Abstract
Infections caused by extended-spectrum β-lactamases (ESBLs) producing bacteria, including Klebsiella pneumoniae have increasingly subjected to therapeutic limitations and patients with these infections are at high risk for treatment failure, long hospital stays, high health care costs, and high mortality. The aim of this study was to screen the prevalence of the blaTEM,blaCTX-M and blaSHV ESBL genes in K. pneumoniae strains isolated from nosocomial urinary tract infections (UTIs). During the March 2016 to December 2017, one hundred isolates of K. pneumoniae were collected from urine specimens of patients suffering from nosocomial UTI referred to Khatam Al-Anbia hospital in Shahrud, north-central Iran. All isolates were identified by standard bacteriological tests. The pattern of antibiotic susceptibility was determined according to the CLSI guidelines. The presence of the ESBLs was investigated using the double-disc synergy test (DDST). Polymerase chain reaction technique was used to detect the blaTEM, blaCTX-M and blaSHV genes in DDST positive isolates. Most isolates showed remarkable resistance to tested antibiotics with highest rate against nitrofurantoin (75%) and trimethoprim/sulfamethoxazole (65%). The imipenem was the most effective antibiotic against K. pneumoniae isolates. ESBL phenotype was detected in 50 (50%) of isolates. The prevalence of blaTEM, blaCTX-M and blaSHV genes among 50 ESBLs-positive isolates was 25 (50%), 15 (30%) and 35 (70%) respectively. The blaTEM and blaSHV genes were seen in 25 isolates (50%) simultaneously. The findings of this study indicated the 50% frequency rate of ESBL-producing K. pneumoniae in our geographic region. Since the treatment of infections caused by this bacterium is associated with many limitations, this high prevalence is a warning sign to adopt new control policies to prevent further spread of this microorganism.
Keywords: Microbiology, Extended spectrum beta-lactamases, ESBL, blaTEM, blaCTX-M, Klebsiella pneumoniae, Multidrug-resistant, Iran
1. Introduction
Urinary tract infection (UTI) is one of the most commonly diagnosed infections in both genders and across all age groups in the world [1]. Uropathogenic Escherichia coli (UPEC) and Klebsiella pneumoniae are classified among the most important microorganisms contributing UTI [2]. As a Gram-negative bacterium, K. pneumoniae is responsible for hospital-acquired UTI, septicemia, pneumonia, and soft tissue infections [3]. In recent decades, due to the excessive and improper use of antibiotics and the spread of organisms that produce extended-spectrum β-lactamases (ESBLs), the emergence of multidrug-resistant uropathogens such as K. pneumoniae has increasingly been raised [4].
Since first described in the 1980s, the ESBL-producing bacteria have been spreading throughout the world, and nowadays, they are frequently isolated from nosocomial and from community-acquired infections [5, 6]. ESBLs are bacterial enzymes have ability to cause resistance against various types of β-lactam antibiotics, including third-generation cephalosporins and monobactams, though inhibited by clavulanic acid. Meanwhile, they do not confer resistance against cephamycins and carbapenems [7]. So far, more than 400 ESBL enzymes have been identified, and most of them have evolved due to mutations in the active center of the classic plasmid β-lactamases, including TEM-1, TEM-2, and SHV-1, with over 150 members. The CTX-M enzymes are the second largest group of ESBLs [8, 9]. Until the late 1990s, the majority of ESBLs (mainly TEM and SHV type) were isolated from K. pneumoniae strains involved in nosocomial outbreaks [10].
Infections caused by ESBL-producing bacteria have increasingly subjected to therapeutic limitations, and patients with these infections are at high risk for treatment failure, long hospital stays, high health care costs, and high mortality [11, 12]. The clinical bacterial isolates all over the world vary in the presence of ESBL enzymes and their pattern is changing rapidly over time [13]. Currently, ESBL-producing Klebsiella species have been categorized as one of the six drug-resistant bacteria that need urgent new therapeutic compounds [14].
Owing to the serious problems in the treatment of ESBL-producing K. pneumoniae infections, identifying the most prevalent ESBL enzymes locally is of great importance for countries, to monitor the changing of antibiotic resistance patterns. These observations afford valuable information on ESBL epidemiology and could aid medical personnel in choosing the right and most effective treatment [14].
By virtue of the scarce data on the frequency rate of the most prevalent ESBL genes (blaTEM, blaCTX-M, and blaSHV) in UTIs-isolated K. pneumoniae strains in our region at this time, our study was conducted to screen the presence of these genes among nosocomial uropathogen isolates of K. pneumoniae in Shahrud City, north-central Iran. Furthermore, the multidrug-resistant (MDR) (resistant to three or more antimicrobial categories) K. pneumoniae isolates were determined according to the most frequently used definition [15].
