Mosquito-borne flaviviruses include a group of pathogenic viruses that cause serious diseases in humans and animals, including dengue, West Nile, and Japanese encephalitis viruses. These flaviviruses are zoonotic and use animals, including birds, as amplifying and reservoir hosts. Avian Tembusu virus (TMUV) is an emerging mosquito-borne flavivirus that is pathogenic for many avian species and can infect cells derived from mammals and humans in vitro. Although not currently pathogenic for primates, the infection of duck industry workers and the potential risk of TMUV infection in immunocompromised individuals have been highlighted. Thus, the prevention of TMUV in flocks is important for both avian and mammalian health. Our study reveals the escape of TMUV from the first line of the host defense system in the arthropod-borne transmission route of arboviruses, possibly helping to extend our understanding of flavivirus infection in birds and refine the design of anti-TMUV therapeutics.
Keywords: : flavivirus, virus-host interactions, monocytes/macrophages, innate immunity
ABSTRACT
Avian Tembusu virus (TMUV) is a newly emerging avian pathogenic flavivirus in China and Southeast Asia with features of rapid spread, an expanding host range, and cross-species transmission. The mechanisms of its infection and pathogenesis remain largely unclear. Here, we investigated the tropism of this arbovirus in peripheral blood mononuclear cells of specific-pathogen-free (SPF) ducks and SPF chickens and identified monocytes/macrophages as the key targets of TMUV infection. In vivo studies in SPF ducks and SPF chickens with monocyte/macrophage clearance demonstrated that the infection of monocytes/macrophages was crucial for viral replication, transmission, and pathogenesis. Further genome-wide transcriptome analyses of TMUV-infected chicken macrophages revealed that host antiviral innate immune barriers were the major targets of TMUV in macrophages. Despite the activation of major pattern recognition receptor signaling, the inductions of alpha interferon (IFN-α) and IFN-β were blocked by TMUV infection on transcription and translation levels, respectively. Meanwhile, TMUV inhibited host redox responses by repressing the transcription of genes encoding NADPH oxidase subunits and promoting Nrf2-mediated antioxidant responses. The recovery of either of the above-mentioned innate immune barriers was sufficient to suppress TMUV infection. Collectively, we identify an essential step of TMUV infection and reveal extensive subversion of host antiviral innate immune responses.
IMPORTANCE Mosquito-borne flaviviruses include a group of pathogenic viruses that cause serious diseases in humans and animals, including dengue, West Nile, and Japanese encephalitis viruses. These flaviviruses are zoonotic and use animals, including birds, as amplifying and reservoir hosts. Avian Tembusu virus (TMUV) is an emerging mosquito-borne flavivirus that is pathogenic for many avian species and can infect cells derived from mammals and humans in vitro. Although not currently pathogenic for primates, the infection of duck industry workers and the potential risk of TMUV infection in immunocompromised individuals have been highlighted. Thus, the prevention of TMUV in flocks is important for both avian and mammalian health. Our study reveals the escape of TMUV from the first line of the host defense system in the arthropod-borne transmission route of arboviruses, possibly helping to extend our understanding of flavivirus infection in birds and refine the design of anti-TMUV therapeutics.
INTRODUCTION
Avian Tembusu virus (TMUV) is a newly emerging avian pathogenic flavivirus, identified first in the People’s Republic of China in 2010, which causes mainly ovarian hemorrhage and a subsequent substantial decrease in egg laying together with a sudden decline in feed uptake and neurological signs (1). To date, TMUV continues to cause massive economic losses in the poultry industry not only in China but also in Southeast Asia, where the mosquito TMUV isolate has been grouped with a duck serum TMUV isolate from China (2). TMUV is a single-stranded positive-sense RNA arbovirus belonging to the genus Flavivirus, family Flaviviridae, and shares many conserved motifs with other flaviviruses (1, 3). Many flaviviruses, including West Nile virus (WNV), dengue virus (DENV), yellow fever virus (YEV), Japanese encephalitis virus (JEV), and Zika virus, are zoonotic. Given that birds serve as amplifying and reservoir hosts of some flaviviruses in nature, such as WNV and Bagaza virus (4–6), the relevance of TMUV to human and animal health has been investigated. A wide range of natural host species of TMUV have been identified, including mosquitos, ducks, chickens, geese, pigeons, and sparrows (2, 7). Although TMUV has been reported to be not pathogenic for primates due to its high sensitivity to mammalian interferon antiviral responses according to an in vivo study in rhesus monkeys (8), TMUV replicates well in many types of nonavian cells, including many human cell lines (i.e., Vero, BHK21, A549, HeLa, HepG2, and SH-SY5Y) in vitro and induces high neurovirulence that is typical of many other encephalitic flaviviruses and even death in mice upon intracerebral inoculation (8–11). The potential transmission from birds to humans has been further demonstrated by an investigation of duck industry workers, which reported that 71.9% of the serum samples tested contained antibodies against TMUV and that the RNA of TMUV was observed in 47.7% of the oral swab samples evaluated (9). Although TMUV did not cause viremia or clinical symptoms in rhesus monkeys, TMUV-specific humoral immune responses were induced, and the potential risk of TMUV infection in immunocompromised individuals was highlighted by the authors (8). Taken together, the rapid spread, expanding host range, and cross-species transmission of TMUV demonstrate the possibility that TMUV might emerge as a zoonotic flavivirus in the future, although the risk is still low, and the prevention of TMUV in flocks now is important for both avian and mammalian health. Further studies on the pathogenesis and host-pathogen interaction of this novel flavivirus are urgently needed.
Tembusu virus, West Nile virus, Usutu virus, Bagaza virus, and Israel turkey encephalitis virus currently constitute the five flaviviruses transmitted by mosquito bites with marked pathogenicity in birds (12). Despite the occurrence of a nonvector transmission strain due to the mutation at position 156 in the envelope protein (13), similar to other mosquito-borne flaviviruses, arthropod-borne transmission via the host blood is still the major route of transmission for TMUV. In this route, blood immune cells constitute the first line of the host antiviral defense system that TMUV must escape at the beginning of infection. However, the interaction between this arbovirus and poultry blood immune cells remains unclear.
Macrophages play a critical role in the induction and regulation of both innate and adaptive immune responses and sometimes act as a double-edged sword during certain viral infections, including flavivirus infections, as macrophages may not only help fight against viral infection but also contribute to virus production and dissemination during viral infections (14–18). The interactions between host macrophages and a number of viruses have been extensively studied in mammal models. However, limited information is known about the interaction between viruses and avian macrophages. Although avian macrophages have been shown to serve as the main target for some avian virus infections (19–22), the exact biological consequences and the underlying mechanisms of the infection of avian macrophages with these viruses are largely uncertain.
In the present study, the TMUV tropism for peripheral blood mononuclear cells (PBMCs) was investigated in specific-pathogen-free (SPF) ducks and SPF chickens, and the infection of monocytes/macrophages has been identified as the essential step of TMUV infection. Extensive subversion of the antiviral innate immune responses of monocytes/macrophages by TMUV was investigated.
RESULTS
Monocytes/macrophages are the primary targets of TMUV in host PBMCs.
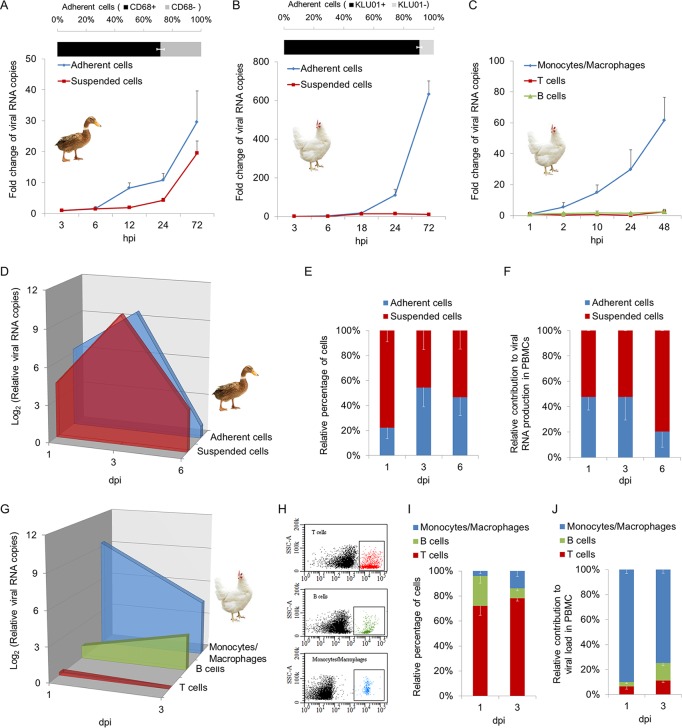
The susceptibilities of host PBMCs to TMUV were detected both in vitro and in vivo. We first isolated monocytes/macrophages and lymphocytes from PBMCs of 45- to 60-week-old female SPF ducks and chickens by two rounds of differential adhesion (23). The purities of monocytes/macrophages in adherent cells were 71.77% in ducks and 90.45% in chickens, as identified by antibodies targeting CD68 and the monocyte/macrophage marker antibody KUL01, respectively (Fig. 1A and B, upper panel). The cells were then infected with TMUV at a multiplicity of infection (MOI) of 0.01. TMUV grew in both adherent cells and suspended cells of duck PBMCs, as evidenced by the detection of viral RNA using reverse transcription-quantitative PCR (RT-qPCR) (Fig. 1A, lower panel). In contrast, for chicken PBMCs, the growth of TMUV was observed only in adherent cells, not in suspended cells (Fig. 1B, lower panel). This finding was further confirmed by TMUV infection of monocytes/macrophages, B lymphocytes, and T lymphocytes isolated from PBMCs of chickens by fluorescence-activated cell sorting (FACS) using antibodies targeting the specific cell surface markers of each type of cell (Fig. 1C). Next, we investigated TMUV infection of monocytes/macrophages and lymphocytes in vivo by detecting cells isolated from PBMCs of TMUV-inoculated SPF ducks and SPF chickens at the time points postinfection indicated in Fig. 1D. Consistent with the above-described in vitro investigations, TMUV infected both adherent cells and suspended cells of duck PBMCs, with peak viral RNA levels at 3 days postinfection (dpi) (Fig. 1D). Although TMUV preferred to replicate in adherent cells, the adherent cells and suspended cells contributed equally to the total viral RNA production in PBMCs at 1 dpi because of the higher proportion of suspended cells in PBMCs, while the contribution of adherent cells decreased to approximately 20% at 6 dpi (Fig. 1E and F). In chicken PBMCs, TMUV grew mainly in monocytes/macrophages, and the level of viral RNA decreased after 1 dpi (Fig. 1G). TMUV infection of B cells was observed in vivo (Fig. 1G). Although the level of viral RNA in B cells increased following the time of infection, due to the reduced proportion of B cells, the total viral RNA production in chicken PBMCs was contributed mainly by monocytes/macrophages, with a weak contribution by B cells at 3 dpi (Fig. 1H to J). In summary, despite the equal contribution of lymphocytes to viral RNA production in PBMCs in ducks, monocytes/macrophages are the major targets of TMUV in host PBMCs of both ducks and chickens.
FIG 1.
