Abstract
Background
Campylobacter is a leading global cause of bacterial gastroenteritis, motivating research to identify sources of human infection. Population genetic studies have been increasingly applied to this end, mainly using multilocus sequence typing (MLST) data.
Objectives
This review aimed to summarise approaches and findings of these studies and identify best practice lessons for this form of genomic epidemiology.
Methods
We systematically reviewed publications using MLST data to attribute human disease isolates to source. Publications were from January 2001, when this type of approach began. Searched databases included Scopus, Web of Science and PubMed. Information on samples and isolate datasets used, as well as MLST schemes and attribution algorithms employed, was obtained. Main findings were extracted, as well as any results’ validation with subsequent correction for identified biases. Meta-analysis is not reported given high levels of heterogeneity.
Results
Of 2,109 studies retrieved worldwide, 25 were included, and poultry, specifically chickens, were identified as principal source of human infection. Ruminants (cattle or sheep) were consistently implicated in a substantial proportion of cases. Data sampling and analytical approaches varied, with five different attribution algorithms used. Validation such as self-attribution of isolates from known sources was reported in five publications. No publication reported adjustment for biases identified by validation.
Conclusions
Common gaps in validation and adjustment highlight opportunities to generate improved estimates in future genomic attribution studies. The consistency of chicken as the main source of human infection, across high income countries, and despite methodological variations, highlights the public health importance of this source.
Keywords: Campylobacter, MLST, source attribution, food-borne infections, bacterial infections, surveillance, epidemiology, molecular methods
Introduction
Campylobacter gastroenteritis is a leading cause of acute bacterial gastroenteritis in high, low, and middle income countries. The number of confirmed cases has continued to increase across countries of the European Union (214,000 in 2013 to 246,000 in 2016 and 2017) [1], and over 800,000 cases are estimated to occur annually in the United States (data from 2000 to 2008) [2]. In low income countries Campylobacter is increasingly implicated in growth faltering among children under 2 years of age [3].
Chicken products have been identified as an important risk factor for human infection by a variety of techniques including natural experiments, case–control studies, and increasingly by the application of genotypic methods [4-10]. Other infection sources identified by observational epidemiological studies include cattle, sheep, pigs, wild birds and the environment [10].
Alongside epidemiological studies there has been an increasing use of population genetic analyses to attribute human cases to likely sources. In these analyses, the genetic diversity of isolates from humans is compared with that of collections of Campylobacter isolates obtained from possible sources of infection, allowing quantitative attribution to these sources.
Multilocus sequence type (MLST) data [8] have become the standard data used in such population genetic analyses, the results of which are generally consistent with the findings from epidemiological analyses [11,12]. Large collections of isolates have been sequenced at the MLST loci from a wide range of sources. The approaches provide a potential means of monitoring change in sources of human infection, for example those that occur as a consequence of public health and food chain interventions [13]. Insights obtained from seven-gene MLST analyses can also inform analyses using more extensive genomic data, as large well sampled datasets of whole genome sequenced (WGS) isolates accumulate from humans and putative sources. Other techniques such as multiplex PCR, PFGE, and comparative genomic fingerprinting have neither been taken up widely nor offer compatibility with whole genome based approaches.
Studies analysing MLST data vary in terms of both the analytical algorithm applied and the reference datasets used [13-18] (‘reference’ data throughout this paper describe data from known reservoirs such as animal species that can act as sources of human infection). Here, our objectives on the use of MLST analysis to attribute infection in human populations to sources are to: (i) summarise the findings from these studies to date; (ii) describe the approaches used; and (iii) identify lessons to guide further genetic source attribution work using these data and more extensive genomic data as they become available.
Methods
Search strategy
The literature search strategy aimed to identify articles attributing Campylobacter isolates from human infections to possible sources using MLST-based attribution algorithms. Systematic searches were performed on the Scopus, Web of Science, and PubMed databases using a search string comprised of the following terms: ‘campylobacter$’ AND ‘multilocus OR genotype OR genotyping’ AND ‘source$ OR assignment OR attribution’ AND ‘human OR clinical OR disease’. These were carried out on 23 November 2017 and limited to items published from January 2001 onwards, as the C. jejuni MLST scheme was first described in this year [8].
The publication lists arising from the three searches were combined and duplicate records removed. Titles and abstracts of the remaining studies were reviewed to ensure that they described the source attribution of clinical C. jejuni and/or C. coli isolates to potential source populations using an MLST-based algorithm, in English. Complete texts of the final list were then considered to identify whether or not they satisfied the inclusion criteria. Reference sections of these papers were also searched for further candidate publications.
Texts of the resulting article list were scrutinised to identify: (i) datasets used, including their size, geographical origin, and year of disease and potential source isolates; (ii) sample types from which isolates were obtained, e.g. retail chicken meat, cattle faecal sample; (iii) attribution method(s) employed; (iv) loci in the typing scheme; (v) validation, such as self-attribution of isolates from known sources; (vi) adjustment of attribution to correct for identified biases; (vii) proportion of Campylobacter clinical isolates attributed to each source.
When relevant details were not in the text, values were calculated from available data or obtained by contacting the authors where possible. In articles that compared results from more than one dataset, for example comparing clinical samples among different years or rural vs urban disease samples, baseline or mean values are reported in this review.