2. Materials and methods
2.1. Ethical consideration
This cross-sectional study was conducted in accordance with the Declaration of Helsinki and written informed consent was obtained from all patients.
2.2. Bacterial isolates
The isolates that were used in this study belonged to different wards (internal, surgery, intensive care unit, emergency, etc.) of Khatam Al-Anbia hospital in Shahrud, north-central Iran, that were collected from urine samples of patients with nosocomial UTI during the March 2016 to December 2017. Totally, 100 isolates of K. pneumoniae were collected from nosocomial UTI. The patients with primary negative urine culture, whose urine culture test showed a positive result after 48–72 hours of hospitalization, were considered as a nosocomial UTI. The 100 K. pneumoniae strains were isolated from 58 females and 42 males, respectively. The age of the patients was between 5-79 years (average 39 years).
2.3. Sample collection
For isolation of the K. pneumoniae, a 10 ml of clean-catch midstream urine specimen was collected in a sterile container. The specimens were cultured on blood agar and MacConkey agar (Quelab, Canada) media. A volume of urine measured using the calibrated loop method was inoculated into blood agar medium for colony counting. Densities of growth of K. pneumoniae greater than or equal to 104 CFU/ml (colony forming units per milliliter) were interpreted as positive for urinary tract infection [16].
2.4. Identification of K. pneumoniae
All isolates were confirmed by standard bacteriological tests, which included Gram staining, lysine iron agar (LIA), triple sugar iron agar (TSI), SIM (sulfide-indole- motility), Simon citrate, MR-VP (Methyl Red-Voges Proskauer), and urea broth [17]. All media were purchased from Merck Co, Germany. The verified K. pneumoniae isolates were suspended in trypticase soy broth (TSB, Merck, Germany) with 20% (v/v) glycerol and frozen at -80 °C for further investigations.
2.5. Antimicrobial susceptibility testing (AST)
Antibiotic susceptibility testing of the isolates was carried out on Mueller-Hinton (MH) agar (Merck, Germany) by the Kirby-Bauer method, according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [18]. The antibiotics were as follows: amikacin (30μg), cefotaxime (30μg), ceftazidime (30 μg), ciprofloxacin (5μg), co-trimoxazole (25μg), gentamicin (10μg), imipenem (10μg), nitrofurantoin (30μg), nalidixic acid (30μg) cefepime (30μg), ceftriaxone (30μg) and norfloxacin (10μg) (Mast Co, Merseyside, UK). The MDR isolates were determined according to previous definition [15]. The Escherichia coli ATCC 25922 was used as a quality control strain.
2.6. Phenotypic detection of ESBL production
All isolates that were resistant to one of the third generation cephalosporins (ceftazidime, cefotaxime, and ceftriaxone) by AST were analyzed for ESBL production [18]. ESBL phenotypic detection was performed using a double-disc synergy test (DDST) standard on MH agar. Briefly, the bacteria with turbidity equivalent to 0.5 McFarland standards were swabbed on to MH agar plates, an amoxicillin/clavulanic acid (20/10μg) was placed in the center of the plate and ceftazidime (30μg), cefotaxime (30μg) and ceftriaxone (30μg) discs were placed 15 mm away from the central disc. The plates were incubated at 37 °C for up to 24 h. An increase of >5 mm inhibition zone for antibiotics around the amoxyclav disc compared to the cephalosporin discs alone was considered ESBL production. ESBL production was further confirmed by the phenotypic confirmatory test (PCT) by using both ceftotaxime/cefotaxime-clavulanic acid (30μg/10μg) and ceftazidime/ceftazidime-clavulanic acid (30μg/10μg) as described previously [9, 19]. K. pneumoniae ATCC 700603 was used as an ESBL-positive control.
2.7. Preparation of DNA template for polymerase chain reaction
Bacterial DNA templates were extracted by suspending some colonies of an overnight growth on Luria–Bertani agar (Merck, Germany) in 500 ml DNase- and RNase-free water. The suspension was boiled at 95 °C for 10 min in a dry bath incubator (Fisher Scientific, USA), then centrifuged at 14000 rpm for 10 min at 4 °C. Finally, 0.5 ml of the supernatant was used as DNA template for PCR [9]. The yielded DNA was stored at -20 °C for molecular screening.
