Hepatitis C virus (HCV) is known for its ability to modulate phosphoinositide signaling pathways for its replication. Elevated levels of phosphatidylinositol 4-phosphate [PI(4)P] in HCV replication organelles (ROs) recruits lipid transfer proteins (LTPs), like oxysterol-binding protein (OSBP). OSBP exchanges PI(4)P with cholesterol, thus removing PI(4)P from the HCV RO. Here, we found that the phosphatidylinositol transfer protein Nir2 acts as an LTP and may replenish PI at the HCV RO by interacting with VAMP-associated proteins (VAPs), enabling continuous viral replication during chronic infection. Therefore, the coordination of OSBP, Nir2, and VAPs completes our understanding of the phosphoinositide cycle between the ER and HCV ROs.
KEYWORDS: hepatitis C virus, lipid transfer protein, membrane contact sites, phosphoinositides, viral replication
ABSTRACT
The endoplasmic reticulum (ER)-resident proteins vesicle-associated membrane protein (VAMP)-associated protein A and B (VAPA and VAPB) have been reported to be necessary for efficient hepatitis C virus (HCV) replication, but the specific mechanisms are not well understood. VAPs are known to recruit lipid transfer proteins to the ER, including oxysterol binding protein (OSBP), which has been previously shown to be necessary for cholesterol delivery to the HCV replication organelle in exchange for phosphatidylinositol 4-phosphate [PI(4)P]. Here, we show that VAPA and VAPB are redundant for HCV infection and that dimerization is not required for their function. In addition, we identify the phosphatidylinositol transfer protein Nir2 as an effector of VAPs to support HCV replication. We propose that Nir2 functions to replenish phosphoinositides at the HCV replication organelle to maintain elevated steady-state levels of PI(4)P, which is removed by OSBP. Thus, Nir2, along with VAPs, OSBP, and the phosphatidylinositol 4-kinase, completes a cycle of phosphoinositide flow between the ER and viral replication organelles to drive ongoing viral replication.
IMPORTANCE Hepatitis C virus (HCV) is known for its ability to modulate phosphoinositide signaling pathways for its replication. Elevated levels of phosphatidylinositol 4-phosphate [PI(4)P] in HCV replication organelles (ROs) recruits lipid transfer proteins (LTPs), like oxysterol-binding protein (OSBP). OSBP exchanges PI(4)P with cholesterol, thus removing PI(4)P from the HCV RO. Here, we found that the phosphatidylinositol transfer protein Nir2 acts as an LTP and may replenish PI at the HCV RO by interacting with VAMP-associated proteins (VAPs), enabling continuous viral replication during chronic infection. Therefore, the coordination of OSBP, Nir2, and VAPs completes our understanding of the phosphoinositide cycle between the ER and HCV ROs.
INTRODUCTION
Like all positive-sense RNA viruses, hepatitis C virus (HCV) induces cytoplasmic membrane alterations in infected cells (1), which have been termed “membranous webs” or replication organelles (ROs) and are believed to be sites of HCV RNA synthesis. ROs may serve to shield viral components from host innate immunity and may also concentrate RNA templates with viral and host replication cofactors to promote efficient replication. Additionally, the lipid composition of ROs may be optimized for optimal viral replication.
The host phosphatidylinositol 4-kinase PI4KA and its downstream effector oxysterol-binding protein (OSBP) are essential for HCV replication and RO integrity (2–8). OSBP is a member of a large class of cellular lipid transfer proteins (LTPs) and is recruited by vesicle-associated membrane protein (VAMP)-associated proteins (VAPs) and phosphatidylinositol 4-phosphate [PI(4)P] to membrane contact sites (MCS) between the endoplasmic reticulum (ER) and other cellular membranes, where it exchanges sterols for PI(4)P between apposed membranes (9). VAPs, which in humans are encoded by the VAPA and VAPB genes, are highly conserved ER-resident transmembrane proteins (10). VAPs interact with OSBP and many other LTPs by binding to an FFAT (two phenylalanine in an acidic tract) motif. This recruitment of LTPs by VAPs to form MCSs has become recognized as a key mechanism of interorganellar nonvesicular transfer of lipids (10). VAPs are necessary for efficient HCV replication (11, 12) and interact with the HCV NS5A and NS5B nonstructural proteins (12–14), but the mechanistic details of how they support HCV replication are not well understood.
There are five known human phosphatidylinositol transfer proteins (PITPs) that are implicated in the nonvesicular trafficking of phosphatidylinositol (PI) between intracellular membranes (15). One of these PITPs is PITPNM1/Nir2, which contains a PI transfer protein domain as well as an FFAT domain for interaction with VAPs (15) and which has been reported to exchange PI for phosphatidic acid (PA) at ER-plasma membrane MCSs (16).
Here, we report that VAPA and VAPB play redundant roles in supporting HCV infection and that efficient VAP dimerization is not required to support HCV replication. Additionally, we find that VAPs interact with Nir2 in HCV-infected cells and that Nir2 is essential for efficient HCV infection as well as for upregulation of PI(4)P levels in HCV-expressing cells. This is consistent with a model in which Nir2 promotes the replenishment of PI(4)P at viral ROs to counteract its removal by OSBP-mediated exchange for sterols.
RESULTS
VAPA and VAPB play redundant roles in HCV replication.
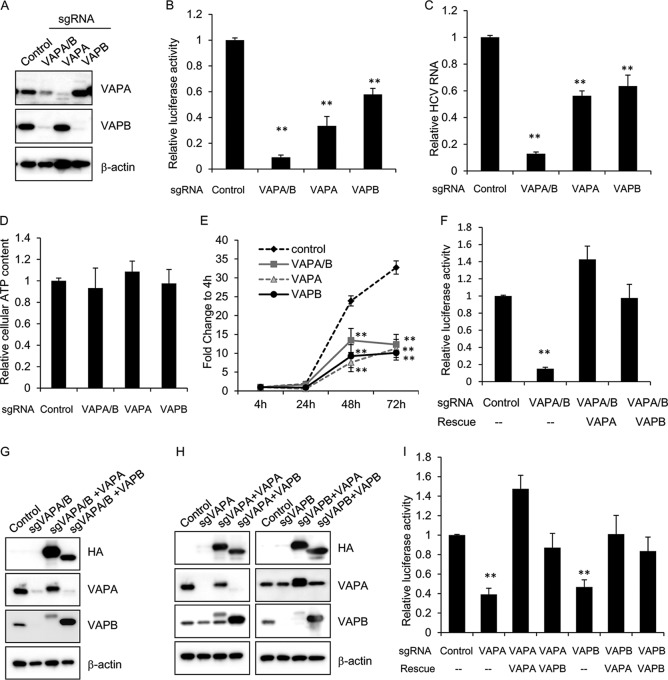
To test the requirement for the two VAP genes in HCV infection, we employed the CRISPR-Cas9 knockout system using single guide RNAs (sgRNAs) targeting VAPA, VAPB, or a conserved sequence in the major sperm protein (MSP) domain of both VAPA and VAPB (sequences are found in Table S1 in the supplemental material). Huh7.5.1 cells were then stably transduced with lentiviral vectors encoding the sgRNA and Streptococcus pyogenes Cas9. After validation of gene targeting (Fig. 1A), we infected the stable cell pools with full-length HCV encoding a NanoLuc reporter, where luciferase activity directly correlates with viral propagation. Three days postinfection, we saw a strong decrease in luciferase activity in VAPA knockout cells and moderate inhibition in VAPB knockout cells (Fig. 1B). Cells with a double knockout of VAPA and VAPB (VAPA/B) had a greater decrease in luciferase activity than single knockouts (Fig. 1B), indicating that both VAPA and VAPB are required for efficient HCV infection. HCV viral RNA quantitation after JFH-1 infection (under multicycle infection conditions) showed similar inhibition in VAP knockout cells (Fig. 1C), and the viability of single- and double-knockout cells was similar to that of control cells (Fig. 1D). We also transfected a subgenomic replicon containing a Renilla luciferase reporter (17) into the knockout cells to assess whether the specific step of viral replication requires VAPA or VAPB. We found significant decreases in luciferase activity in all knockout cell pools compared to levels in the control at both 48 and 72 h posttransfection (Fig. 1E). These data confirm that VAPs are required for efficient HCV replication.
FIG 1.