Statistical analysis
The proportions of human infection with C. jejuni attributed to poultry estimated by the two most commonly used algorithms (Asymmetric Island (AI) [17] and STRUCTURE [19]) were compared by the two-group mean comparison test and variation in this proportion across studies described using the I2 index from the metaprop command using StataIC 15 (StataCorpLP, Texas).
Results
Search results and subsequent exclusions, detailed in the preferred reporting items for systematic reviews and meta-analyses (PRISMA) flow diagram (Figure 1) and Table, resulted in 25 articles [13,14,16,17,20-41].
Figure 1.
Flow diagram of the search strategy used to identify articles for inclusion in the systematic reviewon source attribution of human campylobacteriosis using multilocus sequence typing, since January 2001
MLST: multilocus sequence typing.
a The search terms were broad and brought in diverse articles with a very diverse corresponding set of reasons for exclusion, including articles that had no relation with source attribution as well as those using other approaches. No useful summary of this diversity was possible.
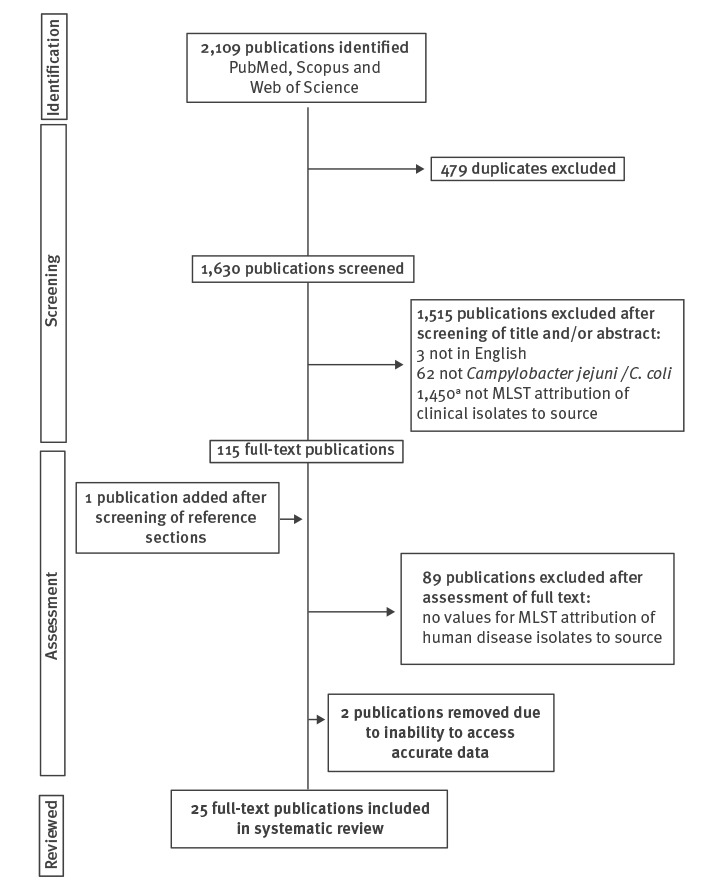
Table. Summary of studies reviewed, indicating composition of reference datasets used for attribution analysis of human Campylobacter infections to animal or environmental sources, 2001−2017 (n = 25 studies).
| Paper (author/year) |
Poultry/chicken | Ruminant/cattle/sheep | Environmental/wild bird dataset(s) | Multiple sample types per source | Number of source populations | Years (max) between clinical and reference isolates | |
|---|---|---|---|---|---|---|---|
| C. jejuni | C. coli | ||||||
| Bessell (2012) [46] | Chicken | Ruminant | Wild bird | Yes | 3 | NA | 16 |
| Boysen (2014) [14] | Poultry | Cattle | NA | Yes | 5 | NA | 1a |
| Cody (2015) [22] | Chicken | Cattle, sheep | Wild bird | No | 4 | NA | 14 |
| Di Giannatale (2016) [23] | Chicken | Cattle, small ruminant | Wild bird, environmental | Yes | 6 | NA | No data |
| French (2008) [24] | Chicken | Cattle, sheep | Wild bird, environmental water | Yes | 5 | NA | 3a |
| Jonas (2015) [25] | Chicken | Cattle | NA | Yes | 3 | 3 | 6 |
| Kittl (2013) [26] | Chicken | NA | NA | Yes | 2 | 2 | 10a |
| Kovac (2018) [27] | Poultry | Cattle | Environmental (inc. wild birds) | Yes | 3 | NA | 12a |
| Levesque (2013) [28] | Chicken | Cattle | Environmental water, wild bird | No | 4 | NA | 2a |
| Mossong (2016) [29] | Poultry | Ruminant | Environmental water | Yes | 4 | 4 | 10 |
| Mughini Gras (2012) [30] | Chicken | Cattle, sheep | Environmental (inc. wild birds) | Yes | 5 | 5 | 13 |
| Mughini Gras (2013) [31] | Chicken | Cattle, sheep | Environmental (inc. wild birds) | Yes | 5 | 5 | 13 |
| Mullner (2009a) [16] | Chicken | Cattle, sheep | Environmental (inc. wild birds) | Yes | 4 | NA | 3a |
| Mullner (2009b) [13] | Chicken | Cattle, sheep | Environmental (inc. wild birds) | Yes | 4 | NA | 3a |
| Nohra (2016) [32] | Poultry | Ruminant | Environmental water | No | NA | 3 | 5 |
| Rosner (2017) [33] | Chicken | Cattle | NA | Yes | 5 | 5 | 10 |
| Roux (2013) [34] | Chicken | Cattle, sheep | NA | Yes | NA | 4 | 1a |
| Sears (2011) [35] | Poultry | Cattle, sheep | Environmental water | Yes | 4 | NA | 3a |
| Sheppard (2009) [36] | Chicken | Ruminant | Environmental (inc. wild birds) | Yes | 3b | 5b | 16 |
| Sheppard (2010) [37] | Chicken | Ruminant | Environmental (inc. wild birds) | Yes | NA | 5 | 6 |
| Smid (2013) [38] | Chicken | Cattle, sheep | Environmental (inc. wild birds) | Yes | 4 | 4 | 28 |
| Strachan (2009) [39] | Chicken | Cattle, sheep | Wild bird | No | 5 | 5 | 6 |
| Strachan (2013) [40] | Chicken | Cattle, sheep | Wild bird | No | 5 | 5 | 2a |
| Thépault (2017) [41] | Chicken | Ruminant | Environmental water | Yes | 3 | NA | 10 |
| Wilson (2008) [17] | Chicken | Cattle and sheep | Wild bird, water, sand | Yes | 8 | NA | 12 |
Inc.: including; NA: not applicable, whereby this species (C. jejuni or C. coli) or possible source was not included in the study.