2.8. Molecular characterization of ESBL genes
The phenotypic ESBLs-positive K. pneumoniae isolates were investigated for presence of blaSHV, blaCTX-M, and blaTEM ESBL genes by polemerase chain reaction (PCR). The primers used in this study are listed in Table 1. The PCR reactions were carried out in an Eppendorf thermal cycler (Germany) in a final volume of 25 μL containing 2.5 μL of 10X PCR buffer, 0.6 mg/μL MgCl2, 200 μM of deoxynucleotide triphosphates (dNTPs), 0.5 units of Taq polymerase, 10 pmol of each primer, and 5 μL of sample DNA. The PCR carried out in the following conditions: initial denaturation at 94 °C (5 min), followed by 35 cycles of denaturation at 95 °C (30 sec), different annealing temperatures (Table 1) (30 sec), and extension at 72 °C (30 sec), with final extension period of 72 °C (5 min). To determination of expected products, the PCR bands were separated by electrophoresis on 1% agarose gel stained with ethidium bromide and evaluated using an ultraviolet transilluminator (ENDURO™ UV, Labnet International, CA, USA). Positive gene controls prepared from the Pasteur Institute of Iran were used in each PCR run.
Table 1.
PCR primers used for detection of ESBL genes in K. pneumoniae isolates.
| Target Genes | Primer Sequence (5′ to 3′) | Amplicon Size, bp | Annealing Temperature, °C |
|---|---|---|---|
| blaCTX-M | F: CGCTTTGCGATGTGCAG | 550 | 60 |
| R: ACCGCGATATCGTTGGT | |||
| blaSHV | F:CGCCTGTGTATTATCTCCCTGTTAGCC | 843 | 62 |
| R:TTGCCAGTGCTCGATCAGCG | |||
| blaTEM | F: CATTTCCGTGTCGCCCTTATTC | 800 | 60 |
| R: CGTTCATCCATAGTTGCCTGAC |
2.9. Statistical analysis
The data analysis was done by using SPSSTM software, version 22.0 (IBM Corporation, Armonk, NY, USA). The results are presented as descriptive statistics in terms of relative frequency. Values were expressed as the percentages of the variables.
3. Results
3.1. Antibiotic resistance patterns
The results of AST of the 100 K. pneumoniae isolates against 12 antibiotics are shown in Table 2. Overall, from 100 K. pneumoniae isolates, 75 (75%) of them were resistant to nitrofurantoin, followed by 65 (65%) to trimethoprim/sulfamethoxazole and norfloxacin, 60 (60%) to ciprofloxacin, 55 (55%) to nalidixic acid and ceftazidime, 50 (50%) to cefepime and amikacin, 40 (40%) to ceftriaxone and cefotaxime, and 30 (30%) to gentamicin. The results revealed that imipenem was the most effective antibiotic against K. pneumoniae isolates with 80% susceptibility. Among the K. pneumoniae isolates studied, 45 (45%) were resistant to at least one of third-generation cephalosporins. In total, 60 (60%) isolates were resistant to third-generation cephalosporins. The antibiotic resistance rate in ESBL-positive isolates was as follows: nitrofurantoin (76%), nalidixic acid (50%), cefepime (58%), gentamicin (22%), ceftazidime (74%), cefotaxime (66%), ciprofloxacin (70%), cefteriaxon (64%), trimethoprim/sulfamethoxaz (78%), norfloxacin (62%), imipenem (16%), and amikacin (58%) (Table 2).
Table 2.
The antibiotic susceptibility testing results of 100 K. pneumoniae isolates based on CLSI 2015.
