Abstract
Many sensory systems use stochastic fate specification to increase their repertoire of neuronal types. How these stochastic decisions are coordinated with the development of their target post-synaptic neurons in processing centers is not understood. In the Drosophila visual system, two subtypes of the UV-sensitive R7 color photoreceptors called yR7 and pR7 are stochastically specified in the retina. In contrast, the target neurons of photoreceptors in the optic lobes are specified through a highly deterministic program. Here, we identify subtypes of the main postsynaptic target of R7, the Dm8 neurons, that are each specific to the different subtypes of R7s. We show that during development the different Dm8 subtypes are produced in excess by distinct neuronal progenitors, independently from R7 subtype specification. Following matching with their respective R7 target, supernumerary Dm8s are eliminated by apoptosis. We show that the two interacting cell adhesion molecules Dpr11, expressed in yR7s, and its partner DIPγ, expressed in yDm8s, are essential for the matching of the synaptic pair. Loss of either molecule leads to the death of yDm8s or their mis-pairing with the wrong pR7 subtype. We also show that competitive interactions between Dm8 subtypes regulate both cell survival and targeting. These mechanisms allow the qualitative and quantitative matching of R7 subtypes with their target in the brain and thus permit the stochastic choice made in R7 to propagate to the deterministically specified downstream circuit to support color vision.
Stochastic specification of neurons is a common features of many sensory systems (1). In the vertebrate olfactory system, it is used to increase the diversity of olfactory sensory neuron types to a repertoire of more than 1400 in mouse (2, 3). In humans and old world monkeys, the stochastic specification of cone cells is the basis of the retinal mosaic responsible for trichromatic color vision (4, 5). A neuron that relies on an initial stochastic decision must stabilize its choice to maintain the proper identity, and then inform its downstream target cells of its choice. The latter is of high importance for neurons as they need to connect to their proper targets to faithfully transmit information to processing centers. The mouse olfactory system offers the most dramatic illustration of this matching problem: The ~1,400 olfactory neuron subtypes are randomly distributed within domains of the olfactory epithelium (6), yet all olfactory neurons of the same subtype project to the exact same glomeruli of the olfactory bulb (7–9).
In the Drosophila retina, a similar stochastic mechanism is employed to ensure the random distribution of photoreceptors with different spectral sensitivity (10, 11). The Drosophila compound eye is composed of ~750 unit eyes called ommatidia, each composed of 8 photoreceptors of two main categories: the six outer photoreceptors R1-6 express the broad spectrum light sensing Rhodopsin 1 (Rh1) and are important for motion and dim-light vision, analogous to vertebrate rods (Figure 1A; reviewed in (12)). The two inner photoreceptors R7 and R8 are responsible for color vision, similar to vertebrate cones. Ommatidia can be classified into different subtypes based on the Rhodopsins with different spectral sensitivity expressed by R7 and R8. The main part of the retina is occupied by two types of ommatidia that are randomly distributed and stochastically specified (Figure 1A). In the yellow (y) type that represents 65% of ommatidia, R7 expresses the UV-sensitive Rh4 whereas the R8 located below R7, and thus seeing the same point in space, always expresses the green-sensitive Rh6. In the remaining 35% of ommatidia of the pale (p) subtype, R7 expresses the shorter UV-sensitive Rh3 and R8 the blue-sensitive Rh5. A third type of ommatidia called Dorsal Rim Area (DRA) is localized in the most dorsal row of ommatidia (13). In this subtype, both R7 and R8 express Rh3 and are responsible for detecting the e-vector of polarized light used for navigation (14).
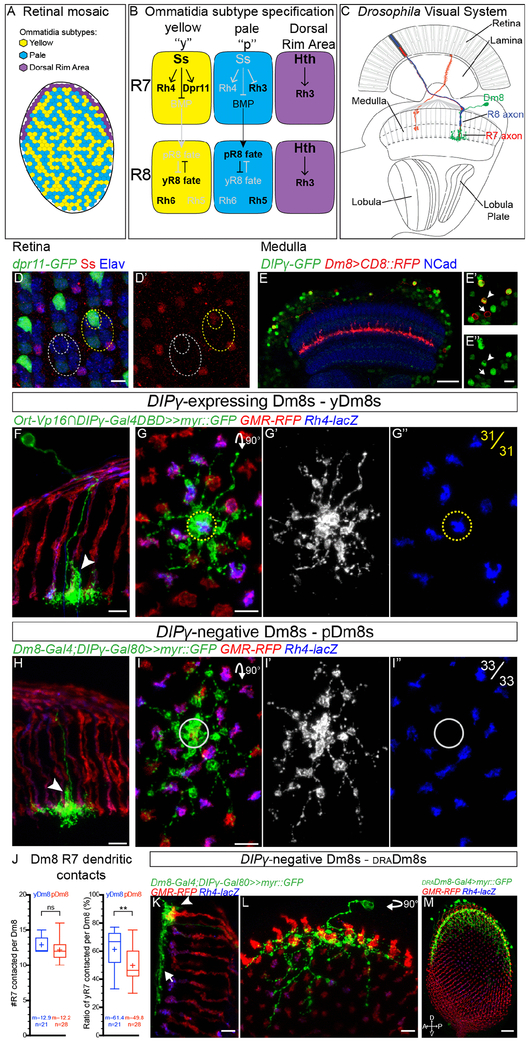
Fig. 1. Identification of three Dm8 subtypes corresponding to the three R7 subtypes:
(A) Schematic representation of the three different subtypes of ommatidia. (B) Regulatory network controlling R7 and R8 fate specification. (C) Schematic of the Drosophila visual system with R7 axons and their postsynaptic target Dm8 neurons in the medulla. (D) Dpr11MI02231 gene-trap expression in retina photoreceptors (Elav, blue) at 25 hours After Puparium Formation (APF). Dpr11-GFP (green) is strongly expressed in yR7, labelled by Ss (red, outline in yellow circles) but absent from pR7 (grey circles). (E) DIPγMI03222 gene-trap drives expression of GFP (green) in the adult medulla (Neuropile labelled using NCad, blue). A subset of Dm8s (labelled by CD8::RFP, in red) expresses DIPγ (DIPγ-expressing Dm8s, arrowhead, DIPγ-negative Dm8s, arrow). (F-G) DIPγ-expressing Dm8s always contact a yR7 in their home column. (F) Dorsoventral view of DIPγ-expressing Dm8 sparsely labeled with myr::GFP (green) extending a single process to the M4 layer in its home column (arrowhead). (G) Proximodistal view of a DIPγ-expressing Dm8. The yellow circle represents the center of the Dm8 dendritic field where the home column is located. yDm8s have a yR7 axon terminal in their home column (31 out 31 clones). Photoreceptors are labeled with GMR-RFP (red) and yR7 with Rh4-lacZ (blue). (H-I) pDm8s don’t express DIPγ and always contact a pR7 in their home-column. (H) Dorsoventral view of DIPγ-negative Dm8 with its single process in its home column (arrowhead). (I) Proximodistal view of a pDm8. pDm8s have a pR7 in their home column (grey circle, 33 out of 33 clones). (J) Tukey boxplots representing the number of R7s contacted per yDm8s and pDm8s outside of their home column, and the percentage of these contacts being with yR7s. Edges of the box indicate the first and third quartiles and the line the median. Mean (m) is represented by a cross. Whiskers represent the highest and lowest data point within 1.5 IQR of the first or third quartile respectively. ns (non-significant), **p>0.005; Student’s t-test. (K-L) A second type of DIPγ-negative Dm8 only contacts draR7s. Unlike y and pDm8s, they do not have a well-defined home-column (arrowhead) and their lateral processes do not contact non-draR7s (arrow). (M) The R13E04-Gal4 line specifically labels draDm8s. Proximodistal view of the entire medulla with draDm8s labelled with myr::GFP (in green), photoreceptors with GMR-RFP (red) and yR7 axons by Rh4-lacZ (blue). Note that draDm8s projections are only located at the edges of the dorsal half the medulla where draR7s axons are. Scale bars: (D), (E’-I) and (K and L) 5 μm, (E and M) 20μm.
Most of the gene regulatory network controlling the establishment of the fly retinal mosaic has been uncovered (11, 15, 16) (Figure 1B). The stochastic fate decision is initially made by R7 and is controlled by the transcription factor Spineless (Ss): Ss is stochastically turned on in 65% of R7s that then adopt the yR7 fate (11, 17). Once this decision is made cell autonomously by R7, it is propagated to R8 in the same ommatidium so that R7 and R8 have coupled Rhodopsin expression. This is achieved through induction of the pR8 fate by pR7s through Activin and BMP signaling while the yR8 fate is specified by default (18, 19). R7 and R8 send their axons to the medulla, the second neuropile of the optic lobe where they make synapses with some of the ~40,000 neurons of more than 80 different cell types that compose the medulla (20, 21)(Figure 1C). The medulla is retinotopically organized in ~750 columns that correspond to the ~750 ommatidia as medulla neurons preserve their spatial relationship to the retina.