a Clinical and attribution datasets from same temporal range.
b C. jejuni results expressed for ruminants; C. coli results expressed for cattle and sheep separately.
Datasets
Twelve papers only investigated C. jejuni [13,14,16,17,21-24,27,28,35,41] and three studied only C. coli [32,34,37]. Of the 10 publications that considered both C. jejuni and C. coli [25,26,29-31,33,36,38,39,42], six expressed results by single species [25,26,29,30,33,36] and four reported attribution for the two species jointly [31,38,39,42]. Studies included human clinical isolates from the United Kingdom (n = 9 studies), New Zealand (n = 5), the Netherlands (n = 3), Germany (n = 2), Luxembourg (n = 2), Switzerland (n = 2), Austria, Canada, Denmark, France, Italy and Slovenia (n = 1 study each) (Supplementary Table).
Fifteen articles reported using reference datasets that combined isolates from more than one potential animal host species or animal species and an environmental reservoir as a single class. Poultry datasets containing predominantly chicken but including other farmed birds were used as an attribution source in five articles [14,27,29,32,35], cattle and sheep isolates combined as ruminants in six reports [21,29,32,36,37,41] and an environmental category comprising at least water and wild bird isolates in 11 [13,16,27,30-32,35-38,43]. A single sample type (e.g. retail meat or faeces) was considered for each host animal species in reference datasets from five studies [22,28,32,39,40] rather than combining isolates across different sample types from the same source.
The highest number of potential source populations to which disease isolates were attributed was eight [17] and the lowest two [26], with reference datasets ranging in size from two [29] to 1,288 [21,36] samples, across the articles reviewed (Supplementary Table). Eleven publications used source isolates from the same time period as human cases [13,16,21,24,26-28,34,35,37,40], with the maximum possible temporal difference between any human and any source isolate ranging between 1 and 12 years in these. The longest time difference between human case and source isolates among the remaining articles was 28 years [38].
One study explicitly considered domestic (with no history of international travel) and travel-associated human cases separately [14] and one compared attribution results of clinical samples from two countries [41]. Reference datasets were limited to the same country as the clinical isolates in 10 studies [13,16,26,28,32,34,35,39,40,43] and an eleventh article compared domestic human case isolates with those of travel-associated cases [14]. Two further publications included reference data from non-domestic sources having established that these countries shared similar disease genotype frequencies as domestic human cases [23,30], and a third compared attribution results using reference MLST data from the same country as the cases and from different countries [38]. The remaining articles used reference isolates from internationally widespread locations.
Attribution models and data
The studies included used one or more of five attribution algorithms: (i) the Asymmetric Island (AI) model (n = 13 reports) [17]; (ii) STRUCTURE (n = 11) [19]; (iii) the Modified Hald (MH) model (n = 4) [16]; (iv) the Dutch model (n = 2) [44]; and (v) the Hald model (n = 1) [45] (Supplementary Table). Twenty-four of the publications reported data based on the seven housekeeping genes originally described by Dingle [7,8] and a single article identified and used 15 novel host-segregating loci [41]. Clinical datasets composed of both C. jejuni and C. coli, and those that considered the two species individually, were analysed using either STRUCTURE or AI analysis, whereas studies using the Dutch, Hald, and MH attribution models were restricted to analyses of C. jejuni (Supplementary Table). Five of the 25 articles used more than one attribution method [13,14,24,35,36], although one of these only reported results from a single model [35].
Self-attribution and other validation
Self-attribution estimates the probability that, for example, a chicken-origin isolate will be attributed back to the chicken host reference sample, and repeats this to measure accuracy. These analyses were performed in five of the articles using the AI, STRUCTURE, or both algorithms, and reported average percentage accuracy values for each source tested [26,36-38,41]. For both C. jejuni [36], and C. coli [26,37,38,41] AI showed greater self-attribution accuracy than the STRUCTURE algorithm. No publication reported adjustment of subsequent attribution of isolates from human cases based on bias identified in self-attribution.
