Abstract
Background and aim
It is commonly noticed that chaotic and inefficient subgenotyping are universally used academically and clinically, a standardized HBV genotype/subgenotype classification criterion is urgently acquired. Sequence similarity, which was commonly used for the last three decades, should be upgraded by phylogenetic analysis in genotyping of recombinant-free HBV strains.
Methods
In this study, 4,429 HBV whole-genome sequences were employed to reconstruct the phylogeny of HBV using Bayesian inference. After excluding recombinant sequences, calculating partitioned evolutionary models, excluding recombinant sequences, reconstructing phylogenetic trees, and performing a correlation analysis of genetic distances, geographical distribution and serotypes, we systematically redefined the genotypes and subgenotypes of HBV.
Results
Compared to previous taxonomy, fourteen subgenotypes (A5-A7; B5-B9; C2-C4, C7; and D6-D7) were revised in the new standard. Now the HBV is divided into ten genotypes (A-J) and 24 subgenotypes (A1-A3; B1-B5; C1-C6; D1-D6; and F1-F4).
Conclusion
Our robust genotype/subgenotype new taxonomy has objectively re-molded the current shape of HBV classification. We believe that all future hepatitis B related researches or diagnosis will be benefited under the new HBV genotyping/subgenotyping standards.
Keywords: Clinical genetics, Infectious disease, Phylogenetic analysis, Genotyping, Taxonomy, Recombination, Hepatitis B virus
1. Introduction
Hepatitis B is second hepatitis to be discovered and hence the name. The disease was discovered in 1947 [1] and confirmed in 1965 [2], and it is one of the leading causes of death in humans in the last two decades [3]. The World Health Organization (WHO) reported that in 2015 there were 240 million people chronically infected with hepatitis B and more than 786,000 mortalities were attributed to it each year [4]. Hepatitis B is caused by the hepatitis B virus (HBV) and can transmit through blood, semen, and other body fluids.
Into the new era of personalized treatment, there are growing evidences which show that the HBV genotype/subgenotype is associated with the HBeAg seroconversion, the severity of liver disease and the response rate to interferon-alpha therapy [5, 6, 7]. As a result, the rational use of antivirus drugs and the precise prognosis is encouraged based on knowing the HBV specific genotyping and subgenotyping in hepatitis patients. However, the current HBV genotype/subgenotype correct classification research seems not so adequate.
The golden standard for classifying HBV used to be the sequence inter-group similarity of the entire viral genome. Okamoto et al. for the first time proposed that sequence divergence in the whole HBV genome exceeding 8% should be categorized as different HBV genotypes after analyzing 18 HBV isolates [8]. Norder et al. then added 4% scalar at S-gene (codes for HBsAg) level in 1992 [9]. These two standards constituted the definition of genotypes of HBV and are still used today [10, 11].
Thanks to the remarkable progress of molecular epidemiology research, more than 4,000 complete genomes of HBV have been investigated. There are ten genotypes (A-J) and more than 30 subgenotypes of human infected HBV that have been discovered and named [7]. As a result, the genotyping methods developed by Okamoto et al. and Norder et al., are nowadays considered as controversial and antiquated because certain phylogenetic inference is lacking [12]. The inconsistency between genotype/subgenotype identification and the clinical outcome had a direct impact on patients’ antivirus therapy [13, 14].
Phylogeny estimation, a technique that has now proved to be the most efficient solution to analyze the divergence of homologous genes, was introduced when more and more researchers realized the defects of the present HBV genotyping system [15, 16]. This significant change in HBV classification is a benefit due to the rapid progress in molecular genetics and bioinformatics. However, the presence of recombination, which is the most significant force of virus evolution especially in high-speed evolved retroviruses, was always being incorrectly applied in phylogenetic analysis [17]. Likewise, alignments including both recombinant and nonrecombinant cases, were used together for phylogeny reconstruction. But in fact, when evolutionary inferences were made from a data set with recombination taking place, the recombinant cases possessing breakpoints could induce an exponential growth of statistics in a test of selection pressure and then a mess of the estimation of substitution rate [17, 18]. The result is an ambiguous relationship that can be obtained between phylogenetic bisected clades which represented different monophyletic genotypes because of the recombinant ones’ swaging between two or more ancestors. Therefore, Huson et al. believed the recombinant ones should be classified in a different system such as network reconstruction (which is not discussed in this study) [19]. Usually, current HBV subgenotyping is directly using phylogenetic tree reconstruction no matter if recombinant occurred in that virus genome. As a result, the abusing of phylogeny has generated the current HBV subgenotype impreciseness. Therefore, in this study, viruses with recombination were carefully excluded, and a more robust HBV genotype/subgenotype frame has been established.
To upgrade the criterion of HBV genotyping/subgenotyping in this study, we reconstructed the phylogenetic tree of HBV genomes, using partitioned Bayesian model and over 4,000 DNA sequences. The partitioned Bayesian model is fully considered and weighted the subset-specific substitution rates of the coding sequence at genomic levels as the best-fit algorithm for phylogenetic analysis [20]. The huge number of sequences applied constructed a comprehensive dataset of HBV genetics which can thoroughly and systematically investigate the current differentiation of HBV. Hence, in this study, a pure non-recombination phylogenetic tree was generated as a basis of HBV classification which could be used as a taxonomy guide. As a major upgrade, this work created a new insight into HBV genotyping and subgenotyping, which would further benefit hepatitis B diagnosis and treatment in the future.
2. Methods
2.1. Sequence acquisition and processing
We acquired 4,429 complete genome sequences of HBV, including 4,186 from humans and 121 from non-human primates from GenBank. Each of the four genes (S, C, P, and X) was extracted using GenScalpel [21] and aligned separately using MEGA 6.0 [22]. The genetic distances between genotypes as well as subgenotypes were estimated under the Kimura 2-parameter model as implemented by MEGA 6.0 (Table 1).
Table 1.
Gene/domain organization and analysis preferences in distance estimation.
|
Gene/domain organization: | ||||
| Gene/Domains | From | To | Sites | Codon Start |
| P/S/X-1 protein | 1 | 2532 | 2532 | 1st site |
| X-2 protein | 2533 | 2747 | 215 | 2nd site |
| C protein |
2748 |
3299 |
552 |
1st site |
|
Analysis Preferences/Distance Estimation: | ||||
| Substitution Type | Nucleotide | |||
| Model/Method | Kimura 2-parameter model | |||
| Substitutions to Include | d: Transitions + Transversions | |||
| Rates among Sites | Uniform rates | |||
| Gaps/Missing Data Treatment | Pairwise deletion | |||
To optimize the following analyses, we reduced the number of sequences by group sequences into the operational taxonomic unit (OTU). We grouped the sequences using a minimum identity of 97% as the “radius” of an OTU, assuming that only sequences belonging to the same genotype/subgenotype would be grouped even in consideration of the possibility of recombination and thus would not affect the accuracy of our following analysis. This was implemented in USEARCH v8.1 [23], and 600 OTUs were obtained and used in the subsequent analyses.
For better accuracy of phylogenetic interfaces, we detected and removed recombinant sequences using what we considered to be a conservative approach as implement in RDP4 [24] accompanied by the use of SplitsTree 4 [25]. RDP4 can determine approximate breakpoint positions and compare phylogenetic signals in opposite sites of recombination breakpoint. SplitsTree is used to analyze phylogeny that provides scenarios of reticulate evolution such as recombination networks. In RDP4, we detected recombination using six algorithms including 3Seq, Chimaera, Geneconv, MaxChi, RDP, and Siscan using the default settings. In case that recombination is observed in four of the six algorithms, we compared the phylogenetic positions of the major and the minor parent sequences estimated in UPGMA trees and inspected whether they were highly distinctive. We also used SplitsTree to determine the recombination network based on an equal angle algorithm to look for a potential recombinant sequence that was unaware by RDP4.
2.2. Phylogenetic inference
We estimated the phylogeny of HBV under Bayesian inference using BEAST v2.2.1 [26]. First, we calculated the best-fit partitioning schemes under the Bayesian information criterion using PartitionFinder v1.1 [27]. We defined the alignment of the complete HBV genome based on the physic positions of the four open reading frames and codon positions into 15 blocks. The gene P is partitioned at two isolated regions of the genome, so we estimated the partition schemes for each region separately. A total of 516 schemes were evaluated using the greedy algorithm. Finally, a partition scheme of 12 partitions was accepted (Table 2). A woolly monkey HBV genome (AF046996) was included in the alignment to root the tree. Each BEAST analysis used 12 partitions, one log-normal relaxed clock model, and a birth-death tree prior. We ran each analysis for 500 million generations and sampled every 50 000 generations. We repeated the analysis four times, calculated the effective sample size of each parameter using Tracer v1.6, and confirmed that all analyses reached similar posterior distribution. The maximum clade credibility tree and the posterior distribution for each node were calculated. Bayesian posterior probabilities (PP)≥0.95 were considered as strong supports for a given relationship [28].