In contrast with the stochastic specification of photoreceptors, the different medulla neurons are formed following a highly stereotypic mode of development (reviewed in (22)). The medulla develops from a neuroepithelium called the Outer Proliferation Center (OPC) during late third instar larval stage and early pupation. A proneuronal wave sequentially converts single rows of neuroepithelial cells into neuroblasts, the Drosophila neural stem cells, until the entire OPC is consumed (23). The medulla is thus sequentially produced, similarly and concomitantly to the retina where single rows of ommatidia are sequentially added in the eye disc at the morphogenetic furrow (24–26). Once specified, medulla neuroblasts sequentially express a series of transcription factors that will command the fate of the neurons produced during each temporal window (27–29). Thus, over time, a single neuroblast is able to generate a wide repertoire of different neurons, including the entire repertoire of uni-columnar neurons that are found in each medulla column with a 1:1 stoichiometry with photoreceptors (30, 31).
Connecting photoreceptors with the correct postsynaptic partners is fundamental to ensure proper color vision. Here we investigate how the stochastic decision made by photoreceptors is propagated to the medulla to instruct the formation of yellow and pale columns in which R7 photoreceptors connect to their proper specific targets. We show that correct matching is achieved through the generation of supernumerary target neurons of each subtype. Neurons that fail to connect to their corresponding R7 photoreceptor are culled by apoptosis. Recognition of future synaptic partners is achieved using a pair of interacting cell adhesion molecules from the Dpr/DIP families expressed in R7 or their Dm8 targets (32). We argue that competition between Dm8 subtypes for the available R7s affects both their survival and their targeting. This mechanism of elimination of supernumerary neurons upon lack of interaction of cell adhesion molecules might be a general mechanism to ensure the quantitative and qualitative matching of synaptic pairs, and to relay the stochastic decisions of sensory neurons to deeper brain region.
Results
Identification of three Dm8 subtypes corresponding to the three R7 subtypes
We first sought to identify the specific target neurons of the distinct R7 subtypes and thus focused on R7s main postsynaptic partner, medulla neuron Dm8s (33, 34). It was shown that the cell adhesion molecule Dpr11 is specifically expressed in yR7 during pupal development and that one of the Dpr11-binding partners, DIPγ, is expressed in a subset of Dm8s (32) (Figure 1D and 1E). Since it was proposed that these molecules play a role in establishing synaptic specificity in the optic lobe (32, 35), we reasoned that the two types of Dm8 neurons that are distinguished by DIPγ expression could correspond to the two R7 subtypes with DIPγ+Dm8s being postsynaptic to Dpr11+yR7s.
To test this, we first developed tools to genetically label the different populations of Dm8s based on DIPγ expression. We took advantage of a MiMIC construct inserted in the first intron of DIPγ (36) (Figure S1A) that faithfully recapitulates DIPγ expression as confirmed by antibody stainings against DIPγ (Figure S1B). We swapped the GFP within the original MiMIC line with the Gal4 DNA binding domain to build a DIPγ split-Gal4 line (DIPγ-Gal4DBD) to label DIPγ-expressing Dm8s, and with Gal80 to generate a DIPγ-Gal80 to label DIPγ-negative Dm8s (Figure S1A). The combination of the DIPγ split-Gal4 line with a hemidriver for the histamine chloride channel ort (ort-C1-3-Vp16) that is expressed in neurons postsynaptic to photoreceptors (33), labels a large subset of Dm8 neurons (Figure S1D). In order to better characterize the DIPγ-expressing Dm8s, and particularly to look at their connectivity, we generated flip-out clones to sparsely label the Dm8 population marked by this split-Gal4 combination (Figure 1F and 1G). We confirmed that Dm8s neurons extend their dendrites in the M6 layer, where R7 projects, each contacting ∼14 columns (34, 37) (Figure 1G and 1J). At the center of their dendritic field, Dm8s extend a much more extensive dendritic branch in their ‘home column’ along the R7 axon, from the M6 to the M4 layer that contains most of their synapses with R7 (33, 38) (Figure 1F). Single cell clonal analysis revealed that DIPγ-expressing Dm8s always have a Rh4-expressing yR7 in their home column (n=31/31 Dm8s; Figure 1F and 1G) but their lateral dendrites contact either pR7s or yR7s (Figure 1G and 1J). Hereafter, we refer to DIPγ-expressing Dm8s as yellow Dm8s (yDm8s).
We next characterized DIPγ-negative Dm8s using two distinct Gal4 lines expressed in Dm8s in combination with DIPγ-Gal80 (Figure S1F), and observed two types of neurons. A first population of Dm8s was morphologically identical to yDm8s (Figure 1H, S1F). However, these neurons always had a pR7 in their home column (n=33/33 Dm8s; Figure 1I), but similarly to yDm8s, they contacted both p and yR7s outside their home column (Figure 1I and J). We will refer to these neurons as pale Dm8s (pDm8s). Although both p and yDm8s show a strict preference for the R7 subtype in their home column and contact on average the same number of R7s outside their home column (Figure 1J), the ratio of R7 subtypes contacted by their lateral dendrites was different: yDm8s connected to yR7 vs. pR7 with the same frequency as the distribution of these photoreceptors (Figure 1J, ratio of yR7 contacted=61.4% vs. yR7=65%) whereas pDm8s had a preference for pR7s (Figure 1J, ratio of pR7 contacted=51.2% vs. pR7=35%). Additionally, around 15% of Dm8s from both populations harbored two main processes and thus had two home columns (Figure S1H and S1J) that were always both occupied by their preferred R7 subtype.
To confirm the strict home column pairing of yDm8s with yR7s and pDm8s with pR7s that we observed in single cell clones, we looked at whole mount stainings of either population (Figure S1E and S1G). We never observed a yDm8 extending its main dendritic branch along a pR7 (Figure S1E and S1I, n=1046) or the reverse for pDm8s (Figure S1G and S1G, n=516). We also quantified the ratio of columns occupied by a Dm8 as a home column. 88% of yR7 columns were occupied by a yDm8 while 97% of pR7 columns were occupied by a pDm8 (Figure S1J). These numbers might be a lower estimate of Dm8s column coverage due to the Gal4 lines not being fully penetrant and not labeling all neurons of a given cell type (see below and (39)).
In addition to pDm8s, we also identified a second type of DIPγ-negative Dm8 that only innervated DRA photoreceptors (Figure 1K and 1L). These Dm8s had a distinct morphology from p and y Dm8s: they did not appear to have any distinctive home column, they only made tight contacts with draR7 termini and did not contact the M6 layer in the main part of the medulla innervated by pale and yellow R7s (Figure 1K and 1L). We also identified a Gal4 line that specifically labeled draDm8s, confirming that these neurons are genetically different from pDm8 neurons and that draDm8s are confined to the outer part of the dorsal half of the medulla where draR7 axons are located (Figure 1M). These neurons correspond to the newly identified Dm-DRA1 neurons that were shown to be postsynaptic to draR7s (40), and thus their connection to draR7 will not be investigated further.
Thus, we have identified three types of Dm8s, corresponding to the three different R7 subtypes. In the main part of the medulla, most columns are occupied by the main process of a single p or yDm8s with a perfect pairing of R7 and Dm8 subtypes. Thus, the topographic organization of R7 subtypes in the retina is propagated to the medulla and mirrored by the mosaic of Dm8 subtypes.
Dm8 subtypes are pre-specified and have distinct lineages
We next sought to identify the mechanisms that lead from the random patterning of photoreceptors to a deterministic output in the medulla, where most of the columns are occupied by a Dm8 with a perfect matching between R7 subtypes and their respective Dm8 subtypes.
Two alternative mechanisms could allow this matching: (i) R7 subtypes could directly coordinate their fate with their Dm8 subtypes by instructing naïve Dm8s during development to adopt the appropriate fate (p vs. y). This would be similar to the coordination between R7 and R8 fates where pR7s signal to R8s within the same ommatidium and instruct them to adopt the pR8 fate (18). (ii) Alternatively, distinct Dm8s subtypes could be specified independently of R7 subtypes, such that matching would occur during later stages in development.
