Abstract
The U.S. Environmental Protection Agency’s ToxCast program has screened thousands of chemicals for biological activity, primarily using high-throughput in vitro bioassays. Adverse outcome pathways (AOPs) offer a means to link pathway-specific biological activities with potential apical effects relevant to risk assessors. Thus, efforts are underway to develop AOPs relevant to pathway-specific perturbations detected in ToxCast assays. Previous work identified a “cytotoxic burst” (CTB) phenomenon wherein large numbers of the ToxCast assays begin to respond at or near test chemical concentrations that elicit cytotoxicity, and a statistical approach to defining the bounds of the CTB was developed. To focus AOP development on the molecular targets corresponding to ToxCast assays indicating pathway-specific effects, we conducted a meta-analysis to identify which assays most frequently respond at concentrations below the CTB. A preliminary list of potentially important, target-specific assays was determined by ranking assays by the fraction of chemical hits below the CTB compared with the number of chemicals tested. Additional priority assays were identified using a diagnostic-odds-ratio approach which gives greater ranking to assays with high specificity but low responsivity. Combined, the two prioritization methods identified several novel targets (e.g., peripheral benzodiazepine and progesterone receptors) to prioritize for AOP development, and affirmed the importance of a number of existing AOPs aligned with ToxCast targets (e.g., thyroperoxidase, estrogen receptor, aromatase). The prioritization approaches did not appear to be influenced by inter-assay differences in chemical bioavailability. Furthermore, the outcomes were robust based on a variety of different parameters used to define the CTB.
Keywords: ToxCast, cytotoxic burst, adverse outcome pathway, computational toxicology, in vitro
In 2007, the U.S. National Research Council advocated that regulatory toxicity testing transition from a system based largely on direct observation of apical outcomes in whole organism toxicity tests, to one in which the impacts of chemicals are predicted based on biological activities measured in high-throughput, primarily in vitro, bioassays (Krewski et al., 2010). A pioneering effort involved in the implementation of that vision has been the U.S. Environmental Protection Agency (EPA)’s ToxCast program (Dix et al., 2007; Judson et al., 2010; Kavlock et al., 2012). This program combines chemical screening from the Toxicity forecaster (Dix et al., 2007, 2012; Kavlock et al., 2012) and Tox21 programs (Attene-Ramos et al., 2013; Tice et al., 2013). Together, these high-throughput screening (HTS) efforts (hereafter collectively referred to as ToxCast) provide a wealth of biological response data for over 9000 chemicals. Details regarding the different assay platforms have been described previously (see Judson et al. [2016] and references therein), and specifics on individual assays, as well as test chemicals, are available on the interactive Chemical Safety and Sustainability (iCSS) ToxCast dashboard (https://actor.epa.gov/dashboard; last accessed March 5, 2018).
Although HTS represents an efficient and standardized approach to chemical testing, there is uncertainty as to applicability of the data for hazard identification and risk assessments. The primary challenge arises from the difficulty in extrapolating in vitro bioactivity to in vivo effects. The adverse outcome pathway (AOP) framework has been proposed as a means to help provide this linkage (Delrue et al., 2016; Kleinstreuer et al., 2016; Villeneuve et al., 2014a,b). AOPs describe the perturbation of a molecular target (the molecular initiating event; MIE), the scientifically credible and defensible effects of that perturbation on measureable downstream key events (KEs) and, ultimately, how sufficiently large perturbations to the pathway may result in an adverse outcome at the individual or population level (Ankley et al., 2010). Numerous publications have already demonstrated the use of in vitro testing and AOPs to screen chemicals for toxic effects (Judson et al., 2014; Knapen et al., 2015; Rotroff et al., 2013). The U.S. EPA’s Endocrine Disruptor Screening Program (EDSP), in particular, exemplifies the marriage of the AOP framework and high-throughput testing (Browne et al., 2017). Examples of how an AOP may be developed in support of an assay target are provided in Russom et al. (2014), Bal-Price et al. (2015), Yauk et al. (2015), and Fay et al. (2017). Given the potential for AOPs to enhance the utility of ToxCast data, the U.S. EPA’s Office of Research and Development has prioritized the development of AOPs for the molecular targets represented by ToxCast assays and supported the entry of these AOPs into an online repository (AOP-wiki; https://aopwiki.org). The open-access AOP-wiki currently contains approximately 200 AOPs in various stages of development, covering 34 different ToxCast targets (as of October 2017). However, given that there are approximately 350 unique molecular targets currently evaluated by ToxCast assays (Assay_Information_Oct_2015 download; https://www.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data; last accessed March 5, 2018), a prioritization scheme for developing AOPs related to the most suitable HTS assays/endpoints is needed.
One prioritization approach is to focus AOP development on those perturbations that reflect target-specific modes of action that may lead to nonlethal chronic effects that may not easily be detected in commonly used toxicity test methods. Chemical interaction with steroidogenic enzymes or neurotransmitter receptors are classic examples of this type of toxicity. Conversely, ToxCast assay targets that reflect more generalized, nonspecific toxicity (e.g., nonpolar narcosis; reactive modes of action) would be lower priority. A recent analysis of the ToxCast dataset by Judson et al. (2016) revealed that test chemicals often activated large numbers of diverse assays over a narrow range of concentrations at which cytotoxicity or cell stress was also observed. The authors concluded that the majority of chemical-assay active calls (hits) actually reflect cytotoxicity, cell stress, or general disruption of molecular machinery, and termed this phenomenon the cytotoxic burst (CTB). They then applied a statistical approach to define the concentration limits of the CTB for each of the 1060 ToxCast chemicals included in their analysis. The previous analysis focused on differentiating those chemicals exhibiting highly specific modes of action from those causing only a more generalized CTB response. In the current analysis, we take advantage of a similar approach to identify the bioactivities associated with ToxCast chemical screening that reflect specifically acting modes of action, as opposed to nonspecific toxicity.
The ToxCast program employs a standardized data analysis pipeline (Filer et al., 2016). Typically, chemicals are tested using a dose-response design incorporating six or more nominal concentrations (usually ranging from 0.1 to 10 μM), with dimethyl sulfoxide as a chemical delivery agent. Several metrics are provided by the data analysis pipeline, including the maximum activity (T, top value, or efficacy), the activity concentration at 50% of maximal activity (AC50), and the activity concentration at cut-off (ACC) (Figure 1). Most applications of the ToxCast data, including the previous CTB analysis, have relied on the AC50 (Filer et al., 2014; Judson et al., 2016; Karmaus et al., 2016; Knudsen et al., 2011). However, as a relative potency metric, the AC50 can be misleading when comparing chemicals with significantly different efficacy in a given bioassay because the 50% maximal activity is based on the chemical-specific maximum response. In contrast, the ACC is indexed to a standard response threshold in each assay (Blackwell et al., 2017). Thus, ACC provides a relative potency estimate that is not biased by differences in the efficacy of individual chemicals in the different systems. Consequently, rather than apply the AC50-based approach of Judson et al. (2016) for CTB definition, we adapted the method to employ the ACC as an alternative estimate of relative effect concentrations, as we have done for other meta-analyses based on the ToxCast dataset (Blackwell et al., 2017; Fay et al., 2017).
Figure 1.
Illustration of a chemical-assay dose-response curve annotated with the ToxCast pipeline metrics: maximal activity or top value (T), concentration at ½ maximal activity (AC50), and concentration at cut-off (ACC).
In our current analysis, the CTB definition was used to distinguish those assays responding to chemicals at concentrations below the CTB lower bound (CTBlb) from those responding predominantly within the CTB region. We hypothesized that this would be a reasonable means to identify assays providing pathway-specific effect information. Using this definition, we applied two simple approaches to prioritize the targets of these assays as a basis for subsequent AOP development. Further, we explored how chemical bioavailability may impact the findings of this work, and evaluated whether the CTB definition can be used for mode of action classification (i.e., to identify chemicals with specific-acting versus nonspecific modes of action in vivo).
MATERIALS AND METHODS
Dataset and definition of the cytotoxic burst
The statistical calculation of the CTB is available as the ToxCast Pipeline R-code (version 3.1.0) package (tcpl; https://www.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data). For the current effort, the code was applied to the October 2015 ToxCast database, containing data for 1198 assays and 8708 chemicals. This dataset contains assays not available in December 2014 download used by Judson et al. (2016) for the original CTB analysis. Additional HTS tests/endpoints include steroidogenesis assays in the CeeTox (CEETOX) platform, CellzDirect (CDL) hepatic transporter and enzyme assays, and zebrafish embryo assays (Tanguay_ZF). Assay endpoints covered in October 2015 versus December 2014 are detailed in Supplementary Table 1.