Table 2.
Best partitioning scheme applied in the phylogenetic inference of the HBV genome.
| Subset | Best model | Subset partitions | Subset sites |
|---|---|---|---|
| 1 | SYM + G | PPa_1st, PPa_2nd | 1-1063∖3, 2-1063∖3 |
| 2 | GTR + G | PPa_3rd | 3-1063∖3 |
| 3 | SYM + G | SP_1st | 1064-1744∖3 |
| 4 | TrNef + G | SP_2nd | 1065-1744∖3 |
| 5 | SYM + G | SP_3rd | 1066-1744∖3 |
| 6 | SYM + G | CP_2nd, PPb_1st | 1745-2282∖3, 2749-3299∖3 |
| 7 | GTR + G | CP_3rd, PPb_2nd | 1746-2282∖3, 2750-3299∖3 |
| 8 | SYM + G | PPb_3rd | 1747-2282∖3 |
| 9 | GTR + G | XP_1st | 2283-2747∖3 |
| 10 | SYM + G | XP_2nd | 2284-2747∖3 |
| 11 | SYM + G | XP_3rd | 2285-2747∖3 |
| 12 | SYM + G | CP_1st | 2748-3299∖3 |
3. Results
3.1. Phylogenetic relationship
Firstly, we removed recombinant sequences recognized by RDP4 and SplitsTree 4, and repeated the analyses three times. The numbers of sequences retained after each run of analyses were 577, 524, and 515, respectively. In total, we removed 85 recombinant OTU consensus sequences from the alignment and the remaining 516 sequences were sent to phylogenetic estimation.
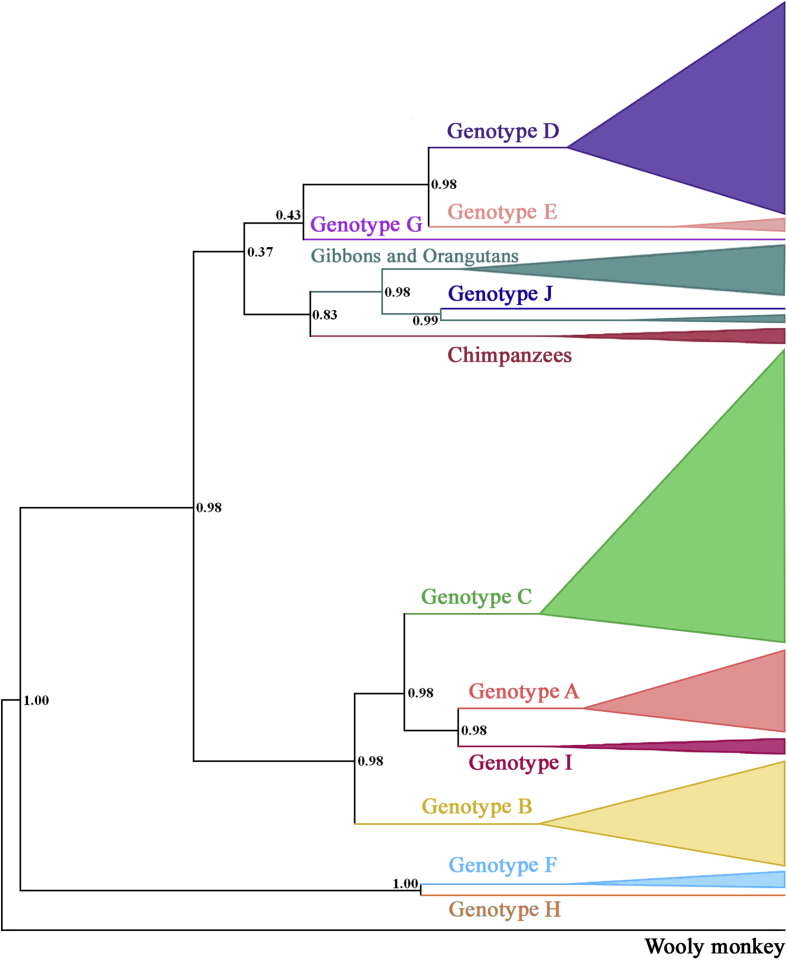
Then, in the phylogenetic tree generated by 516 HBV sequences, eleven clades and two lineages were revealed corresponding to the ten human infected HBV genotypes (A to J) as well as three non-human primates HBV, while the genotypes H and J are represented by a single sequence each (Fig. 1). Genotypes F and H are sister to each other (PP = 1.0) and forms a basal monophyletic group (PP = 0.98). The genotypes A, B, C, and I also develop a strongly supported monophyletic clade (PP = 0.98), and the relationships among them are also well supported (PP ≥ 0.98). Phylogenetic relationships among the rest of the clades and lineages are not fully resolved (PP < 0.85), while the genotype J is strongly supported as being embedded within the non-human primate HBV clades (PP ≥ 0.98).
Fig. 1.
Phylogenetic reconstruction of HBV genomes. The values of Bayesian posterior probability are shown orderly at the nodes.
Based on highly supported phylogenetic relationships within the clades of genotypes, we recognized a series of subgenotypes. Notably, the subgenotypes of A3, D7, B9, etc. are highly incongruent from those reported by original reporters (Tables 3 and 4), while 26 stains failed to genotyping and 136 failed to subgenotyping (Table 5).
Table 3.
The genotype, serotype, and geographical distribution of hepatitis B virus cases reported.
| Genotype | Cases | Serotype‡ | Geographical distribution§ | This study |
|---|---|---|---|---|
| D4 | D(1), D4(3) | ayw(4) | Australia(3), Canada(3), Haiti(3), Papua New Guinea(1) | D4 |
| D7 | D(3), D7(7), Rec(1) | ayw4(1) | Belgium(1), Central African Republic(1), Gabon(1), Tunisia(34) | D6 |
| D2 | D(8), D2(9) | adw3(1), ayw(8), ayw3(5) | Belgium(1), Greenland(1), India(1), Iran(1), Japan(1), Lebanon(3), Poland(3), Russia(1), Serbia(add), Spain(1), Taiwan(2), Turkey(2) | D2 |
| D1 | D(20), D1(50), N/A(2) | ayw(26), ayw2(13), ayw3(3) | China(2), Greece(1), India(2), Indonesia(1), Iran(28), Italy(3), Japan(1), Lebanon(5), Mongolia(2), Pakistan(1), South Africa(1), Syria(10), Tunisia(4), Turkey(11) | D1 |
| D? | D(2) | × | ||
| D3/D6 | D(11), D3(6), D6(1) | adrq+(1), adw1(1), ayw(7), ayw2(15), ayw3(7) | Belarus(3), Belgium(8), Canada(3), China(3), Estonia(2), France(1), Haiti(1), India(15), Indonesia(5), Italy(4), Japan(1), Mongolia(3), Pakistan(1), Russia(1), Serbia(3), South Africa(6), Sweden(3), Turkey(2) | D3 |
| D5 | D(6), D5(1) | ayw(1), ayw2(1), ayw3(11) | India(22), Japan(1) | D5 |
| E | E(9) | ayw4(4) | Argentina(add), Cameroon(add), Central African Republic(add), Colombia(add), Ghana(1), Guinea(2), Namibia(1), Nigeria(5) | E |
| G | G(2), Rec(1) | adw(5), adw2(6) | Argentina(2), Belgium(2), Brazil(4), France(2), Germany(2), Mexico(2), Netherlands(1), Thailand(1), USA(8), | G |
| non-human primates(26) | ||||
| J | J(1) | ayw(1) | Japan(1) | J |
| C2-part1 | B2(1), C(33), C1(2), C2(95), N/A(17) | adr(22), adrq-(1), adrq+(99), adw(2), adw1(10), adw2(2) | Bolivia(1), China(101), Hong Kong(2), Japan(27), Malaysia(4), South Korea(7), Taiwan(6) | C2 |
| C2-part2 | C(4), C2(3), N/A(1) | adr(2), adrq+(6), adw2(1) | China(8), Hong Kong(2), Japan(6), Malaysia(1), South Korea(1), Taiwan(5) | C3 |
| C1-part1 | C(10), C1(22), N/A(1), Rec(4) | adr(11), adrq+(6), adw(1) | Belgium(1), Cambodia(1), China(8), Hong Kong(7), India(2), Indonesia(2), Japan(1), Malaysia(8), Myanmar(1), Taiwan(1), Thailand(8), Tunisia(1), Viet Nam(3) | C1 |
| C1-part2 | C(1), C1(1), C2(2) | adrq+(3) | China(4), Malaysia(2) | C1 |
| C? | C(5), C2(1) | adr(2), adrq-(1), adrq+(13) | China(16), Indonesia(1), Japan(1), Taiwan(1) | C4 |
| C6/C7 | C(2), C6(6), C7(1) | adr(4), ayw(2) | Australia(2), Indonesia(17), South Korea(1) | C6 |