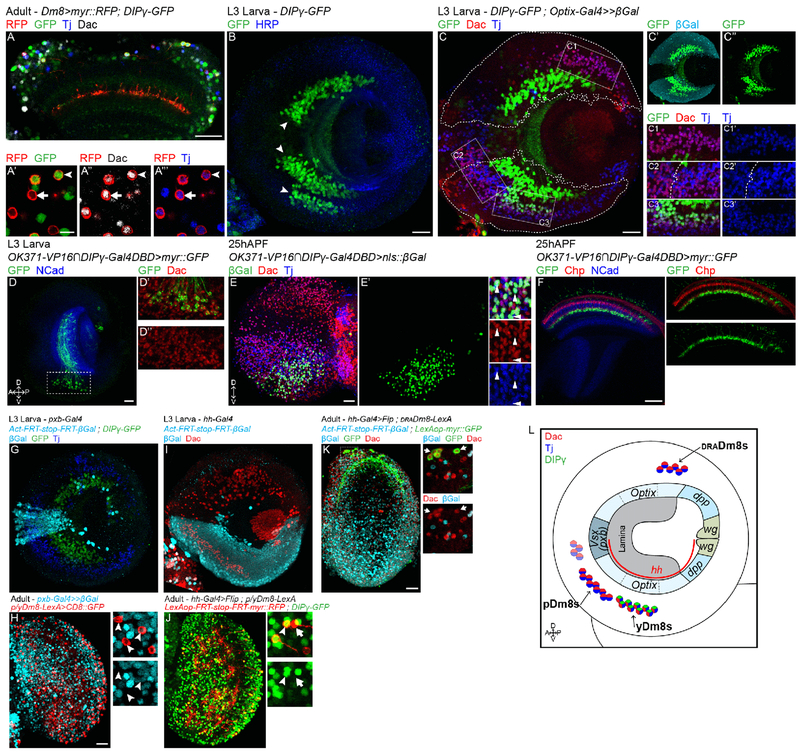
To distinguish between the two models, we sought to identify the origin of the distinct Dm8 subtypes and asked whether one single naïve, or distinct subtypes form during development, before R7 innervation. Since the Gal4 lines used to label Dm8 neurons in adult brains begin expression during late pupal development, we looked for markers expressed by adult Dm8 neurons that may also be expressed during early development. In adults, all three Dm8 subtypes express the transcription factors Dachshund (Dac) and Traffic jam (Tj) (Figure 2A, Figure S2A). We first focused on identifying yDm8s during development and asked when yDm8s adopt their final subtype fate. We looked at the early expression of DIPγ in late L3 larval optic lobes and identified several distinct clusters of cells expressing DIPγ (Figure 2B). One of these clusters also expressed Dac and Tj (Figure 2C) and could represent the yDm8 population. The identification of larval yDm8s based on the markers expressed in adult Dm8s assumes that their expression is maintained throughout development. To confirm that this was indeed the case we followed the Dac+Tj+DIPγ+ cell cluster from L3 until we could identify these neurons as yDm8s based on their morphology. To achieve this, we looked for a split-Gal4 line that would mark this cluster of cells throughout development. The combination of two split Gal4 lines, an enhancer trap for Vesicular glutamate transporter (OK371-VP16) and DIPγ-Gal4DBD specifically labeled the Dac+Tj+DIPγ+ cluster of cells in late L3 stage (Figure 2D). At 25h after puparium formation (APF), the split-Gal4 line remained specific for the same cluster of cells that expressed both Dac and Tj (Figure 2E) and could be identified as yDm8s based on their morphology (Figure 2F). Thus, in late L3 stage, when medulla neurons are just born, yDm8s have already acquired their final identity and express DIPγ.
Fig. 2. Dm8 subtypes are pre-specified and have distinct lineages:
(A) In adult, both p and yDm8s, labeled by myr::RFP (in red), and DIPγ-GFP only for yDm8s (in green), express Dac and Tj (in grey and blue respectively). (B) DIPγ-GFP expression in late L3 optic lobe. DIPγ is expressed in three clusters of neurons (arrowheads). HRP labels neurons membrane (in blue). (C) In L3 stage, only the smaller cluster labelled by DIPγ-GFP also express Tj and Dac (in blue and red). Optix lineage trace with nuclear β-Galactosidase (outlined in C and cyan in C’) revealed that this cluster is coming from the ventral part of the Optix domain (C and C3). Three other clusters of Tj+Dac+ neurons are found in the larval optic lobe: one in the most dorsal part of the Optix region (C1), one in the ventral half of the mOPC (left in C2) and one adjacent in the ventral Optix domain (right in C2). (D) The split-Gal4 line VGlut-VP16∩DIPγ-Gal4DBD specifically labels the DIPγ+Dac+ cell cluster in late L3 optic lobes (labeled with myr::GFP, in green). (E) At 25h APF, VGlut-VP16∩DIPγ-Gal4DBD is still specific to the same cluster of cells expressing Dac (in red) and Tj (in blue). (F) VGlut-VP16∩DIPγ-Gal4DBD driving myr::GFP shows that this cluster of cells are Dm8s based on morphology. Photoreceptors are labelled in red by Chaoptin, and the neuropile in blue by NCad. (G) pxb lineage trace in late L3 optic lobe labeled all mOPC derived neurons with nuclear β-Galactosidase (cyan). Note that none of the DIPγ-GFP neurons are labelled by β-Gal. (H) Same lineage tool in adult in combination with a R24F06-LexA driving myr::GFP in p/yDm8s (red). None of the p/yDm8s express β-Gal (arrowheads). (I) hh lineage trace in late L3 optic lobe labeled all neurons derived from the ventral half of the OPC with nuclear β-Galactosidase (cyan). (J) hh lineage tool in combination with DIPγ-GFP labels both p (arrow) and yDm8s (arrowhead). hh-Gal4 drives the expression of the Flip recombinase that will lead to the excision of a stop cassette within a LexAop-RFP reporter. Thus, only cells coming from the ventral hh+ region and expressing the p/yLexA driver will be labelled by RFP. (K) hh lineage trace in adult does not label draDm8s (lineage: β-Galactosidase in cyan, and draDm8s in green). (L) Schematic of the distinct lineages of the three Dm8 subtypes. Scale bars: (A-J) 20μm, (A’) 5μm.
We then looked for other Tj and Dac double positive cells in the developing larval optic lobe that could represent the other two Dm8 subtypes. We found four other large clusters of Dac+Tj+ neurons (Figure 2C). Unlike for yDm8s, we could not trace p and draDm8s from larva to adult because of the lack of a marker equivalent to DIPγ. Thus, in order to identify which cluster corresponded to which Dm8 subtypes, we used lineage trace experiments. We used the FLEXAMP memory cassette (41), a tool that when used in combination with a Gal4 line immortalizes GFP expression at a given time in development (Figure S2D) and thus allows identification of the neuron types based on their adult morphology. When using the tj-Gal4 line, which faithfully recapitulates tj expression in larvae (Figure S2C), in combination with DIPγ-Gal80, we consistently obtained clones of p and draDm8s among the four neuron types that also expressed Dac (Figure S2E).
The OPC is divided along the dorsoventral axis into compartments based on the expression of spatially restricted factors (30, 41): dpp, Optix and Vsx1 expression define the three major regions of the OPC, Optix is in the two arms of the main OPC (Figure 2C) and dpp in the two lateral parts of the OPC. The ventral half of the OPC can also be defined by its early expression of hedgehog (hh) (Figure 2I). We used lineage tools to identify the neuroepithelium compartments from which the different types of Dm8s originate.
We first used a lineage tool for the main OPC compartment using Optix-Gal4 (Figure 2C) and for the central OPC using pxb-Gal4 (Figure 2G). In larvae, the majority of Dac+Tj+ neurons came from the Optix region: two clusters were present in the ventral half, including the one expressing DIPγ. A third one was in the dorsal half (Figure 2C), whereas a smaller cluster was in the pxb region (Figure 2G). We used these lineage tools to trace y/p Dm8s and draDm8s in adults: all three subtypes were labelled by the Optix lineage tool (Figure S2F and S2G) whereas no Dm8s were labelled by the pxb lineage tool (Figure 2H and S2H). We then traced neurons coming from the ventral half of the OPC by using hh-Gal4 lineage trace (Figure 2I) to identify whether the two Optix-derived Tj+Dac+DIPγ− clusters were the p and draDm8s. pDm8s (and yDm8s) were labelled by the hh lineage trace, but none of the draDm8s were (Figure 2J and 2K). Therefore, pDm8s come from the ventral Optix cluster, next to yDm8s, whereas draDm8s are part of the dorsal Optix-derived cluster (Figure 2L). This shows that the three Dm8 subtypes come from three different neural progenitor domains and thus have distinct lineages (Figure 2L). The distinct fates of Dm8 subtypes are thus pre-established independently of the specification of their presynaptic R7 subtype.
Dm8 subtypes specification is coordinated with R7 stochastic specification
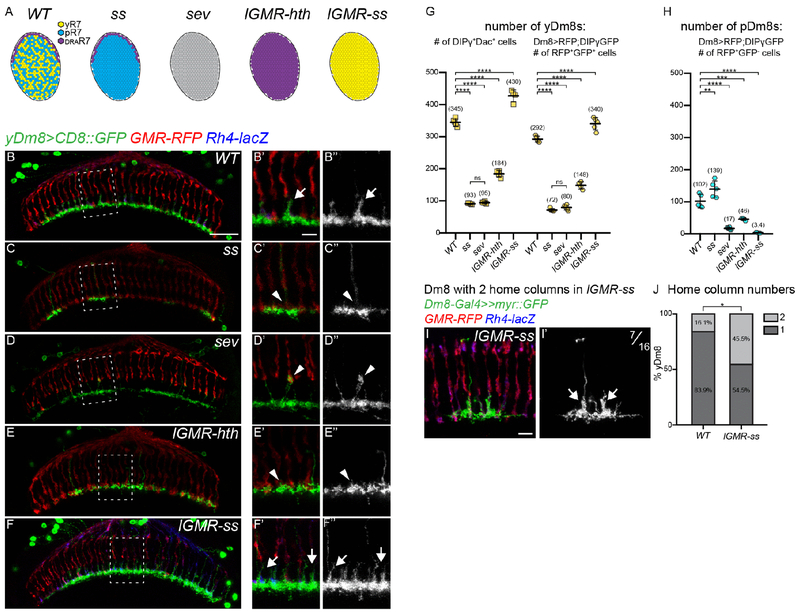
Since the Dm8 subtypes are specified independently of y and pR7, how can the brain accommodate stochastic changes in the ratio of photoreceptor subtypes to ensure that pDm8s always connect to pR7s and yDm8s to yR7s? To address this, we used mutations that affect the specification of the different ommatidial subtypes and looked at the consequence on the formation of Dm8s (Figure 3A). We first focused on yDm8s: In wild type, their arborizations covered almost entirely the M6 layer and their main branch that reached M4 in their home column could be easily identified (Figure 3B). We examined the effect of the absence of yR7s on yDm8s by using a retina-specific allele of spineless in which yR7s are not specified and the main part of the retina is solely composed of pR7s (Robert Johnston, personal communication, Figure 3A). In these ss mutants, large areas of the M6 layer were devoid of yDm8s, suggesting a dramatic decrease in the number of yDm8s (Figure 3C, see next paragraph for quantification). Furthermore, the yDm8s that were present lacked a home column as seen by the absence of the typical main Dm8 arbor reaching the M4 layer (Figure 3C). Similar effects, although varying in magnitude, could be seen in two other genetic backgrounds that lack yR7s: in sevenless (sev) mutants, where R7s are not specified (Figure 3A) there was also a decrease in the innervation of the M6 layer by yDm8s (Figure 3D). However, in contrast with ss mutants, some yDm8s appeared to still have a home column, although their main processes were thinner and reached higher in the medulla to layer M3 where they wrapped around R8 termini (Figure 3D). This might be due to the complete absence of R7s in sev mutants. We also converted the entire retina into DRA ommatidia using homothorax gain-of-function (13) (lGMR-hth, Figure 3A and 3E). In this case we observed a similar, though weaker, decrease in the innervation of the M6 layer by yDm8s and a total absence of yDm8s home column (Figure 3E).