TMUV tropism for peripheral blood mononuclear cells (PBMCs). (A and B) Monocytes/macrophages and lymphocytes were obtained from freshly isolated PBMCs of 45- to 60-week-old female SPF ducks and chickens by two rounds of differential adhesion. The purities of monocytes/macrophages in adherent cells were identified by rabbit anti-CD68 polyclonal antibody in ducks (A) and by mouse anti-chicken monocyte/macrophage-PE clone KUL01 in chickens (B). Separated cells were infected with TMUV at an MOI of 0.01. The level of viral RNA was detected by RT-qPCR at the indicated times postinfection. Data are presented as the mean ± SEM (n = 3). (C) Monocytes/macrophages, T lymphocytes, and B lymphocytes were sorted and recycled by FACS from freshly isolated chicken PBMCs using mouse anti-chicken monocyte/macrophage-PE clone KUL01, mouse anti-chicken CD3-APC clone CT-3, and mouse anti-chicken Bu-1-FITC clone AV20, respectively. Cells were then infected with TMUV at an MOI of 0.01. The level of viral RNA was detected by RT-qPCR at the indicated times postinfection. Data are presented as the mean ± SEM (n = 3). (D to J) SPF ducks (D to F) or chickens (G to J) were inoculated with 100 μl of TMUV specimens at 1 × 105 PFU per ml via intravenous injection. Duck monocytes/macrophages and lymphocytes were isolated as described for panel A at the indicated time points postinfection. The relative percentages of cells were calculated based on cell counting (E). Chicken monocytes/macrophages, T lymphocytes, and B lymphocytes were isolated as described for panel C at the indicated time points postinfection. (H and I) The relative percentages of cells were detected by FACS. Representative flow plots are shown (H). Data are presented as the mean ± SEM (n = 3). (D and G) The level of viral RNA was detected by RT-qPCR. Data are presented as the mean ± SEM (n = 3). (F and J) The relative contribution to the total level of viral RNA in PBMCs by isolated cells was calculated. Data are presented as the mean ± SEM (n = 3).
TMUV infection of monocytes/macrophages is essential for viral replication, dissemination, and pathogenesis in ducks and chickens.
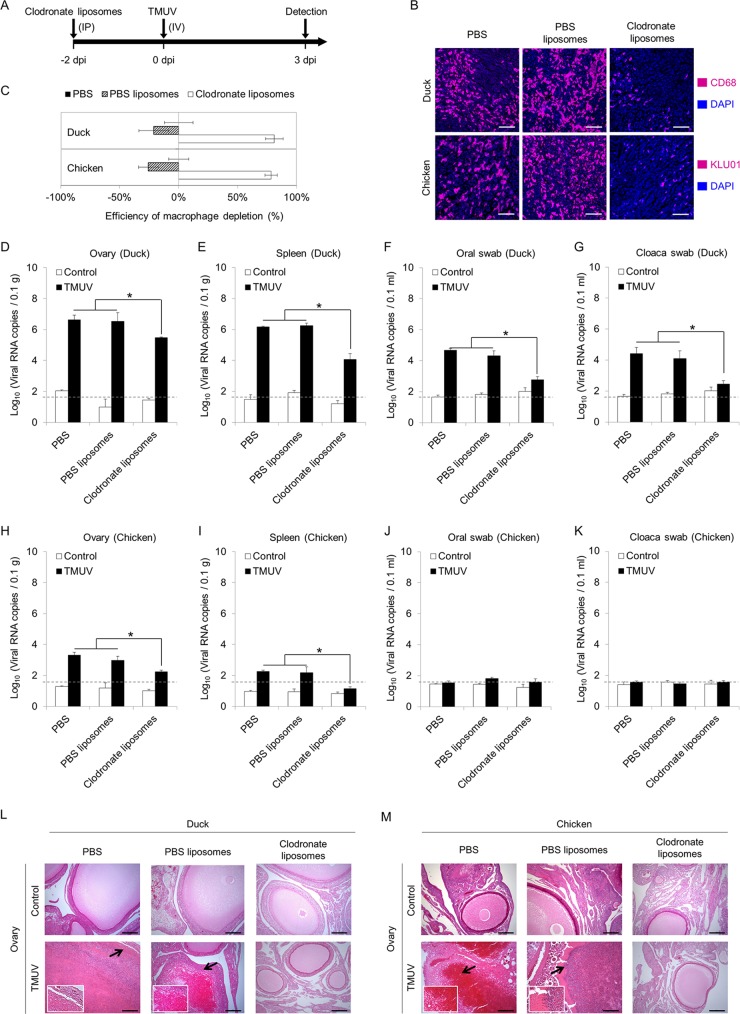
To address the role of monocytes/macrophages in TMUV infection of ducks and chickens, clearance of monocytes/macrophages was performed by injection of clodronate liposomes as described in Fig. 2A. The efficiencies of monocyte/macrophage depletion in ducks and chickens were determined by detecting monocytes/macrophages in the spleen using antibodies targeting CD68 and the monocyte/macrophage marker antibody KUL01, respectively (Fig. 2B). More than 80% of monocytes/macrophages were depleted in both ducks and chickens 2 days after the administration of clodronate liposomes (Fig. 2C). The clearance of monocytes/macrophages in ducks significantly reduced viral RNA production in both the ovary (Fig. 2D) and the spleen (Fig. 2E) and completely prevented oral and fecal shedding of virus, as evidenced by viral RNA detection in oral and cloaca swabs (Fig. 2F and G). The reduction in viral RNA production in the ovary and spleen upon monocyte/macrophage depletion were also observed in chickens (Fig. 2H and I). The level of viral RNA was lower in chickens than in ducks in general, and no viral shedding through the fecal-oral route was observed in inoculated chickens regardless of monocyte/macrophage depletion (Fig. 2H to K). We further investigated the pathogenic damage to the ovary, which is the most typical feature of TMUV infection (1). In line with the decreased level of viral RNA, obvious ovarian hemorrhage with immune cell infiltration appeared at 3 dpi in all infected ducks and chickens pretreated with phosphate-buffered saline (PBS) or PBS liposomes (Fig. 2L and M). Follicle atresia and rupture were observed in the slides of sections from some infected birds. This pathogenic damage was completely prevented by monocyte/macrophage depletion (Fig. 2L and M). These data demonstrate that infection of monocytes/macrophages is a key step of TMUV infection in vivo, which is important for viral replication, dissemination, and pathogenesis.
FIG 2.
Roles of monocytes/macrophages in TMUV infection in vivo. (A) Scheme depicting the design of the experiment. Monocytes/macrophages were depleted by intraperitoneal injection of clodronate liposomes. (B) Representative pictures of immunofluorescence staining for monocyte/macrophage markers. The efficiency of monocyte/macrophage depletion by clodronate liposome injection was examined in sections of spleen samples 2 days after clodronate liposome injection using rabbit anti-CD68 polyclonal antibody for duck samples and mouse anti-chicken monocyte/macrophage-PE clone KUL01 for chickens. Equal volumes of a control liposome suspension and PBS were used as controls. Scale bars, 30 μm. (C) The numbers of monocytes/macrophages were counted, and the reduction rates were calculated. Data are presented as the mean ± SEM (n = 3). (D to M) At 3 days post-intravenous injection of 500 μl of virus specimens at 1 × 105 PFU per ml in ducks and chickens preinjected with clodronate liposomes, control liposome suspension (PBS liposomes), or PBS 2 days prior to infection, ovaries were harvested, and oral swabs and cloaca swabs were collected. The level of viral RNA was detected by RT-qPCR. Samples collected from uninfected animals are indicated as “control.” A dashed line indicates the limit of detection. (L and M) Representative H&E staining of ovary section slides at a higher magnification with arrows depicting areas of damage are shown. The data in panels D to K are presented as the mean ± SEM. Asterisks indicate significant differences (n = 3, P < 0.05). Scale bars in panels L and M, 200 μm.
Genome-wide transcriptional profile analysis of TMUV infection of macrophages.
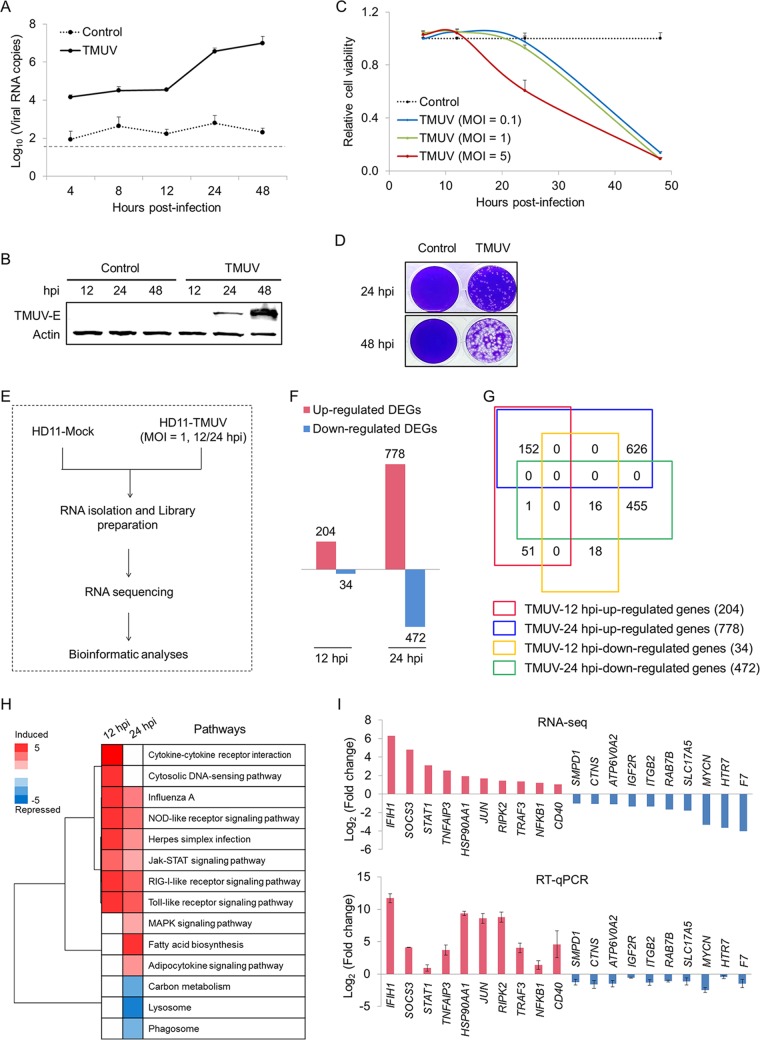
Considering the importance of monocytes/macrophages in TMUV infection, the responses of macrophages to TMUV infection were investigated by genome-wide transcriptional profile analysis to explore the molecular mechanisms of TMUV infection of macrophages. Given the limitations of the purity and long-term in vitro culture of avian primary isolated monocytes/macrophages, TMUV infection of the chicken macrophage cell line HD11 was employed as an experimental model in subsequent mechanistic studies. Similar to TMUV infection of primary isolated monocytes/macrophages and TMUV infection of monocytes/macrophages in vivo, the replication of TMUV in HD11 cells was observed after 12 h postinfection (hpi), as evidenced by the detection of viral RNA and protein using RT-qPCR (Fig. 3A) and Western blotting (Fig. 3B), respectively. Upon TMUV infection, the viability of host cells dropped quickly within 24 hpi at an MOI of 5, at which time no significant change in cell viability was observed at an MOI of 1 (Fig. 3C). Upon TMUV infection at an MOI of 1, the cytopathic effect (CPE) occurred at 24 hpi and became more severe at 48 hpi (Fig. 3D). No significant difference in reduction in viability of host cells was observed between an MOI of 1 and an MOI of 0.1 (Fig. 3C). Taken together, to investigate the initial responses of HD11 to TMUV replication, the transcriptional profiles of HD11 cells were assayed and analyzed at 12 hpi and 24 hpi with an MOI of 1, as illustrated in Fig. 3E. Bioinformatics analysis identified 238 and 1,250 differentially expressed genes (DEGs) at 12 hpi and 24 hpi, respectively, upon TMUV infection (Fig. 3F), based on the following criteria: (i) a P value of <0.001, (ii) a false discovery rate (q value) of <0.001, and (iii) a fold change of >2 (see Table S1 in the supplemental material). Among the 238 genes differentially expressed at 12 hpi, the constant effects of TMUV infection on the transcription of 168 genes were observed at 24 hpi (Fig. 3G). Further pathway analysis of these DEGs was performed to explore the shift of the molecular network of HD11 by TMUV infection. Four of the five major pattern recognition receptor (PRR) pathways, including the cytosolic DNA sensor (CDS) pathway, Toll-like receptor (TLR) signaling pathway, NOD-like receptor (NLR) pathway, and RIG-I-like receptor (RLR) pathway, were induced successfully by TMUV at 12 hpi, and all of these pathways except the CDS pathway were continually activated at 24 hpi (Fig. 3H). The upregulation of several crucial downstream transcriptional factors of PRR pathways, such as interferon (IFN) regulatory factor 7 (IRF7), nuclear factor-κB (NF-κB), and AP-1, was also observed (Table S1). Meanwhile, the suppression of pathways involved in another key antiviral innate immune defense of macrophages, phagocytosis, such as the lysosome and phagosome pathways, was observed at 24 hpi. For validation, the transcript levels of 20 genes randomly selected from these significantly regulated pathways were examined by RT-qPCR analysis. The directions of change determined by RT-qPCR detection and transcriptome sequencing (RNA-seq) analysis are identical in general (Fig. 3I), suggesting a correspondence between these two methods in our study. Our genome-wide transcriptional profile analysis of TMUV infection of HD11 demonstrates antiviral innate immune defenses as the major targets of TMUV infection in macrophages.