| C5 | C5(4) | adw(3), adw2(3) | Indonesia(3), Malaysia(3), Philippines(3), Thailand(2) | C5 |
| A1 | A(5), A1(14), N/A(3) | adw(4), adw2(13), ayw(1), ayw2(1) | Argentina(add), Bangladesh(1), Belgium(1), Central African Republic(add), Colombia(add), Congo(add), Haiti(5), India(3), Malaysia(add), Nepal(add), Philippines(add), Rwanda(add), Somalia(add), South Africa(11), Tanzania(1), Zimbabwe(add) | A1 |
| A3 | A(1), A3(4) | ayw(1) | Cameroon(6), Gabon(3), Nigeria(2) | A3 |
| A5 | A5(5) | Cameroon(2), Haiti(21), Nigeria(3) | A3 | |
| A7 | A7(6) | Cameroon(10) | A3 | |
| A3? | A(1), A3(2) | Cameroon(1), Gabon(1), Guinea(1), Mali(1) | A3 | |
| A2/A6 | A(3), A2(6) | adw(2), adw2(4), ayw1(3) | Argentina(add), Australia(add), Belarus(add), Belgium(3), Canada(add), Estonia(add), France(add), Germany(2), Italy(1), Japan(add), Latvia(add), Poland(2), Russia(add), South Africa(add), Tunisia(1), USA(add), Uzbekistan(add) | A2 |
| I | I(1); N/A(3) | ayw(1), ayw1(1) | China(2), India(1), Laos(1) | I |
| B1 | B(2), B1(8) | adw(3) | Japan(33) | B1 |
| B2 | B(9), B2(9), N/A(1), Rec(2) | adrq+(3), adw(4), adw1(15), adw2(6), ayw(1), ayw2(1) | China(12), Hong Kong(1), Japan(add), Malaysia(8), South Korea(add), Taiwan(add), Thailand(add), Viet Nam(add) | B2 |
| B4 | B4(1) | ayw1(2) | Malaysia(1), Viet Nam(3) | B4 |
| B5 | B5(3) | ayw(3), ayw1(2) | Malaysia(2), Philippines(5) | B3 |
| B3/B7/B8 | B(6), B3(7), B7(1), B8(2), C1(1), N/A(1), Rec(4) | adw(4), ayw(2), ayw1(8) | China(8), Indonesia(36), Malaysia(18), Philippines(1), Taiwan(4), Thailand(1) | B3 |
| B9 | B9(1) | ayw(1), ayw1(1) | China(1), Indonesia(7), Malaysia(2), Philippines(1) | B3 |
| B6 | B(2), B6(3) | Canada(23), Greenland(2) | B5 | |
| F3 | F(1), F3(2) | adw4(2) | Colombia(3), Venezuela(16) | F3 |
| F4 | F4(2), Rec(2) | adw4(14) | Argentina(18), Bolivia(7), Brazil(2) | F4 |
| F2 | F2(2) | adw4(4) | Nicaragua(1), Venezuela(7) | F2 |
| F1 | F1(3) | adw4(29) | Argentina(16), Chile(18), Costa Rica(2), El Salvador(2), Ireland(1), Japan(1), Peru(2), USA: Alaska(9) | F1 |
| H | H(1) | adw4(3), adw(7) | Argentina(2), Japan(5), Mexico(9), Nicaragua(2), Thailand(1), USA(4) | H |
Serotype‡/Geographical distribution§: Except for nine subgenotypes (D2, D1, D?, E, C2-part1, C1-part1, A1, A2/A6 and B2), the serotypes and geographical distributions of other subgenotypes contained all the cases in OTU and expended OTU datasets; Subgenotypes with underlines were recommended in this study but differ from reported. add, the cases reported in expended OTU datasets; N/A, non-available; Rec, recombinant strain; ×, delete for recombinant strain(s) only.
Numbers in parenthesis indicate the sequences reported in GenBank. For the absence of subgenotype/serotype of numbers of sequence records, the numbers in the cases/Serotype column could be lower than the sum of geographical distribution.
Table 4.
516 sequences in Fig. 1.
Table 5.
Statistics of Fig. 1.
| Clade | Number of Sequences | Failed to genotyping | Failed to subgenotyping | Inconsistent subgenotyping within clade |
|---|---|---|---|---|
| D4 | 4 | 1 | ||
| D7 | 11 | 3 | 1(D/E Rec) | |
| D2 | 17 | 8 | ||
| D1 | 72 | 2 | 20 | |
| D? | 2 | 2 | ||
| D3/6 | 18 | 11 | 1(D6) | |
| D5 | 7 | 6 | ||
| E | 9 | |||
| G | 3 | 1(G/C Rec) | ||
| J | 1 | |||
| C2-part1 | 148 | 17 | 33 | 3(B2, C1×2) |
| C2-part2 | 8 | 1 | 4 | |
| C1-part1 | 37 | 1 | 10 | 4(C/A Rec ×2, C/A/B Rec, C/G Rec) |
| C1-part2 | 4 | 1 | 2(C2×2) | |
| C? | 6 | 5 | ||
| C6/C7 | 9 | 2 | 1(C7) | |
| C5 | 4 | |||
| A1 | 22 | 3 | 5 | |
| A3 | 5 | 1 | ||
| A5 | 5 | |||
| A7 | 6 | |||
| A3? | 3 | 1 | ||
| A2/A6 | 9 | 3 | ||
| I | 4 | |||
| B1 | 10 | 2 | ||
| B2 | 21 | 1 | 9 | 2(B/C Rec ×2) |
| B4 | 1 | |||
| B5 | 3 | |||
| B3/B7/B8 | 22 | 1 | 6 | 4(B/C Rec ×4) |
| B9 | 1 | |||
| B6 | 5 | 2 | ||
| F3 | 3 | 1 | ||
| F4 | 4 | 2(F/D Rec, F/G Rec) | ||
| F2 | 2 | |||
| F1 | 3 | |||
| H | 1 |
Rec, recombinant strain.
In order to calibrate the HBV genotype/subgenotype nomenclature and calculate the genetic distances within/between groups, each of HBV genotypes was further analyzed as follows.
3.2. Genotypes of HBV
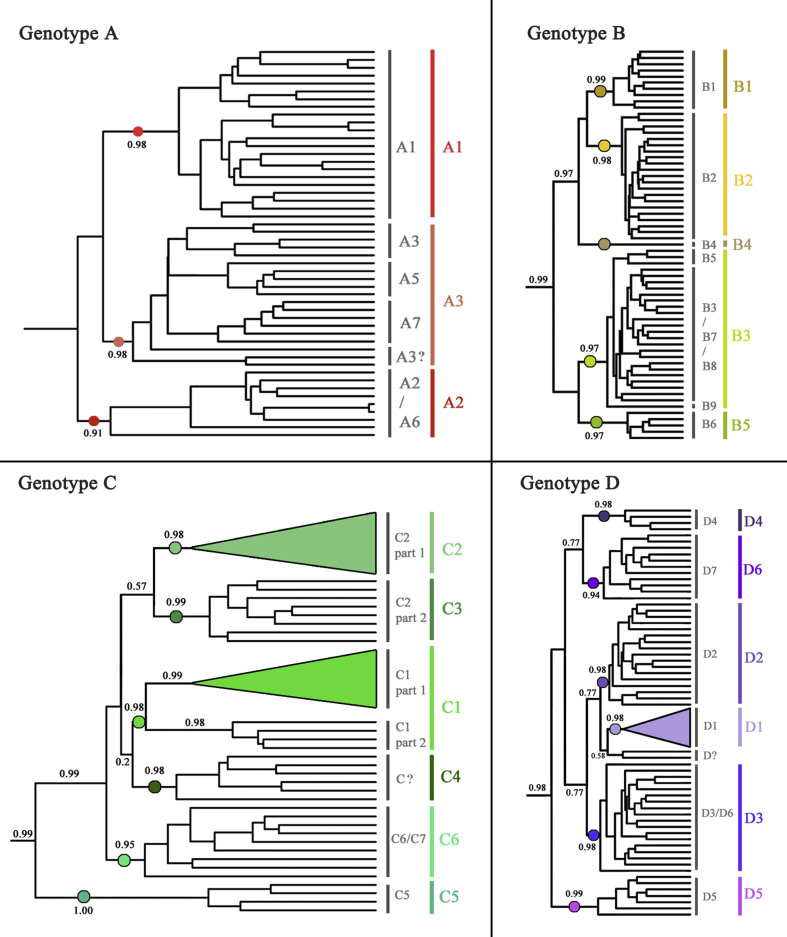
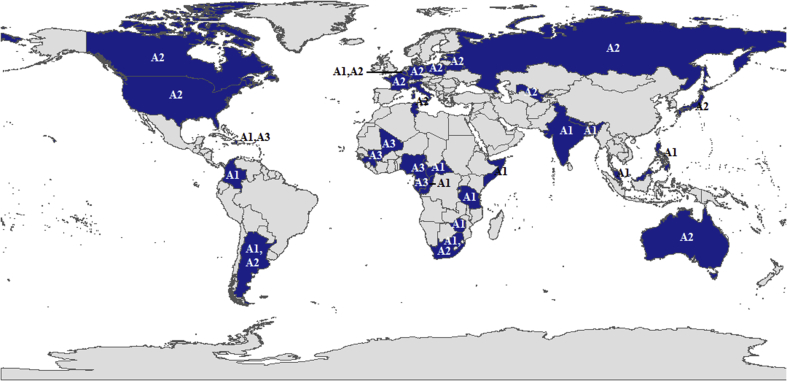
Genotype A. 50 species were recognized as in genotype A and used to assign into seven subgenotypes (A1-A7, Fig. 2A). While A1 (as known as Aa) and A2 (Ae) are widely distributed in Africa/Asia and Europe [29], respectively, A3/A5/A7/A3? were only reported in one case study each (Table 6). Geographically, A3/A5/A7/A3? were from five countries in west-central Africa as well as Haiti, a state in which 90% of the population are descendants of African slaves (Table 3) [30]. The estimated genetic distances between these subgenotypes were only 0.0380–0.0435 (Table 7).