Judson et al. (2016) describe their CTB definition/analysis in detail. Briefly, a subset of assays designed to primarily measure nonspecific cytotoxicity or cell stress as their endpoint were identified (Supplementary Table 2). Only chemicals eliciting hits in two or more of these cytotoxicity assays were considered. Then, the log AC50 values for each chemical assay pair (i.e., −log10(AC50/1 × 106) in the dataset were used to calculate the chemical’s cytotoxicity concentration threshold. Inactive chemicals were assigned AC50 values of 1 × 106 μM, resulting in values of 0 for these chemical assay pairs. For these chemicals, the median log AC50 determined in the cytotoxicity (cytotox) assays and the median absolute deviation (MAD) of log AC50 (cytotox) were calculated. Judson et al. (2016) used the distribution of these values for all chemicals in their dataset to calculate a global cytotoxicity MAD (0.293), equal to the median of the MAD of the log AC50(cytotox assays). Z-scores were then determined for each chemical-assay pair with equation 1:
| (1) |
Using this approach, hits with Z-scores < 3 are considered within the CTB, whereas those with Z-scores > 3 imply activity at concentrations below those at which cytotoxicity is observed. Where a chemical activated fewer than two of the cytotoxicity assays, the chemical was assumed to be noncytotoxic at the concentrations within the testing range (up to 100 μM), and the chemical’s median cytotoxicity concentration was set to 1000 μM, generating a Z-score above 3. Using this definition of the CTB, it follows that the CTBlb can be calculated from equation 1 by setting Z to 3 and AC50 (chemical, assay) to CTBlb, then solving for CTBlb (equation 2):
| (2) |
Small discrepancies in the designated “cytotoxicity assays” and in the criterion for chemical selection exist among the Judson et al. (2016) description, supplemental files, and available R-code. Specifically, 33 or 35 assays were identified as measuring cytotoxicity (Supplementary Table 2) and the criterion for chemical selection was reported as tested in >90% of the assays, but actually coded as tested in >80% of the (33) cytotoxicity assays. Following Judson et al. (2016), in the current study the AC50 metric was used to define MADs, and the effects of these two parameters (cytotoxicity assay designation and chemical selection criterion) on data inclusion and the CTB definition were evaluated. We then repeated these evaluations using the ACC. The majority of analyses reported herein were conducted with the dataset generated using the ACC and chemicals tested in >80% of 33 cytotoxicity assays.
Prioritization of assay targets for AOP development
Only those ToxCast assays with clearly identifiable molecular targets were considered for our analysis. High-throughput in vivo assays (e.g., zebrafish embryo development assays) and in vitro bioassays involving endpoints that were not mapped to discrete molecular targets were not included, although it is acknowledged that some may be KEs relevant to certain AOPs. Assays were ranked according to two evaluations. First, assay scores were determined from the proportion of hits in the assay that were below the CTBlb relative to the total number of chemicals tested in that assay (equation 3)
| (3) |
The second approach used assay sensitivity (sn), determined as the proportion of “true positives” relative to total hits (equation 4) and responsivity (r; equation 5) to calculate a diagnostic odds ratio (DOR; equation 6).
| (4) |
| (5) |
| (6) |
In cases where all hits for an assay were below the CTBlb (sn = 1), the DOR is undefined. Consequently, an arbitrary value of 0.5 was subtracted from the number of hits < CTBlb. For example, a sensitivity value of 3.5/4 = 0.875 would be used for an assay with 4 hits < CTBlb out of 4 hits total so a DOR value could be calculated.
Target functional annotation
Functional annotation information was obtained for the intended targets of the assays in the top 90th percentile of each prioritization approach (50 assay targets; ACC dataset). UniProt Identifications (IDs) corresponding to the targets were obtained from the ToxCast data download (https://www.epa.gov/chemical-research/toxicity-forecaster-toxcasttm-data). Human UniProt IDs were submitted to the Database for Annotation, Visualization and Integrated Discovery (DAVID; Huang et al. [2009]) v6.8 for gene functional analysis using the following options (parameter values provided in parentheses): kappa similarity term overlap (3), similarity threshold (0.35), classification initial group (2), final group (2) and multiple linkages (0.50). For the target CSNK2A1 (casein kinase II subunit alpha 3), the ToxCast dataset identified UniProt ID Q8NEV1, which corresponded to CSNK2A3. Consequently, UniProt IDs for both CSNK2A1 (P68400) and CSNK2A3 were included for functional annotation. Additionally, in four cases, nonhuman proteins (Ptgs1, P05979; Ptgs2, P79208; Scn1a, P04774; Sigmar1, Q60492) were identified as assay targets. For functional grouping, the human orthologs were substituted (PTGS1, P23219; PTGS2, P35354; SCN1A, P35498; SIGMAR1, Q99720). Details on target functions were further obtained from the gene pages provided by the National Center for Biotechnology Information (NCBI).
Chemical bioavailability as a determinant of assay prioritization
Observed potency in an in vitro bioassay can be highly dependent on the free fraction of chemical available in the test system, that is, the amount of chemical not bound to protein or lipid. The degree of this kind of nonspecific binding is dependent on the unique properties of the test chemical (e.g., lipophilicity and charge). Fischer et al. (2017) recently addressed the issue of in vitro chemical bioavailability by modeling the chemical binding of 100 structurally diverse chemicals in 26 cell-based Tox21 assays. These assays were characterized for medium and cellular water volumes as well as lipid and protein concentrations. Free, cellular and membrane chemical concentrations (ECfree, ECcell, ECmem, respectively) corresponding to the nominal 50% effect concentration (EC50nom, equivalent to the AC50) were estimated in silico for the chemical-assay hit pairs. In the present study, we used the Fischer et al. dataset and performed two evaluations to assess whether chemical bioavailability was likely to have a significant influence on assay prioritization by differentially skewing the nominal response concentrations. First, assay rank, as determined by equation 3, was compared with assay lipid and protein composition as a proxy for chemical bioavailability. Second, the modeled data for 25 of the 26 assays (activity data for TOX21_ARE_ BLA_agonist_viability were not provided) were used to evaluate whether the factors involved in estimating chemical free fraction were correlated with assay rank. Fischer et al. (2017) modeled the freely dissolved chemical concentration associated with a hit call as (equation 7)
| (7) |
where ffree, medium is the chemical fraction in the medium, and Vsystem and Vw, medium are the volumes of the in vitro system and the water phase of the medium, respectively. Linear regression was used to evaluate assay rank against the EC50free divided by the AC50, thus isolating the chemical and assay-specific properties assumed to determine the freely dissolved chemical concentration.
Evaluation of the CTB to predict in vivo and nonspecific toxicity
A DOR approach was used to evaluate if chemicals eliciting hits < CTBlb (in noncytotoxicity assays) and those never eliciting hits < CTBlb (or only in cytotoxicity assays) could be classified into specific-acting and nonspecific modes of action, respectively. In vitro classifications based on the CTB analysis were compared with mode of action classifications derived from in vivo effects data. We hypothesized that chemicals which predominantly exhibited nonspecific bioactivity in vitro could be classified as exhibiting “narcosis” (sometimes termed baseline toxicity) in vivo (Meyer, 1937; Overton, 1901; Wezel and Opperhuizen, 1995). This assessment utilized the concordance array (Table 1), where TP, TN, FP, and FN are the number of “true positives,” “true negatives,” “false positives,” and “false negatives,” respectively, and the DOR′ is calculated as (equation 8):
| (8) |
Table 1.
Concordance Table Between Chemical Categorization According to the Hits Relative to the Cytotoxic Burst Lower Bound (CTBlb) and Verhaar Toxicity Classifications Based on In Vivo Effects in Aquatic Species
| Verhaar Toxicity Classification |
|||
|---|---|---|---|
| Specific Acting | Nonspecific Acting | ||
| Chemical group | Group A: hits< CTBlb in at least one noncytotoxicity assay | True positive (TP) | False positive (FP) |
| Group B: no hits < CTBlb or hits < CTBlb only in cytotoxicity assays | False negative (FN) | True negative (TN) | |
The Verhaar chemical classification system broadly categorizes chemicals by their mode of action based on observed general in vivo responses (in aquatic species). Recent work by Kienzler et al. (2017) evaluated various implementations of the Verhaar chemical classification: OECD QSAR toolbox, Toxtree, and Toxtree amended by additional classification rules encoded as a Konstanz Information Miner (KNIME). Specifics of the KNIME amendments are provided in Ellison et al. (2015). The Kienzler datasets were utilized in the DOR′ evaluation, although chemical coverage was limited (<300). The Verhaar classifications include narcosis toxicity (class I), less inert compounds (class II); unspecific reactivity (class III), and specific mechanism (class IV). Chemicals were classified as nonspecific acting if they were designated as class I. However, given that the CTB effect is, as yet, poorly understood and speculated to be the result of various nonspecific physiological disruptions in vitro, DOR′ calculations were also performed by defining nonspecific acting toxicity as class I and II or I, II, and III (narcotic + reactive).