Attribution results
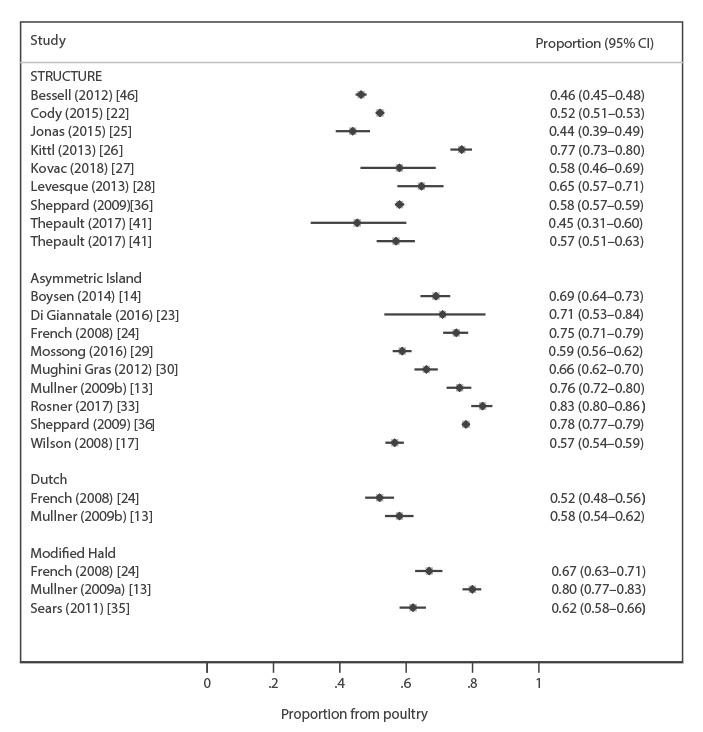
Attribution of human C. jejuni isolates to poultry using seven-locus MLST by the AI model ranged between 57% and 83% [13,14,17,23,24,29,30,33,36]; by STRUCTURE between 44% and 77% [22,25-28,36,41,46]; the Dutch model between 52% and 58% [13,24]; the Hald model 52% [14]; and the MH model between 62% and 80% [13,16,24,35] (Figure 2 and Supplementary Table). In all four studies reporting results from C. jejuni datasets using more than one attribution algorithm, the AI model attributed higher proportions to poultry than other methods [13,14,24,36]. The one study using STRUCTURE analysis of 15 alternative C. jejuni loci attributed 57% to poultry [41]. The variation across estimates for attribution to poultry among studies, as determined by the I2 index, was greater than 90% for both AI and STRUCTURE analyses, showing substantial between-study variation so that a single summary estimate of results by method or overall is not supported (Figure 2).
Figure 2.
Forest plot of the proportion of Campylobacter jejuni clinical isolates attributed to poultry by different studies, and uncertainty around these estimates
CI: confidence interval.
I2 index > 90% for STRUCTURE and Asymmetric Island models so no summary estimates are given.
Between 38.0% and 82.4% of human C. coli were estimated to come from poultry in studies using the AI model [29,30,32,33,36,37], and between 40.0% and 86.4% using STRUCTURE [25,26,34,36].
Where both C. jejuni and C. coli species results were reported together, the AI model attributed between 68% and 77% to poultry [31,38] and STRUCTURE identified a range of between 19% and 48.7% [39,40]. Studies using the AI model were found to attribute significantly more C. jejuni isolates to poultry than STRUCTURE (p = 0.007), but no difference was detected for C. coli isolates attributed to poultry by these two methods.
In all articles that reported the attribution of C. jejuni to poultry, cattle and other sources, using seven-locus MLST, cattle were the second most predominant source identified by all methods with the exception of one STRUCTURE and one AI analysis that identified sheep [22] and pets [33] as the second most prevalent sources, respectively. Where cattle and sheep were considered together they were the second most prevalent C. jejuni source identified regardless of the algorithm used in three seven-locus MLST studies [21,29,36].
Ruminants were identified as the predominant source of clinical C. coli in one AI analysis [32], and sheep in one STRUCTURE analysis [34], while equal proportions of disease were attributed to poultry and sheep in a further STRUCTURE analysis [36]. In the five remaining AI [29,30,33,36,37] studies and one STRUCTURE [25] study considering the sources of human C. coli infection, poultry were the predominant source, followed by either cattle or a combined ruminant class.
The two studies that reported AI analyses of combined C. jejuni and C. coli datasets [31,38] both identified poultry as the primary and cattle as the secondary sources of disease. One STRUCTURE analysis of both species, comparing attribution at three time periods, reported poultry as the main disease source, with the secondary source changing from cattle to sheep in the final study period [40]. A further STRUCTURE analysis investigating the sources of C. jejuni and C. coli in children in urban and rural settings found poultry to be the predominant source followed by cattle in urban areas, but cattle as the prevalent and wild birds as the secondary sources in rural areas [39].