Fig. 2.
Phylogeny of genotype A/B/C/D of HBV. A: Genotype A (upper left); B: Genotype B (upper right); C: Genotype C (lower left); D: Genotype D (lower right). The values of Bayesian posterior probability are shown orderly at the nodes. Nodes of PP > 0.95, which considered as a robust monophyletic group were shown in filled circles. Subgenotype names were shown after the tree while old classification with grey alphabets and new classification with colorful alphabets.
Table 6.
The major reports of subgenotype A3-A7.
| Subgenotype | References |
|---|---|
| A3 | Kurbanov F, Tanaka Y, Fujiwara K, et al. J Gen Virol. 2005; 86(Pt 7):2047–56. |
| A4 | Olinger CM, Venard V, Njayou M, et al. J Gen Virol 2006; 87:1163–1173. |
| A5 | Andernach IE, Nolte C, Pape JW, et al. Emerg Infect Dis 2009; 15:1222–1228. |
| A6 | Pourkarim MR, Lemey P, Amini-Bavil-Olyaee S, et al. J Clin Virol 2010; 47:93–96. |
| A7 | Hübschen JM, Mbah PO, Forbi JC, et al. Clin Microbiol Infect 2011; 17:88–94. |
Table 7.
Estimates of evolutionary divergence over sequences pairs between tentative Genotype A.
| A1 | A3 | A5 | A7 | A3? | A2 | A6 | |
|---|---|---|---|---|---|---|---|
| A1 | |||||||
| A3 | 0.0509 | ||||||
| A5 | 0.0479 | 0.0380 | |||||
| A7 | 0.0551 | 0.0426 | 0.0406 | ||||
| A3? | 0.0496 | 0.0417 | 0.0395 | 0.0435 | |||
| A2 | 0.0558 | 0.0541 | 0.0503 | 0.0574 | 0.0517 | ||
| A6 | 0.0498 | 0.0466 | 0.0451 | 0.0495 | 0.0447 | 0.0494 |
For showing a robust phylogenetic relationship, we adjusted the branches composed by A3/A5/A7/A3? to new subgenotype A3, and A2/A6 to new subgenotype A2 (Table 3 and Fig. 2A). Subgenotype A1 kept the original name. Based on the new subgenotype, evolutionary divergences in Genotype A were analyzed (Table 8), while their epidemiological distribution was also described (Fig. 3).
Table 8.
Estimates of evolutionary divergence between subgenotypes of HBV genotype A.
| A1 | A2 | A3 | |
|---|---|---|---|
| A1 | |||
| A2 | 0.0538 | ||
| A3 | 0.0512 | 0.0514 |
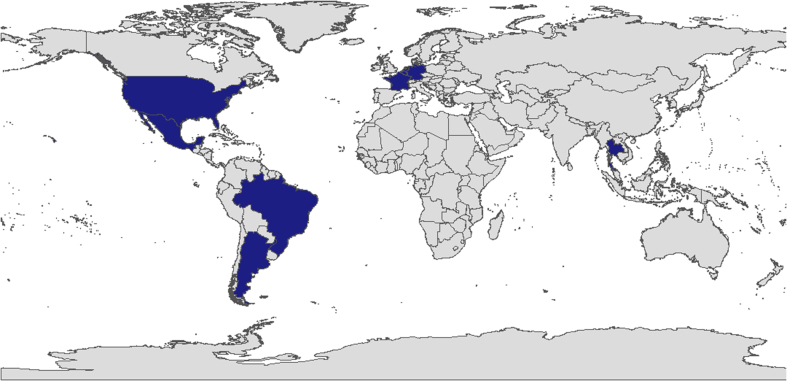
Fig. 3.
Geographical distributions of HBV Genotype A.
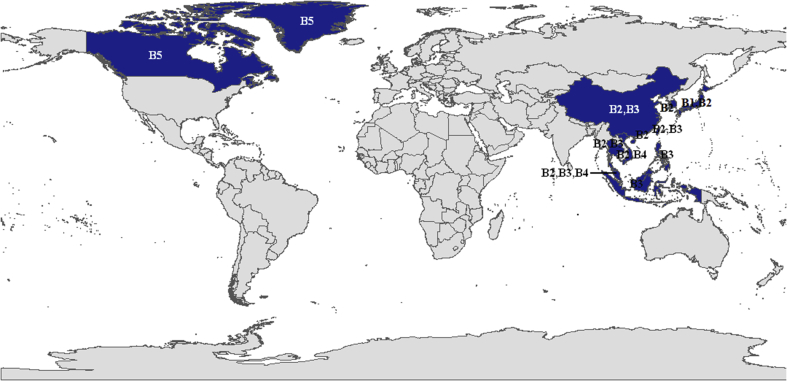
Genotype B. There used to be nine subgenotypes (B1–B9) reported in genotype B, which consists of 63 sequences (Fig. 2B). As a genotype typically distributed in Asia (B1 or Bj in Japan, B2 or Ba in Asia) [31], only B6 were sporadically found in Canada [32] and Greenland [33] (Table 3). Further analysis showed that B5, B7–B9 possess a short distance with B3 and each other (0.0275–0.0398, Table 9).
Table 9.
Estimates of evolutionary divergence over sequences pairs between tentative Genotype B.
| B1 | B2 | B4 | B5 | B3 | B7 | B8 | B9 | B6 | |
|---|---|---|---|---|---|---|---|---|---|
| B1 | |||||||||
| B2 | 0.0548 | ||||||||
| B4 | 0.0572 | 0.0455 | |||||||
| B5 | 0.0686 | 0.0552 | 0.0508 | ||||||
| B3 | 0.0706 | 0.0573 | 0.0536 | 0.0398 | |||||
| B7 | 0.0676 | 0.0538 | 0.0482 | 0.0344 | 0.0314 | ||||
| B8 | 0.0667 | 0.0544 | 0.0486 | 0.0367 | 0.0363 | 0.0305 | |||
| B9 | 0.0650 | 0.0523 | 0.0473 | 0.0316 | 0.0345 | 0.0275 | 0.0310 | ||
| B6 | 0.0646 | 0.0725 | 0.0716 | 0.0636 | 0.0640 | 0.0565 | 0.0633 | 0.0579 |
Along with the phylogenetic relationship showed in Fig. 2B and considering the distribution information [34, 35], we integrated subclade B5, B3/B7/B8, and B9 into new subgenotype B3. Subgenotype B1, B2 and B4 kept their original name. And subgenotype B6 was then numbered to B5 sequentially. So far, genotype B was divided into five new subgenotypes (B1–B5, Table 10), of which B1 is only distributed in Japan, B2–B4 are mainly distributed in Southeast Asia, and B5 is sporadically distributed in Canada and Greenland (Fig. 4).
Table 10.
Estimates of evolutionary divergence between subgenotypes of HBV genotype B.
| B1 | B2 | B3 | B4 | B5 | |
|---|---|---|---|---|---|
| B1 | |||||
| B2 | 0.0548 | ||||
| B3 | 0.0695 | 0.0564 | |||
| B4 | 0.0573 | 0.0474 | 0.0535 | ||
| B5 | 0.0646 | 0.0725 | 0.0631 | 0.0719 |
Fig. 4.
Geographical distributions of HBV Genotype B.
Genotype C. As the biggest group of HBV, genotype C was mainly reported in Asia [36] and believed seven subgenotypes (C1–C7, Fig. 2C). Interestingly, three of five strains that apart from Asia (Table 3) were proved to be caused by transmission from Japanese immigrants [37, 38]. Therefore, we believed that genotype C is predominantly found in Asian populations.