FIG 3.
Genome-wide transcriptome analysis of TMUV infection of HD11 cells. (A and B) The replication of TMUV in HD11 cells was determined by RT-qPCR (A) and Western blotting (B) using an antibody targeting the E protein of TMUV. Samples collected from uninfected cells are indicated as “control.” A dashed line indicates the limit of detection. Actin was used as a loading control. (C) The viability of HD11 cells with or without TMUV infection at the indicated MOI was detected using a CCK-8 kit. (D) The cytopathic effect of TMUV infection on HD11 cells was visualized by crystal violet staining. (E) Workflow of the genome-wide transcriptome analysis. (F) Number of genes that were differentially expressed in HD11 cells at a P of <0.001, a q of 0.001, and a fold change of >2. (G) Venn diagram showing the intersections of genes significantly regulated among subgroups in HD11 cells. (H) Pathway analysis of significantly expressed genes (P < 0.05). (I) The transcription of 20 genes selected for the validation of the RNA sequencing data was assayed by RT-qPCR. Data in panels A, C, and I are presented as the mean ± SEM (n = 3).
TMUV suppresses the expression of type I interferons in macrophages.
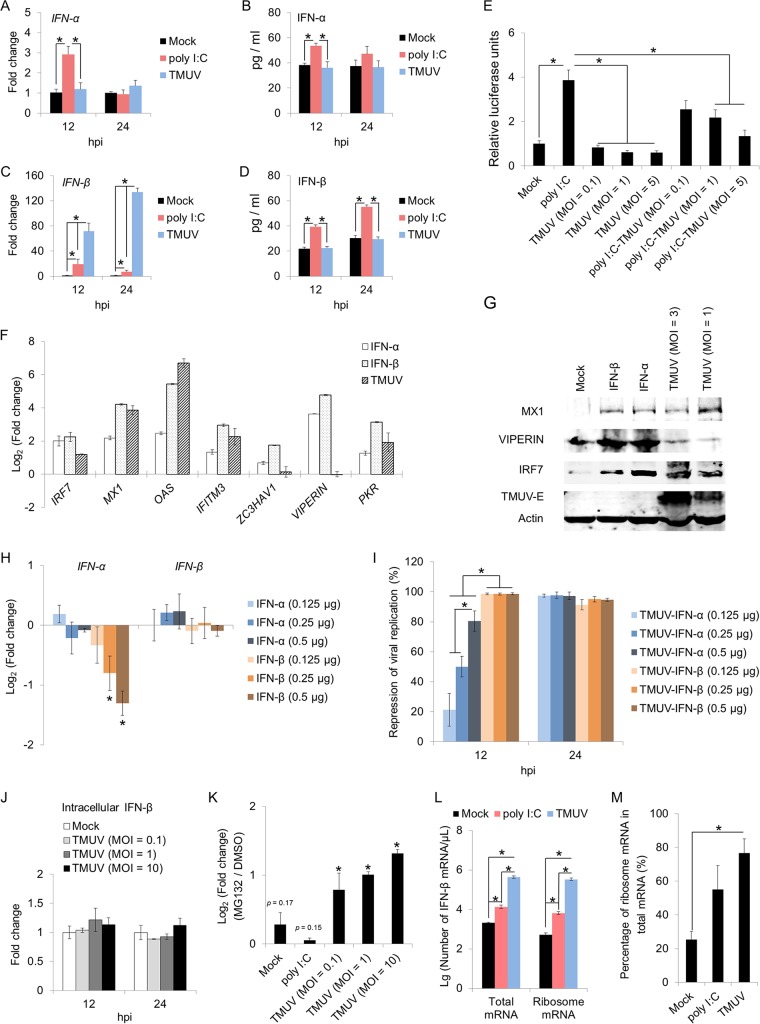
Multiple PRR families, such as TLRs, NLRs, and RLRs, contribute significantly to RNA viral detection, leading to the activation of downstream transcription factors, such as IRF7, NF-κB. and AP-1, and the subsequent induction of cytokines and type I IFNs (24–27). The successful activation of PRR pathways in HD11 cells upon TMUV infection (Fig. 3H) suggested that TMUV might escape PRR-mediated antiviral responses through the direct subversion of type I IFN expression. To address this concept, the expression of the main type I IFNs, alpha IFN (IFN-α) and IFN-β, was detected at both the mRNA and protein levels. The expression of IFN-α and IFN-β was significantly enhanced by poly(I·C) on both the mRNA and protein levels, as assayed by RT-qPCR and ELISA (Fig. 4A to D), suggesting that exogenous RNA was capable of promoting type I IFN expression in HD11 cells. In contrast, no increase in IFN-α expression at either the mRNA or protein level was observed in HD11 cells upon TMUV infection (Fig. 4A and B), indicating that TMUV infection can repress IFN-α expression as early as in the transcription phase. The mRNA level of IFN-β was strongly promoted by TMUV infection in HD11 cells, which was 3.71 times and 18.42 times that induced by poly(I·C) (Fig. 4C). However, the level of extracellular IFN-β was not affected by TMUV infection (Fig. 4D). Further, both the basal transcriptional activity of IFN-β and the enhanced transcriptional activity of IFN-β by poly(I·C) treatment were significantly attenuated by TMUV infection at MOIs of 1 and 5 when assayed by the luciferase assay (P < 0.05) (Fig. 4E), demonstrating an inhibitory effect of TMUV on IFN-β function. In addition, the effect of TMUV infection on the transcription of six well-characterized chicken ISGs (ChISGs), namely, MX1, OAS1, IFITM3, ZC3HAV1, VIPERIN, and PKR, was investigated, and HD11 cells treated with chicken IFN-α and IFN-β were used as a positive control for these ChISGs. All of these ChISGs were significantly induced by both IFN-α and IFN-β, as assayed by RT-qPCR (Fig. 4F), suggesting that these ChISGs can be induced by type I IFNs in macrophages. Four of these ChISGs (MX1, IFITM3, OAS1, and PKR) were also significantly induced by TMUV infection (Fig. 4F), indicating that these four ChISGs might be induced in a type I IFN-independent manner in chicken macrophages during TMUV infection. In agreement with the results of RT-qPCR assay, similar expression patterns of MX1 and viperin were observed at the protein level by a Western blotting assay (Fig. 4G). Different from mammals, chickens lack IRF3 and IRF9 in their type I IFN pathways, and IRF7, which can be induced by IFN-β to promote the expression of IFN-α in mammals, has been found to be constitutively expressed in chicken cells and essential for the regulation of IFN-β expression during viral infection (26). Upon TMUV infection, IRF7 was significantly induced on both transcription and protein levels (Fig. 4F and G), which is relevant to the enhanced transcription of IFN-β in the TMUV-infected cells we observed (Fig. 4C). However, the induction of IRF7 by TMUV infection was insufficient to promote the transcription of IFN-α in HD11 cells (Fig. 4A). Further studies revealed that neither the addition of chicken IFN-α nor the addition of chicken IFN-β could promote the transcription of any type I IFN detected in HD11 cells (Fig. 4H), although both type I IFNs promoted IRF7 expression (Fig. 4F and G). In agreement with a previous study, chicken IRF7 participated in mediating IFN-β, instead of IFN-α, induction upon stimulation (27). To address the biological significance of the repression of IFN-α and IFN-β by TMUV infection in macrophages, the effects of the addition of chicken IFN-α and IFN-β on TMUV infection in HD11 cells were investigated. Both type I IFNs were capable of preventing TMUV infection of HD11 cells, although the antiviral effect of IFN-α was weaker than that of IFN-β at 12 hpi (Fig. 4I), demonstrating the importance of the repression of type I IFNs in TMUV infection of macrophages. Given the significant induction of IFN-β transcription, TMUV might suppress the extracellular level of IFN-β by manipulating the translation, protein stability, and secretion of IFN-β. The involvement of IFN-β secretion was ruled out by the finding that no difference in the intracellular level of IFN-β between cells with or without TMUV infection was observed by ELISA (Fig. 4J). Further investigation using MG132 to prevent protein degradation in HD11 cells observed an ∼2 times greater enhancement in the level of extracellular IFN-β in TMUV-infected cells but not in either noninfected cells or poly(I·C)-treated cells (Fig. 4K), suggesting a virus-induced degradation of IFN-β. However, this inhibitory effect was too weak to counterbalance the dramatic promotion of IFN-β transcription by TMUV (134 times induction) (Fig. 4C), indicating the suppression of IFN-β translation upon TMUV infection. Therefore, the effect of TMUV infection on the translation initiation of IFN-β was assayed by detecting the amount of IFN-β mRNA bound to ribosome by using absolute quantitative real-time PCR. Both the amount of IFN-β mRNA bound to ribosome (Fig. 4L) and its proportion in total mRNA bound to ribosome (Fig. 4M) were significantly increased by TMUV infection, demonstrating that TMUV infection blocked IFN-β translation most likely at the translation elongation stage instead of the translation initiation stage. In summary, TMUV infection activated major PRR pathways successfully in host macrophages but subverted IFN-α transcription and IFN-β translation.
FIG 4.
Effects of TMUV on the expression of type I interferons in HD11 cells. (A to D) The expression of IFN-α (A and B) and IFN-β (C and D) in HD11 cells upon poly(I·C) treatment (1 μM) or TMUV infection (MOI = 1) was detected for both mRNA and protein levels by RT-qPCR (A and C) and ELISA (B and D). (E) Luciferase assay of IFN-β in HD11 cells upon poly(I·C) treatment (1 μM) and/or TMUV infection (MOI = 1). (F and G) The effects of the addition of recombinant chicken IFNs and TMUV infection on the expression of IRF7 and chicken ISGs in HD11 cells were assayed by RT-qPCR (F) and Western blotting (G), respectively. Actin was used as a loading control. (H) The effects of the addition of recombinant chicken IFNs on the expression of IFN-α and IFN-β in HD11 cells were assayed by RT-qPCR. (I) The repression rates of viral replication by the addition of chicken recombinant type IFNs were calculated with the levels of viral RNA detected by RT-qPCR. (J) The intracellular levels of IFN-β in HD11 cells with or without TMUV infection at the indicated MOI were detected by ELISA. (K) The effect of protein degradation on the expression of IFN-β in HD11 cells was determined by calculating the fold change of IFN-β expression levels with or without MG132 pretreatment. The level of IFN-β was detected by ELISA. (L and M) The effects of TMUV infection on the translation of IFN-β in HD11 cells were assayed by detecting the level of IFN-β mRNA bound to ribosomes using RT-qPCR. Data are presented as the mean ± SEM. Asterisks indicate a significant difference (n = 3, P < 0.05).
