Accumulating evidence indicates that RNA viruses use numerous host factors during replication of their genomes. However, the host factors involved in rubella virus (RUBV) genome replication are largely unknown. In this study, we demonstrate that the HSP90 molecular chaperone is needed for the efficient replication of the RUBV genome. Further, we reveal that HSP90 interacts with RUBV nonstructural protein p150 and its precursor polyprotein, p200. HSP90 contributes to the stability of p150 and the processing of p200 via its protease domain in the p150 region. We conclude that the cellular molecular chaperone HSP90 is a key host factor for functional maturation of nonstructural proteins for RUBV genome replication. These findings provide novel insight into this host-virus interaction.
KEYWORDS: HSP90, Matonaviridae, Togaviridae, alphavirus, genome replication, host factor, rubella virus
ABSTRACT
Two viral nonstructural proteins, p150 and p90, are expressed in rubella virus (RUBV)-infected cells and mediate viral genome replication, presumably using various host machineries. Molecular chaperones are critical host factors for the maintenance of cellular proteostasis, and certain viral proteins use this chaperone system. The RUBV p150 and p90 proteins are generated from a precursor polyprotein, p200, via processing by the protease activity of its p150 region. This processing is essential for RUBV genome replication. Here we show that heat shock protein 90 (HSP90), a molecular chaperone, is an important host factor for RUBV genome replication. The treatment of RUBV-infected cells with the HSP90 inhibitors 17-allylamino-17-desmethoxygeldanamycin (17-AAG) and ganetespib suppressed RUBV genome replication. HSP90α physically interacted with p150, but not p90. Further analyses into the mechanism of action of the HSP90 inhibitors revealed that HSP90 activity contributes to p150 functional integrity and promotes p200 processing. Collectively, our data demonstrate that RUBV p150 is a client of the HSP90 molecular chaperone and that HSP90 functions as a key host factor for RUBV replication.
IMPORTANCE Accumulating evidence indicates that RNA viruses use numerous host factors during replication of their genomes. However, the host factors involved in rubella virus (RUBV) genome replication are largely unknown. In this study, we demonstrate that the HSP90 molecular chaperone is needed for the efficient replication of the RUBV genome. Further, we reveal that HSP90 interacts with RUBV nonstructural protein p150 and its precursor polyprotein, p200. HSP90 contributes to the stability of p150 and the processing of p200 via its protease domain in the p150 region. We conclude that the cellular molecular chaperone HSP90 is a key host factor for functional maturation of nonstructural proteins for RUBV genome replication. These findings provide novel insight into this host-virus interaction.
INTRODUCTION
Rubella virus (RUBV) is the causative agent of rubella, and humans are the only natural host of this virus. The clinical symptoms of rubella are a fever, a maculopapular rash, and lymphadenopathy. Rubella is usually a mild disease, but it may cause multiple organ defects known as congenital rubella syndrome (CRS) in babies born from mothers who are infected with the virus at an early stage of pregnancy. Typical complications of CRS are cataracts, deafness, and heart defects (1). Although highly safe and effective vaccines are available for rubella, the immunization coverage is not sufficient in many countries to prevent outbreaks of infection. Understanding of the molecular mechanisms of RUBV infection is essential to unveil RUBV pathology.
In 2019, a new family, Matonaviridae, was created to include the genus Rubivirus, which was removed from the family Togaviridae. RUBV is the sole member in the genus Rubivirus (1, 2). The rest of the genus Alphavirus in the Togaviridae family comprises ∼30 viruses, including Sindbis virus (SINV), chikungunya virus (CHIKV), and Semliki Forest virus, which are mainly maintained in arthropod vectors and mammals (3). RUBV and togaviruses are enveloped, single-stranded, positive-sense RNA viruses with a genome of ∼10 to 12 kb (3). Although RUBV differs greatly from alphaviruses in terms of the nucleotide and amino acid sequences of the genome and viral proteins, respectively, the genome organization of RUBV shows a certain similarity to the genome organizations of the alphaviruses, and RUBV proteins have functional and structural domains similar to those of alphavirus proteins (4). The RUBV genome encodes a polyprotein, p200, which is a precursor of two nonstructural proteins (NSPs), p150 and p90. These proteins are believed to have several enzymatic activities because they share amino acid sequence homology with their counterparts in other positive-sense RNA viruses. p150 possesses methyltransferase and protease motifs, while p90 contains helicase and RNA-dependent RNA polymerase (RdRp) motifs (4–8). In fact, the protease activity in the p150 region participates in the cleavage of p200, generating p150 and p90 (9). The two cleaved proteins are considered to function in virus genome replication (1).
Heat shock protein 90 (HSP90) is one of the most evolutionarily conserved molecular chaperones from bacteria to mammals. In mammals, there are four isoforms of HSP90. Two isoforms are cytoplasmic HSP90s, one of which is the stress-inducible HSP90α and the other of which is the constitutively expressed HSP90β; the third isoform is Grp94, which is localized to the endoplasmic reticulum; and the fourth isoform is TRAP1, which is a mitochondrial HSP90 (10). RUBV replicates in the cytoplasm, and thus, cytoplasmic HSP90s are the focus of the present study. HSP90 contributes to the maintenance of cellular proteostasis by promoting the proper folding and maturation of its client proteins (11). Many essential factors in vital cellular processes, such as steroid hormone receptors, protein kinases, and transcription factors, are HSP90 clients (12). Among this variety of factors, HSP90 specifically recognizes clients with the aid of cochaperones (13–15). HSP90 also leads clients to undergo proteasomal degradation, if the clients are misfolded (16). A variety of DNA and RNA viruses are known to use HSP90 for the proper folding and maturation of their viral proteins (17). For togaviruses, nsP4 of SINV and nsP2, nsP3, and nsP4 of CHIKV have been shown to interact with cytoplasmic HSP90s (18–20). It is clear that HSP90 activity is important for efficient CHIKV propagation in vitro and in vivo, while the significance of HSP90 in SINV infection is still unclear (19, 20).
In this study, we analyzed the role of HSP90 in RUBV infection. Our data demonstrate that RUBV p150 is an HSP90 client and that HSP90 activity is essential for the functional integrity of p150 and for genome replication.
RESULTS
HSP90 supports the genome replication of RUBV and SINV.
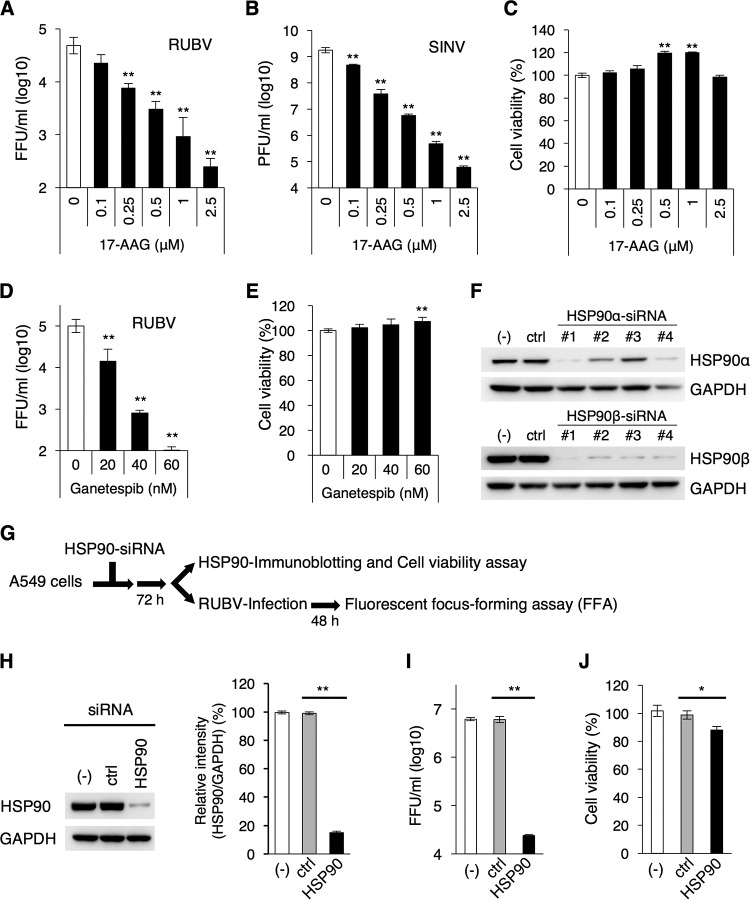
Vero cells were infected with RUBV and SINV at a multiplicity of infection (MOI) of 1.0. In RUBV infection experiments, recombinant RUBV rHS-p150/AG1, which expresses p150 fused with the humanized monomeric Azami green 1 (AG1) fluorescent protein, was used (21, 22). In SINV infection experiments, the isolated MM2215 virus strain was used (23, 24). The infected cells were cultured with various concentrations of 17-allylamino-17-desmethoxygeldanamycin (17-AAG), an inhibitor of HSP90 activity (25). The infectivity titers of RUBV and SINV released into the media were significantly decreased in a dose-dependent manner (Fig. 1A and B), whereas cell viability was not reduced by 17-AAG at these concentrations (Fig. 1C). The titer of RUBV was reduced by ∼100-fold at a concentration of 2.5 μM. The titer of SINV was also reduced by ∼10,000-fold at a concentration of 2.5 μM. When the cells were cultured with ganetespib, another potent HSP90 inhibitor (12, 26), RUBV infectivity titers were again decreased at nontoxic concentrations of ganetespib (Fig. 1D and E). To further demonstrate the role of cytoplasmic HSP90s in RUBV infection, RUBV production was assessed using A549 cells transfected with small interfering RNAs (siRNAs) specific to HSP90α and HSP90β (Fig. 1F). A549 cells have successfully been used in previous siRNA experiments for RUBV infection (27). The reduction in protein levels of HSP90α and HSP90β was assessed in the cells transfected with individual siRNAs. Four individual siRNAs were used for both HSP90α and HSP90β. The amount of HSP90α was decreased most effectively by the treatment of number 1 siRNA specific to HSP90α (Fig. 1F). The amount of HSP90β was also decreased most effectively by the treatment of number 1 siRNA specific to HSP90β (Fig. 1F). In cells treated with a combination of both number 1 siRNAs specific to HSP90α and HSP90β, the total amount of cytoplasmic HSP90 was reduced by ∼85% (Fig. 1H), and the RUBV titer released into the media was decreased by ∼100-fold (Fig. 1I). Under these conditions, cell viability was reduced by only ∼10% (Fig. 1J).