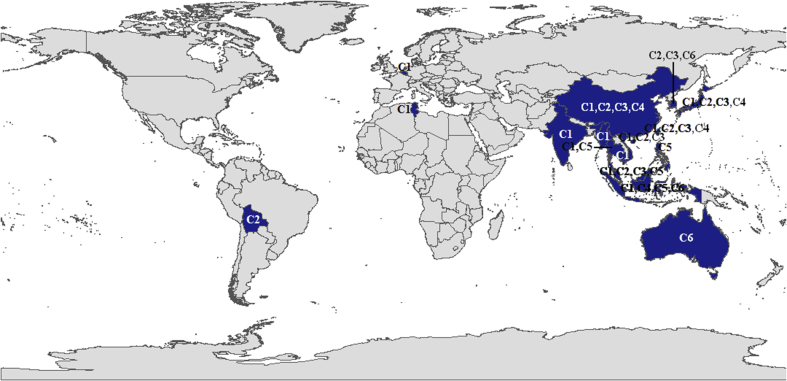
Combining evolutionary distances between groups (Table 11), geographical distribution (Table 3) and robust bootstrap supporting values in the phylogenetic tree, we suggested that C1-part1 and C1-part2 should combine into a new subgenotype C1, C2-part1 should be new subgenotype C2, C2-part2 should be a new subgenotype C3, C? should be a new subgenotype C4, and C6 – C7 should be combined into a new subgenotype C6 (Fig. 2C). Subgenotype C5 kept the original name. Among the six clades, C1–C4 are widely spread in Southeast and East Asia, C5 only in Southeast Asia, and C6 is mainly seen in Indonesia, Australia and scattered in South Korea (Fig. 5). Based on the new taxonomy, genetic variances over all sequence pairs in genotype C was given as 0.0418 to 0.0674 (Table 12).
Table 11.
Estimates of evolutionary divergence over sequences pairs between tentative Genotype C.
| C2-part1 | C2-part2 | C1-part1 | C1-part2 | C? | C6 | C7 | C5 | |
|---|---|---|---|---|---|---|---|---|
| C2-part1 | ||||||||
| C2-part2 | 0.0418 | |||||||
| C1-part1 | 0.0477 | 0.0511 | ||||||
| C1-part2 | 0.0456 | 0.0487 | 0.0473 | |||||
| C? | 0.0430 | 0.0466 | 0.0496 | 0.0488 | ||||
| C6 | 0.0597 | 0.0643 | 0.0665 | 0.0647 | 0.0610 | |||
| C7 | 0.0484 | 0.0520 | 0.0562 | 0.0564 | 0.0501 | 0.0624 | ||
| C5 | 0.0590 | 0.0625 | 0.0649 | 0.0651 | 0.0614 | 0.0734 | 0.0609 |
Fig. 5.
Geographical distributions of HBV Genotype C.
Table 12.
Estimates of evolutionary divergence between subgenotypes of HBV genotype C.
| C1 | C2 | C3 | C4 | C5 | C6 | |
|---|---|---|---|---|---|---|
| C1 | ||||||
| C2 | 0.0475 | |||||
| C3 | 0.0509 | 0.0418 | ||||
| C4 | 0.0495 | 0.0430 | 0.0466 | |||
| C5 | 0.0649 | 0.0590 | 0.0625 | 0.0614 | ||
| C6 | 0.0592 | 0.0513 | 0.0557 | 0.0532 | 0.0674 |
Genotype D. Seven subgenotypes (D1-D7) used to be involved in genotype D, which consisted of 131 sequences (Fig. 2D). Geographical statistics showed that genotype D was prevalent in all continents except South America, but mainly in the Mediterranean area, the Middle East, and India (Table 3) [10, 39]. Further analysis showed that D4-D7 stains were only reported in one or two inter-cited studies, respectively (Table 13); genetic distance between D3 and D6 was only 0.0257 (Table 14); D4 and D7 possess a distinct geographic distribution.
Table 13.
The major reports of subgenotype D4-D7.
| Subgenotype | References |
|---|---|
| D4 | Osiowy C, Larke B, Giles E. J Viral Hepat 2011; 18:e11-e19. |
| D5 | Ghosh S, Banerjee P, RoyChoudhury A, et al. J Clin Microbiol 2010; 48:4063–4071. |
| D6 | Utsumi T, Lusida MI, Yano Y, et al. J Clin Microbiol 2009; 47:1842–1847. |
| D7 | Meldal BH, Moula NM, Barnes IH, et al. J Gen Virol 2009; 90:1622–1628. |
Table 14.
Estimates of evolutionary divergence over sequences pairs between tentative Genotype D.
| D4 | D7 | D2 | D1 | D3 | D6 | D5 | |
|---|---|---|---|---|---|---|---|
| D4 | |||||||
| D7 | 0.0469 | ||||||
| D2 | 0.0530 | 0.0546 | |||||
| D1 | 0.0528 | 0.0529 | 0.0392 | ||||
| D3 | 0.0455 | 0.0494 | 0.0412 | 0.0401 | |||
| D6 | 0.0472 | 0.0509 | 0.0423 | 0.0417 | 0.0257 | ||
| D5 | 0.0549 | 0.0582 | 0.0567 | 0.0560 | 0.0509 | 0.0517 |
Based on the phylogenetic results, we adjusted clade D3/D6 to new subgenotype D3, D7 to new subgenotype D6 and the rest of the subgenotypes remained their old names. Although one of two strains in clade D? was believed to be a recombinant of D1/D7 [40], the sequence successfully survived the recombinant elimination procedure. As a result, we strongly suggested D? should remain as an un-classifiable virus and be re-classified when more genotype D HBV sequences available in the future.
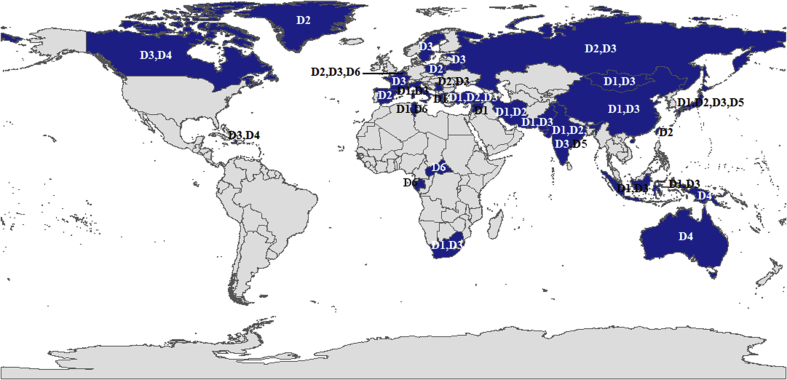
In the updated system of genotype D, D1-D3 has a worldwide distribution but common in the Mediterranean area, the Middle East and India, D4 is scatted across Oceania and Central and North America, D5 in India and D6 mainly in Africa (Fig. 6 and Table 15).
Fig. 6.
Geographical distributions of HBV Genotype D.
Table 15.
Estimates of evolutionary divergence between subgenotypes of HBV genotype D.
| D1 | D2 | D3 | D4 | D5 | D6 | |
|---|---|---|---|---|---|---|
| D1 | ||||||
| D2 | 0.0392 | |||||
| D3 | 0.0422 | 0.0434 | ||||
| D4 | 0.0528 | 0.0530 | 0.0478 | |||
| D5 | 0.0560 | 0.0567 | 0.0531 | 0.0549 | ||
| D6 | 0.0529 | 0.0546 | 0.0516 | 0.0469 | 0.0582 |
Genotype E-H. Although the cases of these four types were mainly reported by few studies, each taxon of genotype E-H apparently formed an independent monophyletic branch in the phylogenetic tree (Fig. 1 and Table 16). By referring to the results of previous reports, the geographical distribution of these HBV strains was updated in this study (Table 17 and Figs. 7, 8, 9, and 10). As to genotyping, however, we tentatively support the present taxonomy that Genotype E, G, H presented independently with no subgenotypes and Genotype F was divided into four subgenotypes (F1–F4, Tables 3 and 18), since the sequence information of those genotype/subgenotype are limited.
Table 16.
The major reports of genotype E-H.
| Genotype | References |
|---|---|
| E | Bekondi C, Olinger CM, Boua N, et al. 2007; J Clin Virol 40:31–7. |
| Garmiri P, Loua A, Haba N, et al. 2009; J Gen Virol 90:2442–51. | |
| Forbi JC, Vaughan G, Purdy MA, et al. 2010; PLoS One 5:e11615. | |
| Hübschen JM, Mbah PO, Forbi JC, et al. 2011; Clin Microbiol Infect 17:88–94. | |
| F | Devesa M, Loureiro CL, Rivas Y, et al. 2008; J Med Virol 80:20–6. |
| Torres C, Piñeiro y Leone FG, et al. 2011; Mol Phylogenet Evol 59:114–22. | |
| Sami H, Rizvi M, Azam M, et al. 2013; Adv Virol 2013:846849. | |
| G | Kato H, Orito E, Gish RG, et al. 2002; J Virol 76:6131–7. |
| Bottecchia M, Souto FJ, Ó KM, et al. 2008; BMC Microbiol 8:11. | |
| Araujo NM, Araujo OC, Silva EM, et al. 2013; J Gen Virol 94:150–8. | |
| H | Arauz-Ruiz P, Norder H, Robertson BH, et al. 2002; J Gen Virol 83:2059–73. |
| Tanaka Y, Sanchez LV, Sugiyama M, et al. 2008; Virology 376:408–15. |
Table 17.