VAPA and VAPB are required for efficient HCV replication. (A) Huh7.5.1 cells were stably transduced with lentivirus encoding a puromycin resistance marker, Cas9, and a single guide RNA (sgRNA) targeting the indicated gene(s). Lysates from the stable knockout cell pools were then immunoblotted for the indicated proteins. (B) Huh7.5.1 knockout cell pools as described in panel A were infected with NanoLuc-HCV for 3 days, and luciferase activity was measured. Values are means ± standard errors of the means of three independent experiments and are normalized to the level of the control. **, P < 0.001, for results compared to the result with the control. (C) Huh7.5.1 knockout cell pools as described in panel A were infected with JFH-1 for 3 days before viral RNA was quantified by quantitative reverse transcription-PCR. Values are normalized to the result with the control. **, P < 0.001, for results compared to the result with the control. (D) Cell viability of Huh7.5.1 cells stably transduced with lentivirus encoding sgRNA targeting the indicated gene was measured by CellTiter-Glo. Values are normalized to the level of the control. (E) Huh7.5.1 knockout cell pools as described in panel A were transfected with a subgenomic replicon encoding a Renilla luciferase reporter. Luciferase activity was measured at 4, 24, 48, and 72 h posttransfection, and data are plotted as fold change over values at 4 h to control for transfection and initial translation. **, P < 0.001, for results compared to the result with the control. (F) VAPA/B double knockout Huh7.5.1 cell pools were transduced with the indicated lentiviral vectors to express HA-tagged sgRNA-resistant VAPA or VAPB and infected with NanoLuc-HCV for 3 days, and luciferase activity was measured. Values are means ± standard errors of the means of three independent experiments and are normalized to the level of the control. **, P < 0.001, for results compared to the result with the control. (G) Stably transduced cells as described in panel F were immunoblotted for the indicated proteins. sgVAPA/B, sgRNA targeting VAPA/B. (H) Indicated knockout cell pools were transduced with the indicated lentiviral vectors to express HA-tagged VAPA or VAPB and immunoblotted for the indicated proteins. (I) Stably transduced cells as described in panel H were infected with NanoLuc-HCV for 3 days, and luciferase activity was measured. Values are means ± standard errors of the means of three independent experiments and are normalized to the level of the control.
In order to confirm the specificity of our knockout cell lines, sgRNA-resistant VAPA and VAPB cDNAs were then introduced by lentiviral transduction into the VAPA/B knockout cell lines to rescue VAPA or VAPB expression (Fig. 1G). Exogenous expression of either VAPA or VAPB fully rescued NanoLuc-HCV infection in VAPA/B knockout cells (Fig. 1F), demonstrating that VAPA and VAPB play redundant roles in supporting HCV infection or that overexpression of one VAP can overcome the requirement of the other. To further confirm this, we performed a reciprocal rescue assay, in which VAPA and VAPB knockout cells were rescued by exogenous expression of sgRNA-resistant VAPA and VAPB, respectively. Immunoblotting confirmed expression of exogenous VAPA and VAPB (Fig. 1H). We found that HCV infection in VAPA knockout cells was rescued by exogenous expression of either VAPA or VAPB, and the same held true for VAPB knockout cells (Fig. 1I).
VAPA dimerization is not required to support HCV infection.
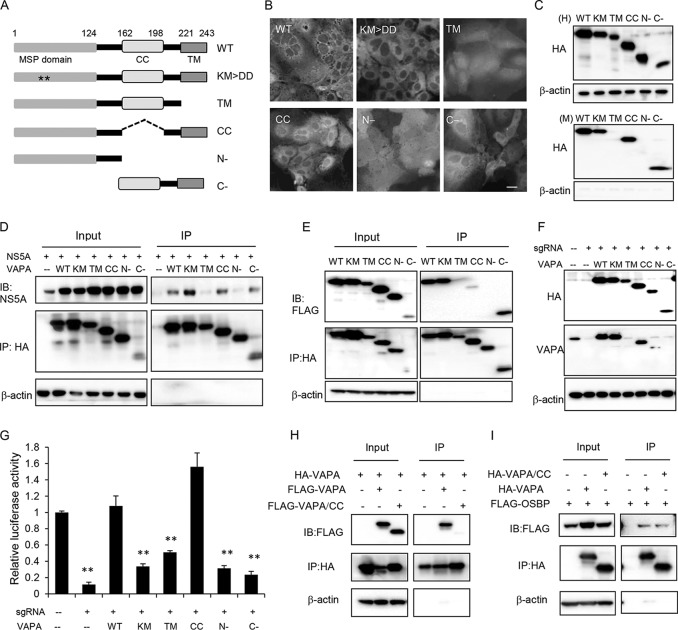
Because VAPA expression could fully rescue the inhibition of HCV replication by VAPB knockout, subsequent characterization of VAP function was performed with VAPA alone in the context of VAPA/B double-knockout cell pools. To elucidate the domain requirement and functional role of VAPA in HCV infection further, we generated five VAPA mutants (Fig. 2A). The mutants featured the following: (i) KM→DD substitutions that ablate its interaction with FFAT domain-containing proteins (18); (ii) a deletion of the transmembrane (TM) domain; (iii) a deletion of the coiled-coil (CC) domain; (iv) the MSP domain alone (N−); and (v) a deletion of the MSP domain (C−). We first overexpressed these hemagglutinin (HA)-tagged mutants in VAPA/B knockout cells and found that wild-type VAPA, the KM→DD mutant, and the CC and C− mutants showed similar reticular distributions in the cell, consistent with ER localization (Fig. 2B). In contrast, the TM mutant and isolated MSP domain (N−) were homogenously distributed in cells, consistent with their expected loss of membrane association due to deletion of the transmembrane domain. Similar results were obtained when the cells were analyzed by a biochemical membrane association assay (Fig. 2C).
FIG 2.
VAPA dimerization is not required to support HCV infection. (A) Schematic diagrams of VAPA domains and mutants. MSP domain, major sperm protein domain; CC, coiled-coil domain; TM, transmembrane domain. Asterisks indicate the lysine and methionine residues in the MSP domain necessary for interaction with FFAT domains. (B) VAPA/VAPB double knockout Huh7.5.1 cell pools were stably transduced with lentiviral vectors expressing the indicated HA-tagged wild-type VAPA or VAPA mutants and then were immunostained with HA antibody. Bar, 20 μm. (C) Membrane association assay. Stably transduced cells as described in panel B were homogenized (H, upper panels), and unbroken cells and cell nuclei were removed by low-speed centrifugation. Membrane fractions (M, lower panels) were spun at 90,000 × average g force for 1 h, and the pellet was resuspended with RIPA buffer containing 0.1% SDS. Samples were immunoblotted with the indicated antibodies. (D) Coimmunoprecipitation of VAPA mutants with HCV NS5A. 293T cells were cotransfected to express HCV NS5A and HA-tagged wild-type VAPA or the indicated VAPA mutants. Forty-eight hours later, cells were lysed and immunoprecipitated with HA antibody, followed by immunoblotting with the indicated antibodies. (E) Homodimerization of VAPA mutants. 293T cells were cotransfected with two expression plasmids encoding wild-type VAPA (lane 1) or the same indicated VAPA mutant (lanes 2 to 6), one with an HA tag and the other with a FLAG tag. Forty-eight hours later, cells were lysed and immunoprecipitated with HA antibody followed by immunoblotting with the indicated antibodies. (F) VAPA/VAPB double knockout Huh7.5.1 cell pools were transduced with lentiviral vectors expressing HA-tagged VAPA or the indicated VAPA mutants before they were immunoblotted with the indicated antibodies. Note that the VAPA antibody was generated against amino acids 121 to 228; thus, it could not detect TM and the C− mutant, and the N− mutant is poorly detected. (G) The stable cell pools described in panel F were infected with NanoLuc-HCV for 3 days, followed by luciferase activity measurement. Values are normalized to the level of the control. **, P < 0.001, for results compared to the result with the control. (H) Heterodimerization of the VAPA CC mutant with wild-type VAPA. 293T cells were cotransfected to express HA-tagged VAPA and FLAG-tagged VAPA or the VAPA CC mutant. Forty-eight hours later, cells were lysed and immunoprecipitated with HA antibody, followed by immunoblotting with the indicated antibodies. (I) VAPA CC interaction with OSBP. 293T cells were cotransfected to express FLAG-tagged OSBP and HA-tagged wild-type VAPA or the VAPA CC mutant. Forty-eight hours later, cells were lysed and immunoprecipitated with HA antibody, followed by immunoblotting with the indicated antibodies. WT, wild type; IB, immunoblotting.