| Antibiotic | Total K. pneumoniae No. (%) |
ESBL-producing K. pneumoniae No. (%) |
||||
|---|---|---|---|---|---|---|
| Resistant | Intermediate | Sensitive | Resistant | Intermediate | Sensitive | |
| Nitrofurantoin | 75 (75%) | 5 (5%) | 20 (20%) | 38 (76%) | 2 (4%) | 10 (20%) |
| Nalidixic acid | 55 (55%) | 5 (5%) | 40 (40%) | 25 (50%) | 2 (4%) | 23 (46%) |
| Cefepime | 50 (50%) | 5 (5%) | 45 (45%) | 29 (58%) | 1 (2%) | 20 (40%) |
| Gentamicin | 30 (30%) | 10 (10%) | 60 (60%) | 11 (22%) | 3 (6%) | 36 (72%) |
| Ceftazidime | 55 (55%) | 5 (5%) | 40 (40%) | 37 (74%) | 3 (6%) | 10 (20%) |
| Cefotaxime | 40 (40%) | 15 (15%) | 45 (45%) | 33 (66%) | 8 (16%) | 9 (18%) |
| Ciprofloxacin | 60 (60%) | 10 (10%) | 30 (30%) | 35 (70%) | 4 (8%) | 11 (22%) |
| Cefteriaxon | 40 (40%) | 15 (15%) | 45 (45%) | 32 (64%) | 8 (16%) | 10 (20%) |
| Trimethoprim/Sulfamethoxazole | 65 (65%) | 5 (5%) | 30 (30%) | 39 (78%) | 0 (0%) | 11 (22%) |
| Norfloxacin | 65 (65%) | 10 (10%) | 25 (25%) | 31 (62%) | 4 (8%) | 15 (30%) |
| Imipenem | 15 (15%) | 5 (5%) | 80 (80%) | 8 (16%) | 0 (0%) | 42 (84%) |
| Amikacin | 50 (50%) | 10 (10%) | 40 (40%) | 29 (58%) | 6 (12%) | 15 (30%) |
3.2. MDR patterns
Regarding the result of AST, all 100 K. pneumoniae isolates were resistant to at least three antibiotics and all of the isolates (n = 100, 100%) were MDR with 19 different patterns (Table 3). The profile II (nitrofurantoin- nalidixic acid- cefepime- norfloxacin- ciprofloxacin) was the most predominant resistance pattern with 10% frequency.
Table 3.
Multidrug-resistance patterns of K. pneumoniae isolates.
| Resistance pattern | Phenotypic resistance | Resistant isolates N (%) |
|---|---|---|
| I | FM-NA-SXT | 5 (5%) |
| II | FM-NA-FEP-NOR-CIP | 10 (10%) |
| III | FM-NA-CAZ-NOR-CIP | 5 (5%) |
| IV | FM-NA-FEP-IPM-NOR-CIP | 5 (5%) |
| V | FM-NA-SXT-CAZ-FEP-CRO-NOR-CIP | 5 (5%) |
| VI | FM-NA-CAZ-FEP-CRO-GM-CTX-NOR-CIP | 5 (5%) |
| VII | FM-NA-SXT-CRO-GM-CIP-CTX-NOR -AN | 5 (5%) |
| VIII | FM-SXT-CTX-AN | 5 (5%) |
| IX | FM-SXT-CAZ-NOR-CIP-AN | 5 (5%) |
| X | FM-SXT- CAZ-CRO-GM-CIP-CTX | 5 (5%) |
| XI | FM-SXT-CAZ-CRO-GM-CTX-AN | 5 (5%) |
| XII | FM-SXT-CAZ-CRO-GM-CTX-AN-NOR-CIP | 5 (5%) |
| XIII | FM-SXT-CAZ- CRO-CTX-AN -FEP- CIP | 5 (5%) |
| XIV | FM-IMP-NOR | 5 (5%) |
| XV | NA-CAZ-FEP-NOR | 5 (5%) |
| XVI | NA-SXT-IPM-NOR | 5 (5%) |
| XVII | NA-SXT-FEP-NOR-AN | 5 (5%) |
| XVIII | SXT-CAZ-FEP-NOR-CIP-AN | 5 (5%) |
| XIX | SXT-CAZ-FEP-CRO-GM-CIP-CTX-AN | 5 (5%) |
Abbreviations: FM, Nitrofurantoin; NA, Nalidixic acid; CTX, Cefotaxime; CRO, Ceftriaxone; CAZ, Ceftazidime; IPM, Imipenem; CIP, Ciprofloxacin; NOR, Norfloxacin; SXT, Trimethoprim/Sulfamethoxazole; FEP, Cefepime; GEN, Gentamicin; AN, Amikacin.
3.3. Phenotypic results for ESBLs
Among the 60 isolates that were resistant to third-generation cephalosporins, ESBL production was detected in 45 and 50 isolates using DDST and PCT, respectively.