Fig. 3. Dm8 subtype specification is coordinated with R7 stochastic specification:
(A) Schematic representing the R7 subtypes in the different mutant conditions. In the spineless (ss) eye specific mutant the retina is only composed of pale and DRA R7s, in sevenless (sev) mutants R7 photoreceptors are not specified and thus absent. lGMR-ss and lGMR-hth are photoreceptors specific gain-of-function where all R7s are either all o›f the yellow or DRA type respectively. (B-F) yDm8s labeled with CD8::GFP in WT (B), ss (C), sev (D), lGMR-hth (E) and lGMR-ss (F). Photoreceptors are labeled with GMR-RFP (red) and yR7 with Rh4-lacZ (blue). (G-H) Quantification of the number of yDm8s and pDm8s per optic lobe in Dm8-LexA>LexAop-RFP;DIPγ-GFP animals. (G) The number of yDm8s is plotted using two different quantification: the number of DIPγ-GFP+Dac+ cells or as the number of RFP+GFP+. (H) Quantification of the number of pDm8s (RFP+GFP−). Bars show the mean +/− SD. ****p<0.0001, one-way ANOVA, Tukey test. (I) Single cell clone of a yDm8 with two home-columns in ss gain-of-function (lGMR-ss). (J) Distribution of yDm8s with 2 home-columns (WT, n=31; lGMR-ss, n=22). p=0.0302, Fischer’s exact test. Scale bars: 20μm in B for (B-F), 5μm in B’ for (B’-F’) and (I) 5μm.
We next performed the reverse experiment and converted the entire retina into yellow ommatidia by overexpressing Ss in photoreceptors (lGMR-ss, Figure 3A). In this case, almost every R7 was occupied by a yDm8 home column (Figure 3F).
Therefore, in all cases, the innervation of the M6 layer by yDm8s appears to correlate with the number of yR7s, suggesting that the number of yDm8s is adjusted to accommodate the number of yR7s.
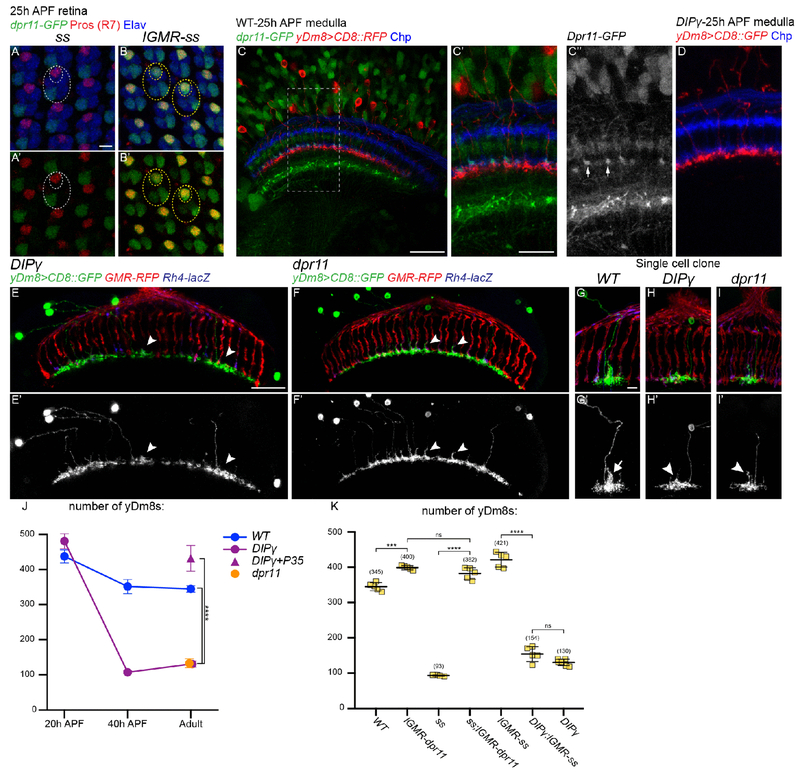
To confirm these changes, we quantified the number of yDm8s using two different methods: (i) we counted the number of Dac+DIPγ+ cells per optic lobe that represents the absolute number of yDm8s; (ii) we also used a Dm8-LexA line to drive an RFP reporter to visualize Dm8s in combination with DIPγ-GFP and quantified the number of RFP+GFP+ cells. Both methods gave similar relative results, with quantification using the Dm8-LexA driver resulting in a lower estimate of the number of yDm8s (Figure 3G). This might be due to the fact that the LexA driver does not label all Dm8s. Thus, we will only discuss the quantification of the Dac+DIPγ+ cells for yDm8s. In ss and sev mutants that lack yR7s, the average number of yDm8s per optic lobe dropped to around 25% of the wild type number (Figure 3G; WT=345, ss=93, sev=95). Converting every R7 to the DRA subtype led to a lower decrease in the number of yDm8s (Figure 3G; WT=345, lGMR-hth=184, discussed below). When every R7 was converted to yR7, the number of yDm8s increased by 25% (WT=345, lGMR-ss=430). We also quantified the number of pDm8s and obtained similar results: in the absence of pR7s (lGMR-ss, sev), the number of pDm8s dropped dramatically (Figure 3H; WT=102, lGMR-ss=3.4, sev=17) whereas it increased in ss mutants (Figure 3H; ss=139). Taken together, these data indicate that the number of y and pDm8s is affected by the number of y/p R7 in the retina.
We noticed that in ss gain-of-function (lGMR-ss), most yR7s were occupied by a yDm8 home column (Figure 3F) while the 25% increase in yDm8 number alone could not account for such a dramatic effect (Figure 3G). We speculated that, in addition to the increase in cell number, an increase in the number of home columns covered by individual yDm8 could explain such an effect. We therefore generated single cell flip-out clones of yDm8s in ss gain-of-function to look at their morphology. In the wild type, 16.1% of yDm8s had two home columns (Figures S1H, S1I) whereas in ss gof, 44.5% of yDm8s had two home columns (Figures 3I and 3J). We never observed yDm8s with more than two home columns. Thus, two mechanisms allow adult yDm8s to accommodate changes in the number of their R7 presynaptic partners: (i) their numbers increase or decrease overall to match the number of yR7s and (ii) when excess yR7 are present, individual yDm8s also increase the number of columns they occupy, such that most yR7s are covered.
Apoptosis of excess Dm8s ensures the numerical matching of R7s and Dm8s
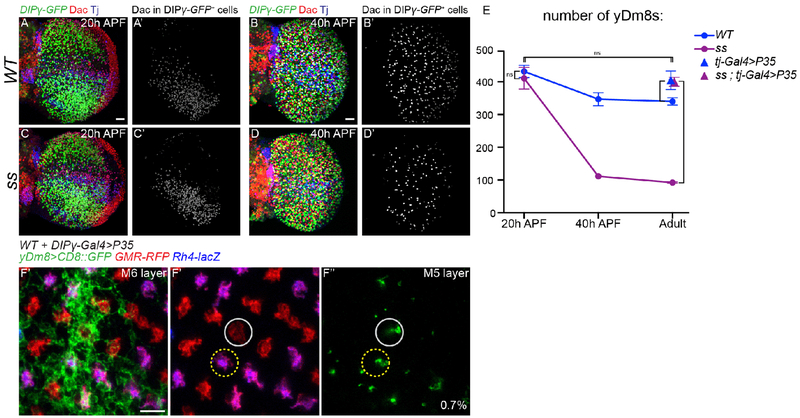
Our data indicate that although the different types of Dm8s are pre-specified, their number can be adjusted to accommodate the ratio of their presynaptic R7s. One hypothesis is that, in ss mutants, yDm8s are produced normally but in the absence of their yR7 partners, they are eliminated during a later stage of development while, when yR7s are in excess (lGMR-ss), all yDm8s are maintained. To test this, we first looked at the number of yDm8s early in development in ss mutants that lack yR7s. At 20h APF, when the neuroblasts no longer divide and thus no more medulla neurons are produced, similar numbers of yDm8s were found in ss mutants and in wild type (Figure 4A, 4C and 4E; WT=437.5, ss=416.5). By 40 APF however, the number of yDm8s decreased to approximately the number observed in the adult in both wild type and ss mutants (Figure 4B, 4D and 4E; P40: WT=351, ss=113, Adult: WT=345, ss=93). This confirms that, in the absence of yR7s, yDm8s are still produced normally but that numerical matching with yR7 happens during pupal development. In ss mutants, the decrease in the number of yDm8s might be due to the death of yDm8s that have failed to find their correct R7 subtype. We confirmed this by inhibiting apoptosis using tj-Gal4 to mis-express the caspase inhibitor P35 in yDm8s. Inhibiting apoptosis restored the number of yDm8s in ss mutants to the number found at 20h APF (Figure 4E; Adult: WT= 345, ss=93, ss+P35=406, P20: ss=416.5). We could also rescue the decreased number of yDm8s in the wild type by mis-expressing P35 (Figure 4E). The final number of adult yDm8s obtained upon cell death inhibition was similar in wild type and in ss mutants and was also similar to the number in the ss gof and in wild type at 20h APF (Figure 3G and 4E; WT-20hAPF=437.5, lGMR-ss=430 and WT+P35=410). This shows that during development, a fixed number of yDm8s are produced in excess but that the relative number of yDm8s surviving depends on the number of their available presynaptic yR7s: Naturally occurring cell death can be rescued by providing more yR7s in ss gof, whereas it can be greatly increased by eliminating yR7s in ss or sev mutants. However about 25% of yDm8s are still found in the absence of any yR7s (Figure 3G), suggesting that some yDm8s that are not connected to yR7s can still survive.