We next assessed the ability of these VAPA mutants to interact with NS5A by immunoprecipitation (IP). We found that the TM mutant and isolated MSP domain (N−) did not interact with NS5A (Fig. 2D), suggesting that membrane interaction is necessary for binding to NS5A. VAPA homodimerizes or heterodimerizes with VAPB (18, 19), and dimerization has been thought to be important for VAP function although this has not been experimentally demonstrated. We next assessed the ability of these mutants to dimerize by cotransfecting 293T cells with HA-tagged and FLAG-tagged VAPA constructs, followed by immunoprecipitation of HA-VAPA and immunoblotting for associated FLAG-VAPA. The TM and N− mutants did not form dimers, while the CC mutant exhibited a significant impairment in dimerization (Fig. 2E). These results suggest that the transmembrane domain and coiled-coil domain are both important for dimerization, consistent with a previous report (18).
Finally, we tested whether these mutants could support HCV infection. VAPA/B double-knockout cell pools were transduced with sgRNA-resistant VAPA mutants and infected with NanoLuc-HCV. Both wild-type VAPA and the CC mutant could fully rescue HCV infection, while other mutants did not (Fig. 2G). Immunoblotting confirmed the expression of individual proteins (Fig. 2F); of note, the levels of CC mutant expression were comparable to those of endogenous VAPA in control cells (lane 1). Surprisingly, the CC mutant, which is defective in interaction with either the CC mutant (Fig. 2E) or with full-length VAPA (Fig. 2H), was able to completely rescue HCV infection in cells depleted of VAPA and VAPB (Fig. 2G) and could also interact with OSBP (Fig. 2I). These results indicate that efficient dimerization is not required for VAPs to interact with OSBP or to support HCV infection.
VAPs support HCV replication through mechanisms different from OSBP.
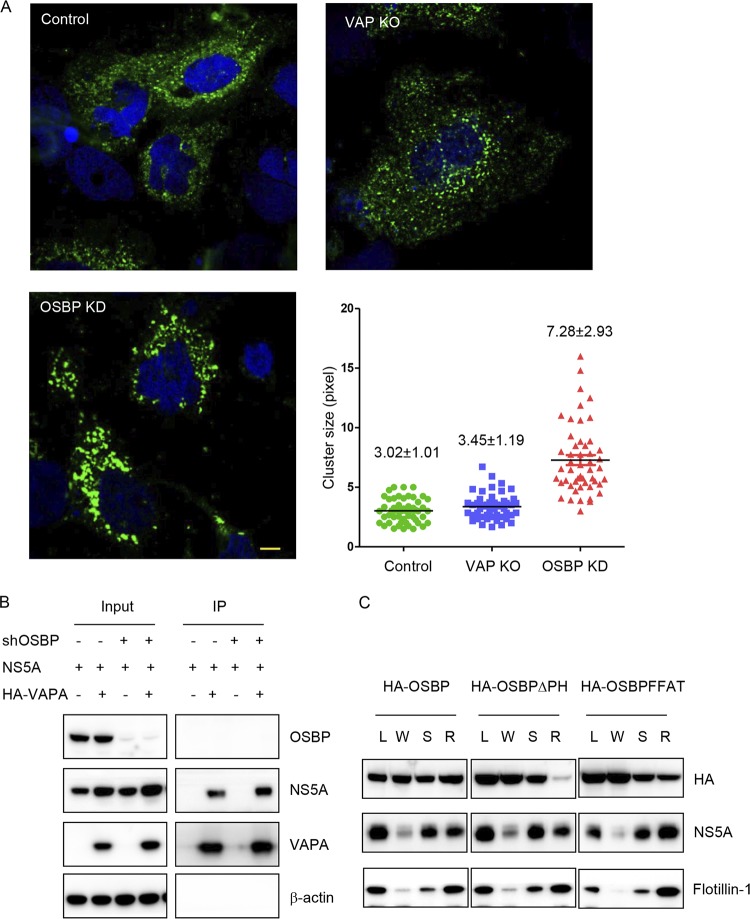
Silencing or pharmacologic inhibition of PI4KA or OSBP induces the accumulation of clusters of small double-membrane vesicles containing HCV NS5A and other viral replication components (3–5, 8). We next tested whether VAP depletion also induced these altered membranous structures using a nonreplicative HCV expression system. Huh7.5.1/T7 cells depleted of VAPs or OSBP were transfected with an NS3-5B/green fluorescent protein (NS3-5B/GFP) polyprotein encoding GFP-tagged NS5A (6). As expected, OSBP silencing induced the formation of NS5A-positive membrane clusters, while no clusters were seen in VAP double-knockout cells (7.28 ± 2.93 versus 3.45 ± 1.19 pixels, respectively) (Fig. 3A). Thus, the NS5A-positive membrane clustering phenotype seen in OSBP-depleted cells is not seen in VAP-depleted cells. This implies that the inhibition of HCV replication in VAP-depleted cells involves one or more mechanisms distinct from that of PI4KA and OSBP.
FIG 3.
VAP function in HCV infection is not solely through OSBP. (A) HCV RO morphology in VAPA/VAPB knockout cells. Huh7.5.1/T7 cells stably expressing T7 RNA polymerase were transduced with the indicated sgRNA with Cas9 or shRNA and then transfected with constructs expressing the NS3-5B/GFP polyprotein encoding NS5A-GFP under the control of the T7 promoter. Forty-eight hours later, cells were immunostained for GFP (green) with DAPI (blue) nuclear counterstaining. The lengths of the NS5A-positive membranous structures in each group were quantitated with ImageJ, and the average length is indicated in pixels as means±SD. Bar, 10 μm. KO, knockout. KD, kinase dead. (B) Coimmunoprecipitation of VAPA with HCV NS5A protein. 293T cells stably expressing a negative-control shRNA or an shRNA targeting OSBP (shOSBP) were cotransfected to express HCV NS5A and HA-tagged VAPA. Forty-eight hours later, cells were lysed and immunoprecipitated with HA antibody, followed by immunoblotting with the indicated antibodies. (C) Huh7.5.1 cells stably transduced with wild-type or mutant OSBP were infected with the JFH-1 strain of HCV. Three days later, cell homogenates (L) were separated into a crude membrane pellet and a water-soluble supernatant (W) by centrifugation. The pellet was treated with cold 1% NP-40 and spun again; the detergent-soluble (S) fraction was removed, and the detergent-resistant membrane (R) pellet was resuspended with RIPA buffer containing 0.1% SDS. Samples were analyzed by SDS-PAGE, followed by immunoblotting for the indicated proteins.
NS5A interacts with both VAPs and OSBP (12, 13, 20). It is not known whether NS5A interaction with VAPs requires OSBP. Here, we found that VAPA could interact with NS5A in both control and OSBP-silenced cells (Fig. 3B), suggesting that the interaction between VAPA and NS5A does not require OSBP expression. Conversely, we tried to test whether NS5A interaction with OSBP requires VAPA. However, in our 293T cotransfection system, we found only very weak coimmunoprecipitation between OSBP and NS5A, preventing us from testing whether this interaction is VAPA dependent. We hypothesize that the low affinity of OSBP for NS5A suggested by coimmunoprecipitation indicates that targeting of OSBP to HCV ROs may not be explained solely by direct interaction with NS5A. For example, HCV ROs have elevated PI(4)P levels (4, 5, 7), and VAPA has been also reported to be enriched at HCV ROs (11, 14, 21). Therefore, OSBP targeting to HCV ROs could be mediated by PI(4)P or VAPA interaction, in addition to direct interaction with NS5A. To distinguish between these two possibilities, we asked whether OSBP interaction with detergent-resistant membranes, which are enriched in HCV ROs (11, 22), requires interaction with PI(4)P or VAPA. We found that both wild-type OSBP and the OSBP FFAT mutant, but not the OSBP pleckstrin homology (PH) domain mutant, cofractionated with NS5A in detergent-resistant membranes from HCV-infected cells (Fig. 3C), suggesting that OSBP localization to NS5A-positive fractions requires PI(4)P but not VAP binding. Collectively, our findings suggest that VAPs and OSBP support HCV infection through distinct mechanisms.
Identification of VAPA-interacting FFAT-containing proteins by mass spectrometry.