Geographical distribution of Genotype E-H.
| Genotype | Previous reports | Extension in this study |
|---|---|---|
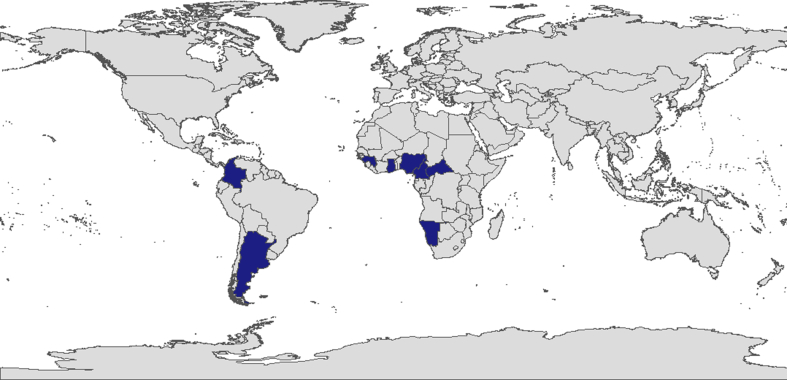
| E | West Coast of Africa, Madagascar | Argentina and Colombia in South America |
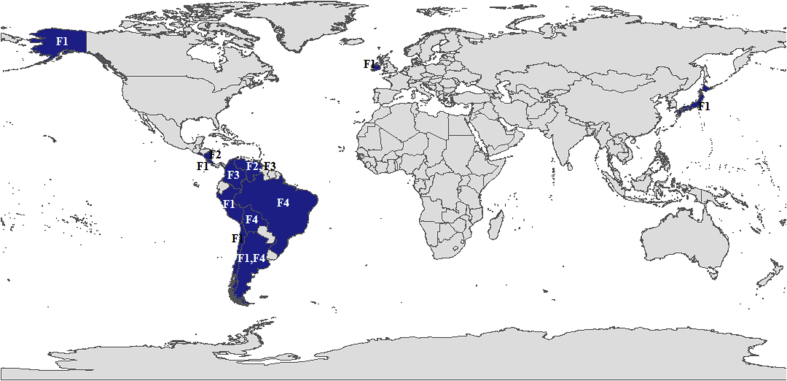
| F | restricted to Central and South America | USA: Alaska; Ireland; Japan |
| G | France, Germany, UK, Italy, USA | Argentina, Belgium, Brasil, Mexico, Netherlands, Tailand |
| H | Nicaragua, Mexico and USA | Argentina, Japan, Tailand |
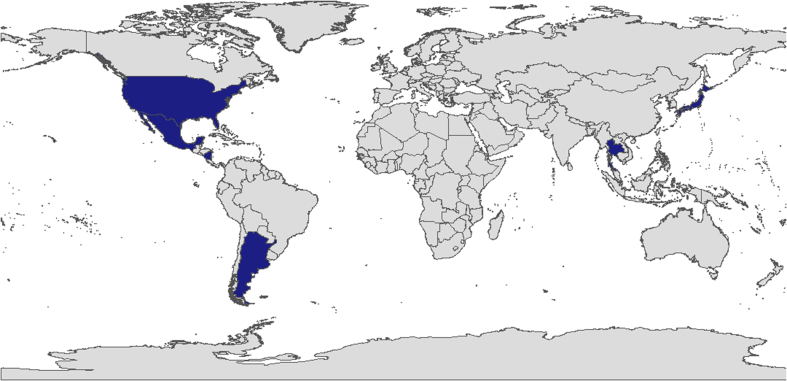
Fig. 7.
Geographical distributions of HBV Genotype E.
Fig. 8.
Geographical distributions of HBV Genotype F.
Fig. 9.
Geographical distributions of HBV Genotype G.
Fig. 10.
Geographical distributions of HBV Genotype H.
Table 18.
Estimates of evolutionary divergence between subgenotypes of HBV genotype F.
| F1 | F2 | F3 | F4 | |
|---|---|---|---|---|
| F1 | ||||
| F2 | 0.0585 | |||
| F3 | 0.0562 | 0.0465 | ||
| F4 | 0.0636 | 0.0504 | 0.0503 |
There is no doubt that more cases of genotype E-H were expected, from which we can better understand evolutionary divergence in these groups.
Genotype I. A previous study indicated that this genotype is derived from the recombination of the genotype C and an unknown genotype [41]. In our analysis using RDP4, recombination is only supported in one of the sequences (FR714499) by all six algorithms, which has an unknown parent sequence from the genotype C. However, there is no evidence to support a scenario of recombination in the other three sequences (EU835240, FJ023667, and FR714496). These results imply that previous studies may have been misled by incomplete sampling (Table 19).
Table 19.
The major reports of genotype I.
| Subgenotype | Country | Region | Reference | Detection of recombination | |
|---|---|---|---|---|---|
| EU835240 | A/G/C | India | South Asia | Unpublished | |
| FJ023667 | I2 | Laos | Southeast Asia | Olinger CM, et al. Emerg Infect Dis 2008; 14:1777–1780. | N/A |
| FR714496 | X/C | China | East Asia | Fang ZL, et al. J Gen Virol 2011; 92:402–411. | RDP2 |
| FR714499 | X/C | China | East Asia | Fang ZL, et al. J Gen Virol 2011; 92:402–411. | RDP2 |
N/A, non-available.
Genotype J. A genetic variant that cannot be ignored was isolated from an 88-year-old Japanese patient with hepatocellular carcinoma. The only human case in the clade of non-human primates was believed to be a new genotype J (AB486012, Fig. 1) [11]. Interestingly, the analysis against its allies seemed to suggest a cross-species transmission of non-human primate HBV to humans (Table 20). The novel genotype derived from Southwest Asia was enlightening and was only reported once so far. Given this, we tentatively support the validation of Genotype J.
Table 20.
The source information of “Clade J”.
4. Discussion
We re-scaled the genotypes and subgenotypes frame of HBV via a Bayesian inference phylogenetic approach. There were totally 14 subgenotypes were either canceled or redefined. Based on the principle of reservation of old names, the monophyletic topology together with genetic variance were considered as the priority criteria of the re-classification.
The amount of HBV subgenotyping conclusion and nomenclature in the pervious studies are considered as inappropriate after our investigation in this study (Tables 3 and 4). At the same time, many works supported our analysis, such as, the phylogenetic position of F2 which was completely consistent with our OTU stains (DQ899142, DQ899144) [42]; the cases of non-typical geographic distribution within clades were screened out by further demographic analysis (AB365453, FJ349225) [38,43] and the researches focusing on origin of HBV constructed evolutionary trees with a topology identical to the ones in our study [44, 45].
Although we would like to insist that the only standard of genotype and subgenotype classification of HBV should be based on the monophyly of composed strains, we also calculate the genetic distance within or between HBV genotypes and subgenotypes. Systematic defects appeared when conducting HBV genotyping using 8% inter-subtypic divergence in genome-scale and 4% in S gene-scale. In this study, genetic differentiation ranges from 7.01% to 16.01% in genotype level, and the average distance reached 12.73% (Table 21), while the maximum variance is 5.16% and the minimum is 0.75% within groups (Table 22). This situation can interpret as the divergence between sequences is dynamically changed and the genotypes could be reassigned even, especially with the rapid growth of HBV genome data quantity. Even though genetic distances were estimated based on new HBV taxonomy, it does not mean that a new taxon should be assigned while values higher than the maximum in a certain group or several types should be integrated while lower than the maximum. Furthermore, since a few subgenotypes were deleted or redefined, it seems that more clinical correlations between HBV genome variation and treatment outcomes could be reviewed in an upgraded angle. Most importantly, we suggested that methods based on partitioned Bayesian Inference to reconstruct phylogeny applied in this study should be followed as a golden standard of HBV genotype/subgenotype classification in the future. With this, the representative sequences of OTUs, which represented each subgenotype of HBV were given in the supplementary data for further study (Table 23). Using phylogenetic analysis, a sequence can be located in a clade with proper classification and that subgenotype of HBV can be precisely medicated more effectively when more knowledge between genetic variation and clinical characteristics is available.
Table 21.