FIG 1.
HSP90 is required for efficient production of RUBV and SINV. (A) Vero cells were infected with RUBV at an MOI of 1 and cultured for 72 h in various concentrations of 17-AAG. The infectivity titers in the media were measured by FFA. (B) Vero cells were infected with SINV at an MOI of 1 and cultured for 18 h in various concentrations of 17-AAG. The infectivity titers in the media were measured by a plaque assay. (C) Vero cells were cultured for 72 h in media containing various concentrations of 17-AAG. After incubation, cell viability was measured by an ATP-based assay and was expressed as a relative value. (D) Vero cells were infected with RUBV at an MOI of 1 and cultured for 72 h in various concentrations of 17-AAG. The infectivity titers in the media were measured by an FFA. (E) Vero cells were cultured for 72 h in media containing various concentrations of ganetespib. After incubation, cell viability was measured by an ATP-based assay and was expressed as a relative value. (F) Immunoblotting of HSP90α and HSP90β in each of four siRNA-transfected cells at 72 h posttransfection (p.t.) (lanes −, untransfected cells; lanes ctrl, control siRNA-transfected cells; lanes #1 to #4, HSP90α or HSP90β siRNA-transfected cells 1 to 4, respectively). (G) Experimental flow for the assays whose results are presented in panels H, I, and J. (H) Immunoblotting of HSP90 in combination with HSP90α number 1 and HSP90β number 1 siRNA-transfected cells (lane −, untransfected cells; lane ctrl, control siRNA-transfected cells; lane HSP90, HSP90α and HSP90β siRNA-transfected cells). The bar graph shows the relative signal intensities of the HSP90-to-GAPDH signals. (I) At 72 h p.t., the transfected cells were infected with RUBV at an MOI of 5 and were cultured for 48 h. After incubation, the infectivity titers in the media were measured by an FFA. (J) The viability of the transfected cells at 72 h p.t. was measured by an ATP-based assay and was expressed as a relative value. (A to E and H to J) Data are representative of those from three independent experiments. (F) Data are representative of those from two independent experiments. (A to E and H to J). Mean values ± standard deviations (SD) for triplicate wells are shown, and the significant differences were determined by two-tailed t tests. *, P < 0.05; **, P < 0.01.
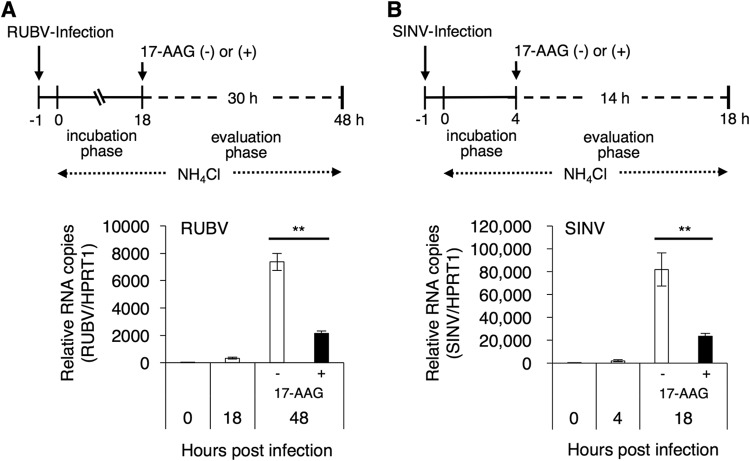
Alphavirus nsPs, which are involved in genome replication, interact with two cytoplasmic HSP90s, as previously demonstrated (18–20). The significance of HSP90 activity in viral genome replication was analyzed. To assess the levels of genome replication in a single round of infection, viruses were allowed to enter the cells for only 1 h. After that, secondary infection was inhibited by NH4Cl throughout the experiment. RUBV, like the togaviruses, enters cells via endocytosis and subsequent low-pH-triggered membrane fusion (28–33). NH4Cl prevents endosomal acidification and thus blocks viral entry (28, 29, 33). At the incubation phase (18 h and 4 h for RUBV and SINV, respectively), the cells were cultured without 17-AAG (Fig. 2A and B) to initiate genome replication. Then, at the evaluation phase, the cells were cultured in the absence or presence of 2.5 μM 17-AAG (30 h and 14 h for RUBV and SINV, respectively). Treatment with 17-AAG inhibited virus genome replication during the evaluation phase of both RUBV and SINV (Fig. 2A and B). These data suggest that HSP90 activity supports virus production and viral genome replication of both RUBV and SINV.
FIG 2.
HSP90 activity is required for efficient genome replication of RUBV and SINV. (A) Vero cells were infected with RUBV for 1 h. After washing, the cells were cultured for 18 h (incubation phase) with NH4Cl. Then, the cells were cultured in the presence or absence of 2.5 μM 17-AAG for 30 h (evaluation phase) with 20 mM NH4Cl to block the secondary infections. The viral genomic RNA and the host HPRT1 mRNA were quantified by real-time RT-PCR at the indicated time points. The graph shows the viral genomic RNA copy numbers relative to those of HPRT1 mRNA. (B) Vero cells were infected with SINV for 1 h. After washing, the cells were cultured for 4 h (incubation phase). Then, the cells were cultured in the presence or absence of 2.5 μM 17-AAG for 14 h (evaluation phase). NH4Cl (20 mM) was used to block secondary infections. The viral RNA and the host HPRT1 mRNA were quantified by real-time RT-PCR at the indicated time points. The graph shows the viral RNA copy numbers relative to those of HPRT1 mRNA. (A, B) Data are representative of those from three independent experiments. Mean values ± SD for four wells are shown. The significant differences were determined by two-tailed t tests. **, P < 0.01.
RUBV p150 and alphavirus nsP4 are HSP90 clients.
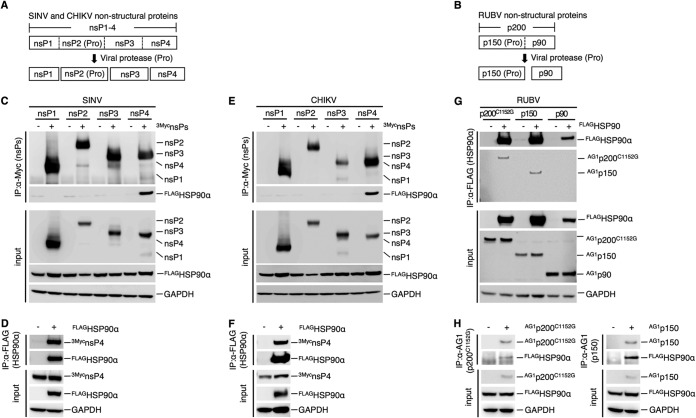
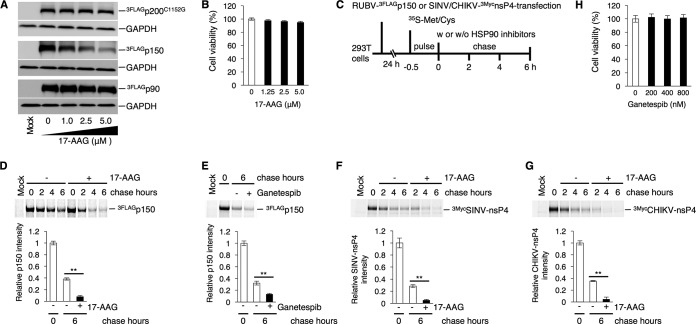
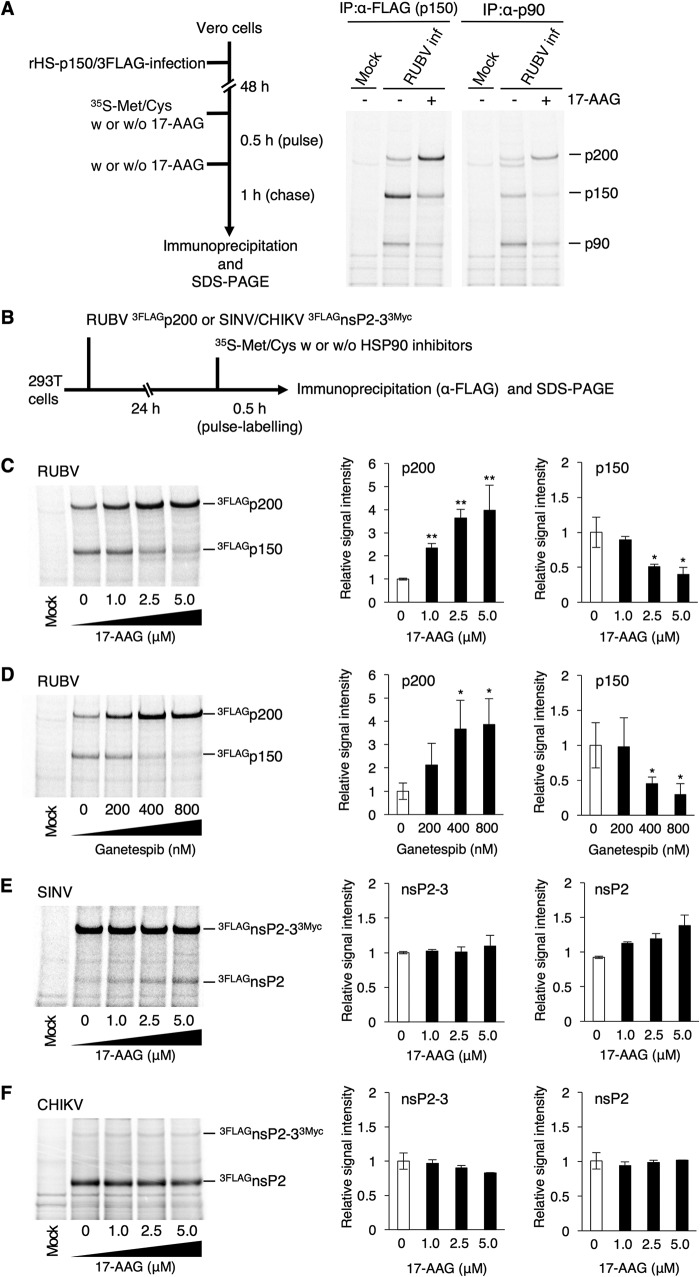
The physical interaction between HSP90 and nonstructural proteins (NSPs) of RUBV or the nsPs of SINV and CHIKV was analyzed by coimmunoprecipitation assays (co-IPAs) using 293T cells, as previously reported (20). The SINV and CHIKV nsP polyprotein nsP1-4 is sequentially cleaved by its own protease activity in the nsP2 region into nsP1, nsP2, nsP3, and nsP4 (34, 35) (Fig. 3A). The RUBV NSP polyprotein p200 is cleaved by its own protease activity into p150 and p90 (9) (Fig. 3B). The co-IPA showed that HSP90 was coimmunoprecipitated with the nsP4 proteins of SINV and CHIKV (Fig. 3C and E). The nsP4 proteins of SINV and CHIKV were also coimmunoprecipitated with HSP90, confirming the interaction of HSP90 with the nsP4 proteins of SINV and CHIKV (Fig. 3D and F) (18, 20). Under our experimental conditions, an interaction of HSP90α with nsP1, nsP2, or nsP3 of SINV or CHIKV was not detected (Fig. 3C and E), although previous studies showed an interaction between HSP90 and CHIKV nsP2 and nsP3 (19, 20). For RUBV, p150 and p200C1152G, a protease-deficient mutant of p200, coimmunoprecipitated with HSP90α, whereas p90 did not (Fig. 3G). Coprecipitation of HSP90 with p150 and p200C1152G was also confirmed (Fig. 