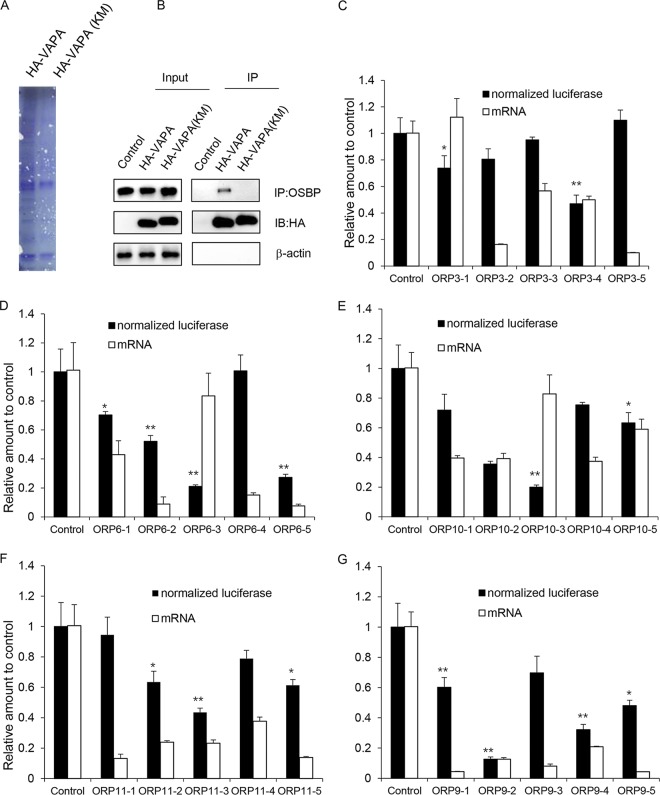
The VAPA KM→DD mutant, which is defective in binding to FFAT motifs, cannot rescue HCV replication in VAP knockout cells, indicating that VAPs support HCV infection by recruiting one or more FFAT-containing proteins other than OSBP to the RO. To identify FFAT-containing proteins interacting with VAPs in HCV-infected cells, VAPA/B knockout cells stably expressing HA-tagged wild-type or KM→DD mutant VAPA were infected with the JFH-1 strain of HCV, and cell lysates were subjected to anti-HA antibody immunoprecipitation. Coomassie staining of the immunoprecipitate demonstrated a number of proteins that specifically copurified with wild-type VAPA (Fig. 4A, left lane), but not with the KM→DD mutant (right lane). Immunoblotting showed that wild-type VAPA coimmunoprecipitated the known FFAT-containing protein OSBP, while the KM→DD mutant did not (Fig. 4B). Immunoprecipitated proteins from both preparations were subjected to mass spectrometry, and several known FFAT-containing proteins were identified (Table S2). These included several members of the OSBP-related protein (ORP) family: ORP3, ORP6, ORP9, ORP10, ORP11, and OSBP; a PI-transfer protein, PITPNM1/Nir2 (henceforth referred to here as Nir2); and the acyl-coenzyme A (CoA) binding protein ACBD5. We next tested the functional role of the ORPs in supporting HCV infection by genetic depletion. We found that ORP3, ORP6, ORP10, or ORP11 was not required to support HCV infection when individually depleted (Fig. 4C to F) though this does not exclude the possibility that HCV infection might be inhibited by depletion of two or more of these ORPs. While knockdown of ORP9 appeared to inhibit HCV infection, (Fig. 4G), it is thought to mediate a similar function as OSBP (8, 20), namely, exchange of cholesterol for PI(4)P at membrane contact sites.
FIG 4.
Identification of FFAT-containing proteins by mass spectrometry. (A) Coomassie blue staining of VAPA or VAPA KM→DD coimmunoprecipitates. VAPA/B knockout Huh7.5.1 cells were stably transduced to express HA-tagged VAPA or the VAPA KM→DD mutant before they were infected with JFH-1. Cells were lysed and immunoprecipitated with HA antibody. Samples were resolved by SDS-PAGE and stained by Coomassie blue. (B) Input and immunoprecipitated (IP) samples as described above were immunoblotted with the indicated antibodies. (C to G) OR6 cells containing a full-length HCV replicon were transduced with a nontargeting shRNA or with five independent shRNAs targeting the indicated gene for 72 h before replication was assessed by Renilla luciferase activity; the mRNA expression level of the indicated gene was quantified by quantitative reverse transcription-PCR. Values were normalized to the level of the control. *, P < 0.05; **, P < 0.01, for results compared to the result with the control.
Nir2 is relevant to HCV replication.
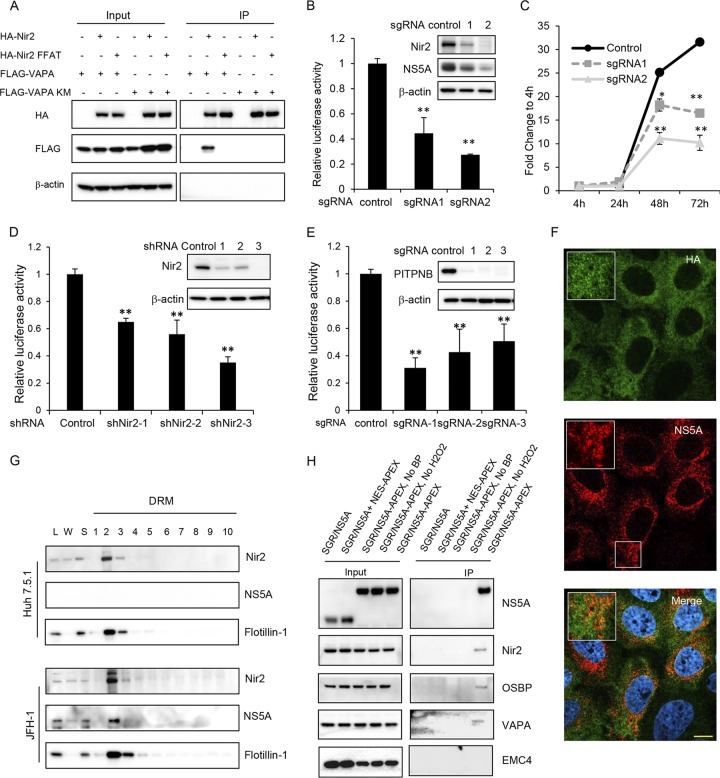
We then turned to Nir2, a membrane-associated phosphatidylinositol transfer domain-containing protein that is a homolog of the Drosophila retinal degeneration B (rdgB) protein (23). We first confirmed its interaction with VAPA by coimmunoprecipitation. Wild-type VAPA interacted with wild-type Nir2, while mutations in either the FFAT motif in Nir2 or the KM motif in VAPA ablated this interaction (Fig. 5A). We next designed two independent sgRNAs targeting Nir2. Nir2 knockout Huh7.5.1 cell pools were then infected with either NanoLuc-HCV (Fig. 5B) or JFH-1 (Fig. 5B, inset). Immunoblotting demonstrated that Nir2 expression was effectively ablated by sgRNA 2 (Fig. 5B, inset). Significant inhibition of viral infection was observed in Nir2-depleted cells compared to that with a control sgRNA. A transient replication assay using transfected replicon RNA showed that viral replication was significantly impaired in Nir2 knockout cells at 48 or 72 h posttransfection (Fig. 5C). There was an association between the efficiency of Nir2 knockout and inhibition of viral replication. Viral replication in OR6 cells containing a full-length genotype 1b HCV replicon expressing a Renilla luciferase reporter gene (24) transduced with three independent short hairpin RNA (shRNA) lentiviral vectors targeting Nir2 was also found to be inhibited in a dose-dependent manner (Fig. 5D), indicating that the dependency of HCV on Nir2 is not genotype specific.
FIG 5.
Nir2 is required for HCV replication and is in close proximity to HCV ROs. (A) Coimmunoprecipitation of VAPA with Nir2 protein. 293T cells were cotransfected to express FLAG-tagged Nir2 or the Nir2 FFAT mutant with HA-tagged VAPA or the VAPA KM→DD mutant. Forty-eight hours later, cells were lysed and immunoprecipitated with HA antibody, followed by immunoblotting with the indicated antibodies. (B) Huh7.5.1 cells stably expressing a negative control or two independent Nir2 sgRNAs with Cas9 were infected with NanoLuc-HCV for 3 days before luciferase activity was measured. Values are normalized to the level of the control. **, P < 0.001, for results compared to the result with the control. Inset, Huh7.5.1 cell pools as described above were infected with the JFH-1 strain of HCV followed by immunoblotting for the indicated proteins. (C) Huh7.5.1 cell pools described as in panel B were transfected with a subgenomic replicon encoding a Renilla luciferase reporter. Luciferase activity was measured at 4, 24, 48, and 72 h posttransfection, and data are plotted as fold change to values at 4 h to control for transfection and RNA translation. *, P = 0.01; **, P < 0.005, for results compared to the result with the control. (D) OR6 replicon cells were transduced with the indicated shRNAs, and replication was assessed at 72 h postransduction by Renilla luciferase activity. Values are normalized to the level of the control. **, P < 0.001, for results compared to the result with the control. Inset, OR6 cell pools as described were immunoblotted for the indicated proteins. (E) Huh7.5.1 cell pools stably expressing a negative control or three independent PITPNB sgRNAs with Cas9 were infected with NanoLuc-HCV for 3 days before luciferase activity was measured. Values are normalized to the level of the control. **, P < 0.001, for results compared to the result with the control. Inset, Huh7.5.1 cell pools as described were immunoblotted for the indicated proteins. (F) Nir2 knockout Huh7.5.1 cell pools stably overexpressing HA-tagged Nir2 were infected with JFH-1. Three days later, cells were fixed and stained with antibodies against NS5A (green) and HA (red) with DAPI nuclear counterstaining (blue). Bar, 10 μm. (G) Uninfected or JFH-1 infected Huh7.5.1 cells were prepared as in the legend of Fig. 3C, and detergent-resistant membranes (DRMs) were fractionated on an iodixanol density gradient. Fractions were analyzed by SDS-PAGE, followed by immunoblotting for the indicated proteins. Fractions are numbered from 1 to 10 in order from top to bottom (light to heavy). L, cell homogenate; W, water-soluble supernatant. (H) Nir2 is in close proximity to NS5A. Huh7.5.1 stably expressing a subgenomic replicon (SGR/NS5A) or coexpressing the SGR/NS5A replicon and NES-APEX2 or a subgenomic replicon with APEX2-tagged NS5A (SGR/NS5A-APEX) were treated, where indicated, with biotin-phenol (BP)-containing medium for 30 min, followed by hydrogen peroxide for 1 min to biotinylate APEX2-proximal proteins prior to quenching and lysis. Biotinylated proteins were affinity isolated with streptavidin beads, and lysates were resolved by SDS-PAGE. Immunoblotting was performed for the indicated proteins for both input and immunoprecipitated (IP) samples.