Membrane partitioning as a determinant of chemical-assay hit data > CTBlb in cell-based assays
Chemicals that elicit toxicity primarily via narcosis are assumed to do so through achieving a chemically independent membrane concentration sufficient to disrupt cellular processes (Di Toro et al., 2000; McGrath et al., 2004; Wezel and Opperhuizen, 1995). Using the cell-based Tox21 assays evaluated by Fischer et al. (2017), the influence of the cellular membrane chemical concentration (EC50mem) on the relationship of a chemical-assay hit relative to the CTBlb (degree above or below the CTBlb) was examined. Fischer and colleagues calculated the EC50mem as (equation 9):
| (9) |
where fmem is the chemical fraction in the membrane, Vsystem and Vlipid, cell are the volumes of the in vitro system and the cellular lipids in the system, respectively (assuming all cellular lipids are membrane lipids). The difference between the AC50 of a chemical-assay hit combination and the chemical’s lower bound of its cytotoxic burst region (AC50-CTBlb) was regressed against the modeled proportion of chemical partitioned to cellular membranes (EC50mem/AC50, from equation 9).
RESULTS
Dataset and Definition of the Cytotoxic Burst
The slight discrepancies in the analysis of Judson et al. (2016) did not cause substantial differences in the final dataset or CTB definition (Supplementary Table 3). Specifically, use of 33 or 35 cytotoxicity assays to define the CTB for each chemical resulted in the same set of chemicals (1366 total) and nearly the same global MAD (0.2710 for 33 cytotoxicity assays, 0.2712 for 35). Similarly, employing a chemical selection criterion of “tested in >80% of (either 33 or 35) cytotoxicity assays” resulted only in four additional chemicals compared with the default criterion of “tested in >90% of cytotoxicity assays.”
A parallel evaluation using the ACC to statistically define the CTB likewise gave results similar to those based on the AC50, although the ACC generated a larger global MAD (0.370 vs. 0.271). Some assays had hits below the burst using one metric (AC50 or ACC) but not the other (Supplementary Table 4). In most cases, this difference was influenced by only one or two chemicals and, hence, would not impact assay prioritization. For the Attagene assays Pax6_CIS_dn and RORg_TRANS_dn, several chemicals caused hits at concentrations below their CTB thresholds using the ACC definition but not the AC50. Examination of the dose-response curves for these chemical-assay pairs shows either a trend of activity above cutoff at the lowest concentration with diminishing activity at higher concentrations, or gain-loss curve shapes, with the maximum response just above cutoff and at relatively low (e.g., < 1 μM) test concentrations (Supplementary Figure 1). Regardless, these assays were not in the top 90th percentile using the ACC evaluation.
Prioritization of Assay Targets for AOP Development
As an initial prioritization approach, assays were ranked according to their proportion of hits below the CTB threshold (<CTBlb) relative to the total number of chemicals tested within the assay. Because we were most concerned with results providing an indication of potential initiation of toxic effects (e.g., MIEs), assays detecting effects on xenobiotic biotransformation enzymes (e.g., some cytochrome P450s) as targets were excluded from this prioritization, as were assays with no identified molecular target. Use of either the ACC or AC50 to statistically define the CTB for each chemical resulted in qualitatively similar prioritizations (Supplementary Table 5). Of the targets represented by these assays, approximately half have already been identified for AOP development and have at least one relevant entry in the AOP-wiki (Table 2). However, the current prioritization suggests that investment in expanding the network of AOPs associated with these assays to provide a broader representation of potential outcomes for additional taxa, lifestages, impacted target organs, etc. may be useful. For example, two assays with the target NFE2L2 (NRF-2) were in the top 12 ranked assays. Two AOPs related to this target are under development (AOP: 61, 232; AOP-wiki), but both describe a similar pathway with the same outcome of liver steatosis. Neither considers the possible taxonomic lifestage nor sex applicability of the response. The remaining assays (Table 2; shown in bold) suggest novel molecular targets for AOP development. Prominent in this group is the translocator protein, TSPO, which is the target of two assays in the top 95th percentile. This protein has traditionally been described as a cholesterol transporter located on the mitochondrial membrane essential to steroidogenesis. However, a recent critical review of the evidence for its essentiality to steroidogenesis concluded TSPO is more likely to be important to immune function (Selvaraj et al., 2015).
Table 2.
Prioritized ToxCast Assay Targets for AOP Development as Identified by Assays Responding With the Greatest Number of Hits Below the Cytotoxic Burst Lower Bound (CTBlb; as Defined by the ACC) Relative to the Total Number of Chemicals Tested
| Assay | Hits< CTBlb/Chemicals | Intended Target (Gene) | Target Name | Extant AOP(s) (AOP-Wiki) |
|---|---|---|---|---|
| NCCT_TPO_GUA_dn | 0.327 | TPO | Thyroperoxidase | 42, 119, 159, 175 |
| ATG_PXRE_CIS_up | 0.327 | NR1I2 | Pregnane X receptor; PXR | 8, 11, 60 |
| ATG_ERE_CIS_up | 0.138 | ESR1 | Estrogen receptor 1 | 29, 30, 146, 165a, 167, 200 |
| NCCT_TPO_AUR_dn | 0.129 | TPO | Thyroperoxidase | 42, 119, 159, 175 |
| ATG_PXR_TRANS_up | 0.119 | NR1I2 | Pregnane X recpetor; PXR | 8, 11, 60 |
| ATG_NRF2_ARE_CIS_up | 0.110 | NFE2L2 | Nuclear factor erythroid 2-related factor 2; NRF2 | 61, 232 |
| ATG_ERa_TRANS_up | 0.106 | ESR1 | Estrogen receptor 1 | 29, 30, 146, 165a, 167, 200 |
| ACEA_T47D_80hr_Positive | 0.103 | ESR1 | Estrogen receptor 1 | 29, 30, 146, 165a, 167, 200 |
| TOX21_ERa_LUC_BG1_Agonist | 0.096 | ESR1 | Estrogen receptor 1 | 29, 30, 146, 165a, 167, 200 |
| ATG_PPARg_TRANS_up | 0.081 | PPARG | Peroxisome proliferator-activated receptor gamma | 72, 163, 206 |
| ATG_VDRE_CIS_up | 0.074 | VDR | Vitamin D receptor | |
| TOX21_ARE_BLA_agonist_ratio | 0.070 | NFE2L2 | Nuclear factor erythroid 2-related factor 2; NRF2 | 61, 232 |
| ATG_RXRb_TRANS_up | 0.070 | RXRB | Retinoid x receptor beta | |
| NVS_NR_hPPARg | 0.067 | PPARG | Peroxisome proliferator-activated receptor gamma | 72, 163, 206 |
| NVS_TR_hDAT | 0.063 | SLC6A3 | Sodium-dependent dopamine transporter | |
| NVS_NR_hAR | 0.060 | AR | Androgen receptor | 19, 23, 111, 117 |
| OT_ER_ERaERb_0480 | 0.056 | ESR1, ESR2 | Estrogen receptor 1 and 2 | 29, 30, 146, 165a, 167, 200 |
| NVS_NR_hER | 0.056 | ESR1 | Estrogen receptor 1 | 29, 30, 146, 165a, 167, 200 |
| OT_AR_ARELUC_AG_1440 | 0.056 | AR | Androgen receptor | 19, 23, 111, 117 |