3H). Experiments using expression plasmids showed that the levels of p150 protein were reduced by 17-AAG treatment in a dose-dependent manner, whereas the levels of p90 and p200C1152G remained unchanged (Fig. 4A). Under these concentrations of 17-AAG, cell viability was not reduced (Fig. 4B). To assess the change in stability of p150 following treatment with HSP90 inhibitors, a pulse-chase experiment was performed. The p150 protein was rapidly decayed in the presence of 5.0 μM 17-AAG or 800 nM ganetespib (Fig. 4D and E). The p150 signal was decreased by ∼92% after 6 h of treatment with 17-AAG, compared with 62% without 17-AAG treatment (Fig. 4D). A similar result was obtained following treatment with 800 nM ganetespib (Fig. 4E). The p150 signal was decreased by ∼87% after 6 h of treatment with ganetespib, compared with 68% without ganetespib treatment (Fig. 4E). The change in the stability of SINV and CHIKV nsP4 with 17-AAG treatment was also assessed. The nsP4 signals of both SINV and CHIKV were decreased by ∼95% after 6 h of 17-AAG treatment, compared with ∼72% and ∼65% for the nsP4 signals of SINV and CHIKV, respectively, without 17-AAG treatment (Fig. 4F and G). Under these concentrations of 17-AAG and ganetespib treatment, cell viability was not affected (Fig. 4B and H). Collectively, these data demonstrate that RUBV p150 and nsP4 of CHIKV and SINV are HSP90 clients, because HSP90 physically interacts with these viral proteins and contributes to their stability.
FIG 3.
HSP90 interacts with RUBV p150. (A) A schematic diagram of SINV and CHIKV nsPs. nsP1, nsP2, nsP3, and nsP4 were processed from the precursor polyprotein, nsP1-4, by the viral protease (Pro), located in the nsP2 region. (B) A schematic diagram of RUBV NSPs. p150 and p90 were processed from the precursor polyprotein, p200, by viral protease, located in the p150 region. (C and D) Co-IPA of FLAGHSP90α with SINV 3MycnsPs (C) and co-IPA of SINV 3MycnsP4 with FLAGHSP90α (D). (C) 293T cells were cotransfected with plasmids expressing FLAG-tagged HSP90α (FLAGHSP90α) and Myc-tagged SINV nsPs (SINV 3MycnsPs). SINV 3MycnsPs were immunoprecipitated with anti-Myc MAb. Proteins in the total cell lysate (input) and in the immunoprecipitated (IP) complexes were detected by immunoblotting using anti-FLAG, anti-Myc, and anti-GAPDH MAbs. (D) For immunoprecipitation with FLAGHSP90α, 293T cells were cotransfected with plasmids expressing FLAGHSP90α and SINV 3MycnsP4. FLAGHSP90α was immunoprecipitated with an anti-FLAG MAb. Proteins in the total cell lysate (input) and in the immunoprecipitated complexes were detected by immunoblotting using anti-FLAG, anti-Myc, and anti-GAPDH MAbs. (E and F) Co-IPA of FLAGHSP90α with CHIKV 3MycnsPs (E) and co-IPA of CHIKV 3MycnsP4 with FLAGHSP90α (F). These co-IPA experiments were performed as described in the legend to panels C and D using CHIKV 3MycnsPs instead of SINV 3MycnsPs. (G and H) Co-IPA of RUBV NSPs with FLAGHSP90α (G) and co-IPA of FLAGHSP90α with RUBV p200 or p150 (H). (G) 293T cells were cotransfected with plasmids expressing AG1-tagged NSP (AG1p200C1152G, AG1p150, or AG1p90) and FLAG-tagged FLAGHSP90α. FLAGHSP90α was immunoprecipitated with an anti-FLAG M2 affinity gel, and the proteins in the total cell lysate (input) and in the immunoprecipitated complexes were detected by immunoblotting using an anti-FLAG pAb, an anti-AG1 pAb, and an anti-GAPDH MAb. (H) For immunoprecipitation with AG1p200C1152G or AG1p150, 293T cells were transfected with plasmids expressing AG1-tagged NSP (AG1p200C1152G or AG1p150) and FLAGHSP90α. AG1p200C1152G or AG1p150 was immunoprecipitated with an anti-AG1 pAb. Proteins in the total cell lysate (input) and in the immunoprecipitated complexes were detected by immunoblotting using an anti-AG1 pAb, an anti-FLAG MAb, and an anti-GAPDH MAb. (C to H) Data are representative of those from three independent experiments.
FIG 4.
HSP90 contributes to RUBV p150 stability. (A) Reduced protein levels of p150 by 17-AAG. 293T cells were transfected with plasmids expressing FLAG-tagged NSPs (3FLAGp200C1152G, 3FLAGp150, or 3FLAGp90). After 24 h of incubation with various concentrations of 17-AAG, the protein amounts of FLAG-tagged NSPs in the cells were analyzed by immunoblotting using an anti-FLAG MAb. GAPDH was detected using an anti-GAPDH MAb as an internal control. (B) 293T cells were cultured for 48 h with the indicated concentrations of 17-AAG. After incubation, cell viability was measured by an ATP-based assay and expressed as a relative value. Error bars indicate standard deviations. (C) Experimental flow for the assays whose results are presented in panels D, E, F, and G. (D to G) Stability assay of RUBV p150 (D, E), SINV nsP4 (F), and CHIKV nsP4 (G) by pulse-chase experiments. 293T cells were transfected with plasmids expressing RUBV 3FLAGp150 (D, E), SINV 3MycnsP4 (F), or CHIKV 3MycnsP4 (G). After 24 h of incubation, the cells were pulse-labeled with [35S]methionine and cysteine (Met and Cys) for 30 min and incubated in the presence or absence of HSP90 inhibitors, 5.0 μM 17-AAG (D, F, G) or 800 nM ganetespib (E). The 17-AAG-treated cells were collected after 0, 2, 4, and 6 h of chasing, while the ganetespib-treated cells were collected after 0 and 6 h of chasing. The pulse-labeled viral proteins in the cell lysate were immunoprecipitated using anti-FLAG or anti-Myc MAbs, subjected to SDS-PAGE, and detected on a Typhoon FLA 7000 image analyzer. The bar graphs show the signal intensity of the viral proteins after 6 h of chasing with or without HSP90 inhibitors relative to the viral protein signals before chasing (0 h). (H) 293T cells were cultured for 6 h with the indicated concentrations of ganetespib. After incubation, cell viability was measured by an ATP-based assay and expressed as a relative value. (A, B, D to H) Data are representative of those from three independent experiments. (B, H) Mean values ± standard deviations (SD) for four wells are shown. (D to G) Mean values ± SD for three wells are shown. Significant differences were determined by two-tailed t tests. **, P < 0.01.
HSP90 is required for RUBV NSP processing.
