaired two-tailed Student’s t test.
Ethics statement.
This study was carried out in accordance with the recommendations for care and use of animals by the Office of Laboratory Animal Welfare, National Institutes of Health. The Institutional Animal Care and Use Committee (IACUC) of University of Texas Medical Branch (UTMB) approved the animal studies under protocols 1711065 and 1707046.
Mice and in vivo infection.
Ten-week-old BALB/c mice were purchased from Charles River Laboratories and maintained in SealSafe HEPA-filtered air in/out units. Animals were anesthetized with isoflurane and infected intranasally (i.n.) with 104 PFU in 50 μl of phosphate-buffered saline (PBS). Infected animals were monitored for weight loss, morbidity, and clinical signs of disease, and lung titers were determined as described previously (62). For experiments involving prophylactic treatment with surfactin, 50 μl of 100 μg/ml surfactin-PBS was administered i.n. to anesthetized animals 18 h prior to infection, with additional treatments on day 0, day 1, and day 2. Infected animals were weighed daily, and lungs were collected 2 and 4 days postinfection for downstream analysis by plaque assay.
ACKNOWLEDGMENTS
Research was supported by grants from NIA and NIAID of the NIH (U19AI100625 and R00AG049092 to V.D.M.; R01AI134907, R21AI126012, and R21AI132479-01 to R.R.; and R24AI120942 to WRCEVA). Research was also supported by a STARs Award provided by the University of Texas System to V.D.M. and by trainee funding provided by the McLaughlin Endowment at UTMB.
REFERENCES
- 1.Man WH, de Steenhuijsen Piters WA, Bogaert D. 2017. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol 15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thaiss CA, Zmora N, Levy M, Elinav E. 2016. The microbiome and innate immunity. Nature 535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 3.Karst SM. 2016. The influence of commensal bacteria on infection with enteric viruses. Nat Rev Microbiol 14:197–204. doi: 10.1038/nrmicro.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger AK, Yi H, Kearns DB, Mainou BA. 2017. Bacteria and bacterial envelope components enhance mammalian reovirus thermostability. PLoS Pathog 13:e1006768. doi: 10.1371/journal.ppat.1006768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rowe HM, Meliopoulos VA, Iverson A, Bomme P, Schultz-Cherry S, Rosch JW. 2019. Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat Microbiol 4:1328. doi: 10.1038/s41564-019-0447-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drexler JF, Corman VM, Drosten C. 2014. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res 101:45–56. doi: 10.1016/j.antiviral.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gralinski LE, Menachery VD, Morgan AP, Totura AL, Beall A, Kocher J, Plante J, Harrison-Shostak DC, Schäfer A, Pardo-Manuel de Villena F, Ferris MT, Baric RS. 2017. allelic variation in the toll-like receptor adaptor protein. G3 (Bethesda) 7:1653–1663. doi: 10.1534/g3.117.041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Totura AL, Whitmore A, Agnihothram S, Schäfer A, Katze MG, Heise MT, Baric RS. 2015. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio 6:e00638. doi: 10.1128/mBio.00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheahan T, Morrison TE, Funkhouser W, Uematsu S, Akira S, Baric RS, Heise MT. 2008. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog 4:e1000240. doi: 10.1371/journal.ppat.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson BA, Hage A, Kalveram B, Mears M, Plante JA, Rodriguez SE, Ding Z, Luo X, Bente D, Bradrick SS, Freiberg AN, Popov V, Rajsbaum R, Rossi S, Russell WK, Menachery VD. 2019. Peptidoglycan associated cyclic lipopeptide disrupts viral infectivity. bioRxiv 10.1101/635854. [DOI] [PMC free article] [PubMed]
- 11.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. doi: 10.1111/j.1574-6976.2007.00094.x. [DOI] [PubMed] [Google Scholar]
- 12.Yang H, Li X, Li X, Yu H, Shen Z. 2015. Identification of lipopeptide isoforms by MALDI-TOF-MS/MS based on the simultaneous purification of iturin, fengycin, and surfactin by RP-HPLC. Anal Bioanal Chem 407:2529–2542. doi: 10.1007/s00216-015-8486-8. [DOI] [PubMed] [Google Scholar]