Fig. 4. Apoptosis of excess Dm8s ensures the numerical matching of R7s and Dm8s:
(A-D) DIPγ-GFP expression in the optic lobe in WT (A,B) and ss mutant (C,D) at 20 hours (A and C) and 40 hours After Pupa Formation (APF) (B and D). yDm8s were labelled by segmenting the Dac staining from the GFP staining (A’-D’, in grey). Scale bar: 20 μm in A for (A-D). (E) Number of yDm8s per optic lobe (DIPγ-GFP+,Dac+ cells, n=4-6 optic lobes per genotype). Bars show the mean +/− SD. ****p<0.0001, one-way ANOVA, Tukey test. (F) Proximodistal view of yDm8s labelled with CD8::GFP in WT upon cell death inhibition by mis-expressing P35 in DIPγ expressing neurons. A single yDm8 mis-paired with a pR7 (circled in grey) showed by dense GFP staining at the level of the M6 layer (E’) and its home column at the level of the M5 layer (F” and F”’) (mis-paired yDm8s=0.7%, n=428). Scale bar: 5 μm.
We propose that this mechanism is sufficient to obtain the perfect matching observed in the wild type, and that cell death plays an essential role in coordinating the size of the Dm8 populations with the ratio of y/p R7 subtypes in the retina.
Physiological apoptosis regulates Dm8 wiring
We next tested whether the physiological cell death might be important for the proper wiring of yDm8s by looking for mis-pairing of yDm8s with pR7s when cell death was abolished in an otherwise wild type background. We did observe a very low but significant frequency of yDm8s mis-paired with pR7s (Figure S1E and 4F; WT+P35=0.7%; n=428). This suggests that the great majority of un-dead yDm8s still manage to integrate the proper circuitry.
yDm8 morphology and survival are affected in DIPγ and dpr11 mutants
We next sought to identify the mechanisms that control the pairing of Dm8s with their specific R7 subtype and to investigate the role of Dpr11 and DIPγ in the process. Dprs and DIPs are two closely related families of Immunoglobulin-containing cell adhesion molecules (42). Each of the 21 Dprs binds to one or several of the 11 DIPs and these interactions are required for their neurogenic function (32, 43, 44). Because of the striking complementary expression pattern of Dpr and DIP pairs in synaptic partners, these families have been proposed to play a role in synaptic partner matching (32, 35, 45). Dpr11 and DIPγ are ideal candidates for the matching of yR7s and yDm8s: dpr11 expression is specific to yR7s (32) (Figure 1D), and depends on ss, as dpr11 is lost from yR7s in ss mutant retinas (Figure 5A) whereas ss overexpression in photoreceptors is sufficient to induce dpr11 expression in all R7s (Figure 5B). dpr11 is widely expressed in the optic lobe, especially in adult brains (Figure S3A). However, at 25h APF, around the time dpr11 expression peaks in yR7s, it is relatively restricted to yR7s in the M6 layer while being still broadly expressed in other medulla layers (Figure 5C). By that time, yDm8s have already reached the M6 layer and have contacted R7s (Figure 5C) but do not have an obvious phenotype in DIPγ mutants (Figure 5D).
Fig. 5. yDm8 morphology and survival are affected in DIPγ and dpr11 mutants:
(A and B) dpr11-GFP expression in 25h APF retinas (green) in ss mutant (A) or ss gain-of-function (B). R7 cells are labelled by Prospero (red) and all photoreceptors by Elav (blue). Large ellipse indicates a single ommatidia and the smaller ellipses the R7 cells. Note that dpr11-GFP expression is not lost in a single outer photoreceptor in ss mutant whereas weak dpr11-GFP is also seen in a few outer photoreceptors in the ss gof. (C) dpr11-GFP expression in 25h APF medulla (green). The split-gal4 line DIPγ-Gal4DBD∩OK371-Vp16 drives CD8::RFP in yDm8s (red). In the M6 layer, dpr11-GFP is mainly seen in a subset of photoreceptors (blue) that corresponds to the yR7s (arrowheads). (D) DIPγ mutant yDm8s labelled with CD8::GFP (red) at 25h APF. (E-F) Dorsoventral view of yDm8s labelled with CD8::GFP in DIPγ (E) and dpr11 (F) mutants. Arrowheads indicate morphological defects in yDm8 home columns. (G-I) Sparsely labeled yDm8s in WT (G), DIPγ (H) and dpr11 whole mutant animals (I). Arrow in G’ indicates the Dm8 process in its home column, and arrowheads in H’ and I’ indicate the defective process in the home column. (J-K) Number of yDm8s per optic lobe (DIPγ-GFP+,Dac+ cells, n=4-7 optic lobes per genotype). Bars show the mean +/− SD. ****p<0.0001, one-way ANOVA, Tukey test. Scale bars: (A and B, C’ and D, G-I) 5μm, (C, E and F) 20μm.
In adults however, the mutant phenotypes for dpr11 or DIPγ were quite obvious: yDm8 innervation of the M6 layer was significantly decreased (Figure 3B, 5E and 5F), suggesting a decrease in their number, as previously shown (32). We quantified the number of yDm8s and confirmed that this number decreased in both dpr11 and DIPγ mutants (Figure 5J; WT=345, DIPγ=120, dpr11=132). This reduction was also due to apoptosis during development (Figure 5J) as the DIPγ phenotype could be rescued by mis-expressing P35 in yDm8s (Figure 5J; DIPγ+P35=432).This phenotype is similar to what was reported for mutants for DIPα and its two Dpr partners, dpr6 and dpr10, where a proportion of the 3 DIPα-expressing Dm neurons (Dm1, Dm4 and Dm12) were shown to undergo increased apoptosis (45).
Additionally, yDm8 morphology was affected in both mutants. DIPγ mutant yDm8s failed to extend a proper process in their home column (Figure 3B and 5E) and only had a short protrusion at the center of their dendritic field (Figure 5G and 5H). In dpr11 mutants, yDm8s had a similar but weaker phenotype and extended a thin process in their home column (Figure 5G and 5I). Rescuing yDm8s cell death by mis-expressing P35 was not sufficient to rescue the morphology of their dendritic extension in their home column (Figure S3B).
Thus, dpr11 and DIPγ mutants phenocopy the loss of yR7s (Figure 3), supporting a model that yDm8s in these mutants are unable to recognize yR7s and thus do not receive the trophic support required for their survival. If this was indeed the case, (i) overexpressing dpr11 in R7s should compensate for the loss of yR7s in ss mutants, and (ii) the loss of DIPγ should be epistatic over the ss gain-of-function. Overexpressing dpr11 in photoreceptors increased the number of yDm8s in wild type and could also rescue yDm8 cell death and morphology in ss mutants (Figure 5K; lGMR-dpr11=400, ss+lGMR-dpr11=382, and S3D). Conversely, increasing the number of yR7s using ss gain-of-function was not sufficient to rescue cell death of yDm8s in a DIPγ mutant background (Figure 5K; DIPγ=130, DIPγ+lGMR-ss=154, and S3C).