3.4. PCR investigation of ESBL genes
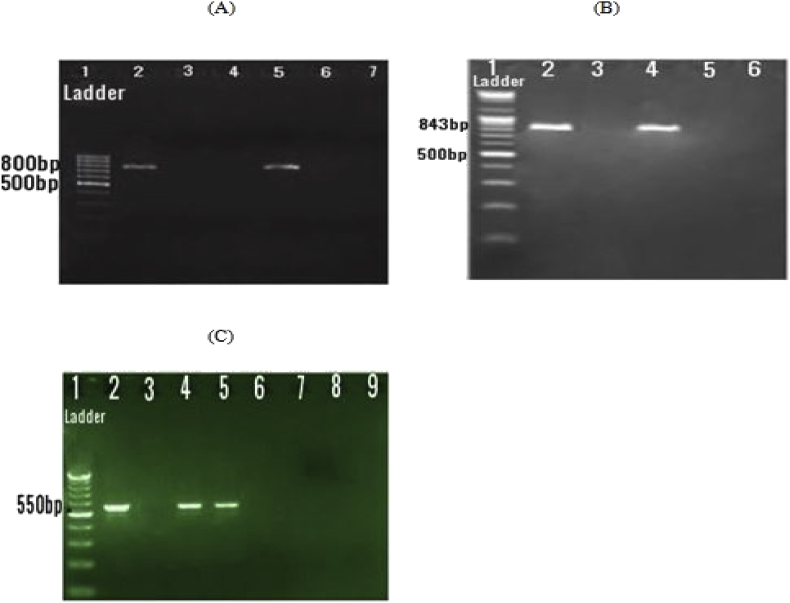
Based on the results of this study, among the phenotypic ESBLs-positive isolates, the PCR assay showed that 25 (50%), 15 (30%) and 35 (70%) of K. pneumoniae isolates were positive for blaTEM, blaCTX-M and blaSHV gene, respectively and 25 (50%) of isolates contained both blaTEM and blaSHV genes (Fig. 1 A,B,C). In the ESBLs-producing K. pneumoniae, 25 isolates (50%) harbored one type of ESBL genes, and 25 isolates (50%) carried two types of ESBL genes.
Fig. 1.
Gel electrophoresis of PCR assays of different ESBL genes. (A) Lane 1: 100 bp DNA marker; lane 2: positive control (blaTEM - 800bp); lane 3: negative control; lane 4,6, and 7: negative isolate; lane 5: positive isolate. (B) Lane 1: 100 bp DNA marker; lane 2: positive control (blaSHV - 843bp); lane 3: negative control; lane 5, and 6: negative isolate; lane 4: positive isolate. (C) Lane 1: 100 bp DNA marker; lane 2: positive control (blaCTX-M - 550bp); lane 3: negative control; lane 4, and 5: positive isolate; lane 6,7,8, and 9: negative isolate.
4. Discussion
This preliminary hospital-based study aimed to identify the ESBL-producing UTI-causing K. pneumoniae isolates and their antibiotic susceptibility pattern. Therefore, our study did not focus on concerns such as incidence and prevalence of other ESBL-UTI causing organisms in north-central Iran. Globally, UTIs afflict 150 million people every year [20]. Available data regarding the UTI-causing K. pneumoniae and its antimicrobial resistance profiles in specific geographical regions may assist physicians in selecting the best empirical antimicrobial therapy. Although K. pneumoniae is the second or third most etiologic agent of UTIs after E. coli, the former creates a problem for clinicians because of the multidrug resistance expressed by this pathogen [21]. The current investigation explored the resistance patterns of K. pneumoniae strains in patients suffered from UTIs in Shahrud, north-central Iran. Moreever, we showed a high resistance rate to antibiotics in patients with nosocomial urinary infections, except for imipenem with the susceptibility rate of 80%. Unlike our study, Latifpour et al. [22] in Sharekord, Iran, reported lower antibiotic resistance rate against norfloxacin (59%), trimethoprim/sulfamethoxazole (61%), and nitrofurantoin (55%). However, the resistance rate to nalidixic acid (72%) was higher, and the ciprofloxacin resistance was the same as the present survey. On the other hand, Najar Peerayeh et al. [23] displayed 30.7% and 51.1% resistance to gentamicin and ceftazidime, respectively, which was in line with our results. Furthermore, consistent with our investigation, they introduced the imipenem as the most effective antibiotic against K. pneumoniae. A previous study by Dallal et al. [24], suggested higher resistance for nalidixic acid and ceftazidime antibiotics [24]. The resistance rate of our isolates against third-generation cephalosporin groups, ceftriaxone and cefotaxime, were 40%. However, some other studies demonstrated a higher resistance rate for cephalosporin antibiotics [24, 25, 26]. The occurrence of MDR K. pneumoniae is a worldwide concern, and many types of research with diverse results have been performed in the recent decade [27].
The current study showed the occurrence of 100% for MDR K. pneumoniae strains, but in other studies conducted in Iran, the frequency of these strains was reported to be 74% and 46.6%, which is less than our results [28, 29]. The possible explanation for this higher rate of MDR characteristic, mainly in developing countries, could be irregular antibiotic prescription, sampling biases, genetic variations, geographic differences, social behaviors, and dissimilar patients' characteristics [28].