The dynamics of RUBV NSPs in virus-infected cells were analyzed by a pulse-chase experiment. In this experiment, recombinant RUBV possessing FLAG-tagged p150 (rHS-p150/3FLAG) was used. At 48 h postinfection (p.i.), rHS-p150/3FLAG-infected cells were radiolabeled for 30 min and incubated for 1 h in the absence or presence of 2.5 μM 17-AAG (Fig. 5A). p150 and p90 were immunoprecipitated and detected by an anti-FLAG mouse monoclonal antibody (MAb) and p90-specific polyclonal antibody (pAb), respectively. The signals for both p150 and p90 were decreased by 17-AAG treatment (Fig. 5A). The decreased level of p150 could be attributed to the reduced stability of p150 (Fig. 4D and E). However, these changes were observed concomitantly with the increased levels of the p200 signals (Fig. 5A). Therefore, our interpretation of these data was that HSP90 activity is important for NSP processing. Both p90 and p150 were detected in these experiments when either p90-specific or FLAG-specific (p150-specifc) antibodies were used, because p90 and p150 interact with each other (36, 37). This hypothesis was also assessed by a radiolabeling experiment using a p200 expression plasmid. 293T cells were transfected with the p200 expression plasmid, and after 24 h of incubation, the cells were radiolabeled and incubated for 30 min in various concentrations of 17-AAG or ganetespib (Fig. 5B). The levels of p200 signals were increased by 17-AAG or ganetespib treatment, while the levels of p150 signals were inversely decreased (Fig. 5C and D). Similar experiments were performed for the nsPs of SINV and CHIKV using expression plasmids encoding nsP2-3, a polyprotein of nsP2 and nsP3 (Fig. 5E and F). The levels of unprocessed (nsP2-3) and processed (nsP2) signals of SINV and CHIKV were unchanged by 17-AAG treatment (Fig. 5E and F). These data show that HSP90 activity is required for RUBV NSP processing.
FIG 5.
HSP90 activity facilitates RUBV p200 processing. (A) A pulse-chase experiment in RUBV-infected cells. Vero cells were infected with rHS-p150/3FLAG, which comprises a p150 protein fused with a 3FLAG tag. After 48 h of incubation, the infected cells were pulse-labeled with [35S]Met and Cys with or without 2.5 μM 17-AAG for 30 min and incubated with or without 2.5 μM 17-AAG for 1 h. The pulse-labeled NSPs (p200, p150, and p90) in the cell lysates were immunoprecipitated with an anti-FLAG MAb or an anti-RUBV p90 pAb, subjected to SDS-PAGE, and detected on a Typhoon FLA 7000 image analyzer. (B) Processing efficiency of RUBV NSPs, SINV nsPs, and CHIKV nsPs, showing the experimental flow for the assays whose results are presented in panels C, D, E, and F. (C to F) 293T cells were transfected with plasmids expressing RUBV 3FLAGp200 (C, D), SINV 3FLAGnsP2-33Myc (E), or CHIKV 3FLAGnsP2-33Myc (F). After 24 h of incubation, the transfected cells were pulse-labeled with [35S]Met and Cys with various concentrations of 17-AAG (C, E, and F) or ganetespib (D) for 30 min and lysed. The RUBV 3FLAGp200, SINV 3FLAGnsP2-33Myc, and CHIKV 3FLAGnsP2-33Myc in the cell lysates were immunoprecipitated with an anti-FLAG MAb, subjected to SDS-PAGE, and detected on a Typhoon FLA 7000 image analyzer. (A, C to F) Data are representative of those from three independent experiments. (C, D) The left bar graphs show the relative signal intensity of 3FLAGp200. The right bar graphs show the relative signal intensity of 3FLAGp150. (E, F) The left bar graphs show the relative signal intensity of 3FLAGnsP2-33Myc. The right bar graphs show the relative signal intensity of 3FLAGnsP2. (C to F) The bar graphs show the signal intensities of viral proteins relative to the nontreatment signals. Mean values ± standard deviations for three wells are shown. Significant differences were determined by two-tailed t tests. *, P < 0.05; **, P < 0.01.
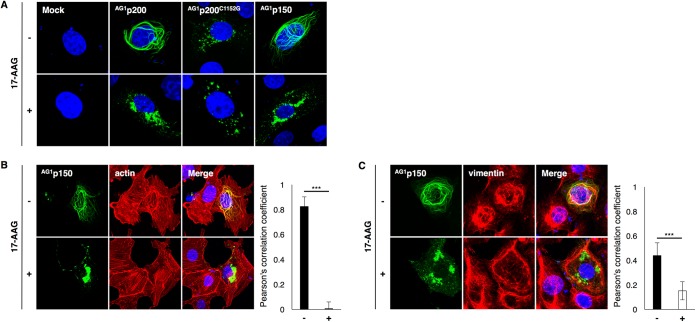
HSP90 is required for the filamentous distribution of RUBV p150.
p150 is distributed among filamentous structures in the cytoplasm (22, 38–40). When p200 was expressed in cells, filament formation was also detected (Fig. 6A), because p200 is processed into p150 and p90. The protease-deficient mutant of p200, p200C1152G, was distributed in dots in the cytoplasm but not in filamentous structures (Fig. 6A). When cells were treated with 2.5 μM 17-AAG, p150 no longer formed filamentous structures and aggregated in the cytoplasm (Fig. 6A). Without 17-AAG treatment, p150 strongly colocalized with actin fibers, forming a filamentous structure (Fig. 6B). This filamentous structure disappeared and p150 no longer colocalized with actin when cells were treated with 17-AAG (Fig. 6B). Vimentin intermediate filaments were partly colocalized with p150-forming filamentous structures (Fig. 6C). This colocalization was also decreased by 17-AAG treatment (Fig. 6C).
FIG 6.
Alteration of the intracellular distribution of RUBV p150 by inhibiting HSP90 activity. (A) Intracellular distribution of RUBV AG1p200, AG1p200C1152G, and AG1p150 in the presence or absence of 2.5 μM 17-AAG. Vero cells were transfected with plasmids expressing AG1-tagged NSPs (AG1p200, AG1p200C1152G, or AG1p150). After 24 h of incubation with or without 2.5 μM 17-AAG, the cells were fixed and the nuclei were counterstained with DAPI (blue). The cellular distributions of NSPs were analyzed by detecting the AG1 fluorescent signals under a confocal microscope. (Top) The intracellular distributions of NSPs in the absence of 2.5 μM 17-AAG; (bottom) the intracellular distributions of NSPs in the presence of 17-AAG. (B and C) Distribution of p150 with cytoskeleton proteins. (B) The actin (F actin) in AG1p150-expressing cells was stained with Alexa Fluor 594-conjugated phalloidin (red), and nuclei were counterstained with DAPI. (C) The intermediate filament vimentin in AG1p150-expressing cells was stained with an anti-vimentin MAb and an Alexa Fluor 594-conjugated secondary antibody (red), and nuclei were counterstained with DAPI. Pearson’s correlation coefficient values between p150 and cytoskeleton proteins were obtained by analyzing 10 cells in each sample. Error bars indicate standard deviations. Significant differences were determined by two-tailed t tests. ***, P < 0.001.
DISCUSSION
In the present study, we investigated the role of HSP90 in RUBV replication. Our data demonstrate that RUBV replicates poorly when HSP90 activity is inhibited by 17-AAG or ganetespib. We conclude that RUBV p150 is a novel client of HSP90, because HSP90 physically interacts with p150 and is required for its stability.
17-AAG inhibits the activity of Grp94 and of cytoplasmic HSP90s (41–44). Ganetespib also strongly inhibits the activity of cytoplasmic HSP90s; however, the inhibitory effect on Grp94 and TRAP1 is unknown, although its binding to all four isoforms of HSP90 has been demonstrated (12, 41, 45). In the present study, we focused only on the roles of cytoplasmic HSP90s. This may be a limitation of this study. However, our data clearly demonstrate that the cytoplasmic HSP90s play key roles in the RUBV life cycle, because the infectivity titers of RUBV were reduced by ∼100-fold following treatment with HSP90α and HSP90β siRNAs. Our data demonstrate that p150 is a viral client protein of HSP90. The RUBV p150 protein has a protease domain, which cleaves p200 into p150 and p90 in cis and trans (9, 46). Our data also demonstrate that the HSP90 inhibitors prevent p200 processing. Since p200 cleavage is critical for RUBV genome replication (46), this may be one of the main reasons why RUBV poorly produced infectious viruses when cells were treated with HSP90 inhibitors. There are two possible reasons for this miscleavage: one is the loss of the protease activities of p200 and p150 due to misfolding of the protease domain located in the p150 region, and the other is a structural change at the self-cleavage site of p200, such that it cannot be recognized by its own protease domain. For most of our experiments, N-terminus-tagged viral proteins and HSP90α were used. We cannot completely rule out the possibility that the tags and the additional N-terminus methionine influence the properties of the proteins. It is predicted that p150 additionally possesses methyltransferase activity (8). This activity might be diminished by inhibition of HSP90, although it remained undetermined in this study. In any case, nonfunctional p150 is the cause of NSP dysfunction, resulting in a reduction in genome replication.
Under normal conditions, RUBV p150 is distributed among filamentous structures, where cellular cytoskeletal actins and intermediate filaments are redistributed to form structures with p150 (38, 40). Only p150 and not either p200 or p90 can be distributed in these structures (39, 40). In this study, the filamentous distribution of p150 in p200-expressing cells disappeared following 17-AAG treatment. The distribution was not mediated by p200, since protease-deficient p200, p200C1152G, localized in dots or aggregates in the cytoplasm. These data suggest that HSP90 supports the filamentous distribution of p150 through p200 processing. The distribution of p150 was also inhibited by the treatment, even when p150 was solely expressed in cells, suggesting that HSP90 also contributes to the inherent intracellular localization of p150 cleaved from p200. The filamentous structures and cytoskeleton rearrangement with p150 were no longer observed following 17-AAG treatment. This may suggest a structural abnormality in p150 caused by misfolding. All our data suggest that without HSP90 activity, the RUBV p200 protein fails to undergo cleavage into p150 and p90. Even when it is cleaved, p150 is distributed aberrantly in cells and forced to undergo degradation or aggregation.