DIPγ and Dpr11 regulate the pairing between yR7s and yDm8s
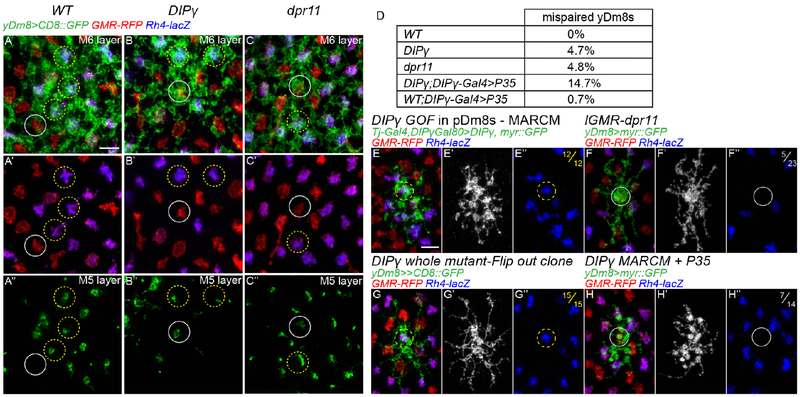
These results imply that Dpr11 and DIPγ mediate the strict matching of yDm8s with yR7s. If true, we would expect to observe mis-pairing of the remaining yDm8s with pR7s in either mutant because of the inability of yDm8 to recognize yR7s. In wild type, yDm8 home columns were always located along yR7 and we never observed mis-pairing with a pR7, either in single cell clones (Figure 1E and 1F) or whole mount (Figure 6A; number of yDm8s, n=1046). We first tested whether DIPγ and dpr11 mutants exhibited defects in yDm8s and yR7s pairing. We did observe mis-pairing in both mutants (Figure 6B and 6C) where around 5% of surviving yDm8s were paired with pR7s (Figure 6D; DIPγ= 4.7%; n=223 and dpr11=4.8%; n=478,). The ratio of yR7s contacted by DIPγ mutant yDm8s lateral dendrites was also decreased (Figure S4B; WT=61.4%, DIPγ=51.1%), without affecting their overall dendritic field size (Figure S4A; WT=12.9, DIPγ=12.5).
Fig. 6. DIPγ and Dpr11 regulate pairing of yR7 and yDm8:
(A-C) Proximodistal view yDm8s labelled with CD8::GFP in WT (A), DIPγ mutant (B) and dpr11 mutant (C), either at the level of the M6 layer (A-C and A’-C’) or at the level of the M5 layer (A”-C”). Some yR7 or pR7 columns were highlight by yellow and grey circles respectively. In WT, yDm8s occupy most yR7 columns but never occupy pR7 columns (A”). In DIPγ and dpr11 mutants many yR7 columns are devoid of yDm8s, and some yDm8s contact pR7s (grey circle in B” and C”). (D) Quantification of yDm8s mis-pairing with pR7s (total number of yDm8s counted: WT; n=1046, DIPγ; n=223, dpr11; n=478, DIPγ;DIPγ-Gal4>P35; n=251 and WT;DIPγ-Gal4>P35; n=428). (E) MARCM clone of a pDm8 overexpressing DIPγ. DIPγ overexpression is sufficient for mis-pairing of pDm8s with yR7s (n=12/12). (F) yDm8 Flip-out clone labelled with myr::GFP with dpr11 overexpressed in all photoreceptors using lGMR-dpr11. Some yDm8s are mis-paired with pR7s (grey circle; n=5/23). (G) yDm8 Flip-out clone expressing CD8::GFP DIPγ background. 100% of the yDm8 clones obtained were paired with yR7s (yellow circle; n=15/15). (H) DIPγ mutant MARCM clone of a yDm8 expressing myr::GFP and P35 in an otherwise heterozygous background. 50% of the yDm8 clones obtained were mis-paired with pR7s (grey circle; n=7/14). Scale bar: 5μm for all micrographs.
We next tested whether DIPγ overexpression in pDm8s or dpr11 in pR7s was sufficient to generate mis-pairing of Dm8s with R7s. When DIPγ was sparsely overexpressed in pDm8 MARCM clones from the late third larval instar stage onward using tj-Gal4, pDm8s were always mis-paired with yR7s (Figure 6E, n=14/14).
We also overexpressed DIPγ in two other medulla neuron types: Dm11 and Dm12. Dm11s project to the M6 layer and have multiple processes going along multiple R7s (Figure S2E) but do not show a preference for a R7 subtype (Figure S4C). Overexpression of DIPγ in Dm11s was sufficient to induce these processes to exclusively occupy yR7 columns (Figure S4D). However, for Dm12 neurons that arborize in the M3 layer, overexpression of DIPγ was not sufficient to make them contact yR7s (Figure S4E and S4F).
We also overexpressed dpr11 in all photoreceptors and looked at whether this was sufficient to create mispairing between yDm8s with pR7s. Indeed, 20% of yDm8s extended their home column in pR7 columns (Figure 6F, n=5/23). Taken together, these data show that the interaction between Dpr11 and DIPγ is absolutely sufficient to promote pairing of Dm8s with yR7s, whereas lack of Dpr11 and DIPγ only causes 5% of yDm8s to mis-pair with pR7s.
Two hypotheses could explain the discrepancy between the requirement and the sufficiency of dpr11 and DIPγ: (i) In the absence of DIPγ, pDm8s are unaffected and thus target pR7s, leaving no space for mutant yDm8s to target pR7s. One could test it by looking at yDm8s pairing in a DIPγ mutant where pDm8s were ablated. As this experiment was not technically feasible, instead of removing the competition with pDm8s we allowed DIPγ mutant yDm8s to compete with both pDm8s and wild type yDm8s. We looked at the pairing of DIPγ homozygous mutant yDm8s in mosaic animal using MARCM where most yDm8s were heterozygous for DIPγ. Since DIPγ mutant yDm8s would normally undergo apoptosis (see below, Figure S6), we rescued cell death by mis-expressing P35 in the mutant clones. In these conditions, half of the DIPγ mutant yDm8s mis-paired with pR7s (Figure 6H, n=7/14) whereas in single cell flip-out clones in whole DIPγ mutants, we did not observe a single mis-paired yDm8 (Figure 6G; n=0/15). Thus, in this experimental setup where p and yR7 columns are equally accessible, yDm8s evenly distribute between the two, suggesting that DIPγ is required for yDm8s to pair with the proper R7 subtype. (ii) A second explanation could be that, as two thirds of yDm8s die during early pupal development in DIPγ mutants because they are unable to find a yR7, they would be able to mistarget if cell death was rescued. Indeed, when we rescued cell death in DIPγ mutants, there was a 3-fold increase in yDm8s mis-pairing (Figure 5D; DIPγ=4.7%; n=223 and DIPγ+P35=14.7%; n=251). However, because not every yDm8s prevented to die mis-paired with a pR7, this suggest that cell death is not the result of the culling of mis-paired yDm8s, but that preventing cell death makes them more competent to compete with pDm8s to occupy pR7 columns.
Taken together, our data indicate that Dpr11 and DIPγ mediate the pairing of yDm8s with their presynaptic partners yR7s.
It is noteworthy that we did not find any other DIPs expressed in pDm8s or pR7s, whereas Dm8s express multiple Dprs based on RNAseq data (21). This suggests that the matching between pR7 and pDm8s uses cell adhesion molecules distinct from Dprs and DIPs.
yDm8s targeting of the M6 layer is not affected in DIPγ mutants and in DIP gain-of-function
Based on their layer-specific expression in the medulla and mis-expression experiments, it was proposed that Dprs and DIPs regulate layer targeting (45). In either dpr11 or DIPγ mutants, yR7s or yDm8s do not mistarget but instead elaborate processes in the appropriate M6 layer (Figure 5E and 5F). Because the mistargeted cells could have been those eliminated by apoptosis, we looked at yDm8s both in DIPγ mutants where cell death was abolished, and during development when apoptosis happens. We did not observe mistargeting to another layer in either case (Figure 5D and S3B). To test whether DIP overexpression was sufficient to mistarget neurons to an improper layer, we performed gain-of-function of DIPs in different neuronal population. As described above, overexpression of DIPγ in Dm12s, that normally innervate the M3 layer, was not sufficient to make them target the M6 layer or other layers where DIPγ is expressed (Figure S4E and S4F). We also tested whether replacing DIPγ by other DIPs normally expressed in different layers would be sufficient to retarget yDm8s to these layers (Figure S5). Overexpression of DIPδ in DIPγ mutant yDm8 had no effect (Figure S5E), but overexpression of DIPα led some yDm8 to send small processes to the M3 and M7 layers (Figure S5C), where DIPα and it’s two ligands are expressed (Figure S5B, S5F and S5G). Thus, yDm8s targeting to the M6 layer is independent of Dpr11/DIPγ interaction and yDm8s cannot be efficiently retargeted to different layers by ectopic expression of other DIPs. However, the extension of their home column requires DIPγ, and ectopic expression of DIPα leads to the formation of small extensions reminiscent of Dm8s home column. Taken together these results suggest that DIPs do not play a significant role in layer targeting in the visual system but instead are involved in the matching of synaptic pairs.
Competitive interactions between Dm8s regulate survival and wiring
yDm8s mis-pairing was drastically enhanced when they had to compete for targeting with wild type yDm8s (See above, Figure 6H). We thus asked whether survival was also affected by competition among Dm8s. We generated DIPγ yDm8s mutant MARCM clones in either a DIPγ heterozygous background, or in a DIPγ mutant background where yDm8s both outside and within the clone had identical genotypes (Figure S6A). If competition among yDm8s played a role in survival, we would expect to see differences in clone size. In the heterozygous background, we obtained small clones of yDm8s (Figure S6B; yDm8s/clone: 2 ± 2.6, mean ± SD) whereas in DIPγ mutant background, we obtained clones of significantly larger size (Figure S6B; yDm8/clone: 10.8 ± 3.5). Because these experiments rely on generating clones of the same size, we controlled for clone size by quantifying the number of pDm8s per clone. In both conditions we obtained similar number of pDm8s (Figure S6B; pDm8/clone: WT background: 29.6 ± 10.3, DIPγ background: 28.6 ± 6.3), confirming that the difference in yDm8 number comes from a difference in survival. Thus, DIPγ mutant yDm8s are much more likely to survive when they compete with DIPγ mutant yDm8s rather than with wild type yDm8s.