Discussion
This review supports poultry and ruminants as the main sources of human campylobacteriosis across the settings investigated, with more than half of human campylobacteriosis cases attributed to poultry. Studies varied in the populations investigated, algorithms used, and approaches to choosing reference datasets for potential sources, but consistently identified the importance of poultry as a source. All studies were from high-income countries, with a substantial evidence gap for low- and middle-income countries.
Between-study comparisons were limited by the wide range of approaches used. Many enhanced the size of reference populations by access to publicly available datasets alongside more limited local data, sometimes using source data distant from the human isolate datasets, while others were limited to smaller reference datasets closer in time and place to the human infections. Although host-associated genetic signal has been shown to be stronger than the effects of geography [20], geographical distance among isolates from a single host source has the potential to cause bias in attribution analyses [20,47]. Smid and colleagues [38] investigated a range of factors affecting the outcome of AI attribution and identified that the inclusion of non-contemporaneous data and data from other countries reduced the attribution of Dutch disease isolates to chicken. Other authors used principal component analysis, to determine the most suitable countries of isolate origin for inclusion in attribution datasets [30,31,33]. Temporal separation between isolates may also create bias, with more pronounced effects reported for C. coli than C. jejuni populations [26]. There is no definitive evidence on the relative benefits of having larger more diverse reference datasets from potential sources of infection, or smaller ones closer to the human case isolates in time and place.
The number and composition of sources considered also varied across studies. Environmental isolates were considered as a proxy for other wildlife sources in some, but frequently included water samples that can be contaminated by farm slurry, agricultural run-off, or the disposal of abattoir effluent, as well as by wild animals [28,30]; indeed, ruminants have been implicated in cases of drinking water contamination by these means [48]. Datasets acquired from publicly available database collections rarely detailed sample type and often included samples from more than one point in the food chain [13,14,17,23,25,26,30,31,33-38,41,46,49,50]. Since all genotypes do not survive food processing procedures equally, broiler farm samples may, for example, represent the exposure of most individuals to Campylobacter from chicken less accurately than samples from retail poultry [51]. A combined ruminant (cattle and sheep) source population rather than separate cattle and sheep datasets was used in some studies but not in others, further limiting comparability among investigations. These ruminant species may host substantially overlapping Campylobacter populations [52].
The seven-locus MLST data used were primarily acquired using techniques described in the original publication of this methodology [8], but which have now been largely superseded as whole genome sequence (WGS) data are more common. Despite this change, the backwards compatibility of the gene-by-gene approach to WGS analysis permits extraction of the relevant internal gene fragment alleles for use in existing attribution methodologies, and also facilitates the identification of additional genes for use in such algorithms [53]. To date there has been only one publication detailing attribution using alternative loci identified from WGS, but as this study did not make any comparison with seven-locus MLST it was not possible to determine whether the novel methodology improved the accuracy of attributing generalist genotypes [41].
Comparison of results from different attribution models analysing the same datasets identified that the choice of model may be important. Results from the AI and STRUCTURE algorithms demonstrated that the AI model attributed more human Campylobacter infection to poultry, whereas the STRUCTURE algorithm attributed a higher proportion to ruminants [36]. These observations were confirmed by assessment of data from across all C. jejuni studies using these two algorithms where AI attributed a larger proportion to poultry.
The same pattern of results was observed when the AI, MH, and Dutch models were used to analyse highly similar C. jejuni datasets in two publications [13,43]; in all analyses poultry was predominant but the extent of this was more extreme with AI than with the MH or Dutch algorithms. In addition, the Dutch model identified environmental and wild-bird sources as causing a greater number of disease cases than either the AI or MH models.
Only in five studies was the accuracy of the attribution approach tested. Some evidence suggested that AI self-attributed poultry more accurately than STRUCTURE, although there is also grey literature reporting the opposite [54]. No study described adjustment of the raw results obtained in attributing human isolates for the biases estimated in self-attribution tests.
This systematic review brings together compelling evidence for poultry as the major source of human campylobacteriosis, with consistent results across several countries, time periods, and using different analytical algorithms and approaches to assembling isolate data from potential sources. The studies were mainly from Europe and New Zealand and highlight the gap in evidence for low and middle-income countries where Campylobacter may have a particularly large health burden [3]. This review also shows marked limitations as regards quality and comparability, with most studies not assessing their own accuracy. Moreover, none of the studies that measured accuracy and bias used this to adjust estimates of the proportion of human infection from each potential source or performing sensitivity analysis. The lack of evolution towards agreed optimal methods in the context of almost all studies using the same MLST data is striking. As WGS data become increasingly available, allowing the use of different genetic data across studies, moving to a consistent or optimum approach may be even more difficult although important to ensure comparability. The performance of tests of accuracy and bias such as self-attribution, and sensitivity analyses to take account of imperfect source attribution will be even more important. We recommend that validations, using approaches such as attribution of isolates from known sources, and adjustment for biases that are identified should be included in future population genetic source attribution studies and reports.
Acknowledgements
This work was supported by the United Kingdom Food Standards Agency [grant number FS101013]; the Wellcome Trust [grant numbers 087622 to M.C.J.M., 072782MA to N.D.M.]; and National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Gastrointestinal Infections at the University of Oxford in partnership with Public Health England (PHE). The views expressed are those of the author(s) and not necessarily those of the FSA, NHS, the NIHR, the Department of Health or Public Health England.