17-AAG treatment had a greater effect on SINV propagation than on RUBV propagation. In alphaviruses, nsP2, nsP3, and nsP4 of CHIKV and nsP4 of SINV interact with cytoplasmic HSP90s (18–20) and HSP90 activity is required for efficient CHIKV propagation in vitro and in vivo (20). The nsP4 proteins of both alphaviruses interacted with cytoplasmic HSP90s, although the significance of the HSP90s for nsP4 is unclear. Our data confirm that HSP90α interacts with nsP4 of CHIKV and SINV. Further, our data reveal that the stability of nsP4 is reduced by 17-AAG treatment, suggesting that HSP90 activity contributes to the folding of nsP4. SINV nsP4 has an RdRp motif and acts as a core component in the viral genome replication machinery (47–49). The translation efficiency of SINV nsP4 is lower than that of the other nsPs, because nsP4 is translated by readthrough of an opal termination codon at the end of nsP3 (50). Even when translated, nsP4 is degraded rapidly by the N-end rule pathway (51). The underexpression of nsP4 was probably accelerated by the inhibition of HSP90 activity, and it could be explained that 17-AAG treatment caused a severe reduction in the infectivity titers in the media of SINV-infected cells. The nsP4 protein is highly conserved in the genus Alphavirus (52), and thus, our findings and those of previous studies suggest that HSP90 participates in the folding and functionality of nsP4 in a variety of alphaviruses.
HSP90 possesses substrate specificity, and the specificity is determined by cochaperones which interact with HSP90 and client proteins (53, 54). In the case of protein kinase clients, the CDC37 cochaperone interacts with certain kinases, and the complexes are recruited into the HSP90 machinery via the interaction of CDC37 with HSP90 (55). In addition to client-recruiting roles, certain cochaperones regulate HSP90 chaperone activities by binding to a specific conformation of HSP90 (53, 56). Despite accumulating evidence for the association of HSP90 with various viral proteins, our understanding of cochaperones of viral proteins is limited (57–61). Further studies are needed to clarify the detailed molecular mechanisms by which cochaperones recruit viral proteins into the HSP90 machinery and regulate HSP90 activity to fold viral proteins.
Viral client proteins are conserved to some extent among related virus family members. P1 structural polyproteins, viral polymerase L proteins, and reverse transcriptase are clients of HSP90 in the families Picornaviridae (59), Paramyxoviridae (62–64), and Hepadnaviridae (65, 66), respectively. We expect that the RUBV p90 protein containing the RNA-dependent RNA polymerase (RdRp) domain is an HSP90 client, because previous studies have shown that HSP90 interacts with the SINV and CHIKV nsP4 proteins, which possess RdRp activity (18, 20). However, our data revealed that p150, but not p90 possessing an RdRp motif, is a client of HSP90 for RUBV. The alphavirus proteins which possess methyltransferase and protease domains are nsP1 and nsP2, respectively, but the interaction of HSP90 with nsP1 and nsP2 was not detected in our study. These findings that viral client proteins of HSP90 differ between RUBV and alphaviruses would be a good opportunity to discuss the new taxonomic categorization of RUBV relative to alphaviruses based on their genetic and molecular backgrounds.
MATERIALS AND METHODS
Chemical reagents and antibodies.
17-AAG and ganetespib were purchased from Focus Biomolecules (Plymouth Meeting, PA, USA) and MedChem Express USA (Monmouth Junction, NJ, USA), respectively. Anti-GAPDH (anti-glyceraldehyde-3-phosphate dehydrogenase; clone 3H12) and anti-Myc (My3) MAbs were purchased from MBL (Nagoya, Japan). Anti-HSP90α (Hyb-K41009), anti-HSP90β (K3701), anti-HSP90α/β (3H3C27), anti-DYKDDDDK (FLAG) (1E6), and anti-vimentin (V9) MAbs were purchased from StressMarq Bioscience (Victoria, Canada), Abcam (Cambridge, United Kingdom), BioLegend (San Diego, CA, USA), Fujifilm Wako Pure Chemical (Tokyo, Japan), and Sigma-Aldrich (St. Louis, MO, USA), respectively. Anti-Azami green 1 (anti-AG1) and anti-FLAG rabbit pAbs were purchased from MBL and Sigma-Aldrich, respectively. Anti-RUBV p150 pAb was a kind gift from T. Ahola (Institute of Biotechnology, University of Helsinki, Helsinki, Finland). Anti-RUBV p90 rabbit pAb was prepared as described previously (22).
Plasmids.
A genomic infectious cDNA clone of the RVi/Hiroshima.JPN/01.03[1J] strain, pHS-p150/AG1, was reported previously (22). pHS-p150/3FLAG was constructed by inserting a three-tandem-FLAG (3FLAG) epitope sequence at the region encoding the amino acid region between positions 717 and 718. Expression plasmids encoding either HS p200, p150, or p90 were constructed based on the pcDNA3.1+ vector (Invitrogen/Thermo Fisher Scientific, Waltham, MA, USA). The AG1 gene or 3FLAG sequence was added at the 5′ terminus of the HS p200 gene (AG1p200 and 3FLAGp200, respectively). A plasmid encoding AG1p150, 3FLAGp150, AG1p90, or 3FLAGp90 was constructed by replacing the HS p200 gene with the HS p150 or p90 gene. Expression plasmids encoding AG1p200C1152G and 3FLAGp200C1152G, which possess an inactivate protease domain, were constructed by introducing a C1152G point mutation into the protease catalytic dyad. Expression plasmids encoding the SINV and CHIKV nsP nsP1, nsP2, nsP3, or nsP4 or polyprotein nsP2-3, which consists of only nsP2 and nsP3, were constructed based on pcDNA3.1+. The cDNAs of SINV nsPs were obtained from a plasmid encoding the subgenomic replicon, pSinRep5 (67). The cDNAs of CHIKV were generated from the genomic RNA of an SL11131 strain as previously described (68). A three-tandem-Myc (3Myc) epitope sequence was added at the 5′ terminus of all SINV and CHIKV nsP genes (SINV 3MycnsP1, 3MycnsP2, 3MycnsP3, and 3MycnsP4 and CHIKV 3MycnsP1, 3MycnsP2, 3MycnsP3, and 3MycnsP4). The 3FLAG sequence was added at the 5′ terminus of SINV nsP2-3, and the 3Myc sequence was added at the 3′ terminus (SINV 3FLAGnsP2-33Myc). CHIKV 3FLAGnsP2-33Myc was constructed using the CHIKV nsP2-3 gene instead of the SINV nsP2-3 gene. An expression plasmid encoding HSP90α tagged with a FLAG epitope at the 5′ terminus (FLAGHSP90α) was constructed using the pcDNA3.1+ vector (Invitrogen/Thermo Fisher Scientific).
Cells and viruses.
Vero, A549, and 293T cells were purchased from the American Type Culture Collection (Manassas, VA, USA) and were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco/Thermo Fisher Scientific) containing 10% fetal bovine serum (FBS) and antibiotics (100 U/ml penicillin and 100 U/ml streptomycin). BHK cells (a preexisting stock) (69) were maintained in DMEM supplemented with 5% FBS and antibiotics. RK13 cells were obtained from the Kitasato Institute and were maintained in minimal essential medium (MEM; Nissui Pharmaceutical, Tokyo, Japan) supplemented with 8% bovine serum and antibiotics. The recombinant virus rHS-p150/AG1, expressing p150 tagged with AG1, was reported previously (22). The recombinant virus rHS-p150/3FLAG, expressing p150 tagged with the 3FLAG epitope, was recovered from the genomic infectious cDNA clone pHS-p150/3FLAG. The MM2215 strain of SINV has been reported previously (23, 24). RUBV and SINV were propagated in BHK cells.
Immunoblotting.
Cells were washed with phosphate-buffered saline (PBS) twice and then lysed with immunoprecipitation (IP) lysis buffer (125 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 5% glycerol, cOmplete protease inhibitor cocktail [Sigma-Aldrich]). The cell debris was removed by centrifugation at 15,000 × g for 5 min at 4°C. Aliquots of the cell lysate were boiled with sodium dodecyl sulfate (SDS) sample buffer for 5 min. Polypeptides in the samples were separated by SDS-polyacrylamide gel electrophoresis (PAGE) using 4% to 20% gradient SDS-PAGE gels (Fujifilm Wako Pure Chemical) and electroblotted onto polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA). After blocking with 5% skim milk in Tris-buffered saline containing 0.5% Tween 20 (TBS-T), the membranes were incubated overnight at 4°C with primary antibodies diluted in TBS-T containing 5% bovine serum albumin. The membranes were then washed three times with TBS-T and incubated with horseradish peroxidase-conjugated secondary antibody diluted with 5% skim milk in TBS-T for 2 h at room temperature. After being washed, the membranes were treated with Signal West Femto maximum-sensitivity substrate (Thermo Fisher Scientific), and the chemiluminescent signals on the membranes were detected with an LAS-3000 luminescent image analyzer (Fujifilm, Tokyo, Japan).
Titration of RUBV and SINV stocks.
The infectivity titers of RUBV in the stocks were determined by a plaque assay using RK13 cells, as described previously (70). Those of SINV were determined by a plaque assay using BHK cells, as reported previously (71). Their infectivity titers were expressed as the numbers of PFU.
Analysis of the effect of HSP90 activity on virus production and genome replication.