- 13.Cochrane SA, Vederas JC. 2016. Lipopeptides from Bacillus and Paenibacillus spp.: a gold mine of antibiotic candidates. Med Res Rev 36:4–31. doi: 10.1002/med.21321. [DOI] [PubMed] [Google Scholar]
- 14.Meena KR, Kanwar SS. 2015. Lipopeptides as the antifungal and antibacterial agents: applications in food safety and therapeutics. Biomed Res Int 2015:473050. doi: 10.1155/2015/473050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seydlová G, Svobodová J. 2008. Review of surfactin chemical properties and the potential biomedical applications. Cent Eur J Med 3:123–133. [Google Scholar]
- 16.Mustafa S, Balkhy H, Gabere MN. 2018. Current treatment options and the role of peptides as potential therapeutic components for Middle East Respiratory Syndrome (MERS): a review. J Infect Public Health 11:9–17. doi: 10.1016/j.jiph.2017.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization. 2004. WHO SARS risk assessment and preparedness framework. WHO Press, Geneva, Switzerland. [Google Scholar]
- 18.World Health Organization. 2017. WHO MERS-CoV global summary and assessment of risk. WHO Press, Geneva, Switzerland. [Google Scholar]
- 19.Yuan L, Zhang S, Wang Y, Li Y, Wang X, Yang Q. 2018. Surfactin inhibits membrane fusion during invasion of epithelial cells by enveloped viruses. J Virol 92:e00809-18. doi: 10.1128/JVI.00809-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoshino N, Takeshita R, Kawamura H, Sasaki Y, Kagabu M, Sugiyama T, Muraki Y, Sato S. 2018. Mast cells partially contribute to mucosal adjuvanticity of surfactin in mice. Immun Inflamm Dis 6:117–127. doi: 10.1002/iid3.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, Hu W, Zhu L, Yang Q. 2017. Bacillus subtilis and surfactin inhibit the transmissible gastroenteritis virus from entering the intestinal epithelial cells. Biosci Rep 37:BSR20170082. doi: 10.1042/BSR20170082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang X, Zhao J, Fang X, Liu H, Zhang Y, Cen S, Yu L. 2017. Surfactin derivatives from Micromonospora sp. CPCC 202787 and their anti-HIV activities. J Antibiot (Tokyo) 70:105–108. doi: 10.1038/ja.2016.63. [DOI] [PubMed] [Google Scholar]
- 23.Miller MA, Stabenow JM, Parvathareddy J, Wodowski AJ, Fabrizio TP, Bina XR, Zalduondo L, Bina JE. 2012. Visualization of murine intranasal dosing efficiency using luminescent Francisella tularensis: effect of instillation volume and form of anesthesia. PLoS One 7:e31359. doi: 10.1371/journal.pone.0031359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Southam DS, Dolovich M, O’Byrne PM, Inman MD. 2002. Distribution of intranasal instillations in mice: effects of volume, time, body position, and anesthesia. Am J Physiol Lung Cell Mol Physiol 282:L833–9. doi: 10.1152/ajplung.00173.2001. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen DS, Shepherd NE, Xu W, Lucke AJ, Stoermer MJ, Fairlie DP. 2017. Orally absorbed cyclic peptides. Chem Rev 117:8094–8128. doi: 10.1021/acs.chemrev.6b00838. [DOI] [PubMed] [Google Scholar]
- 26.Chen H, Langer R. 1998. Oral particulate delivery: status and future trends. Adv Drug Deliv Rev 34:339–350. doi: 10.1016/S0169-409X(98)00047-7. [DOI] [PubMed] [Google Scholar]
- 27.Yuan L, Zhang S, Peng J, Li Y, Yang Q. 2019. Synthetic surfactin analogues have improved anti-PEDV properties. PLoS One 14:e0215227. doi: 10.1371/journal.pone.0215227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carrillo C, Teruel JA, Aranda FJ, Ortiz A. 2003. Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin. Biochim Biophys Acta 1611:91–97. doi: 10.1016/s0005-2736(03)00029-4. [DOI] [PubMed] [Google Scholar]
- 29.Ivanova PT, Myers DS, Milne SB, McClaren JL, Thomas PG, Brown HA. 2015. Lipid composition of viral envelope of three strains of influenza virus—not all viruses are created equal. ACS Infect Dis 1:399–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Meer G, Voelker DR, Feigenson GW. 2008. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harrison MS, Sakaguchi T, Schmitt AP. 2010. Paramyxovirus assembly and budding: building particles that transmit infections. Int J Biochem Cell Biol 42:1416–1429. doi: 10.1016/j.biocel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rossman JS, Lamb RA. 2011. Influenza virus assembly and budding. Virology 411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bavari S, Bosio CM, Wiegand E, Ruthel G, Will AB, Geisbert TW, Hevey M, Schmaljohn C, Schmaljohn A, Aman MJ. 2002. Lipid raft microdomains: a gateway for compartmentalized trafficking of Ebola and Marburg viruses. J Exp Med 195:593–602. doi: 10.1084/jem.20011500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown RS, Wan JJ, Kielian M. 2018. The alphavirus exit pathway: what we know and what we wish we knew. Viruses 10:E89. doi: 10.3390/v10020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor SD, Palmer M. 2016. The action mechanism of daptomycin. Bioorg Med Chem 24:6253–6268. doi: 10.1016/j.bmc.2016.05.052. [DOI] [PubMed] [Google Scholar]
- 36.She AQ, Gang HZ, Mu BZ. 2012. Temperature influence on the structure and interfacial properties of surfactin micelle: a molecular dynamics simulation study. J Phys Chem B 116:12735–12743. doi: 10.1021/jp302413c. [DOI] [PubMed] [Google Scholar]
- 37.Kuhn RJ, Dowd KA, Beth Post C, Pierson TC. 2015. Shake, rattle, and roll: impact of the dynamics of flavivirus particles on their interactions with the host. Virology 479–480:508–517. doi: 10.1016/j.virol.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan Y, Cao D, Zhang Y, Ma J, Qi J, Wang Q, Lu G, Wu Y, Yan J, Shi Y, Zhang X, Gao GF. 2017. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun 8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoefler BC, Gorzelnik KV, Yang JY, Hendricks N, Dorrestein PC, Straight PD. 2012. Enzymatic resistance to the lipopeptide surfactin as identified through imaging mass spectrometry of bacterial competition. Proc Natl Acad Sci U S A 109:13082–13087. doi: 10.1073/pnas.1205586109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, Hooper LV, Dermody TS, Pfeiffer JK. 2011. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science 334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Steenhuijsen Piters WA, Sanders EA, Bogaert D. 2015. The role of the local microbial ecosystem in respiratory health and disease. Philos Trans R Soc Lond B Biol Sci 370:20140294. doi: 10.1098/rstb.2014.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeżewska-Frąckowiak J, Seroczyńska K, Banaszczyk J, Jedrzejczak G, Żylicz-Stachula A, Skowron PM. 2018. The promises and risks of probiotic Bacillus species. Acta Biochim Pol 65:509–519. doi: 10.18388/abp.2018_2652. [DOI] [PubMed] [Google Scholar]
- 43.Burja AM, Abou-Mansour E, Banaigs B, Payri C, Burgess JG, Wright PC. 2002. Culture of the marine cyanobacterium, Lyngbya majuscula (Oscillatoriaceae), for bioprocess intensified production of cyclic and linear lipopeptides. J Microbiol Methods 48:207–219. doi: 10.1016/S0167-7012(01)00324-4. [DOI] [PubMed] [Google Scholar]
- 44.Morikawa M, Daido H, Takao T, Murata S, Shimonishi Y, Imanaka T. 1993. A new lipopeptide biosurfactant produced by Arthrobacter sp. strain MIS38. J Bacteriol 175:6459–6466. doi: 10.1128/jb.175.20.6459-6466.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalili D, Amini M, Faramarzi MA, Fazeli MR, Khoshayand MR, Samadi N. 2015. Isolation and structural characterization of coryxin, a novel cyclic lipopeptide from Corynebacterium xerosis NS5 having emulsifying and anti-biofilm activity. Colloids Surf B Biointerfaces 135:425–432. doi: 10.1016/j.colsurfb.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 46.Kindler E, Jónsdóttir HR, Muth D, Hamming OJ, Hartmann R, Rodriguez R, Geffers R, Fouchier RA, Drosten C, Müller MA, Dijkman R, Thiel V. 2013. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio 4:e00611. doi: 10.1128/mBio.00611-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts A, Deming D, Paddock CD, Cheng A, Yount B, Vogel L, Herman BD, Sheahan T, Heise M, Genrich GL, Zaki SR, Baric R, Subbarao K. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog 3:e5. doi: 10.1371/journal.ppat.