| ATG_Ahr_CIS_up | 0.055 | AHR | Aryl hydrocarbon receptor | 21, 41, 57, 131 150 |
| NVS_MP_rPBR | 0.055 | Tspo | Peripheral benzodiazepine receptor; translocator protein | |
| ATG_PPRE_CIS_up | 0.053 | PPARA, PPARD, PPARG | Peroxisome proliferator-activated receptor, alpha, delta and gamma | 6, 18, 37, 51, 72, 163, 166, 206 |
| OT_ER_ERbERb_0480 | 0.053 | ESR2 | Estrogen receptor 2 | 29a, 30a, 146a, 165a, 167a, 200a |
| BSK_LPS_PGE2_down | 0.052 | PTGER2 | Prostaglandin E receptor 2 | |
| TOX21_AhR_LUC_Agonist | 0.051 | AHR | Aryl hydrocarbon receptor | 21, 41, 57, 131 150 |
| NVS_NR_hGR | 0.047 | NR3C1 | Glucocorticoid receptor | 14, 64, 66, 71 |
| NVS_MP_hPBR | 0.047 | TSPO | Peripheral benzodiazepine receptor; translocator protein | |
| OT_ER_ERaERb_1440 | 0.046 | ESR1, ESR2 | Estrogen receptor 1 and 2 | 29, 30, 146, 165a, 167, 200 |
| OT_ER_ERbERb_1440 | 0.046 | ESR2 | Estrogen receptor 2 | 29a, 30a, 146a, 165a, 167a, 200a |
| NVS_NR_mERa | 0.045 | Esr1 | Estrogen receptor 1 | 29, 30, 146, 165a, 167, 200 |
| NVS_OR_gSIGMA_NonSelective | 0.042 | Sigmar1 | Sigma nonopioid intracellular receptor 1 | |
| NVS_NR_cAR | 0.042 | AR | Androgen receptor | 19, 23, 111, 117 |
| NVS_ENZ_hCK2a2b2 | 0.040 | CSNK2A1 | Casein kinase II alpha 1 | |
| OT_ERa_EREGFP_0120 | 0.039 | ESR1 | Estrogen receptor 1 | 29, 30, 146, 165a, 167, 200 |
| NVS_NR_bER | 0.038 | ESR1 | Estrogen receptor 1 | 29, 30, 146, 165a, 167, 200 |
| ATG_DR5_CIS_up | 0.038 | RARA, RARB, RARG | Retinoic acid receptors alpha, beta, gamma | |
| BSK_LPS_TNFa_down | 0.038 | TNF | Tumor necrosis factor | |
| OT_ERa_EREGFP_0480 | 0.038 | ESR1 | Estrogen receptor 1 | 29, 30, 146, 165a, 167, 200 |
| BSK_3C_HLADR_down | 0.037 | HLA-DRA | Major histocompatability complex, class II, DRalpha | |
| NVS_ENZ_hPDE5 | 0.037 | PDE5A | Phosphodiesterase 5A | |
| NVS_IC_rNaCh_site2 | 0.036 | Scn1a | Sodium-voltage gated channel alpha subunit 1; Nav1.1 | 197 |
| NVS_NR_hPR | 0.035 | PGR | Progesterone receptor | |
| NVS_NR_hCAR_Antagonist | 0.034 | NR1I3 | Constitutive androstane receptor (CAR) | 58, 107 |
| NVS_TR_hNET | 0.033 | SLC6A2 | Norepinephrine transporter | |
| NVS_ENZ_rMAOBC | 0.033 | Maob | Monoamine oxidase B | |
| OT_ER_ERaERa_0480 | 0.033 | ESR1 | Estrogen receptor 1 | 29, 30, 146, 165a, 167, 200 |
| NVS_GPCR_h5HT7 | 0.032 | HTR7 | 5-Hydroxytryptamine receptor 7 | 98a, 97a, 195a, 204a, 203a |
| NVS_ENZ_rMAOAC | 0.031 | Maoa | Monoamine oxidase A | |
| BSK_SAg_Eselectin_down | 0.031 | SELE | Eselectin; endothelial leukocyte adhesion molecule 1 | |
| ATG_PBREM_CIS_up | 0.031 | NR1I3 | Constitutive androstane receptor (CAR) | 58, 107 |
| BSK_hDFCGF_CollagenIII_down | 0.030 | COL3A1 | Collagen type III alpha 1 chain | |
| BSK_hDFCGF_IP10_down | 0.030 | CXCL10 | Chemokine ligand 10 | |
| BSK_hDFCGF_PAI1_down | 0.030 | SERPINE1 | Serpin peptidase inhibitor, clade E | |
| ATG_Oct_MLP_CIS_up | 0.029 | POU2F1 | POU class 2 homeobox | |
| NVS_ENZ_oCOX2 | 0.029 | PTGS2 | Cyclooxygenase 2 | 28, 63, 100–103, 228 |
| TOX21_Aromatase_Inhibition | 0.029 | CYP19A1 | Aromatase | 7, 25, 153 |
| BSK_CASM3C_IL6_down | 0.029 | IL6 | Interleukin 6 | |
| NVS_GPCR_hM4 | 0.028 | CHRM4 | Cholinergic receptor muscarinic 4 | |
| OT_ER_ERaERa_1440 | 0.028 | ESR1 | Estrogen receptor 1 | 29, 30, 146, 165a, 167, 200 |
| ATG_SREBP_CIS_up | 0.028 | SREBF1 | Sterol regulatory element binding transcription factor | |
| NVS_ENZ_oCOX1 | 0.027 | PTGS1 | Cyclooxygenase 1 | 177, 227 |
The top 95th and 90th percentiles of assays are represented along with relevant AOPs in the AOP-Wiki (https:aopkb.org), as of October 2017. Intended targets and target names in bold font indicate potential novel MIE targets for AOP development.
AOP is related to the intended target, but the intended target is not specified in the MIE.
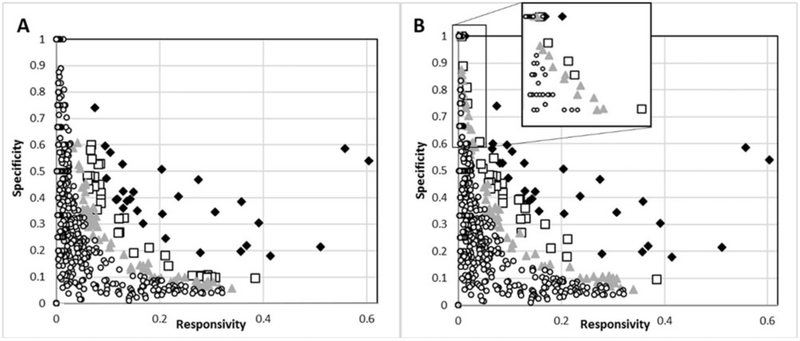
An alternative approach to prioritizing the ToxCast assays for AOP development comes from the field of medical testing. A DOR is an evaluation of the effectiveness of a diagnostic test. From a medical perspective, this calculation requires knowledge of the test’s sensitivity (ability to detect true positives) and specificity (ability to reject true negatives), but an analogous evaluation of the ToxCast assay data can be performed using assay sensitivity (ability to detect pathway-specific hits) and responsivity. The results of the DOR approach to prioritization are very similar to the above evaluation (Supplementary Table 6), except assays with high sensitivity (equation 4) but low responsivity (equation 5) are ranked higher (Figure 2B). Using this approach, additional gene targets in the top 90th percentile included CXCL8, CSNK1D, PTPN2, NR3C2, IRAK4, MapKapk2, and CASP2, none of which have currently been linked to relevant apical outcomes using the AOP framework.
Figure 2.
Distribution of ToxCast assays with biomolecular targets according to assay responsivity (hits/chemicals tested) versus sensitivity (hits < CTBlb/hits). Assays were ranked according to hits < CTBlb/chemicals tested (A) or diagnostic odds ratio (B): top 95th percentile (diamond), 95th > percentile > 90th (square), 90th > percentile >80th (triangle), and < 80th percentile (circle). The two approaches prioritized the assays similarly, although the diagnostic odds ratio favored highly specific, less sensitive assays (inset, B).
Assay targets identified in the top 90th percentile from both ranking approaches (equations 3 and 6; ACC dataset) resulted in 50 unique priority targets (Supplementary Table 7). Examination of these targets indicated several that have a similar function (e.g., MAOA and MAOB, SLC6A2 and SLC6A3) or that operate as part of a common pathway (e.g., inflammatory response: CASP2, SELE, IRAK4, MAPKAP2, TNF, IL6, CXCL8, CXCL10; Table 3). DAVID classification organized the targets into seven functional groups: nuclear receptors (group 1: AR, PGR, PPARA, PPARD, PPARG, NR1I2, NR1I3, NR3C1, ESR1, ESR2, NR3C2, RARA, RARB, RARG, RXRB), cyclooxygenases/peroxidases (group 2: PTGS1, PTGS2, TPO), kinases (group 3: MAPKAPK2, CSNK2A3, IRAK4, CSNK2A1), G-protein-coupled receptors (group 4: CHRM2, CHRM4, HTR7, PTGER2), chemokines (group 5: CXCL8, CXCL10), neurotransmitter transporters (group 6: SLC6A2, SLC6A3), and monoamine oxidases (group 7: MAOA, MAOB). In addition, some targets were not grouped by the DAVID analysis (POUT2F1, AHR, CSNK1D, CASP2, COL3A1, CYP19A1, IL6, HLA-DRA, NFE2L2, PDE5A, PTPN2, SELE, SERPINE1, SIGMAR1, SCN1A, SREBF1, TSPO, TNF). Although the assay ATG_VDRE_CIS_up was ranked highly in both prioritizations, its intended target, the vitamin D receptor (VDR), was excluded from the functional classification as well any further consideration for AOP development because the assay is not specific to VDR activation, but likely reflects PXR or FXR binding to the VDR response element (Supplementary Data).
Table 3.