Vero cells were inoculated with rHS-p150/AG1 or SINV at an MOI of 1.0. After 1 h of incubation at room temperature, the cells were washed with serum-free DMEM twice and cultured with culture medium containing various concentrations of 17-AAG or ganetespib. rHS-p150/AG1-infected cells were cultured at 35°C for 72 h. SINV-infected cells were cultured at 37°C for 18 h. The infectivity titers of rHS-p150/AG1 were measured by a fluorescent focus-forming assay (FFA) as described previously (22). The infectivity titers were expressed as the number of fluorescent focus-forming units (FFUs). The infectivity titers of SINV were measured by a plaque assay as described above. For analysis of the effect of 17-AAG on virus genome replication, Vero cells were infected with RUBV or SINV at an MOI of 1.0 for 1 h at 35°C and 37°C, respectively. After washing with serum-free DMEM twice, the cells were cultured with medium containing 20 mM NH4Cl (DMEM-NH4Cl). NH4Cl was used to prevent secondary infection (28, 29, 33). For RUBV-infected cells, the culture media were replaced with fresh DMEM-NH4Cl in the absence or presence of 2.5 μM 17-AAG at 18 h p.i. Then, the cells were cultured for 30 h. For SINV-infected cells, the culture media were replaced with fresh DMEM-NH4Cl in the absence or presence of 2.5 μM 17-AAG at 4 h p.i. Then, the cells were cultured for 14 h. The total RNA was obtained from RUBV- or SINV-infected cells using a CellAmp Direct RNA preparation kit for reverse transcription (RT)-PCR (TaKaRa Bio, Shiga, Japan), according to the manufacturer’s instructions. The RT reaction was performed to generate the first-strand cDNA from the total RNA using a PrimeScript RT reagent kit (TaKaRa Bio) with a combination of hexamer and oligo(dT) primers. Quantitative PCR was performed using a LightCycler 480 Probe Master instrument (Roche Applied Science, Mannheim, Germany) together with Universal ProbeLibrary probes (Roche Applied Science) or the TaqMan probe (ABI/Thermo Fisher Scientific), in accordance with the manufacturers’ instructions. The copy numbers of the RUBV genomic RNA and SINV genomic/subgenomic RNAs were quantitated with a standard plasmid encoding the target sequence. The data were adjusted using the hypoxanthine phosphoribosyltransferase 1 (HPRT1) mRNA level in the sample as an internal control. The sets of primers and probes for RUBV, SINV, and HPRT1 were reported previously (22, 64, 72).
Analysis of the effects of HSP90 siRNA on RUBV production.
All siRNAs were purchased from Dharmacon (Cambridge, UK). A549 cells were transfected with siRNAs specific for HSP90α (catalog numbers D-005186-01, D-005186-02, D-005186-03, D-005186-05 for HSP90α siRNA numbers 1 to 4, respectively), and HSP90β (catalog numbers D-005187-01, D-005187-02, D-005187-05, and D-005187-18 for HSP90β siRNA numbers 1 to 4, respectively) or with control siRNAs (catalog number D-001210-01-05) using the Lipofectamine RNAi Max reagent (Invitrogen/Thermo Fisher Scientific). After 72 h of incubation at 37°C, the protein levels of HSP90 in the transfected cells were analyzed. The relative intensity of HSP90 was calculated using the signal intensity of GAPDH as an internal control. The transfected cells were infected with rHS-p150/AG1 at an MOI of 5 at 72 h posttransfection (p.t.). After a 1-h incubation at room temperature, the cells were washed with serum-free DMEM twice and then cultured with culture media for 48 h at 35°C. The infectivity titers in the media were measured by an FFA.
Co-IPA.
293T cells were transfected with the plasmid expressing the SINV 3MycnsP1, -P2, -P3, or -P4 protein or the CHIKV 3MycnsP1, -P2, -P3, or -P4 protein, together with the FLAGHSP90α expression plasmid, using branched polyethylenimine (PEI; Sigma-Aldrich). At 6 h p.t., the culture media were replaced with fresh media and the transfected cells were incubated for 24 h at 37°C. Aliquots (5.0% of the total volume) of the cell lysates were used for immunoblotting directly to show the amounts (input) of each protein in the cells. The remaining samples were then preincubated with protein G Sepharose (GE Healthcare, Buckinghamshire, United Kingdom) for 1 h at 4°C. The viral proteins in the samples were bound to an anti-Myc MAb overnight at 4°C. The immune complexes were precipitated with protein G Sepharose for 2 h at 4°C. After being washed with TBS-T five times, the complexes were dissolved from the protein G Sepharose by boiling with SDS sample buffer for 5 min. The FLAGHSP90α proteins that interacted with the viral proteins were then analyzed by immunoblotting using anti-Myc, anti-FLAG, and anti-GAPDH MAbs. 293T cells transfected with SINV or CHIKV 3MycnsP4 together with FLAGHSP90α were also subjected to the following experiments. After preparing the input sample, the remaining samples were preincubated with mouse IgG-agarose (Sigma-Aldrich) for 60 min at 4°C. The FLAGHSP90α protein in each sample was immunoprecipitated with an anti-FLAG M2 affinity gel (Sigma-Aldrich) for 2 h at 4°C. After being washed with TBS-T five times, the proteins bound to the anti-FLAG M2 affinity gel were dissolved by boiling the samples with SDS sample buffer for 5 min. The SINV and CHIKV 3MycnsP4 proteins that interacted with FLAGHSP90α were then analyzed by immunoblotting using anti-Myc, anti-FLAG, and anti-GAPDH MAbs. For analysis of the interaction of RUBV NSPs with HSP90α, 293T cells were transfected with plasmids expressing RUBV NSPs (AG1p200C1152G, AG1p150, or AG1p90) together with the FLAGHSP90α expression plasmid. At 6 h p.t., the culture media were replaced with fresh media and the transfected cells were incubated for 24 h at 37°C. The cells were washed with PBS once and lysed with IP lysis buffer. The cell debris was removed by centrifugation at 15,000 × g for 5 min at 4°C. Aliquots (5.0% of the total volume) of the cell lysates were used for immunoblotting directly to reveal the amounts (input) of each protein in the cells. The remaining samples were preincubated with mouse IgG-agarose (Sigma-Aldrich) for 60 min at 4°C. The FLAGHSP90α protein in each sample was immunoprecipitated with an anti-FLAG M2 affinity gel (Sigma-Aldrich) for 2 h at 4°C. After being washed with TBS-T five times, the proteins bound to the anti-FLAG M2 affinity gel were dissolved by boiling the samples with SDS sample buffer for 5 min. The viral proteins that interacted with FLAGHSP90α were then analyzed by immunoblotting using anti-AG1 and anti-FLAG pAbs and anti-GAPDH MAb. 293T cells transfected with FLAGHSP90α together with either AG1p200C1152G or AG1p150 were also subjected to the following experiments. After input sample preparation, the remaining samples were preincubated with protein G Sepharose (GE Healthcare) for 1 h at 4°C. The viral proteins in the samples were bound to an anti-AG1 pAb overnight at 4°C. The immune complexes were precipitated with protein G Sepharose for 2 h at 4°C. After being washed with TBS-T five times, the complexes were dissolved from protein G Sepharose by boiling with SDS sample buffer for 5 min. The AG1p200C1152G and AG1p150 that interacted with the FLAGHSP90α proteins were then analyzed by immunoblotting using anti-AG1 pAb and anti-FLAG and anti-GAPDH MAbs.
Analysis of the effect of HSP90 activity on the stability of RUBV NSPs and SINV and CHIKV nsP4s.
293T cells were transfected with plasmids expressing RUBV NSPs (3FLAGp200C1152G, 3FLAGp150, or 3FLAGp90) using PEI. At 6 h p.t., the culture media were replaced with fresh culture media containing various concentrations of 17-AAG, and the cells were cultured for 24 h. The proteins levels of RUBV NSPs were analyzed by immunoblotting using anti-FLAG and anti-GAPDH MAbs. For pulse-chase analysis of RUBV p150, 293T cells were transfected with the 3FLAGp150 expression plasmid. After a 24-h incubation period, the cells were preincubated with cysteine- and methionine-depleted DMEM (Gibco/Thermo Fisher Scientific) for 1 h and then pulse-labeled with [35S]methionine and cysteine using an EasyTag Express35S protein labeling mix (PerkinElmer, Waltham, MA, USA) for 30 min. The pulse-labeled cells were cultured with fresh culture media in the presence or absence of 5.0 μM 17-AAG or 800 nM ganetespib. For pulse-chase analysis of SINV or CHIKV nsP4, 293T cells were transfected with the SINV or CHIKV 3MycnsP4 expression plasmids. After a 24-h incubation period, the cells were preincubated with cysteine- and methionine-depleted DMEM (Gibco/Thermo Fisher Scientific) for 1 h and then pulse-labeled with [35S]methionine and cysteine using an EasyTag Express35S protein labeling mix (PerkinElmer) for 30 min. The pulse-labeled cells were cultured with fresh culture media in the presence or absence of 5.0 μM 17-AAG. Every 2 h, the cells were washed with PBS once and lysed with radioimmunoprecipitation assay buffer (Thermo Fisher Scientific) containing cOmplete protease inhibitor cocktail. The cell lysates were centrifuged at 15,000 × g for 1 h at 4°C, and the radiolabeled viral proteins in the supernatants were immunoprecipitated using anti-FLAG or anti-Myc MAbs and protein G Sepharose. The immunoprecipitated samples were subjected to SDS-PAGE, and the radiolabeled viral proteins in the samples were visualized using a Typhoon FLA 7000 image analyzer (GE Healthcare).
Analysis of the effects of HSP90 inhibitors on NSP/nsP processing.
For experiments involving viruses, Vero cells were infected with rHS-p150/3FLAG at an MOI of 10. At 48 h p.i., the culture media were replaced with cysteine- and methionine-depleted DMEM, and the cells were cultured for 1 h. Then, the cells were pulse-labeled with [35S]methionine and cysteine with or without 2.5 μM 17-AAG for 30 min and were cultured with fresh culture media and the same concentration of 17-AAG for 1 h. The pulse-labeled NSPs in the samples were precipitated with an anti-FLAG MAb, an anti-RUBV p90 pAb, and protein G Sepharose. The immunoprecipitated samples were subjected to SDS-PAGE and visualized as described above. When using expression plasmids, 293T cells were transfected with plasmids expressing RUBV 3FLAGp200, SINV 3FLAGnsP2-33Myc, or CHIKV 3FLAGnsP2-33Myc using PEI. At 24 h p.t., the cells were incubated in cysteine- and methionine-depleted DMEM for 1 h. The cells were then pulse-labeled with [35S]methionine and cysteine in various concentrations of 17-AAG or ganetespib for 30 min. The pulse-labeled NSPs/nsPs in the samples were precipitated with an anti-FLAG MAb and protein G Sepharose. The immunoprecipitated samples were subjected to SDS-PAGE and visualized as described above.
Cell viability assay.