Supplementary Data
Conflict of interest: None declared.
Authors’ contributions: Alison Cody undertook the literature searches, analysis and writing of the first draft as well as revisions of the work. Noel McCarthy conceived and led the study, undertook some analyses and edited manuscript drafts and this revision. Norval Strachan and Martin Maiden contributed to planning of the work and reviewed and edited drafts of the manuscript.
References
- 1. European Food Safety Authority (EFSA) The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2017. EFSA J. 2018;16(12):5500 10.2903/j.efsa.2018.5500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, et al. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis. 2011;17(1):7-15. 10.3201/eid1701.P11101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amour C, Gratz J, Mduma E, Svensen E, Rogawski ET, McGrath M, et al. Epidemiology and Impact of Campylobacter Infection in Children in 8 Low-Resource Settings: Results From the MAL-ED Study. Clin Infect Dis. 2016;63(9):1171-9. 10.1093/cid/ciw542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Friesema IH, Havelaar AH, Westra PP, Wagenaar JA, van Pelt W. Poultry culling and Campylobacteriosis reduction among humans, the Netherlands. Emerg Infect Dis. 2012;18(3):466-8. 10.3201/eid1803.111024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stern NJ, Hiett KL, Alfredsson GA, Kristinsson KG, Reiersen J, Hardardottir H, et al. Campylobacter spp. in Icelandic poultry operations and human disease. Epidemiol Infect. 2003;130(1):23-32. 10.1017/S0950268802007914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vellinga A, Van Loock F. The dioxin crisis as experiment to determine poultry-related Campylobacter enteritis. Emerg Infect Dis. 2002;8(1):19-22. 10.3201/eid0801.010129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dingle KE, Colles FM, Falush D, Maiden MC. Sequence typing and comparison of population biology of Campylobacter coli and Campylobacter jejuni. J Clin Microbiol. 2005;43(1):340-7. 10.1128/JCM.43.1.340-347.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dingle KE, Colles FM, Wareing DRA, Ure R, Fox AJ, Bolton FE, et al. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39(1):14-23. 10.1128/JCM.39.1.14-23.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ikram R, Chambers S, Mitchell P, Brieseman MA, Ikam OH. A case control study to determine risk factors for Campylobacter infection in Christchurch in the summer of 1992-3. N Z Med J. 1994;107(988):430-2. [PubMed] [Google Scholar]
- 10. Domingues AR, Pires SM, Halasa T, Hald T. Source attribution of human campylobacteriosis using a meta-analysis of case-control studies of sporadic infections. Epidemiol Infect. 2012;140(6):970-81. 10.1017/S0950268811002676 [DOI] [PubMed] [Google Scholar]
- 11. Dingle KE, Colles FM, Ure R, Wagenaar JA, Duim B, Bolton FJ, et al. Molecular characterization of Campylobacter jejuni clones: a basis for epidemiologic investigation. Emerg Infect Dis. 2002;8(9):949-55. 10.3201/eid0809.02-0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sheppard SK, Dallas JF, MacRae M, McCarthy ND, Sproston EL, Gormley FJ, et al. Campylobacter genotypes from food animals, environmental sources and clinical disease in Scotland 2005/6. Int J Food Microbiol. 2009;134(1-2):96-103. 10.1016/j.ijfoodmicro.2009.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mullner P, Spencer SE, Wilson DJ, Jones G, Noble AD, Midwinter AC, et al. Assigning the source of human campylobacteriosis in New Zealand: a comparative genetic and epidemiological approach. Infect Genet Evol. 2009;9(6):1311-9. 10.1016/j.meegid.2009.09.003 [DOI] [PubMed] [Google Scholar]
- 14. Boysen L, Rosenquist H, Larsson JT, Nielsen EM, Sørensen G, Nordentoft S, et al. Source attribution of human campylobacteriosis in Denmark. Epidemiol Infect. 2014;142(8):1599-608. 10.1017/S0950268813002719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McCarthy ND, Colles FM, Dingle KE, Bagnall MC, Manning G, Maiden MC, et al. Host-associated genetic import in Campylobacter jejuni. Emerg Infect Dis. 2007;13(2):267-72. 10.3201/eid1302.060620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mullner P, Jones G, Noble A, Spencer SE, Hathaway S, French NP. Source attribution of food-borne zoonoses in New Zealand: a modified Hald model. Risk Anal. 2009;29(7):970-84. 10.1111/j.1539-6924.2009.01224.x [DOI] [PubMed] [Google Scholar]
- 17. Wilson DJ, Gabriel E, Leatherbarrow AJH, Cheesbrough J, Gee S, Bolton E, et al. Tracing the source of campylobacteriosis. PLoS Genet. 2008;4(9):e1000203. 10.1371/journal.pgen.1000203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.mlstdbNet Home Page. Available at: http://pubmlst.org/software/database/mlstdbnet/