In addition to Nir2, there are four other known PI transfer proteins in mammalian cells. Of those, PITPNB has been reported to be an essential host factor for rhinovirus infection (25), possibly by shuttling PI between the ER and rhinovirus ROs. We also tested the requirement of PITPNB for HCV infection, and we also found inhibition of HCV infection after PITPNB knockout (Fig. 5E). Whether this and other PI transfer proteins play a role in HCV infection and how they coordinate during viral infection need further investigation.
Because Nir2 is essential for efficient HCV replication, we hypothesized that Nir2 might associate with HCV ROs. However, we did not observe redistribution of endogenous Nir2 in HCV-infected cells. We did observe partial colocalization between Nir2 and HCV NS5A, a viral nonstructural protein found in ROs (Mander’s coefficient M1, 0.694) (Fig. 5F), though this analysis was limited by the rather diffuse reticular staining pattern of Nir2. HCV RO membranes cofractionate with detergent-resistant membranes (DRMs) in HCV-infected cells (11, 22). A membrane flotation assay of DRMs showed that Nir2 cofractionated with NS5A and the DRM marker flotillin-1 (Fig. 5G) though Nir2 was found in DRMs from both HCV-infected and uninfected cells.
To more conclusively demonstrate that Nir2 is in close proximity to HCV NS5A, we conducted a proximity biotinylation assay using the engineered peroxidase APEX2 to label proteins in proximity to NS5A. APEX2 catalyzes the biotinylation of proteins within an ∼10-nm radius in the presence of biotin-phenol and hydrogen peroxide (26). This reaction is performed in live cells over a short labeling period of 1 min. We generated a cell line stably expressing an NS3-5B subgenomic replicon encoding APEX2 at an insertion-tolerant site in NS5A (27). Following APEX2-mediated biotinylation, cells were lysed, and affinity purification of biotinylated proteins was performed using streptavidin beads, followed by SDS-PAGE and immunoblotting.
We found, as expected, that NS5A-APEX2 was specifically and heavily biotinylated. However, NS5A was not biotinylated when APEX2 was instead fused to a nuclear export signal (NES) for cytoplasmic APEX2 localization in a cell line stably expressing a wild-type NS3-5B subgenomic replicon (Fig. 5H). Consistent with our hypothesis, Nir2 as well as OSBP and VAPA were all biotinylated in NS5A-APEX2 replicon cells, while EMC4, an ER membrane protein complex subunit necessary for dengue viral infection (28), was not (Fig. 5H). These results suggest that Nir2 is in close proximity to the HCV NS5A protein.
VAPs and Nir2 are required to maintain PI(4)P upregulation in HCV infection.
A proposed model of lipid trafficking to the HCV RO posits that increased levels of PI(4)P recruit lipid transfer proteins such as OSBP to the viral RO, and OSBP then delivers cholesterol to the HCV RO by exchange for PI(4)P (8). However, under this model phosphoinositide levels must be depleted from the RO by OSBP unless there is a mechanism for phosphoinositide replenishment at the RO during chronic infection. Thus, we hypothesized that the PI transfer protein Nir2 might be necessary to maintain PI levels at the HCV RO for subsequent conversion to PI(4)P by PI4KA.
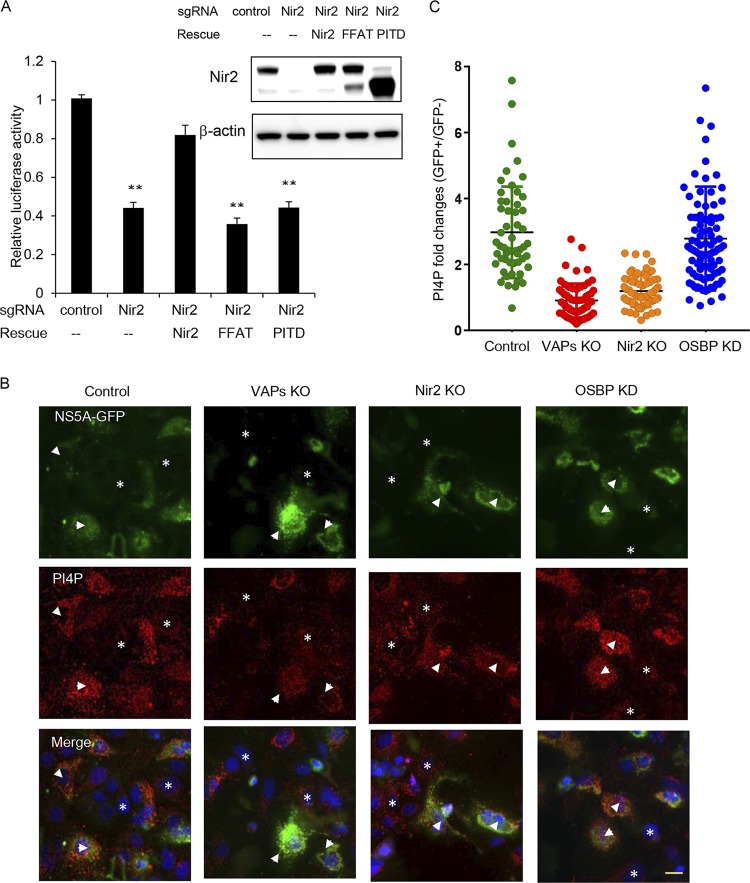
Nir2 has multiple protein domains. Among these are the following: (i) the N-terminal PI transfer domain (PITD) that has been shown to have phosphatidylinositol transfer activity in vitro, (ii) the FFAT motif, and (iii) the C-terminal domain that binds to phosphatidic acid (16, 29). To assess the role of the PITD and the FFAT motif in HCV replication, Nir2 knockout cell pools were transduced with sgRNA-resistant Nir2 mutants and infected with NanoLuc-HCV. Neither PITD deletion nor the FFAT mutant rescued HCV replication (Fig. 6A), suggesting that the PI transfer activity and interaction with VAPs is required for Nir2 to support HCV infection.
FIG 6.
VAPs and Nir2 are required for PI(4)P upregulation in HCV-expressing cells. (A) Control or Nir2 knockout Huh7.5.1 cells were transduced with the indicated lentiviral vectors to express sgRNA-resistant Nir2 or mutants before they were infected with NanoLuc-HCV for 3 days, and luciferase activity was measured. Values are normalized to the level of the control. **, P < 0.001, for results compared to the result with the control. Inset, Huh7.5.1 cell pools as described above were immunoblotted for the indicated proteins. (B) Huh7.5.1/T7 cells stably expressing T7 RNA polymerase were transduced by the indicated sgRNA or shRNA to deplete VAPA/B, Nir2, or OSBP and then transfected with constructs expressing the NS3-5B/GFP polyprotein encoding NS5A-GFP under the control of the T7 promoter. Forty-eight hours later, cells were immunostained for PI(4)P (red) and GFP (green) with DAPI nuclear counterstaining (blue). Arrowheads indicate cells expressing the NS3-5B/GFP polyprotein, while asterisks indicate cells with no NS3-5B/GFP expression. Bar, 20 μm. (C) Quantitation of PI(4)P fluorescence from HCV-expressing cells versus HCV-nonexpressing cells in the same experiment as described for panel B. Each point denotes the fold change of integrated fluorescence signal from a single HCV-expressing cell over the average intensity of HCV-nonexpressing cells, with the mean fold change indicated by the black lines.