General Function and Classifications of the Novel Molecular Targets Identified in ToxCast Assays Prioritized by Their Response Below the CTB
| Intended Target gene | Target Name | Description | General Biological Category |
|---|---|---|---|
| RXRB | Retinoid X receptor beta | This nuclear receptor is involved in mediating the effects of retinoic acid. This receptor forms homodimers with the retinoic acid, thyroid hormone, and vitamin D receptors, increasing both DNA binding and transcriptional function on their respective response elements. The gene lies within the major histocompatibility complex class II region on chromosome 6 | Nuclear receptor involved in hormonal regulation |
| RARA, RARB, RARG | Retinoic acid receptor alpha, beta, gamma | The nuclear retinoic acid receptors dimerize with RXR and bind hormone response elements (retinoic acid response elements), repressing transcription of target genes. Upon ligand binding (e.g., retinoic acid) to the RAR, transcription is activated. RARs are involved in various biological processes, including limb bud development, skeletal growth, and matrix homeostasis. RARA has been implicated in regulation of development, differentiation, apoptosis, granulopoeisis, and transcription of clock genes. RARB is thought to limit growth of many cell types | |
| PGR | Progesterone receptor | The progesterone receptor is a member of the steroid receptor superfamily and mediates the physiological effects of progesterone, which plays a central role in reproductive events associated with the establishment and maintenance of pregnancy | |
| NR3C2 | Mineralocorticoid receptor | The mineralocorticoid receptor mediates aldosterone actions on salt and water balance within restricted target cells. This ligand-dependent transcription factor binds to the mineralocorticoid response element. Mutations in this gene cause autosomal dominant pseudohypoaldosteronism type I, a disorder characterized by urinary salt wasting. Defects in this gene are also associated with early onset hypertension with severe exacerbation in pregnancy | |
| TSPO | Translocator protein; peripheral benzodiazepine receptor | The translocator protein regulates the transport of cholesterol into mitochondria to permit the initiation of steroid hormone synthesis. It also interacts with some benzodiazepines | Transporter involved in hormone regulation |
| HTR7 | 5-Hydroxytryptamine receptor 7 | The serotonin receptor 7 is a G protein-coupled receptor, which mediates the effects of the neurotransmitter serotonin (5-hydroxytryptamine) on cognition and behavior. Mutations in this gene are associated with autistic disorder and other neuropsychiatric disorders | Receptor involved in neurotransmission |
| SIGMAR1 | Sigma nonopioid intracellular receptor 1 | This receptor interacts with a variety of psychotomimetic drugs, including cocaine and amphetamines, and is believed to play an important role in the cellular functions of various tissues associated with the endocrine, immune, and nervous systems. Mutations in this gene have been associated with juvenile amyotrophic lateral sclerosis 16 | |
| CHRM2 | Cholinergic receptor muscarinic 2 | This receptor is a G protein-coupled receptor which binds acetylcholine. Activated muscarinic receptors are inhibitory regulators of acetylcholine transmission. Muscarinic cholinergic receptor 2 is involved in mediation of bradycardia and a decrease in cardiac contractility | |
| CHRM4 | Cholinergic receptor muscarinic 4 | This receptor is a G protein-coupled receptor which binds acetylcholine. Activated muscarinic receptors are inhibitory regulators of acetylcholine transmission and have many effects in the central and peripheral nervous system. Muscarinic acetylcholine receptors also influence dopaminergic neurotransmission | |
| SLC6A2 | Norepinephrine transporter | This gene encodes a member of the sodium: neurotransmitter symporter family. This protein is responsible for reuptake of norepinephrine into presynaptic nerve terminals and is a regulator of norepinephrine homeostasis. Mutations in this gene cause orthostatic intolerance, a syndrome characterized by lightheadedness, fatigue, altered mentation and syncope | Transporter involved in neurotransmission |
| SLC6A3 | Sodium-dependent dopamine transporter | The sodium-dependent dopamine transporter moves the neurotransmitter dopamine from the synaptic cleft back into the cytosol. Genetic polymorphisms are associated with idiopathic epilepsy, attention-deficit hyperactivity disorder, dependence on alcohol and cocaine, susceptibility to Parkinson disease and protection against nicotine dependence | |
| MAOA | Monoamine oxidase A | Monoamine oxidase A is an enzyme, which catalyzes the oxidative deamination of amines, such as dopamine, norepinephrine, and serotonin. Mutation of this gene results in Brunner syndrome. This gene has also been associated with a variety of other psychiatric disorders, including antisocial behavior | Enzyme involved in neurotransmission |
| MAOB | Monoamine oxidase B | Monoamine oxidase B is an enzyme located in the mitochondrial outer membrane. It catalyzes the oxidative deamination of biogenic and xenobiotic amines and plays an important role in the metabolism of neuroactive and vasoactive amines in the central nervous sysytem and peripheral tissues. This protein preferentially degrades benzylamine and phenylethylamine | |
| NFE2L2 | Nuclear factor, erythroid 2 like 2; NRF2 | This transcription factor is a member of a small family of basic leucine zipper (bZIP) proteins. The encoded transcription factor regulates genes, which contain antioxidant response elements (ARE) in their promoters; many of these genes encode proteins involved in response to injury and inflammation which includes the production of free radicals | Transcription factor involved in antioxidant and inflammatory response |
| CSNK2A1 | Casein kinase II alpha 1 | Casein kinase II is a serine/threonine protein kinase that phosphorylates acidic proteins such as casein. It is involved in various cellular processes, including cell cycle control, apoptosis, and circadian rhythm. The kinase exists as a tetramer and is composed of an alpha, an alpha-prime, and two beta subunits. The alpha subunits contain the catalytic activity whereas the beta subunits undergo autophosphorylation | Kinase involved in cell cycle regulation |
| CSNK1D | Casein kinase 1 delta | Members of the casein kinase I family have been implicated in the control of cytoplasmic and nuclear processes, including DNA replication and repair. Casein kinase II alpha 1 may also be involved in the regulation of apoptosis, circadian rhythm, microtubule dynamics, chromosome segregation, and p53-mediated effects on growth | |
| TNF | Tumor necrosis factor | Tumor necrosis factor is a multifunctional proinflammatory cytokine mainly secreted by macrophages. It binds the receptors TNFRSF1A/TNFR1 and TNFRSF1B/TNFBR. This cytokine is involved in the regulation of a wide spectrum of biological processes including cell proliferation, differentiation, apoptosis, lipid metabolism, and coagulation, and has been implicated in a variety of diseases, including autoimmune diseases, insulin resistance, and cancer | Cytokine involved in inflammatory response |
| IL6 | Interleukin 6 | This cytokine functions in inflammation, the maturation of B cells, and is an endogenous pyrogen capable of inducing fever in people with autoimmune diseases or infections. The protein is primarily produced at sites of acute and chronic inflammation, where it is secreted into the serum and induces a transcriptional inflammatory response through interleukin 6 receptor, alpha. The gene is implicated in a wide variety of inflammation-associated disease states, including susceptibility to diabetes mellitus and systemic juvenile rheumatoid arthritis | |
| CXCL8 | C-X-C motif chemokine ligand 8; interleukin 8 | This chemokine is in the CXC family and is one of the major mediators of the inflammatory response. Secreted by several cell types, it functions as a chemoattractant, and is also a potent angiogenic factor. It is also believed to play a role in the pathogenesis of bronchiolitis, a common respiratory tract disease caused by viral infection | |
| CXCL10 | Chemokine ligand 10 | This chemokine belongs to the CXC subfamily and is a ligand for the receptor CXCR3. Binding of this protein to CXCR3 results in pleiotropic effects, including stimulation of monocytes, natural killer and T-cell migration, and modulation of adhesion molecule expression | |
| PDE5A | Phosphodiesterase 5A | Phosphodiesterase 5A is a member of the cyclic nucleotide phosphodiesterase family and specifically hydrolyzes cGMP to 5’-GMP. It is involved in the regulation of intracellular concentrations of cyclic nucleotides and is important for nitric-oxide-mediated smooth muscle relaxation. Important in cardiovascular muscle relaxation and penile erection | Phosphodiesterase involved in signal transduction |
| SELE | Eselectin; endothelial leukocyte adhesion molecule 1 | Eselectin is found in cytokine-stimulated endothelial cells and is thought to be responsible for the accumulation of blood leukocytes at sites of inflammation by mediating the adhesion of cells to the vascular lining. As a member of the selectin family of cell adhesion molecules, it participates in the interaction between leukocytes and the endothelium and appears to be involved in the pathogenesis of atherosclerosis | Protein involved in cell adhesion and inflammatory response |