Estimates of evolutionary divergence over sequences pairs between HBV genotypes.
| A | B | C | D | E | F | G | H | I | |
|---|---|---|---|---|---|---|---|---|---|
| B | 0.1031 | ||||||||
| C | 0.0966 | 0.0965 | |||||||
| D | 0.1068 | 0.1148 | 0.1111 | ||||||
| E | 0.1109 | 0.1250 | 0.1185 | 0.0902 | |||||
| F | 0.1539 | 0.1553 | 0.1502 | 0.1530 | 0.1549 | ||||
| G | 0.1230 | 0.1387 | 0.1360 | 0.1251 | 0.1157 | 0.1594 | |||
| H | 0.1557 | 0.1573 | 0.1525 | 0.1522 | 0.1569 | 0.0865 | 0.1601 | ||
| I | 0.0848 | 0.0938 | 0.0701 | 0.1061 | 0.1118 | 0.1490 | 0.1198 | 0.1506 | |
| J | 0.1277 | 0.1208 | 0.1180 | 0.1337 | 0.1274 | 0.1520 | 0.1364 | 0.1533 | 0.1145 |
Table 22.
Estimates of evolutionary divergence over sequences pairs within HBV genotypes.
| A | B | C | D | E | F | G | H | I | J |
|---|---|---|---|---|---|---|---|---|---|
| 0.0471 | 0.0516 | 0.0405 | 0.0424 | 0.0295 | 0.0502 | 0.0075 | 0.0149 | 0.0463 | n/c |
n/c, not computable for only one sequence in Genotype J.
Table 23.
Reference sequences recommended for phylogenetic analysis of HBV genotyping.
| Genotype A | |
|---|---|
| A1 | AB116082, AF418685, AF418689, FJ692566, AF418683, FJ692570, FJ692583, FJ692586, AB246317, JN182323, JN182326, AF297623, GU563545, AF297625, HQ646555, HQ646556, FJ692573, AY233275, AF297621, U87742, AY233290, AY233279 |
| A2 | AF143304, AF143306, DQ298163, GQ477469, EU859908, EU859927, GQ477500, FJ904411, EU859952 |
| A3 | AM184125, AB194952, HM363613, FN545825, FJ692555, FJ692599, FJ692609, FJ692608, FJ692611, FN545829, FN545840, FN545832, FN545839, FN545833, FN545837, GQ161813, FJ349296, FN545826 |
| Genotype B | |
|---|---|
| B1 | AB073847, AB300371, AB073849, AB073856, AB642101, AB073858, AB302095, AB106884, AB073853, AB642093 |
| B2 | AF461360, HM011504, EU939638, EU939660, GQ377638, HM011475, EU939636, FJ386648, EU579441, EU939675, JQ027313, EU547563, HM011466, FJ518811, FJ562262, GQ924630, DQ995804, FJ899790, EU939661 |
| B3 | AB219427, AB241116, GQ924640, AB219430, GQ924617, JQ027328, JQ429079, EU660230, GQ358140, GQ924628, GQ924639, GQ924641, DQ361535, EU331000, GQ358144, GQ358145, HM011487, GQ924621, GQ924635, AY800392, GQ924656, GQ358150 |
| B4 | GQ924626 |
| B5 | AB287320, DQ463799, DQ463802, JN792899, JN792901 |
| Genotype E | |
|---|---|
| E | AB219533, DQ060829, GQ161805, EU239220, GQ161774, AB219534, HM363611, HM363583, FN545824 |
| Genotype F | |
|---|---|
| F1 | EU670261, HM590472, FJ589065 |
| F2 | DQ899142, DQ899144 |
| F3 | AB036905, FJ589068, DQ899150 |
| F4 | AB214516, JQ272888, AB365453, HE981178 |
| Genotype H | |
|---|---|
| H | AB275308 |
| Genotype J | |
|---|---|
| J | AB486012 |
We believe that in the near future, after correctly genotyping and subgenotyping, more discoveries should be jointly carried out based on the symptoms, diagnosis, drug use, drug resistance feedbacks and prognosis of a certain type of hepatitis B. Only by combining molecular genetics with clinical practice, with the joint efforts of both laboratory scientists and physicians in departments, the work of genotyping can play a more profound role in the treatment of hepatitis B [46, 47, 48, 49].
In conclusion, based on over 4,000 sequences and partitioned Bayesian inference, this study updated the classification system of non-recombinant HBV. That is, phylogenetic analysis rather than sequence divergence should be used in genotyping or subgenotyping of HBV. Compared to previous taxonomy, fourteen subgenotypes (A5–A7, B5–B9, C2–C4, C7, and D6-D7) were revised in the new standard. Now the HBV is divided into ten genotypes (A-J) and 24 subgenotypes (A1–A3, B1–B5, C1–C6, D1–D6 and F1–F4; Fig. 11).
Fig. 11.
Reference topology recommended for phylogenetic analysis of HBV genotyping. For limited space, the subgenotypes of genotype F [F3(3 sequences), F4(2 sequences), F2(2 sequences) and F1(3 sequences) from the top down] were unmarked in the figure.
As a significant change to the criterion that has existed for nearly 30 years, it will trigger a radical rethink about how the development of science promoting human cognition. Accompanied with subsequent clinical investigation, the right solutions will eventually be unraveled.
Declarations
Author contribution statement
Yonghua Yin, Kai He: performed the experiments; analyzed and interpreted the data; wrote the paper.
Wei Liu, Pu Liao, Min Xu, Bingting Wu: performed the experiments; analyzed and interpreted the data.
Lianming Du: contributed reagents, materials, analysis tools or data.
Yu Liu, Miao He: conceived and designed the experiments; wrote the paper.
Funding statement
This work was supported by the CAMS Innovation Fund for Medical Sciences (CIFMS, No.2016-I2M-3-025 to M He, No.2016-I2M-1-018 to Y Liu, No. 2016-I2M-3-024 to Yin), the Fundamental Research Funds for the Central Universities (No. 3332018125 to Wu), National Natural Science Foundation of China (No. 81572089 to Liao), National Key Research and Development Program of China (No. 2018YFE0107500 to M He), and Non-profit Central Research Institute Fund of Chinese Academy of Medical Sciences (No. 2018PT32016 to Yin).
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Not applicable.
References
- 1.MacCallum F.O. Homologous serum hepatitis. Lancet. 1947;250:691–692. [Google Scholar]
- 2.Blumberg B.S., Alter H.J., Visnich S. A “new” antigen in leukemia sera. Jama. 1965;191:541–546. doi: 10.1001/jama.1965.03080070025007. [DOI] [PubMed] [Google Scholar]
- 3.Lozano R., Naghavi M., Foreman K., Lim S., Shibuya K., Aboyans V., Abraham J. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2013;380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hepatitis B. 2016. WHO.http://www.who.int/mediacentre/factsheets/fs204/en/ [Google Scholar]
- 5.Kao J.H., Chen P.J., Lai M.Y., Chen D.S. Hepatitis B genotypes correlate with clinical outcomes in patients with chronic hepatitis B. Gastroenterology. 2000;118:554–559. doi: 10.1016/s0016-5085(00)70261-7. [DOI] [PubMed] [Google Scholar]
- 6.Kao J.H., Wu N.H., Chen P.J., Lai M.Y., Chen D.S. Hepatitis B genotypes and the response to interferon therapy. J. Hepatol. 2000;33:998–1002. doi: 10.1016/s0168-8278(00)80135-x. [DOI] [PubMed] [Google Scholar]
- 7.Kramvis A. Genotypes and genetic variability of hepatitis B virus. Intervirology. 2014;57:141–150. doi: 10.1159/000360947. [DOI] [PubMed] [Google Scholar]
- 8.Okamoto H., Tsuda F., Sakugawa H., Sastrosoewignjo R.I., Imai M., Miyakawa Y., Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J. Gen. Virol. 1988;69:2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 9.Norder H., Hammas B., Löfdahl S., Couroucé A.M., Magnius L.O. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J. Gen. Virol. 1992;73:1201–1208. doi: 10.1099/0022-1317-73-5-1201. [DOI] [PubMed] [Google Scholar]
- 10.Kramvis A., Kew M., François G. Hepatitis B virus genotypes. Vaccine. 2005;23:2409–2423. doi: 10.1016/j.vaccine.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 11.Tatematsu K., Tanaka Y., Kurbanov F., Sugauchi F., Mano S., Maeshiro T., Nakayoshi T. A genetic variant of hepatitis B virus divergent from known human and ape genotypes isolated from a Japanese patient and provisionally assigned to new genotype J. J. Virol. 2009;83:10538–10547. doi: 10.1128/JVI.00462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nei M., Kumar S. first ed. Oxford University press; New York: 2000. Molecular Evolution and Phylogenetics; pp. 23–36. [Google Scholar]
- 13.Kurbanov F., Tanaka Y., Kramvis A., Simmonds P., Mizokami M. When should ‘I’ consider a new hepatitis B virus genotype? J. Virol. 2008;82:8241–8242. doi: 10.1128/JVI.00793-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayerat C., Mantegani A., Frei P.C. Does hepatitis B virus (HBV) genotype influence the clinical outcome of HBV infection? J. Viral Hepat. 1999;6:299–304. doi: 10.1046/j.1365-2893.1999.00174.x. [DOI] [PubMed] [Google Scholar]