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sims AC, Tilton SC, Menachery VD, Gralinski LE, Schäfer A, Matzke MM, Webb-Robertson BJ, Chang J, Luna ML, Long CE, Shukla AK, Bankhead AR, Burkett SE, Zornetzer G, Tseng CT, Metz TO, Pickles R, McWeeney S, Smith RD, Katze MG, Waters KM, Baric RS. 2013. Release of severe acute respiratory syndrome coronavirus nuclear import block enhances host transcription in human lung cells. J Virol 87:3885–3902. doi: 10.1128/JVI.02520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Josset L, Menachery VD, Gralinski LE, Agnihothram S, Sova P, Carter VS, Yount BL, Graham RL, Baric RS, Katze MG. 2013. Cell host response to infection with novel human coronavirus EMC predicts potential antivirals and important differences with SARS coronavirus. mBio 4:e00165. doi: 10.1128/mBio.00165-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bradrick SS, Lieben EA, Carden BM, Romero JR. 2001. A predicted secondary structural domain within the internal ribosome entry site of echovirus 12 mediates a cell-type-specific block to viral replication. J Virol 75:6472–6481. doi: 10.1128/JVI.75.14.6472-6481.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plante KS, Rossi SL, Bergren NA, Seymour RL, Weaver SC. 2015. Extended preclinical safety, efficacy and stability testing of a live-attenuated Chikungunya vaccine candidate. PLoS Negl Trop Dis 9:e0004007. doi: 10.1371/journal.pntd.0004007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Freiberg AN, Worthy MN, Lee B, Holbrook MR. 2010. Combined chloroquine and ribavirin treatment does not prevent death in a hamster model of Nipah and Hendra virus infection. J Gen Virol 91:765–772. doi: 10.1099/vir.0.017269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bente DA, Alimonti JB, Shieh WJ, Camus G, Ströher U, Zaki S, Jones SM. 2010. Pathogenesis and immune response of Crimean-Congo hemorrhagic fever virus in a STAT-1 knockout mouse model. J Virol 84:11089–11100. doi: 10.1128/JVI.01383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Widman DG, Young E, Yount BL, Plante KS, Gallichotte EN, Carbaugh DL, Peck KM, Plante J, Swanstrom J, Heise MT, Lazear HM, Baric RS. 2017. A reverse genetics platform that spans the Zika virus family tree. mBio 8:e02014-16. doi: 10.1128/mBio.02014-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rajsbaum R, Albrecht RA, Wang MK, Maharaj NP, Versteeg GA, Nistal-Villán E, García-Sastre A, Gack MU. 2012. Species-specific inhibition of RIG-I ubiquitination and IFN induction by the influenza A virus NS1 protein. PLoS Pathog 8:e1003059. doi: 10.1371/journal.ppat.1003059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bharaj P, Atkins C, Luthra P, Giraldo MI, Dawes BE, Miorin L, Johnson JR, Krogan NJ, Basler CF, Freiberg AN, Rajsbaum R. 2017. The host E3-ubiquitin ligase TRIM6 ubiquitinates the Ebola virus VP35 protein and promotes virus replication. J Virol 91:e00833-17. doi: 10.1128/JVI.00833-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu YS, Ngai SC, Goh BH, Chan KG, Lee LH, Chuah LH. 2017. Anticancer activities of surfactin and potential application of nanotechnology assisted surfactin delivery. Front Pharmacol 8:761. doi: 10.3389/fphar.2017.00761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim H-S, Yoon B-D, Lee C-H, Suh H-H, Oh H-M, Katsuragi T, Tani Y. 1997. Production and properties of a lipopeptide biosurfactant from Bacillus subtilis C9. J Ferment Bioeng 84:41–46. doi: 10.1016/S0922-338X(97)82784-5. [DOI] [Google Scholar]
- 59.Abdel-Mawgoud AM, Aboulwafa MM, Hassouna NA. 2008. Characterization of surfactin produced by Bacillus subtilis isolate BS5. Appl Biochem Biotechnol 150:289–303. doi: 10.1007/s12010-008-8153-z. [DOI] [PubMed] [Google Scholar]
- 60.Berryman MA, Rodewald RD. 1990. An enhanced method for post-embedding immunocytochemical staining which preserves cell membranes. J Histochem Cytochem 38:159–170. doi: 10.1177/38.2.1688894. [DOI] [PubMed] [Google Scholar]
- 61.Gilling DH, Kitajima M, Torrey JR, Bright KR. 2014. Mechanisms of antiviral action of plant antimicrobials against murine norovirus. Appl Environ Microbiol 80:4898–4910. doi: 10.1128/AEM.00402-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheahan T, Whitmore A, Long K, Ferris M, Rockx B, Funkhouser W, Donaldson E, Gralinski L, Collier M, Heise M, Davis N, Johnston R, Baric RS. 2011. Successful vaccination strategies that protect aged mice from lethal challenge from influenza virus and heterologous severe acute respiratory syndrome coronavirus. J Virol 85:217–230. doi: 10.1128/JVI.01805-10. [DOI] [PMC free article] [PubMed] [Google Scholar]