TMUV maintains intracellular redox homeostasis in macrophages.
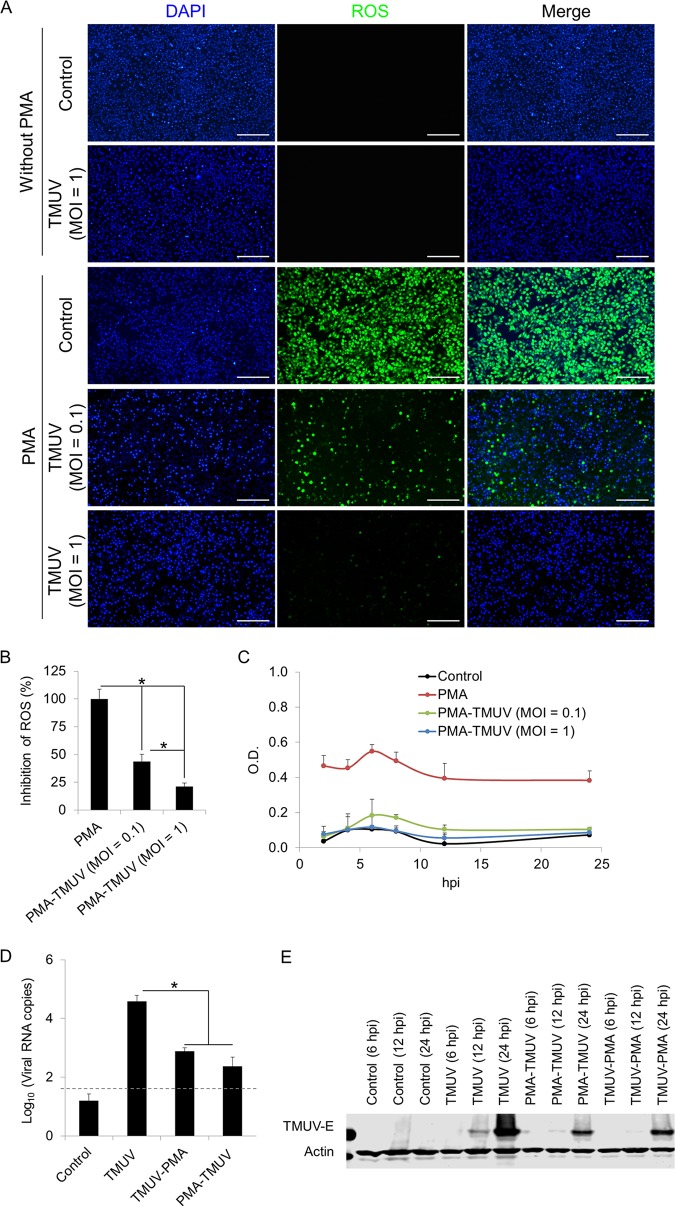
Along with PRR-mediated microbicidal action, the phagocytosis-dependent redox response is another key antiviral innate immune defense of macrophages. In line with the suppression of phagocytosis-related pathways (Fig. 3H), TMUV infection did not trigger the production of reactive oxygen species (ROS) in HD11 cells, as detected with the 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA)-based assay (Fig. 5A). Considering the low primary level of ROS in HD11 cells, we enhanced ROS production by stimulating cells with phorbol-12-myristate-13-acetate (PMA) (5 μg/ml), a protein kinase C (PKC) agonist widely used for ROS induction, to ascertain whether TMUV infection had an inhibitory effect on ROS production. Approximately 56.35% and 78.79% of ROS triggered by PMA were reduced by TMUV infection at MOIs of 0.1 and 1, respectively (Fig. 5A and B). Given that the DCFH-DA-based assay we used for ROS detection is not sensitive for the detection of superoxide, the total superoxide production was determined using a water-soluble tetrazolium salt 1 (WST-1)-based assay. A similar conclusion was obtained in HD11 cells upon PMA prestimulation (Fig. 5C). Hence, our data demonstrate that TMUV maintains the intracellular redox homeostasis in macrophages after infection. Next, we investigated the biological significance of the unaffected intracellular redox state during TMUV infection of macrophages by comparing the levels of viral replication under different superoxide levels. Upon prepromotion of ROS by PMA stimulation, TMUV replication was suppressed significantly regardless of the order of stimuli, as assayed by RT-qPCR (Fig. 5D) and Western blotting (Fig. 5E). Therefore, a stable intracellular redox state is important for TMUV evasion from the innate immune defenses of macrophages.
FIG 5.
Effects of TMUV on the redox state in HD11 cells. HD11 cells were infected with TMUV at the indicated MOI in the presence or absence of PMA pretreatment (5 μg/ml). (A) Cellular ROS were detected using a DCFH-DA-based reactive oxygen species assay kit at 24 h postinfection. All cell nuclei were stained with DAPI. Scale bars, 200 μm. (B) Fluorescent signal was analyzed statistically with ImageJ. Data are presented as the mean ± SEM. Asterisks indicate a significant difference (n = 6, P < 0.05). (C) Cellular superoxide production was detected using a WST-1-based superoxide assay kit. The optical density (OD) was read at 450 nm. Data are presented as the mean ± SEM (n = 3). (D and E) The replication of TMUV in HD11 cells with stimulation of PMA (5 μg/ml) before or after TMUV infection (MOI = 1) was determined by RT-qPCR at 24 h postinfection (D) and Western blotting using antibody targeting E protein of TMUV at the indicated time point (E). Samples collected from uninfected cells are indicated as “control.” A dashed line indicates the limit of detection. Actin was used as a loading control. Data in panel D are presented as the mean ± SEM. Asterisks indicate a significant difference (n = 3, P < 0.05).
TMUV maintains redox homeostasis in macrophages by both direct and indirect control of the intracellular superoxide level.
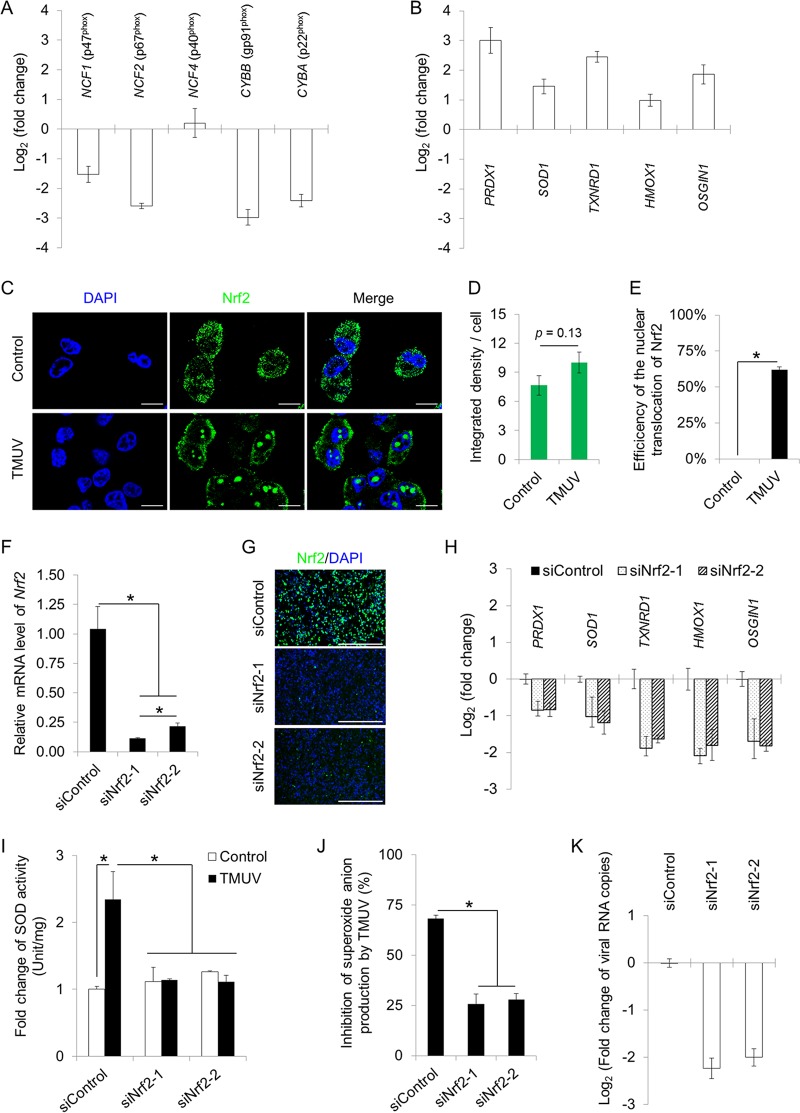
The generation of superoxide in phagocytic cells such as macrophages is mediated mainly by NADPH oxidases on the membrane of phagosomes. The phagocytic NADPH oxidase includes five subunits, namely, p47phox (NCF1), p67phox (NCF2), p40phox (NCF4), p22phox (CYBA), and gp91phox (CYBB). The transcription of four of the five subunits was suppressed by TMUV infection as assayed by RT-qPCR (Fig. 6A). In contrast, the transcription of five well-known antioxidant genes, namely, PRDX1, SOD1, TXNRD1, HMOX1 and OSGIN1, was induced by TMUV infection significantly (Fig. 6B).
FIG 6.
TMUV maintains intracellular redox homeostasis in macrophages. (A) The effects of TMUV infection on the transcription of NADPH oxidase subunits were assayed by RT-qPCR. Data are presented as the mean ± SEM (n = 3). (B) The effects of TMUV infection on the transcription of five antioxidant genes were assayed by RT-qPCR. Data are presented as the mean ± SEM (n = 3). (C) The effects of TMUV infection on Nrf2 expression and cellular location were examined by confocal microscopy using rabbit multiclonal antibodies specifically recognizing the C terminus of Nrf2, followed by an FITC-conjugated anti-rabbit secondary antibody. Cell nuclei were stained with DAPI (blue). Scale bars, 10 μm. (D) The integrated density of the fluorescent signal was quantified statistically with ImageJ. Data are presented as the mean ± SEM (n = 6). (E) The percentage of cells with Nrf2 nuclear translocation per field was quantified statistically by observing 100 cells per slide. Data are presented as the mean ± SEM. Asterisks indicate significant differences (n = 6, P < 0.05). (F and G) The efficiency of Nrf2 knockdown was verified by RT-qPCR (F) and immunofluorescent staining (G). Data in panel F are presented as the mean ± SEM. Asterisks indicate significant differences (n = 3, P < 0.05). Scale bar in panel G, 400 μm. (H) The effects of Nrf2 knockdown on the transcription of five antioxidant genes were assayed by RT-qPCR. Data are presented as the mean ± SEM (n = 3). (I and J) The effects of Nrf2 knockdown on the repression of SOD activity (I) and cellular superoxide production (J) by TMUV infection (MOI = 1) in HD11 cells were detected using a SOD activity assay kit and a WST-1-based superoxide assay kit, respectively. The absorbance was read at 450 nm. The SOD activity and the inhibition rate of superoxide production by TMUV infection were calculated. Data in panels I and J are presented as the mean ± SEM. Asterisks indicate a significant difference (n = 3, P < 0.05). (K) The effect of Nrf2 knockdown on TMUV RNA production was determined by RT-qPCR. Data are presented as the mean ± SEM (n = 3).