- 15.Holder M., Lewis P.O. Phylogeny estimation: traditional and Bayesian approaches. Nat. Rev. Genet. 2003;4:275–284. doi: 10.1038/nrg1044. [DOI] [PubMed] [Google Scholar]
- 16.Bartholomeusz A., Schaefer S. Hepatitis B virus genotypes: comparison of genotyping methods. Rev. Med. Virol. 2004;14:3–16. doi: 10.1002/rmv.400. [DOI] [PubMed] [Google Scholar]
- 17.Posada D., Crandall K.A. The effect of recombination on the accuracy of phylogeny estimation. J. Mol. Evol. 2002;54:396–402. doi: 10.1007/s00239-001-0034-9. [DOI] [PubMed] [Google Scholar]
- 18.Schierup M.H., Hein J. Consequences of recombination on traditional phylogenetic analysis. Genetics. 2000;156:879–891. doi: 10.1093/genetics/156.2.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2005;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 20.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin Y.H., Du L.M., Yue B.S. GenScalpel: an application for sequence retrieval and extraction from the GenBank flatfile. J. Hered. 2012;103:908–911. doi: 10.1093/jhered/ess052. [DOI] [PubMed] [Google Scholar]
- 22.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 24.Martin D.P., Murrell B., Golden M., Khoosal A., Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kloepper T.H., Huson D.H. Drawing explicit phylogenetic networks and their integration into SplitsTree. BMC Evol. Biol. 2008;8:22. doi: 10.1186/1471-2148-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouckaert R., Heled J., Kühnert D., Vaughan T., Wu C.H., Xie D., Suchard M.A. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2014;10 doi: 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanfear R., Calcott B., Ho S.Y., Guindon S. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 2012;29:1695–1701. doi: 10.1093/molbev/mss020. [DOI] [PubMed] [Google Scholar]
- 28.Huelsenbeck J., Rannala B. Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Syst. Biol. 2004;53:904–913. doi: 10.1080/10635150490522629. [DOI] [PubMed] [Google Scholar]
- 29.Sugauchi F., Kumada H., Acharya S.A., Shrestha S.M., Gamutan M.T., Khan M., Gish R.G. Epidemiological and sequence differences between two subtypes (Ae and Aa) of hepatitis B virus genotype A. J. Gen. Virol. 2004;85:811–820. doi: 10.1099/vir.0.79811-0. [DOI] [PubMed] [Google Scholar]
- 30.Andernach I.E., Nolte C., Pape J.W., Muller C.P. Slave trade and hepatitis B virus genotypes and subgenotypes in Haiti and Africa. Emerg. Infect. Dis. 2009;15:1222–1228. doi: 10.3201/eid1508.081642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugauchi F., Orito E., Ichida T., Kato H., Sakugawa H., Kakumu S., Ishida T. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J. Virol. 2002;76:5985–5992. doi: 10.1128/JVI.76.12.5985-5992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowalec K., Minuk G.Y., Børresen M.L., Koch A., McMahon B.J., Simons B., Osiowy C. Genetic diversity of hepatitis B virus genotypes B6, D and F among circumpolar indigenous individuals. J. Viral Hepat. 2013;20:122–130. doi: 10.1111/j.1365-2893.2012.01632.x. [DOI] [PubMed] [Google Scholar]
- 33.Sakamoto T., Tanaka Y., Simonetti J., Osiowy C., Borresen M.L., Koch A., Kurbanov F. Classification of hepatitis B virus genotype B into 2 major types based on characterization of a novel subgenotype in Arctic indigenous populations. J. Infect. Dis. 2007;196:1487–1492. doi: 10.1086/523111. [DOI] [PubMed] [Google Scholar]
- 34.Thedja M.D., Muljono D.H., Nurainy N., Sukowati C.H., Verhoef J., Marzuki S. Ethnogeographical structure of hepatitis B virus genotype distribution in Indonesia and discovery of a new subgenotype. B9. Arch. Virol. 2011;156:855–868. doi: 10.1007/s00705-011-0926-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nurainy N., Muljono D.H., Sudoyo H., Marzuki S. Genetic study of hepatitis B virus in Indonesia reveals a new subgenotype of genotype B in east Nusa Tenggara. Arch. Virol. 2008;153:1057–1065. doi: 10.1007/s00705-008-0092-z. [DOI] [PubMed] [Google Scholar]
- 36.Huy T.T., Ushijima H., Quang V.X., Win K.M., Luengrojanakul P., Kikuchi K., Sata T. Genotype C of hepatitis B virus can be classified into at least two subgroups. J. Gen. Virol. 2004;85:283–292. doi: 10.1099/vir.0.19633-0. [DOI] [PubMed] [Google Scholar]
- 37.Sugauchi F., Mizokami M., Orito E., Ohno T., Kato H., Suzuki S., Kimura Y. A novel variant genotype C of hepatitis B virus identified in isolates from Australian Aborigines: complete genome sequence and phylogenetic relatedness. J. Gen. Virol. 2001;82:883–892. doi: 10.1099/0022-1317-82-4-883. [DOI] [PubMed] [Google Scholar]
- 38.Khan A., Tanaka Y., Saito H., Ebinuma H., Sekiguchi H., Iwama H., Wakabayashi G. Transmission of hepatitis B virus (HBV) genotypes among Japanese immigrants and natives in Bolivia. Virus Res. 2008;132:174–180. doi: 10.1016/j.virusres.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 39.Hannachi N., Fredj N.B., Bahri O., Thibault V., Ferjani A., Gharbi J., Triki H. Molecular analysis of HBV genotypes and subgenotypes in the Central-East region of Tunisia. Virol. J. 2010;7:302. doi: 10.1186/1743-422X-7-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meldal B.H., Moula N.M., Barnes I.H., Boukef K., Allain J.P. A novel hepatitis B virus subgenotype, D7, in Tunisian blood donors. J. Gen. Virol. 2009;90:1622–1628. doi: 10.1099/vir.0.009738-0. [DOI] [PubMed] [Google Scholar]
- 41.Fang Z.L., Hué S., Sabin C.A., Li G.J., Yang J.Y., Chen Q.Y., Fang K.X. A complex hepatitis B virus (X/C) recombinant is common in Long an county, Guangxi and may have originated in southern China. J. Gen. Virol. 2011;92:402–411. doi: 10.1099/vir.0.026666-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devesa M., Loureiro C.L., Rivas Y., Monsalve F., Cardona N., Duarte M.C., Poblete F. Subgenotype diversity of hepatitis B virus American genotype F in Amerindians from Venezuela and the general population of Colombia. J. Med. Virol. 2008;80:20–26. doi: 10.1002/jmv.21024. [DOI] [PubMed] [Google Scholar]
- 43.Pourkarim M.R., Amini-Bavil-Olyaee S., Verbeeck J., Lemey P., Zeller M., Rahman M., Maes P. Molecular evolutionary analysis and mutational pattern of full-length genomes of hepatitis B virus isolated from Belgian patients with different clinical manifestations. J. Med. Virol. 2010;82:379–389. doi: 10.1002/jmv.21726. [DOI] [PubMed] [Google Scholar]
- 44.Kay A., Zoulim F. Hepatitis B virus genetic variability and evolution. Virus Res. 2007;127:164–176. doi: 10.1016/j.virusres.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 45.Littlejohn M., Locarnini S., Yuen L. Origins and evolution of hepatitis B virus and hepatitis D virus. Cold Spring Harb. Perspect. Med. 2016;6:a021360. doi: 10.1101/cshperspect.a021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Märschenz S., Endres A.S., Brinckmann A., Heise T., Kristiansen G., Nürnberg P., Krüger D.H. Functional analysis of complex hepatitis B virus variants associated with development of liver cirrhosis. Gastroenterology. 2006;131:765–780. doi: 10.1053/j.gastro.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 47.Schweitzer A., Horn J., Mikolajczyk R.T., Krause G., Ott J.J. Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013. Lancet. 2015;386:1546–1555. doi: 10.1016/S0140-6736(15)61412-X. [DOI] [PubMed] [Google Scholar]
- 48.Shih H.H., Jeng K.S., Syu W.J., Huang Y.H., Su C.W., Peng W.L., Sheen I.J. Hepatitis B surface antigen levels and sequences of natural hepatitis B virus variants influence the assembly and secretion of hepatitis d virus. J. Virol. 2008;82:2250–2264. doi: 10.1128/JVI.02155-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugiyama M., Tanaka Y., Kato T., Orito E., Ito K., Acharya S.K., Gish R.G. Influence of hepatitis B virus genotypes on the intra- and extracellular expression of viral DNA and antigens. Hepatology. 2006;44:915–924. doi: 10.1002/hep.21345. [DOI] [PubMed] [Google Scholar]