The amounts of ATP in cells were measured as an indicator of cell viability using a CellTiter-Glo luminescent cell viability assay (Promega, Madison, WI, USA) and a Power Scan HT reader (BioTek, Winooski, VT, USA), according to the manufacturer’s instructions.
Indirect immunofluorescent assay.
Vero cells were transfected with the expression plasmid AG1p200C1152G, AG1p200, or AG1p150 using the TransIT-LT1 transfection reagent (Mirus, Madison, WI, USA). After 6 h p.t., the culture media were replaced with fresh media with or without 2.5 μM 17-AAG and the cells were cultured for 24 h. After the 24-h incubation period, the cells were fixed with 4% paraformaldehyde in PBS and permeabilized with 0.5% Triton X-100 in PBS. The F actin in the cells was stained with phalloidin conjugated with Alexa Fluor 594. The vimentin in the cells was immunostained with an anti-vimentin MAb and an anti-mouse immunoglobulin secondary antibody conjugated with Alexa Fluor 594 (Molecular Probes/Thermo Fisher Scientific). The nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI; Lonza Cologne GmbH, Walkersville, MD, USA). The stained cells were observed with an FV1000D spectral-type confocal laser-scanning microscope (Olympus, Tokyo, Japan). Pearson’s correlation coefficient values between p150 and F actin or vimentin were calculated using the Coloc2 plug-in from ImageJ software (National Institutes of Health, Bethesda, MD, USA).
ACKNOWLEDGMENTS
We thank M. Nagai for her technical assistance. We thank Kate Fox from the Edanz Group for editing a draft of the manuscript.
This work was supported by grants from the Japan Society for the Promotion of Science (Grant-in-Aid for Young Scientists [B] 16K19620) and from the Takeda Science Foundation.
REFERENCES
- 1.Hobman TC. 2013. Rubella virus, p 687–711. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 2.Chen R, Mukhopadhyay S, Merits A, Bolling B, Nasar F, Coffey L, Powers A, Weaver S, Smith D, Simmonds P, Siddell S. 2018. Create a new family Matonaviridae to include the genus Rubivirus, removed from the family Togaviridae. ICTV Taxonomy history: Rubivirus. https://talk.ictvonline.org//taxonomy/p/taxonomy-history?taxnode_id=201855114.
- 3.Kuhn RJ. 2013. Togaviridae, p 629–650. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 4.Dominguez G, Wang CY, Frey TK. 1990. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during togavirus evolution. Virology 177:225–238. doi: 10.1016/0042-6822(90)90476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gorbalenya AE, Koonin EV, Lai MM. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett 288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamer G, Argos P. 1984. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res 12:7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gros C, Wengler G. 1996. Identification of an RNA-stimulated NTPase in the predicted helicase sequence of the rubella virus nonstructural polyprotein. Virology 217:367–372. doi: 10.1006/viro.1996.0125. [DOI] [PubMed] [Google Scholar]
- 8.Rozanov MN, Koonin EV, Gorbalenya AE. 1992. Conservation of the putative methyltransferase domain: a hallmark of the ‘Sindbis-like’ supergroup of positive-strand RNA viruses. J Gen Virol 73:2129–2134. doi: 10.1099/0022-1317-73-8-2129. [DOI] [PubMed] [Google Scholar]
- 9.Chen JP, Strauss JH, Strauss EG, Frey TK. 1996. Characterization of the rubella virus nonstructural protease domain and its cleavage site. J Virol 70:4707–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen B, Piel WH, Gui L, Bruford E, Monteiro A. 2005. The HSP90 family of genes in the human genome: insights into their divergence and evolution. Genomics 86:627–637. doi: 10.1016/j.ygeno.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 11.Taipale M, Jarosz DF, Lindquist S. 2010. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol 11:515–528. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 12.Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. 2012. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith DF, Sullivan WP, Marion TN, Zaitsu K, Madden B, McCormick DJ, Toft DO. 1993. Identification of a 60-kilodalton stress-related protein, p60, which interacts with hsp90 and hsp70. Mol Cell Biol 13:869–876. doi: 10.1128/mcb.13.2.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jascur T, Brickner H, Salles-Passador I, Barbier V, El Khissiin A, Smith B, Fotedar R, Fotedar A. 2005. Regulation of p21(WAF1/CIP1) stability by WISp39, a Hsp90 binding TPR protein. Mol Cell 17:237–249. doi: 10.1016/j.molcel.2004.11.049. [DOI] [PubMed] [Google Scholar]
- 15.Pearl LH. 2005. Hsp90 and Cdc37—a chaperone cancer conspiracy. Curr Opin Genet Dev 15:55–61. doi: 10.1016/j.gde.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 16.McClellan AJ, Tam S, Kaganovich D, Frydman J. 2005. Protein quality control: chaperones culling corrupt conformations. Nat Cell Biol 7:736–741. doi: 10.1038/ncb0805-736. [DOI] [PubMed] [Google Scholar]
- 17.Geller R, Taguwa S, Frydman J. 2012. Broad action of Hsp90 as a host chaperone required for viral replication. Biochim Biophys Acta 1823:698–706. doi: 10.1016/j.bbamcr.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cristea IM, Rozjabek H, Molloy KR, Karki S, White LL, Rice CM, Rout MP, Chait BT, MacDonald MR. 2010. Host factors associated with the Sindbis virus RNA-dependent RNA polymerase: role for G3BP1 and G3BP2 in virus replication. J Virol 84:6720–6732. doi: 10.1128/JVI.01983-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Das I, Basantray I, Mamidi P, Nayak TK, Pratheek BM, Chattopadhyay S, Chattopadhyay S. 2014. Heat shock protein 90 positively regulates chikungunya virus replication by stabilizing viral non-structural protein nsP2 during infection. PLoS One 9:e100531. doi: 10.1371/journal.pone.0100531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rathore AP, Haystead T, Das PK, Merits A, Ng ML, Vasudevan SG. 2014. Chikungunya virus nsP3 & nsP4 interacts with HSP-90 to promote virus replication: HSP-90 inhibitors reduce CHIKV infection and inflammation in vivo. Antiviral Res 103:7–16. doi: 10.1016/j.antiviral.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 21.Karasawa S, Araki T, Yamamoto-Hino M, Miyawaki A. 2003. A green-emitting fluorescent protein from Galaxeidae coral and its monomeric version for use in fluorescent labeling. J Biol Chem 278:34167–34171. doi: 10.1074/jbc.M304063200. [DOI] [PubMed] [Google Scholar]
- 22.Sakata M, Otsuki N, Okamoto K, Anraku M, Nagai M, Takeda M, Mori Y. 2014. Short self-interacting N-terminal region of rubella virus capsid protein is essential for cooperative actions of capsid and nonstructural p150 proteins. J Virol 88:11187–11198. doi: 10.1128/JVI.01758-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otsuki N, Sakata M, Saito K, Okamoto K, Mori Y, Hanada K, Takeda M. 2017. Both sphingomyelin and cholesterol in the host cell membrane are essential for rubella virus entry. J Virol 92:e01130-17. doi: 10.1128/JVI.01130-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson K, Trent DW. 1985. Genetic and antigenic variations among geographical isolates of Sindbis virus. J Gen Virol 66:797–810. doi: 10.1099/0022-1317-66-4-797. [DOI] [PubMed] [Google Scholar]
- 25.Schulte TW, Neckers LM. 1998. The benzoquinone ansamycin 17-allylamino-17-demethoxygeldanamycin binds to HSP90 and shares important biologic activities with geldanamycin. Cancer Chemother Pharmacol 42:273–279. doi: 10.1007/s002800050817. [DOI] [PubMed] [Google Scholar]
- 26.Lin TY, Bear M, Du Z, Foley KP, Ying W, Barsoum J, London C. 2008. The novel HSP90 inhibitor STA-9090 exhibits activity against Kit-dependent and -independent malignant mast cell tumors. Exp Hematol 36:1266–1277. doi: 10.1016/j.exphem.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Z, Anderson R, Hobman TC. 2011. The capsid-binding nucleolar helicase DDX56 is important for infectivity of West Nile virus. J Virol 85:5571–5580. doi: 10.1128/JVI.01933-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petruzziello R, Orsi N, Macchia S, Rieti S, Frey TK, Mastromarino P. 1996. Pathway of rubella virus infectious entry into Vero cells. J Gen Virol 77:303–308. doi: 10.1099/0022-1317-77-2-303. [DOI] [PubMed] [Google Scholar]
- 29.Glomb-Reinmund S, Kielian M. 1998. The role of low pH and disulfide shuffling in the entry and fusion of Semliki Forest virus and Sindbis virus. Virology 248:372–381. doi: 10.1006/viro.1998.9275. [DOI] [PubMed] [Google Scholar]
- 30.Cong H, Jiang Y, Tien P. 2011. Identification of the myelin oligodendrocyte glycoprotein as a cellular receptor for rubella virus. J Virol 85:11038–11047. doi: 10.1128/JVI.05398-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose PP, Hanna SL, Spiridigliozzi A, Wannissorn N, Beiting DP, Ross SR, Hardy RW, Bambina SA, Heise MT, Cherry S. 2011. Natural resistance-associated macrophage protein is a cellular receptor for Sindbis virus in both insect and mammalian hosts. Cell Host Microbe 10:97–104. doi: 10.1016/j.chom.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dube M, Rey FA, Kielian M. 2014. Rubella virus: first calcium-requiring viral fusion protein. PLoS Pathog 10:e1004530. doi: 10.1371/journal.ppat.1004530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dube M, Etienne L, Fels M, Kielian M. 2016. Calcium-dependent rubella virus fusion occurs in early endosomes. J Virol 90:6303–6313. doi: 10.1128/JVI.00634-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Groot RJ, Hardy WR, Shirako Y, Strauss JH. 1990. Cleavage-site preferences of Sindbis virus polyproteins containing the non-structural proteinase. Evidence for temporal regulation of polyprotein processing in vivo. EMBO J 9:2631–2638. doi: 10.1002/j.1460-2075.1990.tb07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Solignat M, Gay B, Higgs S, Briant L, Devaux C. 2009. Replication cycle of chikungunya: a re-emerging arbovirus. Virology 393:183–197. doi: 10.1016/j.virol.2009.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews JD, Tzeng WP, Frey TK. 2012. Determinants in the maturation of rubella virus p200 replicase polyprotein precursor. J Virol 86:6457–6469. doi: 10.1128/JVI.06132-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forng RY, Frey TK. 1995. Identification of the rubella virus nonstructural proteins. Virology 206:843–853. doi: 10.1006/viro.1995.1007. [DOI] [PubMed] [Google Scholar]