More than 75% of the studies have reported ESBL-producing infections with K. pneumoniae [30]. Our findings revealed that 50% of the K. pneumoniae isolates were ESBL producers. ESBL producing rate in this study was higher than those reported in the United States (12%) and Europe (33%) [31]. Conversely, higher prevalence rates of ESBL-producing K. pneumoniae isolates were identified in Tehran (69.7%), and Zahedan (66.7%), cities of Iran [29]. In the present study the PCT method detected more ESBL-positive isolates than DDST. DDST found 45% of ESBL producers K. pneumoniae, whereas PCT identified 50%. The results of the PCR assay in this study demonstrated the blaSHV as the most prevalent ESBL gene, a finding which was similar to Khosravi et al. [32] study in Ahvaz, Iran. The prevalence of blaSHV gene in clinical isolates of K. pneumoniae has differently been reported in some previous studies from Iran, which ranged from 43.1% to 67.4% [32, 33, 34, 35].
Nowadays, there are reports presenting the prevalence of other ESBL enzymes in different countries [9, 19, 31]. The co-existence of varied ESBL genes within the same isolate, as detected in this study, has also been reported in other regions of Iran and other countries [9, 29, 32, 36]. Furthermore, most of the blaSHV ESBL positive strains in our study were detected to be blaTEM positive. This investigation has some limitations such as short period time of sample collection, small sample size, single hospital evaluation, and focusing only on blaCTX-M, blaTEM, and blaSHV ESBL genes. However, they could be the subject of ongoing studies.
5. Conclusions
Even with the above-mentioned limitations, this report offers an insight into the current prevalence and molecular types of ESBL-producing K. pneumoniae in Shahrud, north-central Iran, contributing to a better understanding of the epidemiology of these enzymes at local and national levels. In addition, to the best of our knowledge, we have presented the first report regarding the ESBL-producing K. pneumoniae in north-central Iran. The findings of this study indicated the 50% frequency rate of ESBL-producing K. pneumoniae in our geographic region. Since the treatment of infections caused by this bacterium is associated with many limitations, this high prevalence is a warning sign to adopt new control policies to prevent further spread of this microorganism. Our results emphasize the necessity for the adequate screening of ESBL-producing strains in our region.
Declarations
Author contribution statement
Sajjad Yazdansetad, Miaad K. Alkhudhairy: Conceived and designed the experiments; Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Reza Najafpour, Elika Farajtabrizi, Reham M. Al-Mosawi: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Morteza Saki, Elham Jafarzadeh, Farokh Izadpour, Atefeh Ameri: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
We would like to express our gratitude to the personnel of the Microbiology Laboratory of Golestan University of Medical Sciences, Gorgan, Iran, for their cooperation with this study.
References
- 1.Magliano E., Grazioli V., Deflorio L., Leuci A.I., Mattina R., Romano P. Gender and age-dependent etiology of community-acquired urinary tract infections. Sci. World J. 2012;2012:349597. doi: 10.1100/2012/349597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Y.S., Ku C.H., Lin J.C., Shang S.T., Chiu C.H., Yeh K.M. Impact of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae on the outcome of community onset bacteremic urinary tract infections. J. Microbiol. Immunol. Infect. 2010;43(3):194–199. doi: 10.1016/S1684-1182(10)60031-X. [DOI] [PubMed] [Google Scholar]
- 3.Guoa Y., Cenb Z., Zoub Y., Fanga X., Lia T., Wanga J. Whole-genome sequence of Klebsiella pneumoniae strain LCT-KP214. J. Bacteriol. 2012;194(12):3281. doi: 10.1128/JB.00531-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerceo E., Deitelzweig S.B., Sherman B.M., Amin A.N. Multidrug-resistant gram-negative bacterial infections in the hospital setting: overview, implications for clinical practice, and emerging treatment options. Microb. Drug Resist. 2016;22(5):412–431. doi: 10.1089/mdr.2015.0220. [DOI] [PubMed] [Google Scholar]