| COL3A1 | Collagen type III alpha 1 chain | The pro-alpha1 chains of type III collagen contribute to the fibrillar collagen that is found in extensible connective tissues such as skin, lung, uterus, intestine and the vascular system, and frequently in association with type I collagen. Mutations in this gene are associated with Ehlers–Danlos syndrome type IV, and with aortic and arterial aneurysms | Protein involved in connective tissue |
| SERPINE1 | Serpin peptidase inhibitor, clade E | This serine proteinase inhibitor (serpin) is the principal inhibitor of tissue plasminogen activator (tPA) and urokinase (uPA), and hence is an inhibitor of fibrinolysis. Defects in this gene are the cause ofplasminogen activator inhibitor-1 deficiency (PAI-1 deficiency), and high concentrations of the gene product are associated with thrombophilia | Inhibitor inolved the immune response |
| POU2F1 | POU class 2 homeobox | This transcription factor was among the first identified members of the POU transcription factor family. Members of this family contain the POU domain, a 160-amino acid region necessary for DNA binding to the octameric sequence ATGCAAAT. POU2F1 appears to be involved in lens and nasal sensory development | Transcription factor involved in cell cycle and development |
| SREBF1 | Sterol regulatory element binding transcription factor | This transcription factor binds to the sterol regulatory element-1 (SRE1), which flanks the low-density lipoprotein receptor gene and some genes involved in sterol biosynthesis. The protein is synthesized as a precursor that is attached to the nuclear membrane and endoplasmic reticulum. Following cleavage, the mature protein translocates to the nucleus and activates transcription by binding to the SRE1. Sterols inhibit the cleavage of the precursor, and the mature nuclear form is rapidly catabolized, thereby reducing transcription | Transcription factor involved in sterol/cholesterol regulation |
| PTPN2 | Tyrosine-protein phosphatase nonreceptor type 2 | This signaling molecule is a member of the protein tyrosine phosphatase (PTP) family, which regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. Epidermal growth factor receptor and the adaptor protein Shc were reported to be substrates of this PTP, which suggested it plays a role in growth factor-mediated cell signaling | Signaling molecule involved in cell cycle regulation |
| IRAK4 | Interleukin-1 receptor-associated kinase 4 | This kinase activates NF-kappaB in both the Toll-like receptor (TLR) and T-cell receptor (TCR) signaling pathways. The protein is essential for most innate immune responses. Mutations in this gene result in IRAK4 deficiency and recurrent invasive pneumococcal disease | Kinase involved in inflammatory response |
| MAPKAP2 | Mitogen-activated protein kinase-activated protein kinase 2 | This Ser/Thr protein kinase is regulated through direct phosphorylation by p38 MAP kinase. In conjunction with p38 MAP kinase, this kinase is known to be involved in many cellular processes including stress and inflammatory responses, nuclear export, gene expression regulation, and cell proliferation. Heat shock protein HSP27 is one substrate of this kinase. | |
| CASP2 | Caspase 2 | Caspases (cystein-asparctic acid proteases) mediate cellular apoptosis through the proteolytic cleavage of specific protein substrates. Caspase 2 may function in stress-induced cell death pathways, cell cycle maintenance, and the suppression of tumorigenesis. Increased expression of this gene may play a role in neurodegenerative disorders including Alzheimer’s disease, Huntington’s disease, and temporal lobe epilepsy | Protease involved in cell death |
Chemical Bioavailability as a Determinant of Assay Prioritization
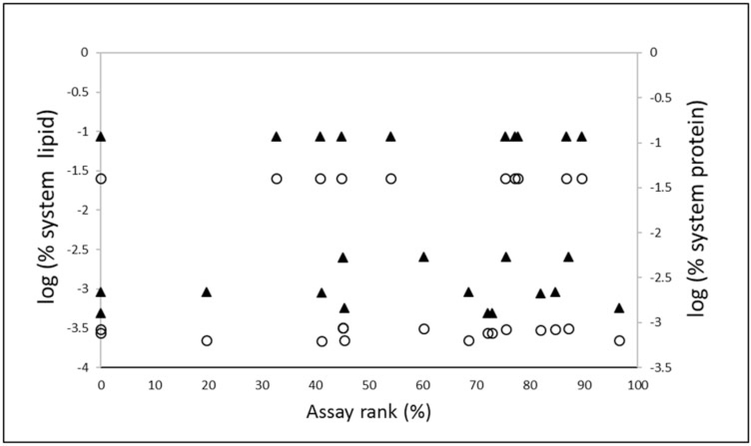
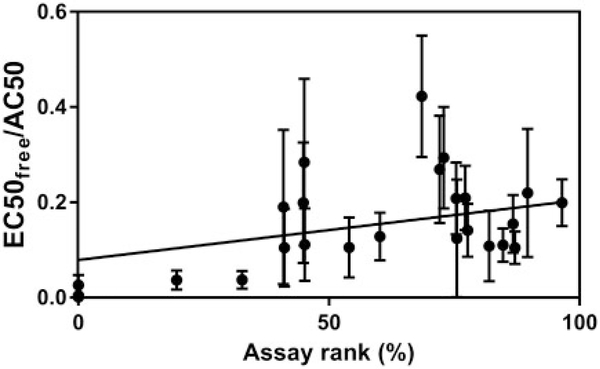
Assay composition and chemical binding were determined by Fisher et al. (2017) for a subset of Tox21 assays with a nearly even distribution across the assay prioritization of hits < CTBlb/total chemicals tested (Figure 3; Supplementary Figure 2). Three assays had no hits < CTBlb and were assigned a rank of 0%. No relationship was apparent between either assay lipid or protein content and assay rank (Figure 3). Further, regression of assay rank against EC50free/AC50 resulted in a positive trend, perhaps suggesting greater chemical bioavailability in higher ranked assays, but the relationship was not significant (Figure 4).
Figure 3.
Assay rank versus assay content in terms of % protein (filled triangles) and % lipid (open circles). Assay rank was determined according to the number of chemical-assay hits < CTBlb/chemicals tested. Percent protein and lipid (v/v) were determined by Fischer et al. (2017) for 25 Tox21 assays and are presented on a logarithmic (base 10) scale. Neither system % lipid nor % protein appeared to influence the assay rank.
Figure 4.
Regression of assay rank (determined as hits < CTBlb/chemicals tested) versus EC50free/AC50 for 25 TOX21 assays modeled by Fischer et al. (2017). Toxicity data for ARE_BLA_agonist_viability were not provided. The slope was not significantly different from zero (95% confidence intervals: −0.01953 to 0.2712), suggesting that chemical bioavailability was not a determinant of the ranking.
Utility of CTB for Mode of Action Classification
An evaluation of the utility of the CTB analysis for differentiating chemicals with nonspecific modes of action (i.e., Verhaar categories I, II, or III) from specifically acting (Verhaar category IV) was conducted using a DOR′ approach (equation 8). The results (Supplementary Table 8) suggest that chemicals which elicited hits < CTBlb in noncytotoxicity assays (Group A) were more often classified as specific acting (Verhaar class IV) compared with chemicals which either never elicited a hit < CTBlb or only did so in the cytotoxicity assays (Group B). Group B chemicals were more often classified as Verhaar narcotics (class I). Predictive accuracy was highest when compared with the Toxtree Verhaar classification with KNIME correction (DOR′ = 12.5) and diminished with the unadulterated Toxtree and the OECD QSAR toolbox Verhaar classifications (DOR′ = 7.8 and 4.6, respectively). When classifications of less inert and reactive chemicals (classes II and III) were combined with class I as nonspecific acting chemicals, there was no clear predictive relationship (DOR′ range: 1.7–0.8).
Given that the above analysis resulted in positive DOR′s only when using the strictest classification of nonspecific toxicity (narcosis; class I), an additional evaluation of cell membrane concentration versus the nominal hit concentration relationship to the CTB threshold (AC50-CTBlb) was performed using the Fischer et al (2017) dataset (i.e., a subset of 23 Tox21 assays). A positive relationship is expected between increasing AC50 concentrations and EC50mem because the latter is dependent on the reported AC50 (equation 5). Indeed, this is the case for chemical-assay pairs with hits above the CTBlb (Supplementary Figure 3). However, the EC50mem/AC50 reflects the chemical (fmem) and assay (Vsystem/Vlipid, cell)-specific attributes which control the modeled chemical partitioning to the cellular membrane. The regression of (AC50-CTBlb) against (EC50mem/AC50) did not result in a significant relationship, suggesting that the CTB definition may not provide a robust basis for identifying baseline toxicity (i.e., chemical disruption of cellular membranes) from ToxCast hit data (Figure 5).
Figure 5.
Regression of EC50mem/AC50 and the nominal hit concentration relation to the burst threshold (AC50-CTBlb) for chemical assay data falling within the CTB (AC50-CTBlb > 0). No significant correlation between the factors driving chemical partitioning to the cellular membrane (EC50mem/AC50) and the relationship of the hit from the CTB threshold (CTBlb) was determined, suggesting that even in cell-based assays, the CTB effect is more complicated than simple baseline toxicity. Slope regression was 0.0016 with a 95% confidence interval of −0.0020 to 0.0051.
DISCUSSION
Prioritization of ToxCast Assay-Related AOP Development
The primary aim of the current study was to prioritize ToxCast assay targets for AOP development based on the assumption that assays for which larger proportions of responding chemicals were doing so at concentrations well below the CTB would be higher priorities from a predictive toxicology perspective. Similar targets were identified regardless of the metric used to define the CTB (AC50 or ACC). This consistent outcome supports the robustness of the CTB definition as well as our target rankings. Further, the two different approaches to assay target prioritization yielded similar results. Ranking by the proportion of hits < CTBlb relative to the total number of chemicals tested highlighted assays that, presumably, reflect a target susceptible to perturbation by a greater chemical space. The DOR-type approach resulted in a similar prioritization, but placed a greater value on highly sensitive assays.