To test whether Nir2 is required to maintain PI(4)P upregulation in HCV infection, we measured the PI(4)P level in Nir2 knockout cells. To avoid confounding effects of Nir2 knockout on HCV replication, this experiment was conducted with Huh7/T7 cells expressing the NS3-5B/GFP polyprotein as described above. PI(4)P was elevated approximately 3-fold in NS3-5B/GFP-expressing cells transduced with a negative-control sgRNA (Fig. 6B and C) compared with the level in HCV-nonexpressing cells. As has been previously reported (8) and also shown in Fig. 6, OSBP silencing did not block the upregulation of intracellular PI(4)P levels in HCV-expressing cells. In contrast, knockout of VAPs or Nir2 blocked the upregulation of intracellular PI(4)P levels by HCV polyprotein expression, indicating that these proteins are required to support elevated steady-state PI(4)P levels in HCV infection.
DISCUSSION
Although VAPA and VAPB have been shown to be necessary for efficient HCV replication (11, 12), their specific mechanisms and effectors in this process have not been defined. In this study, we found that VAPA and VAPB play redundant roles in HCV infection, which explains why inhibition of either VAPA or VAPB alone only moderately inhibits viral infection. Furthermore, VAPs are known to form homodimers or heterodimers (30) though their roles in mediating VAP function have not been clearly elucidated. Here, we found that VAP dimerization is not necessary to support viral replication. Of note, Saccharomyces cerevisiae VAP homologs Scs2p and Scs22p lack the CC domain (31) and are believed to function as monomers (10).
The ER is the major site for biosynthesis of most membrane lipids, including PI, which are then transported to different cellular membrane compartments by both vesicular and nonvesicular transport mechanisms (32). In eukaryotic cells, PI generally constitutes less than 10% of total cellular phospholipid (33). As the HCV RO is believed to be derived from the ER and not in physical continuity with cellular membrane compartments (34), it is likely that the initial mass of PI derived from the ER is consumed by PI4KA to generate PI(4)P. Furthermore, we have previously found that cholesterol is delivered to HCV ROs by OSBP in exchange for PI(4)P (8). This should result in a net decrease in RO phosphoinositide content and, ultimately, a decrease in cholesterol delivery over time if there is no mechanism for phosphoinositide resupply. However, using pulse-chase labeling of HCV NS5A protein, we found that cholesterol trafficking to “old” NS5A-positive membrane structures is not decreased compared to that to “new” ones (17), hinting at the existence of a mechanism for PI replenishment. Here, we identified Nir2 as a VAP effector that we propose functions to transfer PI to ROs, which can then be used as a substrate for PI4KA to generate PI(4)P. The PI(4)P exchanged to the ER by OSBP is then converted by the lipid phosphatase SacI back to PI (9, 35), which can then be used to continue the phosphoinositide cycle at the HCV RO.
In conjunction with VAPs, OSBP tethers membranes from different organelles with its PH domain and FFAT motif to form membrane contact sites where nonvesicular lipid transfer occurs (9, 36). We have previously shown that an OSBP FFAT mutant failed to rescue the viral replication inhibition caused by OSBP depletion (8), suggesting that OSBP requires VAP interaction to support HCV replication. However, VAP depletion does not result in the same membranous web clusters as those seen in OSBP (or PI4KA) depletion, suggesting that VAPs and OSBP mediate overlapping but distinct roles in RO biogenesis. We hypothesize that in VAP-depleted cells, the resulting low levels of PI(4)P generation at the viral RO prevents not only the recruitment of OSBP but also that of other cellular proteins that interact with PI(4)P. Conversely, during OSBP depletion, although OSBP-mediated PI(4)P-cholesterol transfer is inhibited (8), elevated PI(4)P may still recruit other PI(4)P-interacting proteins to the HCV RO, such as other ORPs, FAPP2 (37, 38), and ceramide transfer protein (CERT) (37, 39). In addition, the ER-resident protein VAP has also been reported to be enriched in HCV ROs (11, 14, 21), suggesting the possibility that VAPs are located on both sides of the ER-RO MCS. However, the directionality of cholesterol/PI(4)P exchange by OSBP at this interface is likely determined by PI(4)P enrichment at the HCV RO, and we hypothesize that the directionality of net PI transport by Nir2 from the ER to the HCV RO is determined by the PI concentration gradient between these two membrane compartments.
Wild-type HCV isolates and cell culture-adapted variants have divergent requirements for PI4KA activity and PI(4)P levels to replicate efficiently (40). Specifically, while HCV strains adapted to growth in Huh7 hepatoma cells promote PI4KA activation and require relatively high levels of PI(4)P for optimal replication, wild-type HCV isolates appear to replicate efficiently in hepatoma cells when PI(4)P production is reduced. However, the same study also demonstrated that a PI4KA inhibitor suppressed wild-type HCV replication in cells expressing low levels of PI4KA, such as primary human hepatocytes, indicating that wild-type HCV isolates do require PI4KA activity for replication. Here, we have shown that adapted strains of both genotype 1b and 2a require Nir2 for efficient replication in cell culture. We speculate that wild-type HCV strains may also require some level of Nir2 expression for efficient replication though we have not tested this in this study.
Nir2 has been proposed to exchange PI for PA at ER-plasma membrane contact sites (16), where PA is generated from phosphatidylinositol-3,4-bisphosphate [PI(4,5)P2] by the sequential actions of phospholipase C and diacylglycerol kinase. Interestingly, HCV ROs have also been reported to be enriched in PI(4,5)P2 (41), which could be a potential source of PA generation. However, it is not known whether phospholipase C and diacylglycerol kinase are present at the HCV RO or whether Nir2 might be able to exchange PI for lipid species other than PA.
In addition to the PITPs mentioned above, there is another structurally unrelated family of proteins that has the ability to bind and transfer PI, namely, the Sec14p family (42). Interestingly, SEC14L2 has been shown to support pan-genotype HCV replication in cell culture by enhancing vitamin E-mediated protection against lipid peroxidation (43); whether this protein also involves PI transfer in HCV infection remains to be further studied.
In conclusion, the new insights into VAP function provided by this study advance our knowledge of nonvesicular lipid transfer during viral infection. The identification of Nir2 as a PI transfer protein located in close proximity to the HCV replication complex may answer the question of how high levels of PI(4)P are maintained in cells during chronic HCV infection and helps complete our understanding of the mechanisms that establish a phosphoinositide cycle at the HCV RO.
MATERIALS AND METHODS
Reagents and antibodies.
Antibodies used in this study include the following: VAPA and flotillin-1 (BD Biosciences, San Jose, CA), VAPB, Nir2, PITPNB, and OSBP (Proteintech, Chicago, IL), EMC4 (Life Technologies, Carlsbad, CA), HCV NS5A (monoclonal 9E10; Charles Rice, Rockefeller University, New York, NY). FLAG, β-actin (monoclonal; Sigma-Aldrich, St. Louis, MO), GFP and anti-HA tag (Cell Signaling Technology, Danvers, MA), PI(4)P (mouse IgM monoclonal; Echelon, Salt Lake City, UT). Alexa Fluor- and 4′,6′-diamidino-2-phenylindole (DAPI)-conjugated secondary antibodies for microscopy experiments were purchased from Life Technologies. Puromycin, blasticidin S, and G418 were also purchased from Life Technologies.
Viruses and viral replicons.
The genotype 2a HCV strain JFH-1 (44) was produced and propagated as described in Kato et al. (45). We generated a full-length NanoLuc-tagged virus (Jc1/NanoLuc2A) by replacing the Gaussia luciferase coding sequence in the Jc1/Gluc2A (46) with the NanoLuc (Promega, Madison, WI) coding sequence. Primers are listed in Table S1 in the supplemental material.
We also generated a subgenomic replicon expressing APEX2-tagged NS5A based on the JFH-1 genotype 2a sequence, termed here as pSGR-JFH1(NS5A/APEX2). We subcloned the APEX2 sequence into the unique MluI site in the construct described in Wang and Tai (17) using the primers shown in Table S1. In vitro-transcribed RNA was prepared and transfected into Huh7.5.1 cells as described in Kato et al. (45). A cell line stably harboring pSGR-JFH1(NS5A/APEX2) was isolated by G418 selection.
Luciferase assay.