- 5.Pilmis B., Delory T., Groh M., Weiss E., Emirian A., Lecuyer H. Extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBL-PE) infections: are carbapenem alternatives achievable in daily practice? Int. J. Infect. Dis. 2015;39:62–67. doi: 10.1016/j.ijid.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 6.Padmini N., Ajilda A.A.K., Sivakumar N., Selvakumar G. Extended spectrum β-lactamase producing Escherichia coli and Klebsiella pneumoniae: critical tools for antibiotic resistance pattern. J. Basic Microbiol. 2017;57:460–470. doi: 10.1002/jobm.201700008. [DOI] [PubMed] [Google Scholar]
- 7.Rawat D., Nair D. Extended-spectrum β-lactamases in gram negative bacteria. J. Glob. Infect. Dis. 2010;2(3):263–274. doi: 10.4103/0974-777X.68531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rahman S., Ali T., Ali I., Khan N.A., Han B., Gao J. The growing genetic and functional diversity of extended-spectrum beta-lactamases. BioMed Res. Int. 2018;2018:9519718. doi: 10.1155/2018/9519718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barguigua A., El Otmani F., Talmi M., Bourjilat F., Haouzane F., Zerouali K. Characterization of extended-spectrum beta-lactamase producing Escherichia coli and Klebsiella pneumoniae isolates from the community in Morocco. J. Med. Microbiol. 2011;60(9):1344–1352. doi: 10.1099/jmm.0.032482-0. [DOI] [PubMed] [Google Scholar]
- 10.Mnif B., Harhour H., Jdidi J., Mahjoubi F., Genel N., Arlet G. Molecular epidemiology of extended-spectrum beta-lactamase-producing Escherichia coli in Tunisia and characterization of their virulence factors and plasmid addiction systems. BMC Microbiol. 2013;13:147. doi: 10.1186/1471-2180-13-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiratisin P., Apisarnthanarak A., Laesripa C., Saifon P. Molecular characterization and epidemiology of extended-spectrum beta-lactamase producing Escherichia coli and Klebsiella pneumoniae isolates causing health care-associated infection in Thailand, where the CTX-M family is endemic. Antimicrob. Agents Chemother. 2008;52(8):2818–2824. doi: 10.1128/AAC.00171-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benner K.W., Prabhakaran P., Lowros A.S. Epidemiology of infections due to extended-spectrum beta-lactamase producing bacteria in a pediatric intensive care unit. J. Pediatr. Pharmacol. Ther. 2014;19(2):83–90. doi: 10.5863/1551-6776-19.2.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alipourfard I., Nili N.Y. Antibiogram of extended-spectrum Beta-lactamase (ESBL) producing Escherichia coli and Klebsiella pneumoniae isolated from hospital samples. Bangladesh J. Med. Microbiol. 2010;04(01):32–36. [Google Scholar]
- 14.Ranjbar R., Memariani H., Sorouri R. Molecular epidemiology of extended-spectrum beta-lactamase-Producing Klebsiella pneumoniae strains isolated from children with urinary tract infections. Arch. Pediatr. Infect. 2017;5(2) [Google Scholar]
- 15.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 16.Irajian G., Moghadas A.J. Frequency of extended-spectrum beta lactamase positive and multidrug resistance pattern in Gram-negative urinary isolates, Semnan, Iran. Jundishapur J. Microbiol. 2010;3(3):107–113. [Google Scholar]
- 17.Mac Faddin J.F. third ed. Lippincott Williams & Wilkinson; Philadelphia: 1999. Biochemical Tests for Identification of Medical Bacteria. [Google Scholar]
- 18.Clinical and Laboratory Standards Institute . 2015. Performance Standards for Antimicrobial Susceptibility Testing. Twenty-Fifth informational supplement. Document M100-S25. [Google Scholar]
- 19.Kjerulf A., Hansen D.S., Sandvang D., Hansen F., Frimodt-Moller N. The prevalence of ESBL-producing E. coli and Klebsiella strains in the Copenhagen area of Denmark. Apmis. 2008;116(2):118–124. doi: 10.1111/j.1600-0463.2008.00777.x. [DOI] [PubMed] [Google Scholar]
- 20.Flores-Mireles A.L., Walker J.N., Caparon M., Hultgren S.J. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat. Rev. Microbiol. 2015;13(5):269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paczosa M.K., Mecsas J. Klebsiella pneumoniae: going on the offense with a strong defense. Microbiol. Mol. Biol. Rev. 2016;80(3):629–661. doi: 10.1128/MMBR.00078-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Latifpour M., Gholipour A., Damavandi M.S. Prevalence of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates in nosocomial and community-acquired urinary tract infections. Jundishapur J. Microbiol. 2016;9(3) doi: 10.5812/jjm.31179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Najar Peerayeh S., Rostami E., Eslami M., Ahangarzadeh Rezaee M. High frequency of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae and Escherichia coli isolates from male patients’ urine. Arch. Clin. Infect. Dis. 2016;11(2) [Google Scholar]