From the combined prioritization approaches, several functional groups of potentially important targets were identified. Although the enzymes, transporters, and receptors can readily be understood as targets of chemical perturbation reflecting the MIE in a AOP, other gene targets indicate a more generalized response to insult (e.g., antioxidant, inflammatory, or immune effects). In these instances, the assays likely measure a downstream consequence of a chemical perturbation, that is, a KE that does not result from a direct chemical interaction with the target. Nonetheless, when appropriately linked to downstream apical outcomes, the information could be useful for hazard prediction. For example, many of the targets in the top 90th percentile (Table 2) share a role in inflammatory response. Those assays detecting changes in inflammatory factors at concentrations below the CTBlb are nearly all analyzed in the negative direction (BSK_LPS_TNFa_down, BSK_hDFCGF_IP10_down, BSK_CASM3C_IL6_down, BSK_SAg_Eselectin_down). This trend suggests that, together, these assays may effectively screen chemicals with anti-inflammatory effects. Although anti-inflammatory properties may be useful from a therapeutic perspective, in some cases, such effects may be maladaptive. For example, IL6 and TNFa have known neuroprotective effects, suggesting their downregulation could result in an adverse effect (Barger et al., 1995; Carlson et al., 1999).
The AC50 or ACC values provided by the ToxCast pipeline are based on nominal chemical concentrations only. No measurements of free versus bound fraction of the administered chemicals are available. Consequently, there is some question as to whether differences in chemical bioavailability across assays may influence the number of hit responses at concentrations below the burst. To assess this possibility, we employed a dataset described by Fischer et al. (2017) which provides modeled free and bound chemical concentrations for 26 cell-based Tox21 assays tested with 100 chemicals. Their dataset spanned our assay ranking (Supplementary Table 5) from 0 to 97% with fairly even distribution. Assay lipid and protein content, which presumably could impact the bioavailability of all test chemicals to varying degrees, were not found to be correlated with assay rank. Likewise, chemical and assay-specific determinants of chemical-free fraction were not a significant factor affecting prioritization. These results support the hypothesis that the hits observed below the CTBlb are, indeed, pathway-specific and the corresponding chemical concentrations are probably not artificially potent because of low binding relative to other, lower-ranked assays.
CTB and Mode of Action Classification
A natural extension of using the CTB to identify assays detecting pathway-specific chemical effects is to consider whether chemicals eliciting nonspecific toxicity can likewise be identified, and how well these in vitro effects translate to in vivo observations. This possibility was suggested by the results of Judson et al. (2016) that showed that chemicals designed to interact with specific biological targets (e.g., pesticides, pharmaceuticals) and endogenously active compounds (e.g., steroid hormones) accounted for a disproportionate number of the chemicals eliciting responses below the CTB threshold. In contrast, chemicals associated with industrial activities and nonbiomedical commercial products accounted for greater fractions of those responding only within the CTB concentration range.
In the present study, the DOR′ evaluation between CTB-defined chemical groups and Verhaar toxicity classifications suggested that the CTB definition may be able to distinguish chemicals which cause specific-acting and nonspecific toxicity in vivo. However, the dataset was too small to be conclusive, and the DOR′ was at best 12.5—using the strictest, and arguably most favorable scenario (comparing only class I and class IV compounds of the Toxtree implementation with KNIME post-processing). DOR′ evaluations using other Verhaar implementations as well as other classification schemes such as the Assessment Tool for the Evaluation of Risk (ASTER, USEPA, 2007) or Mode of Action-Aquatic Toxicity Database (MOATox, Barron et al. [2015]) were not as favorable (data not shown). Likewise, our assessment of the Tox21 cell-based assays included in the Fischer et al. (2017) study, did not support that chemical disruption of lipid membranes (narcosis) was correlated with chemical assay hit position relative to the CTB threshold. This finding suggests that the CTB effect, even when limited to cell-based assays, is more complicated than cellular membrane disruption alone.
The limited capability to extrapolate the in vitro hit data to in vivo observations of specific and nonspecific toxicity is not unexpected. For example, biotransformation is known to greatly impact in vitro to in vivo extrapolations and the majority of the current ToxCast in vitro assays likely do not actively metabolize xenobiotics (Dix et al., 2007; Kavlock et al., 2012). The omission of relevant toxicity targets within the ToxCast suite of assays also may contribute to the limited predictive utility of the CTB definition. However, the greatest factor limiting the in vitro to in vivo correlation attempted in the present work may be discrepancies within and limitations of the Verhaar classification scheme itself. In the study by Kienzler et al. (2017), a curated toxicity dataset for 3448 chemicals with confirmed Chemical Abstract Service (CAS) and Simplified molecular-input line entry systems (SMILES) identifiers were evaluated for classification concordance across different mode of action prediction tools. Approximately half of the chemicals were outside of the domain of the Verhaar classification or were not recognized by the implementation rules. Of the remaining chemicals, 38% were classified differently using various Verhaar rule implementations (i.e., OECD QSAR toolbox vs. Toxtree). Further, the proportion of chemicals in the dataset classified as narcosis chemicals by Verhaar (27%) was substantially lower than those determined using MOAtox (50%) or ASTER (59%). These kinds of inconsistencies among in vivo classifications of chemical toxicity diminish confidence in using the data to evaluate the veracity of in vitro chemical predictions.
CONCLUSIONS
Adverse outcome pathway development has been recognized as one way to support the use of HTS for hazard prediction and prioritization of testing resources. The AOP framework is an increasingly important component of predictive approaches to risk assessment, both in tiered testing strategies, and integrated approaches to testing and assessment (IATA; Patlewicz et al. [2014]; Tollefsen et al. [2014]). However, the development of formal AOPs is a resource-intensive endeavor (Fay et al., 2017). The current work used the CTB definition to identify a subset of ToxCast assay targets for which investment in AOP development may have the greatest potential to enhance current hazard prediction approaches. The use of the CTB as a means to evaluate ToxCast assays for pathway-specific activity was supported both by the resilience of the dataset and assay ranking to various approaches as well as our analysis indicating interassay differences in chemical bioavailability was not a likely influence on the prioritization. Overall, these results contribute to on-going efforts to develop both a systematic knowledge-base and analytical tools to better inform how ToxCast HTS data can be used in twenty-first century regulatory decision making. In conclusion, the results of this study can inform future use of ToxCast data, enhance the interpretation of HTS bioactivity and guide the development of needed AOPs.
Supplementary Material
ACKNOWLEDGMENTS
This article has been reviewed in accordance with USEPA guidelines, but does not necessarily reflect the views or policies of the Agency. We thank Richard Judson for comments on an earlier version of the article.
FUNDING
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Footnotes
Disclaimer: This document has been subjected to review by the National Health and Environmental Effects Research Laboratory, U.S. Environmental Protection Agency and approved for publication. Approval does not signify that the contents reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
SUPPLEMENTARY DATA
Supplementary data are available at Toxicological Sciences online.