In mammals, the transcription of these antioxidant genes has been reported to be under the control of transcription factor Nrf2, a central regulator of intracellular redox homeostasis (28). Therefore, we detected the influence of TMUV infection on Nrf2 by immunofluorescence (IF) by using multiclonal antibodies specifically recognizing the C terminus of Nrf2 (Fig. 6C). Upon TMUV infection, the expression of Nrf2 was not affected (Fig. 6D), but the nuclear translocation of Nrf2 was greatly promoted, as the nuclear translocation of Nrf2 was observed in nearly 70% of TMUV-infected cells (Fig. 6E). To address the role of Nrf2 in the repression of the transcription of antioxidants and the subsequent stable redox state, knockdown of the Nrf2 gene using short interfering RNAs (siRNAs) targeting two different sites of the Nrf2 transcript was performed. The efficiency of knockdown was evidenced by both RT-qPCR (Fig. 6F) and IF (Fig. 6G). Knockdown of Nrf2 resulted in transcriptional repression of all antioxidants detected (Fig. 6H) and compromised the superoxide dismutase (SOD) activity elevated by TMUV infection (Fig. 6I). In agreement, the inhibitory effect of TMUV on superoxide anion production was greatly attenuated by Nrf2 knockdown (Fig. 6J), leading to significant repression of viral replication (Fig. 6K). Taken together, our data revealed pan-transcriptional repression of NADPH oxidase subunits and elevation of antioxidant activity through the promotion of nuclear translocation of Nrf2 by TMUV, which was essential for maintaining redox homeostasis in infected macrophages and subsequent viral replication.
DISCUSSION
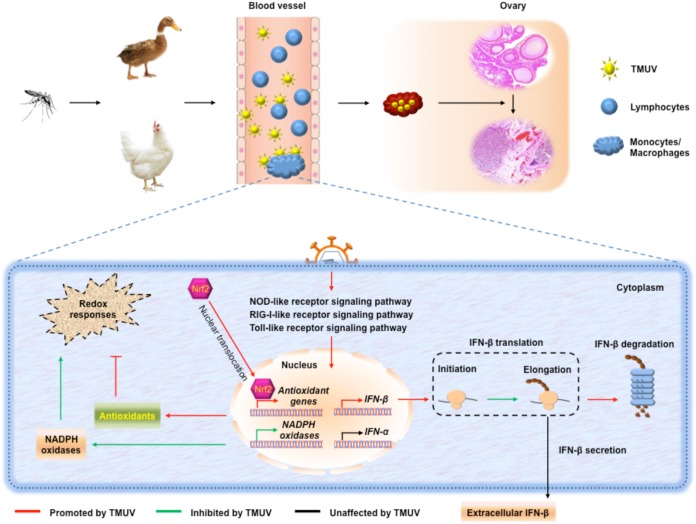
The newly emerging avian pathogenic flavivirus avian TMUV continues to cause massive economic losses in the poultry industry in China and Southeast Asia annually and may be a public health concern due to its potential transmission from birds to human or other nonavian hosts. The interactions between avian TMUV and its host remain mostly unknown. The present study investigated the interaction between this mosquito-borne flavivirus and its host PBMCs, the first line of the antiviral defense system in the arthropod-borne transmission route. Monocytes/macrophages were identified as the key target of TMUV infection, and the infection of monocytes/macrophages was found to be essential for viral replication, transmission, and pathogenesis in both ducks and chickens. Thus, illustration of the escape mechanism by which TMUV survives in monocytes/macrophages is important for developing therapeutics against TMUV infection. As shown by a schematic model (Fig. 7), monocytes/macrophages are successfully alerted upon TMUV infection through the activation of major PRR signaling pathways, which results in dramatic upregulation of IFN-β transcription. To survive in monocytes/macrophages, TMUV attenuates type I IFN-mediated antiviral immune responses by preventing the elevation of IFN-α transcription and reducing the translation of IFN-β (probably translation elongation). Meanwhile, TMUV, on the one hand, inhibits the transcription of NADPH oxidase subunits and, on the other hand, promotes antioxidant responses by promoting the nuclear translocation of Nrf2, which maintains a stable redox state in monocytes/macrophages. Together, the present study reveals TMUV tropism for avian host PBMCs, highlights the importance of monocytes/macrophages in TMUV infection in both ducks and chickens, and uncovers extensive subversion of host antiviral innate immune responses by TMUV in host cells.
FIG 7.
Schematic summary. Monocytes/macrophages are the key targets of TMUV in its arthropod-borne transmission route, which is essential for viral replication, transmission, and pathogenesis. Upon TMUV infection, monocytes/macrophages are successfully alerted through the activation of major PRR signaling pathways, which results in dramatic upregulation of IFN-β transcription. To survive in monocytes/macrophages, TMUV attenuates type I IFN-mediated antiviral immune responses by preventing the elevation of IFN-α transcription and reducing the translation and protein stability of IFN-β. Meanwhile, TMUV inhibits the transcription of NADPH oxidase subunits and enhances the transcription of antioxidant genes by promoting the nuclear translocation of Nrf2 to maintain a stable redox state in monocytes/macrophages.
Macrophages, which are not only members of the innate immune system but also regulators and effectors of the adaptive immune system, play a central role throughout antiviral immune responses (29–31). Subversion of the antiviral activities of macrophages by viruses such as HIV, severe acute respiratory syndrome (SARS) virus, African swine fever virus (ASFV), and porcine reproductive and respiratory syndrome virus (PRRSV) can facilitate viral replication and viral spread and can even enhance the intensity of immune responses, which leads to severe immune-mediated disease (32–35). Recently, the subversion of macrophages by flaviviruses has been reported for tick-borne encephalitis virus (TBEV), WNV, DENV, Zika virus, and classical swine fever virus (CSFV) (36–41). Antibody-dependent enhancement of TMUV infection has been observed in mice, indicating the potential interaction between TMUV and mouse macrophages (42). However, no direct evidence for TMUV infection of immune cells, including monocytes/macrophages, has been reported yet. A recent in vivo investigation published in February 2019 observed multivesicular bodies containing TMUV in macrophages and lymphocytes in the spleens of infected ducks by the use of electron microscopy (43). However, whether these viruses observed in the multivesicular bodies in macrophages and lymphocytes could replicate and transmit and the biological significance of the existence of TMUV in macrophages and lymphocytes in the spleen were not addressed. Our present study provided direct experimental evidence for both in vitro and in vivo infection of host monocytes/macrophages by TMUV (Fig. 1) and revealed that infection of monocytes/macrophages was essential for TMUV replication, transmission, and pathogenesis in ducks and chickens (Fig. 2).
In mammals, a two-step type I IFN gene regulation model has been proposed; in this model, IRF3 is crucial for the initial induction of IFN-β and the induced IFN-β activates IRF7 to promote IFN-α expression. Current knowledge of the avian type I IFN system indicates that these cytokines are functionally, structurally, and evolutionary related to their mammalian counterparts. Therefore, it is important to investigate in birds the potential existence of this two-step regulation model that has been proposed in mammals. Unlike mammals, chickens lack IRF3 and IRF9 in their type I IFN pathways (44). A number of efforts have been made recently to clarify whether the functions of mammalian IRF3 are compensated by IRF7 in birds. Similar to those of mammalian IRF3, constitutive expression of IRF7 and the regulation of IFN-β expression by IRF7 in different types of chicken cells, including DF-1, HD11, and primarily isolated chicken embryo fibroblasts (CEFs), have been revealed (26). The induction of IRF7 by TMUV infection is relevant to the enhanced transcription of IFN-β in TMUV-infected cells that we observed. However, chicken IRF7 seems less like the regulator of IFN-α upon stimulation, since IRF7 has been found to be dispensable for the induction of IFN-α in chicken cells upon stimulation and no effect of its overexpression on IFN-α expression has been observed (27). In agreement with these findings, the induction of IRF7 by TMUV infection is insufficient to promote the transcription of IFN-α in our experimental model (Fig. 4A). Our further studies revealed that neither the addition of chicken IFN-α nor the addition of chicken IFN-β could promote the transcription of any type I IFN detected in HD11 cells (Fig. 4H), although both type I IFNs promoted IRF7 expression (Fig. 4F and G). Neither previous studies nor our present study supports the existence of an IRF7-mediated link between IFN-β expression and IFN-α expression, which enables us to propose that chicken IRF7 participates in mediating IFN-β, instead of IFN-α, induction in macrophages upon stimulation and that the escape of TMUV from host IFN-α and IFN-β pathways in chicken macrophages may be independent events. The precise underlying mechanisms of the escape of TMUV from the type I IFN system of host macrophages needs to be further elucidated.
Currently, 108 ChISGs have been predicted by publicly available transcriptome data of chicken cell line DF-1 upon type I IFN treatment (45), while only a few have been functionally validated in chicken cells. In our present study, the transcription of six validated ChISGs, namely, MX1, IFITM3, OAS1, PKR, ZC3HAV1, and VIPERIN, was detected. Four of these ChISGs (MX1, IFITM3, OAS1, and PKR) were induced by TMUV infection, although host type I IFNs were not induced (Fig. 4F). It is known that to counteract the evasion of pathogens from host IFN responses, a subset of ISGs can be induced directly by IRFs, such as MX1, IFITM3, and OAS1 (46, 47). It is possible that these four ChISGs (MX1, IFITM3, OAS1, and PKR) were induced in a type I IFN-independent manner in chicken macrophages upon TMUV infection, since IRF7 was significantly induced by TMUV infection (Fig. 4F and G). In agreement with the results of the RT-qPCR assay, similar expression patterns of MX1 and viperin were observed at the protein level by a Western blotting assay (Fig. 4G). The enhanced protein levels of IRF7 and MX1 by TMUV infection suggest that the inhibition of IFN-β by TMUV infection at the translation level is more likely an immune evasion strategy specific to IFN-β rather than a global block of host translation by TMUV infection.
Both PRR signaling-triggered type I IFN pathways and phagocytosis-dependent redox responses are the key components of the antiviral innate immune system of macrophages and need to be overcome by viruses. Crosstalk of these two antiviral innate immune barriers has been revealed recently. ROS can initiate inflammatory responses by activating factors such as NF-κB and AP-1 (48, 49). Given that both NF-κB and AP-1 are essential transcription factors for the induction of type I IFNs, it is possible that the production of ROS could induce type I IFN expression. To date, the association of ROS with PRR signaling-triggered type I IFN expression has been validated in both human and porcine cells, which demonstrates that ROS is crucial for triggering the RIG-I-mediated IRF3 activation and subsequent IFN-β expression (26, 50). On another hand, ROS can also be induced by IFN-β, which has been found to be essential for the pathogenicity of some human diseases like dermatomyositis and the activation and proliferation of hematopoietic stem and progenitor cells (51, 52). In our model, TMUV infection induces the major PRR signaling pathways and subsequent transcription of IFN-β without affecting the host redox state, indicating that phagocytosis-dependent redox responses are not required for type I IFN expression in avian macrophages. Besides, no effect of IFN responses on the transcription of genes encoding NADPH oxidases and antioxidases was observed in either the publicly available transcriptome data of chicken cell line DF-1 upon type I IFN treatment (45) or our unpublished transcriptome data of chicken macrophage cell line HD11 upon poly(I·C) treatment. Thus, the escape of TMUV from host PRR signaling-triggered type I IFN responses and the phagocytosis-dependent redox responses that we observed in chicken macrophages are more likely independent events.
The generation of superoxide in phagocytic cells such as macrophages is mediated mainly by NADPH oxidases. The subversion of NADPH oxidase-mediated antiviral oxidative responses by flaviviruses has been well investigated in dengue virus infection, which revealed repression of antioxidant genes by viruses through promotion of the nuclear entry of Nrf2 (53). A similar mechanism was confirmed in avian flavivirus infection by our present study. In addition, our studies also observed an interaction between NADPH oxidases and viral infection, which, to the best of our knowledge, has not been yet reported for any virus. Its underlying mechanisms remain unclear and need to be investigated in the future.