- 24.Dallal M.S., Sabbaghi A., Aghamirzaeie H.M., Lari A.R., Eshraghian M.R., Mehrabad J.F. Prevalence of AmpC and SHV beta-lactamases in clinical isolates of Escherichia coli from Tehran hospitals. Jundishapur J. Microbiol. 2013;6(2):176–180. [Google Scholar]
- 25.Feizabadi M.M., Mahamadi-Yeganeh S., Mirsalehian A., Mirafshar S.M., Mahboobi M., Nili F. Genetic characterization of ESBL producing strains of Klebsiella pneumoniae from Tehran hospitals. J. Infect. Dev. Ctries. 2010;4(10):609–615. doi: 10.3855/jidc.1059. [DOI] [PubMed] [Google Scholar]
- 26.Fernando M.M.P.S.C., Luke W.A.N.V., Miththinda J.K.N.D., Wickramasinghe R.D.S.S., Sebastiampillai B.S., Gunathilake M.P.M.L. Extended spectrum beta lactamase producing organisms causing urinary tract infections in Sri Lanka and their antibiotic susceptibility pattern -A hospital based cross sectional study. BMC Infect. Dis. 2017;17:138. doi: 10.1186/s12879-017-2250-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rezaee M.A., Sheikhalizadeh V., Hasani A. Detection of integrons among multi-drug resistant (MDR) Escherichia coli strains isolated from clinical specimens in Northern West of Iran. Braz. J. Microbiol. 2011;42(4):1308–1313. doi: 10.1590/S1517-838220110004000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moini A.S., Soltani B., Taghavi Ardakani A., Moravveji A., Erami M., Haji Rezaei M. Multidrug-resistant Escherichia coli and Klebsiella pneumoniae isolated from patients in Kashan, Iran. Jundishapur J. Microbiol. 2015;8(10) doi: 10.5812/jjm.27517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maleki N., Tahanasab Z., Mobasherizadeh S., Rezaei A., Faghri J. Prevalence of CTX-M and TEM β-lactamases in Klebsiella pneumoniae isolates from patients with urinary tract infection, Al-Zahra hospital, Isfahan, Iran. Adv. Biomed. Res. 2018;7:10. doi: 10.4103/abr.abr_17_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paterson D.L., Bonomo R.A. Extended-spectrum b-lactamases: a clinical update. Clin. Microbiol. Rev. 2005;18(4):657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gharrah M.M., El-Mahdy A.M., Barwa R.F. Association between virulence factors and extended Spectrum beta-lactamase producing Klebsiella pneumoniae compared to nonproducing isolates. Interdiscip. Perspect Infect. Dis. 2017;2017:7279830. doi: 10.1155/2017/7279830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khosravi A.D., Hoveizavi H., Mehdinejad M. Prevalence of Klebsiella pneumoniae encoding genes for Ctx-M-1, Tem-1 and Shv-1 extended-spectrum beta lactamases (ESBL) enzymes in clinical specimens. Jundishapur J. Microbiol. 2013;6(10) [Google Scholar]
- 33.Eftekhar F., Rastegar M., Golalipoor M., MansourSamaei N. Detection of extended spectrum b-lactamases in urinary isolates of Klebsiella pneumoniae in relation to blaSHV, blaTEM and blaCTX-M gene carriage. Iran. J. Public Health. 2012;41(3):127–132. [PMC free article] [PubMed] [Google Scholar]
- 34.Hashemi A. Detection of blaCTX-M, blaTEM, blaSHV genes in Klebsiella pneumoniae strains isolated from two haspitals of Tehran, Iran. Iran. J. Public Health. 2014;43(2):98. [Google Scholar]
- 35.Feizabadi M.M., Delfani S., Raji N., Majnooni A., Aligholi M., Shahcheraghi F. Distribution of blaTEM, blaSHV, blaCTX-M genes among clinical isolates of Klebsiella pneumoniae at Labbafinejad hospital, Tehran, Iran. Microb. Drug Resist. 2010;16(1):49–53. doi: 10.1089/mdr.2009.0096. [DOI] [PubMed] [Google Scholar]
- 36.Amin M., Sirous M., Javaherizadeh H., Motamedifar M., Saki M., Veisi H. Antibiotic resistance pattern and molecular characterization of extended-spectrum β-lactamase producing enteroaggregative Escherichia coli isolates in children from southwest Iran. Infect. Drug Resist. 2018;11:1097. doi: 10.2147/IDR.S167271. [DOI] [PMC free article] [PubMed] [Google Scholar]