REFERENCES
- Ankley GT, Bennet RS, Erickson RJ, Hoff DJ, Hornung MW, Johnson RD, Mount DR, Nichols JW, Russom CL, Schmieder, et al. (2010). Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem 29, 730–741. [DOI] [PubMed] [Google Scholar]
- Attene-Ramos MS, Miller N, Huang R, Michael S, Itkin M, Kavlock RJ, Austin CP, Shinn P, Simeonov A, Tice RR, et al. (2013). The Tox21 robotic platform for the assessment of environmental chemicals-from vision to reality. Drug Discov. Today 18, 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal-Price A, Crofton KM, Sachana M, Shafer TJ, Behl M, Forsby A, Hargreaves A, Landesmann B, Lein PJ, Louisse J, et al. (2015). Putative adverse outcome pathways relevant to neurotoxicity. Crit. Rev. Toxicol 45, 83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barger SW, Horster D, Furukawa K, Goodman Y, Krieglstein J, and Mattson MP (1995). Tumor necrosis factors alpha and beta protect neurons against amyloid betapeptide toxicity: evidence for involvement of a kappa B-binding factor and attenuation of peroxide and Ca2+ accumulation. Proc. Natl. Acad. Sci. U. S. A 92, 9328–9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barron M, Lilavois C, and Martin T (2015). MOAtox: a comprehensive mode of action and acute aquatic toxicity database for predictive model development. Aquat. Toxicol 161, 102–107. [DOI] [PubMed] [Google Scholar]
- Blackwell BR, Ankley GT, Corsi SR, DeCicco LA, Houck KA, Judson RS, Li S, Martin MT, Murphy E, Schroeder AL, et al. (2017). An “EAR” on environmental surveillance and monitoring: A case study on the use of exposure–activity ratios (EARs) to prioritize sites, chemicals, and bioactivities of concern in Great Lakes waters. Environ. Sci. Technol 51,8713–8724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne P, Noyes PD, Casey WM, and Dix DJ (2017). Application of adverse outcome pathways to US EPA’s Endocrine Disruptor Screening Program. Environ. Health Perspect 96001, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson NG, Wieggel WA, Chen J, Bacchi A, Rogers SW, and Gahring LC (1999). Inflammatory cytokines IL-1α, IL-1β, IL-6, and TNF-α impart neuroprotection to an excitotoxin through distinct pathways. j. Immunol 163, 3963–3968. [PubMed] [Google Scholar]
- Delrue N, Sachana M, Sakuratani Y, Gourmelon A, Leinala E, and Diderich R (2016). The adverse outcome pathway concept: A basis for developing regulatory decision-making tools. Alternat. Lab. Anim 44, 417–429. [DOI] [PubMed] [Google Scholar]
- Di Toro DM, McGrath JA, and Hansen DJ (2000). Technical basis for narcotic chemicals and polycyclic aromatic hydrocarbon criteria. I. Water and tissue. Environ. Toxicol. Chem 19, 1951–1970. [DOI] [PubMed] [Google Scholar]
- Dix DJ, Houck KA, Judson RS, Kleinstreuer NC, Knudsen TB, Martin MT, Reif DM, Richard AM, Shah I, Sipes NS, et al. (2012). Incorporating biological, chemical, and toxicological knowledge into predictive models of toxicity. Toxicol. Sci 130,440–441. [DOI] [PubMed] [Google Scholar]
- Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, and Kavlock RJ (2007). The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci 95, 5–12. [DOI] [PubMed] [Google Scholar]
- Ellison CM, Madden JC, Cronin MTD, and Enoch SJ (2015). Investigation of the Verhaar scheme for predicting acute aquatic toxicity: improving predictions obtained from Toxtree ver. 2.6. Chemosphere 139, 146–154. [DOI] [PubMed] [Google Scholar]
- Fay KA, Villeneuve DL, LaLone CA, Song Y, Tollefsen KE, and Ankley GT (2017). Practical approaches to adverse outcome pathway development and weight-of-evidence evaluation as illustrated by ecotoxicological case studies. Environ. Toxicol. Chem 36,1429–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filer DL, Kothiya P, Setzer RW, Judson RS, and Martin MT (2016). tcpl: the ToxCast pipeline for high-throughput screening data. Bioinformatics. 33, 618–620. [DOI] [PubMed] [Google Scholar]
- Filer D, Patisaul HB, Schug T, Reif D, and Thayer K (2014). Test driving ToxCast: Endocrine profiling for 1858 chemicals included in phase II. Curr. Opin. Pharmacol 19, 145–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer FC, Henneberger L, König M, Bittermann K, Linden L, Goss K-U, and Escher BI (2017). Modeling exposure in the Tox21 in vitro bioassays. Chem. Res. Toxicol 30, 1197–1208. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, and Lempicki RA (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protocols 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Judson R, Houck K, Martin M, Knudsen T, Thomas RS, Sipes N, Shah I, Wambaugh J, and Crofton K (2014). In vitro and modelling approaches to risk assessment from the U.S. Environmental Protection Agency ToxCast programme. Basic Clin. Pharmacol. Toxicol 115, 69–76. [DOI] [PubMed] [Google Scholar]
- Judson R, Houck K, Martin M, Richard AM, Knudsen TB, Shah I, Little S, Wambaugh J, Woodrow Setzer R, Kothya P, et al. (2016). Analysis of the effects of cell stress and cytotoxicity on in vitro assay activity across a diverse chemical and assay space. Toxicol. Sci 152, 323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson RS, Houck KA, Kavlock RJ, Knudsen TB, Martin MT, Mortensen HM, Reif DM, Rotroff DM, Shah I, Richard AM, et al. (2010). In vitro screening of environmental chemicals for targeted testing prioritization: The ToxCast project. Environ. Health Perspect 118, 485–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karmaus AL, Filer DL, Martin MT, and Houck KA (2016). Evaluation of food-relevant chemicals in the ToxCast high-throughput screening program. Food Chem. Toxicol 92, 188–196. [DOI] [PubMed] [Google Scholar]
- Kavlock R, Chandler K, Houck K, Hunter S, Judson R, Kleinstreuer N, Knudsen T, Martin M, Padilla S, Reif D, et al. (2012). Update on EPA’s ToxCast program: Providing high throughput decision support tools for chemical risk management. Chem. Res. Toxicol 25, 1287–1302. [DOI] [PubMed] [Google Scholar]
- Kienzler A, Barron MG, Belanger SE, Beasley A, and Embry MR (2017). Mode of action (MOA) assignment classifications for ecotoxicology: An evaluation of approaches. Environ. Sci. Technol 51, 10203–10211. [DOI] [PubMed] [Google Scholar]
- Kleinstreuer NC, Sullivan K, Allen D, Edwards S, Mendrick DL, Embry M, Matheson J, Rowlands JC, Munn S, Maull E, et al. (2016). Adverse outcome pathways: from research to regulation scientific workshop report. Regul. Toxicol. Pharmacol 76, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapen D, Vergauwen L, Villeneuve DL, and Ankley GT (2015). The potential of AOP networks for reproductive and developmental toxicity assay development. Reprod. Toxicol 56, 52–55. [DOI] [PubMed] [Google Scholar]
- Knudsen TB, Houck KA, Sipes NS, Singh AV, Judson RS, Martin MT, Weissman A, Kleinstreuer NC, Mortensen HM, Reif DM, et al. (2011). Activity profiles of 309 ToxCast™ chemicals evaluated across 292 biochemical targets. Toxicology 282, 1–15. [DOI] [PubMed] [Google Scholar]
- Krewski D, Acosta D Jr., Andersen, Anderson MH, Bailar JC III, Boekelheide, Brent K, Charnley RG, Cheung VG, and Green S Jr. (2010). Toxicity testing in the 21st century: Avision and a strategy. J. Toxicol. Environ. Health Part B 13, 51–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath JA, Parkerton TF, and Di Toro DM (2004). Application of the narcosis target lipid model to algal toxicity and deriving predicted-no-effect concentrations. Environ. Toxicol. Chem 23, 2503–2517. [DOI] [PubMed] [Google Scholar]
- Meyer KH (1937). Contributions to the theory of narcosis. Trans. Faraday Soc 33, 1062–1064. [Google Scholar]
- Overton E (1901). Studien uber die Narkose. Fischer Jena 45. [Google Scholar]
- Patlewicz G, Kuseva C, Kesova A, Popova I, Zhechev T, Pavlov T, Roberts DW, and Mekenyan O (2014). Towards AOP application-implementation of an integrated approach to testing and assessment (IATA) into a pipeline tool for skin sensitization. Regul. Toxicol. Pharmacol 69, 529–545. [DOI] [PubMed] [Google Scholar]
- Rotroff DM, Dix DJ, Houck KA, Knudsen TB, Martin MT, McLaurin KW, Reif DM, Crofton KM, Singh AV, Xia M, et al. (2013). Using in vitro high throughput screening assays to identify potential endocrine-disrupting chemicals. Environ. Health Perspect 121, 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russom CL, LaLone CA, Villeneuve DL, and Ankley GT (2014). Development of an adverse outcome pathway for acetylcholinesterase inhibition leading to acute mortality. Environ. Toxicol. Chem 33, 2157–2169. [DOI] [PubMed] [Google Scholar]
- Selvaraj V, Stocco DM, and Tu LN (2015). Minireview: Translocator protein (TSPO) and steroidogenesis: A reappraisal. Mol. Endocrinol 29, 490–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice RR, Austin CP, Kavlock RJ, and Bucher JR (2013). Improving the human hazard characterization of chemicals: a Tox21 update. Environ. Health Perspect. (Online) 121, 756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefsen KE, Scholz S, Cronin MT, Edwards SW, de Knecht J, Crofton K, Garcia-Reyero N, Hartung T, Worth A, and Patlewicz G (2014). Applying adverse outcome pathways (AOPs) to support integrated approaches to testing and assessment (IATA). Regul. Toxicol. Pharmacol 70, 629–640. [DOI] [PubMed] [Google Scholar]
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, Landesmann B, Lettieri T, Munn S, Nepelska M, et al. (2014a). Adverse outcome pathway (AOP) development I: strategies and principles. Toxicol. Sci 142, 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve DL, Crump D, Garcia-Reyero N, Hecker M, Hutchinson TH, LaLone CA, Landesmann B, Lettieri T, Munn S, Nepelska M, et al. (2014b). Adverse outcome pathway development II: best practices. Toxicol. Sci 142, 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wezel APV, and Opperhuizen A (1995). Narcosis due to environmental pollutants in aquatic organisms: Residue-based toxicity, mechanisms, and membrane burdens. Crit. Rev. Toxicol 25, 255–279. [DOI] [PubMed] [Google Scholar]
- Yauk CL, Lambert IB, Meek M, Douglas GR, and Marchetti F (2015). Development of the adverse outcome pathway “alkylation of DNA in male premeiotic germ cells leading to heritable mutations” using the OECD’s users’ handbook supplement. Environ. Mol. Mutagen 56, 724–750. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.