Ducks are thought to be more resistant to pathogens, such as the highly pathogenic H5N1 strain of avian influenza virus, than chickens (54, 55). However, ducks seemed more sensitive to TMUV than chickens in the present study because the in vivo viral load in ducks was significantly higher than that in chickens, and viral dissemination was observed only in ducks, not in chickens, under our experimental conditions (Fig. 2D to M). This correlated with the different sensitivities of lymphocytes to TMUV between ducks and chickens (Fig. 1), indicating that TMUV infection of lymphocytes might affect viral replication in vivo. Further investigations are needed to validate this indication in the future. No difference in ovarian pathogenesis was observed between ducks and chickens (Fig. 2L and M), which, together with the importance of monocytes/macrophages in TMUV infection (Fig. 2), suggested that the pathogenesis in ovaries in infected birds is most likely determined by TMUV infection of monocytes/macrophages.
The antiviral immune response of host blood immune cells is the first barrier that arboviruses must overcome. Our present study provides the first direct experimental evidence for the infection of host monocytes/macrophages by avian flavivirus TMUV and highlights the essential role of the infection of monocytes/macrophages in TMUV infection, which may contribute to further illustration of the mechanisms underlying the infection and pathogenesis of this newly emerging avian flavivirus. The present study also uncovers extensive subversion of host antiviral innate immune responses by TMUV in host monocytes/macrophages, which may provide potential targets for the development of novel therapeutics to combat TMUV infection.
MATERIALS AND METHODS
Ethics statement.
The animal experimental protocol was approved and performed in accordance with the ethical guidelines of the Animal Ethics Committee of Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences (approval no. SYXK [Hei] 2011022).
Virus, cells, and agents.
The duck Tembusu virus strain Du/CH/LSD/110128 (GenBank accession no. KC136210.1) is stored at the Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences (HVRI, CAAS). This strain can be propagated in duck embryo fibroblasts (DEFs) with clear cytopathic effects (CPEs) observed (56). The DEFs isolated from specific-pathogen-free (SPF) duck embryos and a permanent chicken bone marrow macrophage cell line, HD11, were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. Cell cultures were incubated at 39°C and 5% CO2. Cell viability was determined using a CCK-8 kit (Beyotime Institute of Biotechnology, Beijing, China) or crystal violet staining (Beyotime Institute of Biotechnology) according to the manufacturer’s instructions. Phorbol 12-myristate 13-acetate (PMA), poly(I·C), diapocynin, and MG132 were all purchased from Sigma (Sigma-Aldrich, St. Louis, MO, USA).
Experimental animals.
Female SPF Jinding ducks and female SPF White Leghorn chickens were all obtained from and kept at the Laboratory Animal Science Department of Harbin Weike Biotechnology Development Company (ABSL-3; accredited by the China National Accreditation Service for Conformity Assessment [CNAS]), a state-owned enterprise subordinate to Harbin Veterinary Research Institute of CAAS.
Isolation of PBMCs.
Peripheral blood mononuclear cells (PBMCs) were isolated from 45- to 60-week-old female SPF ducks and SPF chickens using an avian lymphocyte isolation kit (Tianjin Haoyang Biological Technology, Tianjin, China) according to the manufacturer’s instructions. Monocytes/macrophages and lymphocytes were further separated from freshly isolated PBMCs by two rounds of differential adhesion according to a previous description (23). The purity of monocytes/macrophages in adherent cells in ducks was identified by a rabbit anti-CD68 polyclonal antibody (Boster Biological Technology, Wuhan, China), and the purity of monocytes/macrophages in adherent cells in chickens was identified by the mouse anti-chicken monocyte/macrophage-phycoerythrin (PE) clone KUL01 (Southern Biotechnology Associates, Birmingham, AL, USA). Separated cells were cultured in RPMI 1640 supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine at 37°C and 5% CO2.
FACS.
We conducted fluorescence-activated cell sorting (FACS) using a BD FACScan and CellQuest software version 4.0.2 (BD, Mountain View, CA). Monocytes/macrophages, T lymphocytes, and B lymphocytes were sorted and recycled from freshly isolated chicken PBMCs using mouse anti-chicken monocyte/macrophage-PE clone KUL01 (Southern Biotechnology Associates), mouse anti-chicken CD3-allophycocyanin (APC) clone CT-3 (Southern Biotechnology Associates), and mouse anti-chicken Bu-1-fluorescein isothiocyanate (FITC) clone AV20 (Southern Biotechnology Associates), respectively. Cells were then cultured in RPMI 1640 supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine at 37°C and 5% CO2.
In vivo experiments.
For monocyte/macrophage clearance, a clodronate liposome suspension (Vrije Universiteit Amsterdam, Yeasen, Shanghai, China), a control liposome suspension (Vrije Universiteit Amsterdam, Yeasen), or PBS was administered at 500 μl per animal via intraperitoneal (i.p.) injection 2 days prior to infection according to the manufacturer’s instructions. The efficiency of monocyte/macrophage depletion by clodronate liposome injection was examined in sections of spleen samples by using rabbit anti-CD68 polyclonal antibody for duck samples and mouse anti-chicken monocyte/macrophage-PE clone KUL01 for chicken samples. For in vivo infection, 500 μl of virus specimens at 1 × 105 PFU per ml were inoculated into the blood vessels of chickens or ducks. At 3 days postinfection, oral swabs and cloaca swabs were collected. The spleens and ovaries were then harvested, homogenized, and subjected to three cycles of freeze-thawing. Hematoxylin-eosin (H&E) staining was performed on sections of ovary samples fixed with 4% paraformaldehyde according to a previous description (57).
Virus detection and quantification.
Viral titers were determined using a PFU assay on DEFs. Levels of viral replication were determined using Western blot analysis with mouse monoclonal antibody targeting the E protein of TMUV and qPCR assays targeting the M gene of TMUV (56). Cells were infected with Du/CH/LSD/110128 at the multiplicity of infection (MOI) indicated in Results. The indicated MOI was obtained according to the number of cells to be infected and the number of infectious particles determined by PFU assays.
Immunofluorescence.
For immunofluorescent staining, samples were washed with PBS and fixed with 4% paraformaldehyde for 10 min. After quenching of excess aldehyde, the samples were permeabilized with 0.1% Triton X-100. Nonspecific antibody binding was blocked with 2% bovine serum albumin (BSA) for 1 h, and then the samples were incubated with rabbit anti-CD68 polyclonal antibody, mouse anti-chicken monocyte/macrophage-PE clone KUL01, or rabbit polyclonal antibody against chicken Nrf2 (GenScript, Nanjing, China), followed by a secondary goat anti-mouse/rabbit antibody conjugated to FITC (Jackson Laboratory, Bar Harbor, ME, USA). Background was determined using normal rabbit control serum from nonimmunized rabbits or a mouse IgG isotype control (Abcam, Shanghai, China). All cell nuclei were stained with 4′,-6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich). Fluorescent signals were detected with an EVOS FL fluorescence microscope (AMG, Bothell, WA). For confocal imaging, cells were cultured in 35-mm glass-bottom cell culture dishes and examined using a confocal microscope system (LSM880; Zeiss, Oberkochen, Germany).
Histopathological examination.
Ovaries collected from ducks and chickens were fixed in 4% paraformaldehyde. The fixed samples were sent to the Plaques Diagnosis and Technical Service Center of the Harbin Veterinary Research Institute (ABSL-3; accredited by the CNAS) for histopathological examination. Briefly, samples were embedded in paraffin and then sectioned. The sample slides were stained with H&E and observed by light microscopy or used for immunofluorescent staining. Histopathological examination reports were provided by the Plaques Diagnosis and Technical Service Center.
RT-qPCR.
RNA was isolated from cells or tissue samples using the EasyPure RNA purification kit (TransGen Biotech, Beijing, China), according to the manufacturer’s protocol. For ribosome mRNA detection, ribosome-nascent chain complex (RNC) was isolated according to a previous description (58) using sucrose buffer (30% sucrose in RB buffer) with ultracentrifugation at 185,000 × g for 4 h in a Beckman SW41-Ti rotor at 4°C. RNC-RNA was extracted using the TRIzol RNA extraction reagent (Ambion, USA) according to the manufacturer’s instructions. RT-qPCR and absolute RT-qPCR were performed using a SYBR PrimeScript kit (TaKaRa Bio Inc., Tokyo, Japan), as described previously (56). The standard of the absolute RT-qPCR was prepared by cloning the 180-nucleotide PCR product of the Gallus IFN-β gene into the pMD18-T plasmid (TaKaRa-Bio, Shiga, Japan) according to the manufacturer’s instructions. The primer sequences are shown in Table 1.
TABLE 1.
List of RT-qPCR primers
| Gene | Primer directiona | Sequence (5′ to 3′) |
|---|---|---|
| CD40 | F | GGAAACGCAACGCACAAC |
| R | GTCCCTTTCACCTTCACCAC | |
| JUN | F | ACGAGGATGCCCTGAACG |
| R | CCCGTTGCTGGACTGGAT | |
| IFIH1 | F | GATGCCGCCAGAAGAGTAT |
| R | GGAATGTTATTAGTGAAGGGTT | |
| TRAF3 | F | TTTTCAGGGAACAAACCAA |
| R | ATTCCGTAGCATTTCCTTCT | |
| HSP90AA1 | F | TCCTGTCCTCTGGCTTTA |
| R | GTGGCATCTCCTCGGTAA | |
| NFKB1 | F | GCCAACTGGGAGGTGTATG |
| R | CCCAGGGTCATCTTGCTAA | |
| RIPK2 | F | AAACATCCGCTTCAATACA |
| R | GACCACCAGAATCTCCATC | |
| TNFAIP3 | F | TGTGAACACCCAGCCCTAC |
| R | AGCATTGTAGCAGCGTTCA | |
| SOCS3 | F | ATGGTCACCCACAGCAAGTT |
| R | TGACGCTGAGGGTGAAGAAG | |
| STAT1 | F | CCTATGCCTCTGGAACGA |
| R | ATCCGAGATACCTCATCAAACT | |
| CTNS | F | TTGAGAACTGGCGACGAA |
| R | AGGGTTCACTCCGTTGG | |
| IGF2R | F | GAGTGGAGGACCTTTGTTG |
| R | GCTGTGAGTTTGGGACCTG | |
| SLC17A5 | F | GGGTTTGGCATCTTTGGTA |
| R | GCTAACCAGCCACATCCAG | |
| SMPD1 | F | ACTACCGCATCGTGAACAGG |
| R | CTGGGTGCCACGAAAGC | |
| ITGB2 | F | CTGGGCTTCACAGACACG |
| R | CTGGTCTTGCCTTTGGTG | |
| RAB7B | F | AGATTGCCTCTGCCTGGTG |
| R | CTTGCTTGGCGAGCGTCT | |
| F7 | F | CTGCTGGTTCCTCCTTCTC |
| R | CAGCGGCAGACATAATCCT | |
| HTR7 | F | TGGTGGTCATCTCCGTCTG |
| R | GCCATACACTTCCCGTTCT | |
| MYCN | F | AGCGTCAGAGGCGTAATG |
| R | TCTGCTCCTCTGCCTGAA | |
| ATP6V0A2 | F | AAGCCTCCGAATCCATCT |
| R | CCACGGGACACCAAACCT | |
| Nrf2 | F | CAAGCCAGCGGAGATGC |
| R | TGGCTGCTGTCGTCTGG | |
| GAPDH | F | GGCACTGTCAAGGCTGAGAA |
| R | TGCATCTGCCCATTTGATGT | |
| M (TMUV) | F | AGACTGCTGGTGCAATGAGAC |
| R | CGTCGTTCCCAGATTCCA | |
| MX1 | F | AAACGACCTGATGTTGCCTG |
| R | TTACCCCTTTCCATTCCTGC | |
| OASL1 | F | GCCACATCCTCGCCATCA |
| R | CCCAGTGCGTCGTAAGC | |
| IFITM3 | F | TGGTGACGGTGGAGACG |
| R | GGCAACCAGGGCGATGA | |
| ZAP | F | TTCCAAGTCAAGCCTGTCCC |
| R | CTCCGCTCTGCCTCTTCATC | |
| VIPERIN | F | AACGGTGGTTCAAGAAGTATGG |
| R | ACAGCATAATCTCGGCACCA | |
| PKR | F | TGACTTCTGTGACATACAACCCTC |
| R | TTTCAAACCAAATCAATCCC | |
| IRF7 | F | AACGACGACCCGCACAAG |
| R | GCAGCAGGTCCAAATCCA |
F, forward; R, reverse.