- 19. Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sheppard SK, Colles F, Richardson J, Cody AJ, Elson R, Lawson A, et al. Host association of Campylobacter genotypes transcends geographic variation. Appl Environ Microbiol. 2010;76(15):5269-77. 10.1128/AEM.00124-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bessell PR, Rotariu O, Innocent GT, Smith-Palmer A, Strachan NJ, Forbes KJ, et al. Using sequence data to identify alternative routes and risk of infection: a case-study of Campylobacter in Scotland. BMC Infect Dis. 2012;12(1):80. 10.1186/1471-2334-12-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cody AJ, McCarthy ND, Bray JE, Wimalarathna HM, Colles FM, Jansen van Rensburg MJ, et al. Wild bird-associated Campylobacter jejuni isolates are a consistent source of human disease, in Oxfordshire, United Kingdom. Environ Microbiol Rep. 2015;7(5):782-8. 10.1111/1758-2229.12314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Di Giannatale E, Garofolo G, Alessiani A, Di Donato G, Candeloro L, Vencia W, et al. Tracing Back Clinical Campylobacter jejuni in the Northwest of Italy and Assessing Their Potential Source. Front Microbiol. 2016;7:887. 10.3389/fmicb.2016.00887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.French NP. Enhancing Surveillance of Potentially Foodborne Enteric Diseases in New Zealand: Human Campylobacteriosis in the Manawatu. Final report: FDI / 236/2005 2008. [Accessed 19 Nov 2018]. Available from: https://www.foodsafety.govt.nz/elibrary/industry/enhancing-surveillance-potentially-research-projects-2/Campy_Attribution_Manawatu.pdf
- 25. Jonas R, Kittl S, Overesch G, Kuhnert P. Genotypes and antibiotic resistance of bovine Campylobacter and their contribution to human campylobacteriosis. Epidemiol Infect. 2015;143(11):2373-80. 10.1017/S0950268814003410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kittl S, Heckel G, Korczak BM, Kuhnert P. Source attribution of human Campylobacter isolates by MLST and fla-typing and association of genotypes with quinolone resistance. PLoS One. 2013;8(11):e81796. 10.1371/journal.pone.0081796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kovac J, Stessl B, Čadež N, Gruntar I, Cimerman M, Stingl K, et al. Population structure and attribution of human clinical Campylobacter jejuni isolates from central Europe to livestock and environmental sources. Zoonoses Public Health. 2018;65(1):51-8. 10.1111/zph.12366 [DOI] [PubMed] [Google Scholar]
- 28. Lévesque S, Fournier E, Carrier N, Frost E, Arbeit RD, Michaud S. Campylobacteriosis in urban versus rural areas: a case-case study integrated with molecular typing to validate risk factors and to attribute sources of infection. PLoS One. 2013;8(12):e83731. 10.1371/journal.pone.0083731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mossong J, Mughini-Gras L, Penny C, Devaux A, Olinger C, Losch S, et al. Human Campylobacteriosis in Luxembourg, 2010-2013: A Case-Control Study Combined with Multilocus Sequence Typing for Source Attribution and Risk Factor Analysis. Sci Rep. 2016;6(1):20939. 10.1038/srep20939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mughini Gras L, Smid JH, Wagenaar JA, de Boer AG, Havelaar AH, Friesema IH, et al. Risk factors for campylobacteriosis of chicken, ruminant, and environmental origin: a combined case-control and source attribution analysis. PLoS One. 2012;7(8):e42599. 10.1371/journal.pone.0042599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mughini Gras L, Smid JH, Wagenaar JA, Koene MG, Havelaar AH, Friesema IH, et al. Increased risk for Campylobacter jejuni and C. coli infection of pet origin in dog owners and evidence for genetic association between strains causing infection in humans and their pets. Epidemiol Infect. 2013;141(12):2526-35. 10.1017/S0950268813000356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nohra A, Grinberg A, Midwinter AC, Marshall JC, Collins-Emerson JM, French NP. Molecular Epidemiology of Campylobacter coli Strains Isolated from Different Sources in New Zealand between 2005 and 2014. Appl Environ Microbiol. 2016;82(14):4363-70. 10.1128/AEM.00934-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rosner BM, Schielke A, Didelot X, Kops F, Breidenbach J, Willrich N, et al. A combined case-control and molecular source attribution study of human Campylobacter infections in Germany, 2011-2014. Sci Rep. 2017;7(1):5139. 10.1038/s41598-017-05227-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Roux F, Sproston E, Rotariu O, Macrae M, Sheppard SK, Bessell P, et al. Elucidating the aetiology of human Campylobacter coli infections. PLoS One. 2013;8(5):e64504. 10.1371/journal.pone.0064504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sears A, Baker MG, Wilson N, Marshall J, Muellner P, Campbell DM, et al. Marked campylobacteriosis decline after interventions aimed at poultry, New Zealand. Emerg Infect Dis. 2011;17(6):1007-15. 10.3201/eid/1706.101272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sheppard SK, Dallas JF, Strachan NJ, MacRae M, McCarthy ND, Wilson DJ, et al. Campylobacter genotyping to determine the source of human infection. Clin Infect Dis. 2009;48(8):1072-8. 10.1086/597402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sheppard SK, Dallas JF, Wilson DJ, Strachan NJ, McCarthy ND, Jolley KA, et al. Evolution of an agriculture-associated disease causing Campylobacter coli clade: evidence from national surveillance data in Scotland. PLoS One. 2010;5(12):e15708. 