VAPs or Nir2 knockout stable cell lines were either infected with NanoLuc-HCV (Fig. 1B, F, and I, 2G, 5B and E, and 6A) or transfected with a subgenomic replicon encoding Renilla luciferase (Fig. 1E and 5C) (17). Renilla or NanoLuc activity was measured at the indicated time points using a Synergy 2 plate reader (BioTek, Winooski, VT). Cell viability was assessed by cellular ATP content (CellTiter-Glo; Promega).
Immunofluorescence staining.
Immunofluorescence staining was performed as described in Tai and Salloum (6) and Wang et al. (8). PI(4)P staining of intracellular membranes was performed exactly as described in Hammond et al. (47) and Wang et al. (8). Images were taken with a Nikon A1 laser scanning confocal microscope in sequential scanning mode to limit cross talk between fluorochromes. Quantification of PI(4)P fluorescence intensity was determined from multiple random fields obtained with identical acquisition settings using ImageJ software.
Coimmunoprecipitation and mass spectrometry.
293T cells transiently transfected with HA-tagged constructs were lysed with IP buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, and 5% glycerol). HA antibody (0.5 μg) was added to cell lysate and incubated at 4°C for 1 h. Protein G Dynabeads (Life Technologies) were then added to lysate, incubated at 4°C for 1 h, and washed three times in phosphate-buffered saline (PBS) with 0.5% Triton X-100. Immunoprecipitated proteins were then eluted with SDS sample buffer and subjected to SDS-PAGE. For mass spectrometry (MS), VAP knockout cells stably transduced with HA-tagged wild-type VAPA or VAPA KM→DD (107 cells) were infected with JFH-1 before they were lysed and subjected to anti-HA immunoprecipitation as described above. Proteins on beads were washed with PBS once before they were reduced with dithiothreitol (DTT) and digested with sequencing-grade modified trypsin (Promega). Peptides were separated on a SepPak C18 cartridge (Waters Corp.) for liquid chromatography (LC)-tandem MS -. Proteins were identified by searching the data against Homo sapiens (UniProt taxonomy identifier 9606) and JFH-1 using Proteome Discoverer (version 2.1; Thermo Scientific). The false discovery rate (FDR) was determined using Percolator, and proteins/peptides with an FDR of ≤1% were retained for further analysis.
Lentiviral vector preparation and transduction.
Vesicular stomatitis virus G protein (VSV-G) pseudotyped lentiviruses were produced as previously reported (8, 48). The sgRNA sequences used are listed in Table S1. pLKO.1 shRNA constructs (Sigma-Aldrich) targeting the following were used: OSBP (TRCN0000155602 and TRCN0000155752), ORP3 (TRCN0000285469, TRCN0000155275, TRCN0000157183, TRCN0000158226, and TRCN0000275975), ORP6 (TRCN0000159097, TRCN0000159956, TRCN0000161428, TRCN0000161853, and TRCN0000160736), ORP9 (TRCN0000159131, TRCN0000164901, TRCN0000160352, TRCN0000352848, and TRCN0000162447), ORP10 (TRCN0000147030, TRCN0000147511, TRCN0000330427, TRCN0000330426, and TRCN0000149806), ORP11 (TRCN0000161454, TRCN0000158810, TRCN0000160386, TRCN0000343356, and TRCN0000160605), and Nir2 (TRCN0000441337, TRCN0000029760, and TRCN0000029763). Cells were transduced with lentiviral particles for 4 h in the presence of 8 μg/ml Polybrene (Sigma-Aldrich). Cell lines stably transduced with lentiviral vectors were obtained by puromycin selection (for sgRNA or shRNA) or blasticidin selection (for overexpression constructs).
Subcellular fractionation.
Huh7.5.1 cells were infected with JFH-1 at a multiplicity of infection (MOI) of 0.1; uninfected or infected cells were harvested at 4 days postinfection. Detergent-resistant membrane isolation and density gradient fractionation were performed as described previously (8, 49).
APEX labeling and purification of biotinylated proteins.
APEX2-mediated biotinylation and enrichment of biotinylated proteins were performed as previously described (50). Briefly, cells seeded in six-well plates were first labeled with 500 μM biotin-phenol (Iris Biotech, Germany) for 30 min at 37°C, followed by 1 mM H2O2 treatment for 1 min at room temperature. Cells were then washed extensively with quencher buffer containing sodium azide, Trolox, and sodium ascorbate and lysed with radioimmunoprecipitation assay (RIPA) buffer. Portions (10%) of the lysates were reserved to be run as input controls. Five microliters of streptavidin magnetic beads (catalog no. 88817; ThermoFisher) was added to each tube and rotated for 1 h at 4°C. Complexes were washed three times in PBS with 0.1% Triton X-100. Proteins were eluted in SDS sample buffer and then subjected to SDS-PAGE and Western blotting.
Statistics.
Unless otherwise indicated, all values represent means ± standard deviations (SD) and represent the results of a minimum of three independent experiments. A two-tailed Student's t test was used to compare the means of control and experimental groups.
Supplementary Material
ACKNOWLEDGMENTS
Huh7.5.1 cells, the OR6 cell line, the infectious JFH-1 clone, anti-HCV NS5A antibody (clone 9E10), and VAPA, VAPB, and Nir2 expression constructs were generous gifts from Francis Chisari (Scripps Institute, La Jolla, CA), Nobuyuki Kato (Okayama University, Okayama, Japan), Takaji Wakita (National Institute of Infectious Diseases, Tokyo, Japan), Charles Rice (Rockefeller University, New York, NY), Bruno Antonny (Université Nice Sophia Antipolis and CNRS), Pietro De Camilli (Yale University School of Medicine), and Tamas Balla (National Institutes of Health, Bethesda, MD), respectively. The mass spectrometry was carried out by the Proteomics Resource Facility at the Department of Pathology, University of Michigan.
This work was supported by the National Natural Science Foundation of China (grant 81871662 to H.W.), the National Institutes of Health (grant R01DK097374 to A.W.T.), and the University of Michigan Center for Gastrointestinal Research (UMCGR) (NIH 5P30DK034933).
A.W.T. and H.W. conceived and designed the experiments; H.W. performed the experiments; H.W. and A.W.T. analyzed the data; H.W. and A.W.T. wrote the paper.
We have no competing interests to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00742-19.
REFERENCES
- 1.den Boon JA, Diaz A, Ahlquist P. 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8:77–85. doi: 10.1016/j.chom.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. 2009. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A 106:7577–7582. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. 2009. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe 5:298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Bühler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bianco A, Reghellin V, Donnici L, Fenu S, Alvarez R, Baruffa C, Peri F, Pagani M, Abrignani S, Neddermann P, De Francesco R. 2012. Metabolism of phosphatidylinositol 4-kinase IIIα-dependent PI4P is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activity. PLoS Pathog 8:e1002576. doi: 10.1371/journal.ppat.1002576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tai AW, Salloum S. 2011. The role of the phosphatidylinositol 4-kinase PI4KA in hepatitis C virus-induced host membrane rearrangement. PLoS One 6:e26300. doi: 10.1371/journal.pone.0026300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger KL, Kelly SM, Jordan TX, Tartell MA, Randall G. 2011. Hepatitis C virus stimulates the phosphatidylinositol 4-kinase III alpha-dependent phosphatidylinositol 4-phosphate production that is essential for its replication. J Virol 85:8870–8883. doi: 10.1128/JVI.00059-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Perry JW, Lauring AS, Neddermann P, De Francesco R, Tai AW. 2014. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology 146:1373–1385.e11. doi: 10.1053/j.gastro.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B. 2013. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell 155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- 10.Lev S, Ben Halevy D, Peretti D, Dahan N. 2008. The VAP protein family: from cellular functions to motor neuron disease. Trends Cell Biol 18:282–290. doi: 10.1016/j.tcb.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Gao L, Aizaki H, He JW, Lai MM. 2004. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol 78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamamoto I, Nishimura Y, Okamoto T, Aizaki H, Liu M, Mori Y, Abe T, Suzuki T, Lai MM, Miyamura T, Moriishi K, Matsuura Y. 2005. Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J Virol 79:13473–13482. doi: 10.1128/JVI.79.21.13473-13482.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tu H, Gao L, Shi ST, Taylor DR, Yang T, Mircheff AK, Wen Y, Gorbalenya AE, Hwang SB, Lai MM. 1999. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology 263:30–41. doi: 10.1006/viro.1999.9893. [DOI] [PubMed] [Google Scholar]