Protein extraction and Western blot analysis.
Western blot analysis was performed under reduced denaturing conditions, according to previously described procedures (59). Briefly, cells were washed with ice-cold PBS and soluble proteins were extracted with cell lysis buffer (100 mM Tris-HCl [pH 8], 150 mM NaCl, 1% NP-40, phosphatase, and protease inhibitor cocktail tablets) (Abcam) according to the manufacturer’s protocol. The protein concentration was determined using the Bio-Rad Bradford assay (Bio-Rad, Hercules, CA, USA) and BSA standards (Sigma-Aldrich). An equal amount of protein was separated by SDS-PAGE. Mouse monoclonal antibody targeting the E protein of TMUV, rabbit polyclonal antibody targeting MX1 (Proteintech Group, Rosemont, IL, USA), rabbit polyclonal antibody targeting viperin (Bioss, Beijing, China), and mouse monoclonal antibody targeting actin (Sigma-Aldrich) were used. Rabbit polyclonal antibody targeting IRF7 was kindly provided by Kai Li (HVRI, CAAS).
RNA sequencing.
Genome-wide gene expression profiling of HD11 cells was performed via RNA deep sequencing by Annoroad Gene Technology Co., Ltd. (Beijing, China). Three biological repeats were performed. RNA was isolated from cells using an RNeasy Plus minikit (Qiagen, Hilden, Germany). Library construction was performed using the Illumina platform (Illumina, Inc., San Diego, CA, USA), according to the manufacturer’s instructions. Samples were sequenced on an Illumina HiSeq 2500 instrument.
High-throughput data analysis.
RNA sequencing data were analyzed with the Galaxy web-based tool (60). Pathway analysis was performed with DAVID (gene-enrichment analysis using the EASE score, a modified Fisher’s exact P value, as the threshold) (61).
ELISA.
The levels of IFN-α and IFN-β in cell cultures and cells were analyzed using an ELISA kit for chicken IFN-α (USCN Life Science, Wuhan, China) and IFN-β (USCN Life Science) according to the manufacturer’s instructions. Recombinant chicken IFN-α and IFN-β were also purchased from USCN Life Science.
Transfection and dual-luciferase reporter assays.
HD11 cells were cotransfected with the firefly luciferase reporter plasmid IFN-β-luc and the Renilla luciferase reporter pRL-TK, which served as an internal control, using the TransIT-X2 dynamic delivery system (Mirus, Madison, WI, USA) according to the manufacturer’s instructions and a previous description (26). At 24 h posttransfection, cells were inoculated with TMUV at an MOI of 1 with or without pretreatment with poly(I·C). At 24 h postinfection, cells were lysed and subjected to assays for firefly and Renilla luciferase activities using the Dual-Luciferase reporter assay system (Promega, Madison, WI, USA). Relative luciferase activity was normalized to Renilla luciferase activity.
ROS/superoxide detection.
HD11 cells were inoculated with TMUV at an MOI of 0.1 or 1 as indicated in the presence or absence of PMA pretreatment. At 24 h postinfection, cellular reactive oxygen species (ROS) were detected using a DCFH-DA-based reactive oxygen species assay kit (Beyotime Institute of Biotechnology) according to the manufacturer’s instructions. All cell nuclei were stained with DAPI (Sigma-Aldrich). Fluorescent signals were detected with an EVOS FL fluorescence microscope (AMG) and analyzed with ImageJ. Cellular superoxide production was detected using a WST-1-based superoxide assay kit (Beyotime Institute of Biotechnology) according to the manufacturer’s instructions. The absorbance was read at 450 nm using a multifunctional microplate reader (PerkinElmer, Waltham, MA, USA).
RNA interference and transfection.
Short interfering RNAs (siRNAs) that specifically recognize two different sequences of the chicken Nrf2 mRNA (NM_205117.1; siNrf2-1, 5′-GCU GAA UGU GAA CUC UUU ATT-3′; siNrf2-2, 5′-GCA CCA CUC UAA CUA GUU UTT-3′) and a control siRNA (siControl, 5′-GCA CUU GAU ACA CGU GUA A-3′) with no specific target site in chickens were used (GenePharma, Shanghai, China). Transfection of siRNA was conducted using the TransIT-X2 dynamic delivery system (Mirus) according to the manufacturer’s instructions. Twenty-four hours after transfection, cells were harvested or infected with TMUV for further analysis. The knockdown efficiency was verified by RT-qPCR and immunofluorescent staining.
Statistical analysis.
The SPSS software package (SPSS for Windows version 13.0; SPSS Inc., Chicago, IL, USA) was used for all statistical analyses. Data obtained from several experiments are reported as the mean ± standard error of the mean (SEM). The significance of differences between two groups was determined with a two-tailed Student's t test. One-way or two-way analysis of variances with Bonferroni’s correction was employed for multigroup comparisons. For all analyses, a probability (P) value of <0.05 was considered statistically significant.
Data availability.
RNA raw sequencing data were uploaded to the National Center for Biotechnology Information database under accession number GSE127092.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the National Key Research and Development Project of China (no. 2016YFD0500107-4), the China Agriculture Research System (no. CARS-40-K18), the Elite Youth Program of the Chinese Academy of Agricultural Sciences (CAASQNYC-KYYJ-56), and the Agricultural Science and Technology Innovation Program (CAAS-XTCX2016011-04).
This publication reflects only the authors’ views.
We are greatly indebted to our colleagues who shared valuable reagents with us, and we are grateful to Xijun He, Zuo Zhang, Kai Li, and other colleagues for providing materials, technical support, and valuable suggestions.
H.L. and S.L. conceived and designed the experiments, Y.M., Y.L., N.W., Z.C., H.W., C.Z., Z.W., and L.C. performed the experiments, H.L., Y.M., and S.L. analyzed the data, H.L., Y.M., and S.L. contributed reagents/materials/analysis tools, and H.L. and S.L. wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00978-19.
REFERENCES
- 1.Su J, Li S, Hu X, Yu X, Wang Y, Liu P, Lu X, Zhang G, Hu X, Liu D, Li X, Su W, Lu H, Mok NS, Wang P, Wang M, Tian K, Gao GF. 2011. Duck egg-drop syndrome caused by BYD virus, a new Tembusu-related flavivirus. PLoS One 6:e18106. doi: 10.1371/journal.pone.0018106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang W, Chen S, Mahalingam S, Wang M, Cheng A. 2017. An updated review of avian-origin Tembusu virus: a newly emerging avian flavivirus. J Gen Virol 98:2413–2420. doi: 10.1099/jgv.0.000908. [DOI] [PubMed] [Google Scholar]
- 3.Cao Z, Zhang C, Liu Y, Liu Y, Ye W, Han J, Ma G, Zhang D, Xu F, Gao X, Tang Y, Shi S, Wan C, Zhang C, He B, Yang M, Lu X, Huang Y, Diao Y, Ma X, Zhang D. 2011. Tembusu virus in ducks, China. Emerg Infect Dis 17:1873–1875. doi: 10.3201/eid1710.101890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swayne DE, Beck JR, Smith CS, Shieh WJ, Zaki SR. 2001. Fatal encephalitis and myocarditis in young domestic geese (Anser anser domesticus) caused by West Nile virus. Emerg Infect Dis 7:751–753. doi: 10.3201/eid0704.010429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Traore-Lamizana M, Zeller HG, Mondo M, Hervy JP, Adam F, Digoutte JP. 1994. Isolations of West Nile and Bagaza viruses from mosquitoes (Diptera: Culicidae) in central Senegal (Ferlo). J Med Entomol 31:934–938. doi: 10.1093/jmedent/31.6.934. [DOI] [PubMed] [Google Scholar]
- 6.Agüero M, Fernández-Pinero J, Buitrago D, Sánchez A, Elizalde M, San Miguel E, Villalba R, Llorente F, Jiménez-Clavero MA. 2011. Bagaza virus in partridges and pheasants, Spain, 2010. Emerg Infect Dis 17:1498–1501. doi: 10.3201/eid1708.110077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou H, Yan B, Chen S, Wang M, Jia R, Cheng A. 2015. Evolutionary characterization of Tembusu virus infection through identification of codon usage patterns. Infect Genet Evol 35:27–33. doi: 10.1016/j.meegid.2015.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Wang HJ, Li XF, Liu L, Xu YP, Ye Q, Deng YQ, Huang XY, Zhao H, Qin ED, Shi PY, Gao GF, Qin CF. 2016. The emerging duck flavivirus is not pathogenic for primates and is highly sensitive to mammalian interferon antiviral signaling. J Virol 90:6538–6548. doi: 10.1128/JVI.00197-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Y, Diao Y, Chen H, Ou Q, Liu X, Gao X, Yu C, Wang L. 2015. Isolation and genetic characterization of a Tembusu virus strain isolated from mosquitoes in Shandong, China. Transbound Emerg Dis 62:209–216. doi: 10.1111/tbed.12111. [DOI] [PubMed] [Google Scholar]
- 10.Li S, Li X, Zhang L, Wang Y, Yu X, Tian K, Su W, Han B, Su J. 2013. Duck Tembusu virus exhibits neurovirulence in BALB/c mice. Virol J 10:260–267. doi: 10.1186/1743-422X-10-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ti J, Zhang M, Li Z, Li X, Diao Y. 2016. Duck Tembusu virus exhibits pathogenicity to Kunming mice by intracerebral inoculation. Front Microbiol 7:190. doi: 10.3389/fmicb.2016.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benzarti E, Linden A, Desmecht D, Garigliany M. 2019. Mosquito-borne epornitic flaviviruses: an update and review. J Gen Virol 100:119–132. doi: 10.1099/jgv.0.001203. [DOI] [PubMed] [Google Scholar]
- 13.Yan D, Shi Y, Wang H, Li G, Li X, Wang B, Su X, Wang J, Teng Q, Yang J, Chen H, Liu Q, Ma W, Li Z. 2018. A single mutation at position 156 in envelope protein of Tembusu virus is responsible for virus tissue tropism and transmissibility in ducks. J Virol 92:e00427-18. doi: 10.1128/JVI.00427-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray PJ, Wynn TA. 2011. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol 11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haddadi S, Kim DS, Jasmine H, van der Meer F, Czub M, Abdul-Careem MF. 2013. Induction of toll-like receptor 4 signaling in avian macrophages inhibits infectious laryngotracheitis virus replication in a nitric oxide dependent way. Vet Immunol Immunopathol 155:270–275. doi: 10.1016/j.vetimm.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Cline TD, Beck D, Bianchini E. 2017. Influenza virus replication in macrophages: balancing protection and pathogenesis. J Gen Virol 98:2401–2412. doi: 10.1099/jgv.0.000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pawelek KA, Dor D Jr, Salmeron C, Handel A. 2016. Within-host models of high and low pathogenic influenza virus infections: the role of macrophages. PLoS One 11:e0150568. doi: 10.1371/journal.pone.0150568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan SW, Wu-Hsieh BA, Lin YS, Chen WY, Huang Y, Anderson R. 2018. The monocyte-macrophage-mast cell axis in dengue pathogenesis. J Biomed Sci 25:77. doi: 10.1186/s12929-018-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amarasinghe A, Abdul-Cader MS, Nazir S, De Silva Senapathi U, van der Meer F, Cork SC, Gomis S, Abdul-Careem MF. 2017. Infectious bronchitis corona virus establishes productive infection in avian macrophages interfering with selected antimicrobial functions. PLoS One 12:e0181801. doi: 10.1371/journal.pone.0181801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornax I, Diel DG, Rue CA, Estevez C, Yu Q, Miller PJ, Afonso CL. 2013. Newcastle disease virus fusion and haemagglutinin-neuraminidase proteins contribute to its macrophage host range. J Gen Virol 94:1189–1194. doi: 10.1099/vir.0.048579-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrow AD, Burgess SC, Baigent SJ, Howes K, Nair VK. 2003. Infection of macrophages by a lymphotropic herpesvirus: a new tropism for Marek’s disease virus. J Gen Virol 84:2635–2645. doi: 10.1099/vir.0.19206-0. [DOI] [PubMed] [Google Scholar]