- 38.Kujala P, Ahola T, Ehsani N, Auvinen P, Vihinen H, Kaariainen L. 1999. Intracellular distribution of rubella virus nonstructural protein P150. J Virol 73:7805–7811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthews JD, Tzeng WP, Frey TK. 2009. Determinants of subcellular localization of the rubella virus nonstructural replicase proteins. Virology 390:315–323. doi: 10.1016/j.virol.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 40.Matthews JD, Tzeng WP, Frey TK. 2010. Analysis of the function of cytoplasmic fibers formed by the rubella virus nonstructural replicase proteins. Virology 406:212–227. doi: 10.1016/j.virol.2010.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taldone T, Patel PD, Patel M, Patel HJ, Evans CE, Rodina A, Ochiana S, Shah SK, Uddin M, Gewirth D, Chiosis G. 2013. Experimental and structural testing module to analyze paralogue-specificity and affinity in the Hsp90 inhibitors series. J Med Chem 56:6803–6818. doi: 10.1021/jm400619b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang BH, Plescia J, Song HY, Meli M, Colombo G, Beebe K, Scroggins B, Neckers L, Altieri DC. 2009. Combinatorial drug design targeting multiple cancer signaling networks controlled by mitochondrial Hsp90. J Clin Invest 119:454–464. doi: 10.1172/JCI37613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu W, Mimnaugh EG, Kim JS, Trepel JB, Neckers LM. 2002. Hsp90, not Grp94, regulates the intracellular trafficking and stability of nascent ErbB2. Cell Stress Chaper 7:91–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu W, Mimnaugh E, Rosser MF, Nicchitta C, Marcu M, Yarden Y, Neckers L. 2001. Sensitivity of mature Erbb2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90. J Biol Chem 276:3702–3708. doi: 10.1074/jbc.M006864200. [DOI] [PubMed] [Google Scholar]
- 45.Ying W, Du Z, Sun L, Foley KP, Proia DA, Blackman RK, Zhou D, Inoue T, Tatsuta N, Sang J, Ye S, Acquaviva J, Ogawa LS, Wada Y, Barsoum J, Koya K. 2012. Ganetespib, a unique triazolone-containing Hsp90 inhibitor, exhibits potent antitumor activity and a superior safety profile for cancer therapy. Mol Cancer Ther 11:475–484. doi: 10.1158/1535-7163.MCT-11-0755. [DOI] [PubMed] [Google Scholar]
- 46.Liang Y, Gillam S. 2000. Mutational analysis of the rubella virus nonstructural polyprotein and its cleavage products in virus replication and RNA synthesis. J Virol 74:5133–5141. doi: 10.1128/jvi.74.11.5133-5141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hahn YS, Grakoui A, Rice CM, Strauss EG, Strauss JH. 1989. Mapping of RNA- temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J Virol 63:1194–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubach JK, Wasik BR, Rupp JC, Kuhn RJ, Hardy RW, Smith JL. 2009. Characterization of purified Sindbis virus nsP4 RNA-dependent RNA polymerase activity in vitro. Virology 384:201–208. doi: 10.1016/j.virol.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomar S, Hardy RW, Smith JL, Kuhn RJ. 2006. Catalytic core of alphavirus nonstructural protein nsP4 possesses terminal adenylyltransferase activity. J Virol 80:9962–9969. doi: 10.1128/JVI.01067-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strauss EG, Rice CM, Strauss JH. 1983. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc Natl Acad Sci U S A 80:5271–5275. doi: 10.1073/pnas.80.17.5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Groot RJ, Rumenapf T, Kuhn RJ, Strauss EG, Strauss JH. 1991. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc Natl Acad Sci U S A 88:8967–8971. doi: 10.1073/pnas.88.20.8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weston J, Villoing S, Bremont M, Castric J, Pfeffer M, Jewhurst V, McLoughlin M, Rodseth O, Christie KE, Koumans J, Todd D. 2002. Comparison of two aquatic alphaviruses, salmon pancreas disease virus and sleeping disease virus, by using genome sequence analysis, monoclonal reactivity, and cross-infection. J Virol 76:6155–6163. doi: 10.1128/jvi.76.12.6155-6163.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schopf FH, Biebl MM, Buchner J. 2017. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol 18:345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 54.Hartl FU, Bracher A, Hayer-Hartl M. 2011. Molecular chaperones in protein folding and proteostasis. Nature 475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 55.Keramisanou D, Aboalroub A, Zhang Z, Liu W, Marshall D, Diviney A, Larsen RW, Landgraf R, Gelis I. 2016. Molecular mechanism of protein kinase recognition and sorting by the Hsp90 kinome-specific cochaperone Cdc37. Mol Cell 62:260–271. doi: 10.1016/j.molcel.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahasrabudhe P, Rohrberg J, Biebl MM, Rutz DA, Buchner J. 2017. The plasticity of the Hsp90 co-chaperone system. Mol Cell 67:947–961.e945. doi: 10.1016/j.molcel.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 57.Okamoto T, Nishimura Y, Ichimura T, Suzuki K, Miyamura T, Suzuki T, Moriishi K, Matsuura Y. 2006. Hepatitis C virus RNA replication is regulated by FKBP8 and Hsp90. EMBO J 25:5015–5025. doi: 10.1038/sj.emboj.7601367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anderson I, Low JS, Weston S, Weinberger M, Zhyvoloup A, Labokha AA, Corazza G, Kitson RA, Moody CJ, Marcello A, Fassati A. 2014. Heat shock protein 90 controls HIV-1 reactivation from latency. Proc Natl Acad Sci U S A 111:E1528–E1537. doi: 10.1073/pnas.1320178111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Geller R, Vignuzzi M, Andino R, Frydman J. 2007. Evolutionary constraints on chaperone-mediated folding provide an antiviral approach refractory to development of drug resistance. Genes Dev 21:195–205. doi: 10.1101/gad.1505307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang X, Grammatikakis N, Hu J. 2002. Role of p50/CDC37 in hepadnavirus assembly and replication. J Biol Chem 277:24361–24367. doi: 10.1074/jbc.M202198200. [DOI] [PubMed] [Google Scholar]
- 61.Katoh H, Sekizuka T, Nakatsu Y, Nakagawa R, Nao N, Sakata M, Kato F, Kuroda M, Kidokoro M, Takeda M. 2019. The R2TP complex regulates paramyxovirus RNA synthesis. PLoS Pathog 15:e1007749. doi: 10.1371/journal.ppat.1007749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Connor JH, McKenzie MO, Parks GD, Lyles DS. 2007. Antiviral activity and RNA polymerase degradation following Hsp90 inhibition in a range of negative strand viruses. Virology 362:109–119. doi: 10.1016/j.virol.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bloyet LM, Welsch J, Enchery F, Mathieu C, de Breyne S, Horvat B, Grigorov B, Gerlier D. 2016. HSP90 chaperoning in addition to phosphoprotein required for folding but not for supporting enzymatic activities of measles and Nipah virus L polymerases. J Virol 90:6642–6656. doi: 10.1128/JVI.00602-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katoh H, Kubota T, Nakatsu Y, Tahara M, Kidokoro M, Takeda M. 2017. Heat shock protein 90 ensures efficient mumps virus replication by assisting with viral polymerase complex formation. J Virol 91:e02220-16. doi: 10.1128/JVI.02220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu J, Toft DO, Seeger C. 1997. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J 16:59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hu J, Flores D, Toft D, Wang X, Nguyen D. 2004. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J Virol 78:13122–13131. doi: 10.1128/JVI.78.23.13122-13131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bredenbeek PJ, Frolov I, Rice CM, Schlesinger S. 1993. Sindbis virus expression vectors: packaging of RNA replicons by using defective helper RNAs. J Virol 67:6439–6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lim CK, Nishibori T, Watanabe K, Ito M, Kotaki A, Tanaka K, Kurane I, Takasaki T. 2009. Chikungunya virus isolated from a returnee to Japan from Sri Lanka: isolation of two sub-strains with different characteristics. Am J Trop Med Hyg 81:865–868. doi: 10.4269/ajtmh.2009.09-0009. [DOI] [PubMed] [Google Scholar]
- 69.Umino Y. 1995. Improved potency assay of rubella vaccine: parameters for plaque formation. J Virol Methods 51:317–328. doi: 10.1016/0166-0934(94)00127-3. [DOI] [PubMed] [Google Scholar]
- 70.Sakata M, Komase K, Nakayama T. 2009. Histidine at position 1042 of the p150 region of a KRT live attenuated rubella vaccine strain is responsible for the temperature sensitivity. Vaccine 27:234–242. doi: 10.1016/j.vaccine.2008.10.049. [DOI] [PubMed] [Google Scholar]
- 71.Sokoloski KJ, Nease LM, May NA, Gebhart NN, Jones CE, Morrison TE, Hardy RW. 2017. Identification of interactions between Sindbis virus capsid protein and cytoplasmic vRNA as novel virulence determinants. PLoS Pathog 13:e1006473. doi: 10.1371/journal.ppat.1006473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sane J, Kurkela S, Levanov L, Nikkari S, Vaheri A, Vapalahti O. 2012. Development and evaluation of a real-time RT-PCR assay for Sindbis virus detection. J Virol Methods 179:185–188. doi: 10.1016/j.jviromet.2011.10.021. [DOI] [PubMed] [Google Scholar]