- 14.Evans MJ, Rice CM, Goff SP. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc Natl Acad Sci U S A 101:13038–13043. doi: 10.1073/pnas.0405152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saheki Y, De Camilli P. 2017. Endoplasmic reticulum-plasma membrane contact sites. Annu Rev Biochem 86:659–684. doi: 10.1146/annurev-biochem-061516-044932. [DOI] [PubMed] [Google Scholar]
- 16.Kim YJ, Guzman-Hernandez ML, Wisniewski E, Balla T. 2015. Phosphatidylinositol-phosphatidic acid exchange by Nir2 at ER-PM contact sites maintains phosphoinositide signaling competence. Dev Cell 33:549–561. doi: 10.1016/j.devcel.2015.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Tai AW. 2017. Continuous de novo generation of spatially segregated hepatitis C virus replication organelles revealed by pulse-chase imaging. J Hepatol 66:55–66. doi: 10.1016/j.jhep.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiser SE, Brickner JH, Reilein AR, Fenn TD, Walter P, Brunger AT. 2005. Structural basis of FFAT motif-mediated ER targeting. Structure 13:1035–1045. doi: 10.1016/j.str.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Kanekura K, Nishimoto I, Aiso S, Matsuoka M. 2006. Characterization of amyotrophic lateral sclerosis-linked P56S mutation of vesicle-associated membrane protein-associated protein B (VAPB/ALS8). J Biol Chem 281:30223–30233. doi: 10.1074/jbc.M605049200. [DOI] [PubMed] [Google Scholar]
- 20.Amako Y, Sarkeshik A, Hotta H, Yates J III, Siddiqui A. 2009. Role of oxysterol binding protein in hepatitis C virus infection. J Virol 83:9237–9246. doi: 10.1128/JVI.00958-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramage HR, Kumar GR, Verschueren E, Johnson JR, Von Dollen J, Johnson T, Newton B, Shah P, Horner J, Krogan NJ, Ott M. 2015. A combined proteomics/genomics approach links hepatitis C virus infection with nonsense-mediated mRNA decay. Mol Cell 57:329–340. doi: 10.1016/j.molcel.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi ST, Lee KJ, Aizaki H, Hwang SB, Lai MM. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J Virol 77:4160–4168. doi: 10.1128/jvi.77.7.4160-4168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ocaka L, Spalluto C, Wilson DI, Hunt DM, Halford S. 2005. Chromosomal localization, genomic organization and evolution of the genes encoding human phosphatidylinositol transfer protein membrane-associated (PITPNM) 1, 2 and 3. Cytogenet Genome Res 108:293–302. doi: 10.1159/000081519. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda M, Abe K, Dansako H, Nakamura T, Naka K, Kato N. 2005. Efficient replication of a full-length hepatitis C virus genome, strain O, in cell culture, and development of a luciferase reporter system. Biochem Biophys Res Commun 329:1350–1359. doi: 10.1016/j.bbrc.2005.02.138. [DOI] [PubMed] [Google Scholar]
- 25.Roulin PS, Lotzerich M, Torta F, Tanner LB, van Kuppeveld FJ, Wenk MR, Greber UF. 2014. Rhinovirus uses a phosphatidylinositol 4-phosphate/cholesterol counter-current for the formation of replication compartments at the ER-Golgi interface. Cell Host Microbe 16:677–690. doi: 10.1016/j.chom.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 26.Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY. 2015. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods 12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BD, Rice CM. 2004. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J Virol 78:7400–7409. doi: 10.1128/JVI.78.14.7400-7409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Savidis G, McDougall WM, Meraner P, Perreira JM, Portmann JM, Trincucci G, John SP, Aker AM, Renzette N, Robbins DR, Guo Z, Green S, Kowalik TF, Brass AL. 2016. Identification of Zika virus and dengue virus dependency factors using functional genomics. Cell Rep 16:232–246. doi: 10.1016/j.celrep.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 29.Trivedi D, Padinjat R. 2007. RdgB proteins: functions in lipid homeostasis and signal transduction. Biochim Biophys Acta 1771:692–699. doi: 10.1016/j.bbalip.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 30.Murphy SE, Levine TP. 2016. VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim Biophys Acta 1861:952–961. doi: 10.1016/j.bbalip.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Loewen CJ, Levine TP. 2005. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J Biol Chem 280:14097–14104. doi: 10.1074/jbc.M500147200. [DOI] [PubMed] [Google Scholar]
- 32.Lev S. 2012. Nonvesicular lipid transfer from the endoplasmic reticulum. Cold Spring Harb Perspect Biol 4:a013300. doi: 10.1101/cshperspect.a013300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Bankaitis VA. 2010. Phosphoinositide phosphatases in cell biology and disease. Prog Lipid Res 49:201–217. doi: 10.1016/j.plipres.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog 8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Filseck JM, Vanni S, Mesmin B, Antonny B, Drin G. 2015. A phosphatidylinositol-4-phosphate powered exchange mechanism to create a lipid gradient between membranes. Nat Commun 6:6671. doi: 10.1038/ncomms7671. [DOI] [PubMed] [Google Scholar]
- 36.Dong R, Saheki Y, Swarup S, Lucast L, Harper JW, De Camilli P. 2016. Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell 166:408–423. doi: 10.1016/j.cell.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bishe B, Syed G, Siddiqui A. 2012. Phosphoinositides in the hepatitis C virus life cycle. Viruses 4:2340–2358. doi: 10.3390/v4102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan I, Katikaneni DS, Han Q, Sanchez-Felipe L, Hanada K, Ambrose RL, Mackenzie JM, Konan KV. 2014. Modulation of hepatitis C virus genome replication by glycosphingolipids and four-phosphate adaptor protein 2. J Virol 88:12276–12295. doi: 10.1128/JVI.00970-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amako Y, Syed GH, Siddiqui A. 2011. Protein kinase D negatively regulates hepatitis C virus secretion through phosphorylation of oxysterol-binding protein and ceramide transfer protein. J Biol Chem 286:11265–11274. doi: 10.1074/jbc.M110.182097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harak C, Meyrath M, Romero-Brey I, Schenk C, Gondeau C, Schult P, Esser-Nobis K, Saeed M, Neddermann P, Schnitzler P, Gotthardt D, Perez-Del-Pulgar S, Neumann-Haefelin C, Thimme R, Meuleman P, Vondran FW, De Francesco R, Rice CM, Bartenschlager R, Lohmann V. 2016. Tuning a cellular lipid kinase activity adapts hepatitis C virus to replication in cell culture. Nat Microbiol 2:16247. doi: 10.1038/nmicrobiol.2016.247. [DOI] [PubMed] [Google Scholar]
- 41.Cho NJ, Lee C, Pang PS, Pham EA, Fram B, Nguyen K, Xiong A, Sklan EH, Elazar M, Koytak ES, Kersten C, Kanazawa KK, Frank CW, Glenn JS. 2015. Phosphatidylinositol 4,5-bisphosphate is an HCV NS5A ligand and mediates replication of the viral genome. Gastroenterology 148:616–625. doi: 10.1053/j.gastro.2014.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsuan J, Cockcroft S. 2001. The PITP family of phosphatidylinositol transfer proteins. Genome Biol 2:REVIEWS3011. doi: 10.1186/gb-2001-2-9-reviews3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saeed M, Andreo U, Chung HY, Espiritu C, Branch AD, Silva JM, Rice CM. 2015. SEC14L2 enables pan-genotype HCV replication in cell culture. Nature 524:471–475. doi: 10.1038/nature14899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato T, Date T, Murayama A, Morikawa K, Akazawa D, Wakita T. 2006. Cell culture and infection system for hepatitis C virus. Nat Protoc 1:2334–2339. doi: 10.1038/nprot.2006.395. [DOI] [PubMed] [Google Scholar]
- 46.Phan T, Beran RK, Peters C, Lorenz I, Lindenbach BD. 2009. Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-4A enzyme complexes. J Virol 83:8379–8395. doi: 10.1128/JVI.00891-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammond GR, Schiavo G, Irvine RF. 2009. Immunocytochemical techniques reveal multiple, distinct cellular pools of PtdIns4P and PtdIns(4,5)P(2). Biochem J 422:23–35. doi: 10.1042/BJ20090428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin DL, Cherepanova NA, Bozzacco L, MacDonald MR, Gilmore R, Tai AW. 2017. Dengue virus hijacks a noncanonical oxidoreductase function of a cellular oligosaccharyltransferase complex. mBio 8:e00939-17. doi: 10.1128/mBio.00939-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saxena V, Lai CK, Chao TC, Jeng KS, Lai MM. 2012. Annexin A2 is involved in the formation of hepatitis C virus replication complex on the lipid raft. J Virol 86:4139. doi: 10.1128/JVI.06327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hung V, Udeshi ND, Lam SS, Loh KH, Cox KJ, Pedram K, Carr SA, Ting AY. 2016. Spatially resolved proteomic mapping in living cells with the engineered peroxidase APEX2. Nat Protoc 11:456–475. doi: 10.1038/nprot.2016.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.