- 22.Chhabra R, Chantrey J, Ganapathy K. 2015. Immune responses to virulent and vaccine strains of infectious bronchitis viruses in chickens. Viral Immunol 28:478–488. doi: 10.1089/vim.2015.0027. [DOI] [PubMed] [Google Scholar]
- 23.Reddy VR, Trus I, Desmarets LM, Li Y, Theuns S, Nauwynck HJ. 2016. Productive replication of nephropathogenic infectious bronchitis virus in peripheral blood monocytic cells, a strategy for viral dissemination and kidney infection in chickens. Vet Res 47:70. doi: 10.1186/s13567-016-0354-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson AJ, Locarnini SA. 2007. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol 85:435–445. doi: 10.1038/sj.icb.7100100. [DOI] [PubMed] [Google Scholar]
- 25.Bowie AG, Unterholzner L. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol 8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao L, Li K, Zhang Y, Liu Y, Liu C, Zhang Y, Gao Y, Qi X, Cui H, Wang Y, Wang X. 2019. Inhibition of DNA-sensing pathway by Marek’s disease virus VP23 protein through suppression of interferon regulatory factor 7 activation. J Virol 93:e01934-18. doi: 10.1128/JVI.01934-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim TH, Zhou H. 2015. Functional analysis of chicken IRF7 in response to dsRNA analog poly(I:C) by integrating overexpression and knockdown. PLoS One 10:e0133450. doi: 10.1371/journal.pone.0133450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calkins MJ, Jakel RJ, Johnson DA, Chan K, Kan YW, Johnson JA. 2005. Protection from mitochondrial complex II inhibition in vitro and in vivo by Nrf2-mediated transcription. Proc Natl Acad Sci U S A 102:244. doi: 10.1073/pnas.0408487101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. 2010. Development of monocytes, macrophages, and dendritic cells. Science 327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serbina NV, Jia T, Hohl TM, Pamer EG. 2008. Monocyte-mediated defense against microbial pathogens. Annu Rev Immunol 26:421–452. doi: 10.1146/annurev.immunol.26.021607.090326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hume DA. 2006. The mononuclear phagocyte system. Curr Opin Immunol 18:49–53. doi: 10.1016/j.coi.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 32.Mlcochova P, Sutherland KA, Watters SA, Bertoli C, de Bruin RA, Rehwinkel J, Neil SJ, Lenzi GM, Kim B, Khwaja A, Gage MC, Georgiou C, Chittka A, Yona S, Noursadeghi M, Towers GJ, Gupta RK. 2017. A G1-like state allows HIV-1 to bypass SAMHD1 restriction in macrophages. EMBO J 36:604–616. doi: 10.15252/embj.201696025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. 2016. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19:181–193. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reis AL, Netherton C, Dixon LK. 2017. Unraveling the armor of a killer: evasion of host defenses by African swine fever virus. J Virol 91:e02338-16. doi: 10.1128/JVI.02338-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang R, Zhang YJ. 2014. Antagonizing interferon-mediated immune response by porcine reproductive and respiratory syndrome virus. Biomed Res Int 2014:315470. doi: 10.1155/2014/315470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carletti T, Zakaria MK, Marcello A. 2017. The host cell response to tick-borne encephalitis virus. Biochem Biophys Res Commun 492:533–540. doi: 10.1016/j.bbrc.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 37.Halstead SB, O’Rourke EJ. 1977. Dengue viruses and mononuclear phagocytes. I. Infection enhancement by non-neutralizing antibody. J Exp Med 146:201–217. doi: 10.1084/jem.146.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miorin L, Maestre AM, Fernandez-Sesma A, García-Sastre A. 2017. Antagonism of type I interferon by flaviviruses. Biochem Biophys Res Commun 492:587–596. doi: 10.1016/j.bbrc.2017.05.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simoni MK, Jurado KA, Abrahams VM, Fikrig E, Guller S. 2017. Zika virus infection of Hofbauer cells. Am J Reprod Immunol 77:e12613. doi: 10.1111/aji.12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quicke KM, Bowen JR, Johnson EL, McDonald CE, Ma H, O’Neal JT, Rajakumar A, Wrammert J, Rimawi BH, Pulendran B, Schinazi RF, Chakraborty R, Suthar MS. 2016. Zika virus infects human placental macrophages. Cell Host Microbe 20:83–90. doi: 10.1016/j.chom.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong XY, Tang SQ. 2016. Classical swine fever virus NS5A protein changed inflammatory cytokine secretion in porcine alveolar macrophages by inhibiting the NF-κB signaling pathway. Virol J 13:101. doi: 10.1186/s12985-016-0545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Z, Ji Y, Huang X, Fu Y, Wei J, Cai X, Zhu Q. 2013. An adapted duck Tembusu virus induces systemic infection and mediates antibody-dependent disease severity in mice. Virus Res 176:216–222. doi: 10.1016/j.virusres.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 43.Liu E, Sun X, Wang X, Wang T, Li W, Tarique I, Yang P, Chen Q. 2019. In vivo dynamic distribution of multivesicular bodies and exosomes in spleen of DTMUV infected duck. Vet Microbiol 229:138–146. doi: 10.1016/j.vetmic.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 44.Santhakumar D, Rubbenstroth D, Martinez-Sobrido L, Munir M. 2017. Avian interferons and their antiviral effectors. Front Immunol 8:49. doi: 10.3389/fimmu.2017.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Giotis ES, Robey RC, Skinner NG, Tomlinson CD, Goodbourn S, Skinner MA. 2016. Chicken interferome: avian interferon-stimulated genes identified by microarray and RNA-seq of primary chick embryo fibroblasts treated with a chicken type I interferon (IFN-α). Vet Res 47:75. doi: 10.1186/s13567-016-0363-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ashley C, Abendroth A, Mcsharry B, Slobedman B. 2019. Interferon-independent upregulation of interferon-stimulated genes during human cytomegalovirus infection is dependent on IRF3 expression. Viruses 11:246. doi: 10.3390/v11030246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pulit-Penaloza JA, Scherbik SV, Brinton MA. 2012. Type 1 IFN-independent activation of a subset of interferon stimulated genes in West Nile virus Eg101-infected mouse cells. Virology 425:82–94. doi: 10.1016/j.virol.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D, Kim YJ, Koh HS, Jang TY, Park HE, Kim JY. 2010. Reactive oxygen species enhance TLR10 expression in the human monocytic cell line THP-1. Int J Mol Sci 11:3769–3782. doi: 10.3390/ijms11103769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morgan MJ, Liu ZG. 2011. Crosstalk of reactive oxygen species and NF-κB signaling. Cell Res 21:103–115. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soucy-Faulkner A, Mukawera E, Fink K, Martel A, Jouan L, Nzengue Y, Lamarre D, Vande Velde C, Grandvaux N. 2010. Requirement of NOX2 and reactive oxygen species for efficient RIG-I-mediated antiviral response through regulation of MAVS expression. PLoS Pathog 6:e1000930. doi: 10.1371/journal.ppat.1000930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer A, Laverny G, Allenbach Y, Grelet E, Ueberschlag V, Echaniz-Laguna A, Lannes B, Alsaleh G, Charles AL, Singh F, Zoll J, Lonsdorfer E, Maurier F, Boyer O, Gottenberg JE, Nicot AS, Laporte J, Benveniste O, Metzger D, Sibilia J, Geny B. 2017. IFN-β-induced reactive oxygen species and mitochondrial damage contribute to muscle impairment and inflammation maintenance in dermatomyositis. Acta Neuropathol 134:655–666. doi: 10.1007/s00401-017-1731-9. [DOI] [PubMed] [Google Scholar]
- 52.Tasdogan A, Kumar S, Allies G, Bausinger J, Beckel F, Hofemeister H, Mulaw M, Madan V, Scharffetter-Kochanek K, Feuring-Buske M, Doehner K, Speit G, Stewart AF, Fehling HJ. 2016. DNA damage-induced HSPC malfunction depends on ROS accumulation downstream of IFN-1 signaling and bid mobilization. Cell Stem Cell 19:752–767. doi: 10.1016/j.stem.2016.08.007. [DOI] [PubMed] [Google Scholar]
- 53.Olagnier D, Peri S, Steel C, van Montfoort N, Chiang C, Beljanski V, Slifker M, He Z, Nichols CN, Lin R, Balachandran S, Hiscott J. 2014. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog 10:e1004566. doi: 10.1371/journal.ppat.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hulse-Post DJ, Sturm-Ramirez KM, Humberd J, Seiler P, Govorkova EA, Krauss S, Scholtissek C, Puthavathana P, Buranathai C, Nguyen TD, Long HT, Naipospos TS, Chen H, Ellis TM, Guan Y, Peiris JS, Webster RG. 2005. Role of domestic ducks in the propagation and biological evolution of highly pathogenic H5N1 influenza viruses in Asia. Proc Natl Acad Sci U S A 102:10682–10687. doi: 10.1073/pnas.0504662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim JK, Negovetich NJ, Forrest HL, Webster RG. 2009. Ducks: the “Trojan horses” of H5N1 influenza. Influenza Other Respir Viruses 3:121–128. doi: 10.1111/j.1750-2659.2009.00084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun L, Li Y, Zhang Y, Han Z, Xu Y, Kong X, Liu S. 2014. Adaptation and attenuation of duck Tembusu virus strain Du/CH/LSD/110128 following serial passage in chicken embryos. Clin Vaccine Immunol 21:1046–1053. doi: 10.1128/CVI.00154-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu G, Wang Q, Liu N, Xiao Y, Tong T, Liu S, Wu D. 2012. Infectious bronchitis virus nucleoprotein specific CTL response is generated prior to serum IgG. Vet Immunol Immunopathol 148:353–358. doi: 10.1016/j.vetimm.2012.06.028. [DOI] [PubMed] [Google Scholar]
- 58.Wang T, Cui Y, Jin J, Guo J, Wang G, Yin X, He QY, Zhang G. 2013. Translating mRNAs strongly correlate to proteins in a multivariate manner and their translation ratios are phenotype specific. Nucleic Acids Res 41:4743–4754. doi: 10.1093/nar/gkt178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li H, Gao Q, Shao Y, Sun B, Wang F, Qiao Y, Wang N, Liu S. 2018. Gallid herpesvirus 1 initiates apoptosis in uninfected cells through paracrine repression of p53. J Virol 92:e00529-18. doi: 10.1128/JVI.00529-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol Chapter 19:Unit 19.10.1-21. doi: 10.1002/0471142727.mb1910s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA raw sequencing data were uploaded to the National Center for Biotechnology Information database under accession number GSE127092.