We noticed that affecting yDm8s sometimes yielded unexpected effects on pDm8s that could also be explained by competitive interactions among Dm8s: In DIPγ and dpr11 mutants, the number of pDm8s increased (Figure S6D; WT=102, DIPγ=135 and dpr11=136). Because neither DIPγ nor dpr11 are expressed in pDm8s, the increase in pDm8 number might result from the decrease in the number of yDm8s. We thus explored the non-autonomous effects on pDm8 survival. We first asked whether promoting survival of yDm8s would affect pDm8s survival when we increased the number of yDm8s without affecting pR7s specification: in dpr11 gain-of-function (lGMR-dpr11), the number of yDm8s increased with a corresponding >50% decrease in the number of pDm8s (Figure S6D; WT=102, lGMR-dpr11=41). We obtained the same effect in the absence of any yR7s when dpr11 was overexpressed in a ss mutant (Figure S6D; ss=139, ss+lGMR-dpr11=32). We also asked whether decreasing the number of yDm8s (by mutating DIPγ) in a ss gain-of-function, would have an effect in the survival of pDm8s (Figure S6D), which normally almost all die (Figure 3G). The drastically decreased number of yDm8s was accompanied by an increase of pDm8s (Figure S6D; lGMR-ss=3.4, DIPγ+lGMR-ss=29). Taken together these results show that the size of the pDm8 population is affected by the number of yDm8s. Dm8s must compete for targeting, and affecting the size of one population affects the survival of the other. This competition might be regulated by self-avoidance mechanisms (46) and would explain why there is never more than one Dm8 home column per R7.
Discussion:
In Drosophila, patterning of the mosaic of ommatidial subtypes is established by sequential steps and is initiated in R7 photoreceptors by a single transcription factor, Ss. This initial decision is then transmitted to the R8 of the same ommatidium so that R7 and R8 have paired Rhodopsins expression (18). In contrast, we show here that the mechanism responsible for the coordination of R7 subtype specification with their main post-synaptic target in the brain is different. Dm8 subtype specification does not depend on direct induction from R7s and each Dm8 subtype is produced independently from their corresponding R7s. We have shown that the matching occurs during a later stage of circuit formation after the specification of the different components of the circuit. Following matching with their R7 subtypes, controlled by Dpr11/DIPγ for yR7s/yDm8s (discussed below), supernumerary Dm8s are culled by apoptosis. Because the ratio of ommatidial subtypes can vary among individuals (47), this mechanism allows the perfect matching that is observed in adult, where most R7s, if not all, are innervated by a single Dm8 of the proper type. This developmental plasticity provides a powerful mechanism to transmit the stochastic formation of the photoreceptor mosaic to the deterministic patterning of the brain.
DIPγ and Dpr11 role in synaptic partner pairing
The complementary expression of Dpr and DIP binding pairs in synaptic partners raised the exciting possibility that these proteins might be the long sought-after “Sperry molecules” that act as molecular tags to instruct synaptic specificity (32, 35, 48). Here we provide evidence that DIPγ in yDm8 and Dpr11 in yR7 instruct synaptic partner matching. Similar to the genetic removal of yR7s, loss of either DIPγ or Dpr11 leads to apoptosis of yDm8s, suggesting that in their absence yDm8s are unable to connect to yR7s and to receive from yR7s the trophic support required for their survival. In both mutants, this is accompanied by relatively limited mis-pairing of yDm8s with pR7s whereas ectopic expression of these molecules is absolutely sufficient to create mis-pairing between R7 and Dm8 subtypes. However, targeting to the proper M6 layer of the medulla, or the dendritic size of yDm8s are not affected in these mutants. Thus, we propose that Dprs and DIPs act during a later step of circuit formation to allow distinct neurons that project to the same layer to distinguish their appropriate synaptic partners.
Analysis of other Dpr/DIP pairs in and outside the visual system support this view (45, 49–51). In the medulla, DIPα is expressed in three Dm neurons (Dm1, Dm4 and Dm12) whereas its ligands Dpr6 and Dpr10 are expressed in neurons innervating the layers occupied by these Dm neurons (45). Loss of either DIPα or Dpr6/10 results in apoptosis of a proportion of these Dm neurons during development (20% to 40% depending on the neuron type), likely because these neurons are unable to recognize their targets (45). In the olfactory system, loss of DIPs leads to mistargeting of olfactory receptor neurons and disorganization of olfactory glomeruli (51). At the larval and adult neuromuscular junction, DIPα is expressed in a subset of motoneurons whereas its binding partner Dpr10 is in muscles (49, 52). Loss of either leads to the partial loss of the innervation of motoneurons to the muscle. During development, mutant adult motoneurons extend normal filopodia that target the proper muscles. However these filopodia fail to be maintained, likely because they are unable to recognize the proper muscles in the absence of these molecules (49). In the lamina, loss of both DIPγ and DIPβ leads to ectopic synapse formation of L2 and L4 neurons with the wrong partner, whereas overexpression of DIPγ and DIPε in photoreceptors leads to ectopic synapse formation with lamina neurons putatively expressing the corresponding Dprs (50).
Taken together this supports the role of Dprs/DIPs in establishing synaptic specificity, while the difference in phenotypes, e.g. survival, mistargeting or loss of axonal branches, might reveal the different requirements for such molecules in distinct circuits (discussed below).
Apoptosis as a mechanism for numerical matching of neuronal pairs
Programmed cell death has long been proposed to play a role in the quantitative matching of synaptic partners (53). A classic example is the role of target-derived Nerve Growth Factor (NGF) in promoting the survival of sympathetic and sensory neurons (54, 55). This discovery led to the development of the Neurotrophic theory that states that neurons are produced in excess and that competition for limited trophic support allows for the numerical matching of afferents with their targets in the periphery. Here, we provide evidence that a similar phenomenon happens in the Drosophila central nervous system for Dm8 neurons. We show that yDm8s are produced in excess and that around 25% are eliminated by apoptosis during normal development. This cell death could be fully rescued by increasing the number of yDm8 afferents (i.e. in the ss gain-of-function) or conversely aggravated by decreasing the number of yR7s (i.e. in the ss loss-of-function). Because DIPγ and dpr11 mutant phenotypes phenocopy the loss of yR7, uncovering a link between synaptic pairs matching and survival, it argues that the numerical matching of pairs of R7 and Dm8 is obtained by apoptosis of unmatched Dm8s.
It is worthwhile to mention that in other parts of the visual system and of the brain, neuronal survival is not affected in DIPs mutants, e.g. lamina neurons (50), motoneurons (49, 52) and olfactory receptor neurons (51). Thus, cell death might reveal circuit-specific properties of the formation of the visual system. The medulla is composed in ~750 retinotopically organized columns that can be considered as repetitive microcircuits that each compute information from a single point of the visual field. Most Dm neurons are multicolumnar neurons, i.e. neurons that have a receptive field that covers multiple columns. During larval development, multicolumnar neurons are generated from restricted regions of the neuroepithelium but cover the entire receptive field (30, 56). Our data, and others (45) suggest that multicolumnar neurons are generated in excess and that the proportion of neurons dying during development is increased in DIP mutants.
For yDm8s, the dependency on targeting for survival allows the ~1:1 matching of yDm8s and yR7s. For other multicolumnar neurons, one can only speculate about the function of normally occurring cell death. Because the medulla is a repetitive structure composed of ~750 columns that each need to be innervated by the proper neurons in the proper amount, creating more multicolumnar neurons that require target-derived trophic support to survive might allow the complete innervation of the medulla while having the optimal number of neurons.
One supporting evidence for this model is the lack of cell death of lamina neurons in DIP mutants (50). Lamina differentiation is directly induced by photoreceptors R1-6 axons (57), thus induction of the lamina by R1-6 allows the numerical matching of lamina cartridges and ommatidia, and the production of the right number of lamina neurons in each cartridge (58). This might explain why some neurons do not require target-derived trophic supports for their survival.
Supplementary Material
Acknowledgments:
We would to thank the fly community, the Bloomington and Kyoto Stock centers, and the Drosophila Genomics Resource Center for sharing flies and reagents; Mathias Wernet, Lalanti Venkatasubramanian, Kaushiki Menon and Kai Zinn for sharing data prior to publication; Bob Johnston and Larry Zipursky for unpublished data and fly stocks; Chi-Hon Lee, Lalanti Venkatasubramanian, Richard Mann and Hugo Bellen for fly strains; Dorothea Godt and Yuh-Nung Jan for antibodies. We are grateful to Jessica Treisman, Esteban Mazzoni and Niels Ringstad for their input during the investigation. We would like to thank the entire Desplan lab members for their discussion and comments on the manuscript;
Funding: This work was supported by NIH grant R01 EY13010 to C.D;
Footnotes
Competing interests: Authors declare no competing interests;
Data and materials availability: All data is available in the main text or the supplementary materials.