10.1371/journal.pone.0015708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smid JH, Mughini Gras L, de Boer AG, French NP, Havelaar AH, Wagenaar JA, et al. Practicalities of using non-local or non-recent multilocus sequence typing data for source attribution in space and time of human campylobacteriosis. PLoS One. 2013;8(2):e55029. 10.1371/journal.pone.0055029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Strachan NJ, Gormley FJ, Rotariu O, Ogden ID, Miller G, Dunn GM, et al. Attribution of Campylobacter infections in northeast Scotland to specific sources by use of multilocus sequence typing. J Infect Dis. 2009;199(8):1205-8. 10.1086/597417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Strachan NJ, Rotariu O, MacRae M, Sheppard SK, Smith-Palmer A, Cowden J, et al. Operationalising factors that explain the emergence of infectious diseases: a case study of the human campylobacteriosis epidemic. PLoS One. 2013;8(11):e79331. 10.1371/journal.pone.0079331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Thépault A, Méric G, Rivoal K, Pascoe B, Mageiros L, Touzain F, et al. Genome-Wide Identification of Host-Segregating Epidemiological Markers for Source Attribution in Campylobacter jejuni. Appl Environ Microbiol. 2017;83(7):e03085-16. 10.1128/AEM.03085-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Strachan NJ, Rotariu O, Smith-Palmer A, Cowden J, Sheppard SK, O’Brien SJ, et al. Identifying the seasonal origins of human campylobacteriosis. Epidemiol Infect. 2013;141(6):1267-75. 10.1017/S0950268812002063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. French NP, Midwinter A, Holland B, Collins-Emerson J, Pattison R, Colles F, et al. Molecular epidemiology of Campylobacter jejuni isolates from wild-bird fecal material in children’s playgrounds. Appl Environ Microbiol. 2009;75(3):779-83. 10.1128/AEM.01979-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Van Pelt W, Van De Giessen AW, Van Leeuwen WJ, et al. Oorsprung, omvang en kosten van humane salmonellose. Deel 1. Oorsprung van human salmonellose met betrekking tot varken, rund, kip, ei en overige bronnen. Infectiezikten Bulletin. 1999:240-3. [Google Scholar]
- 45. Hald T, Vose D, Wegener HC, Koupeev T. A Bayesian approach to quantify the contribution of animal-food sources to human salmonellosis. Risk Anal. 2004;24(1):255-69. 10.1111/j.0272-4332.2004.00427.x [DOI] [PubMed] [Google Scholar]
- 46. Bessell PR, Rotariu O, Innocent GT, Smith-Palmer A, Strachan NJ, Forbes KJ, et al. Using sequence data to identify alternative routes and risk of infection: a case-study of Campylobacter in Scotland. BMC Infect Dis. 2012;12(1):80. 10.1186/1471-2334-12-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Griekspoor P, Colles FM, McCarthy ND, Hansbro PM, Ashhurst-Smith C, Olsen B, et al. Marked host specificity and lack of phylogeographic population structure of Campylobacter jejuni in wild birds. Mol Ecol. 2013;22(5):1463-72. 10.1111/mec.12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Clark CG, Price L, Ahmed R, Woodward DL, Melito PL, Rodgers FG, et al. Characterization of waterborne outbreak-associated Campylobacter jejuni, Walkerton, Ontario. Emerg Infect Dis. 2003;9(10):1232-41. 10.3201/eid0910.020584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kovac J, Stessl B, Čadež N, Gruntar I, Cimerman M, Stingl K, et al. Population structure and attribution of human clinical Campylobacter jejuni isolates from central Europe to livestock and environmental sources. Zoonoses Public Health. 2018;65(1):51-8. 10.1111/zph.12366 [DOI] [PubMed] [Google Scholar]
- 50. Mossong J, Mughini-Gras L, Penny C, Devaux A, Olinger C, Losch S, et al. Human Campylobacteriosis in Luxembourg, 2010-2013: A Case-Control Study Combined with Multilocus Sequence Typing for Source Attribution and Risk Factor Analysis. Sci Rep. 2016;6(1):20939. 10.1038/srep20939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yahara K, Méric G, Taylor AJ, de Vries SP, Murray S, Pascoe B, et al. Genome-wide association of functional traits linked with Campylobacter jejuni survival from farm to fork. Environ Microbiol. 2017;19(1):361-80. 10.1111/1462-2920.13628 [DOI] [PubMed] [Google Scholar]
- 52. Ogden ID, Dallas JF, MacRae M, Rotariu O, Reay KW, Leitch M, et al. Campylobacter excreted into the environment by animal sources: prevalence, concentration shed, and host association. Foodborne Pathog Dis. 2009;6(10):1161-70. 10.1089/fpd.2009.0327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maiden MCJ, Jansen van Rensburg MJ, Bray JE, Earle SG, Ford SA, Jolley KA, et al. MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol. 2013;11(10):728-36. 10.1038/nrmicro3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Food Standards Agency. Employing Source Attribution and Molecular Epidemiology to measure the impact of interventions on human campylobacteriosis in Scotland. Final Report. 2018. [Accessed 19 Nov 2018]. Available from: https://www.foodstandards.gov.scot/downloads/Campylobacter_Attribution_Extension_-_University_of_Aberdeen_-_FSS00017_-_iCaMPS_Report_-_FINAL_-_13th_Dec_2017.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