References and Notes:
- 1.Johnston RJ, Desplan C, Stochastic Mechanisms of Cell Fate Specification that Yield Random or Robust Outcomes. Annu. Rev. Cell Dev. Biol 26, 689–719 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buck L, Axel R, A Novel Multigene Family May Encode Odrant Receptors: A Molecular Basis for Odor Recognition. Cell. 65, 175–187 (1991). [DOI] [PubMed] [Google Scholar]
- 3.Godfrey PA, Malnic B, Buck LB, The mouse olfactory receptor gene family. Proc. Natl. Acad. Sci 101, 2156–2161 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nathans J, Thomas D, Hogness D, Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science (80-. ). 232, 193–202 (1986). [DOI] [PubMed] [Google Scholar]
- 5.Roorda A, Williams DR, The arrangement of the three cone classes in the living human eye. Nature. 397, 520–522 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Ressler KJ, Sullivan SL, Buck LB, A zonal organization of odorant receptor gene expression in the olfactory epithelium. Cell. 73, 597–609 (1993). [DOI] [PubMed] [Google Scholar]
- 7.Mombaerts P et al. , Visualizing an olfactory sensory map. Cell. 87, 675–686 (1996). [DOI] [PubMed] [Google Scholar]
- 8.Mombaerts P, Axonal Wiring in the Mouse Olfactory System. Annu. Rev. Cell Dev. Biol 22, 713–737 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Ressler KJ, Sullivan SL, Buck LB, Information coding in the olfactory system: Evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 79, 1245–1255 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Franceschini N, Kirschfeld K, Minke B, Fluorescence of photoreceptor cells observed in vivo. Science (80-. ). 213, 1264–1267 (1981). [DOI] [PubMed] [Google Scholar]
- 11.Wernet MF et al. , Stochastic spineless expression creates the retinal mosaic for colour vision. Nature. 440, 174–180 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rister J, Desplan C, Vasiliauskas D, Establishing and maintaining gene expression patterns: insights from sensory receptor patterning. Development. 140, 493–503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wernet MF et al. , Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell. 115, 267–279 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Wernet MF et al. , Genetic dissection reveals two separate retinal substrates for polarization vision in drosophila. Curr. Biol 22, 12–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston RJ et al. , Interlocked feedforward loops control cell-type-specific rhodopsin expression in the drosophila eye. Cell. 145, 956–968 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jukam D et al. , Opposite feedbacks in the Hippo pathway for growth control and neural fate. Science (80-. ). 342 (2013), doi: 10.1126/science.1238016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnston RJ, Desplan C, Interchromosomal communication coordinates intrinsically stochastic expression between alleles. Science (80-. ). 343, 661–665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells BS, Pistillo D, Barnhart E, Desplan C, Parallel activin and BMP signaling coordinates R7/R8 photoreceptor subtype pairing in the stochastic Drosophila retina. Elife. 6, 1–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikeladze-Dvali T et al. , The growth regulators warts/lats and melted interact in a bistable loop to specify opposite fates in Drosophila R8 photoreceptors. Cell. 122, 775–787 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Fischbach KF, Dittrich APM, The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 258, 441–475 (1989). [Google Scholar]
- 21.Konstantinides N et al. , Phenotypic Convergence: Distinct Transcription Factors Regulate Common Terminal Features. Cell. 174, 622–635. e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertet C, in Decoding Neural Circuit Structure and Function: Cellular Dissection Using Genetic Model Organisms, Çelik A, Wernet MF, Eds. (Springer International Publishing, Cham, 2017; 10.1007/978-3-319-57363-2_17), pp. 419–435. [DOI] [Google Scholar]
- 23.Egger B, Boone JQ, Stevens NR, Brand AH, Doe CQ, Regulation of spindle orientation and neural stem cell fate in the Drosophila optic lobe. Neural Dev. 2 (2007), doi: 10.1186/1749-8104-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ngo KT, Andrade I, Hartenstein V, Spatio-temporal pattern of neuronal differentiation in the Drosophila visual system: A user’s guide to the dynamic morphology of the developing optic lobe. Dev. Biol 428, 1–24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hofbauer A, Campos-Ortega JA, Proliferation pattern and early differentiation of the optic lobes in Drosophila melanogaster. Roux’s Arch. Dev. Biol 198, 264–274 (1990). [DOI] [PubMed] [Google Scholar]
- 26.Ready DF, Hanson TE, Benzer S, Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol 53, 217–240 (1976). [DOI] [PubMed] [Google Scholar]
- 27.Li X et al. , Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 498, 456–462 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki T, Kaido M, Takayama R, Sato M, A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev. Biol 380, 12–24 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Holguera I, Desplan C, Neuronal specification in space and time. Science (80-. ). 362, 176–180 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erclik T et al. , Integration of temporal and spatial patterning generates neural diversity. Nature. 541, 365–370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Courgeon M, Desplan C, Coordination of neural patterning in the Drosophila visual system. Curr. Opin. Neurobiol 56, 153–159 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrillo RA et al. , Control of Synaptic Connectivity by a Network of Drosophila IgSF Cell Surface Proteins. Cell. 163, 1770–1782 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao S et al. , The Neural Substrate of Spectral Preference in Drosophila. Neuron. 60, 328–342 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karuppudurai T et al. , A Hard-Wired Glutamatergic Circuit Pools and Relays UV Signals to Mediate Spectral Preference in Drosophila. Neuron. 81, 603–615 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan L et al. , Ig Superfamily Ligand and Receptor Pairs Expressed in Synaptic Partners in Drosophila. Cell. 163, 1756–1769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venken KJT et al. , MiMIC: A highly versatile transposon insertion resource for engineering Drosophila melanogaster genes. Nat. Methods. 8, 737–747 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ting CY et al. , Photoreceptor-derived activin promotes dendritic termination and restricts the receptive fields of first-order interneurons in Drosophila. Neuron. 81, 830–846 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takemura SY et al. , A visual motion detection circuit suggested by Drosophila connectomics. Nature. 500, 175–181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfeiffer BD et al. , Refinement of tools for targeted gene expression in Drosophila. Genetics. 186, 735–755 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sancer G et al. , Modality-specific circuits for skylight orientation in the fly visual system. bioRxiv, 638171 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Bertet C et al. , Temporal patterning of neuroblasts controls notch-mediated cell survival through regulation of hid or reaper. Cell. 158, 1173–1186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Özkan E et al. , An extracellular interactome of immunoglobulin and LRR proteins reveals receptor-ligand networks. Cell. 154, 228–39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cosmanescu F et al. , Neuron-Subtype-Specific Expression, Interaction Affinities, and Specificity Determinants of DIP/Dpr Cell Recognition Proteins. Neuron. 100, 1385–1400. e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng S et al. , Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila. Elife. 8, e41028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu S et al. , Interactions between the Ig-Superfamily Proteins DIP-α and Dpr6/10 Regulate Assembly of Neural Circuits. Neuron. 100, 1369–1384. e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grueber WB, Sagasti A, Self-avoidance and tiling: Mechanisms of dendrite and axon spacing. Cold Spring Harb. Perspect. Biol 2 (2010), pp. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson C et al. , Natural variation in stochastic photoreceptor specification and color preference in Drosophila. Elife. 6, 1–20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sperry RW, Chemoaffinity in the Orderly Growth of Nerve Fiber Patterns and Connections. Proc. Natl. Acad. Sci 50, 703–710 (1963). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Venkatasubramanian L et al. , Stereotyped terminal axon branching of leg motor neurons mediated by IgSF proteins DIP-α and Dpr10. Elife. 8 (2019), doi: 10.7554/eLife.42692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu C et al. , Control of synaptic specificity by limiting promiscuous synapse formation. bioRxiv, 415695 (2018). [Google Scholar]
- 51.Barish S et al. , Combinations of DIPs and Dprs control organization of olfactory receptor neuron terminals in Drosophila. PLoS Genet. 14, 1–33 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashley J et al. , Transsynaptic interactions between IgSF proteins DIP-α and Dpr10 are required for motor neuron targeting specificity. Elife. 8 (2019), doi: 10.7554/eLife.42690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cowan W, Fawcett J, O’Leary D, Stanfield B, Regressive events in neurogenesis. Science (80-. ). 225, 1258–1265 (1984). [DOI] [PubMed] [Google Scholar]
- 54.Hamburger V, Levi-Montalcini R, Proliferation, differentiation and degeneration in the spinal ganglia. J. Exp. Zool 111, 457–501 (1949). [DOI] [PubMed] [Google Scholar]
- 55.Cohen S, Purification of a nerve-growth promoting protein from the mouse salivary gland and its neuro-cytotoxic antiserum. Proc. Natl. Acad. Sci …. 46, 302–311 (1960). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nern A, Pfeiffer BD, Rubin GM, Optimized tools for multicolor stochastic labeling reveal diverse stereotyped cell arrangements in the fly visual system. Proc. Natl. Acad. Sci 112, E2967–E2976 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Z, Kunes S, Hedgehog, transmitted along retinal axons, triggers neurogenesis in the developing visual centers of the Drosophila brain. Cell. 86, 411–422 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Fernandes VM, Chen Z, Rossi AM, Zipfel J, Desplan C, Glia relay differentiation cues to coordinate neuronal development in Drosophila. Science (80-. ). 357, 886–891 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diao F et al. , Plug-and-play genetic access to drosophila cell types using exchangeable exon cassettes. Cell Rep. 10, 1410–1421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.